Abstract
The base excision repair (BER) pathway is an important DNA repair pathway and is essential for immune responses. In fact, it regulates both the antigen-stimulated somatic hypermutation (SHM) process and plays a central function in the process of class switch recombination (CSR). For both processes, a central role for apurinic/apyrimidinic endonuclease 1 (APE1) has been demonstrated. APE1 acts also as a master regulator of gene expression through its redox activity. APE1's redox activity stimulates the DNA-binding activity of several transcription factors, including NF-κB and a few others involved in inflammation and in immune responses. Therefore, it is possible that APE1 has a role in regulating the CSR through its function as a redox coactivator. The present study was undertaken to address this question. Using the CSR-competent mouse B-cell line CH12F3 and a combination of specific inhibitors of APE1's redox (APX3330) and repair (compound 3) activities, APE1-deficient or -reconstituted cell lines expressing redox-deficient or endonuclease-deficient proteins, and APX3330-treated mice, we determined the contributions of both endonuclease and redox functions of APE1 in CSR. We found that APE1's endonuclease activity is essential for IgA-class switch recombination. We provide evidence that the redox function of APE1 appears to play a role in regulating CSR through the interleukin-6 signaling pathway and in proper IgA expression. Our results shed light on APE1's redox function in the control of cancer growth through modulation of the IgA CSR process.
Keywords: base excision repair (BER), DNA endonuclease, DNA transcription, immunoglobulin A (IgA), immunology, apurinic/apyrimidinic endonuclease 1 (APE1), base excision repair (BER), class switch recombination, immunoglobulin A (IgA), redox-inhibitor
Introduction
Antigen-dependent B cell activation is a dynamically integrated cascade of cell-signaling events. It requires accessory signals that can come either from an armed helper T cell or, in some cases, directly from microbial constituents. Downstream consequences of transduced signals are the initiating of genomic rearrangement that induces somatic hypermutation (SHM)4 (1) and a class switch recombination (CSR) that switches the CH region (1). Both immunoglobulin SHM and CSR are essential mechanisms for the generation of a high-affinity, adaptive humoral immune response; require transcription; and are dependent on the activation-induced cytidine deaminase (AID), which converts cytosine to uracil and also contributes to the formation of requisite single-stranded DNA substrates. Both processes engage activities of normal cellular base excision repair (BER) and mismatch repair, converting AID cytidine deamination lesions to mutational and/or double-stranded break outcomes.
The BER pathway is a highly conserved pathway from bacteria to humans and is responsible for the maintenance of genome stability through repairing DNA damage caused by oxidative stress, ionizing radiation (IR), and chemotherapy treatments. These damages are typically nondistorting alkylated and oxidized bases, as well as single-stranded breaks. However, the enzymes involved in BER also take part in a variety of apparently unrelated cellular functions related to gene expression mechanisms (2).
The role of BER enzymes in SHM and CSR strongly depends on cross-talk between each single protein partner and is finely tuned by the expression levels of each protein and by specific regulated post-translational modifications, such as phosphorylation of Ser38 in AID protein (3, 4). APE1, which is the unique apurinic/apyrimidinic endonuclease in mammalian cells, is responsible for the recognition and cleavage of the abasic sites generated spontaneously or by the action of specialized glycosylase enzymes (2). APE1 deficiency, although not affecting AID-induced S-region break formation, impairs both the recruitment of Ku80 to the S region and the synapse formation between Sμ and Sα (5). APE1 endonuclease activity likely removes 3′-tyrosyl residues from the DNA end after topoisomerase 1 cleavage and topoisomerase 1-cc degradation, which is essential to efficient recombination of broken S regions. Additionally, S regions are subjected to a high rate of transcription driven by cytokine-inducible promoters, which regulates the CSR process.
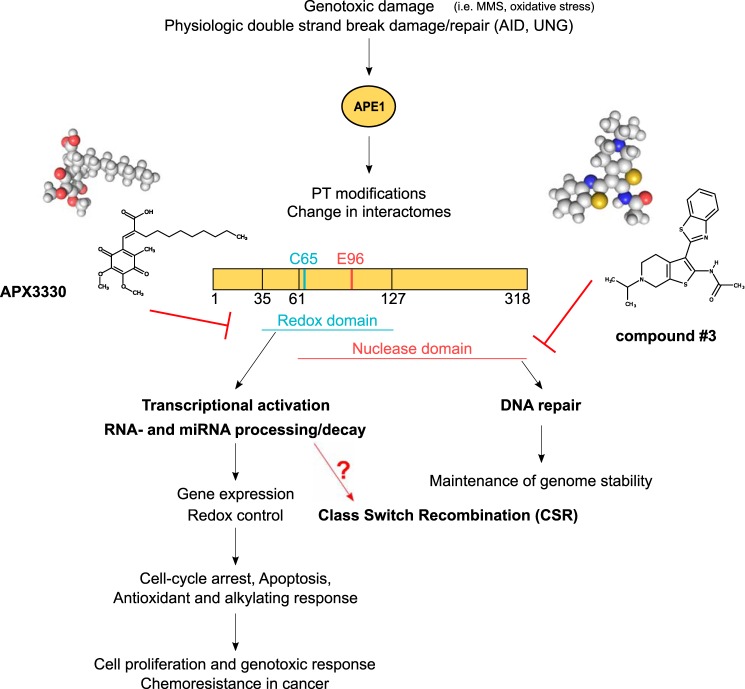
In addition to serving in a crucial role in the maintenance of genome stability through its endonuclease activity, APE1 also acts as a master regulator of cellular response–regulating gene expression through its redox-signaling activity (6–8). APE1 redox mediated stimulation of the DNA-binding activity of several transcription factors (for this reason APE1 is also called Ref-1, for redox effector factor 1) including STAT3, NF-κB, AP-1, HIF-1α, p53, and others affecting the promotion of tumor growth and progression, as well as the inflammation process. APE1 is a unique nuclear redox-signaling factor bearing seven Cys residues. Three of the Cys residues, Cys65, Cys93, and Cys99, are sufficient for its redox activity, and the mechanism for its redox activity has been characterized in detail and involves a redox cycle with potential formation of intermolecular disulfide bonds with the protein target (9–14). Although Cys65 acts as the nucleophilic cysteine, Cys93 and Cys99 likely play roles in resolving disulfide bonds that are formed in APE1 upon oxidation. The redox-active Cys residues are clustered in one of the two β-sheets that make up the β-sandwich fold of the APE1 structure. Cys65 and Cys93 are located within adjacent strands in the middle of one of the β-sheets within APE1, albeit positioned on opposite sides of the sheet, whereas Cys99 is located within a connecting loop region. Structural studies demonstrated that APE1 exists in both native and partially unfolded conformations. Thioredoxin is involved in the redox cycle through the formation of a large complex upon mixed disulfide bond with partially unfolded APE1 (9). Therefore, the partially unfolded state of APE1 represents the redox active intermediate of the enzyme. The unique mechanism of action of the most well-characterized APE1 redox inhibitor, i.e. APX3330 (formerly E3330), is based on APX3330 interacting with the locally unfolded state of APE1. APX3330 stabilizes this unfolded state causing buried Cys residues such as Cys65 and Cys93 to be exposed and facilitates disulfide-bond formation. This disulfide-bond formation results in an inactivation of APE1 and a decrease in interaction with downstream transcriptor factors, effectively causing them to be inactive (10, 12, 13). Fine-tuning of the different APE1 functions is affected by numerous post-translational modifications, as well as by the protein interactome (15).
