Abstract
The vast majority of eukaryotic messenger RNAs (mRNAs) initiate translation through a canonical, cap-dependent mechanism requiring a free 5′ end and 5′ cap and several initiation factors to form a translationally active ribosome. Stresses such as hypoxia, apoptosis, starvation, and viral infection down-regulate cap-dependent translation during which alternative mechanisms of translation initiation prevail to express proteins required to cope with the stress, or to produce viral proteins. The diversity of noncanonical initiation mechanisms encompasses a broad range of strategies and cellular cofactors. Herein, we provide an overview and, whenever possible, a mechanistic understanding of the various noncanonical mechanisms of initiation used by cells and viruses. Despite many unanswered questions, recent advances have propelled our understanding of the scope, diversity, and mechanisms of alternative initiation.
Cap-dependent translation relies on recognition of the 5′ cap by the eukaryotic initiation factor (eIF)4F complex (eIF4E, eIF4G, and eIF4A), which recruits the 43S preinitiation complex ([PIC], 40S, eIF3, eIF1, eIF1A, eIF5, and ternary complex [eIF2•Met-tRNAi•GTP]) to the 5′ end of the messenger RNA (mRNA). Following recruitment, the 43S ribosomal subunit scans the mRNA in a 5′-to-3′ direction until the AUG start codon is recognized and 60S ribosomal subunit joining occurs to make an 80S elongation-competent ribosome (see Merrick and Pavitt 2018). Cap-dependent initiation is down-regulated under conditions of cellular stress (see Wek 2018) or during some viral infections (see Stern-Ginossar et al. 2018). Therefore, viral proteins and cellular proteins that are required to survive or adapt to the cellular stress are synthesized using alternative mechanisms of translation initiation (Fig. 1).
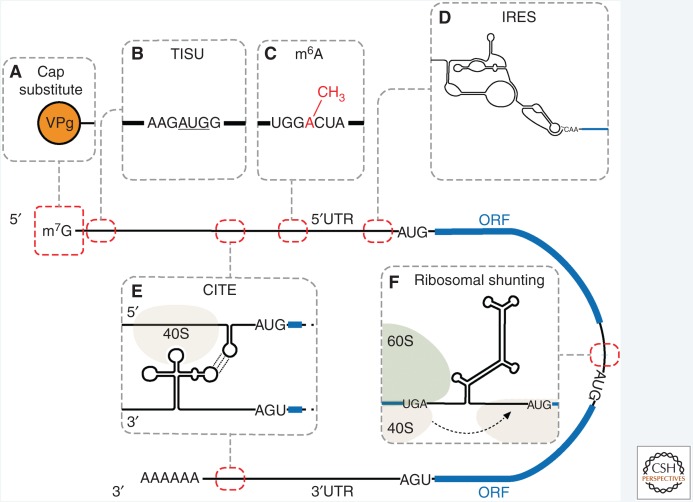
Figure 1.
A model conveying the different mechanisms of noncanonical translation initiation. The locations of the cis-acting elements (red dashed lines), the open reading frames ([ORFs], blue rectangles), and 5′ and 3′ untranslated regions ([UTRs], black lines) are indicated. (A) Viral protein linked to the genome (VPg) rather than a 5′ cap. (B) Translation initiator of short 5′UTR (TISU) element with the start codon underlined. (C) m6A modification. (D) Internal ribosomal entry site (IRES): the intergenic region (IGR) IRES is shown. (E) 3′ Cap-independent translation element (CITE): barley yellow dwarf virus-like translation element (BTE) CITE in the 3′UTR is shown forming a kissing-loop (dotted line) interaction with a stem loop in the 5′UTR. (F) Ribosomal shunting: cauliflower mosaic virus (CaMV) ribosomal shunt is shown; the highly structured region promotes shunting of the 40S from the “donor” site to a downstream “acceptor” site for initiation.
Noncanonical mechanisms of initiation vary with respect to which eIFs are required, whether ribosome scanning and cap recognition are used, and the conditions in which they are favored. For example, ribosomal shunting requires a 5′ cap and all of the canonical initiation factors to load the 40S subunit onto the mRNA before translocating the 40S subunit to a downstream location to initiate translation (Morley and Coldwell 2008). In contrast, the initiation factor requirements for internal ribosome entry sites (IRESs), which require neither a 5′ cap nor a free 5′ end to recruit a ribosome internally to the mRNA, have a range of initiation factor requirements dependent on the type of IRES. For instance, the Dicistroviridae intergenic region (IGR) IRES is the simplest studied IRES, requiring no initiation factors (Jan et al. 2003), whereas the hepatitis A viral IRES uses all of the factors needed for canonical translation initiation including eIF4E (Avanzino et al. 2017). The mechanisms of noncanonical translation initiation that are known are very diverse and differ significantly with respect to how the ribosome is recruited and the mechanism of initiation. Our understanding of these noncanonical mechanisms of initiation has lagged behind our knowledge of cap-dependent translation, largely because of the lack of good genetic and biochemical systems, as most of these noncanonical mechanisms function only in mammalian cells, but not in yeast.
Although many of the mechanisms for noncanonical translation initiation are not well understood, recent advances in the IRES field have shed light on these mechanisms. For example, the first structure of an IRES bound to a ribosome was revealed using cryoelectron microscopy (cryo-EM) (Spahn et al. 2004). Soon afterward, the ribosome-binding domain of an IRES was crystalized, revealing the three-dimensional fold of an IRES (Pfingsten et al. 2006). Previously, the identification of mRNAs that harbor IRESs was cumbersome because of the fact that IRESs must be identified using functional assays to show that translation is cap-independent (Spriggs et al. 2010; Thompson 2012). However, this limitation was largely overcome by using microarrays and next-generation sequencing to identify mRNAs that remained polysome-associated when cap-dependent translation was down-regulated (Johannes et al. 1999; Coldwell et al. 2001; Bushell et al. 2006; Thomas and Johannes 2007). Although these large-scale analyses are not sufficient to attribute IRES activity to any of the mRNAs, a recent high-throughput approach validated most of these candidate mRNAs as having bona fide IRES activity in the 5′ untranslated region (5′UTR) (Weingarten-Gabbay et al. 2016). IRESs were first discovered in viruses in 1988 (Jang et al. 1988; Pelletier and Sonenberg 1988), and minimal factors that were required for the activity of the encephalomyocarditis virus (EMCV) IRES were identified in vitro about 8 years later (Pestova et al. 1996a). However, reconstitution of the poliovirus (PV) IRES was more enigmatic because it required additional RNA-binding proteins that were not initiation factors (Sweeney et al. 2014). mRNA modifications have also been implicated in directing noncanonical initiation (see Meyer et al. 2015; Zhou et al. 2015; Peer et al. 2018). Furthermore, the ribosome, which was once considered to be a large protein synthesis machine that merely translated mRNAs into proteins, is becoming better appreciated for its roles in regulation of initiation and selection of mRNAs based on ribosomal RNA modifications and nonessential ribosomal proteins, such as eS25, eL38, and eL40 (Jack et al. 2011; Hertz et al. 2013; Lee et al. 2013; Ban et al. 2014; Meyer et al. 2015; Xue et al. 2015; Yang et al. 2017). This review focuses on how these discoveries have contributed to our understanding of noncanonical mechanisms of translation initiation.
INTERNAL RIBOSOME ENTRY SITES (IRESs)
IRESs are RNA elements that can recruit a ribosome internally to the mRNA rather than at the 5′ end. IRESs are diverse, ranging in size from 9 to >1000 nucleotides and can be unstructured or contain stable secondary and tertiary structures. IRESs were first discovered in PV and EMCV, members of the Picornaviridae family (Jang et al. 1988; Pelletier and Sonenberg 1988). Later, IRESs were identified in cellular mRNAs that encode proteins related to cellular stress, survival, angiogenesis, and mitosis (Yang and Sarnow 1997; Walters and Thompson 2016). In 1995, Chen and Sarnow presented compelling evidence that the ribosome is recruited internally to the mRNA by showing that a circular RNA (no 5′ end or 5′ cap) containing the EMCV IRES but no stop codons could initiate translation and translate around the circular RNA multiple times. Inclusion of a thrombin cleavage site encoded by the circular RNA showed that on addition of thrombin the polypeptide product that was over 106 kDa was reduced to a 40 kDa protein, demonstrating that the circular RNA was intact and that the large protein generated was due to the ribosome transiting around the circular RNA multiple times and was not a protein aggregate (Chen and Sarnow 1995). This also was one of the first studies to challenge the idea that the 5′ end of the mRNA had to be threaded through the 40S subunit. Together, these results showed that ribosomes can be recruited onto mRNAs internally and do not require a free 5′ end or 5′ cap. Since then, the field has continued to amass a large body of evidence to show that both viral and cellular mRNAs can contain IRESs and they are now widely recognized as a cap-independent mechanism of initiation (Carter et al. 2000; Schneider et al. 2001; Ruggero 2013). The significance of IRESs and other noncanonical mechanisms of eukaryotic translation initiation during viral infections, cellular stress, and survival has suggested a major role for translational control during tumorigenesis and viral infection (Walters and Thompson 2016; Lacerda et al. 2017).
