SUMMARY
Group II introns, self-splicing retrotransposons, serve as both targets of investigation into their structure, splicing, and retromobility and a source of tools for genome editing and RNA analysis. Here, we describe the first cryo-electron microscopy (cryo-EM) structure determination, at 3.8–4.5 Å, of a group II intron ribozyme complexed with its encoded protein, containing a reverse transcriptase (RT), required for RNA splicing and retromobility. We also describe a method called RIG-seq using a retrotransposon indicator gene for high-throughput integration profiling of group II introns and other retrotransposons. Targetrons, RNA-guided gene targeting agents widely used for bacterial genome engineering, are described next. Finally, we detail thermostable group II intron RTs, which synthesize cDNAs with high accuracy and processivity, for use in various RNA-seq applications and relate their properties to a 3.0-Å crystal structure of the protein poised for reverse transcription. Biological insights from these group II intron revelations are discussed.
1. INTRODUCTION
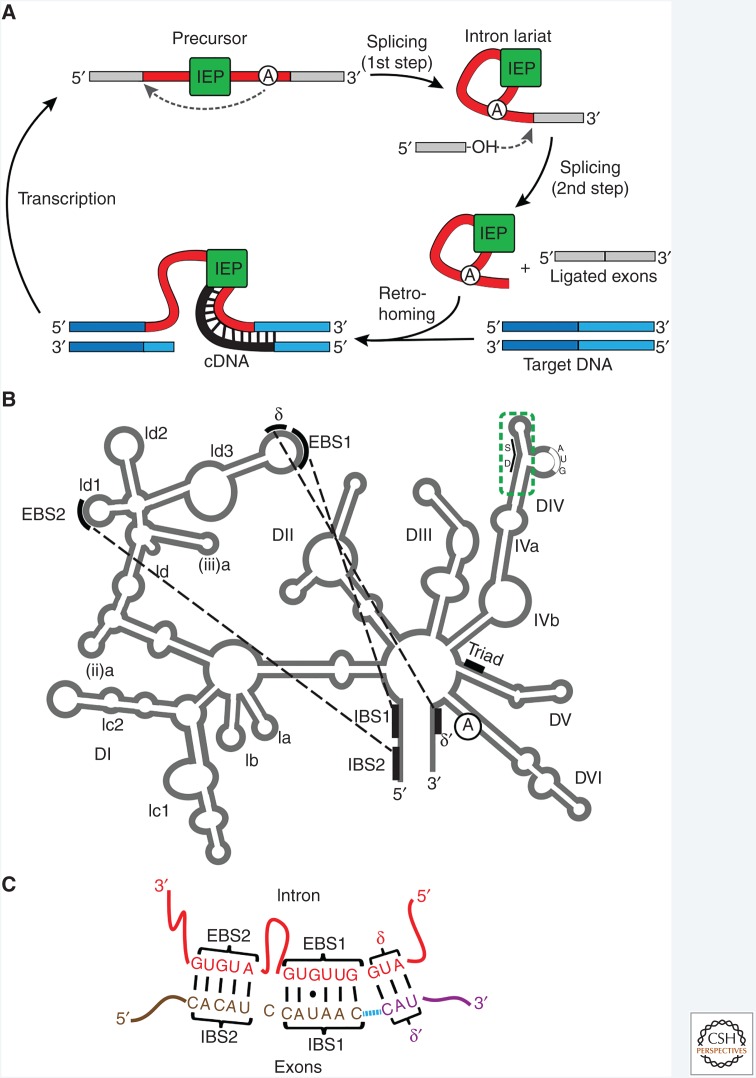
Group II introns are self-splicing RNAs and mobile retroelements that occupy a pivotal evolutionary niche as putative ancestors of spliceosomal introns and the spliceosome, retrotransposons, telomerase, and retroviruses (Lambowitz and Belfort 2015; Novikova and Belfort 2017). They also are important genome editing tools (Enyeart et al. 2014) and the source of reverse transcriptases (RTs) that have broad application in high-throughput characterization of RNAs (Mohr et al. 2013b). These group II intron ribozymes catalyze their self-excision from RNA precursors, and ligation of the flanking exons, with the aid of an intron-encoded protein (IEP), which is an ancient form of RT3 (Fig. 1A) (Lambowitz and Zimmerly 2011). The ribozyme comprises a conserved, six-domain structure (DI–DVI) (Fig. 1B). Splicing occurs via two transesterification reactions, which are initiated by the 2′-OH of a bulged adenosine nucleophile in DVI and result in ligated exons and an excised intron lariat, in precise analogy to spliceosomal intron splicing (Fig. 1A). The IEP functions as a “maturase” to facilitate RNA splicing and reverse splicing of the excised intron RNA into a DNA site on the top strand. After endonuclease cleavage of the bottom strand, retrohoming proceeds when the RT reverse transcribes the reverse-spliced intron RNA into an intron cDNA that is incorporated into the host genome. Group II intron RTs have a DNA-binding domain (denoted D) and many also have a DNA endonuclease (EN) domain, both of which are involved in retromobility.
Figure 1.
Group II intron structure and function. (A) Intron life cycle. Splicing is initiated by attack on the 5′ splice site by the 2′-OH of the bulged adenosine (circled A) in DVI to form a lariat intermediate (first step). Attack on the 3′ splice site by the 3′-OH of the upstream exon results in ligated exons and the intron lariat (second step). The intron lariat can reverse splice into target DNA (retrohoming). The endonuclease of the intron-encoded protein (IEP) then nicks the opposite DNA strand, and the reverse transcriptase (RT) uses the cleaved DNA strand as a primer to make a cDNA copy of the integrated intron. After degradation of the intron RNA, second-strand DNA synthesis and repair, leading to a fully integrated DNA copy of the intron in the genome (not shown), transcription can ensue to complete the cycle. (B) Secondary structure of a group II intron. The six domains (DI–DVI) radiating from a central hub are labeled along with various subdomains, the “exon-binding sequence”–“intron-binding sequence” (EBS–IBS) pairings, the IEP anchor site in DIV (dashed green box), the catalytic triad in DV, and the bulged adenosine in DVI. SD, Shine–Dalgarno sequence. (C) EBS–IBS pairings. EBS1–IBS1, EBS2–IBS2, and δ-δ′ are shown. Blue dashed line denotes ligation site.
Structural studies have provided insight into the RNA component of the ribonucleoprotein (RNP) complex (Toor et al. 2008; Robart et al. 2014; Costa et al. 2016). DI, the largest intron RNA domain, folds first. DI docks the active site domain, DV, and also base-pairs its exon-binding sequences (EBSs) with the intron-binding sequences (IBSs) in the exons (Fig. 1B,C). These base-pairings determine the specificity of forward and reverse splicing reactions (Lambowitz and Zimmerly 2011; Marcia and Pyle 2012). DII and DIII contribute structurally to the catalytic complex, whereas DIV encompasses an unstructured open reading frame (ORF) that encodes the RT, which in turn binds tightly to a stem-loop preceding the ORF (DIVa for the Ll.LtrB intron shown in Fig. 1B). How the IEP interacts with the intron RNA remained elusive until a 3.8–4.5 Å cryo-electron microscopy (cryo-EM) structure of the RNP was solved (Qu et al. 2016). The RNP complex, purified from native host cells, revealed its major structural features including that of the RT and also underscored the relationship of the group II intron RNPs to the spliceosome (Fig. 2). High-resolution crystal structures of amino-terminal proteolytic fragments of group II intron RTs have also been reported, but with the RT active site region improperly folded (Zhao and Pyle 2016). A recent 3.0-Å crystal structure of a full-length group II intron RT in complex with an RNA-template–DNA primer substrate and incoming dNTP revealed the active structure of the enzyme for reverse transcription and a host of novel structural features that may contribute to the distinctive enzymatic properties of group II intron RTs (Stamos et al. 2017).
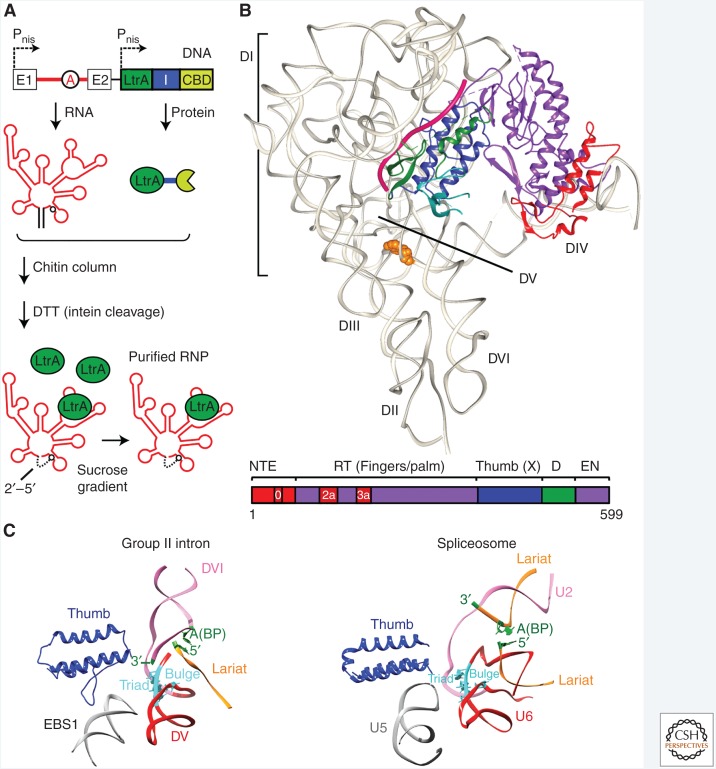
Figure 2.