It can be speculated that a role for APE1 in regulating the CSR through its function as a transcriptional activator could account for the remaining CSR activity observed in APE1-endonuclease activity–deficient cells (16). Although APE1 endonuclease activity is important for CSR, it is dispensable for SHM and AID-induced DNA breaks and may function as a DNA end-processing enzyme to facilitate the joining of broken ends during CSR (5). Notably, the mechanism by which AID is targeted to its substrates is of great interest, given the potentially deleterious consequences of AID's mutagenic activity (17). Off-target AID activities can activate oncogenes via mutations or translocations and thereby contribute to B cell as well as in other cancers. Cumulating evidence demonstrates that APE1 may represent a new signaling node in cancer and other diseases through a function in RNA metabolism, including micr0RNA and RNA processing (6).
Because APE1 has emerged as a promising target for cancer therapy, great efforts have been directed, by different groups including ours, to develop specific small molecules inhibitors of the different APE1 functions. Actually, DNA repair (compound 3) (18), redox (i.e. APX3330) (10, 11, 19–22), and inhibitors of the APE1/NPM1 interaction (i.e. spiclomazine, fiduxosin, SB206553) (23) are all available to dissect the specific role of each function in controlling multiple biological processes by APE1 (6). APX3330 has been shown to be specific for inhibition of APE1 redox function and blocking the activity of redox-regulated transcription factors. Additionally, APX3330 has entered phase I clinical trials, which are concluding (NCT03375086) (24–26). Furthermore, the blockade of APE1's redox activity has been shown to reduce growth-promoting, inflammatory, and anti-apoptotic activities in cells (27–29).
Importantly, the SHM apparatus, as well as the rate of mutation, can be influenced by the impact of radio- and chemotherapeutic regimens on DNA damage, genome stability, and gene expression. The maintenance of DNA integrity through repair is critical for genetic stability and limiting the deleterious effects on the immune system (30). Increased expression of the central BER protein APE1 confers resistance to some oxidative and alkylating agents, as well as IR (7). Therefore, the role of BER in controlling the physiologic function of the immune system is central, under normal and under genotoxic conditions, such as those represented by chemotherapy and radiotherapy regimens.
In the present study, we analyzed the contribution of both the DNA repair endonuclease and redox-signaling functions of APE1 in CSR using different APE1 inhibitors and APE1 DNA-repair and redox-defective mutants. We confirmed that the endonuclease function of APE1 is essential for IgA-class switch recombination. Interestingly, and for the first time, we provide evidence that the redox function of the protein also plays an important role in regulating CSR through the IL-6 signaling pathway and proper IgA expression.
Results
APE1 redox and endonuclease inhibitors reduce class switch recombination in CH12F3 cells
In these studies, we took advantage of the CSR-competent mouse B-cell line CH12F3, which undergoes robust IgA switching in response to appropriate stimuli. To elucidate which function of APE1 is important for CSR, CH12F3 cells were pretreated with specific APE1 inhibitors and then induced to CSR. Two different inhibitors were employed (Fig. 1): APX3330, a well-known inhibitor of APE1 redox signaling activity that does not affect the DNA repair activity, and the small molecule inhibitor compound 3 (18) that inhibits the APE1 endonuclease activity without hampering APE1 redox function (19). Both inhibitors were initially evaluated for toxicity to determine the appropriate subtoxic concentrations and to exclude off-target effects (Fig. S1). Cell viability was determined to be unaffected by after treating the cells up to 50 μm of APX3330, whereas some cell toxicity was observed at the highest tested dose of compound 3 (1 μm). The efficacy of 0.2 μm of compound 3 inhibitor, as well as the expression levels of APE1, were also tested (Fig. S2). The doses of 0.2 μm of compound 3 and 50 μm of APX3330 were subsequently used in which APE1 protein expression remained unaffected.
Figure 1.
Different APE1 functions in mammalian cells involve independent subdomains.
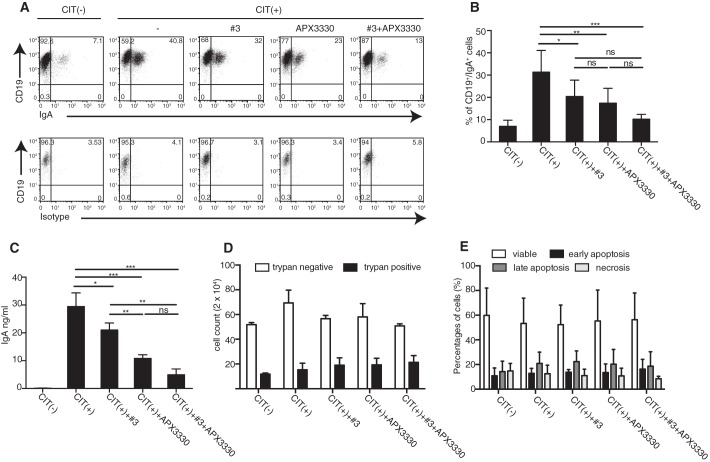
For the CSR experiment, CH12F3 were preincubated with 50 μm APX3330, 0.2 μm of compound 3, and a combination of both drugs for 30 min and then induced to CSR through the addition of the anti-CD40 antibody and recombinant IL-4 and TGF-β (CIT). After 72 h, the cells were harvested and analyzed for surface IgA expression (Fig. 2, A and B). Unstimulated CH12F3 cells, cultured in a normal medium, were used as negative control (CIT(−)). Under CSR conditions, and in the absence of the inhibitors, the percentage of IgA-positive cells reached 32% versus 7% for the unstimulated CH12F3, whereas the overall IgA-class switching in cells treated with compound 3 and APX3330 was reduced with respect to untreated cells (20 and 17% of IgA+ cells in presence of compound 3 and APX3330, respectively). Lower doses of APX3330 and compound 3 were also tested, but as shown in Table 1, the observed reduction of IgA expression was less pronounced than that obtained with 50 μm APX3330 and 0.2 μm of compound 3. The higher dose of compound 3 (1 μm) demonstrated the best results in terms of IgA reduction (Table 1) but is associated with a toxic effect (Fig. S1A).