Classification of Viral IRESs
Viral IRESs are classified based on their structure, initiation factor requirements, and mechanism of initiation (Table 1). Picornaviral IRESs comprise five of the eight classes of viral IRESs. Type I IRESs were the first to be discovered and are present in enteroviruses (e.g., PV, enterovirus 71) and rhinoviruses (human rhinovirus type 2). Type I IRESs are approximately 490 nucleotides long (Pelletier and Sonenberg 1988) and require eIFs, eIF2, eIF3, eIF4A, and the central domain of eIF4G, as well as the IRES trans-acting factor (ITAF) poly(C)-binding protein 2 (PCBP2) (Sweeney et al. 2014). eIF1A and eIF4B are not required, but strongly stimulate type I IRES activity (Sweeney et al. 2014). For type I IRESs, the 40S ribosomal subunit is recruited to an upstream AUG before scanning the RNA in a 5′-to-3′ direction to initiate translation at a downstream start codon. eIF1 facilitates correct start codon recognition for type I IRESs, consistent with its role in cap-dependent initiation (Kaminski et al. 2010; Sweeney et al. 2014; see Merrick and Pavitt 2018).
Table 1.
Classification of viral internal ribosome entry sites (IRESs)
| Requirements | |||||
|---|---|---|---|---|---|
| eIFs | ITAFs | Examples | Notable features | ||
| Picornaviral IRES subtypes | |||||
Type I (entero-/rhinovirus)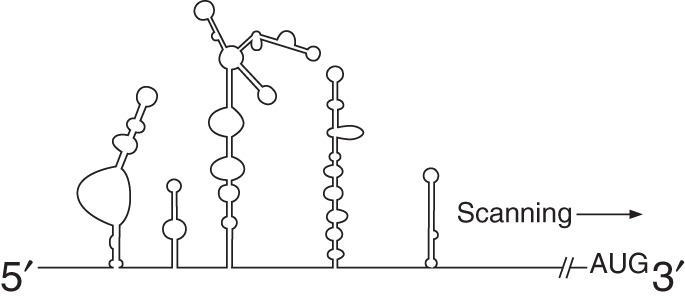
|
eIF2 eIF3 eIF4A eIF4Gm |
PTB PCBP1 PCBP2 |
Poliovirus EV71 BEV Coxsackievirus B3 HRV2 |
Ribosomal scanning downstream of ribosome binding to the AUG | |
Type II (cardio-/apthovirus)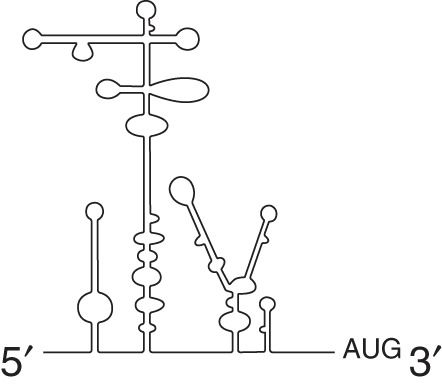 |
eIF2 eIF3 eIF4A eIF4B eIF4Gm |
PTB ITAF45 |
EMCV FMDV TMEV |
Initiates translation at the site of 40S recruitment, 40S does not scan | |
|
Type IV (HAV-like) 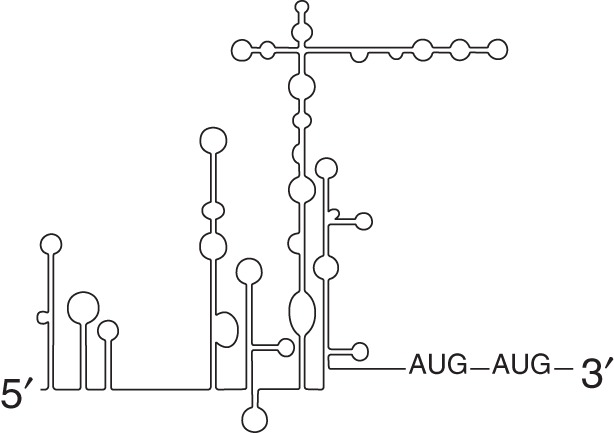 |
eIF2 eIF3 eIF4A eIF4B eIF4E eIF4G |
PTB PCBP2 La |
HAV |
Requires eIF4E but not binding to the 5′ cap |
|
Type V (Aichivirus)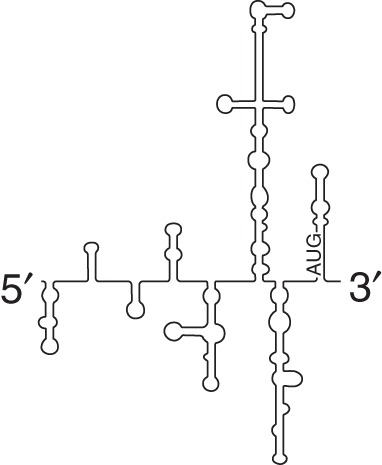 |
eIF2 eIF3 eIF4A eIF4B eIF4Gm |
PTB DHX29 |
Aichivirus | Does not require eIF1 and eIF1A | |
| Type III (HCV-like) HCV-like 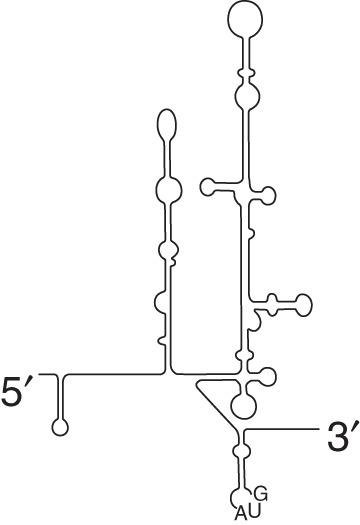
|
eIF2 eIF3 |
None | AEV PTV-1 SPV-9 |
||
| eIF2 eIF3 |
None | HCV CSFV |
Direct binding to 40S ribosomal subunit; translates under conditions with low ternary complex | ||
Dicistrovirus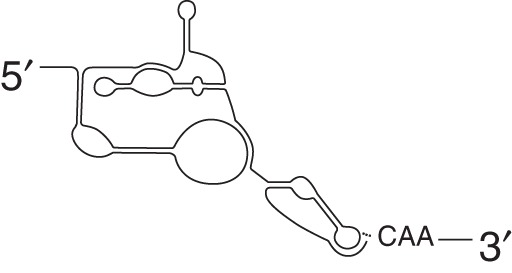
|
None | None | CrPV DCV IAV PSIV |
Initiates translation in the absence of any eIFs in the A-site at a non-AUG codon Can translate in a +1 reading frame (ORFx) |
|
Retrovirus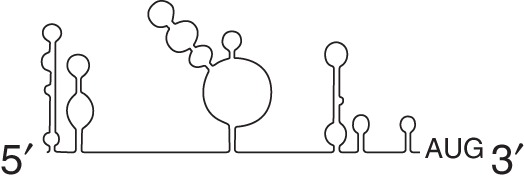
|
eIF4A eIF4G eI5A |
hnRNPA1 hRIP DDX3 |
HIV-1 HTLV-1 SIV FIV |
||
Herpesvirus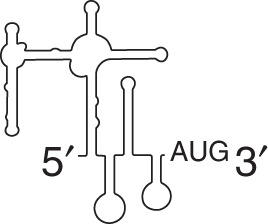
|
eIF4F | PTB PCBP1 KSHV-ORF57 |
KSHV-vFLIP MDV |
vFLIP IRES located in ORF of vCyclin | |
eIF, Eukaryotic initiation factor; ITAF, IRES trans-acting factor; BEV, bovine enterovirus; HRV2, human rhinovirus serotype 2; TMEV, Theiler’s murine encephalymyelitis virus; HAV, hepatitis A virus; SPV-9, simian picornavirus 9; CSFV, classical swine fever virus; DCV, drosophila C virus; IAV, influenza A virus; PSIV, Plautia stali intestine virus; HIV, human immunodeficiency virus; HTLV-1, human T-cell leukemia virus type 1; SIV, simian immunodeficiency virus; FIV, feline immunodeficiency virus; KSHV, Kaposi sarcoma herpesvirus; MDV, Marek’s disease virus; CrPV, cricket paralysis virus.
Type II IRESs, found in cardio- and aphthoviruses (e.g., EMCV and foot and mouth disease virus [FMDV]), are structurally different from type I IRESs, yet both classes have similar eIF requirements (Pestova et al. 1996a,b; Lomakin et al. 2000). A defining difference between type I and type II IRESs is that no scanning occurs downstream from the type II IRES despite recruitment of the ribosome to a similar AUG motif (Sweeney et al. 2014). Instead, this AUG serves as the authentic start codon for type II IRESs and neither scanning nor eIF1 is required (Pestova et al. 1996a). However, the FMDV IRES is an exception in that protein synthesis can also occur at a downstream start codon, AUG2, which is dependent on eIF1 and likely involves scanning, meaning that this IRES has characteristics of both type I and type II IRESs (Andreev et al. 2007). This is an example of the complexity of IRES-mediated translation and how the presence or absence of an initiation factor can modulate initiation codon selection for an IRES.
