Abstract
Heat shock factors (HSFs) are the main transcriptional regulators of the heat shock response and indispensable for maintaining cellular proteostasis. HSFs mediate their protective functions through diverse genetic programs, which are composed of genes encoding molecular chaperones and other genes crucial for cell survival. The mechanisms that are used to tailor HSF-driven proteostasis networks are not yet completely understood, but they likely comprise from distinct combinations of both genetic and proteomic determinants. In this review, we highlight the versatile HSF-mediated cellular functions that extend from cellular stress responses to various physiological and pathological processes, and we underline the key advancements that have been achieved in the field of HSF research during the last decade.
Since its initial discovery by the Italian geneticist Ferruccio Ritossa (Ritossa 1962), the evolutionarily conserved heat shock response (HSR), has been the focus of intense investigation. The HSR exists in all organisms and provides an essential survival mechanism against intracellular and extracellular challenges. HSR is characterized by extremely rapid activation of transcription, leading to massive increase in the synthesis of molecular chaperones, such as the heat shock proteins (HSPs). These proteins enable the maintenance of cellular proteostasis, and thereby ensure the survival of the cell. The rapid transcriptional induction of genes encoding HSPs is regulated by a family of transcription factors called the heat shock factors (HSFs). On activation, HSFs oligomerize, localize to the nucleus, and activate the transcription of their target genes. Albeit originally identified as key factors for cell protection during acute stress, HSFs are now known to respond to a variety of extrinsic and intrinsic signals and the repertoire of HSF target genes has expanded significantly. Consequently, the current view presents HSFs as versatile regulators of many cellular and organismal processes, ranging from stress protection to gametogenesis and neurogenesis. Moreover, misregulated HSF function is tightly linked to diseases originating from disrupted proteostasis, such as neurodegenerative disorders, metabolic diseases, and different forms of cancers, highlighting the importance of understanding the precise context-dependent regulation of HSFs’ activity.
HEAT SHOCK TRANSCRIPTION FACTOR FAMILY
The HSR is an evolutionarily well-conserved stress protective pathway that has been identified in every living organism, all the way from unicellular bacteria and yeasts to multicellular plants and animals. A key feature of the HSR is the rapid transcriptional activation of genes, including molecular chaperones, which is achieved through specific stimulus-dependent transcription factors. In Escherichia coli, the stress-inducible transcription factor is called σ32, which on exposure to heat activates transcription of DnaK, a prokaryotic homologue of HSP70 (Grossman et al. 1984). In eukaryotes, HSR is regulated by a family of transcription factors, the heat shock transcription factors, HSFs. In the yeast Saccharomyces cerevisiae, the nematode Caenorhabditis elegans, and the fruit fly Drosophila melanogaster, only a single HSF is expressed (ScHSF, CeHSF, and DmHSF, respectively), whereas multiple HSFs exist in fish, birds, mammals, and plants (Fig. 1).
Figure 1.
Domain organization of heat shock factor (HSF) family members. In mammals, six HSFs are expressed (HSF1–4, HSFX, and HSFY), which differ in their tissue expression patterns and biological functions. HSF3 is a pseudogene in humans, whereas functional HSF3 protein has been found in mouse (Mus musculus). Four HSFs have been identified in chicken (Gallus gallus). In contrast to vertebrates, a single HSF is expressed in fruit fly (Drosophila melanogaster), nematode (Caenorhabditis elegans), and yeast (Saccharomyces cerevisiae). The evolutionarily conserved DNA-binding domain (DBD) is present in all HSFs. Hydrophobic heptad repeat domain HR-A/B mediates oligomerization. Carboxy-terminal heptad repeat (HR-C) provides an intramolecular interaction site that can repress HSF oligomerization by interacting with HR-A/B. Transactivation domain (TAD) is required to enhance the transcriptional activation of certain HSFs. The mammalian HSF1 contains also a regulatory domain (RD) that is subjected to many posttranslational modifications (see Fig. 2). The numbers indicate amino acids.
The vertebrate HSF family consists of six members (HSF1–4, HSFX, and HSFY), of which HSF1 and HSF2 are most extensively studied. HSF1 is the functional counterpart of ScHSF, CeHSF, and DmHSF, and because of its fundamental role in chaperone gene expression during acute heat stress, it is generally accepted as the master regulator of HSR (McMillan et al. 1998). HSF1 is ubiquitously expressed in mammalian tissues (Fiorenza et al. 1995) and it is essential for protection against heat-induced protein damage and for acquisition of thermotolerance (McMillan et al. 1998). HSF2 also binds DNA stress inducibly, but its role in the acute stress response is diminutive (Östling et al. 2007). Instead, HSF2 appears to be more important during febrile-range thermal stress (Shinkawa et al. 2011) and in various differentiation and developmental processes (Sistonen et al. 1992; Mezger et al. 1994; Rallu et al. 1997; Kallio et al. 2002; Wang et al. 2003; Chang et al. 2006).
The third member of the vertebrate HSF family, HSF3, is expressed in both chicken and mouse cells, cHSF3 and mHSF3, respectively (Nakai and Morimoto 1993; Fujimoto et al. 2010); but in humans, HSF3 is a pseudogene. Interestingly, the mammalian and avian HSF3s seem to be functionally distinct, as cHSF3 is the main factor regulating HSR and thermotolerance (Nakai and Morimoto 1993; Tanabe et al. 1998), whereas mHSF3 participates in the regulation of nonclassical heat shock genes (Fujimoto et al. 2010). However, overexpression of mHSF3 in Hsf1−/− MEFs (mouse embryonic fibroblasts) does partially restore the cellular resistance to heat shock, suggesting that mHSF3 might have a unique, yet undefined, role during acute stress (Fujimoto et al. 2010).
HSF4 does not show stress-inducible DNA-binding activity. HSF4 is highly expressed in both embryonic and adult eye tissue, where it regulates fiber cell differentiation and lens development (Fujimoto et al. 2004; Min et al. 2004). Mutations in the HSF4 DNA-binding domain (DBD) lead to severe cataracts, linking HSF4 directly to human diseases (Bu et al. 2002; Anand et al. 2018). The biological functions of HSF5, as well as the sex-chromosomal HSFX and HSFY are poorly understood, but they are suggested to regulate gametogenesis (Tessari et al. 2004; Widlak and Vydra 2017).
Structural Features of HSFs
All HSF family members are characterized by functional domains that share both common and unique features. The most conserved domain is the amino-terminal, winged helix–turn–helix DBD (Fig. 2), which is present in all eukaryotic HSFs (Vuister et al. 1994). The core structure of DBD consists of a four-stranded antiparallel β-sheet and three α-helices, of which the third helix inserts into the major groove of DNA (Harrison et al. 1994). The wing structure is located between the third and fourth β-sheet and it differs from equivalent structures in other winged helix–turn–helix transcription factors, as it is not in contact with DNA (Littlefield and Nelson 1999). Recently, HSF1 and HSF2 DBDs were cocrystallized with DNA, uncovering a novel topology of DNA-bound HSF oligomers (Jaeger et al. 2016; Neudegger et al. 2016). In contrast to the old view, the new model positions DBD and the remaining parts of HSF on the opposite sites of the DNA strand in a so-called DNA-embracing structure (Fig. 2). X-ray crystallography analyses of the HSF2 DBD wing domain further revealed that HSF1 and HSF2 wing domains differ both structurally and functionally, providing a platform for HSF-specific regulation on diverse cellular states (Jaeger et al. 2016).
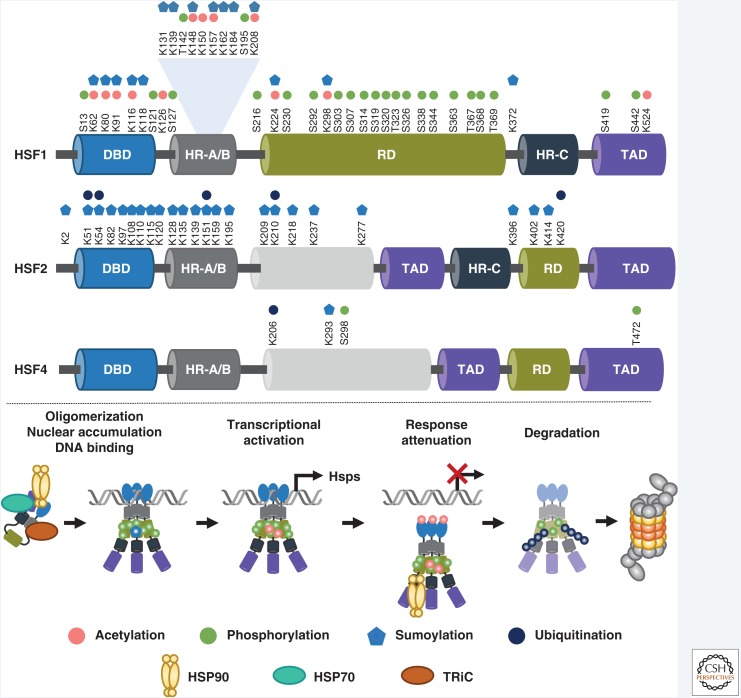
Figure 2.