Figure 2.
APE1 inhibitors APX3330 and compound 3 reduced class switch recombination in CH12F3-treated cells. A–C, representative dot plots (A), percentages of CD19/IgA double-positive cells (B), and amounts of released IgA (C) of CH12F3 under CIT(−) or CIT(+) conditions in absence or in presence of 50 μm APX3330, 0.2 μm compound 3, or both. The data are representative of three independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.001; ns, not significant. D, live (trypan negative) and dead (trypan positive) cells evaluated by trypan blue exclusion after 72 h of culture under CIT(−) or CIT(+) condition in the presence of indicated inhibitors. E, percentages of viable, early apoptotic, late apoptotic, and necrotic CH12F3 cells measured by annexin/PI staining. The data are representative of two independent experiments performed in triplicate.
Table 1.
Effectiveness of different doses of compound 3 and APX3330 in reducing IgA switching of CH12F3 cells.
| Compound 3 |
APX3330 |
|||||
|---|---|---|---|---|---|---|
| Dose (μm) | 0.04 | 0.2 | 1 | 2.5 | 12.5 | 50 |
| IgA reduction (%) | 5 ± 2.6 | 12 ± 1.9 | 25 ± 5.9 | 0.5 ± 2.5 | 7 ± 3.5 | 31 ± 8 |
Interestingly, when APE1 inhibitors were used in combination, a greater inhibitory effect was observed, resulting in less than 10% of CH12F3 cells expressing IgA. This inhibitory effect was confirmed by evaluation of IgA released in the cells supernatants especially when APX3330 and compound 3 were used in combination (Fig. 2C and Fig. S3A). The effect of both APE1 inhibitors on CSR was not due to toxicity or hampered proliferation at the doses employed in the CSR experiments, as determined by viable cell counting (Fig. 2D) and by evaluation of viable and dead cells following 72 h of a challenge with both inhibitors (Fig. 2E). Together, these data indicate that both the endonuclease and the redox activities of APE1 exert a role in CSR.
Expression of APE1-redox and -endonuclease defective mutants differently affect class switch recombination in CH12F3 cells
To investigate the dual role of APE1 through an alternative approach and to exclude possible off-target effects by the two inhibitor compounds, we expressed APE1 WT and its loss-of-function mutant forms (either redox- or DNA repair–deficient) in the CH12F3 cells genetically ablated of Ape1 gene, previously generated by Masani et al. (16). These experiments allow dissecting the dual role of APE1 in CSR, avoiding the confounding contribution of the endogenous APE1 protein. In particular, we employed CH12F3 APE1+/+/Δ and CH12F3 APE1Δ/Δ/Δ cells containing two and zero copies of APE1 gene, resulting from one and three cycles of gene targeting, respectively (Fig. S4). As expected, the absence of APE1 completely impaired the capacity of the CH12F3 cells to undergo CSR, whereas the CH12F3 APE1+/+/Δ cells were still able to switch toward IgA (Fig. 3, A and B) and to secrete the immunoglobulin in the cell supernatant (Fig. 3C).
Figure 3.
Re-expression of APE1WT but not APE1C65S or APE1E96A mutant forms restores the ability of APE1-null CH12F3 cells to undergo isotype switching. A–C, representative dot plots (A), percentages of CD19/IgA double-positive cells (B), and amounts of released IgA (C) by switching CH12F3 APE1+/+/Δ and CH12F3 APE1Δ/Δ/Δ cells under CIT(−) and CIT(+) conditions. D–F, representative dot plots (D), percentages of GFP/IgA double-positive cells (E), and amounts of released IgA (F) by CH12F3 APE1+/+/Δ transfected with empty vector or with GFP-APE1WT, APE1C65S, or APE1E96A mutant forms and induced toward IgA switching. G–I, representative dot plots (G), percentages of GFP/IgA double-positive cells (H), and amounts of released IgA (I) by CH12F3 APE1Δ/Δ/Δ transfected with empty vector, GFP-APE1WT, APE1C65S, or APE1E96A mutant forms and induced toward IgA switching. The data are representative of four independent experiments. *, p < 0.05; **, p < 0.005; ***, p < 0.001; ns, not significant.
Under CSR condition, the exposure to APX3330 and the compound 3 inhibitors reduced the expression of IgA in CH12F3 APE1+/+/Δ, whereas the CH12F3 APE1Δ/Δ/Δ cells were still negative for IgA expression (Fig. S5, A and B). Annexin/PI staining ruled out a negative effect on the cellular viability by the inhibitors (Fig. S5, C and D).
Subsequently, CH12F3 APE1+/+/Δ and CH12F3Δ/Δ/Δ cells were transfected with a nuclease-defective (APE1E96A) (31) and a redox-defective (APE1C65S) APE1 (32). To minimize the potential variation among samples, WT and mutant APE1 constructs were cloned in frame with a green fluorescent protein sequence to allow expression of the GFP-tagged protein. In this way, when performing FACS analysis, IgA-positive cells were investigated among the whole GFP-positive population (Fig. S6A). Cytofluorimetric analysis of the GFP-positive cells showed that, upon transient transfection, WT APE1 (APE1WT) and mutant transgenes were similarly expressed in both CH12F3 APE1+/+/Δ and CH12F3 APE1Δ/Δ/Δ cells (Fig. S6, B and C).
Data in Fig. 2D show that transfection with plasmids containing the APE1WT as well as APE1C65S and APE1E96A mutants did not significantly modify the CSR response of the CH12F3 APE1+/+/Δ cells, being similar to that observed with the empty vector both in terms of surface IgA expression (Fig. 3, D and E) and of IgA secretion (Fig. 3F).
On the other hand, re-expression of APE1WT in CH12F3 APE1Δ/Δ/Δ restored their IgA switching ability, as shown by increased IgA positivity from 0.4 to 3.98%, a 10-fold increase. Notably, transfections with both APE1 mutant forms only mildly restored the CSR capacity of APE1-null cells (Fig. 3, G and H). The ELISA of supernatant-released IgA confirmed the cytofluorimetric data obtained (Fig. 3I). These results demonstrate that the redox-signaling activity of APE1 is required for optimal CSR.
APX3330 impairs the production of IL-6 that is crucial for the efficient class switch recombination
APE1 redox activity is fundamental in immune cells for the production of several cytokines, whose expression is regulated by redox-sensitive transcription factors, such as NF-κB and STAT3 (24). Among these cytokines, IL-6 is primarily involved in B cell proliferation and cell function (33).
