Abstract
Prions are infectious protein aggregates that cause several fatal neurodegenerative diseases. Prion research has been hindered by a lack of cellular paradigms for studying the replication of prions from different species. Although hamster prions have been widely used to study prion replication in animals and within in vitro amplification systems, they have proved challenging to propagate in cultured cells. Because the murine catecholaminergic cell line CAD5 is susceptible to a diverse range of mouse prion strains, we hypothesized that it might also be capable of propagating nonmouse prions. Here, using CRISPR/Cas9-mediated genome engineering, we demonstrate that CAD5 cells lacking endogenous mouse PrP expression (CAD5-PrP−/− cells) can be chronically infected with hamster prions following stable expression of hamster PrP. When exposed to the 263K, HY, or 139H hamster prion strains, these cells stably propagated high levels of protease-resistant PrP. Hamster prion replication required absence of mouse PrP, and hamster PrP inhibited the propagation of mouse prions. Cellular homogenates from 263K-infected cells exhibited prion seeding activity in the RT-QuIC assay and were infectious to naïve cells expressing hamster PrP. Interestingly, murine N2a neuroblastoma cells ablated for endogenous PrP expression were susceptible to mouse prions, but not hamster prions upon expression of cognate PrP, suggesting that CAD5 cells either possess cellular factors that enhance or lack factors that restrict the diversity of prion strains that can be propagated. We conclude that transfected CAD5-PrP−/− cells may be a useful tool for assessing the biology of prion strains and dissecting the mechanism of prion replication.
Keywords: prion, cell culture, neurodegeneration, protein aggregation, CRISPR/Cas, neuron, bovine spongiform encephalopathy, Creutzfeldt-Jakob disease, heterologous system
Introduction
Prions are pathogenic and infectious protein aggregates that cause a variety of fatal neurodegenerative illnesses, including Creutzfeldt-Jakob disease (CJD)2 in humans, chronic wasting disease in cervids, scrapie in sheep, and bovine spongiform encephalopathy (“mad cow disease”) in cattle (1). Prions are composed exclusively of a single protein, the prion protein (PrP), which is encoded by the prion gene (Prnp in rodents, PRNP in humans) (2). The conformational conversion of cellular PrP (PrPC) to an abnormally folded and toxic “prion” state (PrPSc) is central to all forms of prion disease (3–5). PrPC is a glycophosphatidylinositol-anchored protein that is predominantly expressed on the surface of central nervous system (CNS) cells (6). Although the normal function of PrPC within the brain is debated, there is strong evidence that PrPC participates in myelin maintenance within the peripheral nervous system (7–9). PrPC possesses a mainly α-helical structure and is highly sensitive to protease digestion (10). In contrast, PrPSc is enriched in β-sheet content and polymerizes into large, insoluble, and protease-resistant aggregates that deposit within the CNS (4, 11).
Prions are believed to replicate via a template-directed refolding mechanism in which PrPSc guides the conformational conversion of PrPC into additional copies of PrPSc. This ability to self-propagate underlies the ability of prions to transmit disease within a given species. The transmission of prions between different species is typically inefficient or restricted because of the “species barrier” (12), a phenomenon thought to partially arise because of amino acid mismatches between orthologous prion sequences. For instance, WT mice that express endogenous mouse PrP are resistant to hamster prions whereas transgenic mice that also express hamster PrP are highly susceptible (13, 14). The simultaneous presence of multiple PrPC orthologs can also affect prion transmission. For example, transgenic mice expressing human PrP only become susceptible to human prions upon ablation of endogenous mouse PrP (15, 16). A further complication is the existence of “strain barriers” for prion replication. Prion strains are conformational variants of PrPSc aggregates that exhibit distinct biochemical and pathological properties (17, 18). For efficient prion replication to occur, there must be structural compatibility between the PrPC and PrPSc molecules involved (19).
Although studies in animals have undoubtedly advanced our knowledge of prion disease (20), replication of prions in cultured cells offers several advantages, including the ability to rapidly identify anti-prion compounds. Although several murine cell lines that express endogenous mouse PrP can replicate mouse prions, differences exist between lines with regard to the breadth of strains that can be propagated (21–30). For example, N2a neuroblastoma cells are susceptible to the RML and 22L strains, but not the 301C or Me7 strains (27). In contrast, CAD5 cells are permissive to a greater range of mouse strains, including a drug-resistant strain of mouse prions that could not be reliably propagated in N2a cells (27, 31, 32). CAD5 cells are a subclone of the CAD (Cath.a-differentiated) line, which was derived from the immortalized CNS catecholaminergic cell line Cath.a (27, 33, 34). Certain cell lines that lack endogenous PrP can be rendered susceptible to mouse prions by expression of mouse PrP (35, 36), but very few paradigms exist for the propagation of nonmouse prion strains in cultured cells. Rabbit RK13 cells expressing sheep, bank vole, or elk PrPC can propagate prions from the corresponding species (35, 37, 38), but this susceptibility does not extend to all types of prions, including human prions (39).
Hamster prions have largely remained refractory to propagation in cultured cells, despite having played a critical role in the original discovery of prions (3), in the development of in vitro prion replication techniques (40, 41), and in expanding our understanding of prion strains (42) and the molecular basis of the species barrier (13, 14). Given that mouse CAD5 cells can replicate a wide range of mouse prion strains, we hypothesized that they might also represent a suitable paradigm for propagating hamster prions. Here, we show that CAD5 cells expressing hamster PrPC are able to chronically replicate hamster prions, but only after ablation of endogenous mouse PrP. In contrast, N2a cells lacking endogenous PrP were susceptible to mouse but not hamster prions upon transfection of mouse or hamster PrPC, respectively, indicating that cell type–specific factors govern the ability of cells to replicate hamster prions.
Results
Generation of CAD5-PrP−/− cells
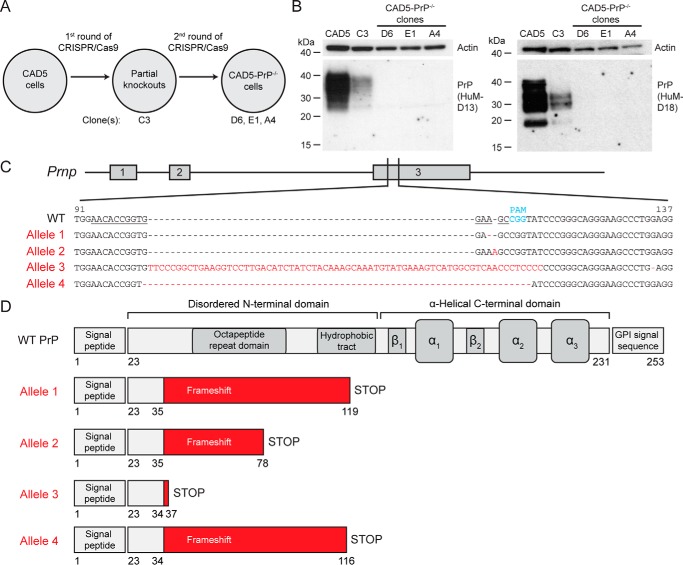
Because the presence of mouse PrP can interfere with the replication of nonmouse prions in transgenic mice (16), we decided to knock out the endogenous mouse PrP in CAD5 cells prior to attempting transmissions with hamster prions. For this purpose, we utilized CRISPR/Cas9-mediated genome engineering (43) to target Prnp exon 3, comprising the entire coding sequence of mature PrP, at an N-terminal site (Fig. 1A). The same ablation strategy had previously been used to delete PrP expression in several cell lines relevant to prion biology, including N2a cells, because it was predicted to pose a low risk for generating off-target effects (44, 45). After transfection of CAD5 cells with the CRISPR/Cas9 reagents, monoclonal lines were isolated and their PrP levels in cell lysates were analyzed by immunoblotting. We surveyed 22 clonal lines and found none that were completely devoid of PrP expression. However, several clones exhibited reduced PrP levels, and we selected one of them, termed C3, for a second round of CRISPR/Cas9 genome engineering (Fig. 1, A and B). Of 35 clones screened, we obtained three putative lines of CAD5-PrP−/− cells, termed D6, E1, and A4, that were robust enough to permit further analysis. Immunoblot analysis of cell lysates from these three clones using the anti-PrP antibodies HuM-D13 (epitope: residues 95–105) and HuM-D18 (epitope: residues 132–156) revealed the complete absence of PrP expression (Fig. 1B).
Figure 1.
Generation of CAD5-PrP−/− cells. A, schematic of the procedure used to isolate CAD5-PrP−/− cells. Four monoclonal lines were kept for further analysis: The partial knockout clone C3 as well as CAD5-PrP−/− clones D6, E1, and A4. B, immunoblots of PrP levels in cell lysates from WT CAD5 cells, the partial knockout clone C3, and the CAD5-PrP−/− lines D6, E1, and A4. PrP was detected using the antibodies HuM-D13 (left blot) or HuM-D18 (right blot). The lower molecular weight PrP bands visible with the HuM-D18 antibody represent a physiological cleavage product termed C1 which is generated by endoproteolysis in the vicinity of PrP residue 110 (74). Blots were reprobed with an antibody to actin (20–33) to compare total protein levels. Molecular mass measurements are indicated in kDa. C, characterization of mutant Prnp alleles in CAD5-PrP−/− clone D6. The numbering is with respect to the ORF of WT Prnp. The targeting sequence of the gRNA is underlined, and the protospacer-adjacent motif (PAM) is indicated in blue. Four mutant alleles were identified: An adenine deletion (Allele 1), an adenine insertion (Allele 2), a large insertion (Allele 3), and a 10-bp deletion (Allele 4). The mutated residues present in the mutant alleles are indicated in red. D, schematic of truncated protein products predicted to be produced by the four mutant alleles. Beginning at PrP residues 34–35, frameshift mutations result in the incorporation of various amounts of non-PrP sequence (red) as well as a premature STOP codon. The domain structure of WT PrPC is shown for comparison.
