Abstract
TraB is an FtsK-like DNA translocase responsible for conjugative plasmid transfer in mycelial Streptomyces. Unlike other conjugative systems, which depend on a type IV secretion system, Streptomyces requires only TraB protein to transfer the plasmid as dsDNA. The γ-domain of this protein specifically binds to repeated 8-bp motifs on the plasmid sequence, following a mechanism that is reminiscent of the FtsK/SpoIIIE chromosome segregation system. In this work, we purified and characterized the enzymatic activity of TraB, revealing that it is a DNA-dependent ATPase that is highly stimulated by dsDNA substrates. Interestingly, we found that unlike the SpoIIIE protein, the γ-domain of TraB does not confer sequence-specific ATPase stimulation. We also found that TraB binds G-quadruplex DNA structures with higher affinity than TraB-recognition sequences (TRSs). An EM-based structural analysis revealed that TraB tends to assemble as large complexes comprising four TraB hexamers, which might be a prerequisite for DNA translocation across cell membranes. In summary, our findings shed light on the molecular mechanism used by the DNA-translocating motor TraB, which may be shared by other membrane-associated machineries involved in DNA binding and translocation.
Keywords: DNA binding protein, bacterial conjugation, ATPase, G-quadruplex, antibiotics, DNA segregation, DNA translocation, DNA-dependent ATPase, FtsK-like ATPase, Streptomyces, DNA translocase, conjugal plasmid transfer, motor protein, TRS sequence
Introduction
Most membrane-associated motors involved in macromolecular transport across bacterial cell membranes belong to the superfamily of hexameric P-loop ATPases (1). These motors are able to couple the chemical energy provided by ATP hydrolysis to the transport of DNA and/or protein effectors through biological membranes. The FtsK/SpoIIIE family of translocases include proteins involved in the transfer of the bacterial chromosome between spatially separated compartments and are the most representative members of this family (2). FtsK and SpoIIIE proteins both play an important role during chromosomal segregation in bacteria, in cell division and sporulation processes, respectively. They are membrane-anchored proteins that have an N-terminal domain with several transmembrane helices (3); a motor domain, common to all RecA-like hexameric ATPases; and a C-terminal γ-domain that confers specificity in DNA binding and dictates the directionality of DNA transport. The γ-domain recognizes specific 8-bp DNA motifs, named KOPS for FtsK (FtsK orienting/polarizing sequence) and SRS (SpoIIIE recognition sequence) for SpoIIIE (4, 5).
A close homolog of these proteins involved in the conjugative transfer of plasmids in Streptomyces was recently discovered (6). This FtsK-like homolog, TraB, mediates the transfer of the plasmid as dsDNA (7). The protein directs plasmid transfer by binding to a specific plasmid region named cis-acting locus of transfer (clt) (8). The clt regions of different plasmids contain direct 8-bp repeats, termed TraB-recognition sequence (TRS),3 which is recognized by the γ-domain of its corresponding TraB protein (6, 9). The predicted structure, domain organization, and DNA-binding characteristics of TraB suggest that the TraB conjugation system is derived from an FtsK-like ancestor (6).
This conjugative mechanism is different from other plasmid-encoded conjugation complexes that translocate DNA through a type IV secretion system (T4SS). Conjugative T4SS consists of at least 12 proteins involved in the processing of the plasmid, the formation of a core channel complex, the assembly of a pilus for cell-to-cell contacts, and also in supplying energy for pilus biogenesis and substrate transport (10, 11). One of these proteins, known as coupling protein because it connects the DNA processing machinery to the secretion channel, is an ATPase that belongs to the FtsK-like family of proteins. The prototype for the T4SS coupling proteins is TrwB, from plasmid R388. TrwB is a hexameric DNA-dependent ATPase implicated in the transfer of the plasmid as an ssDNA molecule (12). The diameter of the central pore in TrwB is 20 Å, which is large enough to accommodate ssDNA. In contrast, the FtsK pore is 30 Å wide, which allows for the passage of dsDNA (13, 14). Contrary to FtsK-like motors, TrwB-like proteins do not contain a γ-domain involved in the specific recognition of a plasmid sequence. Instead, substrate binding is mediated by the interaction with accessory proteins (15), and also by recognizing G-quadruplex structures on the DNA with high affinity (16). These secondary DNA structures have been proposed to act as loading sites for the motor. G-quadruplex structures are formed in G-rich DNA sequences by the pairing of four guanines in a planar array (17). These structural motifs are widespread in genomes and act as control elements in essential biological processes, such as replication or transcription (18). Interestingly, KOPS, SRS, and TRS sequences are also G-rich sequences and, therefore, might form G-quadruplex structures that could act as loading sites.
In this work, we have observed that TraB from plasmid pSVH1 binds G-quadruplex structures with higher affinity than TRS sequences. The γ-domain of TraB is not required for the recognition of these secondary structures. Therefore, the motor domain, common to TrwB-like proteins, is responsible for this DNA-binding mode. Comparative genomic analysis of the motor domain of FtsK-, SpoIIIE-, TraB-, and TrwB-like proteins revealed that these proteins are closely related, sharing an evolutionary common ancestor (19). It is tempting to speculate that they also share a similar mechanism of loading, recognizing secondary DNA structures to establish the first DNA contacts. We have also characterized the enzymatic activity of the protein, showing that TraB is a DNA-dependent ATPase, highly stimulated by dsDNA substrates. The structural analysis of the protein by EM showed the existence of high-order oligomeric complexes that might be formed by the assembly of four TraB hexamers. Although at present is not possible to know if the formation of these high-order oligomeric structures is relevant for TraB function in vivo, all together, these results shed light on the mechanism used by this family of membrane-associated motors for DNA binding and translocation.
Results
TraBs exhibits DNA-dependent ATPase activity
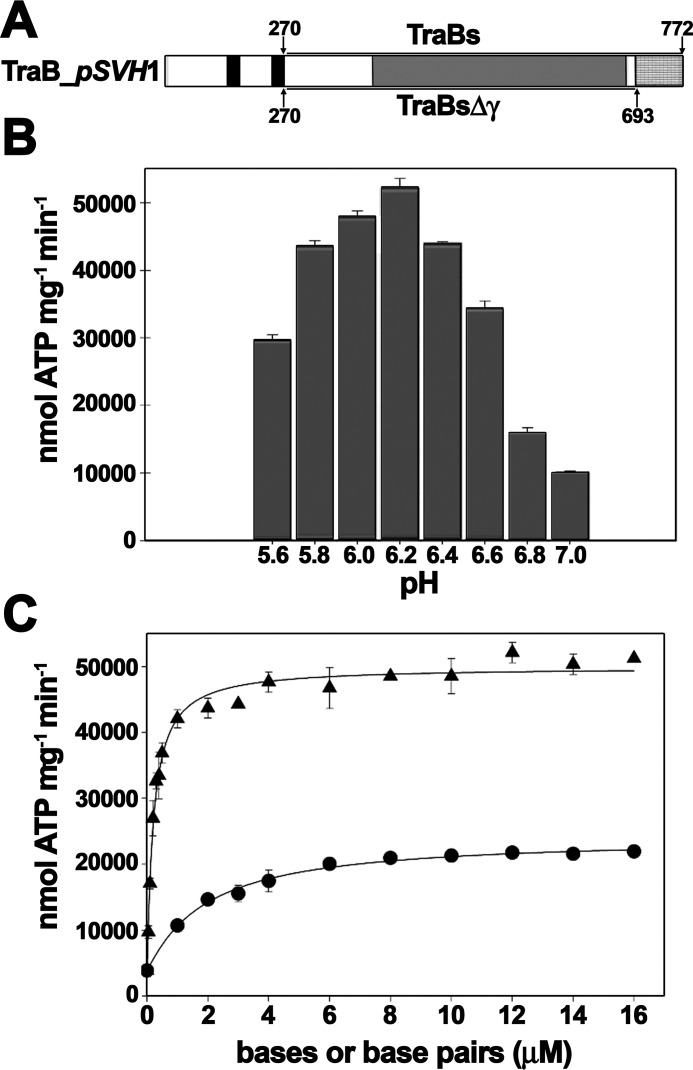
The soluble domain of TraB protein (TraBs), attached to a His-tag at the N terminus (Fig. 1A), was purified to homogeneity and analyzed for ATPase activity in the presence and absence of different DNA substrates under various conditions. The activity was very sensitive to pH, with an optimal range between 5.8 and 6.4 (Fig. 1B). In the absence of DNA, the activity of the protein was ∼2000 nmol ATP min−1 mg−1. ATP hydrolysis was stimulated 35-fold in the presence of dsDNA (Vmax = 72,950 ± 2360 nmol ATP min−1 mg−1), whereas ssDNA caused a 12-fold stimulation (Vmax = 24,870 ± 2380 nmol ATP min−1 mg−1) (Fig. 1C). These values were obtained with DNA substrates lacking the specific TRS recognition sequence.