Heat shock factors (HSFs) undergo a selection of posttranslational modifications (PTMs) during activation and attenuation. HSF1 is phosphorylated on multiple serine (S) and threonine (T) residues, mainly located in the regulatory domain (RD). The DNA-binding domain (DBD) and the oligomerization domain (HR-A/B) harbor lysine (K) residues that are subjected to both acetylation and sumoylation. HSF1 is also ubiquitinated, but the exact ubiquitination target lysines are not known. To date, no phosphorylatable residues have been identified in HSF2. Instead, HSF2 is sumoylated at lysine residues in DBD and HR-A/B regions. Ubiquitination of five lysine residues on HSF2 have been reported. HSF4 is also subjected to phosphorylation, sumoylation, and ubiquitination. In the absence of stimulus, HSF1 is located in the cytosol in a complex that contains chaperones (HSP90, HSP70, TRiC). Steps of activation and attenuation are accompanied by a selection of PTMs. On attenuation, HSF1 is degraded by the ubiquitin-proteasome system. HR-C, carboxy-terminal heptad repeat; TAD, transactivation domain.
Unlike many transcription factors that act as dimers, HSFs form trimers on activation. This oligomerization is mediated by hydrophobic leucine-zipper-like heptad repeats (HR-A/B) that are located directly adjacent to DBD (Fig. 1; Sorger and Nelson 1989). The HR-A/B domains are also highly conserved between the HSF family members. Yet another heptad repeat (carboxy-terminal heptad repeat [HR-C]) resides in the carboxy terminus and has been found only in specific HSFs (Fig. 1). This domain confers an intramolecular interaction site and represses spontaneous oligomerization when interacting with the HR-A/B. Temperature increase results in HR-C unfolding, allowing HR-A/B stabilization and subsequent oligomerization of HSFs (Hentze et al. 2016). Neither mammalian HSF4 nor ScHSF contains HR-C, which results in constitutive DNA-binding competence of these factors (Sorger et al. 1987; Nakai et al. 1997) and likely explains the essential role of ScHSF and HSF4 in cell viability (Sorger and Pelham 1988) and normal tissue development (Fujimoto et al. 2004), respectively.
Formation of DNA-bound HSF trimers does not necessarily induce their target gene expression (Hensold et al. 1990; Jurivich et al. 1992). For that they require the carboxy-terminal transactivation domain (TAD), an interaction site for cofactors and chromatin remodelers (Sullivan et al. 2001; Boellmann et al. 2004). The ScHSF1 is unique among the HSFs in containing both amino- and carboxy-terminal TADs (Nieto-Sotelo et al. 1990). In normal conditions, the TAD is negatively regulated by the centrally located regulatory domain (RD) (Shi et al. 1995; Zuo et al. 1995), which provides the factor with heat-sensing capability (Newton et al. 1996; Hentze et al. 2016). TAD and RD are also present in human HSF2 and HSF4, whereas analogous domains are yet to be identified in murine HSF2 and HSF4.
POSTTRANSLATIONAL REGULATION OF HSFs
Similarly to many other stimulus-dependent transcription factors, all HSFs show transient activation profiles that are achieved through specific posttranslational regulators. Such regulators include both protein-interacting partners and posttranslational modifications (PTMs). The posttranslational regulators have been shown to contribute to literally every step of the activation pathway (i.e., oligomerization, nuclear accumulation, DNA-binding, transcriptional activation, response attenuation, and degradation of HSFs) (Fig. 2).
In the absence of stress, HSF1 interacts with molecular chaperones, including HSP70, HSP90, and the chaperonin TRiC, which maintain repression and localization to the cytosol (Fig. 2; Shi et al. 1998; Guo et al. 2001; Neef et al. 2010; Zheng et al. 2016; for a review, see Gomez-Pastor et al. 2018). In response to stimuli leading to disrupted proteostasis, the chaperones are titrated away from HSF1, which enables oligomerization, nuclear accumulation, and transcriptional activation. Recently, HSP90 was shown to participate also in the removal of HSF1 trimers from DNA (Fig. 2; Kijima et al. 2018), suggesting that chaperones can modulate HSF activity at various phases of the response. In addition to chaperones, HSFs are known to interact with an extensive repertoire of other proteins that among other things increase the access of HSF1 to nucleosomal DNA (Fujimoto et al. 2012), facilitate its DNA-binding activity (Fujimoto et al. 2018), assist in the establishment of the active chromatin state (Takii et al. 2015), and potentiate the transactivation capacity of HSF1 (Tan et al. 2015).
HSFs are subjected to many PTMs, including phosphorylation, acetylation, ubiquitination, and sumoylation. Altogether, 23 phosphorylatable residues have been identified on HSF1, which mainly reside within the RD (Fig. 2; Guettouche et al. 2005). Hyperphosphorylation of HSF1 is strongly induced on heat shock, coinciding with its DNA-binding and transcriptional activities (Fig. 2; Cotto et al. 1996). However, a complete abolishment of phosphorylation does not cause transcriptional incompetence and, thus, phosphorylation is currently considered as a fine-tuning mechanism for HSF1 activity (Budzyński et al. 2015; Zheng et al. 2016). Intriguingly, phosphorylation of HSF1 was recently shown to mediate cell-to-cell variation and phenotypic plasticity in heat-treated S. cerevisiae (Zheng et al. 2018), emphasizing the role of phosphorylation as a modification adjusting HSF1-mediated transcriptional response.
Acetylation has been shown to have a variable role in regulating HSF1 activity (Westerheide et al. 2009; Raychaudhuri et al. 2014). In nonstressed cells, acetylation of K208 and K298 mediates HSF1 stabilization, although the mechanistic details of this stabilization are yet to be determined (Raychaudhuri et al. 2014). During acute stress, HSF1 is acetylated on DBD (K80, K118), which contributes to its release from the DNA and subsequent attenuation of the HSR (Fig. 2; Westerheide et al. 2009; Raychaudhuri et al. 2014; Zelin and Freeman 2015).
The expression levels and activity of HSFs are regulated also by the ubiquitin-proteasome system. Two ubiquitin ligases, NEDD4 and FBXW7, have been shown to directly interact with HSF1 (Kourtis et al. 2015; Kim et al. 2016; Gomez-Pastor et al. 2017). Interaction between HSF1 and FBXW7 depends on the phosphorylation status of S303 and S307 and leads to HSF1 degradation by the proteasome. Interestingly, mutation of S303/307 was shown to prolong the HSR, proposing for the first time that HSF1 degradation is an important step in the attenuation phase of the HSR (Kourtis et al. 2015). Given that the transient HSF activity is often presented as a cyclic process, where DNA-released HSF trimers revert back to monomers and return to the cytosol, these results change the current view by showing that HSF1 is ubiquitinated and degraded during stress recovery (Fig. 2). Moreover, as abnormal regulation of HSF1 stability was associated with disease propagation in both melanoma and Huntington's disease mouse models and patient samples (Kourtis et al. 2015; Gomez-Pastor et al. 2017), these observations underscore the biological relevance of HSF1 ubiquitination. In addition to HSF1, the levels of HSF2 and HSF4 are regulated by ubiquitination. Ubiquitination of HSF2 by E3-ligase APC/C occurs during acute stress and results in proteasomal degradation of the factor (Ahlskog et al. 2010). The consequence of HSF2 ubiquitination is not fully understood, but it seems that HSF2 degradation is required to direct target gene expression on specific cellular states (Ahlskog et al. 2010; Elsing et al. 2014). In unbiased mass spectrometric (MS) screens, five ubiquitination target lysines on HSF2 have been identified (Kim et al. 2007; Wagner et al. 2011), whereas ubiquitination of HSF4 is mainly targeted to K206 and appears to inhibit its transcriptional activity (Fig. 2; Liao et al. 2015).
PTM by sumoylation has been reported for all human HSFs. HSF1 is primarily sumoylated on K298, although a minor sumoylation site on K126 has also been experimentally validated (Fig. 2; Hong et al. 2001; Hietakangas et al. 2003). HSF1 sumoylation is stress-inducible, but it is dispensable for the induction of HSR and rather functions as a negative regulator of HSF1 transactivation capacity (Hietakangas et al. 2003, 2006). K298 resides within an extended sumoylation consensus motif, called phosphorylation-dependent sumoylation motif (PDSM), where phosphorylation of the nearby S303 is a prerequisite for the conjugation of SUMO (Hietakangas et al. 2006). A similar motif has been identified in human HSF4, which undergoes stress-inducible phosphorylation of S298 and sumoylation of K293 (Fig. 2; Hietakangas et al. 2006). Sumoylation of HSF4 also functions as a repressive modification, inhibiting its transcriptional activity (Hietakangas et al. 2006). In HSF2, the main sumoylation target K82 resides within the DBD and inhibits HSF2 DNA-binding activity (Goodson et al. 2001; Anckar et al. 2006; Tateishi et al. 2009; Feng et al. 2016). Additional sumoylation sites on HSF1 and HSF2 were recently identified in an unbiased MS-screen, but the functional importance of these novel sites is unknown (Hendriks et al. 2017). However, considering their accumulation on DBD and HR-A/B (Fig. 2), it is possible that sumoylation functions as a more versatile regulator of HSF oligomerization and DNA-binding activity than previously assumed.
DIVERSITY OF HSF TRANSCRIPTIONAL PROGRAMS: HOW IS THE SPECIFICITY DEFINED?
Despite the central role of HSFs in coordinating stress-induced transcription, it has become well acknowledged that HSFs direct also additional genetic programs that differ extensively from the classical HSR (for detailed description of transcriptional stress responses, see Vihervaara et al. 2018). Multiple genome-wide sequencing studies have identified a great number of novel target sites that display specific HSF-binding profiles depending on cell cycle phase, developmental stage, cell type, stress exposure, and the type of organism (Hahn et al. 2004; Birch-Machin et al. 2005; Åkerfelt et al. 2008, 2010; Vihervaara et al. 2013, 2017; Li et al. 2016; Mahat et al. 2016). The occupancy of HSF1 and HSF2 varies also on malignant transformation (Mendillo et al. 2012), aggregate accumulation (Riva et al. 2012), and fetal alcohol exposure (El Fatimy et al. 2014), altogether indicating that the HSF-driven transcriptional reprogramming is a highly dynamic process that can be regulated by multiple upstream signaling pathways. Although it is not yet fully established how the program diversity is determined in a given state, some common nominators, including promoter architecture and protein-interacting partners, are emerging.
