Abstract
The physiological consequences of the unfolded protein response (UPR) are mediated by changes in gene expression. Underlying them are rapid processes involving preexisting components. We review recent insights gained into the regulation of the endoplasmic reticulum (ER) Hsp70 chaperone BiP, whose incorporation into inactive oligomers and reversible AMPylation and de-AMPylation present a first line of response to fluctuating levels of unfolded proteins. BiP activity is tied to the regulation of the UPR transducers by a recently discovered cycle of ER-localized, J protein–mediated formation of a repressive IRE1–BiP complex, whose working we contrast to an alternative model for UPR regulation that relies on direct recognition of unfolded proteins. We conclude with a discussion of mechanisms that repress messenger RNA (mRNA) translation to limit the flux of newly synthesized proteins into the ER, a rapid adaptation that does not rely on new macromolecule biosynthesis.
In eukaryotes, the synthesis of most proteins destined for secretion or membrane insertion occurs at the endoplasmic reticulum (ER), where their folding and maturation is assisted by compartment-specific molecular chaperones. Thus, protein homeostasis in the secretory pathway relies on the ability of cells to continuously adjust their complement of chaperones to match the load of newly synthesized proteins. An imbalance between folding load and capacity due to excess of non-native ER clients—a condition known as ER stress—can cause proteotoxic misfolding and aggregation, whereas a surplus of chaperones may slow down productive folding and favor aberrant degradation.
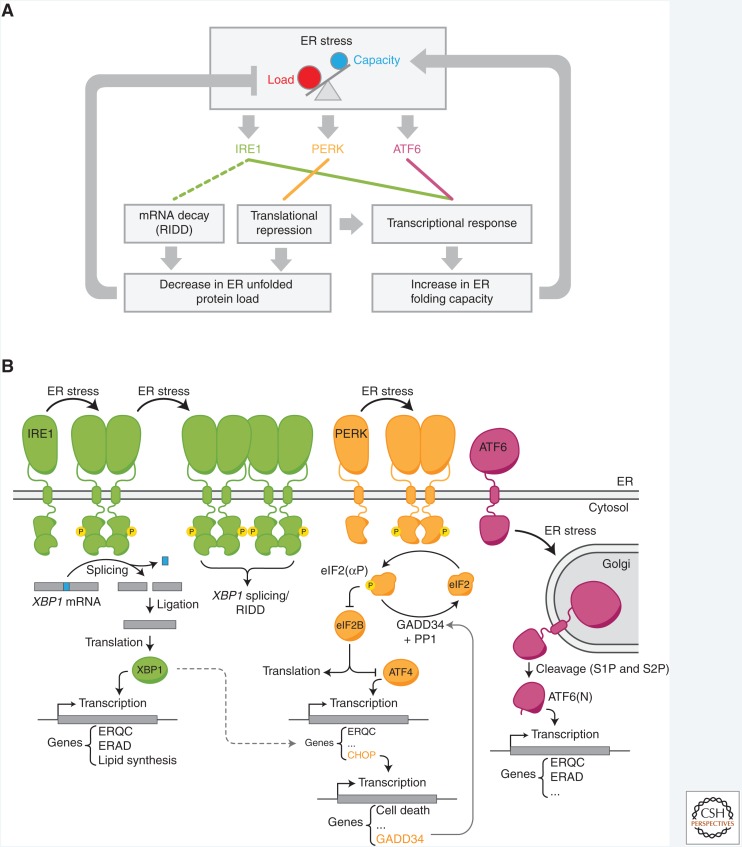
The dynamic adjustments required to defend ER protein homeostasis are governed by a negative feedback–regulated signaling pathway, the unfolded protein response (UPR) (Kozutsumi et al. 1988). The UPR depends on the activation of transmembrane signal transducers during ER stress to evoke mainly two downstream effects: (1) limiting the influx of proteins into the ER to cap the folding load, and (2) increasing the expression of effector molecules with functions in ER protein quality control (ERQC) to expand the folding capacity (Fig. 1).
Figure 1.
Overview of the mammalian unfolded protein response (UPR). (A) The central elements of the UPR signaling network. Each of the three signal transducers: IRE1, protein kinase R-like endoplasmic reticulum (ER) kinase (PERK), and activating transcription factor 6 (ATF6) constitutes a separate branch of the UPR with a specific output to counteract proteostatic perturbations in the ER. (B) Basic components and downstream events of the UPR signaling pathways. ER stress induces dimerization/oligomerization of IRE1 and its trans-autophosphorylation, which leads to nonconventional XBP1 messenger RNA (mRNA) splicing and production of the functional XBP1 transcription factor to induce expression of genes involved in ER protein quality control (ERQC) and ER-associated degradation (ERAD) of misfolded proteins. Activated, oligomeric IRE1 may also degrade ER-targeted mRNAs (via regulated Ire1-dependent decay [RIDD]). PERK-mediated phosphorylation of the α subunit of the eukaryotic initiation factor 2 (eIF2α) causes general translation attenuation, to relieve the load of newly synthesized proteins, and preferred synthesis of ATF4 (see Fig. 5). Production of the downstream transcription factor C/EBP homologous protein (CHOP) feeds back negatively on the PERK pathway (by enhancing expression of the phosphorylated eIF2 [eIF2(αP)]-specific regulatory phosphatase subunit growth arrest and DNA damage 34 [GADD34]) and induction of proapoptotic genes to favor cell death at later stages of unrectified stress. ER stress also induces ATF6 translocation to the Golgi where it is processed by S1P and S2P proteases to liberate the ATF6(N) transcription factor element. At the transcriptional level, all branches primarily facilitate expansion of the ER folding capacity and crosstalk exists between them (e.g., dotted line).