The type III class of IRESs consists of the flavivirus IRESs, the hepatitis C virus (HCV) IRES, and picornaviral IRESs such as porcine teschovirus 1 (PTV-1) and avian encephalomyelitis virus (AEV) (Pisarev et al. 2004; Bakhshesh et al. 2008). The HCV IRES is the prototype for this class of IRESs and initiates translation by binding to the solvent side of the 40S ribosomal subunit. This binding is stabilized by initiation factor eIF3 and base-pairing between the domain IIId region of the HCV IRES and the loop region of helix 26 of the 18S rRNA (Fraser and Doudna 2007; Matsuda and Mauro 2014). Although methionyl-transfer RNA (Met-tRNAi), is required for HCV start codon recognition (Pestova et al. 1998; Fraser and Doudna 2007) in vitro, the HCV IRES has been shown to function under conditions where the ternary complex is limiting, such as when eIF2 is inactivated via phosphorylation (Gonzalez-Almela et al. 2018). In addition, it has been shown that eIF2 is displaced from the initiating complex on HCV, suggesting that HCV does not use it even if it is available (Jaafar et al. 2016). The initiating Met-tRNAi can be brought in independently of eIF2, but how this occurs has been very controversial (Terenin et al. 2008; Jaafar et al. 2016; Gonzalez-Almela et al. 2018). This illustrates the importance of confirming in vitro findings in vivo, because in vitro experiments allow a great deal of control over the components present but may not reflect what is occurring under physiological conditions. It now appears that eIF2D or eIF2A are not required for HCV initiation in vivo as previous findings had suggested, but rather that eIF1A and possibly eIF5B are important for selecting (by codon–anticodon base pairing) and stabilizing Met-tRNAi binding to the HCV 48S complex in Huh-7 cells under conditions where eIF2α is phosphorylated (Jaafar et al. 2016; Gonzalez-Almela et al. 2018). It has been proposed that under stress conditions when eIF2α is phosphorylated, excess levels of Met-tRNAi would be sufficient for recruitment to the HCV 48S complexes in the absence of a dedicated delivery factor (Jaafar et al. 2016). Consistent with this model, the 40S subunit has been reported to bind to the HCV IRES with a dissociation rate that is insignificant on the time scale of initiation (Fuchs et al. 2015).
The most elegant viral IRESs are the IGR IRESs found in the Dicistroviridae family (Sasaki et al. 1998; Sasaki and Nakashima 1999; Wilson et al. 2000; Nakashima and Uchiumi 2009). These IRESs are <200 nucleotides in length and consist of a three-domain, multiple stem-loop structure containing three pseudoknots. The defining mechanistic feature of the IGR-IRES in dicistroviruses is that they can initiate translation in the absence of other initiation factors and also do not make use of a canonical AUG start codon (Sasaki and Nakashima 2000; Wilson et al. 2000; Jan and Sarnow 2002; Deniz et al. 2009). This shows that an RNA structure can independently recruit and manipulate ribosomes to initiate translation, as described next.
Mechanism of Internal Initiation of the Dicistroviridae IGR IRESs
The molecular mechanisms for how a cis-acting RNA element recruits a 40S subunit internally to the mRNA, loads the mRNA into the mRNA channel, recognizes the start codon, and promotes 60S subunit joining is not understood for most IRESs. After all, 10 to 13 initiation factors are required for cap-dependent initiation, yet the IGR IRES is able to bind directly to 40S subunits in the absence of any initiation factors and form translationally competent 80S complexes on addition of 60S subunits (Jan et al. 2003; Pestova and Hellen 2003).
The ∼190 nucleotide cricket paralysis virus (CrPV) IGR IRES forms an RNA structure with three pseudoknots (PKI, PKII, and PKIII) that span all three tRNA-binding sites of the ribosome (Fernandez et al. 2014). PKI mimics an anticodon stem loop bound to a codon and occupies the A site, which is recognized by the universally conserved nucleotides in the decoding center that monitor accurate tRNA selection (G530, A1492, and A1493 in Escherichia coli 16S rRNA) (Schuler et al. 2006; Fernandez et al. 2014). PKII and PKIII extend out toward the E site and are required for affinity of the IRES to the 40S ribosomal subunit (Jan and Sarnow 2002; Pestova and Hellen 2003; Murray et al. 2016). The IGR IRES binds to the intersubunit surface of the 40S subunit with high affinity (5 to 20 nm), resembling a pretranslocation conformation of the ribosome (Spahn et al. 2004; Schuler et al. 2006; Jang et al. 2009; Landry et al. 2009; Jang and Jan 2010; Jack et al. 2011; Fernandez et al. 2014).
For the first amino acid to be decoded, PKI must be translocated to the P site by eukaryotic elongation factor 2 (eEF2), resulting in the first codon that is decoded being positioned in the A site (Fernandez et al. 2014; Koh et al. 2014). After aminoacyl-tRNA (aa-tRNA) binding to the A-site codon, a second translocation event must occur (without peptidyl transfer) to move PKI to the E site and the A site aa-tRNA to the P site (Fernandez et al. 2014; Muhs et al. 2015). The resulting complex resembles the normal 80S initiation complex that contains Met-tRNAi in the P site and an empty A site. Thus, it appears that the CrPV IGR IRES manipulates the ribosome to forego classical initiation and begin with elongation.
Although the CrPV IGR IRES has many properties that make it unique, it provides insights into mechanisms that are applicable to other IRESs. For example, the HCV IRES can also bind to 40S subunits in the absence of any initiation factors. Both HCV and the IGR IRES induce a similar conformational change in the 40S subunit on binding that resembles an “open” conformation for mRNA loading (Spahn et al. 2001, 2004). This conformational change resembles the eIF1- and eIF1A-induced open conformation of the 43S PIC, whereby the mRNA channel is open to allow for mRNA loading (Passmore et al. 2007). Another common requirement for structurally and functionally diverse IRESs is ribosomal protein S25 (eS25/RPS25). Although the role of eS25 is not known, its requirement by both viral and cellular IRESs suggests that they may share a common eS25-dependent mechanism (Hertz et al. 2013). Given the structural differences and eIF requirements, there will likely be some differences in how diverse IRESs initiate translation; however, what is interesting is how these seemingly diverse IRESs appear to have some common mechanisms for initiating translation.
Cellular IRESs
An estimated 10% of mammalian mRNAs have IRESs (Martinez-Salas et al. 2012; Walters and Thompson 2016; Weingarten-Gabbay et al. 2016). These mRNAs encode proteins important during early development, mitosis, and cellular stresses such as hypoxia, apoptosis, starvation, heat shock, tumorigenesis, and viral infection (Table 2) (Johannes and Sarnow 1998; Johannes et al. 1999; Coldwell et al. 2001; Holcik 2004; Bushell et al. 2006; Dresios et al. 2006; Xue et al. 2015; Vaklavas et al. 2016; Walters and Thompson 2016). Our understanding of cellular IRESs lags behind that of viral IRESs because of the fact that many of these IRES-containing mRNAs can be translated cap-dependently but switch to IRES-mediated initiation under cellular stress. To accommodate the switch from cap-dependent to cap-independent initiation, cellular IRESs are generally less structured than viral IRESs and can even be modular, meaning that the IRES activity is distributed throughout the 5′UTR. In fact, some cellular IRESs, such as the Gtx IRES, base pair with a region on the 18S ribosomal RNA reminiscent of Shine–Dalgarno sequences (Dresios et al. 2006). The complementary ribosomal sequence is located in the proximity of the classical Shine–Dalgarno sequence. However, unlike the anti-Shine–Dalgarno sequence, which is single-stranded, this sequence is in a region of helix 26 that is predicted to be base-paired (Deforges et al. 2015). More studies are required to understand the mechanism (Chappell et al. 2000; Dresios et al. 2006; Panopoulos and Mauro 2008). Conversely, some cellular IRESs (such as Bag-1 and c-myc) are structurally complex like viral IRESs (Le Quesne et al. 2001; Pickering et al. 2004; Baird et al. 2007; Spriggs et al. 2008).
Table 2.