Figure 1.
Analysis of TraBs ATPase activity. A, schematic representation of Streptomyces TraB of plasmid pSVH1. Black bars represent the N-terminal transmembrane helices, as predicted with TMHMM (www.cbs.dtu.dk/services/TMHMM-2.0).4 The motor domain is depicted in gray and the C-terminal γ-domain in gray squares. TraBs consists of a deletion of the first 270 residues that contain the transmembrane helices, whereas TraBsΔγ also contains a deletion of the C-terminal γ-domain. B, pH-dependence of TraBs ATPase activity. ATP turnover was measured using a coupled spectrophotometric assay. Reactions contained TraBs (50 nm), 5 mm ATP, and 45 bp dsDNA (250 nm), buffered at the corresponding pH value. C, effect of ssDNA and dsDNA on TraBs ATPase activity. Reactions were carried out at increasing concentrations of ssDNA or dsDNA substrates, represented as micromolar concentrations of bases (for ssDNA) or bp (for dsDNA). Black triangles, TraBs + dsDNA, formed by the annealing of 45-mer nucleotides A and B; black dots, TraBs + ssDNA (oligonucleotide A). Data were fit to a Hill equation and represent the average of at least three different experiments. Error bars: S.D.
It is important to note that these values were much higher than previously reported data for a full-length strepII-TraB fusion protein (Vmax = 800 nmol ATP min−1 mg−1) (8). Moreover, in that previous work the described ATPase activity was not DNA-dependent. The ATPase assays were performed at pH 8, which is not an optimal pH value for measuring this activity. As shown in Fig. 1B, TraBs activity shows a high dependence on low pH values. Because in the previous report a full-length strepII-TraB protein was used, we cannot exclude the possibility that the N-terminal domain had an effect on the DNA-dependent ATPase activity. More likely, the pH dependence observed here might explain the absence of DNA-dependent ATPase activity reported in those previous experiments.
TraB γ-domain does not confer sequence-specific ATPase stimulation
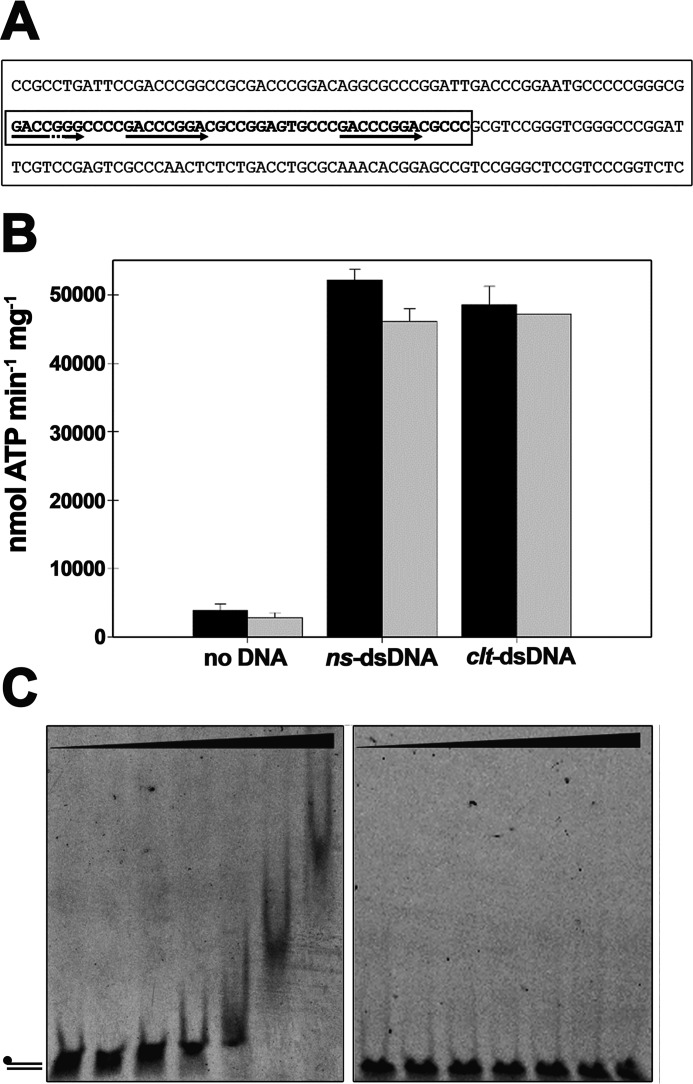
TraB has been reported to have a specific DNA-binding activity, recognizing 8-bp TRS motifs within the clt region of the pSVH1 plasmid (6). This mode of interaction with the DNA is reminiscent of the 8-bp KOPS and SRS motifs recognized by FtsK or SpoIIIE, respectively. In these proteins, the interaction with DNA motifs involves the γ-domain of the protein (5, 20). In an attempt to determine whether the recognition of TRS motifs affected the activity of TraBs, ATP turnover was analyzed using complementary 45-mer oligonucleotides containing part of the clt sequence of the pSVH1 plasmid, which includes three copies of the 8-bp TRS sequence (Fig. 2A) (oligonucleotides C and D from Table 1). As shown in Fig. 2B (black bars), no significant differences were found in ATP turnover when compared with nonspecific complementary 45-mer oligonucleotides (oligonucleotides A and B from Table 1). Independently of the presence or absence of the specific clt sequence and TRS repeats, TraBs basal ATPase activity was always increased by 30-fold.
Figure 2.
TraB does not show sequence-specific ATPase stimulation. A, nucleotide sequence of the TraB-binding region of plasmid pSVH1. 45-base clt sequence is within a box, where the two perfect and one imperfect TRS sequences are marked with arrows. B, the ATP turnover of TraBs (black) and TraBsΔγ (gray) was measured in the absence of any DNA substrate, in the presence of nonspecific dsDNA (ns-dsDNA), formed by the annealing of 45-mer nucleotides A and B from Table 1, and in the presence of dsDNA with the clt sequence (clt-dsDNA), formed by the annealing of 45-mer nucleotides C and D from Table 1. Reactions contained either TraBs or TraBsΔγ (50 nm), 5 mm ATP, and 260 nm DNA substrate. Data represent the average of three different experiments. C, gel mobility shift assays. dsDNA with the clt sequence (5 nm) was formed as above, but with 5′-fluorescein–labeled oligonucleotide C (left panel). The mobility shift with nonspecific DNA (oligonucleotides A and B) is shown on the right panel. DNA was incubated with increasing concentrations of TraBs or TraBsΔγ proteins at 0, 0.05, 0.2, 0.5, 1, 2, and 5 μm, as described under “Experimental procedures.” Error bars: S.D.
Table 1.