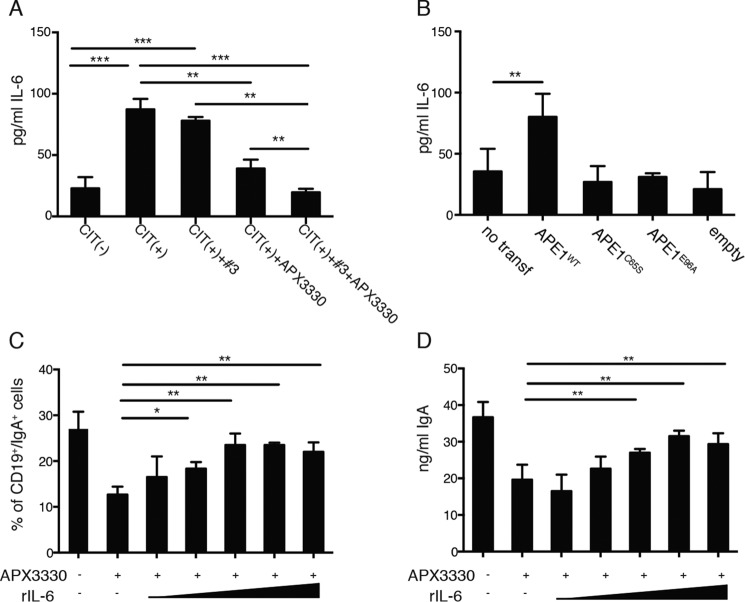
To test whether the redox activity of APE1 in CSR was mediated by IL-6, we assessed the amounts of IL-6 released by CH12F3 treated with the APE1 inhibitors with particular attention to the APX3330 inhibitor. As shown in Fig. 4A, CH12F3 releases a discrete amount of IL-6 both under unstimulated or stimulated conditions. Although pretreatment with compound 3 was ineffective on IL-6 expression, incubation with APX3330 inhibitor strongly reduced the levels of IL-6 detected in the cell supernatants (Fig. 4A and Fig. S3B). Use of the inhibitors in combination did not show any additive effects (Fig. 4A and Fig. S3B). Moreover, as a further confirmation of the important role of APE1 in IL-6 production, an increased release of IL-6 by CH12F3 transfected with APE1WT expression plasmid was determined (Fig. 4B).
Figure 4.
APX3330 impairs the production of IL-6 that is crucial for the optimal CSR. A, levels of secreted IL-6 in the supernatants of CH12F3 cells under CIT(−) or CIT(+) conditions in presence or absence of APE1 inhibitors. B, IL-6 detection in supernatants of not transfected or CH12F3 cells transfected with empty vector or with GFP-APE1WT, APE1C65S, or APE1E96A undergoing CSR. The data in A and B are representative of three independent experiments. C and D, percentages of CD19/IgA double-positive cells (C) and ng/ml of released IgA (D) by switching CH12F3 cells untreated, pretreated with APX3330, or pretreated with APX3330 and induced to CSR in presence of increasing amounts of exogenous recombinant IL-6 (rIL-6) ranging from 0.1 to 10,000 pg/ml. The data are representative of three independent experiments, and comparisons were performed between cells treated with APX3330 versus cells treated with APX330 in presence of IL-6. *, p < 0.05; **, p < 0.005; ***, p < 0.001.
To investigate whether APX3330 reduces IL-6 expression and impact on CSR efficacy, CH12F3 cells were induced to switch in the presence of increasing amounts of recombinant IL-6 added exogenously (doses ranging from 0.1 to 10,000 pg/ml). The percentage of IgA-positive cells (Fig. 4C) and the concentration (ng/ml) of secreted IgA in APX3330-treated CH12F3 (Fig. 4D) were restored by addition of recombinant IL-6, reaching the same values as untreated cells. Thus, exogenously added recombinant IL-6 restored the ability of APX3330-treated cells to switch toward IgA. These data demonstrate that the role of APE1 endonuclease activity in CSR is mainly associated with the DNA recombination process, whereas the redox function plays an essential, although indirect, role through modulation of IL-6 expression.
In vivo effect of APX3330 on T-independent B-cell response to TNP–Ficoll immunization
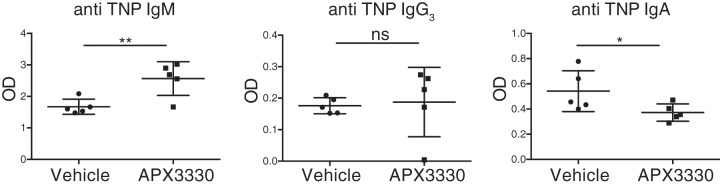
IgA class switching occurs via both T-cell–dependent and T-cell–independent pathways, and the antibody targets both pathogenic and commensal microorganisms. To investigate the in vivo effect of APE1 inhibition on T-cell–independent IgA class switching, we treated mice with 50 mg/kg of APX3330 the day before the immunization with TNP–Ficoll and every day for 7 days. At day 8, the blood levels of anti-TNP IgG, IgM, and IgA were evaluated by ELISA. As shown in Fig. 5, treatment with the APX3330 did not affect the serum level of IgG3 but increased the amount of anti-TNP specific IgM and reduced the amount of anti-TNP–specific IgA.
Figure 5.
In vivo effect of APX3330 on T-independent B-cell response to TNP–Ficoll immunization. APX3330-treated mice (50 mg/Kg) and control mice were immunized with TNP–Ficoll, and the levels of specific anti-TNP IgM, IgG3, and IgA were evaluated in the peripheral blood 1 week after immunization. *, p < 0.05; **, p < 0.005; ns, not significant.
Discussion
This work was undertaken to evaluate the relative contribution of the endonuclease and redox activities of APE1, a central player in the BER pathway, in CSR. BER enzymes play a central and critical role in controlling the physiologic function of the immune system under normal and genotoxic conditions, such as those represented by chemotherapy and radiotherapy regimens. In antigen-stimulated B cells, both SHM and CSR provide diversification of the variable regions of immunoglobulin heavy and light chain genes and diversification of the heavy chain constant region. These two antibody maturation mechanisms require AID, which deaminates DNA cytosines to uracils at transcribed V and switch S regions. Uracil in DNA can be detected and repaired by the glycosylase UNG2-dependent BER pathway. APE1, through its DNA repair AP-endonuclease activity, is responsible for the introduction of DNA nicks, a prerequisite for the repair of the abasic sites generated by glycosylase action on damaged DNA. Despite several uracil glycosylases being present in mammals, APE1 is the only AP-endonuclease, although a weak endonuclease activity has been recently identified for APE2 (16, 34, 35). Although APE1 endonuclease activity is important for CSR, it is dispensable for SHM and AID-induced DNA breaks (5). In addition to APE1 and UNG2, all the other BER enzymes effectively respond to the uracil removal process (36). The role of BER enzymes in SHM and CSR strongly depends on cross-talk between each single protein partner and is finely tuned by the expression level of each partner and by specific regulated post-translational modifications, such as phosphorylation of Ser38 in AID protein (3, 4). Emerging evidence demonstrates that the protein interactome and different post-translational modifications occurring on the APE1 N-terminal domain are a major mechanism to fine-tune and differentiate APE1 activities (7, 15). These properties will deserve additional studies to fully understand the role of BER in CSR and SHM.