To identify the CRISPR/Cas9-induced mutations underlying PrP ablation in the selected clones, genomic DNA was isolated from clones C3 and D6 and the region of Prnp flanking the target site was amplified by PCR. Sequencing of clone D6 revealed the presence of four distinct insertion and deletion (indel) mutations, each of which is predicted to cause a frameshift within the PrP coding region (Fig. 1C). Consistent with the immunoblot results, all four mutant alleles are predicted to produce a nonfunctional PrP because of the presence of premature STOP codons and the incorporation of exogenous sequence beginning at residue 34/35 (Fig. 1D). Sequencing of clone C3 revealed the presence of mutant alleles 2 and 3 (Fig. 1C) as well as the WT allele, which explains the partial reduction of PrP expression observed in this clone (Fig. 1B). Consistent with the results from a separate line of CAD5-PrP−/− cells (46), we concluded that CAD5 cells possess an aneuploid number of four copies of the Prnp gene and selected CAD5-PrP−/− clone D6 for use in subsequent experiments.
Transfected CAD5-PrP−/− cells are susceptible to mouse and hamster prions
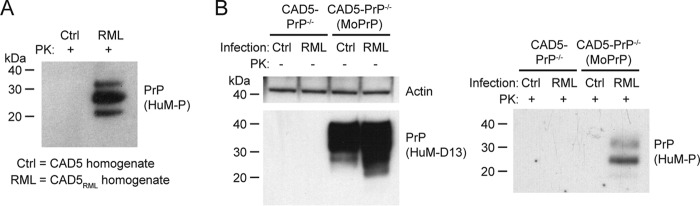
Before moving to hamster prion infection studies, we wanted to verify whether CAD-PrP−/− cells retained their vulnerability to mouse prions. Thus, CAD5-PrP−/− cells were stably transfected with a plasmid that encodes mouse PrP to generate a polyclonal pool of CAD5-PrP−/−(MoPrP) cells. We then challenged CAD5-PrP−/− and CAD5-PrP−/−(MoPrP) cells with cellular homogenate from either uninfected CAD5 cells or CAD5 cells chronically infected with the RML strain of mouse-adapted scrapie prions (CAD5RML cells). As expected, only the cellular homogenate from the CAD5RML cells contained detergent-insoluble and proteinase K (PK)–resistant PrP, a biochemical indicator of PrPSc (Fig. 2A). Following prion challenge of the CAD5-PrP−/− and CAD5-PrP−/−(MoPrP) lines, cells were passaged for seven generations to ensure the complete removal of any residual inoculum. Lysates were collected from cells and then total PrP levels were analyzed by immunoblotting. As expected, only the CAD5-PrP−/−(MoPrP) cells contained detectable levels of PrP expression (Fig. 2B). RML-exposed CAD5-PrP−/−(MoPrP) cells exhibited additional lower molecular weight PrP signal that was absent in the control-inoculated cells, implying the presence of PrPSc that had been partially digested by endogenous cellular proteases. Insoluble PK-resistant PrP was observed in cell lysates from CAD5-PrP−/−(MoPrP) cells, but not CAD5-PrP−/− cells, that had been challenged with RML-infected cell homogenate, confirming the presence of PrPSc (Fig. 2B). No PK-resistant PrP was present in lysates from cells challenged with uninfected cell homogenate. As with the RML inoculum, the monoglycosylated PrPSc glycoform was dominant in RML-infected CAD5-PrP−/−(MoPrP) cells, arguing for the maintenance of strain fidelity. These results demonstrate that, like CAD5 cells, CAD5-PrP−/−(MoPrP) cells are susceptible to infection with the RML strain of mouse prions.
Figure 2.
Replication of mouse RML prions in CAD5-PrP−/−(MoPrP) cells. A, immunoblot of PK-digested cellular homogenates from either uninfected CAD5 cells (Ctrl) or RML prion–infected CAD5 cells (RML). B, infection of CAD5-PrP−/− and CAD5-PrP−/−(MoPrP) cells with RML prions. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to homogenates from either uninfected CAD5 cells or RML prion-infected CAD5 cells. In both panels, PK-resistant PrP was visualized using the antibody HuM-P, and total (undigested) PrP was detected using the antibody HuM-D13. The undigested blot was also reprobed with an antibody to actin (20–33). Molecular mass measurements are indicated in kDa.
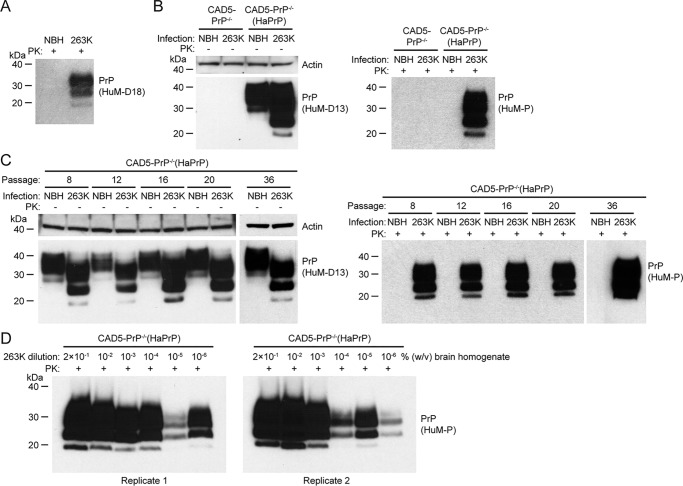
We next decided to test whether CAD5-PrP−/− cells stably transfected with hamster PrP, termed CAD5-PrP−/−(HaPrP), were susceptible to hamster prions. For this purpose, we utilized the 263K strain of hamster-adapted scrapie prions (47), which produces short disease incubation periods and accumulates to high levels in the brains of prion-infected hamsters. As expected, 263K-infected hamster brain homogenate contained abundant amounts of PK-resistant PrP, whereas normal brain homogenate (NBH) from healthy hamsters contained none (Fig. 3A). CAD5-PrP−/− cells and a polyclonal pool of CAD5-PrP−/−(HaPrP) cells were challenged with 263K brain homogenate or NBH and then passaged for seven generations. In undigested cell lysates, high levels of truncated PrP species were observed in the CAD5-PrP−/−(HaPrP) cells exposed to 263K prions, but not in the NBH-treated cells, suggestive of successful prion infection (Fig. 3B). Likewise, insoluble, PK-resistant PrP was only present in the lysates from CAD5-PrP−/−(HaPrP) challenged with 263K prions. We performed nine independent replications of this experiment and observed PK-resistant hamster PrP in each instance. Similar to the PrPSc in 263K brain homogenate, the diglycosylated PrPSc glycoform was predominant in the CAD5-PrP−/−(HaPrP) cells, signifying prion strain maintenance. PK-resistant PrPSc levels in 263K-infected CAD5-PrP−/−(HaPrP) cells remained stable for at least 36 passages, indicating that the cells can be chronically infected with 263K prions (Fig. 3C). The sensitivity of CAD5-PrP−/−(HaPrP) cells to 263K prions was assessed by challenging them with serial 10-fold dilutions of 263K brain homogenate. After seven passages of each 263K prion-exposed culture, PK-resistant PrP was observed with all dilutions tested up to and including 2 × 10−6 % (w/v) brain homogenate, suggesting that CAD5-PrP−/−(HaPrP) cells are highly susceptible to 263K prions (Fig. 3D).
Figure 3.
Replication of hamster 263K prions in CAD5-PrP−/−(HaPrP) cells. A, immunoblot of PK-digested NBH or 263K prion-infected brain homogenate from hamsters. PK-resistant PrP was visualized using the antibody HuM-D18. B, infection of CAD5-PrP−/− and CAD5-PrP−/−(HaPrP) cells with 263K prions. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to NBH or 263K-infected brain homogenate. C, chronic replication of 263K prions in CAD5-PrP−/−(HaPrP) cells. Immunoblots of either undigested (left blots) or PK-digested (right blots) lysates from cells at passage 8, 12, 16, 20, or 36 following exposure to NBH or 263K-infected brain homogenate. D, immunoblots of PK-digested lysates from CAD5-PrP−/−(HaPrP) cells at passage seven following exposure to 10-fold serial dilutions of 0.2% (w/v) 263K-infected brain homogenate. Two independent replicates are shown. B–D, PK-resistant PrP was visualized using the antibody HuM-P, and total (undigested) PrP was detected using the antibody HuM-D13. The undigested blots were also reprobed with an antibody to actin (20–33). Molecular mass measurements are indicated in kDa.