Sequence of oligonucleotides used for substrate preparation
| Oligo | Sequence (5′–3′) |
|---|---|
| A | AAGGACGAAAACCTGTGTAGTGTTATGCCACTACAATATTGCCGC |
| B | GCGGCAATATTGTAGTGGCATAACACTACACAGGTTTTCGTCCTT |
| Ca | GACCGGGCCCCGACCCGGACGCCGGAGTGCCCGACCCGGACGCCC |
| Db | GGGCGTCCGGGTCGGGCACTCCGGCGTCCGGGTCGGGGCCCGGTC |
| Ea | ATGCCACTACAATATTGCCGCGACCCGGAGACCCGGAGACCCGGA |
| F | TCCGGGTCTCCGGGTCTCCGGGTCGCGGCAATATTGTAGTGGCAT |
| G | ATGCCACTACAATATTGCCGCTCCGGGTCTCCGGGTCTCCGGGTC |
| Ha | GACCCGGAGACCCGGAGACCCGGAGCGGCAATATTGTAGTGGCAT |
| Ib | TCGCCACGTTTCGCCGTTTGCGGGGGTTTCTGCGAGGAACTTTGG |
a TRS sequences are underlined.
b G-clusters are indicated by bold letters.
Next, we analyzed the effect of the γ-domain on the ATPase activity of the protein. Previous data obtained with SpoIIIE protein indicated that the γ-domain was inhibiting the ATPase activity of the protein in the absence of DNA (21). Therefore, we purified TraBsΔγ (Fig. 1A and Fig. S1) and the activity of the two proteins was compared. Contrary to what was observed with SpoIIIE, TraBs and TraBsΔγ presented a similar ATP turnover (black and gray bars in Fig. 2B), both in the absence and in the presence of the two types of dsDNA substrates. Although the removal of the γ-domain did not affect the ATPase activity of the protein, previous studies had shown that the TraB γ-domain binds preferentially to TRS sequences over random DNA (6). Therefore, we performed electrophoretic mobility shift assays (EMSA) with the same 45-bp dsDNA fragments that were used in the ATPase assays, with and without the specific clt sequence. Fig. 2C shows that only the dsDNA fragment containing the specific sequence is retarded, with an apparent Kd value of 0.9 μm. Deletion of the γ-domain results in a DNA-binding defect, as previously reported (6).
Stimulation of TraBs ATPase activity is independent of the presence or orientation of the TRS motifs
KOPS and SRS motifs are involved in directing motion during chromosome segregation of FtsK and SpoIIIE translocases, respectively (5, 22). Both translocases move toward the terminus and, therefore, the motors can find these asymmetric motifs in two different orientations: permissive orientation (pointing to the terminus) and nonpermissive orientation (pointing to the origin). At present, there are different models that try to explain how the motors deal with these motifs (20, 23–25). A recent work on SpoIIIE has shown that permissive SRS sequences increase the ATPase activity more than 20-fold above the values obtained with random DNA or nonpermissive SRS (21).
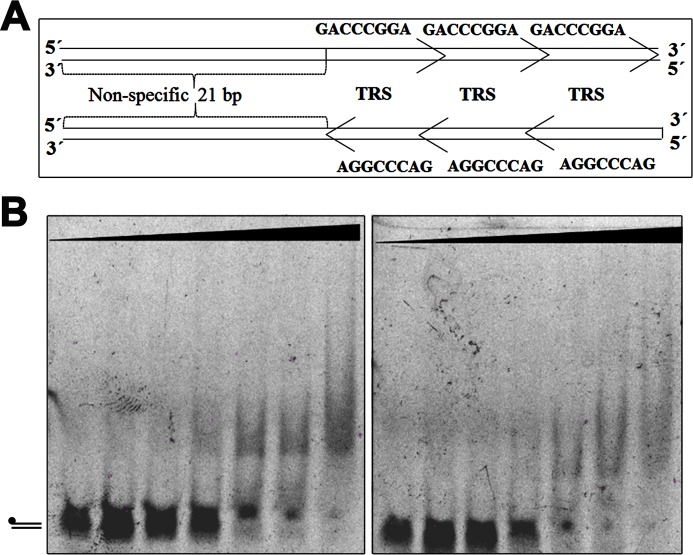
In an attempt to determine whether the stimulation of TraB ATPase activity could be linked to the sensing of the TRS motif in a particular orientation or to the existence of sufficient DNA for a correct loading, we analyzed the turnover of the protein in the presence of two types of synthetic dsDNA. These new dsDNA fragments were long enough to accommodate the assembly of the protein in any orientation and were designed in the same way as those used in the work with SpoIIIE (21). The substrates contained either three overlapping TRS sequences in a “permissive” orientation (3 × TRS) (oligonucleotides E and F from Table 1), or three overlapping TRS sequences in the reverse orientation (3 × revTRS) (oligonucleotides G and H from Table 1) (Fig. 3A). However, it is important to note that the termini “permissive” and “nonpermissive” are not related to the origin of the plasmid but to the two possible orientations that the sequences can acquire in the plasmid. As shown in Fig. 3B, the ATPase activity of the protein in the presence of the three overlapping TRS sequences, in any orientation, was similar to that obtained with random DNA. The values obtained for TraBs and TraBsΔγ proteins were very similar with all the dsDNA substrates tested. Therefore, we can conclude that the ATPase activity of TraBs is independent of the TRS sequence and its orientation.
Figure 3.
TraBs does not require a specific orientation of the TRS motifs for DNA binding. A, schematic representation of the two dsDNA substrates used in the assay (3 × TRS, and 3 × reverse TRS). B, native EMSAs using overlapping TRS sequences in the two different orientations. Substrates (dsDNA) were formed by the annealing of oligonucleotides E to F (left panel) and G to H (right panel), respectively. DNA samples (5 nm) were incubated with increasing concentrations of TraBs protein at 0, 0.05, 0.2, 0.5, 1, 2, and 5 μm, as described under “Experimental procedures.”
To analyze the binding preferences of TraB, we performed EMSA with the two types of substrates. The affinity was similar for both substrates (Kd values of 1.8 and 2.2 μm, respectively), but lower than that obtained for the substrate that contains three TRS sequences together with part of the clt sequence (Kd = 0.9 μm). This result might indicate that the affinity of TraBs for the clt sequence is not only determined by TRS motifs and it might need a larger DNA fragment.
TraBs binds to G-quadruplex structures with higher affinity than to specific TRS sequences
Streptomyces species and their plasmids have a high G + C content in their DNA. More specifically, the GC content of the clt region of plasmid pSVH1 containing the 8-bp TRS repeats is 80% (Fig. 2A). It is well-known that G-rich sequences are involved in the formation of G-quadruplex structures on DNA (G4 DNA) (17). Moreover, it has been shown that TrwB, a motor that belongs to the FtsK-like family which is involved in DNA transfer in Gram-negative bacteria, presents a high affinity for G-quadruplex structures (16), suggesting that this type of secondary DNA structures might act as loading sites for this motor. These data, together with the fact that TraBs presents higher affinity for the clt sequence than for just overlapping TRS sequences, prompted us to analyze the interaction of TraBs with intermolecular G-quadruplex structures. To this end, G4 DNA was formed with the 45-mer oligonucleotide I (Table 1), following a specific protocol to obtain this type of DNA secondary structures, as reported previously (16) (see also “Experimental procedures”).