Cryo-electron microscopy (cryo-EM) structure of the Ll.LtrB group II intron. (A) Intein-based purification. The intron (red) and the intron-encoded protein (IEP) (termed LtrA, green) were expressed independently under nisin-inducible promoters (Pnis), with the IEP fused via an intein to a chitin-binding domain (CBD). The IEP promotes RNA splicing, and the resulting ribonucleoprotein (RNP) complex containing the IEP bound to excised intron lariat RNA was purified from lysed cells by binding to a chitin column. After dithiothreitol treatment, which induces intein cleavage, the released RNP was further purified on a sucrose gradient (Gupta et al. 2014; Qu et al. 2016). (B) Structure of the group II intron RNP determined by cryo-EM. The structure of the RNA (gray) and IEP (colored) was solved at a resolution of 3.8–4.5 Å. Cryo-EM was conducted by the laboratory of Dr. Hongwei Wang (Tsinghua University) and molecular interpretation of the cryo-EM map was a three-way collaboration among the Belfort and Wang laboratories and that of Dr. Raj Agrawal (Wadsworth Center) (Qu et al. 2016). RNA domains I–VI (DI–DVI) are shown (gray), along with the branch point adenosine (orange, space-filling) and trapped ligated exons (magenta). Below is a linear representation of the IEP, color-coded as in the cryo-EM structure, with amino-terminal extension (NTE), RT (fingers-palm), and thumb (X) domains, DNA-binding domain (D) and DNA endonuclease domain (EN). (C) Similarity of group II intron and spliceosome active centers. The group II intron (left) is shown alongside the Schizosaccharomyces pombe spliceosome (right) (Yan et al. 2015), illustrating the spatial similarity between the two. DV of the intron RNP and U6 of the U2/U6 spliceosome RNP (red) are aligned at their triad and bulge regions (cyan). The analogous structures of the group II intron and spliceosome are labeled: thumb domain in IEP and Prp8 (blue), the EBS1 loop of DI and U5 loop I (gray), DV and U6 of U2/U6 (red), DVI and U2 of U2/U6 (mauve), the lariat structures (orange), and their branch-point (BP) adenosines (green). (Adapted from Qu et al. 2016.)
As progress was being made on discerning the biochemical reactions and different functionalities of group II introns, they began to be exploited as genome editing tools (Guo et al. 2000; Mohr et al. 2000; Karberg et al. 2001; Enyeart et al. 2014). The specificity of the IBS–EBS interactions was harnessed to target introns to preprogrammed positions in a wide variety of bacterial genomes. These “targetrons” were then used for a range of genome editing applications in bacteria. Subsequently, the robust properties of thermostable group II intron RTs (TGIRTs) were exploited for many different high-throughput RNA characterization purposes (Mohr et al. 2013b). Thus, these key elements in evolution also provide a toolbox for genetic manipulation and biochemical analysis.
2. TECHNIQUE 1. ISOLATION OF NATIVE RNPS FOR CRYO-EM STRUCTURE DETERMINATION
Cryo-EM structures were determined for the group II Ll.LtrB intron from the ltrB gene of Lactococcus lactis. To isolate RNPs from the native host, an elaborate expression vector was constructed, whereby the intron RNA and the IEP were expressed under separate nisin-inducible promoters (Fig. 2A) (Gupta et al. 2014). The IEP (denoted LtrA protein) was fused to an affinity tag, a chitin-binding domain (CBD), via an intein, to allow cleavage of the IEP from a chitin-affinity column. After induction by nisin, a polycyclic peptide, the L. lactis host cells containing the plasmid were lysed and the lysate applied to a chitin resin. To release the bound IEP and its associated intron RNA, the column was immersed in the reducing agent and nucleophile, dithiothreitol. For cryo-EM analysis, after purification by sucrose gradient sedimentation, residual genomic DNA was digested with DNase and the pure RNP was recovered by gel filtration (Qu et al. 2016).
For EM, the RNP complex was prepared for negative staining or frozen-hydrated for cryo-EM (Qu et al. 2016). In determining the structure of the RNP, negative staining was followed by random conical tilt reconstruction, in which two major class averages were established. Then micrographs of frozen-hydrated intron RNP particles were generated from a 300-kV Titan Krios microscope. A 3D classification of the selected particles yielded three classes with strong LtrA protein occupancy (approximately 450,000 particles) and one class lacking density for the LtrA protein (approximately 100,000 particles). Image processing was then conducted to obtain a 3.8-Å resolution cryo-EM map of the group II intron RNP complex and a 4.5-Å resolution map of the IEP-depleted intron RNA. Soft masks around the most rigid portions of the RNP complex and IEP-depleted form were used for refinement to generate the final 3D reconstruction at the above average resolutions, followed by molecular interpretation of the cryo-EM map.
2.1. Biological Insights and Conclusions from Technique 1
The structures provide the first near-atomic model of the architecture of a group II intron RNP and the first structure of a group II intron RT and its interaction sites with the intron RNA. The six RNA domains form a Y-shaped structure with the IEP making contacts with the terminal stem-loop of DIVa and specific sites in DI (Fig. 1B). The Ll.LtrB group IIA intron RNA shows an overall similarity in structure to that seen in crystal structures of group IIB and IIC intron RNAs, which were derived from in vitro transcripts and lack the IEP (Fig. 2B, top) (Toor et al. 2008; Robart et al. 2014; Costa et al. 2016; Qu et al. 2016). The active-site center around DV, with its AGC catalytic triad, is the most highly conserved region across the different classes of group II introns and also the best-defined structure in the cryo-EM map. The 5′ and 3′ ends of the intron are proximal to the catalytic core, and the bulged A in DVI, which forms the intron lariat with the 5′ end, is adjacent. Remarkably, the structure of the spliced lariat contains the ligated-exon segment of the mRNA, highlighting its interactions with EBS1 and EBS2. The mRNA segment is absent from the IEP-depleted samples with concomitant conformational differences in EBS1 and EBS2, suggesting that the IEP contributes to trapping the mRNA in the RNP (Qu et al. 2016).
The structure gives us the first glimpse of an intact group II intron IEP RT, which has a typical polymerase “right-hand” organization. The IEP has four domains: an RT domain comprised of fingers and palm regions, a thumb domain (previously also referred to as domain X or the maturase domain), a DNA-binding domain (D), and a DNA endonuclease domain (EN), all of which are required for intron mobility (Fig. 2B, bottom). The RT domain contains the catalytic center for reverse transcription. It also has an amino-terminal extension (NTE), with a conserved sequence RT0, and two distinctive “insertion” regions RT2a and RT3a, which are present in group II intron RTs but not in retroviral RTs (Fig. 2B and see below). Protein density revealed some bulky amino acid side-chains but the D and EN domains appear to be dynamic and were less well resolved (Qu et al. 2016). The RT and thumb domains bind the intron RNA to promote both splicing and reverse splicing for retromobility in agreement with previous biochemical analyses (Matsuura et al. 2001; Cui et al. 2004; Gu et al. 2010). The NTE anchors the IEP to DIVa with contributions by RT3a, and the thumb and D domains bind to DI, stabilizing the EBS–IBS interactions for splicing and reverse splicing.
A conundrum raised by the structure is that biochemical and biophysical studies suggest that the IEP is a monomer in solution but binds precursor RNA and functions in splicing at a stoichiometry of ∼2:1, raising the possibility that it functions in splicing as a dimer (Saldanha et al. 1999; Rambo and Doudna 2004). However, the IEP is a monomer in the cryo-EM reconstruction (Agrawal et al. 2016; Qu et al. 2016), and the GsI-IIC group II intron RT, described below, also binds an artificial template-primer substrate as a monomer (Stamos et al. 2017). Although biochemical and structural studies with the proteolytic fragments containing the fingers-palm domain of an IEP of Roseburia intestinalis (Ri) and Eubacterium rectale (Er) suggested dimerization, the dimer interface in those structures would be occluded by the thumb in the full-length protein and presents significant steric clashes when docked into the Ll.LtrB intron RNA (Agrawal et al. 2016; Zhao and Pyle 2016). Recent studies show temperature-dependent dimerization of Ll.LtrB precursor RNPs, although the spliced intron RNPs remained monomeric (Dong et al. 2018). Thus, the monomer–dimer discrepancy may relate to different stoichiometries in different states and will require further experimentation to resolve.