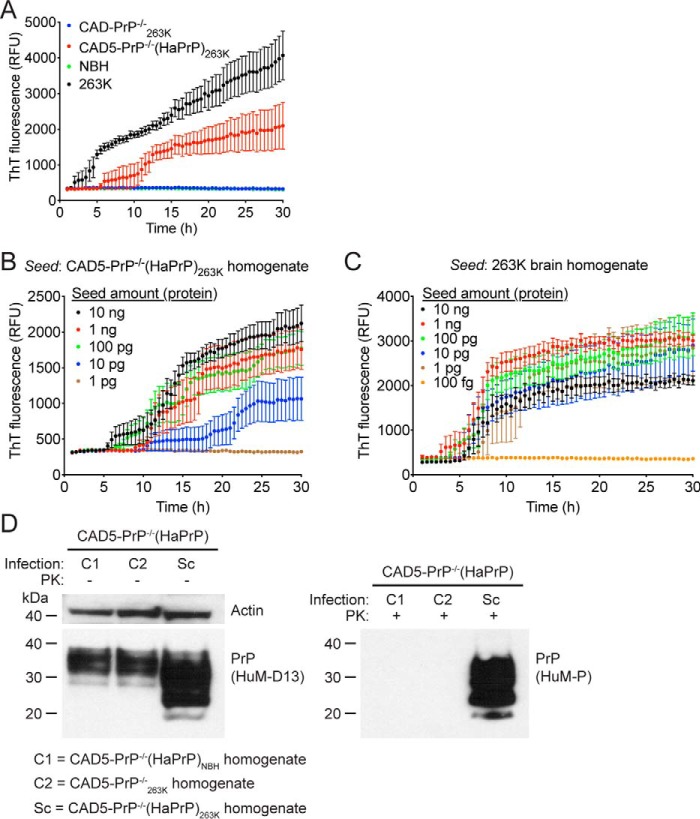
To compare the seeding activities of brain-derived and cell-passaged 263K prions, hereafter referred to as CAD5-PrP−/−(HaPrP)263K, we performed RT-QuIC assays using recombinant hamster PrP(23–231) as the substrate (41, 48). When 100 ng of total protein from either brain or cellular homogenate was used as a seed, amplification was detected with the 263K-infected brain and CAD5-PrP−/−(HaPrP)263K samples, as visualized by an increase in ThT fluorescence (Fig. 4A). In contrast, no seeding activity was detected when NBH or homogenate from 263K-treated CAD5-PrP−/− cells (CAD5-PrP−/−263K) was used (Fig. 4A). This indicates that by passage seven, prion seeding activity from the 263K brain homogenate inoculum had been removed or diluted to a level below the detection threshold of the assay. Serial dilution of CAD5-PrP−/−(HaPrP)263K cell homogenate revealed that seeding activity could be detected when as little as 10 pg of total protein was used as a seed in RT-QuIC reactions (Fig. 4B). When 263K brain homogenate was subjected to an analogous analysis, only 1 pg of total protein was required to observe seeding activity (Fig. 4C). Thus, like brain-derived 263K prions, CAD5-PrP−/−(HaPrP)263K prions exhibit prion seeding activity, although the specific seeding activity is ∼10-fold lower in the cells. To check whether 263K prions can be serially transmitted in cultured cells, CAD5-PrP−/−(HaPrP) cells were exposed to homogenate prepared from either CAD5-PrP−/−(HaPrP)263K, CAD5-PrP−/−263K, or NBH-treated CAD5-PrP−/−(HaPrP) cells, designated CAD5-PrP−/−(HaPrP)NBH, each at passage eight. Treatment with CAD5-PrP−/−(HaPrP)263K homogenate, but not CAD5-PrP−/−263K or CAD5-PrP−/−(HaPrP)NBH homogenate, produced PK-resistant PrP in CAD5-PrP−/−(HaPrP) cells after seven passages, indicating that cell-passaged 263K prions remain infectious to naïve cells (Fig. 4D).
Figure 4.
Prion seeding activity and cellular infectivity of 263K prions passaged in CAD5-PrP−/−(HaPrP) cells. A, RT-QuIC using recombinant HaPrP(23–231) as a substrate and 100 ng of NBH (green), 263K-infected hamster brain homogenate (black), or homogenate from either 263K-exposed CAD5-PrP−/− (blue) or CAD5-PrP−/−(HaPrP) (red) cells (passage seven post infection) as a seed. Each data point represents the mean ± S.E. of quadruplicate reactions. B, RT-QuIC on 10-fold serial dilutions of CAD5-PrP−/−(HaPrP)263K homogenate (passage eight). Each data point represents the mean ± S.E. of eight independent reactions. C, RT-QuIC on 10-fold serial dilutions of 263K-infected hamster brain homogenate. Each data point represents the mean ± S.E. of four independent reactions. D, infection of CAD5-PrP−/−(HaPrP) cells with cell-passaged 263K prions. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to homogenate from either CAD5-PrP−/−(HaPrP) cells treated with NBH (C1), CAD5-PrP−/− cells treated with 263K brain homogenate (C2), or CAD5-PrP−/−(HaPrP) cells treated with 263K brain homogenate (Sc), each at passage eight. PK-resistant PrP was visualized using the antibody HuM-P, and total (undigested) PrP was detected using the antibody HuM-D13. The undigested blot was also reprobed with an antibody to actin (20–33). Molecular mass measurements are indicated in kDa.
CAD5-PrP−/−(HaPrP) cells can propagate multiple hamster prion strains
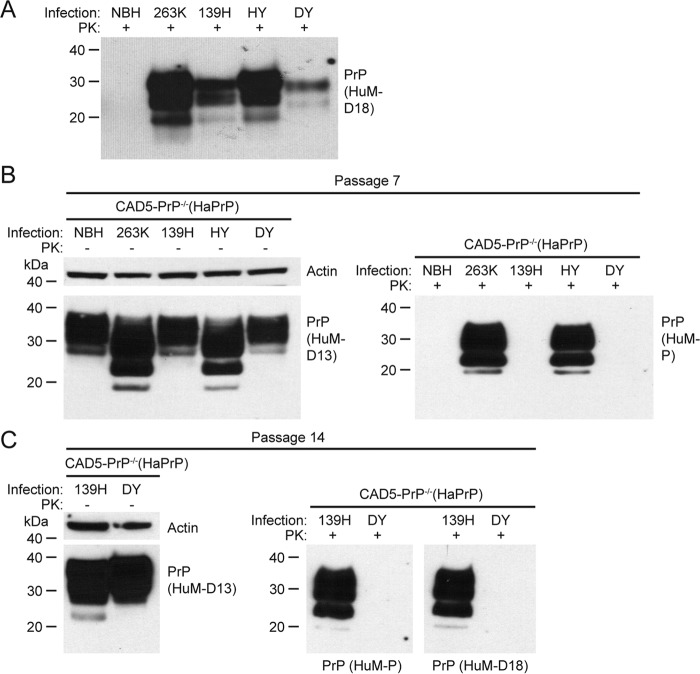
Having established that hamster 263K prions can replicate efficiently in CAD5-PrP−/−(HaPrP) cells, we next tested whether these cells are also vulnerable to other strains of hamster prions. Accordingly, we challenged CAD5-PrP−/−(HaPrP) cells with three additional hamster prion strains: 139H, which was adapted from a strain of mouse-adapted scrapie prions (49), as well as the hamster-adapted hyper (HY) and drowsy (DY) strains (42), both of which originated from strains of transmissible mink encephalopathy prions. Abundant levels of PK-resistant PrP were present in the brain homogenates from hamsters infected with each of the different strains (Fig. 5A). Like cells treated with the 263K strain, cells treated with the HY strain displayed faster-migrating PrP bands as well as PK-resistant PrP following seven passages (Fig. 5B), indicating that CAD5-PrP−/−(HaPrP) can also replicate the HY strain. At passage seven, no PK-resistant PrP was observed in cells challenged with the 139H or DY strains. However, at passage 14, PK-resistant PrP was readily detected in cells exposed to 139H prions, but not in cells challenged with DY prions (Fig. 5C). This implies that many, but not all, strains of hamster prions can be propagated in CAD5-PrP−/−(HaPrP) cells.
Figure 5.
Challenge of CAD5-PrP−/−(HaPrP) cells with different hamster prion strains. A, immunoblot of PK-digested NBH or brain homogenate from hamsters infected with the 263K, 139H, Hyper (HY), or Drowsy (DY) strains. B, challenge of CAD5-PrP−/−(HaPrP) cells with various hamster prion strains. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to NBH or hamster brain homogenate containing the indicated prion strains. C, immunoblots of either undigested (left blot) or PK-digested (right blots) lysates from cells at passage 14 following exposure to DY or 139H brain homogenate. PK-resistant PrP was visualized using the antibodies HuM-D18 (A) or HuM-P (B and C), and total (undigested) PrP was detected using the antibody HuM-D13. The undigested blots were also reprobed with an antibody to actin (20–33). Molecular mass measurements are indicated in kDa.
Assessment of cross-species prion transmission and interference
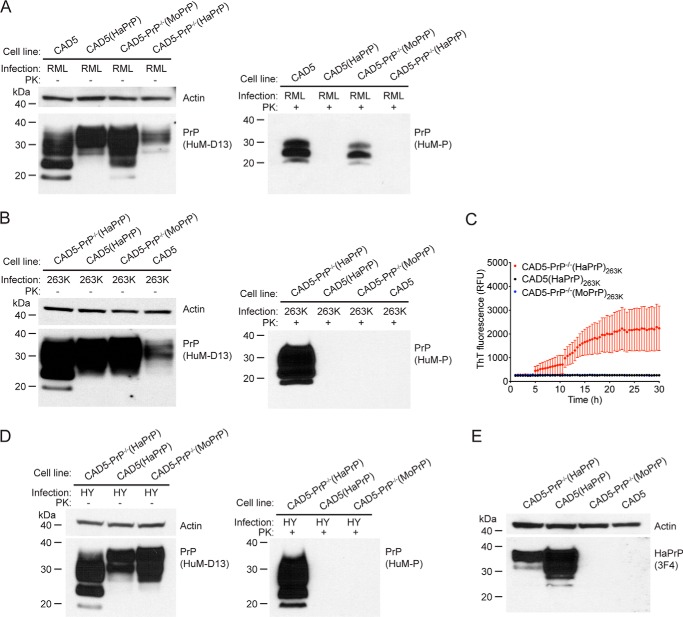
Because previous studies in transgenic mice have demonstrated that co-expression of multiple PrP orthologs can delay or prevent the transmission of prion disease (14, 16), we decided to investigate whether co-expression of mouse PrP has any effect on the propagation of hamster prions in cells and vice versa. For this purpose, we utilized four different cell lines: CAD5 and CAD5-PrP−/−(MoPrP), both of which only express MoPrP; CAD5-PrP−/−(HaPrP), which only expresses HaPrP; and CAD5(HaPrP), which expresses both endogenous MoPrP and stably transfected HaPrP. Each cell line was challenged with either mouse (RML) or hamster (263K) prions. With mouse prion exposure experiments, PK-resistant PrP was observed in the cells that express solely MoPrP but not in the cells that express solely HaPrP or in the cells that express both MoPrP and HaPrP (Fig. 6A). Similarly, in cells exposed to 263K hamster prions, PK-resistant PrP and prion seeding activity in the RT-QuIC assay were observed in the cells that express solely HaPrP, but not in the cells that express solely MoPrP or in the cells that express both MoPrP and HaPrP (Fig. 6, B and C). We repeated this experiment using the HY strain of hamster prions and obtained identical results; PK-resistant PrP was only present in cells that express solely HaPrP (Fig. 6D). Levels of HaPrP were higher in CAD5(HaPrP) cells than in CAD5-PrP−/−(HaPrP) cells, suggesting that the inability of CAD5(HaPrP) cells to be infected by hamster prions was not because of insufficient HaPrP expression (Fig. 6E). Thus, cross-species prion transmission does not occur in transfected CAD5-PrP−/− cells, and the simultaneous presence of different PrPC orthologs completely blocks prion replication.