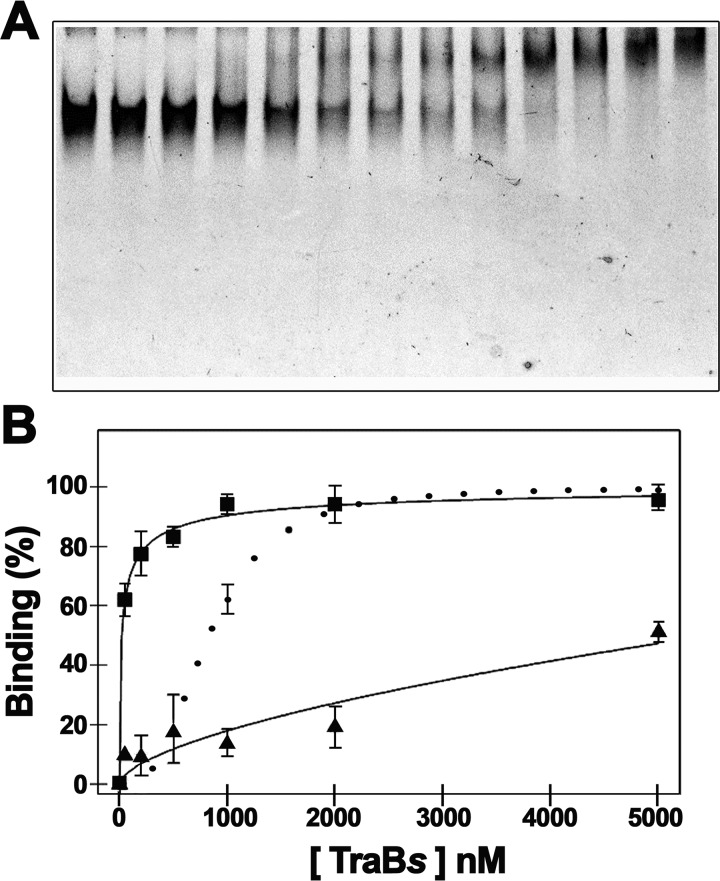
Gel shift assays indicated that TraBs binds G4 DNA with higher affinity than TRS sequences (Fig. 4A). Moreover, TraBsΔγ protein was able to bind G4 DNA almost to the same extent as TraBs, which indicates that the γ-domain is not required for the recognition of this type of structures on the DNA. The analysis and quantification of the TraBs gel shift assays with different DNA substrates showed an apparent Kd value of 23 nm for G4 DNA, 900 nm for dsDNA with the clt sequence, and 1.9 μm for dsDNA with three overlapping TRS sequences (Fig. 4B). As mentioned, TraBsΔγ protein was only able to bind G4 DNA, with a Kd value of 190 nm. All together, these results suggest that TraB might have an initial contact with the DNA by recognizing G-quadruplex structures, without the involvement of the γ-domain.
Figure 4.
Gel mobility shift assays of TraBs and TraBsΔγ with G-quadruplex DNA. A, G-quadruplex DNA substrate, formed with oligonucleotide I as described under “Experimental procedures,” was incubated with increasing concentrations of TraBs or TraBsΔγ proteins at 0, 0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.8, 1, 2, 3, 4, and 5 μm. B, affinity curves obtained from the DNA-binding shift assays in the presence of G-quadruplex DNA (black squares), dsDNA with the clt sequence (black dots), and dsDNA with three overlapping TRS sequences (black triangles). Data represent the average of at least three different experiments. Error bars: S.D.
TraBs forms higher-order structures
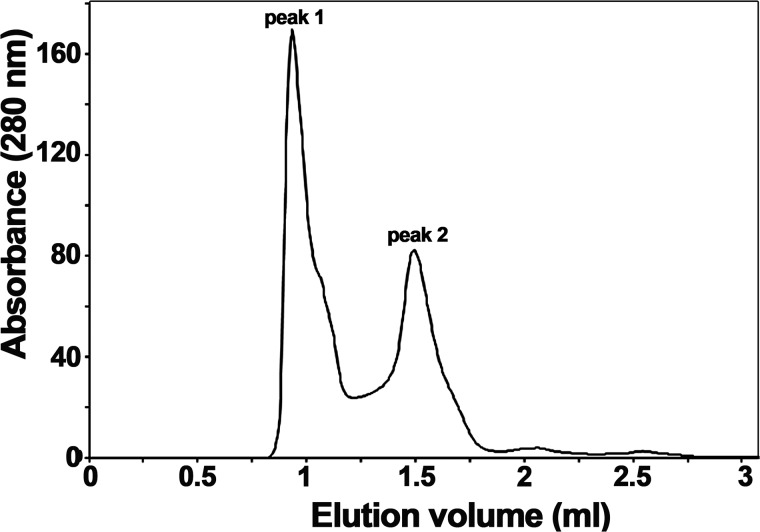
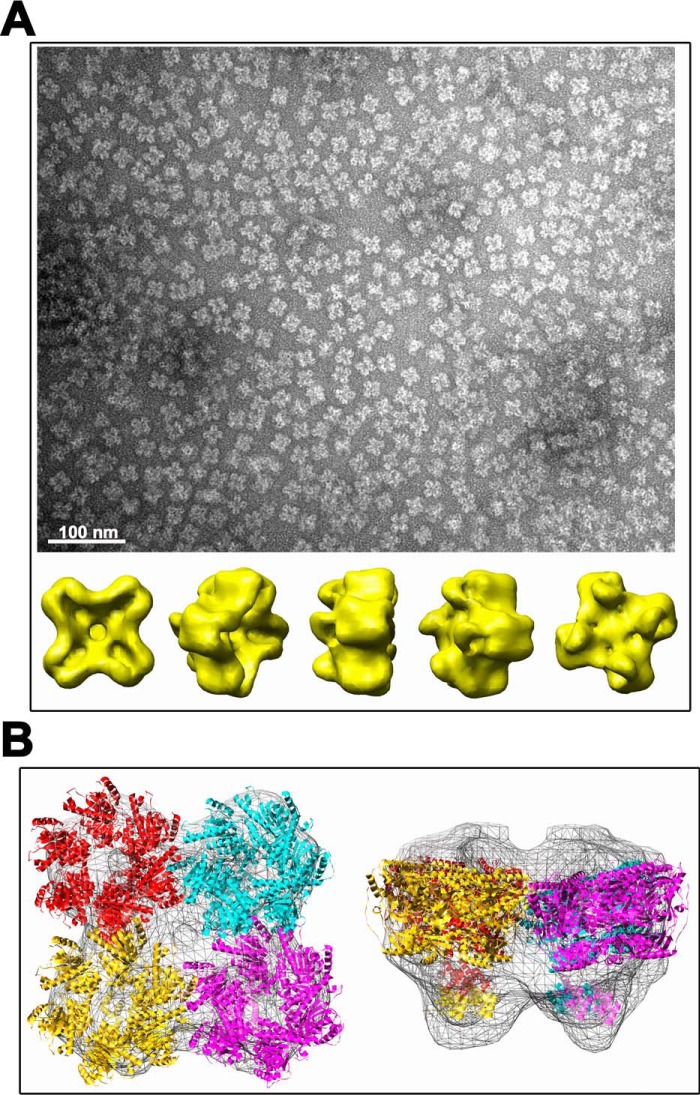
The last purification step of TraBs, consisting of a size exclusion chromatography, showed two different oligomerization states of the protein (Fig. 5). The elution profile exhibited two major protein peaks: peak 1, corresponding to the void volume of the column, and peak 2, corresponding to an apparent molecular mass of 70 kDa (compatible with the predicted molecular mass of the monomer). The eluted fractions were analyzed by SDS-PAGE, confirming that both peaks corresponded to different forms of TraBs protein (data not shown). Variations on the pH value or on the glycerol content of the buffer shifted the balance in favor of a particular oligomeric state, suggesting there was an equilibrium between the two distinct oligomeric species (Fig. S2). EM analysis of fractions from peak 1 revealed a monodisperse sample of single particles, with an apparent diameter of ∼200 Å (Fig. 6A). A 3D reconstruction obtained after classification of these particles revealed a high-order structure, compatible with the assembly of four hexamers. A rotational symmetry analysis of the particles supported the existence of a 4-fold symmetry along the z axis.