Based on similarities in RNA catalysis and structure, the hypothesis that the group II intron is the progenitor of the spliceosomal intron is widely accepted (reviewed in Lambowitz and Belfort 2015; Novikova and Belfort 2017). The cryo-EM structure provides supporting evidence by virtue of LtrA and Prp8, two related proteins, both directing 5′-splice-site recognition. The thumb domain of spliceosomal Prp8 interacts with the U5 snRNP in analogy to the thumb domain of the IEP interacting with the EBSs (Fig. 2C). Likewise, the branch-points are similarly disposed, where DVI presents the bulged A to the DV active site with its catalytic triad, much as U2 of the U2/U6 snRNP pair interacts with the intron to bring the bulged A to the catalytic triad (Agrawal et al. 2016; Piccirilli and Staley 2016; Qu et al. 2016).
The recent revolution in cryo-EM has made possible the close-to-atomic resolution structures of difficult-to-crystallize RNPs, such as the group II intron RNPs described here, and the spliceosome in various states (reviewed in Fica and Nagai 2017; Shi 2017; Galej et al. 2018). The cryo-EM structure of native telomerase holoenzyme has also been determined, but at somewhat lower resolution (Jiang et al. 2015; Nguyen et al. 2018). These cryo-EM studies have revealed the structural and functional relationships of these RNP assemblies.
3. TECHNIQUE 2. RETROMOBILITY INDICATOR GENES AND RIG-SEQ FOR HIGH-THROUGHPUT INTEGRATION PROFILING
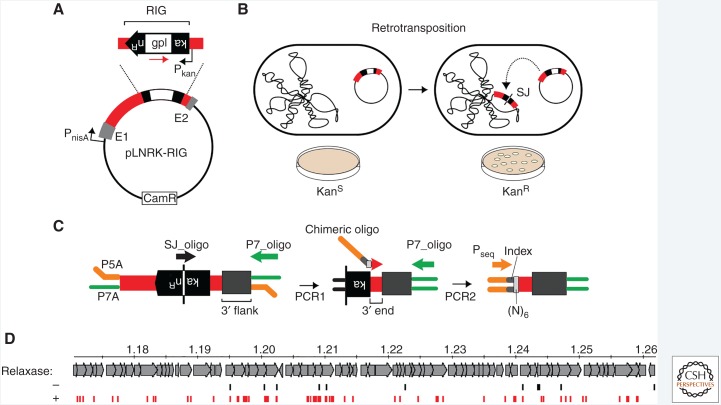
Mechanisms of retrohoming and retrotransposition of group II introns have been well established. The frequency and diversity of retromobility events can be determined by using a retromobility indicator gene (RIG), patterned after that used to measure Ty1 retrotransposition (Fig. 3A) (Curcio and Garfinkel 1991). Refined analysis is facilitated by a high-throughput genomic retrotransposition detection system based on RIG, called RIG-seq. In L. lactis, the group II intron donor plasmid carries the intron (red) flanked by exons (gray) and under a nisin-inducible promoter (PnisA) (Fig. 3A). The intron is interrupted by a RIG with the kanamycin resistance (kanR) gene inserted in the antitranscriptional orientation to the intron and carrying its own promoter (Pkan). The kanR gene is interrupted by a self-splicing group I intron (gpI) in the same orientation as Ll.LtrB (red arrow). Reconstitution of the kanR gene with formation of a characteristic splice junction (SJ) sequence and kanamycin resistance is possible only through reverse transcription of an RNA intermediate that had lost the group I intron during retrotransposition (Fig. 3B).
Figure 3.
RIG-seq for high-throughput retrotransposon profiling. (A) RIG assay. The intron donor plasmid pLNRK-RIG carries the Ll.LtrB intron (red) flanked by exons (gray) under a nisin-inducible promoter (PnisA). The intron is interrupted by a retromobility indicator gene (RIG) with the kanR gene inserted in the anti-transcriptional orientation to the intron and carrying its own promoter (Pkan). The kanR gene is interrupted by a self-splicing group I intron (gpI) in the same orientation as Ll.LtrB (red arrow). (B) Reconstitution of the kanR gene with formation of a characteristic splice junction (SJ) sequence and kanamycin resistance is possible only through reverse transcription of an RNA intermediate that lost the group I intron during retrotransposition. Wavy line to the left is chromosomal DNA, whereas plasmid to the right is the RIG donor. (C) RIG-seq amplification scheme. High-throughput targeted sequencing of insertion loci was based on the generation of the SJ sequence of the kanR gene during retrotransposition. After ligation of Illumina-specific adapters, P5A and P7A, two tandem polymerase chain reactions (PCRs) were used to amplify flanks of retrotransposition events in Illumina libraries: A specific SJ primer (SJ_oligo) with the P7_oligo is used in the first PCR; then a library-specific primer, the chimeric oligo, carrying library-specific indices, is used with the P7_oligo in the second PCR. Pseq is the sequencing primer. (D) Density of intron-insertions in part of the Lactococcus lactis chromosome is shown, in which numbers correspond to mbp from the origin of replication, in the presence (+, red) or absence (−, black) of relaxase. (Adapted from Novikova et al. 2014.)
High-throughput targeted sequencing of group II intron insertion loci is based on generation of the SJ sequence of the kanR gene of RIG during retrotransposition (Fig. 3B,C). After ligation of Illumina-specific adapters (P5A and P7A), two tandem polymerase chain reactions (PCRs) are used to amplify 3′ flanks of retrotransposition events in Illumina libraries. The key to the method is the use of a SJ-specific primer (SJ_oligo) with a P7 primer (P7_oligo) in the first PCR. Then a chimeric library-specific primer (chimeric oligo) carrying library-specific indices and an (N)6 sequence is used with the P7_oligo in the second PCR. The Pseq sequencing primer is then used to obtain Illumina single-end reads of group II intron integration junctions (Fig. 3C).
3.1. Biological Insights and Conclusions from Technique 2
Using the RIG construct, it was shown that retrotransposition of the Ll.LtrB intron to ectopic sites proceeds via reverse splicing into DNA targets (Ichiyanagi et al. 2002), that plasmid targets are favored (Ichiyanagi et al. 2003), and that pathway choice is dictated by cellular environment (Coros et al. 2005). A RIG was also useful for host mutant screens, which revealed both silencers and global stimulators of intron mobility (Coros et al. 2008, 2009). Thus, RNase E, an endoribonuclease that cleaves RNA in single-stranded A- and U-rich regions, was shown to depress retromobility. Conversely, amino acid and glucose starvation, acting through the alarmones ppGpp and cAMP, respectively, were shown to act as stimulators of intron mobility. Thereby, nutritional stress leads to bursts of intron integration.
To provide a comprehensive landscape of integration-site preferences, randomly sampled libraries of the RIG-seq reads were mapped along the L. lactis IL1403 chromosome (Novikova et al. 2014). Mapping of Illumina sequencing reads again showed preference for intron insertion into the intron donor plasmid (pLNRK-RIG). The relationship between retrotransposition of the group II intron and the conjugative plasmid, pRS01, on which it naturally resides, was then probed. This question was of particular interest because the intron is housed within the pRS01 ltrB gene, which encodes the relaxase required for specifically nicking pRS01 to facilitate transfer. Nicking occurs at the transfer origin (oriT), which allows conjugal transmission of pRS01 into the recipient cell. Using RIG-seq, we showed that relaxase introduces off-target nicks in the chromosomal DNA, stimulating both the frequency and density of retromobility events (Fig. 3D). Thus, relaxase, which is rendered functional through group II intron splicing, not only promotes horizontal conjugal transfer but also stimulates intron spread in donor and recipient cells (Belhocine et al. 2005).
The RIG-seq approach adds to the burgeoning high-throughput sequencing technologies for studying mobile-genetic element biology (Xing et al. 2013) and is uniquely and broadly applicable to performing retrotransposition profiling in diverse organisms.
4. TECHNIQUE 3. TARGETRONS FOR BACTERIAL GENE TARGETING APPLICATIONS
The finding that mobile group II introns recognize DNA target sites for retrohoming largely by base-pairing of the intron RNA led to their development into the first RNA-guided gene targeting tools, called targetrons (Guo et al. 2000; Mohr et al. 2000; Karberg et al. 2001; Perutka et al. 2004). Initial targetrons were based on the aforementioned Ll.LtrB group IIA intron, but additional introns, including group IIB introns EcI5 and RmIntI, and TeI3c from a bacterial thermophile, have also been used, as could any group II intron with suitably stringent DNA target-site specificity (Zhuang et al. 2009; Mohr et al. 2013a; Garcia-Rodriguez et al. 2014).