Figure 6.
Evaluation of cross-species prion seeding and interference. A, immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from CAD5, CAD5(HaPrP), CAD5-PrP−/−(MoPrP), or CAD5-PrP−/−(HaPrP) cells at passage seven following exposure to cell homogenate containing mouse RML prions. B, immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from CAD5-PrP−/−(HaPrP), CAD5(HaPrP), CAD5-PrP−/−(MoPrP), or CAD5 cells at passage seven following exposure to hamster brain homogenate containing 263K prions. C, RT-QuIC using recombinant HaPrP(23–231) as a substrate and lysates (10−3 dilution, passage seven post infection) from 263K-exposed CAD5-PrP−/−(HaPrP) (red), CAD5(HaPrP) (black), or CAD5-PrP−/−(MoPrP) (blue) cells as a seed. Each data point represents the mean ± S.E. of quadruplicate reactions. D, immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from CAD5-PrP−/−(HaPrP), CAD5(HaPrP), or CAD5-PrP−/−(MoPrP) cells at passage seven following exposure to hamster brain homogenate containing HY prions. E, HaPrP levels in lysates from uninfected CAD5, CAD5(HaPrP), CAD5-PrP−/−(MoPrP), or CAD5-PrP−/−(HaPrP) cells, as assessed by immunoblotting with the antibody 3F4, which recognizes HaPrP but not MoPrP. A, B, and D, PK-resistant PrP was visualized using the antibody HuM-P, and total (undigested) PrP was detected using the antibody HuM-D13. The undigested blots were also reprobed with an antibody to actin (20–33). Molecular mass measurements are indicated in kDa.
N2a-PrP−/−(HaPrP) cells are resistant to hamster prions
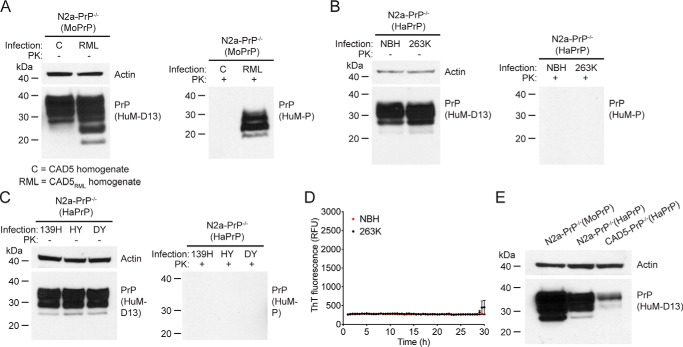
We were curious whether hamster prions could be propagated in other prion-susceptible cell lines following the removal of endogenous PrPC. To address this point, we tested whether a monoclonal N2a-PrP−/− cell line becomes susceptible to hamster prions following the stable transfection of HaPrP. Notably, the N2a-PrP−/− cells were engineered from RML prion-susceptible N2a cells using identical CRISPR/Cas9 reagents as the CAD5-PrP−/− cells (44). Because subclones of N2a cells can exhibit resistance to prion infection (27), we first transfected N2a-PrP−/− cells to generate a polyclonal pool of cells stably expressing MoPrP (N2a-PrP−/−(MoPrP)) and then tested whether they retained their susceptibility to mouse RML prions. Indeed, challenge of N2a-PrP−/−(MoPrP) cells with CAD5RML homogenate, but not CAD5 homogenate, produced a robust prion infection, visualized either in the absence or presence of PK digestion (Fig. 7A). In contrast, when N2a-PrP−/− cells were transfected to generate a polyclonal pool of cells stably expressing HaPrP (N2a-PrP−/−(HaPrP)) and then challenged with 263K prions, no PK-resistant PrP or prion seeding activity was observed in cell lysates at seven passages post exposure (Fig. 7, B and D). Moreover, PK-resistant PrP was also not observed in N2a-PrP−/−(HaPrP) cells challenged with the 139H, HY, or DY hamster prion strains (Fig. 7C). The inability of N2a-PrP−/−(HaPrP) cells to be infected by hamster prions cannot be explained by insufficient expression of HaPrP, because levels of HaPrP were higher in N2a-PrP−/−(HaPrP) cells than in CAD5-PrP−/−(HaPrP) cells (Fig. 7E). This suggests that the expression of HaPrP in N2a cells lacking endogenous PrP is insufficient to permit the replication of hamster prions.
Figure 7.
Challenge of transfected N2a-PrP−/− cells with prions. A, challenge of N2a-PrP−/−(MoPrP) cells with mouse RML prions. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to homogenates from either uninfected CAD5 cells (C) or RML prion-infected CAD5 cells (RML). B, challenge of N2a-PrP−/−(HaPrP) cells with hamster 263K prions. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to NBH or 263K-infected brain homogenate. C, challenge of N2a-PrP−/−(HaPrP) cells with hamster 139H, HY, and DY prion strains. Immunoblots of either undigested (left blot) or PK-digested (right blot) lysates from cells at passage seven following exposure to 139H-, HY-, or DY-infected brain homogenate. D, RT-QuIC using recombinant HaPrP(23–231) as a substrate and lysates (10−3 dilution, passage seven post infection) from N2a-PrP−/−(HaPrP) cells exposed to either 263K homogenate (black) or NBH (red) as a seed. Each data point represents the mean ± S.E. of quadruplicate reactions. E, PrPC levels in lysates from uninfected N2a-PrP−/−(MoPrP), N2a-PrP−/−(HaPrP), and CAD5-PrP−/−(HaPrP) cells, as detected by immunoblotting. A–C and E, PK-resistant PrP was visualized using the antibody HuM-P, and total (undigested) PrP was detected using the antibody HuM-D13. The undigested blots were also reprobed with an antibody to actin (20–33). Molecular mass measurements are indicated in kDa.
Discussion
Here, we have demonstrated that CAD5-PrP−/− cells are capable of stably propagating both mouse and hamster prion strains following transfection with cognate PrPC. CAD5-PrP−/−(HaPrP)263K prions were similar to brain-derived 263K prions in that the dominant PrPSc glycoform was diglycosylated PrP. Conversely, monoglycosylated PrPSc was the dominant glycoform in CAD5-PrP−/−(MoPrP) cells infected with RML prions, suggesting that prion strain properties were at least partially maintained upon propagation in the cultured cells. To the best of our knowledge, this is the first example of chronic and stable replication of hamster prions in an immortalized cell line. A spontaneously immortalized cell line (HaB) isolated from the brain of prion-infected hamsters had previously been used to study the propagation of hamster prions in culture (50). Despite having been isolated from prion-infected animals, HaB cells did not exhibit any evidence of prion infection. Challenge of HaB cells with Sc237 hamster prions occasionally resulted in PrPSc-positive cells, but only when the cells were grown at a lower temperature (34 °C). Moreover, HaB cells expressed very low levels of PrPC (51), and prion infection was inevitably lost during passaging of prion-infected subclones (50, 52), rendering them nonideal for probing the replication behavior of hamster prions. In contrast, CAD5-PrP−/−(HaPrP) cells infected with the 263K strain of hamster prions remained stably infected for at least 36 passages (∼5 months) post infection with no evidence for the gradual loss of PK-resistant PrP. Thus, we contend that prion-infected CAD5-PrP−/−(HaPrP) cells represent a superior cultured cell paradigm for interrogating the biology of hamster prions. Hamster prion replication has also been reported in cultured primary cerebellar granular neurons from transgenic mice expressing hamster PrP, but the nondividing nature of these cells makes scalability difficult (53).
We were able to propagate the 263K, HY, and 139H strains but not the DY strain of hamster prions in CAD5-PrP−/−(HaPrP) cells. This result was unexpected because CAD5 cells appear to be susceptible to all strains of mouse prions tested thus far (27, 31, 32). There are several possible explanations for the partial strain selectively of CAD5-PrP−/−(HaPrP) cells. First, the 139H and DY strains require longer incubation periods to cause disease in hamsters and exhibit lower amplification coefficients in protein misfolding cyclic amplification assays than the 263K and HY strains, suggesting that they replicate more slowly (54). Because the rate of cell division is the primary factor governing the replication of prions in cultured cells (55), CAD5-PrP−/−(HaPrP) cells may divide too rapidly to permit accumulation of the DY strain within a limited number of passages. Indeed, for the 139H strain, additional passaging was necessary to facilitate the detection of PK-resistant PrP by immunoblotting. Second, we did not perform subcloning of our stably transfected CAD5-PrP−/−(HaPrP) cells, so it is expected that variable levels of PrPC expression are present within the polyclonal cell population. It is conceivable that uniform or higher levels of PrPC expression may be necessary for efficient replication of the DY strain. Third, it is possible that the cellular infection paradigm employed, including the composition of the medium, may need to be optimized for each distinct prion strain. Fourth, compared with other hamster prion strains, DY is more conformationally labile and susceptible to protease digestion (54, 56), which could impede cellular infection. Finally, it is conceivable that for reasons not yet understood, CAD5 cells are simply intrinsically resistant to the DY strain.