Figure 5.
TraBs forms high-ordered structures. Elution profile from size-exclusion chromatography of TraBs on a HiLoad Superdex 200 3.2/300 column. Two peaks with an estimated molecular mass of >800 kDa (peak 1) and ∼70 kDa (peak 2) can be distinguished. Samples from peak 1 were loaded into a Superose 6 column to estimate the size (1.15 MDa) (data not shown).
Figure 6.
3D reconstruction of TraBs oligomer. A, TraBs from peak 1 was negatively stained with uranyl acetate and analyzed by EM. Particles were selected, aligned, and classified by maximum likelihood methods. A 3D reconstruction was obtained by projection matching, with imposed C4 symmetry in the final iterations. Scale bar represents 100 nm. B, fitting of TraBs in the EM maps. An atomic model of TraBs was generated by using atomic coordinates from FtsK of P. aeruginosa as template (2iuu.pdb and 2ve9.pdb coordinates for the motor domain and the γ-domain, respectively). The atomic coordinates were docked into the EM maps using SITUS package software.
To get a better estimation of the molecular mass of these high-order structures, samples were also loaded onto a Superose 6 column. Samples from peak 1 eluted with an estimated molecular mass of 1.15 MDa, which is compatible with an assembly of 24 TraBs monomers. Further, an hexameric molecular model of TraBs was generated by molecular threading using as template the atomic coordinates of the motor domain (PDB ID: 2IUU) (14) and the γ-domain of Pseudomonas aeruginosa FtsK (PDB ID: 2VE9) (20). Atomic coordinates were then fitted into the 3D EM map of TraBs. Interestingly, up to four hexamers of FtsK could be docked into the EM map (Fig. 6B).The biological significance of this finding remains unclear.
Discussion
The Streptomyces TraB protein closely resembles DNA translocases of the FtsK/SpoIIIE family (6). Domain organization, predicted structure, and DNA-binding mode of TraB is similar to that of FtsK and SpoIIIE, recognizing a specific 8-bp sequence on the DNA through a γ-domain at the C-terminal domain of the protein. However, TraB is not involved in chromosome segregation but in conjugal plasmid transfer. This unique conjugative system only requires TraB and a specific cis-acting sequence that includes several 8-bp repeats (TRS motifs), which are recognized by the DNA translocase. In contrast to most common conjugative systems, which are involved in the transfer of the plasmid as ssDNA, TraB transfers a dsDNA plasmid molecule to the recipient (7). Conjugative T4SSs also need DNA-dependent ATPases for plasmid transfer, such as TrwB from plasmid R388 (26). These TrwB-like conjugative transporters have a membrane domain and a motor domain, but lack a γ-domain and, hence, they are supposed to translocate plasmid DNA without specific sequence recognition. Instead, it has been shown that TrwB is a structure-specific DNA-binding protein that recognizes G-quadruplex DNA structures with very high affinity, suggesting that these structures might act as loading sites for the motor (16). Interestingly, a phylogenetic analysis of this family of proteins revealed that the motor domain of TraB had diverged from that of TrwB-like proteins after the diversification between FtsK- and TrwB-like families (19, 27). On these bases, we decided to better characterize the activity of TraB, looking for those mechanistic aspects that might be shared by the two DNA transfer systems: chromosome segregation and bacterial conjugation.
Whereas FtsK/SpoIIIE and TrwB proteins have been extensively analyzed, little is known about the activity of TraB. Previous studies showed that the NTP-binding site of TraB was essential for conjugative DNA transfer (28). A more detailed characterization of the full-length strepII-TraB fusion protein, showed a maximal ATPase activity of ∼800 nmol of ATP min−1 mg−1 (8). The same study concluded that the ATPase activity of the protein was not stimulated by DNA. In the work presented here, we have demonstrated that the activity of the protein is highly dependent on the pH value, with a maximal activity at pH 6, which could explain the lower activity values obtained in previous reports, conducted at pH 8. Moreover, here we show that the activity of the protein is highly stimulated with dsDNA substrates, up to a Vmax value of ∼73,000 nmol ATP min−1 mg−1 of protein. It is possible that in the above-mentioned previous report (8), the presence of the membrane anchor domain of the protein had an effect on the ATPase activity. Nonetheless, the pH dependence observed here might also explain the absence of DNA-dependent ATPase activity reported in that prior work. At the conditions tested in these studies the motor can work at a rate of approximately ∼70 ATP s−1 per monomer of protein, which is an order of magnitude lower than the ATPase activity of SpoIIIE (∼800 ATP s−1 per monomer) (21). These differences can be explained on the basis of the biological activities that both proteins perform. SpoIIIE motor must transport the complete chromosome (1.5 megabases of DNA) across the septum in ∼20 min, at a rate of 2 bp of DNA per ATP turned over (21, 29). On these premises, Besprozvannaya et al. (21) estimated that the minimum ATPase rate necessary to support DNA transport in vivo was ∼100 ATP s−1. Taking into account that TraB does not transfer the chromosome, but a single plasmid of 12.6 kb in size, the ATPase rate of TraBs measured in vitro would be sufficient to support plasmid DNA transfer in vivo.
FtsK and SpoIIIE proteins require KOPS and SRS motifs, respectively, for directional DNA translocation. A recent work on SpoIIIE revealed that the γ-domain acts as an allosteric regulator of the ATPase activity of the motor (21). In the absence of an SRS DNA motif, the motor presents a low basal ATPase activity, but upon sensing the recognition motif in a permissive orientation the motor increases the ATPase rates. According to these studies, the γ-domain would play a dual role. On the one hand it would confer a high affinity for the SRS motif and, in addition, it would modulate the directionality by stimulating ATPase activity only when it encounters the recognition motif in a permissive orientation (21). In an attempt to determine whether the γ-domain of TraB plays a similar role in modulating the ATPase activity of the protein, we measured the ATP turnover in the presence and absence of the γ-domain, and also in the presence and absence of DNA substrates with the specific SRS motifs. Contrary to what was observed in the SpoIIIE protein (21), deletion of the γ-domain in TraBs does not abolish the DNA-dependent ATPase activity. Moreover, the ATPase activities of the two proteins, TraBs and TraBsΔγ, were similar in the presence of specific DNA substrates with TRS motifs. Therefore, we can conclude that TraB γ-domain does not confer sequence-specific ATPase stimulation. Interestingly, a recent publication by Chara et al. (30) is in perfect agreement with our results, showing that SpoIIIE has an equivalent specific ATPase activity for both types of DNA substrates (with or without SRS sequences).
Although the removal of the γ-domain does not affect the ATPase activity of the protein, previous studies have shown that TraB γ-domain binds preferentially to TRS sequences over random DNA (6). EMSAs performed in this work corroborate those previous results. TraBsΔγ is not able to bind the DNA fragment with the specific clt sequence at the tested conditions. Therefore, although TraB shares with FtsK/SpoIIIE translocases a similar DNA recognition system by means of the γ-domain, TRS recognition is not coupled to motor activation. The C-terminal γ-domain of FtsK/SpoIIIE translocases is also involved in the recruitment of XerCD DNA recombinases to the dif site, and it is essential for activation of DNA cleavage and rejoining, which results in chromosome resolution (14, 31–33). So far, such a role has not been associated to TraB protein and, therefore, it is not surprising to observe mechanistic differences between these motors. In addition, the amino acid sequence of the γ-domain is highly conserved between FtsK and SpoIIIE proteins and differs quite significantly from that of the TraB homologs (Fig. S3). TraB proteins lack two conserved arginine residues (Arg-745 and Arg-756 in SpoIIIE) that have been shown to play an essential role in DNA-dependent ATPase stimulation of SpoIIIE associated with SRS motifs (21).