Mammalian cellular internal ribosomal entry site (IRES)
| Cellular pathway | Name | References |
|---|---|---|
| Amino acid starvation | Cationic amino acid transporter 1 (CAT-1) | Fernandez et al. 2001 |
| Sodium-coupled amino acid transporter (SNAT2) | Gaccioli et al. 2006 | |
| Nutrient signaling | Human insulin receptor (HIR) | Spriggs et al. 2009 |
| hypoxia | Hypoxia inducible factor 1α (HIF-1α) | Schepens et al. 2005 |
| Vascular endothelial growth factor (VEGF) | Morris et al. 2010 | |
| Fibroblast growth factor 2 (FGF2) | Bonnal et al. 2003 | |
| Apoptosis survival | Apoptotic protease activating factor 1 (Apaf-1) | Sella et al. 1999 |
| B-cell lymphoma 2 (Bcl-2) | Marash et al. 2008 | |
| BAG family molecular chaperone regulator 1 (Bag1) | Pickering et al. 2004 | |
| Cellular inhibitor of apoptosis 1 (cIAP1) | Graber et al. 2010 | |
| X-linked inhibitor of apoptosis (XIAP) | Riley et al. 2010 | |
| Oncogene | c-myc | Le Quesne et al. 2001 |
| c-Jun | Blau et al. 2012 | |
| Mitosis | p58 PITSLRE | Cornelis et al. 2000 |
| Cyclin-dependent kinase 1 (CDK1) | Marash et al. 2008 | |
| p120 catenin (p120) | Silvera et al. 2009 | |
| DNA damage response | p53 | Yang et al. 2006 |
| Serine hydroxymethyltransferase 1 (SHMT1) | Fox et al. 2009 | |
| Replication protein A2 (RPA2) | Yin et al. 2013 | |
| Differentiation | Homeobox transcription factor (Hox) | Xue et al. 2015 |
| Platelet-derived growth factor 2 (PDGF2) | Sella et al. 1999 | |
| Runt-related transcription factor 1 (Runx1) | Pozner et al. 1999 | |
| Lymphoid enhancer-binding factor 1 (LEF-1) | Jimenez et al. 2005 | |
| Cold shock | Cold inducible RNA-binding protein (CIRP) | Al Fageeh and Smales 2009 |
An example of an extensively studied cellular protein that is translated by an IRES-dependent mechanism of initiation is the apoptotic protease activating factor 1 (Apaf-1) protein. Upon binding cytochrome c and dATP, Apaf-1 forms an oligomeric apoptosome that cleaves the procaspase 9 protein to stimulate the caspase cascade that commits the cell to apoptosis. Apaf-1 is important for programmed cell death during early development and knockout mice die early in embryogenesis because of a malformation of the brain caused by reduced apoptosis (Cecconi et al. 1998; Yoshida et al. 1998). As expected, the Apaf-1 IRES is most active in neuronal cells (25-fold over reporter activity with no IRES activity). This is a result, in part, of the higher affinity of the nPTB (neuronal polypyrimidine tract-binding protein 1) ITAF compared with the nonneuronal PTB found in other cell types (i.e., HEK293T cells, which show only sixfold more activity than no-IRES controls) (Coldwell et al. 2000; Mitchell et al. 2003; Andreev et al. 2009). Studies that questioned whether Apaf-1 has an IRES were all performed in a cell line known not to support robust Apaf-1 IRES activity (Andreev et al. 2009). The Apaf-1 mRNA is expressed in all tissues except skeletal muscle, but it is poorly translated through a cap-dependent mechanism of initiation, suggesting that most of the Apaf-1 initiation occurs through the IRES (Burgess et al. 1999; Coldwell et al. 2000). In a bicistronic reporter assay (described below) the Apaf-1 IRES was more active than the human rhinovirus IRES but less active than the c-myc IRES (Coldwell et al. 2000). Preventing expression of the first cistron by inserting a hairpin upstream of it did not diminish IRES activity, demonstrating that expression of the second cistron was not caused by readthrough from the first cistron (Coldwell et al. 2000). In fact, insertion of a stable hairpin upstream of the 5′UTR in a monocistronic reporter reduced Apaf-1 mRNA translation only 1.5-fold compared with cap-dependent translation, which was inhibited 200-fold (Coldwell et al. 2000). We note that studies disputing the existence of the Apaf-1 IRES are not inconsistent with data from other groups demonstrating its IRES activity; rather, it is the interpretation of the data that differs. Thus, it is important for researchers to publish their raw data, not just normalized data, and to assay IRES activity in correct cell types and under conditions in which the endogenous protein is made.
IRES Trans-Acting Factors
ITAFs bind to IRESs and function as RNA chaperones to remodel the RNA to enhance or repress IRES activity. Both viral and cellular IRESs require ITAFs, excluding the Dicistroviridae IGR IRESs (Jan et al. 2003; Pestova and Hellen 2003; Deniz et al. 2009). Interestingly, although translation occurs in the cytoplasm, most ITAFs are either nuclear proteins or proteins that cycle between the nucleus and cytoplasm. The first identified ITAF was La autoantigen (La), which prevents initiation at aberrant start codons and enhances translation of the PV IRES (Meerovitch et al. 1993).
La enhances the activity of the HCV, coxsackievirus, and several cellular IRESs, such as binding immunoglobulin protein (BiP) and X-linked inhibitor of apoptosis (XIAP) (Holcik and Korneluk 2000; Kim et al. 2001; Ray and Das 2002; Costa-Mattioli et al. 2004). Some ITAF mRNAs, such as those encoding La and upstream of N-ras (Unr), use IRESs for their translation (Carter and Sarnow 2000; Cornelis et al. 2005). Other notable ITAFs reported to interact with multiple IRESs include poly-C-binding protein 2 (PCBP2), and the heterogeneous nuclear ribonucleoprotein (hnRNP) family of RNA-binding proteins (Hunt et al. 1999; Fujimura et al. 2008). Whereas the majority of ITAFs are reported to stimulate IRES-mediated translation, the RNA-binding proteins AU-rich-binding factor (AUF1), KH-type splicing regulatory protein (KHSRP), and double-stranded RNA-binding nuclear protein 76 (DRBP76) negatively modulate IRES activity (Merrill and Gromeier 2006; Lin et al. 2009, 2014; Cathcart et al. 2013; Kung et al. 2017). Taken together, ITAFs play an important role in tissue and host tropism for viruses as well as a regulatory role for cellular IRESs, which may only be active in certain cell types or under certain conditions, such as hypoxia or apoptosis (Coldwell et al. 2000; Mitchell et al. 2003; Jahan et al. 2013).
Identification of Putative IRESs
As has been described above, viral and cellular IRESs are tremendously diverse in their sequence, size, structure and mechanism of initiation, and the trans-acting factors used. This requires validation on an individual basis rather than identification bioinformatically (Baird et al. 2006). Potential IRESs have been identified by determining which mRNAs remain or become associated with polysomes under conditions in which cap-dependent translation is down-regulated (Johannes et al. 1999; Qin and Sarnow 2004; Bushell et al. 2006). A recent study took a high-throughput approach by synthesizing 55,000, 210-nucleotide long oligos from cellular and viral mRNAs that have been previously reported to either have IRES activity or be polysome-associated in one of the stress conditions known to down-regulate cap-dependent initiation. The results confirmed that many of the mRNAs that remain polysome-associated under stress have IRESs (Weingarten-Gabbay et al. 2016).
The most important consideration for identifying and validating putative IRESs is that there is no perfect assay. Rather, establishing a new IRES requires a preponderance of evidence to rule out the possibility that translation could be occurring in any other fashion. The assays and controls have been discussed at length previously (Thompson 2012) and only a short summary will be provided here. The bicistronic reporter assay is commonly used as the first assay toward evaluating a putative IRES. Briefly, a DNA plasmid encoding a bicistronic reporter in which the putative IRES sequence has been inserted between two reporter genes (most commonly Renilla and firefly luciferase) is transfected into cells followed by monitoring of the encoded reporter activity. Expression of the first cistron uses a cap-dependent mechanism of initiating translation, whereas expression of the second cistron is driven by IRES-mediated translation. IRES activity is indicated by expression of the second cistron relative to positive and negative controls and corrected for transfection differences by cotransfection of a separate cap-dependent reporter plasmid. Assays to establish IRES activity should be performed in vivo in cells because translation extracts such as the rabbit reticulocyte lysate can translate promiscuously. Whereas the bicistronic assay itself is simple, the major challenge in the field is that a series of stringent controls are required to show that expression of the second cistron is not dependent on a cryptic promoter, cryptic splicing, or readthrough from the upstream cistron (Bert et al. 2006; Baranick et al. 2008; Thompson 2012). In addition, if studies are performed in vitro there need to be controls for RNA cleavage events that would lead to cap-independent initiation because some in vitro translation extracts can translate indiscriminately. As important as the controls are for identifying an IRES, the same stringency needs to be applied to studies that have claimed that previously identified IRESs have cryptic promoter activity. Specifically, controls need to be in place to show that the promoter activity is coming from the IRES and not the vector backbone (Cobbold et al. 2008). It is also important to note that the bicistronic assay is an artificial setting and that in this context IRESs may have cryptic promoter or splicing activity, but this does not show that they are not IRESs, only that the bicistronic IRES assay cannot be used to establish IRES activity. After all, if an endogenous mRNA has this RNA element in the 5′UTR, then the question still remains as to whether it can function as an IRES even if it cannot be shown in a bicistronic context.
Transfection of a bicistronic reporter as an mRNA to bypass transcription in the nucleus avoids issues such as cryptic promoters and splice sites that might produce false positives. However, transfection of an RNA reporter only works for a subset of cellular IRESs, suggesting that a nuclear experience may be required for assembling ITAFs or adding RNA modifications (Stoneley et al. 2000; Semler and Waterman 2008). The greater dependence of cellular IRESs on ITAFs compared with viral IRESs may be caused by their functional differences, because most viral IRESs are highly structured and are solely translated by an IRES-dependent mechanism, rather than switching back and forth between cap-dependent and IRES-mediated initiation. In contrast, many cellular mRNAs that have an IRES need to be translated both cap-dependently and cap-independently, depending on the cellular conditions (Walters and Thompson 2016).