In unicellular eukaryotes, such as yeast, the adaptation to the burden of unfolded proteins in the ER is dominated by a transcriptional program that centers on the regulated activity of a single nuclear protein, HAC1 (Cox and Walter 1996). HAC1 abundance is coordinated with the level of ER stress by the activity of the transmembrane signaling protein IRE1 via a linear pathway (Karagöz et al. 2018). The targets of the yeast UPR include ER-localized chaperones, such as the Hsp70 BiP, which directly bind and buffer unfolded proteins, but also genes that operate throughout the secretory pathway by enhancing membrane lipid synthesis and the ability to degrade unfolded ER proteins (Cox et al. 1997; Travers et al. 2000).
These features of the UPR as a latent transcriptional program by which cells gradually develop the capacity to cope with the demands of protein secretion is conserved in higher eukaryotes: the animal counterpart to HAC1, XBP1, was originally identified as a developmental gene required for immunoglobulin secretion from plasma cells (Reimold et al. 2001). Animal-cell-specific UPR-regulated, activating transcription factor 6 (ATF6) and BBF2H7 are required for the developmental-stage-specific secretion of type II collagen (Ishikawa et al. 2017). However, animals have also acquired reversible mechanisms that rapidly match the ER's capacity to physiological fluctuations in ER stress, independent of transcription and new protein synthesis. The context for this is likely to be the highly dynamic nature of secretion in animals. For example, in response to fluctuating blood glucose levels, the quantity of proinsulin entering the ER of a pancreatic β cell can change by tenfold over a time frame of <1 h (Itoh and Okamoto 1980). A purely yeast-like transcriptional UPR is ill-equipped to cope with such dynamism.
This article will address the early posttranslational events in the animal cell UPR. We will begin by discussing AMPylation, an animal-specific mechanism for rapid, reversible inactivation of the major ER chaperone BiP. This will lead to a critical discussion of BiP's role in regulating the UPR transducers. We shall end by discussing another animal-specific adaptation to ER stress—control over the rate of new protein synthesis.
EMERGING ROLE OF POSTTRANSLATIONAL MECHANISMS FOR SHORT-TERM REGULATION OF CHAPERONE AVAILABILITY IN THE ER
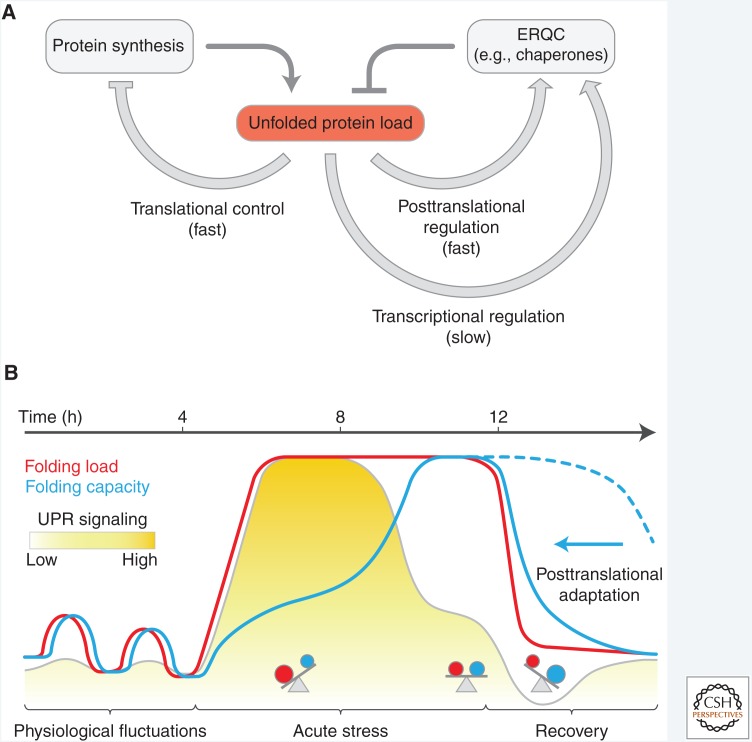
The mammalian UPR is multibranched and depends mainly on the activity of three transmembrane signaling molecules IRE1, ATF6, and protein kinase R–like ER kinase (PERK). The transcriptional output of each is mediated through the pathway-specific transcription factors XBP1, ATF6(N), and ATF4, respectively (Fig. 1). The inherent latency of the UPR limits its responsiveness to changes in the burden of unfolded proteins. In particular, dedicated secretory cells experience recurring physiological fluctuations in the unfolded protein load that appear too transient for alterations of the transcriptional program alone to handle, hinting at the need for other compensatory mechanisms to adjust the folding capacity (Fig. 2).
Figure 2.