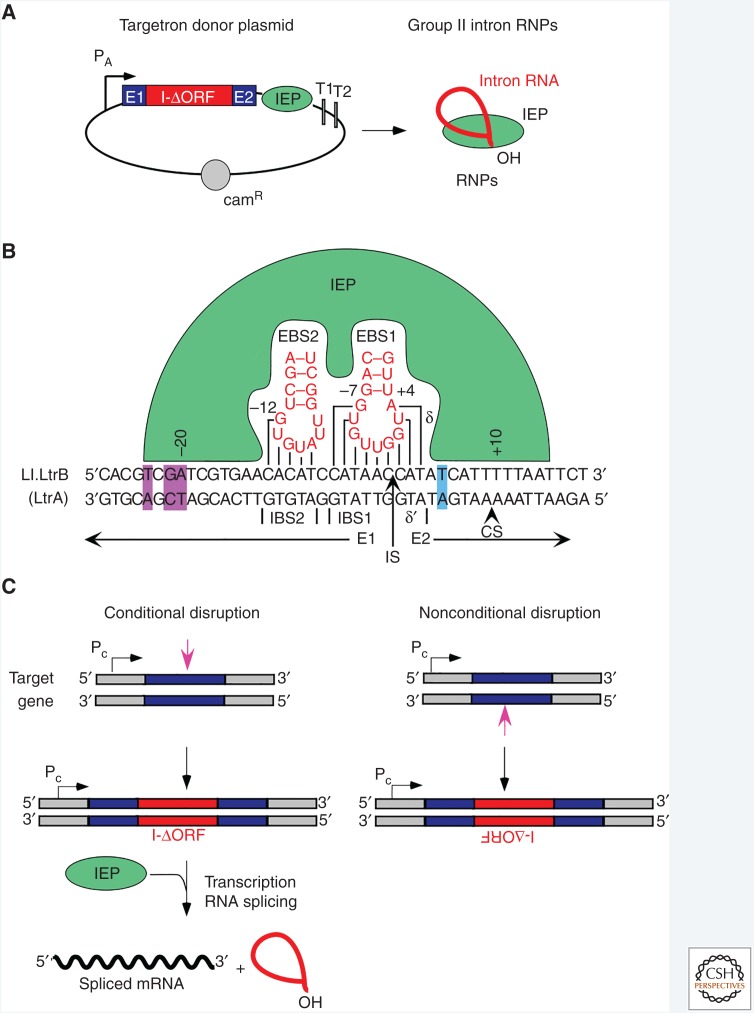
For gene targeting in bacteria, targetrons are typically transcribed from a plasmid expression vector that uses a strong and preferably inducible promoter (Fig. 4A). The targetron cassette expresses a precursor RNA containing a group II intron from which the RT ORF has been deleted (I-ΔORF) flanked by short 5′ and 3′ exons (E1 and E2, respectively), with the RT expressed in tandem downstream from the 3′ exon. The RT promotes splicing of the intron, forming lariat RNPs that recognize and integrate site-specifically into a DNA target site. Group IIA and IIB intron RNPs typically recognize DNA target sites via three base-pairing interactions (EBS1/IBS1, EBS2/IBS2, and δ-δ′ [IIA] or EBS3/IBS3 [IIB]), which provide most of the DNA target specificity (Figs. 1C and 4B). The group II intron RT contributes by recognizing a small number of specific bases in the distal 5′- and 3′-exon regions of the DNA target site to help promote local DNA melting for base-pairing of the intron RNA and bottom-strand cleavage (Singh and Lambowitz 2001; Perutka et al. 2004; Zhuang et al. 2009).
Figure 4.
Group II introns as targetrons for bacterial gene targeting. (A) Targetron donor plasmid. The plasmid has a strong and preferably inducible active promoter (PA) to express a targetron cassette consisting of a group II intron ribozyme with the reverse transcriptase (RT) open reading frame (ORF) deleted (I-ΔORF) flanked by short 5′ and 3′ exons (E1 and E2, respectively) and followed by an ORF encoding the intron-encoded protein (IEP). The IEP splices the group II intron RNA and remains bound to the excised intron lariat RNA in an RNP that can be preprogrammed to insert into desired target sites. (B) DNA target site interactions for the Ll.LtrB group IIA intron used for gene targeting involve base-pairing of EBS1, EBS2, and δ sequences in DI with IBS1, IBS2, and δ′ sequences flanking the intron insertion site (IS). The IEP recognizes specific nucleotides (purple) in the distal 5′-exon region upstream of IBS2 and promotes local DNA melting, enabling base-pairing of the intron RNA to the DNA target sequence. Additional interactions between the IEP and nucleotides in the 3′ exon (blue) are required for bottom-strand cleavage by the EN domain to generate the primer for reverse transcription of the intron RNA, leading to intron insertion via the retrohoming pathway. CS, bottom-strand cleavage site. (C) Use of targetrons for conditional or nonconditional disruptions. Targetrons can be made to insert in either orientation relative to transcription of the target gene (dark blue) by targeting a sequence (magenta arrow) in the top or bottom strand. A targetron that inserts in the antisense orientation yields an unconditional disruption, whereas a targetron that inserts in the sense orientation can yield a conditional disruption by linking splicing of the intron from the pre-mRNA to expression of the IEP from a separate construct. Pc, host chromosomal gene promoter. (Adapted from Enyeart et al. 2014.)
Targetrons are programmed to insert into desired sites with the aid of a commercially available computer algorithm that identifies optimal matches for the small number of nucleotide positions recognized by the IEP and then designs PCR primers for modifying the sequence elements in the intron RNA to base-pair to the IBS and δ sequences into the DNA target site (Perutka et al. 2004; Zhuang et al. 2009; Garcia-Rodriguez et al. 2014). Ll.LtrB and EcI5 targetrons designed by using such algorithms typically insert into DNA target sites with high efficiency (1%–100% without selection), with nonspecific integrations rarely detected. The insertion frequency of targetrons is usually high enough to detect desired integration events by PCR screening of colonies without selection, but if needed, a retrotransposition-activated genetic marker (RAM), equivalent to a RIG, can be incorporated into the intron RNA for genetic selections (Zhong et al. 2003). Polar effects on the expression of downstream genes can be mitigated by inserting a separate promoter near the 3′ end of the targetron.
Broad host-range vectors and promoters enable the use of the same targetron construct in a wide variety of bacteria (Yao and Lambowitz 2007). Targetrons can be made to insert in either the sense or antisense orientation relative to target gene transcription, depending on whether they are targeted to a sequence in the sense or antisense strands (Fig. 4C). A targetron that inserts in the antisense orientation yields an unconditional gene disruption, whereas a targetron that inserts in the sense orientation can be used to obtain a conditional disruption by linking splicing of the intron to expression of the IEP from a separate construct (Frazier et al. 2003; Yao et al. 2006). Targetrons can also be used to introduce a targeted double-strand break that can be repaired by homologous recombination with a cotransformed DNA fragment, enabling the seamless introduction of point mutations (Karberg et al. 2001; Mastroianni et al. 2008). A thermotargetron derived from a thermostable mobile group II intron from a bacterial thermophile was developed to enable gene targeting in thermophilic bacteria and archaea, which include biologically and commercially important species that had been recalcitrant to genetic manipulation (Mohr et al. 2013a; Hong et al. 2014). In recent applications, targetrons have been used to site-specifically position recombinase (lox) sites at two or more desired genomic regions, which can be used to obtain large deletions, insertions, and chromosome inversions after supplying Cre from a separate construct (Enyeart et al. 2013). Although targetrons afford the same benefits of largely RNA-guided design as clustered regularly interspersed short tandem repeat (CRISPR)–Cas gene-targeting systems, they work only inefficiently in eukaryotes because of the high Mg2+ requirements of the group II intron ribozyme (Mastroianni et al. 2008; Truong et al. 2015). Nevertheless, targetrons are useful for gene targeting in virtually any bacterium, and particularly those in which homology-based DNA repair outcomes are challenging (Enyeart et al. 2014).
4.1. Biological Insights and Conclusions from Technique 3
Targetrons have been sold commercially for 15 years and have been used in a wide variety of bacteria, including medically and commercially important species that were previously intractable to detailed genetic analysis or facile genetic engineering (reviewed in Enyeart et al. 2014). These include Pseudomonas aeruginosa, Staphylococcus aureus, Francisella tularensis, and a large variety of clostridial species. Recently, targetrons have also enabled the first directly targeted chromosomal mutations in the intracellular bacterial pathogen Chlamydia trachomatis (Johnson and Fisher 2013). Targetrons continue to be used in diverse bacteria for analysis of pathogenicity determinants (Buchan et al. 2009; Cole et al. 2016; Herrera et al. 2016; Kint et al. 2017), identification of new antibiotic targets (Zoraghi et al. 2011; Donnelly et al. 2017), construction of attenuated vaccine strains (Harper et al. 2016), and the engineering of industrially important bacteria for increased biofuel and chemical production (Hong et al. 2014; Dai et al. 2016; Xue et al. 2016).
5. TECHNIQUE 4. RTS WITH NOVEL ENZYMATIC PROPERTIES AS POTENT RESEARCH TOOLS
RTs are widely used for research and biotechnological applications that require cDNA synthesis, including RT-qPCR and high-throughput RNA sequencing (RNA-seq). Until recently, however, the only RTs available for such applications were retroviral RTs, and these have inherently low fidelity and processivity, presumably to help retroviruses evade host defenses by introducing and propagating mutational variations (Hu and Hughes 2012). In contrast, group II intron RTs function in retrohoming, which requires reverse transcription of a long, highly structured intron RNA with high fidelity and processivity (Cousineau et al. 1998; Conlan et al. 2005), properties that are desirable for biotechnological applications, particularly RNA-seq (Mohr et al. 2013b). Group II intron RTs also have a robust end-to-end template-switching activity, which enables RNA-seq adapter addition to target RNAs without RNA tailing or ligation, inefficient steps in many RNA-seq protocols (Mohr et al. 2013b). The development of new general methods for the bacterial expression of group II intron RTs with high yield and activity in a form that remains soluble when removed from the intron RNA have enabled their large-scale commercial production and use for biotechnological applications (Mohr et al. 2013b).