CAD5-PrP−/−(HaPrP) cells, but not CAD5(HaPrP) cells, were capable of propagating 263K prions, suggesting that the presence of endogenous MoPrP interfered with the replication of hamster prions. This is similar to what has been observed in transgenic mice expressing human PrP, which were only susceptible to human prions following the ablation of MoPrP (16). In contrast, transgenic mice expressing HaPrP as well as endogenous MoPrP were susceptible to hamster prions (14), although the effect of ablating MoPrP on the disease incubation period was not investigated. The incubation periods for mouse prions in these transgenic mice were slightly prolonged, indicating that HaPrP partially interfered with the propagation of mouse prions. In agreement with this observation, we found that CAD5(HaPrP) cells were resistant to infection with mouse prions. The reason(s) why cross-species prion interference for hamster and mouse prions was more pronounced in our cellular models than in transgenic mice remains to be determined. The molecular basis of cross-species prion interference has yet to be deciphered, but it was originally postulated that these dominant-negative effects were mediated by a hypothetical prion replication co-factor termed protein X (16, 57). Although co-factors that influence prion replication in vitro have been identified (58, 59), a chaperone-like molecule that modulates prion replication in vivo has not been found (60). It was hypothesized that mouse protein X binds more strongly to MoPrP than to other PrP orthologs, explaining why the removal of MoPrP is necessary to obtain efficient replication of nonmouse PrPSc. The lack of interference of endogenous mouse PrP on hamster prion replication in transgenic mice would argue that mouse protein X binds equally well to MoPrP and HaPrP. However, given that 263K prion propagation was completely inhibited by the presence of MoPrP in cells and that dominant-negative inhibition of prion formation can be achieved in vitro using purified PrPC and PrPSc (61), our results are more consistent with cross-species prion interference being governed solely by interactions between structural isoforms of PrP orthologs.
CAD5-PrP−/− cells, but not N2a-PrP−/− cells, transfected with HaPrP were susceptible to hamster 263K prions. However, both lines were susceptible to mouse RML prions following transfection with MoPrP. This is consistent with the supposition that CAD5 cells possess cellular factors that enhance the diversity of prion strains that can be propagated, including nonmouse strains. It is not yet clear what determines whether a cell line is susceptible to prion infection. Prion-resistant and prion-susceptible subclones of N2a cells have been isolated (27, 62), which have revealed that prion susceptibility cannot solely be explained on the basis of PrPC expression levels. Instead, prion-resistant subclones show alterations in the transcription of genes related to the remodeling of the extracellular matrix (63). Interestingly, cell lysates from prion-susceptible and prion-resistant cell lines function as equally good substrates for the in vitro replication of prions, suggesting that prion susceptibility versus resistance is a property of the intact cell (64). This could indicate that the relative rates of PrPSc formation and clearance may govern prion susceptibility in cell lines, which is consistent with clearance rates of the DY strain controlling prion tropism in hamsters (65). Recent work comparing the PrP interactomes in various cell lines has found that PrP is exposed to distinct cellular milieus in N2a and CAD5 cells (45). Thus, interaction of PrP with a protein expressed in CAD5 but not N2a cells may result in an expanded repertoire of strains that can be replicated. Alternatively, N2a cells may possess inhibitory factors that limit the number of strains that can be propagated.
We believe that CAD5-PrP−/− cells represent a versatile cellular paradigm for probing the biology of prions. Here, we have shown that, like RK13 cells, CAD5-PrP−/− cells can be rendered susceptible to prions upon transfection of cognate PrPC. Unlike RK13 cells, CAD5 cells are CNS-derived, express many neuronal proteins, and can be differentiated into a neuron-like state (34). Moreover, there has been no report describing the successful propagation of hamster prions in RK13 cells transfected with HaPrP, although lysates from such cells can be used as a substrate to amplify hamster prions by protein misfolding cyclic amplification (66, 67). Given that CAD5-PrP−/−(HaPrP) cells became infected even when challenged with very low doses of hamster prions, it is likely that they can be adapted for use as a cellular bioassay for quantifying hamster prion titers in biological samples, as has been done for mouse RML and cervid chronic wasting disease prions (38, 62). Such a paradigm would permit hamster prion infectivity to be measured more rapidly and in a more high-throughput fashion than possible with traditional end point titration bioassays in animals (20).
Whether CAD5-PrP−/− cells become susceptible to other types of nonmouse prions upon transfection of the appropriate PrPC remains to be determined. Of particular importance are human prions, which have remained refractory to propagation within immortalized cultured cells. A recent study has described the successful replication of CJD prions in differentiated primary human astrocytes (68). However, an immortalized cell line that can be chronically infected with human prions would be preferable for discovering anti-prion compounds active against CJD prions. The discovery of a cell line capable of stably propagating human prions may be paramount to the development of CJD therapeutics, because small molecules that are efficacious against mouse prions in cultured cells and mice can fail to inhibit the replication of human prions in transgenic mice (32, 69–71). Small molecules that are active against both mouse and hamster prion strains may be more likely to exhibit activity against a broad range of prion strains from different species. Consequently, it will be interesting to assess whether anti-prion small molecules that are active against mouse prion strains can also reduce the levels of hamster prions in chronically infected CAD5-PrP−/−(HaPrP) cells.
Experimental procedures
Cell lines
Murine neuro2a (N2a) neuroblastoma cells (ATCC no. CCL-131) were cultured in DMEM (Thermo Fisher Scientific, no. 11965) containing 10% (v/v) fetal bovine serum, 1% GlutaMAX, and 0.2× penicillin-streptomycin. N2a-PrP−/− cells (clone 9.21) generated by CRISPR/Cas9-mediated gene editing (44) were cultured in the same medium. Murine CAD5 cells were a generous gift from Charles Weissmann. CAD5 cells, which are a subclone of CAD cells enriched for prion susceptibility (27, 34), and their derivatives were cultured in Opti-MEM medium (Thermo Fisher Scientific) containing 10% fetal bovine serum and 0.2× penicillin-streptomycin. All lines were maintained in a humidified 37 °C/5% CO2 environment. N2a-derived lines were routinely passaged every 3–5 days by trypsinization, whereas CAD5-derived lines were passaged using enzyme-free cell dissociation buffer (EMD Millipore).
Generation of CAD5-PrP−/− cells
CAD-PrP−/− cells were generated by CRISPR/Cas9-mediated gene editing (43) using reagents identical to those that were previously used to create N2a-PrP−/− cells (44). CAD5 cells were plated at a density of 5 × 105 cells/well in 6-well plates and allowed to grow for 1 day. Cells were then transiently transfected with plasmids encoding Streptococcus pyogenes Cas9 and a customized gRNA targeted to the coding exon 3 of Prnp (3:1 ratio by plasmid weight) using Lipofectamine 2000 in Opti-MEM according to the manufacturer's instructions. Following transfection, cells were grown for 2 days before being subjected to limiting dilution subcloning. Transfected cells were incubated in enzyme-free cell dissociation buffer (EMD Millipore) then serially diluted in growth medium to a concentration of 10–100 cells/ml. Diluted preparations were plated onto 10-cm dishes and left to grow for ∼3–4 weeks to obtain monoclonal colonies. Individual clones were expanded and then PrP levels in cell lysates were analyzed by immunoblotting. To isolate CAD5-PrP−/− clones, a second round of CRISPR/Cas9 editing was necessary, which was performed identically to the first round.
To characterize the null alleles in CAD-PrP−/− clones, confluent cells were washed twice with phosphate-buffered saline (PBS) and then lysed with ice-cold lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.5% (w/v) sodium deoxycholate, and 0.5% (v/v) Nonidet P-40 containing cOmplete Protease Inhibitor mixture (Roche)). Lysate was incubated on ice for 20 min with periodic vortexing and then centrifuged at 1000 × g to remove insoluble debris. Genomic DNA was isolated using the QIAamp DNA Mini Kit (Qiagen) and then the Prnp region flanking the gRNA target sites was amplified by PCR using the primers 5′-TCTTTGTGACTATGTGGACTG-3′ and 5′-TGCCACATGCTTGAGGTTGGTT-3′. PCR conditions were set at 94 °C for 5 min followed by 40 cycles of 94 °C for 45 s, 52 °C for 45 s, and 72 °C for 60 s, and finally a 7-min incubation at 72 °C. Amplified DNA was then cloned into pCR4-TOPO vectors and transformed into TOP10 Escherichia coli cells to isolate individual clones. Plasmid DNA Minipreps (Qiagen) were generated and sent for DNA sequence analysis (ACGT Corp, Toronto, Canada).
Generation of stable cell lines
The coding sequences of Syrian hamster or mouse PrP were cloned into the NheI and BamHI sites of the plasmid pIRESneo3 (Clontech). CAD5-PrP−/− or N2a-PrP−/− cells were plated at a density of 5 × 105 cells/well in 6-well plates and then transfected using Lipofectamine 2000 according to the manufacturer's instructions. The next day, cells were passaged into growth medium containing 1 mg/ml G418 and transferred to 10-cm dishes to allow for the selection of stably transfected cells. Cells were selected and expanded over a period of 3–4 weeks at which point the concentration of G418 was lowered to 0.2 mg/ml for routine stable cell maintenance. No clonal selection was performed on any of the stably transfected cell lines.