Better assays are needed to identify and validate IRESs. One possible mechanism that needs to be explored further stems from the observation that viral and cellular IRESs depend on eS25, whereas cap-dependent translation does not (Hertz et al. 2013). Thus, if expression of a downstream open reading frame (ORF) from a bicistronic reporter were caused by a cryptic promoter, splicing, or readthrough, then its expression would not be affected by the loss of eS25 from the cells. Moreover, this also provides us with another way to examine whether endogenous messages are using IRES-mediated translation under a given cellular condition by determining whether polysome association of an mRNA depends on eS25, as shown for p53 (Hertz et al. 2013).
CAP-INDEPENDENT TRANSLATION ELEMENTS (CITEs)
The majority of plant viral RNAs lack a 5′-cap and poly(A) tail but are nevertheless translated efficiently by the host cell translational machinery. A major mechanism behind this phenomenon is the CITE, which was first discovered in satellite tobacco necrosis virus (STNV). CITEs are located in the 3′UTR and recruit the 40S subunit by binding some component of the eIF4F complex while using RNA–RNA interactions between the 5′UTR and 3′UTR that circularize the viral RNA. Ribosomes assemble at or near the 5′ end and scan to the start codon (Simon and Miller 2013; Miras et al. 2017). Although CITEs have only been found in the 3′UTR, they can still promote initiation when they are moved to the 5′UTR or into the coding region (Meulewaeter et al. 1998).
CITEs vary widely in sequence and structure and have been classified into seven categories: translation enhancer domain (TED), barley yellow dwarf virus-like translation element (BTE), Panicum mosaic virus-like translation enhancer (PTE), I-shaped structures (ISS), Y-shaped structures (YSS), T-shaped structures (TSS), and CABYV-Xinjiang-like translation element (CXTE) (Table 3). These have been described in detail previously (Simon and Miller 2013; Miras et al. 2017). Almost all classes of 3′-CITEs bind to some component of the eIF4F complex and circularize with the 5′UTR. For example, BTE binds preferentially to eIF4G and recruits the 40S subunit to the 3′UTR before transferring it to the 5′UTR where translation initiates (Treder et al. 2008; Sharma et al. 2015). However, PTE binds preferentially to eIF4E and TED, ISS and YSS CITEs require intact eIF4F for translation (Gazo et al. 2004; Truniger et al. 2008; Nicholson et al. 2010; Wang et al. 2011). An unusual case is the turnip crinkle virus that has a TSS CITE in the 3′UTR that interacts with the 60S ribosomal subunit instead of forming RNA–RNA interactions with the 5′UTR. Circularization of the RNA occurs through 60S binding with the 40S subunit that is bound to the 5′UTR (Stupina et al. 2008). In the case of pea enation mosaic virus 2 (PEMV2), there are three CITE structures in the 3′UTR: kissing loop (kl)-TSS, PTE, and TSS. The kl-TSS is responsible for the RNA–RNA interaction with the 5′UTR, whereas the PTE binds eIF4E to facilitate translation initiation (Du et al. 2017). More studies are required to understand the mechanism of initiation for CITEs and whether they share similarities or not. Also, it is unknown how CITEs compete with cap-dependent initiation.
Table 3.
Classification of cap-independent translation elements (CITEs)
| Type | Examples | Notable features |
|---|---|---|
TED (STNV)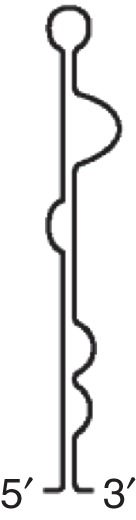 |
STNV PLPV PCLPV |
Binds eIF4F Apical loop interacts with hairpin in 5′UTR |
BTE (BYDV)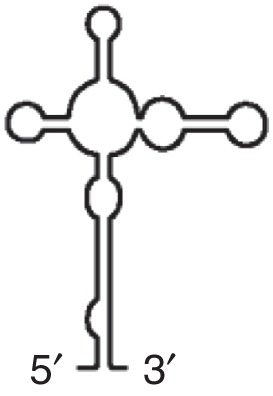
|
Alphanecrovirus Betanecrovirus Dianthovirus Luteovirus Umbravirus |
Binds eIF4G, eIF4E-independent Up to six stem loops can radiate from central hub |
|
PTE (PMV) 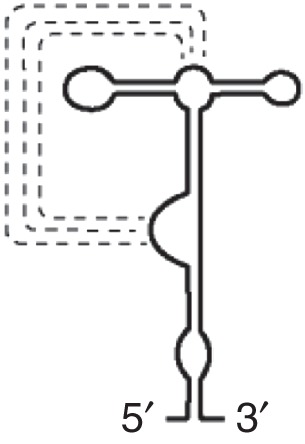
|
PMV PEMV2 SCV |
Binds eIF4E Base-pairing interactions (dotted lines) form pseudoknot |
|
ISS (MNSV) 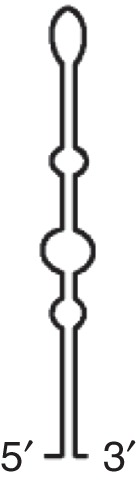 |
MNSV MNeSV |
Binds intact eIF4F Apical loop interacts with hairpin in 5′UTR |
YSS (TBSV)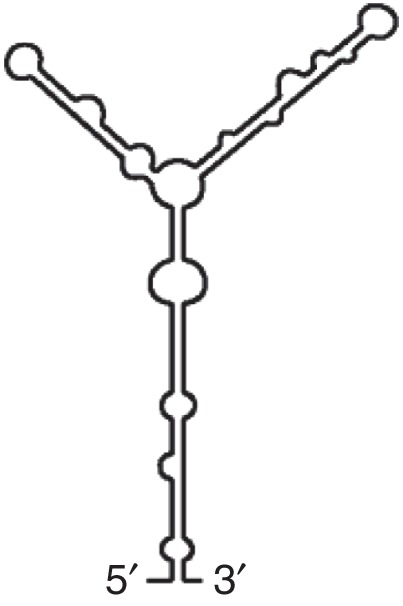
|
TBSV CIRV |
Requires eIF4F/eIFis04F 5′ hairpin interacts with hairpin in 5′UTR |
|
TSS (TCV) 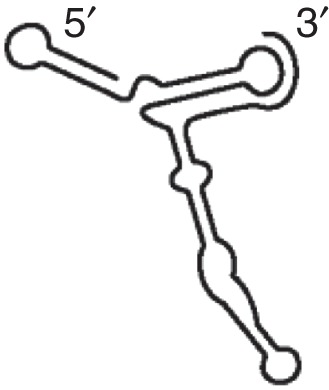
|
TCV PEMV2 |
TCV TSS competes with tRNA for P-site binding in 60S |
|
CXTE (MNSV) 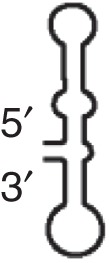
|
MNSV |
eIF4E independent |
PLPV, Pelargonium line pattern virus; PCLPV, pelargonium chlorotic ring pattern virus; BYDV, barley yellow dwarf virus; PMV, panicum mosaic virus; SCV, strawberry crinkle virus; MNSV, melon necrotic spot virus; MNeSV, maize necrotic streak virus; TBSV, tomato bushy stunt virus; CIRV, carnation Italian ringspot virus; TCV, turnip crinkle virus.
N6-METHYLADENOSINE
The most common posttranscriptional modification, methylation of adenine residues to generate N6-methyladenosine (m6A), affects mRNA turnover and translation efficiency (see Peer et al. 2018). Although the majority of m6A modifications are localized to the coding region and 3′UTR, it was recently discovered that modifications located in the 5′UTR can mediate cap-independent translation initiation, although a free 5′ end is still required (Meyer et al. 2015). Importantly, 5′UTR m6A modifications are up-regulated in response to heat shock by relocalization of the reader protein, YTHDF2, to the nucleus upon heat shock, which prevents the eraser protein, FTO, from removing m6A modifications (Meyer et al. 2015; Zhou et al. 2015). m6A promotes selective cap-independent initiation on certain mRNAs, such as HSP70, which depends on eIF3 binding to m6A residues to recruit the 40S subunit (Meyer et al. 2015). In contrast, another study showed that m6A can promote internal initiation of endogenous circular RNAs (circRNA) in cells and that the RNAs are polysome-associated (Yang et al. 2017). m6A-mediated translation is still a relative newcomer to the field (see Chekulaeva and Rajewsky 2018), and what is known has only been characterized in single reports that need to be reproduced. Additional studies are required to show that m6A modifications are required for noncanonical mechanisms of initiation and to reveal the mechanism for m6A-mediated initiation.
RIBOSOMAL SHUNTING
Ribosomal shunting occurs when ribosomes bypass or shunt over parts of the 5′UTR before reaching the start codon. Ribosomes are loaded onto the mRNA in a cap-dependent manner with recruitment of 40S subunits to the 5′ end of a capped mRNA followed by limited scanning to reach a “donor” site at which point the 40S jumps or “shunts” over major parts of a complex leader and lands at a downstream start codon (acceptor site) to initiate translation. Ribosomal shunting has been studied in cauliflower mosaic virus (CaMV) and adenovirus and is also used by a number of other plant and animal viruses (Stern-Ginossar et al. 2018). Additionally, cellular mRNAs such as BACE1, cIAP2, and HSP70 use ribosomal shunting to initiate translation under stress (Yueh and Schneider 2000; Rogers et al. 2004; Sherrill and Lloyd 2008; Koh et al. 2013).