Different models have been proposed to explain how FtsK and SpoIIIE load onto the DNA and keep directionality. Most of the FtsK studies point to a preferential loading model, where the translocase would assemble on the KOPS sequence and begin translocation in the direction dictated by KOPS, ignoring all subsequent sequences (20, 23, 24). In contrast, SpoIIIE hexamer formation is SRS-independent (34). Recent reports on this protein suggest a target search and activation model. Accordingly, preformed SpoIIIE hexamers would bind nonspecifically to DNA and find SRS by an ATP-independent target search mechanism, with ensuing sequence-specific activation of the motor mediated by binding and oligomerization of the γ-domain on SRS (25, 34). This mechanism implies that the 8-bp recognition motifs do not have a role in protein assembly. In fact, equilibrium-binding studies revealed that SpoIIIE binds SRS with a relatively low specificity (25). The experiments carried out in this work also show a low affinity of TraBs for TRS motifs. In contrast, TraBs presents a higher affinity for G-quadruplex DNA. Both, TraBs and TraBsΔγ proteins bind these secondary DNA structures with high affinity, revealing that the γ-domain of the protein does not play an essential role in this binding. TrwB from plasmid R388, involved in bacterial conjugation through a T4SS, also binds G-quadruplex structures with high affinity (16). These DNA structures were suggested to act as loading sites for the motor. Given the high degree of homology between the motor domains of these translocases, it is tempting to propose a similar mechanism for DNA loading, in which the initial nonspecific DNA-binding step would consist of the recognition of G4 DNA by the motor domain.
TRS, KOPS, and SRS are G-rich sequences with the potential to form G-quadruplex structures but it is unknown whether the motor domains of FtsK and SpoIIIE proteins exhibit a high affinity for these types of secondary DNA structures. Whereas KOPS sequences are distributed over the whole chromosome, with a strong bias from the origin toward the terminus region, repeated TRS sequences are exclusively found within the clt region. The GC content of this region is as high as 80% and, therefore, it is likely to be involved in the formation of G-quadruplex structures in vivo. Moreover, although two copies of TRS were sufficient for TraB binding in vitro (8), binding of TraB to a larger clt fragment containing additional TRS copies was more efficient and required lower protein concentrations, indicating that the complete clt region is more effective in vivo (8) (this work). A 45-mer oligonucleotide with the clt DNA sequence (oligonucleotide D from Table 1) was analyzed in nondenaturing polyacrylamide gels, showing an increased mobility compared with other oligonucleotides of the same length (data not shown). These data suggest the formation of intramolecular G-quadruplex, because this type of DNA substrates fold into a more compact structure than the ssDNA, and have a faster migration rate than ssDNA in a native gel (35). Interestingly, FtsK also shows a higher effectiveness with a DNA substrate that contains an overlapping triple KOPS sequence (20). The four clusters of G-residues in this substrate perfectly match the consensus signature to form a G4 DNA structure (G3–5, N1–3, G3–5, N1–3, G3–5, N1–3, G3–5) (18).
The role of G-quadruplex structures in vivo as loading sites remains unclear and needs further investigation, just as the oligomeric state of the protein. Our data point out to the formation of a high-order protein assembly, compatible with the association of four TraB hexamers. A previous structural analysis of TraB by EM revealed hexameric ring-shaped structures with a central pore (6). In that previous report, TraB was purified as a strepII-TraB fusion protein, under different experimental conditions. Although we cannot exclude the possibility that the formation of protein assemblies of higher order is an artifact, the fact that these assemblies show ATPase activity ratios and DNA affinity Kd values similar to those of the monomeric protein suggests that they are an active form of the protein. To our knowledge, there is not any report on this family of hexameric motors showing these high-order oligomeric structures and, at present, it is not possible to explain why an assembly of four hexamers would be required in vivo. Different models exist to explain how FtsK or SpoIIIE translocate the DNA across the septum. Superresolution microscopy studies showed that SpoIIIE assembles into 45-nm complexes that are recruited to nascent sites during septum formation (36). Such SpoIIIE complexes contain 47 ± 20 SpoIIIE molecules after the induction of sporulation, a majority of which are assembled into hexamers. Clusters in division septa of exponentially growing cells displayed 20 ± 4 SpoIIIE molecules. Their results support a model in which DNA translocation occurs through an aqueous DNA-conducting pore, where SpoIIIE prevents membrane fusion until completion of chromosome segregation (36). In view of these results, an assembly of four hexamers for each chromosomal arm might be possible. Each hexamer is ∼12 nm in size (14) and the assembly of the four hexamers observed in our work has a size of ∼24 nm. A single PALM-limited cluster of SpoIIIE, located at the septum, is 45 nm in size, as mentioned above, which reinforces the idea that DNA-translocating complexes might assemble in a compartment-specific fashion.
In summary, we report here a combined structural and functional analysis of TraB. Even though many questions still remain open, the work presented here sheds light on a mechanism that might be shared by complex machineries involved in DNA transport across biological membranes.
Experimental procedures
Cloning and protein purification
The DNA of traBs gene from plasmid pSVH1 was amplified by PCR and cloned into a pET28a expression vector (Novagen, Madison, WI), using the forward primer 5′-ATTAAACATATGGGGGGCGCGTTCGTCGGTCGC, and the reverse primer 5′-ATTAAGAATTCTCAGGCGGTAAGCCCGAATCGGTC. The resultant DNA fragments were digested with NdeI and HindIII restriction enzymes and ligated into the corresponding sites in the MCS of vector pET28a. The truncated traBsΔγ gene was cloned in the same way, but using the reverse primer 5′-ATTAAGAATTCACTCGAGGACGGCGGCCAGGAGCAGCGCGTCGTC. Plasmid DNAs were then used to transform Escherichia coli strain C41 (DE3) (37).
Overexpression of the N-terminal histidine-tagged TraBs and TraBsΔγ proteins was induced by the addition of 1 mm isopropyl-β-d-thiogalactopyranoside (IPTG). After 3-h induction at 37 °C, cells were harvested and suspended in a buffer consisting of 50 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5% glycerol, and 0.001% PMSF. Thawed cells were disrupted by French press and lysates collected by centrifugation and loaded onto a HiTrap HP (5 ml) column (GE Healthcare). Protein was eluted from the column in a 500 mm imidazole linear gradient. Selected fractions were applied to a HiTrap Q Sepharose (1 ml) column (GE Healthcare), and protein was eluted with 5 ml of buffer 50 mm Tris-HCl, pH 7.5, 400 mm NaCl, 5% glycerol, and 0.001% PMSF. Fractions were pooled and stored at −20 °C.
Preparation of DNA substrates
Oligonucleotides were purchased from Sigma-Genosys. G4 DNA was prepared as described previously (16). Oligonucleotide D from Table 1 (300 μm) was incubated overnight at 60 °C in buffer 50 mm Pipes, pH 6.2, 50 mm NaCl, 5% glycerol, and 100 mm sodium acetate. Then, the mixture was incubated at 37 °C for 1 day and cooled down at room temperature for 2 h. Formation of the G-quadruplex was confirmed by gel filtration chromatography and PAGE. To prepare duplex DNA substrate, complementary 45-mer oligonucleotides (85 μm) were incubated at 95 °C for 3 min in buffer (50 mm Pipes, pH 6.2, 50 mm NaCl, and 5% glycerol). Then, the mixture was cooled down at room temperature for 2 h. Formation of both substrates was confirmed by gel filtration chromatography. For DNA retardation assays, 5′-fluorescein–labeled oligonucleotides were used (Sigma-Genosys).