As mentioned, although the role of APE1 endonuclease activity in the CSR process is established, a role for APE1 in regulating the CSR through its function as a redox-transcriptional coactivator is still unknown. Therefore, we here evaluated the role of the APE1 redox activity in CSR of B cells compared with the DNA repair endonuclease activity by using a combination of specific small molecules inhibition and genetic approaches.
We confirmed that the endonuclease function of APE1 is an important, but not unique, determinant for IgA-class switch recombination. We also provided evidence that the redox function of the protein plays an important role in regulating CSR through the IL-6 signaling pathway and proper IgA expression. A relationship between APE1 and IL-6 secretion has been previously established through APE1's redox regulation of NF-κB and AP1 transcription factors, promoting IL-6 expression (24, 37). Furthermore, IL-6 itself was reported to induce cytoplasmic translocation of APE1 followed by its secretion and subsequent feedforward loop as a mediator of the cellular response by inducing IL-6 production and secretion (38). However, we cannot exclude that the redox function of APE1 may influence the CSR of CH12F3 cells through additional IL-6–independent mechanisms, and further studies are required to address this issue.
In conclusion, we determined that both the endonuclease and redox activities are required for the optimal antibody diversification of the CH12F3 and, more importantly, those specific APE1 inhibitors can influence the isotype switching both in in vitro and in vivo settings. Our findings open new clinical perspectives for the use of APE1 redox inhibitors as novel anti-inflammatory compounds, in light of the fact that APX3330 is currently completing phase I clinical trials (39). In fact, antibody diversification is essential for the immune system to mount protective humoral response through the generation of secondary IgG, IgA, and IgE isotypes that have the same antigen specificity as IgM and IgD but different effector functions. Indeed, by binding to specific Fc receptors, secondary isotypes can differently activate innate immune effector cells promoting pathogens opsonization and activation of the complement system with a different extent.
Clinically, the levels of serum Igs are often increased in cancer patients, mirroring specific pro- or anti-inflammatory response (40). In particular, the precise role of IgA in cancer is still controversial. It is commonly believed that the IgA switching assumes a pro-tumor connotation because of the intrinsic immunosuppressive activities of IgA. Indeed, this class of Ig is a less potent opsonin and a weak activator of complement cascade, as well as of the antibody-dependent cellular cytotoxicity-mediated activity of the NK cells compared with IgG. Therefore, it can be speculated that the skew toward IgA weakens of the anti-tumor immune response against a specific antigen compared with that induced by IgGs. Several reports document that increased concentrations of IgA in the secretion, as well as a higher expression of IgA in serum from patients with cancer, correlates with a poor prognosis (41). Very recently, high frequency of IgA1-positive tumor cells was found in tissue microarrays of esophagus, colon, testis, lung, breast, bladder, and ovarian cancer. IgA1 was observed in the cytoplasm and the plasma membrane. A correlation was found between intratumor IgA1 and poor overall survival in a large cohort of bladder cancer patients (42). For these reasons, the potential to control the skewing toward IgA could represent a novel and intriguing pharmacological approach to influence the humoral response in the management of the disease.
The inhibitor of the APE1 redox activity, APX3330, is currently completing phase I clinical trials for safety and recommended phase II dose finding for future oncological clinical trials. Our results shed light on a novel effect of APX3330 in the control of cancer growth by potentially modulating the IgA CSR process. Further studies will therefore be of great interest in providing avenues for targeting APE1 in many diseases that are linked to inflammation.
Experimental procedures
Reagents
Anti-mouse CD40 and anti-mouse IgA-FITC–conjugated reagents were from Becton Dickinson. Anti-mouse IgA-PE–conjugated, anti-mouse CD19-APC, and anti-mouse IgG2a reagents were from eBiosciences. APX3330 was from Sigma–Aldrich (E8534) (21), and compound 3 was a kind gift from (David Maloney from NIH) (18). Recombinant murine IL-4, TGF-β, and IL-6 were from Peprotech.
Cell culture and CSR
CH12F3 cells, gently gifted by T. Honjo, were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, glutamine, penicillin/streptomycin, and 50 μm β-mercaptoethanol. CH12F3 containing two (+/+/Δ) and zero copies of APE1 (Δ/Δ/Δ) have been described previously (16). To induce class switch recombination, 5 × 104 cell/ml were seeded in presence of 1 μg/ml anti-CD40 antibody (Becton Dickinson, clone HM40-3), 5 ng/ml of IL-4, and 0.5 ng/ml di-TGF-β (CIT) and grown for 72 h. The cells cultured in both CIT(−) and CIT(+) conditions were stained with fluorochrome-conjugated anti-mouse CD19 and anti-mouse IgA antibody and analyzed on FACScalibur. CSR efficiency was determined as the percentage of CD19/IgA-double positive cells. In experiments with the inhibitors, CH12F3 cells were preincubated with 50 μm or 0.2 μm of compound 3 for 1 h, and then switching stimuli were added to the cells without washing.
Cell viability and proliferation assay
Live and dead CH12F3 cells were evaluated by trypan blue exclusion test. Trypan blue–excluding cells were counted in a hemocytometer in four randomly selected fields, and averaged counts were reported. The cellular death was detected using the annexin V/PI double staining kit (eBiosciences). 1 × 105 cells were first stained with 2.5 μl of annexin V–FITC, and then 5 μl of propidium iodide was added into cell suspension in turn. The cell suspension was incubated at room temperature for 10 min, and the apoptosis rate was detected by flow cytometry on FACScalibur.
Cell viability was measured by using the CellTiter-Glo® luminescent cell viability assay (Promega) on cells grown in 96-well plates after different treatments. Upon treatment, the CellTiter-Glo® reagent was added to each well, and the plates were incubated for an additional 2 h at 37 °C. Then absorbance was measured at 490 nm by using a multiwell plate reader. The values were standardized to wells containing medium alone.
AP-site incision assays
APE1 endonuclease activity was monitored using CH123F3 cell extracts as follows (6, 23). Briefly, enzymatic reactions were carried out in a final volume of 10 μl using 8 ng of cell extracts in a buffer containing 50 mm Tris-HCl, pH 7.5, 50 mm KCl, 10 mm MgCl2, 1 μg ml−1 BSA, and 1 mm DTT. Extracts were incubated for 15 min, at 37 °C, with 100 nm of double-stranded 26-mer abasic DNA substrate containing a single tetrahydrofuranyl artificial AP site at position 14, which is cleaved to a 14-mer in the presence of AP endonuclease activity (23). The double-stranded DNA was obtained by annealing a 5′-DY-782-labeled oligonucleotide 5′-AATTCACCGGTACCFTCTAGAATTCG-3′ (where F indicates the THF residue), with an unlabeled complementary sequence 5′-CGAATTCTAGAGGGTACCGGTGAATT-3′. The reactions were halted by addition of formamide buffer (96% formamide, 10 mm EDTA, and 6× gel loading buffer (Fermentas)), separated onto a 20% (w/v) denaturing polyacrylamide gel, and analyzed on an Odyssey CLx scanner (Li-Cor Biosciences). The percentage of substrate converted to the product was determined using the ImageStudio software (Li-Cor Biosciences).