Ribosome shunting allows viruses to expand their coding capacity by translating downstream ORFs, whereas both viral and cellular mRNAs use shunting to selectively translate mRNAs when cap-dependent translation is down-regulated. It has been proposed that shunting proceeds when the helicase of eIF4F is inactive because little to no unwinding is required (Yueh and Schneider 1996). The criteria for establishing that an mRNA is using ribosomal shunting is that its translation is cap-dependent, yet upstream AUGs and stem-loop structures have minimal or no effect on translation and its 5′UTR does not contain an IRES. Interestingly, many but not all mRNAs that use ribosomal shunting have a short ORF (sORF) that is translated; on termination, instead of the ribosome disassociating from the mRNA, it remains associated and proceeds to shunt or bypass other sORFs and secondary structures to reinitiate at a downstream ORF (Latorre et al. 1998; Yueh and Schneider 2000; Racine and Duncan 2010). Ribosomal bypass or shunting over entire regions of the 5′UTR was shown by insertion of a start codon in a good Kozak consensus in the bypassed sequences. This did not result in translation from this start codon, suggesting that the ribosome was not scanning through this region (Futterer et al. 1993). Additionally, two different RNAs were used to encode the donor site (where the ribosome shunts from) and the pyrimidine-rich acceptor site (where the ribosome lands); when these RNAs were annealed together through complementary sequences, translation still occurred at the downstream ORF, suggesting that the ribosome did not scan through the bypassed region because scanning would have separated the two RNAs (Futterer et al. 1993).
For those mRNAs that have an sORF that is translated, the 40S subunit is retained following termination through base-pairing of sequences in the 5′UTR (adenovirus and hsp70) with the 18S rRNA 3′ hairpin or through binding to eIF3 (CaMV), which remains associated with the 40S after initiation (Ryabova and Hohn 2000; Yueh and Schneider 2000; Morley and Coldwell 2008). Furthermore, the 40S ribosomal protein, eS25, which is required for IRES-mediated initiation, is also required for ribosomal shunting, although its role in shunting is not understood (Hertz et al. 2013). Many questions still remain about how ribosomes land and reinitiate downstream of the donor sites and which initiation factors or trans-acting factors are required for reinitiation.
TRANSLATION INITIATOR OF SHORT 5′UTR (TISU)
A cap-dependent, scanning-free mechanism of translation initiation in mRNAs with extremely short 5′UTRs (<30 nucleotides) requires a TISU element. TISUs are located within 5 to 30 nucleotides of the cap and have a median length of 12 nucleotides (Elfakess and Dikstein 2008). The TISU consensus sequence, SAASAUGGCGGC (S = C or G), differs from the Kozak consensus sequence (RCCAUGG, R = A or G) for start codon recognition (Elfakess and Dikstein 2008). Approximately 4% of mRNAs contain TISUs and encode proteins with housekeeping functions such as mitochondrial activities, energy metabolism, and protein synthesis (Elfakess and Dikstein 2008). TISU-containing mRNAs are specifically resistant to inhibition of protein synthesis caused by energy stress during glucose deprivation (Sinvani et al. 2015).
TISUs require eIF4F, which seems problematic given that recognition of an AUG close to the cap would clash with 43S PIC recruitment proximal to the cap. However, a key step in TISU initiation is release of eIF4F upon AUG recognition (Sinvani et al. 2015). During canonical cap-dependent initiation, eIF1 discriminates against AUGs proximal to the 5′ end, favoring 48S complex assembly on an AUG further downstream (Pestova and Kolupaeva 2002). However, eIF1 is not required for TISU start codon recognition as it did not inhibit translation for a TISU AUG located close to the 5′ cap, nor did it induce leaky scanning to bypass the TISU AUG (Elfakess et al. 2011; Dikstein 2012). Yet, eIF1 and eIF1A are required for TISU initiation. Most likely they are required to induce an open conformation (disruption of h18 and h34 and formation of a new interaction between h16 and uS3) of the 43S PIC for mRNA loading (Haimov et al. 2015; Sharifulin et al. 2016).
The ability of TISUs to prevent scanning of the 48S subunit following cap-dependent recruitment to the mRNA is caused by sequence-specific interactions with ribosomal proteins (uS3 and eS10) positioned in or near the entry channel (Haimov et al. 2017). The interactions between eIF1A and uS3 and eS10 have been suggested to arrest or prevent scanning by displacing helix 16 (h16), which induces the closed conformation and arrests scanning of the 40S. Taken together, this suggests that a TISU element near the 5′ end of the mRNA can recruit a 43S PIC in close proximity to the cap by displacing the cap-binding complex (eIF4F) and immediately arrest scanning on mRNA binding and AUG recognition.
CONCLUSION
Noncanonical translation plays an important role in gene expression during cellular stress. Transcriptomic analysis of mRNA levels correlates poorly with proteomic analysis of their expression, suggesting that there is a large amount of regulation at the posttranscriptional level (Gygi et al. 1999; Ideker et al. 2001; Griffin et al. 2002). Likely, the role of noncanonical translation has been underappreciated in cancer, early development, and cellular stress conditions. A better understanding of the contributions and mechanisms of noncanonical translation is critical for understanding gene expression. Overall, researchers in the translation field have described the various mechanisms of initiation as different or unique. However, given some of the similarities between ribosomal shunting and IRES-mediated translation, such as ribosome landing occurring at a pyrimidine-rich site, base-pairing with the 18S rRNA, and the requirement for eS25, it is tempting to speculate that there could be some common mechanisms shared between IRESs and ribosomal shunts (Morley and Coldwell 2008; Hertz et al. 2013). In addition, some noncanonical mechanisms use a tRNA-like structure to recruit or assemble ribosomes (Colussi et al. 2014; Butcher and Jan 2016). The mechanisms for how CITES, ribosomal shunting, TISU, IRESs, and m6A manipulate ribosomes to load the mRNA and initiate translation are largely unknown. As the mechanisms underlying noncanonical initiation are elucidated, both differences as well as similarities will be revealed.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Al-Fageeh MB, Smales CM. 2009. Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA 15: 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, Fernandez-Miragall O, Ramajo J, Dmitriev SE, Terenin IM, Martinez-Salas E, Shatsky IN. 2007. Differential factor requirement to assemble translation initiation complexes at the alternative start codons of foot-and-mouth disease virus RNA. RNA 13: 1366–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. 2009. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res 37: 6135–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avanzino BC, Fuchs G, Fraser CS. 2017. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc Natl Acad Sci 114: 9611–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SD, Turcotte M, Korneluk RG, Holcik M. 2006. Searching for IRES. RNA 12: 1755–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SD Baird, SM Lewis, Turcotte M, Holcik M. 2007. A search for structurally similar cellular internal ribosome entry sites. Nucleic Acids Res 35: 4664–4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhshesh M, Groppelli E, Willcocks MM, Royall E, Belsham GJ, Roberts LO. 2008. The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J Virol 82: 1993–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, Logg CR. 2008. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc Natl Acad Sci 105: 4733–4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bert AG, Grepin R, Vadas MA, Goodall GJ. 2006. Assessing IRES activity in the HIF-1α and other cellular 5′ UTRs. RNA 12: 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau L, Knirsh R, Ben-Dror I, Oren S, Kuphal S, Hau P, Proescholdt M, Bosserhoff AK, Vardimon L. 2012. Aberrant expression of c-Jun in glioblastoma by internal ribosome entry site (IRES)-mediated translational activation. Proc Natl Acad Sci 109: E2875–E2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnal S, Schaeffer C, Creancier L, Clamens S, Moine H, Prats AC, Vagner S. 2003. A single internal ribosome entry site containing a G quartet RNA structure drives fibroblast growth factor 2 gene expression at four alternative translation initiation codons. J Biol Chem 278: 39330–39336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess DH, Svensson M, Dandrea T, Gronlund K, Hammarquist F, Orrenius S, Cotgreave IA. 1999. Human skeletal muscle cytosols are refractory to cytochrome c-dependent activation of type-II caspases and lack APAF-1. Cell Death Differ 6: 256–261. [DOI] [PubMed] [Google Scholar]
- Bushell M, Stoneley M, Kong YW, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. 2006. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apoptosis. Mol Cell 23: 401–412. [DOI] [PubMed] [Google Scholar]
- Butcher SE, Jan E. 2016. tRNA-mimicry in IRES-mediated translation and recoding. RNA Biol 13: 1068–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter MS, Sarnow P. 2000. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J Biol Chem 275: 28301–28307. [DOI] [PubMed] [Google Scholar]
- Carter MS, Kuhn KM, Sarnow P. 2000. Cellular internal ribosome entry site elements and the use of cDNA microarrays in their investigation. In Translational control of gene expression (ed. Sonenberg N, et al. ), pp. 615–635. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Cathcart AL, Rozovics JM, Semler BL. 2013. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol 87: 10423–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F, Alvarez-Bolado G, Meyer BI, Roth KA, Gruss P. 1998. Apaf1 (CED-4 homolog) regulates programmed cell death in mammalian development. Cell 94: 727–737. [DOI] [PubMed] [Google Scholar]
- Chappell SA, Edelman GM, Mauro VP. 2000. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci 97: 1536–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Chekulaeva M, Rajewsky N. 2018. Roles of IncRNAs and circRNAs in translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Sarnow P. 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. [DOI] [PubMed] [Google Scholar]
- Cobbold LC, Spriggs KA, Haines SJ, Dobbyn HC, Hayes C, de Moor CH, Lilley KS, Bushell M, Willis AE. 2008. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol 28: 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coldwell MJ, Mitchell SA, Stoneley M, MacFarlane M, Willis AE. 2000. Initiation of Apaf-1 translation by internal ribosome entry. Oncogene 19: 899–905. [DOI] [PubMed] [Google Scholar]