Latency of the negative feedback-regulated transcriptional unfolded protein response (UPR). (A) Adaptation of the UPR by negative feedback regulation via endoplasmic reticulum (ER) protein quality control (ERQC) components. Apart from stress-denatured proteins, newly synthesized secretory proteins constitute the majority of unfolded species in the ER and their accumulation activates the UPR transducers. The response entails rapid attenuation of translation to alleviate the unfolded protein load (see Fig. 5), rapid posttranslational regulation of the pool of active chaperones in the ER (see Fig. 3), and the slower induction of ERQC genes to expand the folding capacity and mediate negative feedback regulation of the UPR (see Fig. 4). (B) Theoretical, time-resolved response profiles to illustrate the inherent latency of the UPR. Recurring physiological short-term fluctuations in the unfolded protein load do not cause a significant transcriptional response, suggesting that they are buffered by rapid, nontranscriptional adjustments of the folding capacity. During acute stress the fast rise of the folding load induces UPR signaling but expansion of the folding capacity is delayed as a result of the inherent latency of transcriptional reprogramming. Negative feedback inhibition attenuates the transcriptional response as the folding capacity builds up to establish homeostasis at a new level. During the recovery from ER stress the folding load drops rapidly. Were the adjustment in ERQC capacity to rely exclusively on the decay of the ERQC messenger RNAs (mRNAs) and encoded proteins, ERQC capacity would likely exceed the unfolded protein load in this recovery phase (dotted blue line). However, posttranslational mechanisms that inactivate chaperones rapidly adjust the effective folding capacity and prevent over-chaperoning (arrow).
Among the effector proteins that are targets of the UPR is the Hsp70-type chaperone BiP, an essential ERQC component involved in protein import, guiding polypeptide folding, assembly of protein complexes, and channeling misfolded polypeptides to degradation (for review, see Behnke et al. 2015). BiP also has important roles as a folding load sensor and negative feedback regulator of UPR signaling (see below), making it a key player in the control of ER protein homeostasis.
BiP, which has a relatively long half-life in cultured cells (≥24 h) (Satoh et al. 1993) and in tissues (≥3 days) (Price et al. 2010; Lau et al. 2016), is present at high concentrations in the ER basally (∼0.5 mm) and can increase several fold during prolonged ER stress (Bakunts et al. 2017). Shortage of BiP corrupts central aspects of ERQC, whereas excess of BiP also impairs secretion (Dorner et al. 1992). The cost of excess BiP suggests that a UPR based solely on changes in levels of gene expression would be ill-equipped to cope with short-term physiological changes in the demand for a highly abundant chaperone with slow turnover rates.
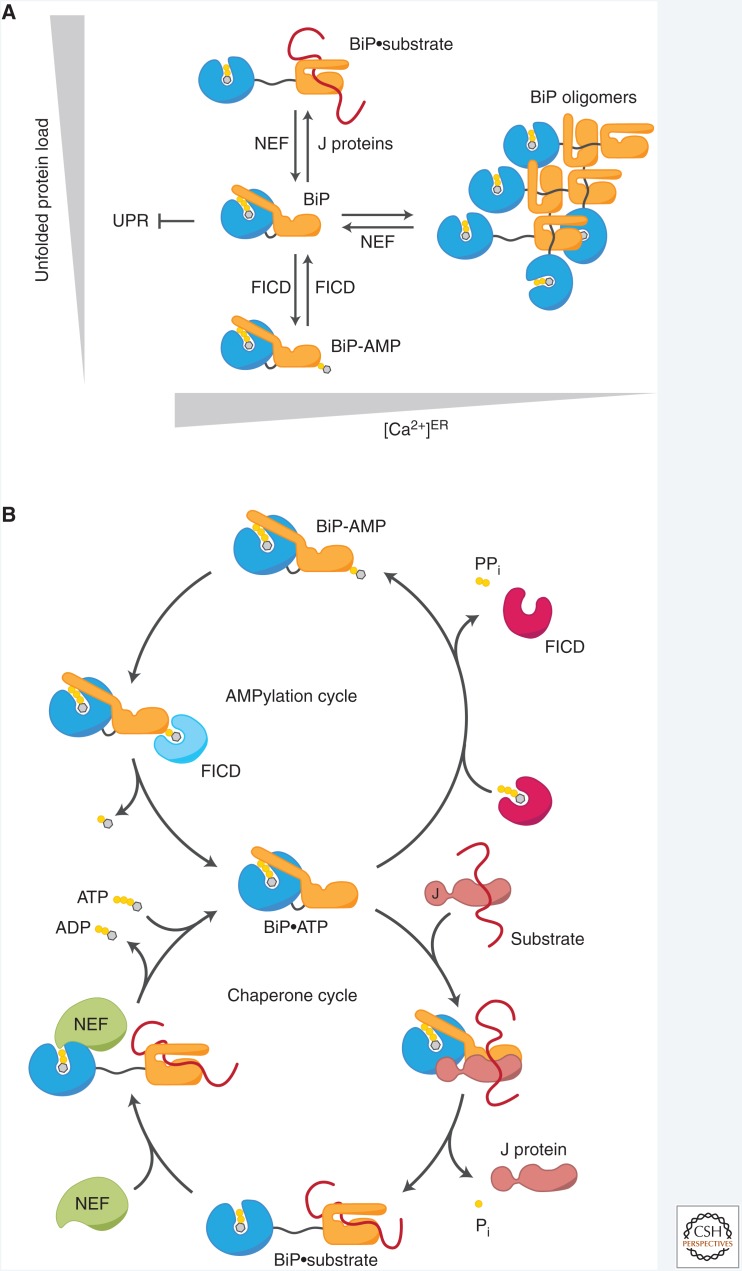
Recent discoveries brought to attention two reversible, posttranslational mechanisms that regulate the quantity of active BiP in response to changes in the concentration of unfolded substrates. The first entails the sequestration of client-free BiP into homomeric complexes. That BiP forms oligomers has been long known (Carlino et al. 1992; Freiden et al. 1992; Blond-Elguindi et al. 1993), but recently their potential physiological role came into focus. In vitro experiments revealed that oligomerization is based on canonical substrate interactions between individual BiP molecules and thus in direct competition with binding to unfolded clients (Blond-Elguindi et al. 1993; Preissler et al. 2015a). Furthermore, BiP oligomers diminish in cells treated with ER stress-inducing agents (Freiden et al. 1992; Preissler et al. 2015a). Together, these observations suggest that BiP oligomers constitute a dynamic inactive pool into which free BiP is deposited to prevent nonproductive interactions with native proteins and from which active chaperone can be recruited when the concentration of unfolded proteins rises (Fig. 3A).