Nucleotide hydrolysis assays
ATP hydrolysis was analyzed by a coupled enzyme assay as described previously (16). TraBs and TraBsΔγ proteins were pre-incubated with different DNA substrates for 10 min at 37 °C in 150 μl of an ATPase assay mixture consisting of 50 mm Pipes-NaOH, pH 6.2, 50 mm NaCl, 5 mm Mg-acetate, 5% glycerol, 0.5 mm phosphoenolpyruvate, 0.25 mm NADH, 60 μg/ml pyruvate kinase, and 60 μg/ml lactate dehydrogenase (Roche Applied Science). Reactions were started by the addition of ATP (5 mm).
Gel filtration analysis
TraBs protein was chromatographed at 25 °C at a flow rate of 30 μl/min on a Superdex 200 PC 3.2/30 column (Amersham Biosciences SMART system) and on a Superose 6 HR 10/30 (Äkta chromatography systems). After isocratic elution in a buffer consisting of 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm MgCl2, 5% glycerol, and 0.001% PMSF, fractions were analyzed by gel electrophoresis.
Electrophoretic mobility shift assays
TraBs and TraBsΔγ DNA-binding activity was measured using a gel shift assay, as described previously (16). Fluorescein-labeled DNA (5 nm) was incubated with increasing concentrations of TraBs or TraBsΔγ proteins in a final volume of 20 μl. Buffer consisted of 50 mm Tris-HCl, pH 7, 50 mm NaCl, 100 μg/μl BSA, 5% glycerol, and the indicated quantities of DNA and protein. After incubation at 37 °C for 30 min, reaction mixtures were loaded onto an 8% native PAGE gel. Free nucleic acids and nucleoprotein complexes were resolved by electrophoresis at 200 V for 30 min at room temperature. Gels were analyzed using a Molecular Imager FX ProSystem (Bio-Rad Laboratories). For quantification, the intensity of bands corresponding to free DNA and TraBs-DNA complex was determined using ImageJ software, as described previously (16), where the percentage of bound DNA was calculated as follows: Binding DNA(%) = 100 × (TraBs-DNA/(TraBs-DNA + Free DNA)).
Electron microscopy and image analysis
Aliquots of TraBs (5 μl at 0.1 mg/ml of protein concentration) were applied onto freshly glow-discharged carbon-coated grids. Samples were negatively stained with 2% (w/v) uranyl acetate. Images were recorded at 200,000 × nominal magnification in a JEM 1011 microscope (JEOL) equipped with an Orius SC1000 CCD (Gatan) at the EM path between the objective and projection lenses. Image acquisition was performed at low dose conditions (0.4 s of exposition) at a sampling rate of 4 Å pixel/size and demagnified twice by linear interpolation. Contrast transfer function was estimated (38) and corrected using cctfind software (39). Alignment and classification was performed by maximum likelihood methods (40). 3D reconstructions were performed using Xmipp software (41) that is part of the Scipion package (42). The initial refinements were carried out without any symmetry imposed. Final reconstructions were done by projection matching after introducing a C4-symmetry in the last refinement steps. Volumes were rendered by using UCSF Chimera (43).
Molecular modeling and fitting into EM maps
An atomic model of TraBs from Streptomyces pSHV1 plasmid was generated by molecular threading (44), using FtsK protein from P. aeruginosa as a template (2iuu.pdb and 2ve9.pdb atomic coordinates for the motor domain and the γ-domain, respectively). Fitting of the atomic coordinates into the EM map was done using SITUS package software (45) and refined by using UCSF Chimera.
Author contributions
E. A. and I. A. investigation; G. M. and E. C. conceptualization; G. M., I. A., and E. C. writing-review and editing; I. A. and E. C. formal analysis; I. A. and E. C. supervision; I. A. and E. C. funding acquisition; I. A. and E. C. project administration; E. C. methodology; E. C. writing-original draft.
Supplementary Material
This work was supported by Spanish Ministerio de Economia y Competitividad (MINECO) Grants BFU2016-78521-R (to E. C. and I. A.) and Deutsche Forschungsgemeinschaft SFB766 (to G. M). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- TRS
- TraB-recognition sequence
- T4SS
- type IV secretion system.
References
- 1. Iyer L. M., Makarova K. S., Koonin E. V., and Aravind L. (2004) Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 32, 5260–5279 10.1093/nar/gkh828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Demarre G., Galli E., and Barre F. X. (2013) The FtsK Family of DNA Pumps. Adv. Exp. Med. Biol. 767, 245–262 10.1007/978-1-4614-5037-5_12 [DOI] [PubMed] [Google Scholar]
- 3. Berezuk A. M., Goodyear M., and Khursigara C. M. (2014) Site-directed fluorescence labeling reveals a revised N-terminal membrane topology and functional periplasmic residues in the Escherichia coli cell division protein FtsK. J. Biol. Chem. 289, 23287–23301 10.1074/jbc.M114.569624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bigot S., Saleh O. A., Cornet F., Allemand J. F., and Barre F. X. (2006) Oriented loading of FtsK on KOPS. Nat. Struct. Mol. Biol. 13, 1026–1028 10.1038/nsmb1159 [DOI] [PubMed] [Google Scholar]
- 5. Ptacin J. L., Nollmann M., Becker E. C., Cozzarelli N. R., Pogliano K., and Bustamante C. (2008) Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat. Struct. Mol. Biol. 15, 485–493 10.1038/nsmb.1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vogelmann J., Ammelburg M., Finger C., Guezguez J., Linke D., Flötenmeyer M., Stierhof Y. D., Wohlleben W., and Muth G. (2011) Conjugal plasmid transfer in Streptomyces resembles bacterial chromosome segregation by FtsK/SpoIIIE. EMBO J. 30, 2246–2254 10.1038/emboj.2011.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Possoz C., Ribard C., Gagnat J., Pernodet J. L., and Guérineau M. (2001) The integrative element pSAM2 from Streptomyces: Kinetics and mode of conjugal transfer. Mol. Microbiol. 42, 159–166 [DOI] [PubMed] [Google Scholar]
- 8. Reuther J., Gekeler C., Tiffert Y., Wohlleben W., and Muth G. (2006) Unique conjugation mechanism in mycelial streptomycetes: A DNA-binding ATPase translocates unprocessed plasmid DNA at the hyphal tip. Mol. Microbiol. 61, 436–446 10.1111/j.1365-2958.2006.05258.x [DOI] [PubMed] [Google Scholar]
- 9. Franco B., González-Cerón G., and Servín-González L. (2003) Direct repeat sequences are essential for function of the cis-acting locus of transfer (clt) of Streptomyces phaeochromogenes plasmid pJV1. Plasmid 50, 242–247 10.1016/S0147-619X(03)00063-5 [DOI] [PubMed] [Google Scholar]
- 10. Cabezón E., Ripoll-Rozada J., Peña A., de la Cruz F., and Arechaga I. (2015) Toward an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39, 81–95 10.1111/1574-6976.12085 [DOI] [PubMed] [Google Scholar]
- 11. Zechner E. L., Lang S., and Schildbach J. F. (2012) Assembly and mechanisms of bacterial type IV secretion machines. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 1073–1087 10.1098/rstb.2011.0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tato I., Zunzunegui S., de la Cruz F., and Cabezon E. (2005) TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc. Natl. Acad. Sci. U.S.A. 102, 8156–8161 10.1073/pnas.0503402102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gomis-Rüth F. X., Moncalián G., Pérez-Luque R., González A., Cabezón E., de la Cruz F., and Coll M. (2001) The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409, 637–641 10.1038/35054586 [DOI] [PubMed] [Google Scholar]
- 14. Massey T. H., Mercogliano C. P., Yates J., Sherratt D. J., and Löwe J. (2006) Double-stranded DNA translocation: Structure and mechanism of hexameric FtsK. Mol. Cell 23, 457–469 10.1016/j.molcel.2006.06.019 [DOI] [PubMed] [Google Scholar]
- 15. Tato I., Matilla I., Arechaga I., Zunzunegui S., de la Cruz F., and Cabezon E. (2007) The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: Implications for a common assembly mechanism of DNA translocating motors. J. Biol. Chem. 282, 25569–25576 10.1074/jbc.M703464200 [DOI] [PubMed] [Google Scholar]