- Coldwell MJ, deSchoolmeester ML, Fraser GA, Pickering BM, Packham G, Willis AE. 2001. The p36 isoform of BAG-1 is translated by internal ribosome entry following heat shock. Oncogene 20: 4095–4100. [DOI] [PubMed] [Google Scholar]
- Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. 2014. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 511: 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. 2000. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell 5: 597–605. [DOI] [PubMed] [Google Scholar]
- Cornelis S, Tinton SA, Schepens B, Bruynooghe Y, Beyaert R. 2005. UNR translation can be driven by an IRES element that is negatively regulated by polypyrimidine tract binding protein. Nucleic Acids Res 33: 3095–3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Svitkin Y, Sonenberg N. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol Cell Biol 24: 6861–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deforges J, Locker N, Sargueil B. 2015. mRNAs that specifically interact with eukaryotic ribosomal subunits. Biochimie 114: 48–57. [DOI] [PubMed] [Google Scholar]
- Deniz N, Lenarcic EM, Landry DM, Thompson SR. 2009. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA 15: 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikstein R. 2012. Transcription and translation in a package deal: The TISU paradigm. Gene 491: 1–4. [DOI] [PubMed] [Google Scholar]
- Dresios J, Chappell SA, Zhou W, Mauro VP. 2006. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat Struct Mol Biol 13: 30–34. [DOI] [PubMed] [Google Scholar]
- Du Z, Alekhina OM, Vassilenko KS, Simon AE. 2017. Concerted action of two 3′ cap-independent translation enhancers increases the competitive strength of translated viral genomes. Nucleic Acids Res 45: 9558–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Dikstein R. 2008. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PLoS ONE 3: e3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. 2011. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 39: 7598–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez J, Yaman I, Mishra R, Merrick WC, Snider MD, Lamers WH, Hatzoglou M. 2001. Internal ribosome entry site-mediated translation of a mammalian mRNA is regulated by amino acid availability. J Biol Chem 276: 12285–12291. [DOI] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JT, Shin WK, Caudill MA, Stover PJ. 2009. A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. J Biol Chem 284: 31097–31108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Doudna JA. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol 5: 29–38. [DOI] [PubMed] [Google Scholar]
- Fuchs G, Petrov AN, Marceau CD, Popov LM, Chen J, O’Leary SE, Wang R, Carette JE, Sarnow P, Puglisi JD. 2015. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc Natl Acad Sci 112: 319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Kano F, Murata M. 2008. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA 14: 425–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futterer J, Kiss-Laszlo Z, Hohn T. 1993. Nonlinear ribosome migration on cauliflower mosaic virus 35S RNA. Cell 73: 789–802. [DOI] [PubMed] [Google Scholar]
- Gaccioli F, Huang CC, Wang C, Bevilacqua E, Franchi-Gazzola R, Gazzola GC, Bussolati O, Snider MD, Hatzoglou M. 2006. Amino acid starvation induces the SNAT2 neutral amino acid transporter by a mechanism that involves eukaryotic initiation factor 2α phosphorylation and cap-independent translation. J Biol Chem 281: 17929–17940. [DOI] [PubMed] [Google Scholar]
- Gazo BM, Murphy P, Gatchel JR, Browning KS. 2004. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem 279: 13584–13592. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Almela E, Williams H, Sanz MA, Carrasco L. 2018. The initiation factors eIF2, eIF2A, eIF2D, eIF4A, and eIF4G are not involved in translation driven by hepatitis C virus IRES in human cells. Front Microbiol 9: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber TE, Baird SD, Kao PN, Mathews MB, Holcik M. 2010. NF45 functions as an IRES trans-acting factor that is required for translation of cIAP1 during the unfolded protein response. Cell Death Differ 17: 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin TJ, Gygi SP, Ideker T, Rist B, Eng J, Hood L, Aebersold R. 2002. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1: 323–333. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol 19: 1720–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Dikstein R. 2015. Cap-dependent, scanning-free translation initiation mechanisms. Biochim Biophys Acta 1849: 1313–1318. [DOI] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Martin F, Ulitsky I, Emmanuel R, Tamarkin-Ben-Harush A, Vardy A, Dikstein R. 2017. Efficient and accurate translation initiation directed by TISU involves RPS3 and RPS10e binding and differential eukaryotic initiation factor 1A regulation. Mol Cell Biol 37: e00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. 2013. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol 33: 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holcik M. 2004. Targeting translation for treatment of cancer—A novel role for IRES? Curr Cancer Drug Targets 4: 299–311. [DOI] [PubMed] [Google Scholar]
- Holcik M, Korneluk RG. 2000. Functional characterization of the X-linked inhibitor of apoptosis (XIAP) internal ribosome entry site element: Role of La autoantigen in XIAP translation. Mol Cell Biol 20: 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt SL, Hsuan JJ, Totty N, Jackson RJ. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev 13: 437–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideker T, Thorsson V, Ranish JA, Christmas R, Buhler J, Eng JK, Bumgarner R, Goodlett DR, Aebersold R, Hood L. 2001. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science 292: 929–934. [DOI] [PubMed] [Google Scholar]
- Jaafar ZA, Oguro A, Nakamura Y, Kieft JS. 2016. Translation initiation by the hepatitis C virus IRES requires eIF1A and ribosomal complex remodeling. eLife 5: e21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack K, Bellodi C, Landry DM, Niederer RO, Meskauskas A, Musalgaonkar S, Kopmar N, Krasnykh O, Dean AM, Thompson SR, et al. 2011. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan N, Wimmer E, Mueller S. 2013. Polypyrimidine tract binding protein-1 (PTB1) is a determinant of the tissue and host tropism of a human rhinovirus/poliovirus chimera PV1(RIPO). PLoS ONE 8: e60791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, Sarnow P. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci 100: 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CJ, Jan E. 2010. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA 16: 1182–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang CJ, Lo MC, Jan E. 2009. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J Mol Biol 387: 42–58. [DOI] [PubMed] [Google Scholar]
- Jimenez J, Jang GM, Semler BL, Waterman ML. 2005. An internal ribosome entry site mediates translation of lymphoid enhancer factor-1. RNA 11: 1385–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Sarnow P. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4: 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannes G, Carter MS, Eisen MB, Brown PO, Sarnow P. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci 96: 13118–13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski A, Poyry TA, Skene PJ, Jackson RJ. 2010. Mechanism of initiation site selection promoted by the human rhinovirus 2 internal ribosome entry site. J Virol 84: 6578–6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YY, Back SH, Rho J, Lee SH, Jang SK. 2001. La autoantigen enhances translation of BiP mRNA. Nucleic Acids Res 29: 5009–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DC, Edelman GM, Mauro VP. 2013. Physical evidence supporting a ribosomal shunting mechanism of translation initiation for BACE1 mRNA. Translation (Austin) 1: e24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CS, Brilot AF, Grigorieff N, Korostelev AA. 2014. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA-like structural element in the ribosomal decoding center. Proc Natl Acad Sci 111: 9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung YA, Hung CT, Chien KY, Shih SR. 2017. Control of the negative IRES trans-acting factor KHSRP by ubiquitination. Nucleic Acids Res 45: 271–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacerda R, Menezes J, Romao L. 2017. More than just scanning: The importance of cap-independent mRNA translation initiation for cellular stress response and cancer. Cell Mol Life Sci 74: 1659–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre P, Kolakofsky D, Curran J. 1998. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol 18: 5021–5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Burdeinick-Kerr R, Whelan SP. 2013. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci 110: 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quesne JP, Stoneley M, Fraser GA, Willis AE. 2001. Derivation of a structural model for the c-myc IRES. J Mol Biol 310: 111–126. [DOI] [PubMed] [Google Scholar]
- Lin JY, Li ML, Shih SR. 2009. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res 37: 47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JY, Li ML, Brewer G. 2014. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS ONE 9: e103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Hellen CU, Pestova TV. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol Cell Biol 20: 6019–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marash L, Liberman N, Henis-Korenblit S, Sivan G, Reem E, Elroy-Stein O, Kimchi A. 2008. DAP5 promotes cap-independent translation of Bcl-2 and CDK1 to facilitate cell survival during mitosis. Mol Cell 30: 447–459. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, Pineiro D, Fernandez N. 2012. Alternative mechanisms to initiate translation in eukaryotic mRNAs. Comp Funct Genomics 2012: 391546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Mauro VP. 2014. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc Natl Acad Sci 111: 15385–15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerovitch K, Svitkin YV, Lee HS, Lejbkowicz F, Kenan DJ, Chan EK, Agol VI, Keene JD, Sonenberg N. 1993. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol 67: 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill MK, Gromeier M. 2006. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J Virol 80: 6936–6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulewaeter F, Van Montagu M, Cornelissen M. 1998. Features of the autonomous function of the translational enhancer domain of satellite tobacco necrosis virus. RNA 4: 1347–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Miller WA, Truniger V, Aranda MA. 2017. Non-canonical translation in plant RNA viruses. Front Plant Sci 8: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SA, Spriggs KA, Coldwell MJ, Jackson RJ, Willis AE. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol Cell 11: 757–771. [DOI] [PubMed] [Google Scholar]