Promoter Architecture
HSFs are sequence-specific transcription factors that act through a conserved DNA element, heat shock element (HSE), consisting of inverted repeats of a pentameric nGAAn consensus sequence (Amin et al. 1988). Nevertheless, HSEs across the genomes display extensive variation in their primary sequence, length, and orientation (Mendillo et al. 2012; Riva et al. 2012; Vihervaara et al. 2013; Mahat et al. 2016). Interestingly, HSFs are able to distinguish specific properties of the HSEs, such as the length or the orientation of the repeats (Bonner et al. 1994; Jaeger et al. 2014), suggesting that the DNA sequence itself can function as a determinant of HSF target selection. Accordingly, heat-responsive and developmentally important HSF target promoters were recently shown to differ significantly in their HSE architecture (Li et al. 2016), which confirms that the specific composition of HSEs impact the HSF-driven transcriptional response.
In addition to HSEs, other promoter cis elements have also been identified as significant determinants of HSF target site selection. In the developing C. elegans larvae, CeHSF binds specifically to promoters that contain GC-rich motives adjacent to the HSE sequence (Li et al. 2016). The GC motives serve as binding sites for transcriptional cofactors, including E2F, which is indispensable for the developmental activation of CeHSF (Li et al. 2016). In human K562 erythroleukemia cells, specific-chromatin-binding factors (e.g., SP2 and GATA factors) preoccupy HSF1 target sites that differ extensively from the classical heat-inducible genes (Vihervaara et al. 2017). Excitingly, a co-occupancy of oncogenic NOTCH1 and HSF1 was recently observed on the promoters of many HSF1 target genes in human T-cell acute lymphoblastic leukemia cells (Kourtis et al. 2018). Taken together, there is increasing evidence that the chromatin architecture and cross talk with other transcriptional regulators determine the transcriptional outcome of HSF DNA-binding activity.
Functional Interplay between HSFs
Because of the significant homology of the DBDs, all HSFs recognize similar cis elements and thereby can compete or cooperate for the regulation of a given gene. For example, in mouse lens tissue and olfactory epithelia, HSF1 and HSF4 occupy the same target gene promoters but have opposite effects on their expression (Fujimoto et al. 2004; Takaki et al. 2006), whereas in mitotic and heat-shocked cells, HSF2 interferes with HSF1 binding to the HSP70 promoter, resulting in reduced HSP70 expression (Elsing et al. 2014). HSF1 and HSF2 bind to the same target gene promoters also in mouse testis (Korfanty et al. 2014), where their functional cooperation seems to be required for normal sperm production (Wang et al. 2004). HSF1 and HSF2 can also form heterotrimers via their conserved HR-A/B domains, and experimental manipulation of HSF1 and HSF2 stoichiometry impacts the transcriptional properties of the heterotrimers (Sandqvist et al. 2009; El Fatimy et al. 2014; Jaeger et al. 2016). These observations suggest that heterotrimerization can function as a determinant of context-dependent HSF activation. To date, no mechanisms underlying the heterotrimerization of HSF1 and HSF2 have been established and it is unclear whether similar cross talk can occur among other HSF family members. Thus, it remains to be determined whether heterotrimerization is a common mechanism of HSF-mediated transcription and target gene selection.
HSFs AS ENVIRONMENTAL SENSORS: FROM HSR TO PHYSIOLOGICAL PROGRAMS
Developmental Role of HSFs
Besides regulating the HSR, HSFs are vital for other physiological processes, particularly those associated with organismal development (Fig. 3). The single ScHSF is crucial for yeast cell viability and cells lacking ScHSF are unable to produce viable spores (Sorger and Pelham 1988; Gallo et al. 1993). Although ScHSF binds multiple gene loci in nonstressed conditions (Hahn et al. 2004), the lethal knockout phenotype can be rescued with overexpression of HSP70 and HSP90, implying that the main role of ScHSF is to maintain proteostasis (Solís et al. 2016). In fruit fly, DmHSF is not required for adult viability, but it is essential for larval development and oogenesis (Jedlicka et al. 1997). These properties are not, however, caused by impaired HSP expression, indicating that HSFs in multicellular organisms regulate additional genetic programs essential for survival. Supporting this view, CeHSF was recently shown to coordinate a genetic program distinct from the HSR during nematode larval development (Li et al. 2016).
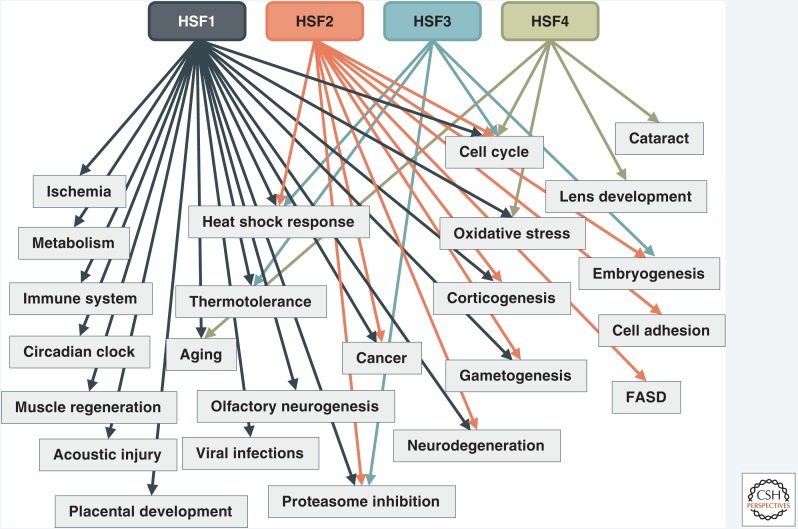
Figure 3.
Vertebrate heat shock factors (HSFs) are linked to a diverse array of biological processes. Although originally identified as the main factors regulating cellular responses to acute heat stress, HSFs are currently linked to an extensive array of different physiological and pathological processes. Most of the processes have been studied in the context of HSF1, and much less is currently known about the biological relevance of HSF2, HSF3, or HSF4. References for the processes that are not discussed in the main text include HSF1, ischemia (Higashi et al. 1995; Nishizawa et al. 1996); HSF1, muscle regeneration (Nishizawa et al. 2013); HSF1, acoustic injury (Sugahara et al. 2003); HSF1, immune system (Inouye et al. 2004); HSF1, viral infections (Filone et al. 2014); HSF1, circadian rhythm (Reinke et al. 2008); HSF1, oxidative stress (Ahn and Thiele 2003); HSF2, embryogenesis (Mezger et al. 1994); HSF3, cell cycle (Nakai and Ishikawa 2001); HSF3, embryogenesis (Nakai and Morimoto 1993); HSF4, oxidative stress (Liao et al. 2018); HSF4, cell cycle (Tu et al. 2006); HSF4, aging (Shi et al. 2008); and HSF1, HSF2, and HSF3, proteasomal inhibition (Kawazoe et al. 1998). The arrows depict the association between a distinct HSF family member and the process. FASD, fetal alcohol spectrum disorder.
In mammals, HSF1 is expressed in a variety of tissues, and although it is not directly required for the survival of the organism, Hsf1−/− mice show many significant developmental defects (McMillan et al. 1998; Xiao et al. 1999). One of the most visible phenotypes is the reduced body and organ size, which has been associated with impaired translation in HSF1-deficient cells (Xiao et al. 1999; Su et al. 2016). In addition, HSF1-null female mice suffer from multiple reproductive abnormalities, such as placental insufficiency and infertility (McMillan et al. 1998; Xiao et al. 1999; Christians et al. 2000). These deficiencies are caused by alterations in the basal HSP levels (Metchat et al. 2009) and decreased expression of distinct meiotic genes (Le Masson et al. 2011). Spermatogenesis is also regulated by HSF1, and HSF1-deficient mice produce less sperm and show disorganized layering of the seminiferous epithelium (Salmand et al. 2008; Åkerfelt et al. 2010). HSF1 is required for the developing brain, because HSF1-null mice display enlarged ventricles, neurodegeneration, demyelination, astrogliosis, and neuronal accumulation of ubiquitinated proteins (Fig. 3; Santos and Saraiva 2004; Homma et al. 2007).
Similarly to HSF1, lack of HSF2 does not compromise organismal viability but impairs corticogenesis and gametogenesis in both genders (Kallio et al. 2002; McMillan et al. 2002; Wang et al. 2003). During mouse embryogenesis, the expression pattern of HSF2 fluctuates (Mezger et al. 1994), and HSF2 is particularly abundant in the central nervous system (Rallu et al. 1997; Kallio et al. 2002; Wang et al. 2003). In adult Hsf2−/− mice, both lateral and third ventricles are profoundly dilated (Kallio et al. 2002; Wang et al. 2003), indicating that HSF2 mediates tissue homeostasis during ventricle formation. Interestingly, HSPs are equally expressed during embryogenesis of Hsf2−/− and wild-type mice, suggesting that HSPs are not the main HSF2 targets in the developing brain (Wang et al. 2003). Hsf2−/− mice also display neuronal mispositioning, which has been attributed to a reduced number of cells supporting neuronal migration (Chang et al. 2006). Importantly, HSF2 activity is misregulated in mice during fetal alcohol exposure, which results in neuronal migratory defects similar to those detected in children diagnosed with fetal alcohol spectrum disorder (FASD) (El Fatimy et al. 2014).