Figure 3.
Posttranslational regulation of BiP. (A) Dynamic partitioning of BiP among different pools. Unfolded substrate polypeptides directly compete with oligomerization for a limited pool of free, ATP-bound BiP. Free BiP also interacts with unfolded protein response (UPR) signal transducers and contributes to repression of the UPR (see Fig. 4). J proteins promote both high-affinity substrate interactions and competing BiP oligomerization (with yet-to-be-characterized relative kinetics), whereas nucleotide exchange factors (NEFs) induce substrate release and oligomer disassembly. When the folding load declines, ATP-bound BiP becomes reversibly modified and inactivated by FICD-mediated AMPylation. Calcium depletion from the endoplasmic reticulum (ER) strongly induces BiP oligomerization by an unknown mechanism. (B) Chaperone and AMPylation cycles of BiP. ATP-bound BiP can be recruited to its substrates by J proteins, which stimulate ATP hydrolysis by BiP to achieve high-affinity interactions with its substrates. NEFs promote ADP release from BiP to enable ATP rebinding and dissociation of substrates. When the folding load is low, ATP-bound BiP becomes AMPylated by FICD. This locks BiP in a low-affinity state for substrates and renders the chaperone insensitive to stimulation by J proteins, thereby causing its functional inactivation. The same enzyme, FICD, demodifies BiP when the unfolded protein load increases to rapidly recruit preexisting inactive BiP into the pool of active chaperones. The different functional states of FICD, default de-AMPylation and yet-to-be-defined AMPylation, are indicated in light blue and magenta, respectively.
A second mechanism of BiP regulation involves its reversible inactivation by posttranslational modification. As the modification entailed labeling of BiP from both cellular phosphate and adenosine pools, it was long believed to represent ADP-ribosylation (Carlsson and Lazarides 1983; Ledford and Jacobs 1986; Hendershot et al. 1988; Chambers et al. 2012). However, the discovery of an ER-localized enzyme, FICD (or HYPE), that uses ATP to catalyze the covalent attachment of AMP via a phosphodiester bond to BiP, suggested an alternative scenario—AMPylation (Ham et al. 2014; Sanyal et al. 2015). Indeed, disruption of the FICD gene in mammalian cells abolishes all detectable BiP modification, indicating that AMPylation (not ADP ribosylation) is the most prominently detectable modification of BiP (Preissler et al. 2015b).
Early observations that BiP modification correlates inversely with the unfolded protein load in the ER (Carlsson and Lazarides 1983; Ledford and Jacobs 1986; Laitusis et al. 1999) and that only unmodified BiP binds substrates (Hendershot et al. 1988; Freiden et al. 1992) suggested an inhibitory effect on BiP's chaperone activity. AMPylation occurs at a single residue in BiP's substrate binding domain, threonine 518. Reconstitution of BiP AMPylation with purified components revealed that BiP, in its ATP-bound conformation, is the favored substrate for AMPylation (Preissler et al. 2015b). Furthermore, AMPylation biases BiP conformationally toward the ATP-bound state (Preissler et al. 2017b; Wieteska et al. 2017), which is characterized by fast substrate dissociation rates. AMPylation also impairs stimulation of BiP's ATPase activity by J protein cofactors (Preissler et al. 2017b), which is required for Hsp70s to attain high affinity for their substrates. Thus, like oligomerization, AMPylation creates a dynamic pool of inactive BiP to match the concentration of active chaperone to the amount of unfolded proteins (Fig. 3B).
Because AMPylation affects BiP's ability to engage in substrate interactions, modified BiP is excluded from oligomers. The two mutually exclusive inactivating processes regulate BiP with different kinetics. Oligomerization is in direct competition with substrate binding and thus is a mass action-driven, fast process, whereas enzyme-catalyzed BiP modification (and demodification) is slower (Ham et al. 2014; Preissler et al. 2015a,b). Indeed, physiological changes of modified BiP levels occur over a few hours in the murine pancreas and correlate with the rates of secreted protein synthesis during alternating fasting and feeding intervals (Chambers et al. 2012). Therefore, it is plausible that both mechanisms operate side by side to regulate BiP availability on short (seconds to few minutes) and intermediate (several minutes to hours) timescales.
BiP AMPylation is especially prominent during the recovery after prolonged ER stress when, as a result of its long half-life, BiP levels remain elevated, while unfolded proteins decrease (Preissler et al. 2015b). BiP inactivation by modification restores ER functionality and secretory efficiency without degrading excess BiP. Indeed, the FICD gene itself is transcriptionally upregulated during the UPR (Ham et al. 2014; Preissler et al. 2015b; Sanyal et al. 2015), which presumably enhances the capacity for BiP inactivation upon ER stress resolution. Thus, posttranslational regulation of BiP activity provides a means to fine-tune the chaperone–client protein balance, obviating a costly and slow adjustment of BiP protein levels by de novo synthesis and degradation (Fig. 2B).