- 16. Matilla I., Alfonso C., Rivas G., Bolt E. L., de la Cruz F., and Cabezon E. (2010) The conjugative DNA translocase TrwB is a structure-specific DNA-binding protein. J. Biol. Chem. 285, 17537–17544 10.1074/jbc.M109.084137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williamson J. R. (1994) G-quartet structures in telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 23, 703–730 10.1146/annurev.bb.23.060194.003415 [DOI] [PubMed] [Google Scholar]
- 18. London T. B., Barber L. J., Mosedale G., Kelly G. P., Balasubramanian S., Hickson I. D., Boulton S. J., and Hiom K. (2008) FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 283, 36132–36139 10.1074/jbc.M808152200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cabezon E., Lanza V. F., and Arechaga I. (2012) Membrane-associated nanomotors for macromolecular transport. Curr. Opin. Biotechnol. 23, 537–544 10.1016/j.copbio.2011.11.031 [DOI] [PubMed] [Google Scholar]
- 20. Löwe J., Ellonen A., Allen M. D., Atkinson C., Sherratt D. J., and Grainge I. (2008) Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell 31, 498–509 10.1016/j.molcel.2008.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Besprozvannaya M., Pivorunas V. L., Feldman Z., and Burton B. M. (2013) SpoIIIE protein achieves directional DNA translocation through allosteric regulation of ATPase activity by an accessory domain. J. Biol. Chem. 288, 28962–28974 10.1074/jbc.M113.484055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sivanathan V., Allen M. D., de Bekker C., Baker R., Arciszewska L. K., Freund S. M., Bycroft M., Löwe J., and Sherratt D. J. (2006) The FtsK γ domain directs oriented DNA translocation by interacting with KOPS. Nat. Struct. Mol. Biol. 13, 965–972 10.1038/nsmb1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graham J. E., Sherratt D. J., and Szczelkun M. D. (2010) Sequence-specific assembly of FtsK hexamers establishes directional translocation on DNA. Proc. Natl. Acad. Sci. U.S.A. 107, 20263–20268 10.1073/pnas.1007518107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee J. Y., Finkelstein I. J., Crozat E., Sherratt D. J., and Greene E. C. (2012) Single-molecule imaging of DNA curtains reveals mechanisms of KOPS sequence targeting by the DNA translocase FtsK. Proc. Natl. Acad. Sci. U.S.A. 109, 6531–6536 10.1073/pnas.1201613109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cattoni D. I., Chara O., Godefroy C., Margeat E., Trigueros S., Milhiet P. E., and Nöllmann M. (2013) SpoIIIE mechanism of directional translocation involves target search coupled to sequence-dependent motor stimulation. EMBO Rep. 14, 473–479 10.1038/embor.2013.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cabezon E., and de la Cruz F. (2006) TrwB: An F(1)-ATPase-like molecular motor involved in DNA transport during bacterial conjugation. Res. Microbiol. 157, 299–305 10.1016/j.resmic.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 27. Peña A., Matilla I., Martín-Benito J., Valpuesta J. M., Carrascosa J. L., de la Cruz F., Cabezón E., and Arechaga I. (2012) The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J. Biol. Chem. 287, 39925–39932 10.1074/jbc.M112.413849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kosońo S., Kataoka M., Seki T., and Yoshida T. (1996) The TraB protein, which mediates the intermycelial transfer of the Streptomyces plasmid pSN22, has functional NTP-binding motifs and is localized to the cytoplasmic membrane. Mol. Microbiol. 19, 397–405 10.1046/j.1365-2958.1996.379909.x [DOI] [PubMed] [Google Scholar]
- 29. Grainge I. (2008) Sporulation: SpoIIIE is the key to cell differentiation. Curr. Biol. 18, R871–R872 10.1016/j.cub.2008.07.047 [DOI] [PubMed] [Google Scholar]
- 30. Chara O., Borges A., Milhiet P. E., Nöllmann M., and Cattoni D. I. (2018) Sequence-dependent catalytic regulation of the SpoIIIE motor activity ensures directionality of DNA translocation. Sci. Rep. 8, 5254 10.1038/s41598-018-23400-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Barre F. X., Aroyo M., Colloms S. D., Helfrich A., Cornet F., and Sherratt D. J. (2000) FtsK functions in the processing of a Holliday junction intermediate during bacterial chromosome segregation. Genes Dev. 14, 2976–2988 10.1101/gad.188700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zawadzki P., May P. F., Baker R. A., Pinkney J. N., Kapanidis A. N., Sherratt D. J., and Arciszewska L. K. (2013) Conformational transitions during FtsK translocase activation of individual XerCD-dif recombination complexes. Proc. Natl. Acad. Sci. U.S.A. 110, 17302–17307 10.1073/pnas.1311065110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Diagne C. T., Salhi M., Crozat E., Salomé L., Cornet F., Rousseau P., and Tardin C. (2014) TPM analyses reveal that FtsK contributes both to the assembly and the activation of the XerCD-dif recombination synapse. Nucleic Acids Res. 42, 1721–1732 10.1093/nar/gkt1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cattoni D. I., Thakur S., Godefroy C., Le Gall A., Lai-Kee-Him J., Milhiet P. E., Bron P., and Nöllmann M. (2014) Structure and DNA-binding properties of the Bacillus subtilis SpoIIIE DNA translocase revealed by single-molecule and electron microscopies. Nucleic Acids Res. 42, 2624–2636 10.1093/nar/gkt1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oganesian L., Moon I. K., Bryan T. M., and Jarstfer M. B. (2006) Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 25, 1148–1159 10.1038/sj.emboj.7601006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fiche J. B., Cattoni D. I., Diekmann N., Langerak J. M., Clerte C., Royer C. A., Margeat E., Doan T., and Nöllmann M. (2013) Recruitment, assembly, and molecular architecture of the SpoIIIE DNA pump revealed by superresolution microscopy. PLoS Biol. 11, e1001557 10.1371/journal.pbio.1001557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miroux B., and Walker J. E. (1996) Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 10.1006/jmbi.1996.0399 [DOI] [PubMed] [Google Scholar]
- 38. Mindell J. A., and Grigorieff N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 10.1016/S1047-8477(03)00069-8 [DOI] [PubMed] [Google Scholar]
- 39. Heymann J. B. (2001) Bsoft: Image and molecular processing in electron microscopy. J. Struct. Biol. 133, 156–169 10.1006/jsbi.2001.4339 [DOI] [PubMed] [Google Scholar]
- 40. Scheres S. H., Valle M., Nuñez R., Sorzano C. O., Marabini R., Herman G. T., and Carazo J. M. (2005) Maximum-likelihood multi-reference refinement for electron microscopy images. J. Mol. Biol. 348, 139–149 10.1016/j.jmb.2005.02.031 [DOI] [PubMed] [Google Scholar]
- 41. Sorzano C. O., Marabini R., Velázquez-Muriel J., Bilbao-Castro J. R., Scheres S. H., Carazo J. M., and Pascual-Montano A. (2004) XMIPP: A new generation of an open-source image processing package for electron microscopy. J. Struct. Biol. 148, 194–204 10.1016/j.jsb.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 42. de la Rosa-Trevín J. M., Quintana A., Del Cano L., Zaldívar A., Foche I., Gutiérrez J., Gómez-Blanco J., Burguet-Castell J., Cuenca-Alba J., Abrishami V., Vargas J., Otón J., Sharov G., Vilas J. L., Navas J., et al. (2016) Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 10.1016/j.jsb.2016.04.010 [DOI] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 44. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wriggers W., Milligan R. A., and McCammon J. A. (1999) Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 125, 185–195 10.1006/jsbi.1998.4080 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.