Among all studied tissue types, HSF2 is most prominently expressed in adult testes (Sarge et al. 1994; Fiorenza et al. 1995) and Hsf2−/− mice display multiple testicular defects, including reduced testis size and increased apoptosis of the spermatocytes (Kallio et al. 2002; Wang et al. 2003). HSF2-deficient female mice show increased embryonic lethality and reduced fertility caused by defective ovulation, meiotic problems, and diverse hormonal abnormalities (Kallio et al. 2002). During spermatogenesis, HSF2 occupies the Y-chromosomal multicopy genes, which have been associated with sperm head development (Åkerfelt et al. 2008). Interestingly, combined disruption of HSF1 and HSF2 exacerbates the Hsf2−/− phenotype and the double knockout mice are completely infertile (Wang et al. 2004). ChIP-seq analyses of HSF1 and HSF2-binding sites in mouse testes have revealed that these factors share the target binding sites during male gametogenesis (Korfanty et al. 2014), indicating that HSF1 and HSF2 cooperatively regulate gene expression during sperm development.
The biological role of HSF4 has been mainly shown in the context of lens development (Fujimoto et al. 2004), although it is expressed also in other mouse tissues (Jin et al. 2011). HSF4-deficient mice display abnormal lens epithelial cell proliferation and premature differentiation, which is at least partially caused by decreased expression of γ-crystallins and increased expression of fibroblast growth factors (FGFs), leading to accumulation of inclusion-like structures (Fujimoto et al. 2004). Intriguingly, altogether, 18 point mutations in the HSF4 gene have been linked to generation of severe cataracts in humans, highlighting the functional relevance of HSF4 in maintaining lens tissue homeostasis (Bu et al. 2002; Anand et al. 2018).
HSF1 in Metabolism
During the last few years, it has become evident that HSF1 acts as a critical sensor for cellular metabolic state and its activity is modulated by a selection of nutrient-sensing pathways. For example, a key transcriptional coactivator of metabolic genes, PGC-1α, directly interacts with HSF1 and represses its transactivation capacity (Minsky and Roeder 2015). Induction of gluconeogenesis in liver during fasting results in PGC-1α activation and subsequent inhibition of HSF1, which has been suggested to mediate some of the antiproliferative effects of nutrient deprivation (Minsky and Roeder 2015). Interestingly, PGC-1α can also promote the HSF1-mediated HSR (Xu et al. 2016), proposing that the impact of PGC-1α on HSF1 activity is context-dependent and sensitive to other regulatory inputs. HSF1 regulates energy expenditure also by binding and activating transcription of the PGC-1α gene, which promotes PGC-1α-dependent white fat browning and mitochondrial function (Ma et al. 2015). Pharmacological stimulation of HSF1 by celastrol, a potent inducer of HSR (Westerheide et al. 2004), enhances PGC-1α-mediated energy consumption and protects against diet-induced obesity and insulin resistance (Ma et al. 2015). These findings suggest that systemic activation of HSF1 might promote the metabolic health of an organism (for a detailed description of HSF1 and organismal metabolism, see Su and Dai 2016).
HSFs IN PATHOPHYSIOLOGICAL CONTEXTS: THE PARADOX OF PROTEOSTASIS
Cancer
Malignant cells are constantly exposed to proteotoxic damage induced by intrinsic and extrinsic factors as they characteristically harbor multiple genetic mutations and maintain growth in adverse environments. Because of their unstable proteome, abnormally high expression of HSPs has been observed in a wide range of human cancers and is currently considered as a general feature of neoplastic cells (Whitesell and Lindquist 2005). Consequently, the importance of HSF1 in cancer progression has been extensively studied and it is now well established that lack of HSF1 protects against malignancy irrespective of the oncogenic driver identity (Dai et al. 2007; Min et al. 2007; Kourtis et al. 2018). In addition to HSPs, HSF1 supports the malignant phenotype by maintaining the expression of multiple other genes, such as those associated with DNA repair, cellular energy metabolism, and translation (Mendillo et al. 2012). This is referred to as the HSF1 cancer program and it has been identified from various human cancer cell lines and patient samples (Mendillo et al. 2012). HSF1 is also required for signaling between cancer cells and tumor stroma (Scherz-Shouval et al. 2014), indicating that the HSF1-driven cell survival pathway is comprehensively tailored to allow malignant progression.
What then causes pronounced HSF1 activation in a given cancer cell? The current literature describes several different oncogenic pathways that alter HSF1 activity, indicating that HSF1 activity is so crucial to tumorous growth that it allows utilization of diverse signaling mechanisms (for detailed description of the molecular pathways, see Dai 2018). Furthermore, HSF1 protein levels are stabilized in some cancer types, which have been shown to contribute to activation of the HSF1-driven gene program (Kourtis et al. 2015). Of note, high HSF1 levels are known to correlate with poor prognosis in both breast and prostate cancer patients (Santagata et al. 2011; Björk et al. 2018), showing that increased HSF1 stability is likely a key feature permitting elevated HSF1 activity in cancer.
In contrast to developmental processes, such as spermatogenesis or corticogenesis, the mammalian HSFs are not known to functionally cooperate in cancer. HSF1 and HSF2 seem to have opposing roles in cancer progression, as lack of HSF2 was associated with enhanced prostate tumor growth and increased invasion (Björk et al. 2016). HSF2 depletion resulted in misregulation of genes linked to GTPase signaling, cell adhesion, and actin cytoskeleton, and down-regulated HSF2 messenger RNA (mRNA) levels were observed in a diverse array of human cancers (Björk et al. 2016). Considering the notion that both HSF1 and HSF2 show altered expression levels in cancer cells, it will be interesting to elucidate how these factors are regulated at gene, mRNA, and protein levels. To this end, oncogenic NOTCH1 was recently reported to bind to the HSF1 promoter and induce its expression, thereby showing for the first time HSF1 regulation at the gene level (Kourtis et al. 2018). Moreover, because HSF1, HSF2, and HSF4 have been reported to display functional cooperativity in specific physiological processes, the importance of cross talk between different HSF family members in cancer should be investigated.
Degeneration and Aging
A great majority of the degenerative diseases are characterized by accumulation of misfolded proteins and imbalances in the protein folding and degradation machineries, leading to proteostasis collapse. The protein quality control becomes limited also during aging and several lines of evidence suggest that enhancing the protein-folding capacity via increased expression of molecular chaperones can prevent the proteostasis collapse (Cummings et al. 2001; Klucken et al. 2004; Magrane et al. 2004; Nagy et al. 2016). Because of the key regulatory role of HSF1 in chaperone gene expression, its involvement in neurodegenerative diseases has been extensively studied. One of the first evidences for HSF1 acting as a suppressor of aggregation was provided by a genome-wide RNAi screen in polyQ-expressing C. elegans, which identified CeHSF1 as a factor required to suppress age-related protein aggregation (Nollen et al. 2004). In Hsf1−/− MEFs, lack of HSF1 results in increased accumulation of ubiquitinated proteins and protein aggregates, and similar observations have been obtained from Hsf1−/− mice with a plethora of degenerative diseases (Fujimoto et al. 2005; Homma et al. 2007; Kondo et al. 2013). Recently, HSF1 expression levels were found to be specifically down-regulated during aggregate accumulation, indicating that abnormal regulation of HSF1 stability, rather than its activity, is a hallmark of neurodegeneration (Kim et al. 2016; Gomez-Pastor et al. 2017).
Although HSF1 has a neuroprotective effect during aging and degeneration, it is not clear how it promotes cell survival in those contexts. Active HSF1 suppresses polyQ-aggregation more efficiently than any combination of HSPs (Hayashida et al. 2010), emphasizing that other HSF1 targets are also essential in preventing disease progression. Accumulation of polyQ-aggregates alters HSF1 binding, particularly at genes linked to cytoskeleton, focal adhesion, and GTPase activity (Riva et al. 2012), but their role in disease propagation is unknown. HSF1 counteracts proteostasis collapse also by maintaining mitochondrial homeostasis (Tan et al. 2015). In aging C. elegans, CeHSF1 promotes stress resistance through PAT-10, a gene specifically required to maintain cytoskeletal integrity (Baird et al. 2014), altogether proposing that the protective functions of HSF1 during degeneration indeed extend beyond the HSPs.
HSF2 has been implicated in the maintenance of proteostasis against polyQ-aggregation and regulation of mouse life span (Shinkawa et al. 2011), but the molecular details of its protective functions are scarce. In cellular model systems, HSF2 is activated during proteasome inhibition and it participates in target gene regulation together with HSF1 (Mathew et al. 1998; Lecomte et al. 2010; Rossi et al. 2014). Moreover, lack of HSF2 has been shown to compromise cell survival during proteasome inhibition (Lecomte et al. 2010), suggesting that HSF2 might have a unique role in proteotoxic stress. Nevertheless, it remains to be established how HSF2 protects cells against protein misfolding and whether the functional interplay between HSFs impacts the degenerative disease progression.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
During the past years, the research focusing on HSFs in acute stress responses has offered numerous key discoveries illuminating the mechanisms of stress-inducible transcription, posttranslational regulation of transcription factors as well as the physiological and pathological relevance of proteostasis. Genomic and epigenomic analyses of HSFs have contributed significantly to the development of new genome-wide methods and provided molecular details that apply generally to chromatin-associated responses. Identification of distinct PTMs has revealed novel insights into how stimulus-specific transcription factors acquire their transient activation profiles. Moreover, the tight link between HSFs and disease processes is a prime example of how the synergistic and antagonistic actions of proteostasis determinants impact the health of an organism. Although it is indisputable that HSFs serve as essential gatekeepers integrating internal and external signals into diverse transcriptional programs, there are still many fundamental questions to be addressed. It is noteworthy that a majority of previous studies have examined HSFs in acute heat stress, which limits understanding of their functions in other biological processes, such as organismal development, prolonged and chronic stress conditions, as well as progressive diseases. Therefore, it would be important to use the already achieved milestones to further explore HSFs in their diverse physiological and pathological working environments.