How is AMPylated BiP reactivated? Surprisingly, it was found that FICD is a bifunctional enzyme that uses the same active site both to modify BiP and to catalyze the hydrolytic removal of AMP from the chaperone (Casey et al. 2017; Preissler et al. 2017a). In fact, while the AMPylation activity of FICD is strongly autoinhibited (Engel et al. 2012; Bunney et al. 2014), the default activity is de-AMPylation (Casey et al. 2017; Preissler et al. 2017a). This implies that AMPylation of BiP in vivo requires a functional switch in FICD that is subordinate to changes in the unfolded protein load. The molecular basis of this switch remains to be uncovered.
THE EARLY STEPS IN UPR ACTIVATION
Downstream events in signaling by UPR transducers and the resultant changes in gene expression have been studied intensely (Karagöz et al. 2018). However, the upstream molecular mechanisms that monitor the balance between folding load and capacity in the ER are less well understood. By definition these processes constitute early events in the UPR.
Any information about proteostatic imbalance must ultimately converge on the signal transducers in the ER membrane and evoke a response proportional to the magnitude of stress. In principle, activation (or repression) of the signal transducers can involve the physical recognition of unfolded polypeptides or either free or occupied effector molecules that interact with the unfolded proteins (i.e., ERQC components such as chaperones).
The UPR signal transducers IRE1 and PERK are both ER-localized type I transmembrane proteins. They consist of a structurally (Credle et al. 2005; Zhou et al. 2006; Carrara et al. 2015a; Wang et al. 2018) and functionally related (Bertolotti et al. 2000; Liu et al. 2000) stress-sensing luminal domain (LD), a transmembrane domain, and a functionally divergent cytosolic kinase/effector domain. It is widely accepted that dimerization/oligomerization in the plane of the membrane, which brings together two or more effector domains, initiates IRE1 and PERK activation. However, the molecular principles that govern this dimerization remain unresolved with two models receiving experimental attention: activation by direct binding of unfolded proteins and chaperone inhibition. Much of the research in this area has been carried out on IRE1, which is conserved among all eukaryotes; however, it is assumed that the metazoan-restricted PERK functions similarly.
DIRECT ACTIVATION OF IRE1 BY UNFOLDED POLYPEPTIDES
The direct binding model was boosted by the crystal structure of the yeast IRE1 (yIRE1) LD. It revealed a symmetric dimer traversed by a groove with similarity to the peptide-binding groove of major histocompatibility complexes (MHCs) (Credle et al. 2005), suggesting that binding of extended polypeptides (i.e., unfolded proteins) stabilizes the LD dimer and favors IRE1 activation (Fig. 4A)—a model supported by the observation that mutations in residues that line the groove adversely affect the ability of yIRE1 to respond to stress in vivo (Credle et al. 2005) and by the observation that the yIRE1-LD possesses chaperone activity, indicative of its ability to bind unfolded proteins in vitro (Kimata et al. 2007). Indeed, the yIRE1-LD interacts with peptides in a ligand-blot assay and a binding motif could be extracted from analyzing their sequences (Gardner and Walter 2011). A solution-phase assay was developed for the highest affinity peptides yielding a dissociation rate in the micromolar range and addition of peptide to dilute solutions of yIRE1-LD enhanced the population of higher order species, although a clear shift from monomers to dimers, predicted by the engagement of peptide in MHC-like groove, was not readily apparent.
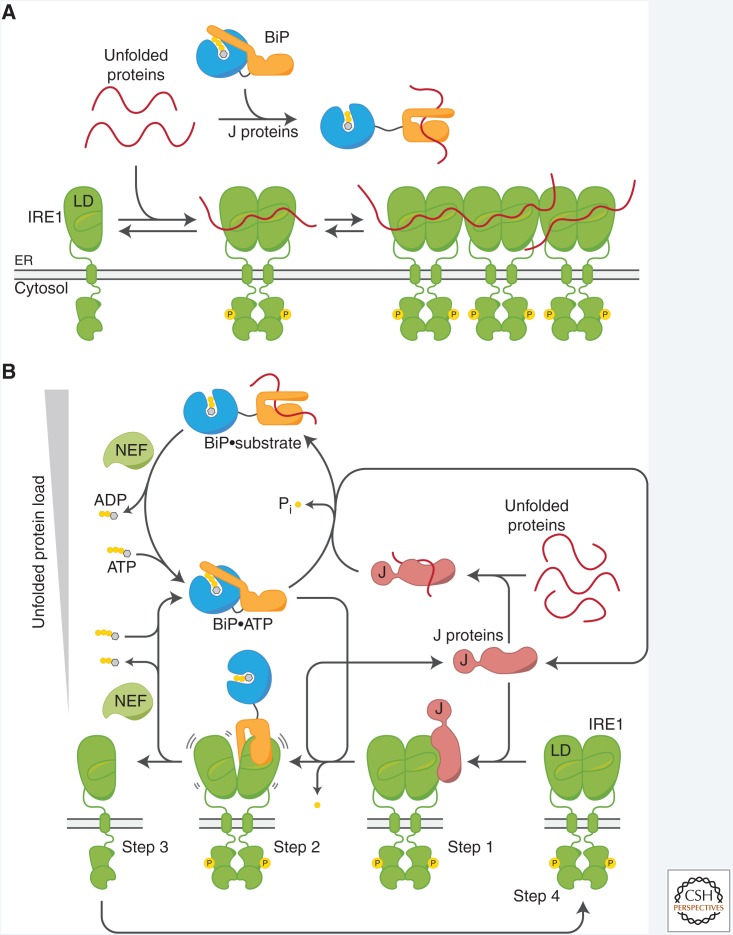
Figure 4.