Cell lysis and immunoblotting
Cells were lysed in lysis buffer, incubated on ice for 20 min, and then centrifuged at 1000 × g to remove insoluble debris. Protein concentrations were determined using the bicinchoninic acid assay. Samples were prepared in 1× LDS loading buffer containing 2.5% (v/v) β-mercaptoethanol, boiled, and then loaded onto 10% Bolt Bis-Tris Plus gels (Thermo Fisher Scientific). Following SDS-PAGE, proteins were transferred to 0.45-micron PVDF membranes, and then membranes were incubated in blocking buffer (5% (w/v) skim milk in Tris-buffered saline containing 0.05% (v/v) Tween 20 (TBST)). Membranes were then incubated with primary antibodies diluted in blocking buffer for 1–2 h at room temperature or overnight at 4 °C. Membranes were washed three times with TBST and then incubated with horseradish peroxidase–conjugated secondary antibodies (Bio-Rad or Thermo Fisher Scientific) diluted in blocking buffer for 1–2 h. Blots were washed three times with TBST and then developed using Western Lightning ECL Pro (PerkinElmer) and exposed to HyBlot CL X-ray film (Denville Scientific). PrP was detected using the recombinant human/mouse chimeric Fabs HuM-D18 (72), HuM-D13 (72), or HuM-P (73), which were a gift from Stanley Prusiner. HaPrP was detected using the antibody 3F4 (BioLegend). For the detection of actin, blots previously probed for PrP were rinsed twice with TBST, incubated with 0.05% (w/v) sodium azide diluted in blocking buffer for 1 h, rinsed five times with TBST, and then reprobed with the actin 20–33 antibody (Sigma Aldrich).
Prion strains
Syrian hamster prion inocula were derived from 10% (w/v) brain homogenates of terminally ill hamsters infected with either the 263K, 139H, HY, or DY strains (54). Brain homogenate from healthy, noninfected hamsters was used as a control. For preparation of inocula containing the RML strain of mouse-adapted scrapie prions, confluent dishes of RML-infected CAD5 cells were scraped into PBS, rinsed in ice-cold PBS, and then homogenized for 30 s using a Minilys bead beater homogenizer (Bertin) and CK14 homogenizing tubes followed by incubation on ice for 5 min. These homogenization steps were repeated an additional three times. Cellular homogenates were then incubated with benzonase (Sigma-Aldrich) at a concentration of 50 units/ml for 30 min at 37 °C with shaking (300 rpm) and then stored at −80 °C. Cell homogenates were plated in tissue culture dishes and incubated for several days to ensure that no viable cells remained. For serial transmission studies, homogenates from cells infected with 263K hamster prions were generated in an identical fashion.
Cellular prion infections
Brain and cellular homogenates were centrifuged at 700 × g for 5 min to remove insoluble debris and then diluted into the appropriate growth medium. For infection of polyclonal pools of stably transfected cells, medium lacking penicillin-streptomycin but containing 0.2 mg/ml G418 was used. Cells were plated in 12-well dishes at a density of 3 × 105 cells/well and allowed to grow for 1 day. Growth medium was then replaced with media containing either 100 μg of cellular homogenate or 0.2% (w/v) brain homogenate. For the serial dilution experiments, prion-containing growth medium (0.2% brain homogenate) was subjected to further 10-fold dilutions in growth medium. Cells were exposed to the inocula for 2 days and were then passaged at a 1:5 dilution every 3–5 days. Cells were passaged a minimum of seven times prior to analysis of prion infection status. Cell lysates were generated in lysis buffer as described above, and then lysates were digested with 50 μg/ml PK for 1 h at 37 °C with 300 rpm shaking, for a PK:protein ratio of 1:50. Digestions were stopped by the addition of PMSF to a final concentration of 4 mm, and then sarkosyl was added to a final concentration of 2% (v/v). Samples were then ultracentrifuged at 100,000 × g for 1 h at 4 °C in a TLA-55 rotor (Beckman Coulter), and pellets were resuspended in 50 μl of 1× LDS loading buffer containing 2.5% (v/v) β-mercaptoethanol and then boiled for 10 min at 95 °C. Digested samples were then analyzed by immunoblotting as described above.
Real-time quaking-induced conversion (RT-QuIC) assays
Recombinant hamster PrP was generated essentially as has been described previously (41). Briefly, a pET41 plasmid encoding Syrian hamster PrP residues 23–231 without a His-tag was transformed into Rosetta2(DE3) E. coli and PrP expression was induced using an overnight autoinduction protocol at 37 °C. Cells were lysed using BugBuster Master Mix followed by the isolation of inclusion bodies, which were solubilized using 8 m guanidine hydrochloride (GdnHCl). Recombinant PrP was purified by binding to nickel-nitrilotriacetic acid Superflow beads (Qiagen) followed by on-column refolding using a 4-h gradient of 6 to 0 m GdnHCl. PrP was eluted in a buffer containing 100 mm sodium phosphate, 10 mm Tris, and 500 mm imidazole, pH 5.8. PrP-containing fractions were pooled and then dialyzed overnight against 10 mm sodium phosphate, pH 5.8. Following filtration through a 0.22-micron membrane and determination of protein concentration by measuring absorbance at 280 nm using a NanoDrop spectrophotometer, recombinant PrP was aliquoted and stored at −80 °C.
The concentration of brain and cellular homogenates was determined using the BCA assay and then homogenates were serially diluted in RT-QuIC dilution buffer (PBS containing 0.1% (w/v) SDS and 1× N-2 Supplement) (Thermo Fisher). For cell lysate RT-QuIC, lysates were diluted 100-fold in dilution buffer. Immediately prior to use, thawed recombinant PrP was centrifuged at 20,000 × g for 10 min at 4 °C to remove any aggregates. The RT-QuIC reaction mixture consisted of PBS, 300 mm NaCl (total concentration including the NaCl present in the PBS), 1 mm EDTA, 0.1 mg/ml recombinant PrP, and 10 μm thioflavin T (ThT). To each well of a black, clear bottom 96-well plate (Nunc), 98 μl of reaction mixture and 2 μl of diluted sample were added. The plate was incubated at 55 °C in a BMG CLARIOstar microplate reader and subjected to alternating cycles of shaking (700 rpm, double orbital) for 1 min and rest for 1 min. ThT fluorescence (excitation: 444 ± 5 nm; emission: 485 ± 5 nm) was measured every 2 min (during the rest periods). A gain setting of 1600 was used. Plates were incubated for a total of 30 h. After this point, control samples would occasionally exhibit increased ThT fluorescence, which we interpreted as spontaneous aggregation of recombinant PrP.
Author contributions
M. E. C. B., G. S.-U., and J. C. W. conceptualization; M. E. C. B. and H. A. data curation; M. E. C. B. and J. C. W. formal analysis; M. E. C. B., H. A., Z. A. M. A.-A., O. H., R. A. S., and M. M. investigation; M. E. C. B., H. A., M. M., and G. S.-U. methodology; M. E. C. B. and J. C. W. writing-original draft; M. E. C. B., H. A., Z. A. M. A.-A., O. H., R. A. S., M. M., G. S.-U., J. C. B., and J. C. W. writing-review and editing; Z. A. M. A.-A., O. H., R. A. S., M. M., G. S.-U., and J. C. B. resources; G. S.-U., J. C. B., and J. C. W. supervision; J. C. W. funding acquisition; J. C. W. project administration.
Acknowledgments
We thank Sukhvir Mahal and Charles Weissmann (Scripps Florida) for providing the CAD5 cells, and Andrew Hughson and Byron Caughey (National Institutes of Health Rocky Mountain Laboratories) for providing the recombinant PrP expression plasmid.
This work was supported by Natural Sciences and Engineering Research Council of Canada Grant RGPIN-2015–05112 (to J. C. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- CJD
- Creutzfeldt-Jakob disease
- CNS
- central nervous system
- PK
- proteinase K
- NBH
- normal brain homogenate
- ThT
- thioflavin T
- gRNA
- guide RNA
- TBST
- Tris-buffered saline with Tween 20
- RT-QuIC
- real-time quaking-induced conversion.