Thermostable group II intron RTs (TGIRTs) from bacterial thermophiles, which can carry out cDNA synthesis at high temperatures (≥60°C) that help melt out stable RNA secondary and tertiary structures, were the first to be adapted for biotechnological applications. Initial work focused on two TGIRTs, TeI4c RT from the thermophilic cyanobacterium Thermosynechococcus elongatus and GsI-IIC RT from the thermophilic soil bacterium Geobacillus stearothermophilus (Mohr et al. 2013b). These TGIRTs were found to have two- to fourfold higher fidelity in vitro than a retroviral RT assayed in parallel, as well as very high processivity, enabling them to continuously copy long and/or structured RNA templates without falling off (macroscopic processivity values defined as the average length of cDNA synthesized in the presence of a trap, 714 and 708 nt for TeI4c and GsI-IIC RT, respectively, fourfold higher than for SuperScript III, a widely used thermostable retrovirus-derived RT [Mohr et al. 2013b]). Nonthermostable group II intron RTs also have high-fidelity and processivity (macroscopic processivity value 616 nt [Zhao et al. 2018]), and it is likely that other group II intron RTs yet to be sampled will have similar properties.
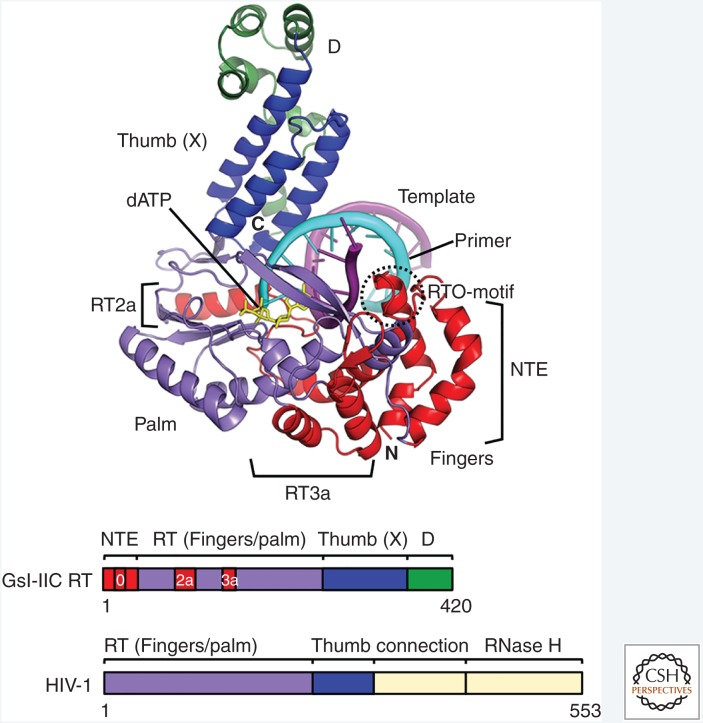
A 3.0-Å X-ray crystal structure of a full-length TGIRT (GsI-IIC RT, a form of which is sold commercially as TGIRT-III) in complex with template-primer substrate and incoming dNTP has provided insight into structural features that may contribute to the beneficial enzymatic properties of group II intron RTs (Fig. 5) (Stamos et al. 2017). Thus, the group II intron RT-specific insertion regions NTE/RT0 and RT2a, which are lacking in retroviral RTs, contribute multiple additional interactions with the template-primer likely accounting for increased processivity. Additionally, the group II intron RT active site has more constrained binding pockets than in retroviral RTs for the templating base, the 3′ end of the primer, and incoming dNTP, potentially increasing fidelity. The RT3a insertion (also known as the insertion-in-fingers domain, IFD) contains a loop (also known as the α-loop), which occupied the RT active site in the misfolded RT fragment structures (Zhao and Pyle 2016; Zhao et al. 2018). Although also suggested to contribute to high processivity, this loop occupies a different position in the structure of the full-length GsI-IIC RT and does not contact the bound template-primer as would be expected for a direct role in processivity (Stamos et al. 2017). Notably, many of the distinctive structural features of group II intron RTs, including homologs of the NTE, RT2a, and RT3a and details of the RT active site, are also found in viral RNA-dependent RNA polymerases (RdRPs), which are potential evolutionary ancestors of RTs. In contrast, retroviral RTs, which are descended from these ancestral proteins, appear to have lost entire regions and active-site features that contribute to the high processivity and fidelity of group II intron RTs (Stamos et al. 2017). The crystal structure of a full-length group II intron RT in complex with template-primer substrate will enable detailed structure–function analysis and the engineering of group II intron RTs for improved performance in biotechnological applications.
Figure 5.
Crystal structure of the thermostable GsI-IIC reverse transcriptase (RT) bound to template-primer and incoming dNTP. The 3.0 Å structure shows full-length GsI-IIC RT bound to an 11-bp RNA template-DNA primer duplex with a single-stranded 5′ overhang (3 nt) on the RNA template strand and a dideoxynucleotide at the 3′ end of the DNA primer. The incoming dATP is bound at the RT active site poised for polymerization. The amino-terminal extension (NTE) and insert regions RT2a and RT3a not present in retroviral RTs are demarcated with brackets. N and C indicate the amino- and carboxyl-termini of the protein. The schematics below show the domain organization of GsI-IIC RT compared with that of HIV-1 RT, with regions color-coded as shown in the figure. The connection and RNase H domains of HIV-1 RT not present in group II intron RTs are shown in yellow. (Adapted from Stamos et al. 2017.)
The beneficial properties of TGIRT enzymes have been exploited in multiple ways for different applications, including three different methods for RNA-seq library construction (Fig. 6). In one method, cDNAs of human cell mRNAs were synthesized by initiating reverse transcription from an oligo(dT)42 primer annealed to the 3′ poly(A) tail (Fig. 6A) (Mohr et al. 2013b). The cDNAs were then converted to dsDNAs by using a commercial kit, and the dsDNAs were fragmented by a transposon with simultaneous addition of RNA-seq adapters for Illumina sequencing. Because of the high processivity of TGIRTs, the resulting RNA-seq data sets showed dramatically more uniform 5′-to-3′ read coverage than those obtained in parallel with SuperScript III RT, which over-represented reads near the 3′ end of the mRNAs (Mohr et al. 2013b). Other applications in which TGIRTs have been used for reverse transcription from an annealed DNA primer include (1) synthesis of long ssDNAs from RNA templates for CRISPR genome editing (Li et al. 2017a); (2) reverse transcription and quantitation of GC-rich repeat expansions, which are clinically important in diseases such as myotonic dystrophy and some forms of amyotrophic lateral sclerosis (ALS) (Carrell et al. 2017); (3) RNA-structure mapping by DMS modification and SHAPE (Wu and Bartel 2017; Zubradt et al. 2017; Mohr et al. 2018); and (4) IR-CLIP for the identification of protein-bound RNAs, in which TGIRT gave four- to eightfold higher cDNA yields than SuperScript III (Zarnegar et al. 2016). TGIRT has also been used for in vitro evolution of the binding specificity of structured RNA aptamers, in which it was able to maintain the stable RNA scaffold structure through multiple rounds of selection, in contrast to retroviral RTs whose low fidelity and inability to reverse transcribe through structured RNAs resulted in the accumulation of mutations that destabilized the RNA scaffold structure, thereby compromising the selections (Porter et al. 2017).
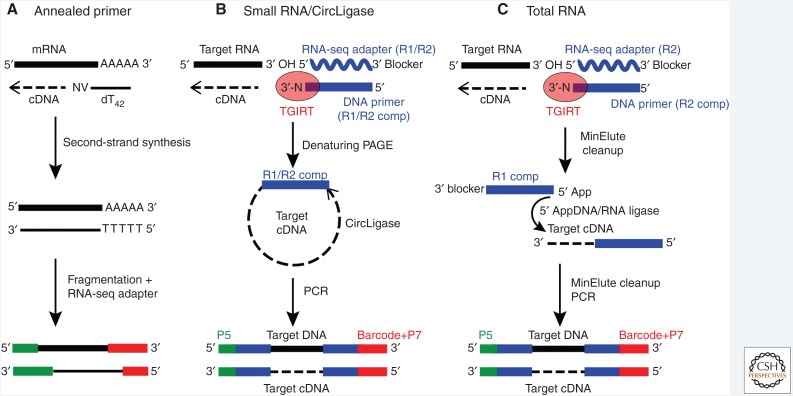
Figure 6.