Role of BiP in regulating IRE1. (A) Direct binding model. Accumulating unfolded proteins bind directly to a groove on the luminal domain (LD) of IRE1 to promote its dimerization/oligomerization and activation. In this model, sequestration of unfolded proteins by chaperones such as BiP counteracts their binding to IRE1, repressing the unfolded protein response (UPR) and establishing a measure for the balance between unfolded proteins and available chaperones. (B) Chaperone inhibition model. Step 1: Endoplasmic reticulum (ER)-localized J proteins (exemplified by ERdj4) associate with the LD of (dimeric) IRE1. Step 2: The J protein recruits BiP and stimulates its ATPase activity to promote a canonical substrate interaction between BiP and IRE1. Step 3: BiP binding triggers J protein release and disrupts IRE1 dimers, causing IRE1 inactivation. BiP dissociates from IRE1 upon nucleotide exchange. Step 4: Chaperone-free IRE1 has an intrinsic propensity to dimerize and become active. J protein–mediated loading of BiP onto IRE1 competes with loading of BiP onto unfolded substrates. The availability of BiP (and J proteins) establishes a dynamic pool of monomeric, inactive IRE1.
Human IRE1α-LD also crystalized as a dimer, with structural features similar to yIRE1 (Zhou et al. 2006). However, the MHC-like groove is less hydrophobic and appears too narrow to accommodate a peptide (although it was argued that flexibility in solution may allow peptide binding). Furthermore, the purified human IRE1α-LD does not possess chaperone activity in vitro (Kimata et al. 2007; Oikawa et al. 2009). Nevertheless, peptides have recently been identified that bind the human IRE1α-LD with micromolar affinity and a nuclear magnetic resonance (NMR) analysis revealed structural rearrangements to the IRE1α-LD following exposure to such peptides (Karagöz et al. 2017). Unfortunately, unlike MHC molecules that invariably co-crystalize with a bound peptide, no such structure of the IRE1-LD is available. Although the recent detailed NMR study did show peptide-induced perturbation of residues near the MHC-like groove, this cannot be unambiguously interpreted as peptide engagement inside the groove. Moreover, although the binding of peptide was apparently saturable, it is unclear whether it featured competitive binding at a single site, consistent with the MHC groove, or if the effects of high concentration of peptide observed in the NMR spectra of the IRE1α-LD arose through multiple, less specific interactions (Karagöz et al. 2017). Resolving this issue seems a paramount consideration for the direct binding model.
Unlike the chaperone inhibition model, described below, the direct binding model lacks an explicit kinetic component (beyond the association and dissociation parameters of peptide binding to the LD). Does peptide release from the LD, which is critical for the system to respond dynamically to changes in ER stress, occur passively or is it an assisted process requiring other agents? This seems especially important for peptides that associate cooperatively with several IRE1 molecules and are predicted to bind tightly.
REGULATION OF IRE1 BY CHAPERONES
The chaperone inhibition model for ER stress sensing is based on an inverse correlation between the amount of BiP recovered by coimmunoprecipitation with IRE1 and PERK and the level of ER stress (Bertolotti et al. 2000; Kimata et al. 2003, 2004). In the simplest scenario, the interaction between BiP and the LDs occurs by the same mechanism governing BiP's interaction with unfolded clients. The model posits that BiP association somehow favors the monomeric, inactive state of IRE1 (and PERK) and that competition for BiP by client proteins titrates BiP away from the LDs, favoring dimerization and activation. This model is analogous to the cytosolic counterpart of the UPR, the heat shock response, in which chaperones associate with the transcription factor Hsf1, in eukaryotes, and σ32, in bacteria, and interfere with their activity (Abravaya et al. 1992; Shi et al. 1998a; Tomoyasu et al. 1998).
Indeed, typical substrate-like interactions between BiP and IRE1 (i.e., via BiP's substrate-binding domain and with affinities dictated by its nucleotide-dependent conformational cycle) have been observed (Bertolotti et al. 2000; Kimata et al. 2003; Liu et al. 2003). A different type of interaction (involving BiP's ATPase domain) and a noncompetitive mechanism of BiP-mediated IRE1 regulation has also been proposed, according to which binding of unfolded proteins to LD-associated BiP allosterically triggers BiP dissociation (Todd-Corlett et al. 2007; Carrara et al. 2015b). However, this mechanism appears to operate independently of BiP's nucleotide state and its acceptance is marred by the difficulty of reconciling it to fundamental principles of nucleotide-dependent allosteric regulation of Hsp70-type chaperones.
Repression of Hsf1 by cytosolic Hsp70 and σ32 by bacterial Hsp70, DnaK, is directed by J-domain co-chaperones, Hdj1 and DnaJ, respectively (Shi et al. 1998a; Tomoyasu et al. 1998). The ER-localized J protein, ERdj4, was recently found to contribute to IRE1 repression in mammalian cells. In vitro characterization indicated that ERdj4 promotes substrate interactions between BiP and the IRE1α-LD that disrupt LD dimers (Amin-Wetzel et al. 2017). J proteins act as adaptors that facilitate high-affinity binding of Hsp70s to specific substrates by recruiting the chaperone in the ATP-bound state with high substrate association rates and converting it to the ADP state with low substrate dissociation rates by stimulation of ATP hydrolysis. Consistent with this canonical J protein function, ERdj4-mediated binding of BiP to IRE1 was strictly dependent on stimulation of BiP's ATPase activity. Furthermore, in the absence of BiP/ERdj4/ATP activity the IRE1-LD was mostly dimeric, a feature emphasized by early work from the Kaufman lab (Liu et al. 2003; Zhou et al. 2006). These observations imply a chaperone inhibition model whereby a nonequilibrium cycle of ERdj4-mediated, ATP hydrolysis-driven BiP–IRE1 complex formation and ATP-binding-induced BiP–IRE1 complex dissociation dynamically establishes a pool of inactive, monomeric IRE1 against its intrinsic propensity to dimerize (Fig. 4B).