References
- 1. Watts J. C., Balachandran A., and Westaway D. (2006) The expanding universe of prion diseases. PLoS Pathog. 2, e26 10.1371/journal.ppat.0020026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oesch B., Westaway D., Wälchli M., McKinley M. P., Kent S. B. H., Aebersold R., Barry R. A., Tempst P., Teplow D. B., Hood L. E., Prusiner S. B., and Weissmann C. (1985) A cellular gene encodes scrapie PrP 27–30 protein. Cell 40, 735–746 10.1016/0092-8674(85)90333-2 [DOI] [PubMed] [Google Scholar]
- 3. Prusiner S. B. (1982) Novel proteinaceous infectious particles cause scrapie. Science 216, 136–144 10.1126/science.6801762 [DOI] [PubMed] [Google Scholar]
- 4. Colby D. W., and Prusiner S. B. (2011) Prions. Cold Spring Harb Perspect Biol. 3, a006833 10.1101/cshperspect.a006833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Collinge J. (2016) Mammalian prions and their wider relevance in neurodegenerative diseases. Nature 539, 217–226 10.1038/nature20415 [DOI] [PubMed] [Google Scholar]
- 6. Stahl N., Borchelt D. R., Hsiao K., and Prusiner S. B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240 10.1016/0092-8674(87)90150-4 [DOI] [PubMed] [Google Scholar]
- 7. Watts J. C., Bourkas M. E. C., and Arshad H. (2018) The function of the cellular prion protein in health and disease. Acta Neuropathol. 135, 159–178 10.1007/s00401-017-1790-y [DOI] [PubMed] [Google Scholar]
- 8. Bremer J., Baumann F., Tiberi C., Wessig C., Fischer H., Schwarz P., Steele A. D., Toyka K. V., Nave K. A., Weis J., and Aguzzi A. (2010) Axonal prion protein is required for peripheral myelin maintenance. Nat. Neurosci. 13, 310–319 10.1038/nn.2483 [DOI] [PubMed] [Google Scholar]
- 9. Küffer A., Lakkaraju A. K., Mogha A., Petersen S. C., Airich K., Doucerain C., Marpakwar R., Bakirci P., Senatore A., Monnard A., Schiavi C., Nuvolone M., Grosshans B., Hornemann S., Bassilana F., Monk K. R., and Aguzzi A. (2016) The prion protein is an agonistic ligand of the G protein-coupled receptor Adgrg6. Nature 536, 464–468 10.1038/nature19312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Riek R., Hornemann S., Wider G., Billeter M., Glockshuber R., and Wüthrich K. (1996) NMR structure of the mouse prion protein domain PrP(121–231). Nature 382, 180–182 10.1038/382180a0 [DOI] [PubMed] [Google Scholar]
- 11. Pan K.-M., Baldwin M., Nguyen J., Gasset M., Serban A., Groth D., Mehlhorn I., Huang Z., Fletterick R. J., Cohen F. E., and Prusiner S. B. (1993) Conversion of α-helices into β-sheets features in the formation of the scrapie prion proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 10962–10966 10.1073/pnas.90.23.10962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pattison I. H. (1965) Experiments with scrapie with special reference to the nature of the agent and the pathology of the disease. in Slow, Latent and Temperate Virus Infections, NINDB Monograph 2 (Gajdusek D. C., Gibbs C. J. Jr., and Alpers M. P., eds) pp 249–257, U.S. Government Printing, Washington, D.C. [Google Scholar]
- 13. Scott M., Foster D., Mirenda C., Serban D., Coufal F., Wälchli M., Torchia M., Groth D., Carlson G., DeArmond S. J., Westaway D., and Prusiner S. B. (1989) Transgenic mice expressing hamster prion protein produce species-specific scrapie infectivity and amyloid plaques. Cell 59, 847–857 10.1016/0092-8674(89)90608-9 [DOI] [PubMed] [Google Scholar]
- 14. Prusiner S. B., Scott M., Foster D., Pan K.-M., Groth D., Mirenda C., Torchia M., Yang S.-L., Serban D., Carlson G. A., Hoppe P. C., Westaway D., and DeArmond S. J. (1990) Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63, 673–686 10.1016/0092-8674(90)90134-Z [DOI] [PubMed] [Google Scholar]
- 15. Telling G. C., Scott M., Hsiao K. K., Foster D., Yang S.-L., Torchia M., Sidle K. C. L., Collinge J., DeArmond S. J., and Prusiner S. B. (1994) Transmission of Creutzfeldt-Jakob disease from humans to transgenic mice expressing chimeric human-mouse prion protein. Proc. Natl. Acad. Sci. U.S.A. 91, 9936–9940 10.1073/pnas.91.21.9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Telling G. C., Scott M., Mastrianni J., Gabizon R., Torchia M., Cohen F. E., DeArmond S. J., and Prusiner S. B. (1995) Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein. Cell 83, 79–90 10.1016/0092-8674(95)90236-8 [DOI] [PubMed] [Google Scholar]
- 17. Telling G. C., Parchi P., DeArmond S. J., Cortelli P., Montagna P., Gabizon R., Mastrianni J., Lugaresi E., Gambetti P., and Prusiner S. B. (1996) Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science 274, 2079–2082 10.1126/science.274.5295.2079 [DOI] [PubMed] [Google Scholar]
- 18. Bessen R. A., and Marsh R. F. (1994) Distinct PrP properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 68, 7859–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collinge J., and Clarke A. R. (2007) A general model of prion strains and their pathogenicity. Science 318, 930–936 10.1126/science.1138718 [DOI] [PubMed] [Google Scholar]
- 20. Watts J. C., and Prusiner S. B. (2014) Mouse models for studying the formation and propagation of prions. J. Biol. Chem. 289, 19841–19849 10.1074/jbc.R114.550707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Butler D. A., Scott M. R., Bockman J. M., Borchelt D. R., Taraboulos A., Hsiao K. K., Kingsbury D. T., and Prusiner S. B. (1988) Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol. 62, 1558–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schätzl H. M., Laszlo L., Holtzman D. M., Tatzelt J., DeArmond S. J., Weiner R. I., Mobley W. C., and Prusiner S. B. (1997) A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol. 71, 8821–8831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nishida N., Harris D. A., Vilette D., Laude H., Frobert Y., Grassi J., Casanova D., Milhavet O., and Lehmann S. (2000) Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 74, 320–325 10.1128/JVI.74.1.320-325.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Birkett C. R., Hennion R. M., Bembridge D. A., Clarke M. C., Chree A., Bruce M. E., and Bostock C. J. (2001) Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J. 20, 3351–3358 10.1093/emboj/20.13.3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vorberg I., Raines A., Story B., and Priola S. A. (2004) Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J. Infect. Dis. 189, 431–439 10.1086/381166 [DOI] [PubMed] [Google Scholar]
- 26. Baron G. S., Magalhães A. C., Prado M. A., and Caughey B. (2006) Mouse-adapted scrapie infection of SN56 cells: Greater efficiency with microsome-associated versus purified PrP-res. J. Virol. 80, 2106–2117 10.1128/JVI.80.5.2106-2117.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mahal S. P., Baker C. A., Demczyk C. A., Smith E. W., Julius C., and Weissmann C. (2007) Prion strain discrimination in cell culture: the cell panel assay. Proc. Natl. Acad. Sci. U.S.A. 104, 20908–20913 10.1073/pnas.0710054104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herbst A., Banser P., Velasquez C. D., Mays C. E., Sim V. L., Westaway D., Aiken J. M., and McKenzie D. (2013) Infectious prions accumulate to high levels in non proliferative C2C12 myotubes. PLoS Pathog. 9, e1003755 10.1371/journal.ppat.1003755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vilette D. (2008) Cell models of prion infection. Vet. Res. 39, 10 10.1051/vetres:2007049 [DOI] [PubMed] [Google Scholar]
- 30. Grassmann A., Wolf H., Hofmann J., Graham J., and Vorberg I. (2013) Cellular aspects of prion replication in vitro. Viruses 5, 374–405 10.3390/v5010374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oelschlegel A. M., Fallahi M., Ortiz-Umpierre S., and Weissmann C. (2012) The extended cell panel assay characterizes the relationship of prion strains RML, 79A, and 139A and reveals conversion of 139A to 79A-like prions in cell culture. J. Virol. 86, 5297–5303 10.1128/JVI.00181-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berry D. B., Lu D., Geva M., Watts J. C., Bhardwaj S., Oehler A., Renslo A. R., DeArmond S. J., Prusiner S. B., and Giles K. (2013) Drug resistance confounding prion therapeutics. Proc. Natl. Acad. Sci. U.S.A. 110, E4160–E4169 10.1073/pnas.1317164110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suri C., Fung B. P., Tischler A. S., and Chikaraishi D. M. (1993) Catecholaminergic cell lines from the brain and adrenal glands of tyrosine hydroxylase-SV40 T antigen transgenic mice. J. Neurosci. 13, 1280–1291 10.1523/JNEUROSCI.13-03-01280.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qi Y., Wang J. K., McMillian M., and Chikaraishi D. M. (1997) Characterization of a CNS cell line, CAD, in which morphological differentiation is initiated by serum deprivation. J. Neurosci. 17, 1217–1225 10.1523/JNEUROSCI.17-04-01217.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Courageot M. P., Daude N., Nonno R., Paquet S., Di Bari M. A., Le Dur A., Chapuis J., Hill A. F., Agrimi U., Laude H., and Vilette D. (2008) A cell line infectible by prion strains from different species. J. Gen. Virol. 89, 341–347 10.1099/vir.0.83344-0 [DOI] [PubMed] [Google Scholar]
- 36. Maas E., Geissen M., Groschup M. H., Rost R., Onodera T., Schätzl H., and Vorberg I. M. (2007) Scrapie infection of prion protein-deficient cell line upon ectopic expression of mutant prion proteins. J. Biol. Chem. 282, 18702–18710 10.1074/jbc.M701309200 [DOI] [PubMed] [Google Scholar]
- 37. Vilette D., Andreoletti O., Archer F., Madelaine M. F., Vilotte J. L., Lehmann S., and Laude H. (2001) Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc. Natl. Acad. Sci. U.S.A. 98, 4055–4059 10.1073/pnas.061337998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bian J., Napier D., Khaychuck V., Angers R., Graham C., and Telling G. (2010) Cell-based quantification of chronic wasting disease prions. J. Virol. 84, 8322–8326 10.1128/JVI.00633-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lawson V. A., Vella L. J., Stewart J. D., Sharples R. A., Klemm H., Machalek D. M., Masters C. L., Cappai R., Collins S. J., and Hill A. F. (2008) Mouse-adapted sporadic human Creutzfeldt-Jakob disease prions propagate in cell culture. Int. J. Biochem. Cell Biol. 40, 2793–2801 10.1016/j.biocel.2008.05.024 [DOI] [PubMed] [Google Scholar]