Antibodies and Western blotting analysis
For Western blotting analyses, whole cell lysates were prepared, and 20 μg of proteins were resolved on 12% SDS-PAGE, transferred onto nitrocellulose membranes (Sigma), and probed with antibodies for APE1 (13B8E5C2, Novus) (1:1000). Data normalization was performed by using monoclonal anti-actin (Sigma–Aldrich) as indicated. The corresponding secondary antibodies labeled with IR-Dye (anti-rabbit IgG IRDye 680 and anti-mouse IgG IRDye 800) were used. Detection and quantification were performed with the Odyssey CLx IR imaging system (LI-COR GmbH). The membranes were scanned in two different channels using an Odyssey IR imager; protein bands were quantified using Odyssey software (Image Studio 5.0).
ELISAs
The concentration of IgA in cell supernatants was assessed by a homemade sandwich ELISA. Briefly, 96-well flat-bottomed0 polystyrene plates (Corning) were coated with affinity-purified anti-mouse IgA (SouthernBiotech) at the final concentration of 2 mg/ml. After 1 h of incubation at 37 °C, the plates were washed with 0.05% Tween 20 in PBS and blocked with 1% BSA in PBS for 1 h at room temperature. 100 ml of cell supernatants or of opportunely diluted mouse sera were added to Ab-coated wells. Purified mouse IgA (BD PharMingen) was used as a standard. After overnight incubation at 4 °C, the plates were washed and optimal concentration of horseradish peroxidase–conjugated goat anti-mouse IgA (Southern Biotech; 1:2000) was added. Next, the plates were incubated for 1 h at room temperature and washed before the addition of tetramethylbenzidine substrate solution (Sigma–Aldrich). The reaction was stopped with 2 mol/liter sulfuric acid, and absorbance was measured at 450 nm. The levels of IL-6 in the supernatants were measured using a commercially available ELISA kit (Biosciences) following the manufacturer's instructions.
Transient transfection and cellular treatments
CH12F3, CH12F3 APE1+/+/Δ, and CH12F3 APE1Δ/Δ/Δ cells were transfected with pEGFP-N2 plasmids encoding for the APE1 WT or mutant protein using the neon transfection system (Invitrogen). Briefly, 1.5 × 105 cells mixed with 0.5 ng of the plasmid of interested in 10 μl of buffer R were electroporated at 1500 V and 30 ms. The cells were firstly recovered in antibiotic-free medium and after 2 h were induced to CSR in presence of G418 selection. After 72 h, the cells were harvested, and CSR efficiency was determined as the percentage of GFP/IgA-double positive cells.
In vivo T-independent immunization assay
All animal experiments were approved by the Indiana University School of Medicine Institutional Animal Care and Use Committee. The animals were maintained under pathogen-free conditions and a 12-h light-dark cycle. WT C57BL/6 mice, 6–8 weeks of age, were purchased from The Jackson Laboratory (Bar Harbor, ME).
Mice were immunized intraperitoneally with 25 μg of TNP(65)-AECM-Ficoll (Biosearch Technologies, Petaluma, CA) at day 1. Antigen was diluted in PBS and injected in a final volume of 200 μl of PBS. On days 0–7 mice received 50 mg/kg APX3330 intraperitoneally. Control mice were injected with the same volume of PBS. On day 8 after immunization, the mice were sacrificed. Blood was taken by cardiac puncture, and separated sera were frozen at −20 °C until use. Four 2-month-old mice were used per group.
For serum Ig detection, ELISA plate was coated overnight at 4 °C with 5 μg/ml TNP(30)-BSA (Biosearch Technologies) in 0.05 m carbonate buffer (pH 9.6). The plate was washed three times with 0.05% Tween 20 in PBS and quenched for 1 h with PBS–BSA (1%) at 37 °C.
Each serum obtained from immunized and control mice was diluted in PBS with 1% BSA (2 μl of sera in 1 ml of PBS). 100 μl of diluted sera were plated for 1 h at 37 °C, and then after washing, horseradish peroxidase–conjugated anti-mouse IgM, IgA, or IgG3 (Biolegend) diluted in PBS with 1% BSA was added for 1 h at 37 °C. The plate was developed by adding 100 μl of room temperature TMB Sure Blue reagent (Sigma) per well, and the reaction was stopped by adding 100 μl of TMB stop solution. Absorbance was read at 450 nm
Statistical analyses
The results are presented as means ± S.D., and data analysis was performed with the Prism GraphPad software. For comparisons between two groups, unpaired and paired Student's t tests were used. When multiple comparisons were necessary, the data were analyzed with the one-way analysis of variance test, and the Bonferroni correction was used as post hoc analysis. In all tests, p values < 0.05 were considered statistically significant. *, p < 0.05; **, p < 0.005; ***, p < 0.001.
Author contributions
B. F., M. H. K., M. R. K., G. T., and C. E. M. P. conceptualization; B. F., G. A., and G. T. data curation; B. F. and G. A. software; B. F., G. A., G. T., and C. E. M. P. formal analysis; B. F., G. A., and N. A. validation; B. F., G. A., G. T., and C. E. M. P. investigation; B. F., G. A., N. A., M. H. K., and M. R. K. visualization; B. F., G. A., K. Y., N. A., G. T., and C. E. M. P. methodology; B. F., M. H. K., M. R. K., G. T., and C. E. M. P. writing-original draft; G. A., K. Y., M. H. K., M. R. K., G. T., and C. E. M. P. writing-review and editing; M. H. K., M. R. K., G. T., and C. E. M. P. resources; M. H. K., M. R. K., G. T., and C. E. M. P. supervision; M. H. K., M. R. K., and G. T. funding acquisition.
Supplementary Material
Acknowledgments
We thank the Tell and Pucillo laboratories for constructive feedbacks during the development of this work.
This work was supported by Associazione Italiana per la Ricerca sul Cancro Grant IG19862 (to G. T.), by National Institutes of Health Grants R01CA205166 and R01CA167291 (to M. R. K.), and by funds granted the Earl and Betty Herr Professor in Pediatric Oncology Research and the Riley Children's Foundation (to M. R. K.). Mark R. Kelley is Chief Scientific Officer of Apexian Pharmaceuticals, the biotech company that has licensed APX3330 used in these studies. Apexian Pharmaceuticals had neither control nor oversight of the studies, interpretation, or presentation of the data in this manuscript. They did not have to approve the manuscript in any way prior to its submission. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S6.