Many details remain to be worked out: although ERdj4 can stimulate BiP binding to IRE1-LD dimers, it is unclear whether the target of binding is the dimeric or the monomeric species, which co-exist in some equilibrium. Likewise, if BiP binds the dimer, it is not known whether J protein–mediated loading of BiP onto IRE1 dimers causes their dissociation entropically or allosterically. The biophysical details, in terms of the structural changes to the IRE1-LD remain to be determined. They may resemble auxillin/Hsc70-driven disassembly of clathrin coats (Xing et al. 2010), or the Hsp40/Hsp70-mediated partial unfolding of the glucocorticoid receptor (Kirschke et al. 2014). The apparent parallels to repression of the bacterial heat shock response, which relies on destabilization and degradation of the σ32 transcription factor by J protein–mediated interaction with the Hsp70 chaperone DnaK (Rodriguez et al. 2008), suggest that mechanisms for sensing unfolded protein stress evolved several times independently exploiting the same functional principles of the highly conserved Hsp70 system.
BiP's concentration vastly exceeds the UPR transducers (Kim et al. 2014; Kulak et al. 2014); an observation seemingly at odds with the sensitivity of UPR signaling to small changes in client protein abundance. However, the fraction of ATP-bound free BiP, which can effectively respond to J proteins, is likely rather small (Freiden et al. 1992; Preissler et al. 2015a,b): most BiP molecules either engage substrates, are sequestered in oligomers, or are inactivated by AMPylation. Indeed, the full functional scope of BiP AMPylation may be revealed in the context of UPR regulation by chaperone inhibition: consistent with its inactivating character, modification of BiP abolishes ERdj4-dependent disruption of IRE1-LD dimers in vitro (Amin-Wetzel et al. 2017). J protein (e.g., ERdj4)–mediated loading onto IRE1 (leading to UPR repression) thus faces fierce competition for the small pool of active BiP by J proteins directing the formation of client–BiP complexes. The constant cycling of BiP onto and off the IRE1-LD provides a sensitive system that can respond rapidly and bidirectionally to changes in the unfolded protein load. The transient accumulation of AMPylated BiP during recovery from ER stress may thus not only counteract over-chaperoning of secretory proteins but also set the tone of the UPR to enable longer-term gene expression programs that build secretory capacity.
ERdj4's role in IRE1 regulation is partially redundant, as knockout cells retain a measure of responsiveness to ER stress (despite a higher basal level of IRE1 signaling). Recent studies implicated a physical interaction between mammalian IRE1α and Sec63 (ERdj2) in IRE1 signaling (Plumb et al. 2015; Adamson et al. 2016; Sundaram et al. 2017). Sec63 is a J protein that associates with the Sec61 translocon channel in all eukaryotes and loads BiP onto polypeptides to support their unidirectional import into the ER (Matlack et al. 1999; Misselwitz et al. 1999). By analogy with ERdj4, Sec63/ERdj2 may promote inhibitory IRE1–BiP complexes when translocation rates are low, to establish an antagonistic crosstalk between protein import and IRE1 repression.
It has been noted that yIRE1 mutants and human-yeast chimeras, that associate with less BiP in coimmunoprecipitation assays, were not constitutively activated and retained responsiveness to ER stress (Kimata et al. 2004; Pincus et al. 2010; Mai et al. 2018). In contrast, mutating the corresponding region in IRE1α (which also affects BiP association) strongly impairs repression in mammalian cells (Oikawa et al. 2009). However, we believe conclusions based on quantifying the association of BiP with IRE1 by coimmunoprecipitation must be drawn with caution. Such studies tend to ignore that most physiological BiP interactions are kinetically driven by its nucleotide-dependent chaperone cycle and thus highly dynamic and prone to postlysis-binding artifacts. The distinction between regulatory and nonregulatory interactions may be further confounded by association of BiP with nascent IRE1 when the latter is overexpressed.
These mechanisms proposed for IRE1 regulation are not mutually exclusive and, despite the overall structural conservation of IRE1-LD's architecture, the contribution of different mechanisms to IRE1 regulation may vary between organisms and cell types. In vitro, mammalian J protein–mediated chaperone inhibition targets the IRE1 monomer–dimer equilibrium to control dimerization and thus the initial step of IRE1 activation (Amin-Wetzel et al. 2017), while high concentration of unfolded proteins promotes further oligomerization (Karagöz et al. 2017). A similar two-pronged regulatory mechanism was also proposed in yeast (Kimata et al. 2007), but the relative contribution of BiP repression and unfolded protein binding remains to be established experimentally (Pincus et al. 2010).
TRANSLATIONAL REPRESSION AS AN EARLY RESPONSE TO ER STRESS
In animal cells, most secreted proteins are translocated into the ER cotranslationally as unfolded nascent polypeptides. It follows therefore that the rate of messenger RNA (mRNA) translation influences the burden of unfolded proteins in the ER. Concordantly, in animal cells a conserved mechanism is found that attenuates protein synthesis in response to ER stress. Its short latency and inherent reversibility qualify this adaptation as an early event in the UPR.