- Morley SJ, Coldwell MJ. 2008. A cunning stunt: An alternative mechanism of eukaryotic translation initiation. Sci Signal 1: pe32. [DOI] [PubMed] [Google Scholar]
- Morris MJ, Negishi Y, Pazsint C, Schonhoft JD, Basu S. 2010. An RNA G-quadruplex is essential for cap-independent translation initiation in human VEGF IRES. J Am Chem Soc 132: 17831–17839. [DOI] [PubMed] [Google Scholar]
- Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol Cell 57: 422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernandez IS. 2016. Structural characterization of ribosome recruitment and translocation by type IV IRES. eLife 5: e13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima N, Uchiumi T. 2009. Functional analysis of structural motifs in dicistroviruses. Virus Res 139: 137–147. [DOI] [PubMed] [Google Scholar]
- Nicholson BL, Wu B, Chevtchenko I, White KA. 2010. Tombusvirus recruitment of host translational machinery via the 3′ UTR. RNA 16: 1402–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panopoulos P, Mauro VP. 2008. Antisense masking reveals contributions of mRNA-rRNA base pairing to translation of Gtx and FGF2 mRNAs. J Biol Chem 283: 33087–33093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50. [DOI] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN. 1996a. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Hellen CU. 1996b. Functional dissection of eukaryotic initiation factor 4F: The 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol Cell Biol 16: 6870–6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev 12: 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfingsten JS, Costantino DA, Kieft JS. 2006. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314: 1450–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickering BM, Mitchell SA, Spriggs KA, Stoneley M, Willis AE. 2004. Bag-1 internal ribosome entry segment activity is promoted by structural changes mediated by poly(rC) binding protein 1 and recruitment of polypyrimidine tract binding protein 1. Mol Cell Biol 24: 5595–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol 78: 4487–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozner A, Goldenberg D, Negreanu V, Le SY, Elroy-Stein O, Levanon D, Groner Y. 2000. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol Cell Biol 20: 2297–2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin X, Sarnow P. 2004. Preferential translation of internal ribosome entry site-containing mRNAs during the mitotic cycle in mammalian cells. J Biol Chem 279: 13721–13728. [DOI] [PubMed] [Google Scholar]
- Racine T, Duncan R. 2010. Facilitated leaky scanning and atypical ribosome shunting direct downstream translation initiation on the tricistronic S1 mRNA of avian reovirus. Nucleic Acids Res 38: 7260–7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray PS, Das S. 2002. La autoantigen is required for the internal ribosome entry site-mediated translation of Coxsackievirus B3 RNA. Nucleic Acids Res 30: 4500–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley A, Jordan LE, Holcik M. 2010. Distinct 5′ UTRs regulate XIAP expression under normal growth conditions and during cellular stress. Nucleic Acids Res 38: 4665–4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr., Edelman GM, Mauro VP. 2004. Differential utilization of upstream AUGs in the β-secretase mRNA suggests that a shunting mechanism regulates translation. Proc Natl Acad Sci 101: 2794–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D. 2013. Translational control in cancer etiology. Cold Spring Harb Perspect Biol 5: a012336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabova LA, Hohn T. 2000. Ribosome shunting in the cauliflower mosaic virus 35S RNA leader is a special case of reinitiation of translation functioning in plant and animal systems. Genes Dev 14: 817–829. [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. 1999. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol 73: 1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci 97: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N, Saito H, Noda H. 1998. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology 244: 50–58. [DOI] [PubMed] [Google Scholar]
- Schepens B, Tinton SA, Bruynooghe Y, Beyaert R, Cornelis S. 2005. The polypyrimidine tract-binding protein stimulates HIF-1α IRES-mediated translation during hypoxia. Nucleic Acids Res 33: 6884–6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Agol VI, Andino R, Bayard F, Cavener DR, Chappell SA, Chen JJ, Darlix JL, Dasgupta A, Donze O, et al. 2001. New ways of initiating translation in eukaryotes. Mol Cell Biol 21: 8238–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. 2006. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13: 1092–1096. [DOI] [PubMed] [Google Scholar]
- Sella O, Gerlitz G, Le SY, Elroy-Stein O. 1999. Differentiation-induced internal translation of c-sis mRNA: Analysis of the cis elements and their differentiation-linked binding to the hnRNP C protein. Mol Cell Biol 19: 5429–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semler BL, Waterman ML. 2008. IRES-mediated pathways to polysomes: Nuclear versus cytoplasmic routes. Trends Microbiol 16: 1–5. [DOI] [PubMed] [Google Scholar]
- Sharifulin DE, Bartuli YS, Meschaninova MI, Ven’yaminova AG, Graifer DM, Karpova GG. 2016. Exploring accessibility of structural elements of the mammalian 40S ribosomal mRNA entry channel at various steps of translation initiation. Biochim Biophys Acta 1864: 1328–1338. [DOI] [PubMed] [Google Scholar]
- Sharma SD, Kraft JJ, Miller WA, Goss DJ. 2015. Recruitment of the 40S ribosome subunit to the 3′-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J Biol Chem 290: 11268–11281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrill KW, Lloyd RE. 2008. Translation of cIAP2 mRNA is mediated exclusively by a stress-modulated ribosome shunt. Mol Cell Biol 28: 2011–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. 2009. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol 11: 903–908. [DOI] [PubMed] [Google Scholar]
- Simon AE, Miller WA. 2013. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 67: 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. 2015. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21: 479–492. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291: 1959–1962. [DOI] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475. [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Stoneley M, Bushell M, Willis AE. 2008. Re-programming of translation following cell stress allows IRES-mediated translation to predominate. Biol Cell 100: 27–38. [DOI] [PubMed] [Google Scholar]
- Spriggs KA, Cobbold LC, Ridley SH, Coldwell M, Bottley A, Bushell M, Willis AE, Siddle K. 2009. The human insulin receptor mRNA contains a functional internal ribosome entry segment. Nucleic Acids Res 37: 5881–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. 2010. Translational regulation of gene expression during conditions of cell stress. Mol Cell 40: 228–237. [DOI] [PubMed] [Google Scholar]
- *.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2018. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. 2000. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res 28: 687–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE. 2008. The 3′ proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA 14: 2379–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. 2014. The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J 33: 76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat Struct Mol Biol 15: 836–841. [DOI] [PubMed] [Google Scholar]
- Thomas JD, Johannes GJ. 2007. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA 13: 1116–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR. 2012. So you want to know if your message has an IRES? Wiley Interdiscip Rev RNA 3: 697–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. 2008. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 14: 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger V, Nieto C, Gonzalez-Ibeas D, Aranda M. 2008. Mechanism of plant eIF4E-mediated resistance against a Carmovirus (Tombusviridae): Cap-independent translation of a viral RNA controlled in cis by an (a)virulence determinant. Plant J 56: 716–727. [DOI] [PubMed] [Google Scholar]
- Vaklavas C, Grizzle WE, Choi H, Meng Z, Zinn KR, Shrestha K, Blume SW. 2016. IRES inhibition induces terminal differentiation and synchronized death in triple-negative breast cancer and glioblastoma cells. Tumour Biol 37: 13247–13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters B, Thompson SR. 2016. Cap-independent translational control of carcinogenesis. Front Oncol 6: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Parisien M, Scheets K, Miller WA. 2011. The cap-binding translation initiation factor, eIF4E, binds a pseudoknot in a viral cap-independent translation element. Structure 19: 868–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. 2016. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 351: aad4939. [DOI] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. [DOI] [PubMed] [Google Scholar]
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. 2015. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Sarnow P. 1997. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: Evidence for specific RNA-protein interactions. Nucleic Acids Res 25: 2800–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DQ, Halaby MJ, Zhang Y. 2006. The identification of an internal ribosomal entry site in the 5′-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene 25: 4613–4619. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. 2017. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 27: 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JY, Dong ZZ, Liu RY, Chen J, Liu ZQ, Zhang JT. 2013. Translational regulation of RPA2 via internal ribosomal entry site and by eIF3a. Carcinogenesis 34: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Kong YY, Yoshida R, Elia AJ, Hakem A, Hakem R, Penninger JM, Mak TW. 1998. Apaf1 is required for mitochondrial pathways of apoptosis and brain development. Cell 94: 739–750. [DOI] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ. 1996. Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev 10: 1557–1567. [DOI] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14: 414–421. [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]