As of today, it is not known how different cells and tissues sense specific stimuli and transform them to the actions of distinct HSF family members. Arrays of posttranslational regulators, including PTMs and protein-interacting partners, orchestrate the activity and stability of HSFs, but their interdependence and combinatorial actions require further investigation. Along the same lines, it will be important to examine the cooperativity between HSF family members, as the interplay likely functions as a more sophisticated rheostat than originally anticipated. Overall, there is a clear demand for a more comprehensive understanding on how HSFs are regulated at the gene, mRNA, and protein levels and how the complex and dynamic chromatin architecture impacts the activity of HSFs. HSFs ultimately configure cellular physiology through transcriptional reprogramming. However, the mechanisms underlying the regulation of their subcellular or subnuclear localization and trafficking remain to be uncovered. The fundamental knowledge obtained from HSF-driven transcriptional networks will allow us to develop advanced molecular strategies to target HSFs in diseases associated with disrupted proteostasis.
ACKNOWLEDGMENTS
We apologize to those investigators whose work could not be cited because of space limitations. The members of the Sistonen laboratory are acknowledged for helpful discussions. Our work is supported by The Academy of Finland, Sigrid Juselius Foundation, Magnus Ehrnrooth Foundation, Cancer Foundation Finland, and the Maud Kuistila Foundation.
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
- Ahlskog JK, Björk JK, Elsing AN, Aspelin C, Kallio M, Roos-Mattjus P, Sistonen L. 2010. Anaphase-promoting complex/cyclosome participates in the acute response to protein-damaging stress. Mol Cell Biol 30: 5608–5620. 10.1128/MCB.01506-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn SG, Thiele DJ. 2003. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev 17: 516–528. 10.1101/gad.1044503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerfelt M, Henriksson E, Laiho A, Vihervaara A, Rautoma K, Kotaja N, Sistonen L. 2008. Promoter ChIP-chip analysis in mouse testis reveals Y chromosome occupancy by HSF2. Proc Natl Acad Sci 105: 11224–11229. 10.1073/pnas.0800620105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerfelt M, Vihervaara A, Laiho A, Conter A, Christians ES, Sistonen L, Henriksson E. 2010. Heat shock transcription factor 1 localizes to sex chromatin during meiotic repression. J Biol Chem 285: 34469–34476. 10.1074/jbc.M110.157552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin J, Ananthan J, Voellmy R. 1988. Key features of heat shock regulatory elements. Mol Cell Biol 8: 3761–3769. 10.1128/MCB.8.9.3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand D, Agrawal SA, Slavotinek A, Lachke SA. 2018. Mutation update of transcription factor genes FOXE3, HSF4, MAF, and PITX3 causing cataracts and other developmental ocular defects. Hum Mutat 39: 471–494. 10.1002/humu.23395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L. 2006. Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol 26: 955–964. 10.1128/MCB.26.3.955-964.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird NA, Douglas PM, Simic MS, Grant AR, Moresco JJ, Wolff SC, Yates JR III, Manning G, Dillin A. 2014. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science 346: 360–363. 10.1126/science.1253168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birch-Machin I, Gao S, Huen D, McGirr R, White RA, Russell S. 2005. Genomic analysis of heat-shock factor targets in Drosophila. Genome Biol 6: R63 10.1186/gb-2005-6-7-r63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk JK, Åkerfelt M, Joutsen J, Puustinen MC, Cheng F, Sistonen L, Nees M. 2016. Heat-shock factor 2 is a suppressor of prostate cancer invasion. Oncogene 35: 1770–1784. 10.1038/onc.2015.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk JK, Ahonen I, Mirtti T, Erickson A, Rannikko A, Bützow A, Nordling S, Lundin J, Lundin M, Sistonen L, et al. 2018. Increased HSF1 expression predicts shorter disease-specific survival of prostate cancer patients following radical prostatectomy. Oncotarget 9: 31200–31213. 10.18632/oncotarget.25756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boellmann F, Guettouche T, Guo Y, Fenna M, Mnayer L, Voellmy R. 2004. DAXX interacts with heat shock factor 1 during stress activation and enhances its transcriptional activity. Proc Natl Acad Sci 101: 4100–4105. 10.1073/pnas.0304768101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JJ, Ballou C, Fackenthal DL. 1994. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol 14: 501–508. 10.1128/MCB.14.1.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L, Jin Y, Shi Y, Chu R, Ban A, Eiberg H, Andres L, Jiang H, Zheng G, Qian M, et al. 2002. Mutant DNA-binding domain of HSF4 is associated with autosomal dominant lamellar and Marner cataract. Nat Genet 31: 276–278. 10.1038/ng921 [DOI] [PubMed] [Google Scholar]
- Budzyński MA, Puustinen MC, Joutsen J, Sistonen L. 2015. Uncoupling stress-inducible phosphorylation of heat shock factor 1 from its activation. Mol Cell Biol 35: 2530–2540. 10.1128/MCB.00816-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Östling P, Åkerfelt M, Trouillet D, Rallu M, Gitton Y, El Fatimy R, Fardeau V, Le Crom S, Morange M, et al. 2006. Role of heat-shock factor 2 in cerebral cortex formation and as a regulator of p35 expression. Genes Dev 20: 836–847. 10.1101/gad.366906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christians E, Davis AA, Thomas SD, Benjamin IJ. 2000. Maternal effect of Hsf1 on reproductive success. Nature 407: 693–694. 10.1038/35037669 [DOI] [PubMed] [Google Scholar]
- Cotto JJ, Kline M, Morimoto RI. 1996. Activation of heat shock factor 1 DNA binding precedes stress-induced serine phosphorylation. Evidence for a multistep pathway of regulation. J Biol Chem 271: 3355–3358. 10.1074/jbc.271.7.3355 [DOI] [PubMed] [Google Scholar]
- Cummings CJ, Sun Y, Opal P, Antalffy B, Mestril R, Orr HT, Dillmann WH, Zoghbi HY. 2001. Over-expression of inducible HSP70 chaperone suppresses neuropathology and improves motor function in SCA1 mice. Hum Mol Genet 10: 1511–1518. 10.1093/hmg/10.14.1511 [DOI] [PubMed] [Google Scholar]
- Dai C. 2018. The heat-shock, or HSF1-mediated proteotoxic stress, response in cancer: From proteomic stability to oncogenesis. Philos Trans R Soc Lond B Biol Sci 373: 20160525 10.1098/rstb.2016.0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. 2007. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell 130: 1005–1018. 10.1016/j.cell.2007.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Fatimy R, Miozzo F, Le Mouel A, Abane R, Schwendimann L, Saberan-Djoneidi D, de Thonel A, Massaoudi I, Paslaru L, Hashimoto-Torii K, et al. 2014. Heat shock factor 2 is a stress-responsive mediator of neuronal migration defects in models of fetal alcohol syndrome. EMBO Mol Med 6: 1043–1061. 10.15252/emmm.201303311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsing AN, Aspelin C, Björk JK, Bergman HA, Himanen SV, Kallio MJ, Roos-Mattjus P, Sistonen L. 2014. Expression of HSF2 decreases in mitosis to enable stress-inducible transcription and cell survival. J Cell Biol 206: 735–749. 10.1083/jcb.201402002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Wang S, Guo L, Punekar AS, Ladenstein R, Wang DC, Liu W. 2016. MD simulation of high-resolution X-ray structures reveals post-translational modification dependent conformational changes in HSF-DNA interaction. Protein Cell 7: 916–920. 10.1007/s13238-016-0331-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filone CM, Caballero IS, Dower K, Mendillo ML, Cowley GS, Santagata S, Rozelle DK, Yen J, Rubins KH, Hacohen N, et al. 2014. The master regulator of the cellular stress response (HSF1) is critical for orthopoxvirus infection. PLoS Pathog 10: e1003904 10.1371/journal.ppat.1003904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorenza MT, Farkas T, Dissing M, Kolding D, Zimarino V. 1995. Complex expression of murine heat shock transcription factors. Nucleic Acids Res 23: 467–474. 10.1093/nar/23.3.467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Izu H, Seki K, Fukuda K, Nishida T, Yamada S, Kato K, Yonemura S, Inouye S, Nakai A. 2004. HSF4 is required for normal cell growth and differentiation during mouse lens development. EMBO J 23: 4297–4306. 10.1038/sj.emboj.7600435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A. 2005. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem 280: 34908–34916. 10.1074/jbc.M506288200 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Hayashida N, Katoh T, Oshima K, Shinkawa T, Prakasam R, Tan K, Inouye S, Takii R, Nakai A. 2010. A novel mouse HSF3 has the potential to activate nonclassical heat-shock genes during heat shock. Mol Biol Cell 21: 106–116. 10.1091/mbc.e09-07-0639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Takii R, Tan K, Prakasam R, Hayashida N, Iemura SI, Natsume T, Nakai A. 2012. RPA assists HSF1 access to nucleosomal DNA by recruiting histone chaperone FACT. Mol Cell 48: 182–194. 