- SHM
- somatic hypermutation
- APE1
- apurinic/apyrimidinic endonuclease 1
- AID
- activation-induced cytidine deaminase
- BER
- base excision repair
- CSR
- class switch recombination
- IL
- interleukin
- IR
- ionizing radiation
- PI
- propidium iodide
- STAT
- signal transducers and activators of transcription
- TGF
- transforming growth factor.
References
- 1. Stavnezer J., Guikema J. E., and Schrader C. E. (2008) Mechanism and regulation of class switch recombination. Annu. Rev. Immunol. 26, 261–292 10.1146/annurev.immunol.26.021607.090248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wallace S. S. (2014) Base excision repair: a critical player in many games. DNA Repair 19, 14–26 10.1016/j.dnarep.2014.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vuong B. Q., Herrick-Reynolds K., Vaidyanathan B., Pucella J. N., Ucher A. J., Donghia N. M., Gu X., Nicolas L., Nowak U., Rahman N., Strout M. P., Mills K. D., Stavnezer J., and Chaudhuri J. (2013) A DNA break- and phosphorylation-dependent positive feedback loop promotes immunoglobulin class-switch recombination. Nat. Immunol. 14, 1183–1189 10.1038/ni.2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vuong B. Q., and Chaudhuri J. (2012) Combinatorial mechanisms regulating AID-dependent DNA deamination: interacting proteins and post-translational modifications. Semin. Immunol. 24, 264–272 10.1016/j.smim.2012.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu J., Husain A., Hu W., Honjo T., and Kobayashi M. (2014) APE1 is dispensable for S-region cleavage but required for its repair in class switch recombination. Proc. Natl. Acad. Sci. U.S.A. 111, 17242–17247 10.1073/pnas.1420221111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Antoniali G., Serra F., Lirussi L., Tanaka M., D'Ambrosio C., Zhang S., Radovic S., Dalla E., Ciani Y., Scaloni A., Li M., Piazza S., and Tell G. (2017) Mammalian APE1 controls miRNA processing and its interactome is linked to cancer RNA metabolism. Nat. Commun. 8, 797 10.1038/s41467-017-00842-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tell G., Fantini D., and Quadrifoglio F. (2010) Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol. Life Sci. 67, 3589–3608 10.1007/s00018-010-0486-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Antoniali G., Lirussi L., D'Ambrosio C., Dal Piaz F., Vascotto C., Casarano E., Marasco D., Scaloni A., Fogolari F., and Tell G. (2014) SIRT1 gene expression upon genotoxic damage is regulated by APE1 through nCaRE-promoter elements. Mol. Biol. Cell 25, 532–547 10.1091/mbc.e13-05-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luo M., Zhang J., He H., Su D., Chen Q., Gross M. L., Kelley M. R., and Georgiadis M. M. (2012) Characterization of the redox activity and disulfide bond formation in apurinic/apyrimidinic endonuclease. Biochemistry 51, 695–705 10.1021/bi201034z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J., Luo M., Marasco D., Logsdon D., LaFavers K. A., Chen Q., Reed A., Kelley M. R., Gross M. L., and Georgiadis M. M. (2013) Inhibition of apurinic/apyrimidinic endonuclease I's redox activity revisited. Biochemistry 52, 2955–2966 10.1021/bi400179m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Luo M., Delaplane S., Jiang A., Reed A., He Y., Fishel M., Nyland R. L. 2nd, Borch R. F., Qiao X., Georgiadis M. M., and Kelley M. R. (2008) Role of the multifunctional DNA repair and redox signaling protein Ape1/Ref-1 in cancer and endothelial cells: small-molecule inhibition of the redox function of Ape1. Antioxid. Redox. Signal. 10, 1853–1867 10.1089/ars.2008.2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Georgiadis M., Luo M., Zhang J., Basavarajappa H., Mahalingan K., Chen Q., Reed A., Gross M., and Kelley M. (2012) An uncharged amide derivative of E3330 inhibits the redox but not the DNA-binding activity of APE. Bioorg. Med. Chem. [Google Scholar]
- 13. Su D., Delaplane S., Luo M., Rempel D. L., Vu B., Kelley M. R., Gross M. L., and Georgiadis M. M. (2011) Interactions of apurinic/apyrimidinic endonuclease with a redox inhibitor: evidence for an alternate conformation of the enzyme. Biochemistry 50, 82–92 10.1021/bi101248s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Georgiadis M. M., Luo M., Gaur R. K., Delaplane S., Li X., and Kelley M. R. (2008) Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease. Mutat. Res. 643, 54–63 10.1016/j.mrfmmm.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tell G., and Demple B. (2015) Base excision DNA repair and cancer. Oncotarget 6, 584–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masani S., Han L., and Yu K. (2013) Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol. Cell Biol. 33, 1468–1473 10.1128/MCB.00026-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gostissa M., Alt F. W., and Chiarle R. (2011) Mechanisms that promote and suppress chromosomal translocations in lymphocytes. Annu. Rev. Immunol. 29, 319–350 10.1146/annurev-immunol-031210-101329 [DOI] [PubMed] [Google Scholar]
- 18. Rai G., Vyjayanti V. N., Dorjsuren D., Simeonov A., Jadhav A., Wilson D. M. 3rd, Maloney D. J. (2012) Synthesis, biological evaluation, and structure-activity relationships of a novel class of apurinic/apyrimidinic endonuclease 1 inhibitors. J. Med. Chem. 55, 3101–3112 10.1021/jm201537d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson D. M. 3rd, and Simeonov A. (2010) Small molecule inhibitors of DNA repair nuclease activities of APE1. Cell Mol. Life Sci. 67, 3621–3631 10.1007/s00018-010-0488-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kelley M. R., Wikel J. H., Guo C., Pollok K. E., Bailey B. J., Wireman R., Fishel M. L., and Vasko M. R. (2016) Identification and characterization of new chemical entities targeting apurinic/apyrimidinic endonuclease 1 for the prevention of chemotherapy-induced peripheral neuropathy. J. Pharmacol. Exp. Ther. 359, 300–309 10.1124/jpet.116.235283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kelley M. R., Luo M., Reed A., Su D., Delaplane S., Borch R. F., Nyland R. L. 2nd, Gross M. L., and Georgiadis M. M. (2011) Functional analysis of novel analogues of E3330 that block the redox signaling activity of the multifunctional AP endonuclease/redox signaling enzyme APE1/Ref-1. Antioxid. Redox. Signal. 