Three methods of using thermostable group II intron reverse transcriptase (TGIRT) enzymes for RNA-seq library construction (TGIRT-seq). (A) TGIRT-seq of eukaryotic mRNAs using an anchored oligo(dT)42 primer for initiation of cDNA synthesis. The primer of sufficient length to stably anneal to mRNA poly(A) tails at high temperature is extended by TGIRTs at 60°C to synthesize cDNA copies of the mRNAs. After second-strand synthesis, dsDNAs are fragmented and RNA-seq adapters are added by conventional methods, such as the transposon-based Illumina Nextera method. (B) TGIRT-seq Small RNA/CircLigase method. The TGIRT initiates by template switching from an initial template-primer substrate comprised of an RNA oligonucleotide containing Illumina R1 and R2 adapter sequences annealed to a 5′-labeled DNA primer that leaves a single-nucleotide 3′ overhang (N, an equimolar mixture of A, C, G, and T) that can base-pair to the 3′ nucleotide of target RNAs. The single base-pair is sufficient to direct TGIRT template switching, even at 60°C (Mohr et al. 2013b; Qin et al. 2016). After reverse transcription, cDNAs are purified on a denaturing polyacrylamide gel to select specific size classes, circularized with CircLigase (Epicentre), and minimally polymerase chain reaction (PCR) amplified with primers that add capture sites and barcodes for Illumina sequencing. (C) TGIRT Total RNA-seq method for construction of comprehensive RNA-seq libraries without size selection. The TGIRT initiates from an initial template-primer substrate similar to that in panel B but containing only an Illumina R2 adapter sequence. After reverse transcription, cDNAs are cleaned up by using a MinElute column to remove unincorporated adapters and a second oligonucleotide containing the reverse complement of an Illumina R1 adapter is ligated to the 3′ end of the cDNA using thermostable 5′ AppDNA/RNA ligase (New England Biolabs). After an additional MinElute cleanup, the cDNAs with adapters on both ends are amplified by PCR with primers that add Illumina capture sites and barcodes. Before sequencing, the libraries are cleaned up using Ampure beads to remove adapter dimers (not shown).
The template switching activity of TGIRTs, which enables facile attachment of RNA-seq adapters to target RNA sequences (Mohr et al. 2013b), has been used to develop two new methods of RNA-seq library construction for Illumina sequencing, the Small RNA/CircLigase method and the Total RNA-seq method (Fig. 6B and C, respectively). In both methods, the TGIRT starts from a synthetic RNA template-DNA primer substrate that contains an RNA-seq adapter sequence and has a single nucleotide 3′ DNA overhang that directs template switching by base-pairing to the 3′ nucleotide of the target RNA (Mohr et al. 2013b). For RNA-seq library construction from a pool of RNAs in which minimal bias is required, the template-primer substrate is added in excess to the target RNA and the 3′ DNA overhang is an equimolar mix of A, C, G, and T (denoted N) that can base-pair to target RNAs in the pool having different 3′ termini. After template switching to the 3′ end of the target RNA, the TGIRT synthesizes a full-length cDNA in which the RNA-seq adapter is seamlessly linked to the target RNA sequence (Mohr et al. 2013b).
In the TGIRT Small RNA/CircLigase method, the initial RNA template/DNA primer substrate contains both Illumina Read 1 and Read 2 (R1 and R2) adapter sequences, and after template switching, the cDNAs with the attached adapter are size-selected on a gel and circularized with CircLigase (Epicentre) followed by PCR with primers that add Illumina barcodes and capture sites for sequencing (Fig. 6B). This variation of the method has been used for miRNA profiling (Mohr et al. 2013b), RIP-seq of protein-bound RNAs (Katibah et al. 2014), and quantitative tRNA sequencing (Katibah et al. 2014; Shen et al. 2015; Zheng et al. 2015). Recently, a further adaptation of the method was developed to measure tRNA aminoacylation levels by reverse transcription with TGIRT before and after removal of the attached amino acid, whose presence blocks template switching (Evans et al. 2017).
The second TGIRT-seq method, referred to as the Total RNA-seq method, enables the construction of comprehensive RNA-seq libraries from pools of RNAs without size selection (Fig. 6C). In this variation, the initial template/primer substrate used for template switching contains only an Illumina R2 sequence, and after cDNA synthesis a second adapter containing the reverse complement of an Illumina R1 sequence is ligated to the 3′ end of the cDNA by an efficient single-stranded DNA ligation (Nottingham et al. 2016; Qin et al. 2016). The resulting cDNAs, with different sizes, are then PCR amplified with primers that anneal to the RNA-seq adapters and add barcodes and capture sites for sequencing. Validation of the method using human reference RNA samples with External RNA Control Consortium (ERCC) spike-ins compared with the widely used strand-specific TruSeq v3 method for transcriptome profiling showed better quantitation of the relative abundance of cellular mRNAs and ERCC spike-ins, more uniform 5′-to-3′ coverage, even for fragmented RNAs, and larger numbers of detected SJs, particularly near the 5′ ends of mRNAs (Nottingham et al. 2016; Qin et al. 2016). Additionally, the TGIRT-seq method had higher strand specificity than TruSeq v3 and eliminated biases because of random hexamer primers, which are inherent in TruSeq. Finally, a major advantage of the Total RNA-seq method is that it enables simultaneous quantitation of mRNAs and structured small noncoding RNAs (ncRNAs) in the same RNA-seq instead of requiring separate preparations for long and short RNAs (Nottingham et al. 2016; Boivin et al. 2018).
In both the Small RNA/CircLigase and Total RNA-seq methods, the high processivity and strand displacement activities of TGIRTs combined with their ability to initiate directly at the 3′ nucleotide of RNA templates by template switching provide the ability to obtain full-length reads of tRNAs and other structured small ncRNAs (Katibah et al. 2014; Shen et al. 2015), which is not possible for retroviral RTs. This was first shown in Katibah et al. (2014) for RIP-seq of RNAs bound to the human interferon–induced protein IFIT5, which were found to include full-length mature and pre-tRNAs beginning at the PolIII initiation site. The method enabled detection of the 3′ CCA at the end of mature tRNAs, as well as poly(U) tails at the 3′ end of RNAs targeted for degradation. Further, the ability to reverse transcribe through tRNA structure is critical for resolving full-length tRNAs from tRNA fragments, which have biological and medical importance, but are difficult to distinguish from strong stops because of RNA secondary structures that impede conventional RTs (Qin et al. 2016). Additionally, in contrast to retroviral RTs, which tend to dissociate at RNA base modifications that affect base-pairing, TGIRTs pause at such modifications but eventually read through by misincorporation, with the spectrum of misincorporated nucleotides being distinctive for different modifications (Katibah et al. 2014). This ability of TGIRTs to read through posttranscriptional modifications that affect base-pairing by misincorporation has been used for high-throughput mapping of posttranscriptional modifications at single-nucleotide resolution in both tRNAs and mRNAs (Clark et al. 2016; Li et al. 2017b; Safra et al. 2017), as well as DMS-induced A and C modifications for RNA structure mapping in DMS-MaPseq (Wu and Bartel 2017; Zubradt et al. 2017). Because these methods map mutations rather than reverse transcription stops they can detect multiple modifications in the same RNA molecule.
The ability of the TGIRT template-switching activity to enable construction of comprehensive RNA-seq libraries from small amounts of starting material has been used for the analysis of extracellular RNAs present in human plasma or packaged in secreted membrane vesicles termed exosomes (Qin et al. 2016; Shurtleff et al. 2017). In both cases, TGIRTs revealed the presence of full-length tRNAs and other structured small ncRNAs, which could not be detected previously by retroviral RTs. Although highly purified exosomes contained only small amounts of mRNAs, the more complex plasma RNAs contain RNA fragments that map to large numbers of protein-coding and long ncRNAs (lncRNAs). These findings suggest that TGIRTs may provide advantages for liquid biopsy, including the ability to identify mRNA, lncRNA, and small ncRNA biomarkers in the same RNA-seq reaction and to capture RNA species that are refractory to retroviral RTs.
Finally, in another liquid biopsy application, a method has been developed for using TGIRT-template-switching activity for ssDNA-seq of human plasma DNAs, which are released into blood by apoptosis of blood lymphoid and myeloid cells or by necrosis of tumors and damaged tissues in cancer and other diseases (Wu and Lambowitz 2017). This TGIRT-ssDNA-seq method uses a workflow similar to the TGIRT-seq Total RNA method, but is optimized for copying of DNA templates, which under appropriate conditions TGIRTs can do robustly and with error rates after corrections approaching those for a conventional DNA-seq method (see Wu and Lambowitz 2017). In addition to a more streamlined workflow than other ssDNA-seq methods, by avoiding tailing steps, this method facilitates precise mapping of DNA ends for analysis of nucleosome positioning and bisulfite mapping of DNA methylation sites. The analysis of nucleosome positioning and mapping of transcription factor-binding and DNA methylation sites of human plasma DNAs can be used to identify their tissues of origin and to follow disease progression and response to treatment (Sun et al. 2015; Snyder et al. 2016; Wu and Lambowitz 2017).