10.1016/j.molcel.2012.07.026 [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Takii R, Katiyar A, Srivastava P, Nakai A. 2018. Poly(ADP-ribose) polymerase 1 promotes the human heat shock response by facilitating heat shock transcription factor 1 binding to DNA. Mol Cell Biol 14: e00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo GJ, Prentice H, Kingston RE. 1993. Heat shock factor is required for growth at normal temperatures in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 13: 749–761. 10.1128/MCB.13.2.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Neef DW, Jaeger AM, Cabiscol E, McKinstry SU, Doss A, Aballay A, Lo DC, Akimov SS, et al. 2017. Abnormal degradation of the neuronal stress-protective transcription factor HSF1 in Huntington's disease. Nat Commun 8: 14405 10.1038/ncomms14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pastor R, Burchfiel ET, Thiele DJ. 2018. Regulation of heat shock transcription factors and their roles in physiology and disease. Nat Rev Mol Cell Biol 19: 4–19. 10.1038/nrm.2017.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson ML, Hong Y, Rogers R, Matunis MJ, Park-Sarge OK, Sarge KD. 2001. SUMO-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem 276: 18513–18518. 10.1074/jbc.M008066200 [DOI] [PubMed] [Google Scholar]
- Grossman AD, Erickson JW, Gross CA. 1984. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38: 383–390. 10.1016/0092-8674(84)90493-8 [DOI] [PubMed] [Google Scholar]
- Guettouche T, Boellmann F, Lane WS, Voellmy R. 2005. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem 6: 4 10.1186/1471-2091-6-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R. 2001. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem 276: 45791–45799. 10.1074/jbc.M105931200 [DOI] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24: 5249–5256. 10.1128/MCB.24.12.5249-5256.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison CJ, Bohm AA, Nelson HC. 1994. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 263: 224–227. 10.1126/science.8284672 [DOI] [PubMed] [Google Scholar]
- Hayashida N, Fujimoto M, Tan K, Prakasam R, Shinkawa T, Li L, Ichikawa H, Takii R, Nakai A. 2010. Heat shock factor 1 ameliorates proteotoxicity in cooperation with the transcription factor NFAT. EMBO J 29: 3459–3469. 10.1038/emboj.2010.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, Lyon D, Young C, Jensen LJ, Vertegaal ACO, Nielsen ML. 2017. Site-specific mapping of the human SUMO proteome reveals co-modification with phosphorylation. Nat Struct Mol Biol 24: 325–336. 10.1038/nsmb.3366 [DOI] [PubMed] [Google Scholar]
- Hensold J, Hunt C, Calderwood S, Housman D, Kingston R. 1990. DNA binding of heat shock factor to the heat shock element is insufficient for transcriptional activation in murine erythroleukemia cells. Mol Cell Biol 10: 1600–1608. 10.1128/MCB.10.4.1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze N, Le Breton L, Wiesner J, Kempf G, Mayer MP. 2016. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. eLife 5: e11576 10.7554/eLife.11576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, Sistonen L. 2003. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol Cell Biol 23: 2953–2968. 10.1128/MCB.23.8.2953-2968.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci 103: 45–50. 10.1073/pnas.0503698102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi T, Nakai A, Uemura Y, Kikuchi H, Nagata K. 1995. Activation of heat shock factor 1 in rat brain during cerebral ischemia or after heat shock. Brain Res Mol Brain Res 34: 262–270. 10.1016/0169-328X(95)00163-M [DOI] [PubMed] [Google Scholar]
- Homma S, Jin X, Wang G, Tu N, Min J, Yanasak N, Mivechi NF. 2007. Demyelination, astrogliosis, and accumulation of ubiquitinated proteins, hallmarks of CNS disease in hsf1-deficient mice. J Neurosci 27: 7974–7986. 10.1523/JNEUROSCI.0006-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Rogers R, Matunis MJ, Mayhew CN, Goodson M, Park-Sarge OK, Sarge KD. 2001. Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276: 40263–40267. 10.1074/jbc.M104714200 [DOI] [PubMed] [Google Scholar]
- Inouye S, Izu H, Takaki E, Suzuki H, Shirai M, Yokota Y, Ichikawa H, Fujimoto M, Nakai A. 2004. Impaired IgG production in mice deficient for heat shock transcription factor 1. J Biol Chem 279: 38701–38709. 10.1074/jbc.M405986200 [DOI] [PubMed] [Google Scholar]
- Jaeger AM, Makley LN, Gestwicki JE, Thiele DJ. 2014. Genomic heat shock element sequences drive cooperative human heat shock factor 1 DNA binding and selectivity. J Biol Chem 289: 30459–30469. 10.1074/jbc.M114.591578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger AM, Pemble CW, Sistonen L, Thiele DJ. 2016. Structures of HSF2 reveal mechanisms for differential regulation of human heat-shock factors. Nat Struct Mol Biol 23: 147–154. 10.1038/nsmb.3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C. 1997. Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J 16: 2452–2462. 10.1093/emboj/16.9.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Eroglu B, Moskophidis D, Mivechi NF. 2011. Targeted deletion of Hsf1, 2, and 4 genes in mice. Methods Mol Biol 1709: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurivich DA, Sistonen L, Kroes RA, Morimoto RI. 1992. Effect of sodium salicylate on the human heat shock response. Science 255: 1243–1245. 10.1126/science.1546322 [DOI] [PubMed] [Google Scholar]
- Kallio M, Chang Y, Manuel M, Alastalo TP, Rallu M, Gitton Y, Pirkkala L, Loones MT, Paslaru L, Larney S, et al. 2002. Brain abnormalities, defective meiotic chromosome synapsis and female subfertility in HSF2 null mice. EMBO J 21: 2591–2601. 10.1093/emboj/21.11.2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazoe Y, Nakai A, Tanabe M, Nagata K. 1998. Proteasome inhibition leads to the activation of all members of the heat-shock-factor family. Eur J Biochem 255: 356–362. 10.1046/j.1432-1327.1998.2550356.x [DOI] [PubMed] [Google Scholar]
- Kijima T, Prince TL, Tigue ML, Yim KH, Schwartz H, Beebe K, Lee S, Budzynski MA, Williams H, Trepel JB, et al. 2018. HSP90 inhibitors disrupt a transient HSP90-HSF1 interaction and identify a noncanonical model of HSP90-mediated HSF1 regulation. Sci Rep 8: 6976 10.1038/s41598-018-25404-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386. 10.1074/jbc.M609659200 [DOI] [PubMed] [Google Scholar]
- Kim E, Wang B, Sastry N, Masliah E, Nelson PT, Cai H, Liao FF. 2016. NEDD4-mediated HSF1 degradation underlies α-synucleinopathy. Hum Mol Genet 25: 211–222. 10.1093/hmg/ddv445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klucken J, Shin Y, Masliah E, Hyman BT, McLean PJ. 2004. Hsp70 reduces α-synuclein aggregation and toxicity. J Biol Chem 279: 25497–25502. 10.1074/jbc.M400255200 [DOI] [PubMed] [Google Scholar]
- Kondo N, Katsuno M, Adachi H, Minamiyama M, Doi H, Matsumoto S, Miyazaki Y, Iida M, Tohnai G, Nakatsuji H, et al. 2013. Heat shock factor-1 influences pathological lesion distribution of polyglutamine-induced neurodegeneration. Nat Commun 4: 1405 10.1038/ncomms2417 [DOI] [PubMed] [Google Scholar]
- Korfanty J, Stokowy T, Widlak P, Gogler-Piglowska A, Handschuh L, Podkowiński J, Vydra N, Naumowicz A, Toma-Jonik A, Widlak W. 2014. Crosstalk between HSF1 and HSF2 during the heat shock response in mouse testes. Int J Biochem Cell Biol 57: 76–83. 10.1016/j.biocel.2014.10.006 [DOI] [PubMed] [Google Scholar]
- Kourtis N, Moubarak RS, Aranda-Orgilles B, Lui K, Aydin IT, Trimarchi T, Darvishian F, Salvaggio C, Zhong J, Bhatt K, et al. 2015. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat Cell Biol 17: 322–332. 10.1038/ncb3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Lazaris C, Hockemeyer K, Balandrán JC, Jimenez AR, Mullenders J, Gong Y, Trimarchi T, Bhatt K, Hu H, et al. 2018. Oncogenic hijacking of the stress response machinery in T cell acute lymphoblastic leukemia. Nat Med 24: 1157–1166. 10.1038/s41591-018-0105-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecomte S, Desmots F, Le Masson F, Le Goff P, Michel D, Christians ES, Le Dréan Y. 2010. Roles of heat shock factor 1 and 2 in response to proteasome inhibition: Consequence on p53 stability. Oncogene 29: 4216–4224. 10.1038/onc.2010.171 [DOI] [PubMed] [Google Scholar]
- Le Masson F, Razak Z, Kaigo M, Audouard C, Charry C, Cooke H, Westwood JT, Christians ES. 2011. Identification of heat shock factor 1 molecular and cellular targets during embryonic and adult female meiosis. Mol Cell Biol 31: 3410–3423. 10.1128/MCB.05237-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chauve L, Phelps G, Brielmann RM, Morimoto RI. 2016. E2F coregulates an essential HSF developmental program that is distinct from the heat-shock response. Genes Dev 30: 2062–2075. 10.1101/gad.283317.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S, Du R, Wang L, Qu Z, Cui X, Li C, Liu F, Huang M, Wang J, Chen J, et al. 2015. BCAS2 interacts with HSF4 and negatively regulates its protein stability via ubiquitination. Int J Biochem Cell Biol 68: 78–86. 10.1016/j.biocel.2015.08.016 [DOI] [PubMed] [Google Scholar]