14, 1387–1401 10.1089/ars.2010.3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nyland R. L., Luo M., Kelley M. R., and Borch R. F. (2010) Design and synthesis of novel quinone inhibitors targeted to the redox function of apurinic/apyrimidinic endonuclease 1/redox enhancing factor-1 (Ape1/ref-1). J. Med. Chem. 53, 1200–1210 10.1021/jm9014857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poletto M., Malfatti M. C., Dorjsuren D., Scognamiglio P. L., Marasco D., Vascotto C., Jadhav A., Maloney D. J., Wilson D. M. 3rd, Simeonov A., and Tell G. (2016) Inhibitors of the apurinic/apyrimidinic endonuclease 1 (APE1)/nucleophosmin (NPM1) interaction that display anti-tumor properties. Mol. Carcinog. 55, 688–704 10.1002/mc.22313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cesaratto L., Codarin E., Vascotto C., Leonardi A., Kelley M. R., Tiribelli C., and Tell G. (2013) Specific inhibition of the redox activity of ape1/ref-1 by e3330 blocks TNF-α-induced activation of IL-8 production in liver cancer cell lines. PLoS One 8, e70909 10.1371/journal.pone.0070909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Biswas A., Khanna S., Roy S., Pan X., Sen C. K., and Gordillo G. M. (2015) Endothelial cell tumor growth is Ape/ref-1 dependent. Am. J. Physiol. Cell Physiol. 309, C296–C307 10.1152/ajpcell.00022.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Logsdon D. P., Grimard M., Luo M., Shahda S., Jiang Y., Tong Y., Yu Z., Zyromski N., Schipani E., Carta F., Supuran C. T., Korc M., Ivan M., Kelley M. R., and Fishel M. L. (2016) Regulation of HIF1α under hypoxia by APE1/Ref-1 impacts CA9 expression: dual targeting in patient-derived 3D pancreatic cancer models. Mol. Cancer Ther. 15, 2722–2732 10.1158/1535-7163.MCT-16-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zaky A., Bouali-Benazzouz R., Favereaux A., Tell G., and Landry M. (2018) APE1/Ref-1 redox function contributes to inflammatory pain sensitization. Exp. Neurol. 307, 1–11 10.1016/j.expneurol.2018.05.014 [DOI] [PubMed] [Google Scholar]
- 28. Montaldi A. P., Godoy P. R., and Sakamoto-Hojo E. T. (2015) APE1/REF-1 down-regulation enhances the cytotoxic effects of temozolomide in a resistant glioblastoma cell line. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 793, 19–29 10.1016/j.mrgentox.2015.06.001 [DOI] [PubMed] [Google Scholar]
- 29. Guan Z., Basi D., Li Q., Mariash A., Xia Y.-F., Geng J.-G., Kao E., and Hall J. L. (2005) Loss of redox factor 1 decreases NF-kappaB activity and increases susceptibility of endothelial cells to apoptosis. Arterioscler. Thromb. Vasc. Biol. 25, 96–101 10.1161/01.ATV.0000150418.14698.75 [DOI] [PubMed] [Google Scholar]
- 30. Mantha A. K., Sarkar B., and Tell G. (2014) A short review on the implications of base excision repair pathway for neurons: relevance to neurodegenerative diseases. Mitochondrion 16, 38–49 10.1016/j.mito.2013.10.007 [DOI] [PubMed] [Google Scholar]
- 31. Barzilay G., Walker L. J., Robson C. N., and Hickson I. D. (1995) Site-directed mutagenesis of the human DNA repair enzyme HAP1: identification of residues important for AP endonuclease and RNase H activity. Nucleic Acids Res. 23, 1544–1550 10.1093/nar/23.9.1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walker L. J., Robson C. N., Black E., Gillespie D., and Hickson I. D. (1993) Identification of residues in the human DNA repair enzyme HAP1 (Ref-1) that are essential for redox regulation of Jun DNA binding. Mol. Cell Biol. 13, 5370–5376 10.1128/MCB.13.9.5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Akira S., Hirano T., Taga T., and Kishimoto T. (1990) Biology of multifunctional cytokines: IL 6 and related molecules (IL 1 and TNF). FASEB J. 4, 2860–2867 10.1096/fasebj.4.11.2199284 [DOI] [PubMed] [Google Scholar]
- 34. Schrader C. E., Guikema J. E., Wu X., and Stavnezer J. (2009) The roles of APE1, APE2, DNA polymerase β and mismatch repair in creating S region DNA breaks during antibody class switch. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 364, 645–652 10.1098/rstb.2008.0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guikema J. E., Linehan E. K., Tsuchimoto D., Nakabeppu Y., Strauss P. R., Stavnezer J., and Schrader C. E. (2007) APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J. Exp. Med. 204, 3017–3026 10.1084/jem.20071289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akbari M., Otterlei M., Peña-Diaz J., Aas P. A., Kavli B., Liabakk N. B., Hagen L., Imai K., Durandy A., Slupphaug G., and Krokan H. E. (2004) Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res. 32, 5486–5498 10.1093/nar/gkh872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie J.-Y., Li M.-X., Xiang D.-B., Mou J.-H., Qing Y., Zeng L.-L., Yang Z.-Z., Guan W., and Wang D. (2010) Elevated expression of APE1/Ref-1 and its regulation on IL-6 and IL-8 in bone marrow stromal cells of multiple myeloma. Clin. Lymphoma Myeloma Leuk. 10, 385–393 10.3816/CLML.2010.n.072 [DOI] [PubMed] [Google Scholar]
- 38. Nath S., Roychoudhury S., Kling M. J., Song H., Biswas P., Shukla A., Band H., Joshi S., and Bhakat K. K. (2017) The extracellular role of DNA damage repair protein APE1 in regulation of IL-6 expression. Cell Signal. 39, 18–31 10.1016/j.cellsig.2017.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jiang S., Zhu L., Tang H., Zhang M., Chen Z., Fei J., Han B., and Zou G.-M. (2015) Ape1 regulates WNT/β-catenin signaling through its redox functional domain in pancreatic cancer cells. Int. J. Oncol. 47, 610–620 10.3892/ijo.2015.3048 [DOI] [PubMed] [Google Scholar]
- 40. Lee Y. T. (1977) Quantitative change of serum protein and immunoglobulin in patients with solid cancers. J. Surg. Oncol. 9, 179–187 10.1002/jso.2930090212 [DOI] [PubMed] [Google Scholar]
- 41. Roberts M. M., Bathgate E. M., and Stevenson A. (1975) Serum immunoglobulin levels in patients with breast cancer. Cancer 36, 221–224 [DOI] [PubMed] [Google Scholar]
- 42. Welinder C., Jirström K., Lehn S., Nodin B., Marko-Varga G., Blixt O., Danielsson L., and Jansson B. (2016) Intra-tumour IgA1 is common in cancer and is correlated with poor prognosis in bladder cancer. Heliyon 2, e00143 10.1016/j.heliyon.2016.e00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.