5.1. Biological Insights and Conclusions from Technique 4
The novel properties of TGIRTs expand and improve RNA-seq in ways that are difficult or impossible for retroviral RTs and have provided a more complete picture of the RNA composition of human plasma, exosomes, and cells (Qin et al. 2016; Shurtleff et al. 2017; Boivin et al. 2018). The use of TGIRT for quantitative tRNA-seq of a yeast ribosome quality control complex identified tRNAAla and tRNAThr isoacceptor species that are recruited by a protein component to stalled ribosomes, in which they function in conjunction with free 60S subunits to promote non-mRNA-mediated addition of carboxy-terminal Ala and Thr extensions (CAT tails) helping to target nascent polypeptides for degradation (Shen et al. 2015). The method also contributed to establishing that codon optimality dictated by tRNA abundance regulates mRNA stability and translation during the maternal-to-zygote transition in zebrafish and likely across vertebrates (Bazzini et al. 2016). The ability of TGIRTs to read through RNA posttranscriptional modifications by misincorporation enabled high-throughput mapping and functional analysis of m1A base methylations in mammalian tRNAs and mRNAs, establishing roles for this modification in translational and developmental regulation (Clark et al. 2016; Li et al. 2017b; Safra et al. 2017). Likewise, the new methods for in vivo RNA structure mapping based on the ability of TGIRTs to read through and mutationally map DMS modifications enabled the first analysis of RNA structure within an animal tissue and revealed the widespread influence of 3′ end structures that juxtapose poly(A) signals and 3′ cleavage sites on mammalian mRNA processing and stability (Wu and Bartel 2017; Zubradt et al. 2017).
6. CONCLUDING REMARKS
Fundamental studies into group II intron structure and function have provided insight into the evolution of retroelements and formed the basis for applications in gene targeting and editing, as well as RT-based high-throughput RNA analysis. The group II intron RNP cryo-EM structure provided a number of “firsts,” including an impression of group II RNP architecture at 3.8–4.5 Å resolution and a global view of similarities to the spliceosome. The RIG-seq methodology showed integration at spurious nicks in DNA, providing novel insights into intron spread. Targetrons, the first RNA-guided gene targeting system, have enabled genetic analysis and genome engineering in a wide variety of bacteria. Finally, group II intron RTs have enabled the development of new methods for RNA-seq, with unprecedented capabilities for comprehensive transcriptome profiling, RNA structure mapping, analysis of tRNAs and other small ncRNAs, mapping of posttranscriptional modifications, and potentially liquid biopsy.
Competing Interest Statement
Targetrons and TGIRT enzymes and methods for their use are the subject of patents and patent applications that have been exclusively licensed by the Ohio State University, the University of Texas, and East Tennessee State University to InGex, LLC. A.M.L., M.B., the Ohio State University, and the University of Texas are minority equity holders in InGex, and A.M.L., the Ohio State University, the University of Texas, and some present and former Lambowitz laboratory members receive royalty payments from the sale of targetrons, TGIRT enzymes, and kits and the sublicensing of the intellectual property by InGex to other companies.
ACKNOWLEDGMENTS
We thank Matt Stanger, Olga Novikova, Eren (Xiaolong) Dong, Rebecca McCarthy, and Jennifer Stamos for help with the figures and manuscript and members of the Belfort and Lambowitz laboratories for their comments on the manuscript. Work in our laboratories is supported by National Institutes of Health (NIH) Grants GM39422 and GM44844 to M.B. and NIH Grant GM37949 and Welch Foundation Grant F-1607 to A.M.L.
We use RT and IEP interchangeably, in keeping with nomenclature for other RTs that have multiple activities (e.g., RNase H).
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Agrawal RK, Wang HW, Belfort M. 2016. Forks in the tracks: Group II introns, spliceosomes, telomeres and beyond. RNA Biol 13: 1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno-Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ. 2016. Codon identity regulates mRNA stability and translation efficiency during the maternal-to-zygotic transition. EMBO J 35: 2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhocine K, Yam KK, Cousineau B. 2005. Conjugative transfer of the Lactococcus lactis chromosomal sex factor promotes dissemination of the Ll.LtrB group II intron. J Bacteriol 187: 930–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin V, Deschamps-Francoeur G, Couture S, Nottingham RM, Bouchard-Bourelle P, Lambowitz AM, Scott MS, Abou-Elela S. 2018. Simultaneous sequencing of coding and non-coding RNAs reveals a human transcriptome dominated by a small number of highly expressed non-coding genes. RNA 10.1261/rna.064493.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan BW, McCaffrey RL, Lindemann SR, Allen LA, Jones BD. 2009. Identification of migR, a regulatory element of the Francisella tularensis live vaccine strain iglABCD virulence operon required for normal replication and trafficking in macrophages. Infect Immun 77: 2517–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell ST, Tang Z, Mohr S, Lambowitz AM, Thornton CA. 2017. Detection of expanded RNA repeats using thermostable group II intron reverse transcriptase. Nucleic Acids Res 46: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. 2016. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22: 1771–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole AL, Muthukrishnan G, Chong C, Beavis A, Eade CR, Wood MP, Deichen MG, Cole AM. 2016. Host innate inflammatory factors and staphylococcal protein A influence the duration of human Staphylococcus aureus nasal carriage. Mucosal Immunol 9: 1537–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan LH, Stanger MJ, Ichiyanagi K, Belfort M. 2005. Localization, mobility and fidelity of retrotransposed group II introns in rRNA genes. Nucleic Acids Res 33: 5262–5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros CJ, Landthaler M, Piazza CL, Beauregard A, Esposito D, Perutka J, Lambowitz AM, Belfort M. 2005. Retrotransposition strategies of the Lactococcus lactis Ll.LtrB group II intron are dictated by host identity and cellular environment. Mol Microbiol 56: 509–524. [DOI] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Belfort M. 2008. A mutant screen reveals RNase E as a silencer of group II intron retromobility in Escherichia coli. RNA 14: 2634–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coros CJ, Piazza CL, Chalamcharla VR, Smith D, Belfort M. 2009. Global regulators orchestrate group II intron retromobility. Mol Cell 34: 250–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Walbott H, Monachello D, Westhof E, Michel F. 2016. Crystal structures of a group II intron lariat primed for reverse splicing. Science 354. [DOI] [PubMed] [Google Scholar]
- Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller JE, Yang J, Mills D, Manias D, Dunny G, Lambowitz AM, Belfort M. 1998. Retrohoming of a bacterial group II intron: Mobility via complete reverse splicing, independent of homologous DNA recombination. Cell 94: 451–462. [DOI] [PubMed] [Google Scholar]
- Cui X, Matsuura M, Wang Q, Ma H, Lambowitz AM. 2004. A group II intron-encoded maturase functions preferentially in cis and requires both the reverse transcriptase and X domains to promote RNA splicing. J Mol Biol 340: 211–231. [DOI] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ. 1991. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci 88: 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Dong H, Zhang Y, Li Y. 2016. Elucidating the contributions of multiple aldehyde/alcohol dehydrogenases to butanol and ethanol production in Clostridium acetobutylicum. Sci Rep 6: 28189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Ranganathan S, Qu G, Piazza CL, Belfort M. 2018. Structural accommodations accompanying splicing of a group II intron RNP. Nucleic Acids Res doi: 10.1093/nar/gky416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly ML, Li W, Li YQ, Hinkel L, Setlow P, Shen A. 2017. A Clostridium difficile-specific, gel-forming protein required for optimal spore germination. mBio 8: e02085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyeart PJ, Chirieleison SM, Dao MN, Perutka J, Quandt EM, Yao J, Whitt JT, Keatinge-Clay AT, Lambowitz AM, Ellington AD. 2013. Generalized bacterial genome editing using mobile group II introns and Cre-lox. Mol Syst Biol 9: 685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyeart PJ, Mohr G, Ellington AD, Lambowitz AM. 2014. Biotechnological applications of mobile group II introns and their reverse transcriptases: Gene targeting, RNA-seq, and non-coding RNA analysis. Mob DNA 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans ME, Clark WC, Zheng G, Pan T. 2017. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res 45: e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Nagai K. 2017. Cryo-electron microscopy snapshots of the spliceosome: Structural insights into a dynamic ribonucleoprotein machine. Nat Struct Mol Biol 24: 791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier CL, San Filippo J, Lambowitz AM, Mills DA. 2003. Genetic manipulation of Lactococcus lactis by using targeted group II introns: Generation of stable insertions without selection. Appl Environ Microbiol 69: 1121–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Toor N, Newman AJ, Nagai K. 2018. Molecular mechanism and evolution of nuclear pre-mRNA and group II intron splicing: Insights from cryo-electron microscopy structures. Chem Rev 118: 4156–4176. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez FM, Hernandez-Gutierrez T, Diaz-Prado V, Toro N. 2014. Use of the computer-retargeted group II intron RmInt1 of Sinorhizobium meliloti for gene targeting. RNA Biol 11: 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu SQ, Cui X, Mou S, Mohr S, Yao J, Lambowitz AM. 2010. Genetic identification of potential RNA-binding regions in a group II intron-encoded reverse transcriptase. RNA 16: 732–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Karberg M, Long M, Jones JP III, Sullenger B, Lambowitz AM. 2000. Group II introns designated to insert into therapeutically-relevant DNA target sites in human cells. Science 289: 452–457. [DOI] [PubMed] [Google Scholar]