- Liao S, Qu Z, Li L, Zhou B, Gao M, Huang M, Li D. 2018. HSF4 transcriptional regulates HMOX-1 expression in HLECs. Gene 655: 30–34. 10.1016/j.gene.2018.02.033 [DOI] [PubMed] [Google Scholar]
- Littlefield O, Nelson HCM. 1999. A new use for the “wing” of the “winged” helix-turn-helix motif in the HSF-DNA cocrystal. Nat Struct Biol 6: 464–470. 10.1038/8269 [DOI] [PubMed] [Google Scholar]
- Ma X, Xu L, Alberobello AT, Gavrilova O, Bagattin A, Skarulis M, Liu J, Finkel T, Mueller E. 2015. Celastrol protects against obesity and metabolic dysfunction through activation of a HSF1-PGC1α transcriptional axis. Cell Metab 22: 695–708. 10.1016/j.cmet.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Magrane J, Smith RC, Walsh K, Querfurth HW. 2004. Heat shock protein 70 participates in the neuroprotective response to intracellularly expressed β-amyloid in neurons. J Neurosci 24: 1700–1706. 10.1523/JNEUROSCI.4330-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Salamanca HH, Duarte FM, Danko CG, Lis JT. 2016. Mammalian heat shock response and mechanisms underlying its genome-wide transcriptional regulation. Mol Cell 62: 63–78. 10.1016/j.molcel.2016.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A, Mathur SK, Morimoto RI. 1998. Heat shock response and protein degradation: Regulation of HSF2 by the ubiquitin-proteasome pathway. Mol Cell Biol 18: 5091–5098. 10.1128/MCB.18.9.5091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan DR, Xiao X, Shao L, Graves K, Benjamin IJ. 1998. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J Biol Chem 273: 7523–7528. 10.1074/jbc.273.13.7523 [DOI] [PubMed] [Google Scholar]
- McMillan DR, Christians E, Forster M, Xiao X, Connell P, Plumier JC, Zuo X, Richardson J, Morgan S, Benjamin IJ. 2002. Heat shock transcription factor 2 is not essential for embryonic development, fertility, or adult cognitive and psychomotor function in mice. Mol Cell Biol 22: 8005–8014. 10.1128/MCB.22.22.8005-8014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. 2012. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 150: 549–562. 10.1016/j.cell.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metchat A, Åkerfelt M, Bierkamp C, Delsinne V, Sistonen L, Alexandre H, Christians ES. 2009. Mammalian heat shock factor 1 is essential for oocyte meiosis and directly regulates Hsp90α expression. J Biol Chem 284: 9521–9528. 10.1074/jbc.M808819200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezger V, Renard JP, Christians E, Morange M. 1994. Detection of heat shock element-binding activities by gel shift assay during mouse preimplantation development. Dev Biol 165: 627–638. 10.1006/dbio.1994.1281 [DOI] [PubMed] [Google Scholar]
- Min JN, Zhang Y, Moskophidis D, Mivechi NF. 2004. Unique contribution of heat shock transcription factor 4 in ocular lens development and fiber cell differentiation. Genesis 40: 205–217. 10.1002/gene.20087 [DOI] [PubMed] [Google Scholar]
- Min JN, Huang L, Zimonjic DB, Moskophidis D, Mivechi NF. 2007. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 26: 5086–5097. 10.1038/sj.onc.1210317 [DOI] [PubMed] [Google Scholar]
- Minsky N, Roeder RG. 2015. Direct link between metabolic regulation and the heat-shock response through the transcriptional regulator PGC-1α. Proc Natl Acad Sci 112: E5669–E5678. 10.1073/pnas.1516219112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy M, Fenton WA, Li D, Furtak K, Horwich AL. 2016. Extended survival of misfolded G85R SOD1-linked ALS mice by transgenic expression of chaperone Hsp110. Proc Natl Acad Sci 113: 5424–5428. 10.1073/pnas.1604885113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Ishikawa T. 2001. Cell cycle transition under stress conditions controlled by vertebrate heat shock factors. EMBO J 20: 2885–2895. 10.1093/emboj/20.11.2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Morimoto RI. 1993. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol 13: 1983–1997. 10.1128/MCB.13.4.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto RI, Nagata K. 1997. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol 17: 469–481. 10.1128/MCB.17.1.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neef DW, Turski ML, Thiele DJ. 2010. Modulation of heat shock transcription factor 1 as a therapeutic target for small molecule intervention in neurodegenerative disease. PLoS Biol 8: e1000291 10.1371/journal.pbio.1000291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neudegger T, Verghese J, Hayer-Hartl M, Hartl FU, Bracher A. 2016. Structure of human heat-shock transcription factor 1 in complex with DNA. Nat Struct Mol Biol 23: 140–146. 10.1038/nsmb.3149 [DOI] [PubMed] [Google Scholar]
- Newton EM, Knauf U, Green M, Kingston RE. 1996. The regulatory domain of human heat shock factor 1 is sufficient to sense heat stress. Mol Cell Biol 16: 839–846. 10.1128/MCB.16.3.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto-Sotelo J, Wiederrecht G, Okuda A, Parker CS. 1990. The yeast heat shock transcription factor contains a transcriptional activation domain whose activity is repressed under nonshock conditions. Cell 62: 807–817. 10.1016/0092-8674(90)90124-W [DOI] [PubMed] [Google Scholar]
- Nishizawa J, Nakai A, Higashi T, Tanabe M, Nomoto S, Matsuda K, Ban T, Nagata K. 1996. Reperfusion causes significant activation of heat shock transcription factor 1 in ischemic rat heart. Circulation 94: 2185–2192. 10.1161/01.CIR.94.9.2185 [DOI] [PubMed] [Google Scholar]
- Nishizawa S, Koya T, Ohno Y, Goto A, Ikuita A, Suzuki M, Ohira T, Egawa T, Nakai A, Sugiura T, et al. 2013. Regeneration of injured skeletal muscle in heat shock transcription factor 1-null mice. Physiol Rep 1: e00071 10.1002/phy2.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EAA, Garcia SM, van Haaften G, Kim S, Chavez A, Morimoto RI, Plasterk RHA. 2004. Genome-wide RNA interference screen identifies previously undescribed regulators of polyglutamine aggregation. Proc Natl Acad Sci 101: 6403–6408. 10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L. 2007. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem 282: 7077–7086. 10.1074/jbc.M607556200 [DOI] [PubMed] [Google Scholar]
- Rallu M, Loones M, Lallemand Y, Morimoto R, Morange M, Mezger V. 1997. Function and regulation of heat shock factor 2 during mouse embryogenesis. Proc Natl Acad Sci 94: 2392–2397. 10.1073/pnas.94.6.2392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Loew C, Körner R, Pinkert S, Theis M, Hayer-Hartl M, Buchholz F, Hartl FU. 2014. Interplay of acetyltransferase EP300 and the proteasome system in regulating heat shock transcription factor 1. Cell 156: 975–985. 10.1016/j.cell.2014.01.055 [DOI] [PubMed] [Google Scholar]
- Reinke H, Saini C, Fleury-Olela F, Dibner C, Benjamin IJ, Schibler U. 2008. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev 22: 331–345. 10.1101/gad.453808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritossa F. 1962. A new puffing pattern induced by temperature shock and DNP in Drosophila. Experientia 18: 571–573. 10.1007/BF02172188 [DOI] [Google Scholar]
- Riva L, Koeva M, Yildirim F, Pirhaji L, Dinesh D, Mazor T, Duennwald ML, Fraenkel E. 2012. Poly-glutamine expanded huntingtin dramatically alters the genome wide binding of HSF1. J Huntingtons Dis 1: 33–45. [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Riccio A, Coccia M, Trotta E, La Frazia S, Santoro MG. 2014. The proteasome inhibitor bortezomib is a potent inducer of zinc finger an1-type domain 2A gene expression: Role of heat shock factor 1 (HSF1)- heat shock factor 2 (HSF2) heterocomplexes. J Biol Chem 289: 12705–12715. 10.1074/jbc.M113.513242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmand PA, Jungas T, Fernandez M, Conter A, Christians ES. 2008. Mouse heat-shock factor 1 (HSF1) is involved in testicular response to genotoxic stress induced by doxorubicin. Biol Reprod 79: 1092–1101. 10.1095/biolreprod.108.070334 [DOI] [PubMed] [Google Scholar]
- Sandqvist A, Björk JK, Åkerfelt M, Chitikova Z, Grichine A, Vourc'h C, Jolly C, Salminen TA, Nymalm Y, Sistonen L. 2009. Heterotrimerization of heat-shock factors 1 and 2 provides a transcriptional switch in response to distinct stimuli. Mol Biol Cell 20: 1340–1347. 10.1091/mbc.e08-08-0864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S, Hu R, Lin NU, Mendillo ML, Collins LC, Hankinson SE, Schnitt SJ, Whitesell L, Tamimi RM, Lindquist S, et al. 2011. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc Natl Acad Sci 108: 18378–18383. 10.1073/pnas.1115031108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos SD, Saraiva MJ. 2004. Enlarged ventricles, astrogliosis and neurodegeneration in heat shock factor 1 null mouse brain. Neuroscience 126: 657–663. 10.1016/j.neuroscience.2004.03.023 [DOI] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK, Kirby JD, Mayo KE, Morimoto RI. 1994. Expression of heat shock factor 2 in mouse testis: Potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod 50: 1334–1343. 10.1095/biolreprod50.6.1334 [DOI] [PubMed] [Google Scholar]