- 40. Saborio G. P., Permanne B., and Soto C. (2001) Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813 10.1038/35081095 [DOI] [PubMed] [Google Scholar]
- 41. Wilham J. M., Orrú C. D., Bessen R. A., Atarashi R., Sano K., Race B., Meade-White K. D., Taubner L. M., Timmes A., and Caughey B. (2010) Rapid end-point quantitation of prion seeding activity with sensitivity comparable to bioassays. PLoS Pathog. 6, e1001217 10.1371/journal.ppat.1001217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bessen R. A., and Marsh R. F. (1992) Identification of two biologically distinct strains of transmissible mink encephalopathy in hamsters. J. Gen. Virol. 73, 329–334 10.1099/0022-1317-73-2-329 [DOI] [PubMed] [Google Scholar]
- 43. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehrabian M., Brethour D., MacIsaac S., Kim J. K., Gunawardana C. G., Wang H., and Schmitt-Ulms G. (2014) CRISPR-Cas9-based knockout of the prion protein and its effect on the proteome. PLoS One 9, e114594 10.1371/journal.pone.0114594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ghodrati F., Mehrabian M., Williams D., Halgas O., Bourkas M. E. C., Watts J. C., Pai E. F., and Schmitt-Ulms G. (2018) The prion protein is embedded in a molecular environment that modulates transforming growth factor β and integrin signaling. Sci. Rep. 8, 8654 10.1038/s41598-018-26685-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bardelli M., Frontzek K., Simonelli L., Hornemann S., Pedotti M., Mazzola F., Carta M., Eckhardt V., D'Antuono R., Virgilio T., González S. F., Aguzzi A., and Varani L. (2018) A bispecific immunotweezer prevents soluble PrP oligomers and abolishes prion toxicity. PLoS Pathog. 14, e1007335 10.1371/journal.ppat.1007335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kimberlin R. H., and Walker C. (1977) Characteristics of a short incubation model of scrapie in the golden hamster. J. Gen. Virol. 34, 295–304 10.1099/0022-1317-34-2-295 [DOI] [PubMed] [Google Scholar]
- 48. Orru C. D., Hughson A. G., Groveman B. R., Campbell K. J., Anson K. J., Manca M., Kraus A., and Caughey B. (2016) Factors that improve RT-QuIC detection of prion seeding activity. Viruses 8, E140 10.3390/v8050140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kimberlin R. H., Walker C. A., and Fraser H. (1989) The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J. Gen. Virol. 70, 2017–2025 10.1099/0022-1317-70-8-2017 [DOI] [PubMed] [Google Scholar]
- 50. Taraboulos A., Serban D., and Prusiner S. B. (1990) Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol. 110, 2117–2132 10.1083/jcb.110.6.2117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Blochberger T. C., Cooper C., Peretz D., Tatzelt J., Griffith O. H., Baldwin M. A., and Prusiner S. B. (1997) Prion protein expression in Chinese hamster ovary cells using a glutamine synthetase selection and amplification system. Protein Eng. 10, 1465–1473 10.1093/protein/10.12.1465 [DOI] [PubMed] [Google Scholar]
- 52. Kristensson K., Feuerstein B., Taraboulos A., Hyun W. C., Prusiner S. B., and DeArmond S. J. (1993) Scrapie prions alter receptor-mediated calcium responses in cultured cells. Neurology 43, 2335–2341 10.1212/WNL.43.11.2335 [DOI] [PubMed] [Google Scholar]
- 53. Cronier S., Beringue V., Bellon A., Peyrin J. M., and Laude H. (2007) Prion strain- and species-dependent effects of antiprion molecules in primary neuronal cultures. J. Virol. 81, 13794–13800 10.1128/JVI.01502-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ayers J. I., Schutt C. R., Shikiya R. A., Aguzzi A., Kincaid A. E., and Bartz J. C. (2011) The strain-encoded relationship between PrP replication, stability and processing in neurons is predictive of the incubation period of disease. PLoS Pathog. 7, e1001317 10.1371/journal.ppat.1001317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ghaemmaghami S., Phuan P. W., Perkins B., Ullman J., May B. C., Cohen F. E., and Prusiner S. B. (2007) Cell division modulates prion accumulation in cultured cells. Proc. Natl. Acad. Sci. U.S.A. 104, 17971–17976 10.1073/pnas.0708372104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bessen R. A., and Marsh R. F. (1992) Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J. Virol. 66, 2096–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaneko K., Zulianello L., Scott M., Cooper C. M., Wallace A. C., James T. L., Cohen F. E., and Prusiner S. B. (1997) Evidence for protein X binding to a discontinuous epitope on the cellular prion protein during scrapie prion propagation. Proc. Natl. Acad. Sci. U.S.A. 94, 10069–10074 10.1073/pnas.94.19.10069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Deleault N. R., Lucassen R. W., and Supattapone S. (2003) RNA molecules stimulate prion protein conversion. Nature 425, 717–720 10.1038/nature01979 [DOI] [PubMed] [Google Scholar]
- 59. Deleault N. R., Piro J. R., Walsh D. J., Wang F., Ma J., Geoghegan J. C., and Supattapone S. (2012) Isolation of phosphatidylethanolamine as a solitary cofactor for prion formation in the absence of nucleic acids. Proc. Natl. Acad. Sci. U.S.A. 109, 8546–8551 10.1073/pnas.1204498109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tamgüney G., Giles K., Glidden D. V., Lessard P., Wille H., Tremblay P., Groth D. F., Yehiely F., Korth C., Moore R. C., Tatzelt J., Rubinstein E., Boucheix C., Yang X., Stanley P., et al. (2008) Genes contributing to prion pathogenesis. J. Gen. Virol. 89, 1777–1788 10.1099/vir.0.2008/001255-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Geoghegan J. C., Miller M. B., Kwak A. H., Harris B. T., and Supattapone S. (2009) Trans-dominant inhibition of prion propagation in vitro is not mediated by an accessory cofactor. PLoS Pathog. 5, e1000535 10.1371/journal.ppat.1000535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Klöhn P. C., Stoltze L., Flechsig E., Enari M., and Weissmann C. (2003) A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc. Natl. Acad. Sci. U.S.A. 100, 11666–11671 10.1073/pnas.1834432100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marbiah M. M., Harvey A., West B. T., Louzolo A., Banerjee P., Alden J., Grigoriadis A., Hummerich H., Kan H. M., Cai Y., Bloom G. S., Jat P., Collinge J., and Klöhn P. C. (2014) Identification of a gene regulatory network associated with prion replication. EMBO J. 33, 1527–1547 10.15252/embj.201387150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Herva M. E., and Weissman C. (2012) Cell-specific susceptibility to prion strains is a property of the intact cell. Prion 6, 371–374 10.4161/pri.20198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shikiya R. A., Langenfeld K. A., Eckland T. E., Trinh J., Holec S. A. M., Mathiason C. K., Kincaid A. E., and Bartz J. C. (2017) PrPSc formation and clearance as determinants of prion tropism. PLoS Pathog. 13, e1006298 10.1371/journal.ppat.1006298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mays C. E., Yeom J., Kang H. E., Bian J., Khaychuk V., Kim Y., Bartz J. C., Telling G. C., and Ryou C. (2011) In vitro amplification of misfolded prion protein using lysate of cultured cells. PLoS One 6, e18047 10.1371/journal.pone.0018047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Moudjou M., Chapuis J., Mekrouti M., Reine F., Herzog L., Sibille P., Laude H., Vilette D., Andréoletti O., Rezaei H., Dron M., and Béringue V. (2016) Glycoform-independent prion conversion by highly efficient, cell-based, protein misfolding cyclic amplification. Sci. Rep. 6, 29116 10.1038/srep29116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Krejciova Z., Alibhai J., Zhao C., Krencik R., Rzechorzek N. M., Ullian E. M., Manson J., Ironside J. W., Head M. W., and Chandran S. (2017) Human stem cell-derived astrocytes replicate human prions in a PRNP genotype-dependent manner. J. Exp. Med. 214, 3481–3495 10.1084/jem.20161547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Giles K., Berry D. B., Condello C., Dugger B. N., Li Z., Oehler A., Bhardwaj S., Elepano M., Guan S., Silber B. M., Olson S. H., and Prusiner S. B. (2016) Optimization of aryl amides that extend survival in prion-infected mice. J. Pharmacol. Exp. Ther. 358, 537–547 10.1124/jpet.116.235556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Giles K., Berry D. B., Condello C., Hawley R. C., Gallardo-Godoy A., Bryant C., Oehler A., Elepano M., Bhardwaj S., Patel S., Silber B. M., Guan S., DeArmond S. J., Renslo A. R., and Prusiner S. B. (2015) Different 2-aminothiazole therapeutics produce distinct patterns of scrapie prion neuropathology in mouse brains. J. Pharmacol. Exp. Ther. 355, 2–12 10.1124/jpet.115.224659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lu D., Giles K., Li Z., Rao S., Dolghih E., Gever J. R., Geva M., Elepano M. L., Oehler A., Bryant C., Renslo A. R., Jacobson M. P., DeArmond S. J., Silber B. M., and Prusiner S. B. (2013) Biaryl amides and hydrazones as therapeutics for prion disease in transgenic mice. J. Pharmacol. Exp. Ther. 347, 325–338 10.1124/jpet.113.205799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., and Burton D. R. (1998) Mapping the prion protein using recombinant antibodies. J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Safar J. G., Scott M., Monaghan J., Deering C., Didorenko S., Vergara J., Ball H., Legname G., Leclerc E., Solforosi L., Serban H., Groth D., Burton D. R., Prusiner S. B., and Williamson R. A. (2002) Measuring prions causing bovine spongiform encephalopathy or chronic wasting disease by immunoassays and transgenic mice. Nat. Biotechnol. 20, 1147–1150 10.1038/nbt748 [DOI] [PubMed] [Google Scholar]
- 74. Altmeppen H. C., Puig B., Dohler F., Thurm D. K., Falker C., Krasemann S., and Glatzel M. (2012) Proteolytic processing of the prion protein in health and disease. Am. J. Neurodegener. Dis. 1, 15–31 [PMC free article] [PubMed] [Google Scholar]