- Gupta K, Contreras LM, Smith D, Qu G, Huang T, Spruce LA, Seeholzer SH, Belfort M, Van Duyne GD. 2014. Quaternary arrangement of an active, native group II intron ribonucleoprotein complex revealed by small-angle X-ray scattering. Nucleic Acids Res 42: 5347–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper M, John M, Edmunds M, Wright A, Ford M, Turni C, Blackall PJ, Cox A, Adler B, Boyce JD. 2016. Protective efficacy afforded by live Pasteurella multocida vaccines in chickens is independent of lipopolysaccharide outer core structure. Vaccine 34: 1696–1703. [DOI] [PubMed] [Google Scholar]
- Herrera A, Vu BG, Stach CS, Merriman JA, Horswill AR, Salgado-Pabon W, Schlievert PM. 2016. Staphylococcus aureus β-toxin mutants are defective in biofilm ligase and sphingomyelinase activity, and causation of infective endocarditis and sepsis. Biochemistry 55: 2510–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Zhang J, Feng Y, Mohr G, Lambowitz AM, Cui GZ, Liu YJ, Cui Q. 2014. The contribution of cellulosomal scaffoldins to cellulose hydrolysis by Clostridium thermocellum analyzed by using thermotargetrons. Biotechnol Biofuels 7: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu WS, Hughes SH. 2012. HIV-1 reverse transcription. Cold Spring Harbor Perspect Med 2: a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K, Beauregard A, Lawrence S, Smith D, Cousineau B, Belfort M. 2002. Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol Microbiol 46: 1259–1272. [DOI] [PubMed] [Google Scholar]
- Ichiyanagi K, Beauregard A, Belfort M. 2003. A bacterial group II intron favors retrotransposition into plasmid targets. Proc Natl Acad Sci 100: 15742–15747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O’Brien Johnson R, Collins K, Loo JA, et al. 2015. Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 350: aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CM, Fisher DJ. 2013. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PLoS ONE 8: e83989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM. 2001. Group II introns as controllable gene targeting vectors for the genetic manipulation of bacteria. Nature Biotechnol 19: 1162–1167. [DOI] [PubMed] [Google Scholar]
- Katibah GE, Qin Y, Sidote DJ, Yao J, Lambowitz AM, Collins K. 2014. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc Natl Acad Sci 111: 12025–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kint N, Janoir C, Monot M, Hoys S, Soutourina O, Dupuy B, Martin-Verstraete I. 2017. The alternative sigma factor σB plays a crucial role in adaptive strategies of Clostridium difficile during gut infection. Environ Microbiol 19: 1933–1958. [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. 2015. Mobile bacterial group II introns at the crux of eukaryotic evolution. Microbio Spectr 3: MDNA3-0050-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S. 2011. Group II introns: Mobile ribozymes that invade DNA. Cold Spring Harbor Perspect Biol 3: a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Beckman KA, Pessino V, Huange B, Weissman JS, Leonetti MD. 2017a. Design and specificity of long ssDNA donors for CRISPR-based knock-in. BioRxiv 10.1101/178905. [DOI] [Google Scholar]
- Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, Mao Y, Lv J, Yi D, Chen XW, et al. 2017b. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell 68: 993–1005.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastroianni M, Watanabe K, White TB, Zhuang F, Vernon J, Matsuura M, Wallingford J, Lambowitz AM. 2008. Group II intron-based gene targeting reactions in eukaryotes. PLoS ONE 3: e3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Noah JW, Lambowitz AM. 2001. Mechanism of maturase-promoted group II intron splicing. EMBO J 20: 7259–7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Smith D, Belfort M, Lambowitz AM. 2000. Rules for DNA target site recognition by a Lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev 14: 559–573. [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Hong W, Zhang J, Cui GZ, Yang Y, Cui Q, Liu YJ, Lambowitz AM. 2013a. A targetron system for gene targeting in thermophiles and its application in Clostridium thermocellum. PLoS ONE 8: e69032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr S, Ghanem E, Smith W, Sheeter D, Qin Y, King O, Polioudakis D, Iyer VR, Hunicke-Smith S, Swamy S, et al. 2013b. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. RNA 19: 958–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G, Kang SY, Park SK, Qin Y, Grohman J, Yao J, Stamos JL, Lambowitz AM. 2018. A highly proliferative group IIC intron from Geobacillus stearothermophilus reveals new features of group II intron mobility and RNA splicing. J Mol Biol 430: 2760–2783. 10.1016/j.jmb.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K. 2018. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 557: 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham RM, Wu DC, Qin Y, Yao J, Hunicke-Smith S, Lambowitz AM. 2016. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA 22: 597–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova O, Belfort M. 2017. Mobile group II introns as ancestral eukaryotic elements. Trends Genet 33: 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova O, Smith D, Hahn I, Beauregard A, Belfort M. 2014. Interaction between conjugative and retrotransposable elements in horizontal gene transfer. PLoS Genet 10: e1004853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perutka J, Wang W, Goerlitz D, Lambowitz AM. 2004. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336: 421–439. [DOI] [PubMed] [Google Scholar]
- Piccirilli JA, Staley JP. 2016. Reverse transcriptases lend a hand in splicing catalysis. Nat Struct Mol Biol 23: 507–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter EB, Polaski JT, Morck MM, Batey RT. 2017. Recurrent RNA motifs as scaffolds for genetically encodable small-molecule biosensors. Nat Chem Biol 13: 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Yao J, Wu DC, Nottingham RM, Mohr S, Hunicke-Smith S, Lambowitz AM. 2016. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22: 111–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu G, Kaushal PS, Wang J, Shigematsu H, Piazza CL, Agrawal RK, Belfort M, Wang HW. 2016. Structure of a group II intron in complex with its reverse transcriptase. Nat Struct Mol Biol 23: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Doudna JA. 2004. Assembly of an active group II intron–maturase complex by protein dimerization. Biochemistry 43: 6486–6497. [DOI] [PubMed] [Google Scholar]
- Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. 2014. Crystal structure of a eukaryotic group II intron lariat. Nature 514: 193–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, Erlacher M, Rossmanith W, Stern-Ginossar N, Schwartz S. 2017. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 551: 251–255. [DOI] [PubMed] [Google Scholar]
- Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz AM. 1999. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry 38: 9069–9083. [DOI] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, Cox J, Cheng Y, Lambowitz AM, Weissman JS, et al. 2015. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347: 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2017. The spliceosome: A protein-directed metalloribozyme. J Mol Biol 429: 2640–2653. [DOI] [PubMed] [Google Scholar]
- Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, Lambowitz AM. 2017. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci 114: E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Lambowitz AM. 2001. Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J Mol Biol 309: 361–386. [DOI] [PubMed] [Google Scholar]
- Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. 2016. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 164: 57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamos JL, Lentzsch AM, Lambowitz AM. 2017. Structure of a thermostable group II intron reverse transcriptase with template-primer and its functional and evolutionary implications. Mol Cell 68: 926–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, Chan WK, Ma ES, Chan SL, Cheng SH, et al. 2015. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci 112: E5503–E5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong DM, Hewitt FC, Hanson JH, Cui X, Lambowitz AM. 2015. Retrohoming of a mobile group II intron in human cells suggests how eukaryotes limit group II intron proliferation. PLoS Genet 11: e1005422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bartel DP. 2017. Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell 169: 905–917. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu DC, Lambowitz AM. 2017. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching. Sci Rep 7: 8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Jorde LB. 2013. Mobile element biology: New possibilities with high-throughput sequencing. Trends Genet 29: 280–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Q, Yang Y, Chen J, Chen L, Yang S, Jiang W, Gu Y. 2016. Roles of three AbrBs in regulating two-phase Clostridium acetobutylicum fermentation. Appl Microbiol Biotechnol 100: 9081–9089. [DOI] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yao J, Lambowitz AM. 2007. Gene targeting in gram-negative bacteria by use of a mobile group II intron (“Targetron”) expressed from a broad-host-range vector. Appl Environ Microbiol 73: 2735–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Zhong J, Fang Y, Geisinger E, Novick RP, Lambowitz AM. 2006. Use of targetrons to disrupt essential and nonessential genes in Staphylococcus aureus reveals temperature sensitivity of Ll.LtrB group II intron splicing. RNA 12: 1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. 2016. irCLIP platform for efficient characterization of protein-RNA interactions. Nat Methods 13: 489–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Pyle AM. 2016. Crystal structures of a group II intron maturase reveal a missing link in spliceosome evolution. Nat Struct Mol Biol 23: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Liu F, Pyle AM. 2018. An ultraprocessive, accurate reverse transcriptase encoded by a metazoan group II intron. RNA 24: 183–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong J, Karberg M, Lambowitz AM. 2003. Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res 31: 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang F, Karberg M, Perutka J, Lambowitz AM. 2009. EcI5, a group IIB intron with high retrohoming frequency: DNA target site recognition and use in gene targeting. RNA 15: 432–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoraghi R, Worrall L, See RH, Strangman W, Popplewell WL, Gong H, Samaai T, Swayze RD, Kaur S, Vuckovic M, et al. 2011. Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities. J Biol Chem 286: 44716–44725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubradt M, Gupta P, Persad S, Lambowitz AM, Weissman JS, Rouskin S. 2017. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nat Methods 14: 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]