- Scherz-Shouval R, Santagata S, Mendillo ML, Sholl LM, Ben-Aharon I, Beck AH, Dias-Santagata D, Koeva M, Stemmer SM, Whitesell L, et al. 2014. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 158: 564–578. 10.1016/j.cell.2014.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kroeger PE, Morimoto RI. 1995. The carboxyl-terminal transactivation domain of heat shock factor 1 is negatively regulated and stress responsive. Mol Cell Biol 15: 4309–4318. 10.1128/MCB.15.8.4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. 1998. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12: 654–666. 10.1101/gad.12.5.654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Shi X, Jin Y, Miao A, Bu L, He J, Jiang H, Lu Y, Kong X, Hu L. 2008. Mutation screening of HSF4 in 150 age-related cataract patients. Mol Vis 14: 1850–1855. [PMC free article] [PubMed] [Google Scholar]
- Shinkawa T, Tan K, Fujimoto M, Hayashida N, Yamamoto K, Takaki E, Takii R, Prakasam R, Inouye S, Mezger V, et al. 2011. Heat shock factor 2 is required for maintaining proteostasis against febrile-range thermal stress and polyglutamine aggregation. Mol Biol Cell 22: 3571–3583. 10.1091/mbc.e11-04-0330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen L, Sarge KD, Phillips B, Abravaya K, Morimoto RI. 1992. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol 12: 4104–4111. 10.1128/MCB.12.9.4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solís EJ, Pandey JP, Zheng X, Jin DX, Gupta PB, Airoldi EM, Pincus D, Denic V. 2016. Defining the essential function of yeast Hsf1 reveals a compact transcriptional program for maintaining eukaryotic proteostasis. Mol Cell 63: 60–71. 10.1016/j.molcel.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Nelson HCM. 1989. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell 59: 807–813. 10.1016/0092-8674(89)90604-1 [DOI] [PubMed] [Google Scholar]
- Sorger PK, Pelham HRB. 1988. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell 54: 855–864. 10.1016/S0092-8674(88)91219-6 [DOI] [PubMed] [Google Scholar]
- Sorger PK, Lewis MJ, Pelham HRB. 1987. Heat shock factor is regulated differently in yeast and HeLa cells. Nature 329: 81–84. 10.1038/329081a0 [DOI] [PubMed] [Google Scholar]
- Su KH, Dai C. 2016. Metabolic control of the proteotoxic stress response: Implications in diabetes mellitus and neurodegenerative disorders. Cell Mol Life Sci 73: 4231–4248. 10.1007/s00018-016-2291-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su KH, Cao J, Tang Z, Dai S, He Y, Sampson SB, Benjamin IJ, Dai C. 2016. HSF1 critically attunes proteotoxic stress sensing by mTORC1 to combat stress and promote growth. Nat Cell Biol 18: 527–539. 10.1038/ncb3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara K, Inouye S, Izu H, Katoh Y, Katsuki K, Takemoto T, Shimogori H, Yamashita H, Nakai A. 2003. Heat shock transcription factor HSF1 is required for survival of sensory hair cells against acoustic overexposure. Hear Res 182: 88–96. 10.1016/S0378-5955(03)00180-1 [DOI] [PubMed] [Google Scholar]
- Sullivan EK, Weirich CS, Guyon JR, Sif S, Kingston RE. 2001. Transcriptional activation domains of human heat shock factor 1 recruit human SWI/SNF. Mol Cell Biol 21: 5826–5837. 10.1128/MCB.21.17.5826-5837.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaki E, Fujimoto M, Sugahara K, Nakahari T, Yonemura S, Tanaka Y, Hayashida N, Inouye S, Takemoto T, Yamashita H, et al. 2006. Maintenance of olfactory neurogenesis requires HSF1, a major heat shock transcription factor in mice. J Biol Chem 281: 4931–4937. 10.1074/jbc.M506911200 [DOI] [PubMed] [Google Scholar]
- Takii R, Fujimoto M, Tan K, Takaki E, Hayashida N, Nakato R, Shirahige K, Nakai A. 2015. ATF1 modulates the heat shock response by regulating the stress-inducible heat shock factor 1 transcription complex. Mol Cell Biol 35: 11–25. 10.1128/MCB.00754-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Fujimoto M, Takii R, Takaki E, Hayashida N, Nakai A. 2015. Mitochondrial SSBP1 protects cells from proteotoxic stresses by potentiating stress-induced HSF1 transcriptional activity. Nat Commun 6: 6580 10.1038/ncomms7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe M, Kawazoe Y, Takeda S, Morimoto RI, Nagata K, Nakai A. 1998. Disruption of the HSF3 gene results in the severe reduction of heat shock gene expression and loss of thermotolerance. EMBO J 17: 1750–1758. 10.1093/emboj/17.6.1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi Y, Ariyoshi M, Igarashi R, Hara H, Mizuguchi K, Seto A, Nakai A, Kokubo T, Tochio H, Shirakawa M. 2009. Molecular basis for SUMOylation-dependent regulation of DNA binding activity of heat shock factor 2. J Biol Chem 284: 2435–2447. 10.1074/jbc.M806392200 [DOI] [PubMed] [Google Scholar]
- Tessari A, Salata E, Ferlin A, Bartoloni L, Slongo ML, Foresta C. 2004. Characterization of HSFY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol Hum Reprod 10: 253–258. 10.1093/molehr/gah036 [DOI] [PubMed] [Google Scholar]
- Tu N, Hu Y, Mivechi NF. 2006. Heat shock transcription factor (Hsf)-4b recruits Brg1 during the G1 phase of the cell cycle and regulates the expression of heat shock proteins. J Cell Biochem 98: 1528–1542. 10.1002/jcb.20865 [DOI] [PubMed] [Google Scholar]
- Vihervaara A, Sergelius C, Vasara J, Blom MAH, Elsing AN, Roos-Mattjus P, Sistonen L. 2013. Transcriptional response to stress in the dynamic chromatin environment of cycling and mitotic cells. Proc Natl Acad Sci 110: E3388–E3397. 10.1073/pnas.1305275110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Mahat DB, Guertin MJ, Chu T, Danko CG, Lis JT, Sistonen L. 2017. Transcriptional response to stress is pre-wired by promoter and enhancer architecture. Nat Commun 8: 255 10.1038/s41467-017-00151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihervaara A, Duarte FM, Lis JT. 2018. Molecular mechanisms driving transcriptional stress responses. Nat Rev Genet 19: 385–397. 10.1038/s41576-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuister GW, Kim SJ, Orosz A, Marquardt J, Wu C, Bax A. 1994. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Mol Biol 1: 605–614. 10.1038/nsb0994-605 [DOI] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C. 2011. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 10: M111.013284 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhang J, Moskophidis D, Mivechi NF. 2003. Targeted disruption of the heat shock transcription factor (hsf)-2 gene results in increased embryonic lethality, neuronal defects, and reduced spermatogenesis. Genesis 36: 48–61. 10.1002/gene.10200 [DOI] [PubMed] [Google Scholar]
- Wang G, Ying Z, Jin X, Tu N, Zhang Y, Phillips M, Moskophidis D, Mivechi NF. 2004. Essential requirement for both hsf1 and hsf2 transcriptional activity in spermatogenesis and male fertility. Genesis 38: 66–80. 10.1002/gene.20005 [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Bosman JD, Mbadugha BNA, Kawahara TLA, Matsumoto G, Kim S, Gu W, Devlin JP, Silverman RB, Morimoto RI. 2004. Celastrols as inducers of the heat shock response and cytoprotection. J Biol Chem 279: 56053–56060. 10.1074/jbc.M409267200 [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM Jr, Sistonen L, Morimoto RI. 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase sirt1. Science 323: 1063–1066. 10.1126/science.1165946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L, Lindquist SL. 2005. HSP90 and the chaperoning of cancer. Nat Rev Cancer 5: 761–772. 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- Widlak W, Vydra N. 2017. The role of heat shock factors in mammalian spermatogenesis. Adv Anat Embryol Cell Biol 222: 45–65. 10.1007/978-3-319-51409-3_3 [DOI] [PubMed] [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J 18: 5943–5952. 10.1093/emboj/18.21.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ma X, Bagattin A, Mueller E. 2016. The transcriptional coactivator PGC1α protects against hyperthermic stress via cooperation with the heat shock factor HSF1. Cell Death Dis 7: e2102 10.1038/cddis.2016.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelin E, Freeman BC. 2015. Lysine deacetylases regulate the heat shock response including the age-associated impairment of HSF1. J Mol Biol 427: 1644–1654. 10.1016/j.jmb.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Krakowiak J, Patel N, Beyzavi A, Ezike J, Khalil AS, Pincus D. 2016. Dynamic control of Hsf1 during heat shock by a chaperone switch and phosphorylation. eLife 5: e18638 10.7554/eLife.18638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Beyzavi A, Krakowiak J, Patel N, Khalil AS, Pincus D. 2018. Hsf1 phosphorylation generates cell-to-cell variation in Hsp90 levels and promotes phenotypic plasticity. Cell Rep 22: 3099–3106. 10.1016/j.celrep.2018.02.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Rungger D, Voellmy R. 1995. Multiple layers of regulation of human heat shock transcription factor 1. Mol Cell Biol 15: 4319–4330. 10.1128/MCB.15.8.4319 [DOI] [PMC free article] [PubMed] [Google Scholar]