The PERK Arm of the UPR
The ER-localized transmembrane protein PERK is upstream of another UPR pathway found in metazoans. Though PERK's stress-sensing LD is only distantly related in primary sequence to IRE1, it assumes a very similar fold (Carrara et al. 2015a; Wang et al. 2018), suggesting that similar principles govern its activation. The cytosolic, effector domain of PERK resembles kinases that phosphorylate the α subunit of translation initiation factor 2α (eIF2α) on serine 51 (Shi et al. 1998b). This phosphorylation event negatively affects the rate at which translation is initiated on most mRNAs (Wek 2018). Thus, PERK couples ER stress to repression of global protein synthesis (Harding et al. 1999).
Such repression develops rapidly. Within minutes of exposing cells to agents that cause ER stress, PERK is autophosphorylated, levels of phosphorylated eIF2α increase, and global rates of protein synthesis decline. PERK has a nonredundant role in this chain of events. Mammalian cells that lack PERK (Harding et al. 2000b) or cells treated with the potent PERK kinase inhibitor, GSK2606414 (Axten et al. 2012), maintain normal levels of protein synthesis in the face of ER stress. PERK's role in modulating the rate of protein synthesis is observed not only under severe, pharmacologically imposed ER stress, but also in the context of physiological signals that affect the rate of ER client protein synthesis. In cultured insulin-secreting β cells, acute inhibition of PERK signaling deregulates metabolically entrained proinsulin biosynthesis (Harding et al. 2012) and in the tissues of PERK-deficient mice heightened activity of the parallel IRE1 pathway was observed, suggesting loss of important feedback (Fig. 1A; Harding et al. 2001).
In animal cells, IRE1's RNAse activity is not limited to the XBP1 mRNA. Rather IRE1 has the capacity to degrade other ER membrane-associated mRNAs in a process known as Ire1-dependent decay (RIDD) (Fig. 1A; Hollien and Weissman 2006; Hollien et al. 2009). The quantitative impact of RIDD on protein-folding homeostasis in the ER remains to be determined. Confounding this issue is the observation that PERK-mediated translational repression contributes to RIDD (Moore and Hollien 2015). Thus, the roles of PERK-mediated eIF2α phosphorylation and IRE1-mediated RIDD cannot be readily apportioned by genetic criteria. Similar considerations apply to other processes that may influence ER client protein synthesis, such as the observed ER stress-induced mRNA dissociation from the ER (Reid et al. 2014).
These observations are consistent with a PERK-imposed restraint on the rate of ER client protein synthesis, contributing to ER protein-folding homeostasis under physiological circumstances. PERK deficiency, which in mammals is associated with rapid decline in the function of major secretory tissues (Delepine et al. 2000; Harding et al. 2001; Zhang et al. 2002), may be understood to exert its consequences through the disruption of this short-term adaptation to ER stress. However, this might be an oversimplification (Cavener et al. 2010).
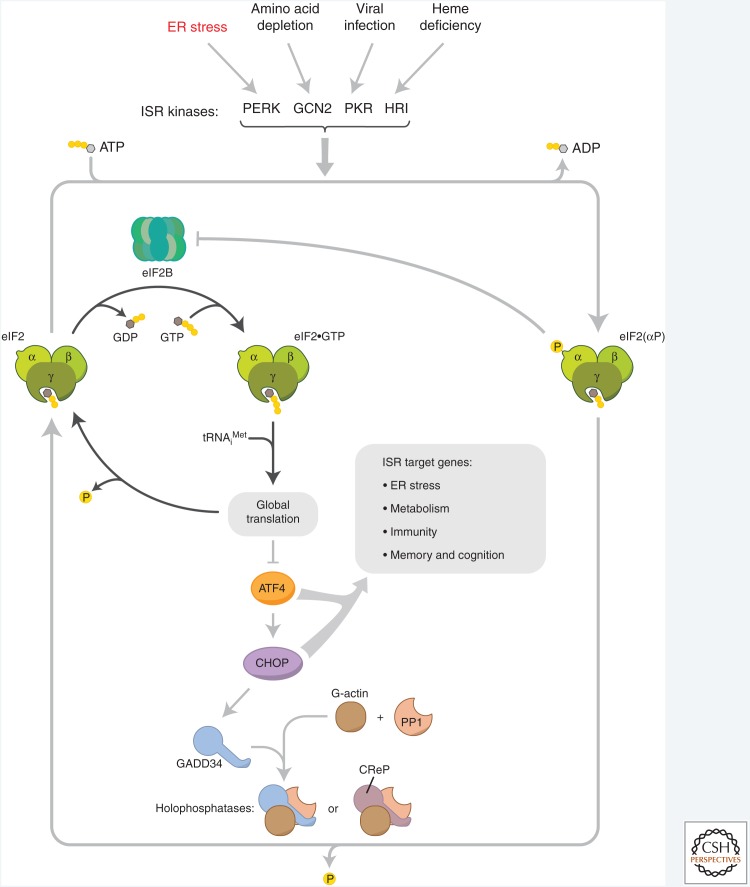
Phosphorylated eIF2 (eIF2(αP)) attenuates the initiation step in mRNA translation indirectly by inhibiting eIF2B, the guanine nucleotide exchange factor (GEF) that recycles eIF2 from its inactive GDP bound to its active GTP-bound form (Fig. 5; Panniers and Henshaw 1983). PERK and other eIF2α kinases limit the stock of active eIF2 in the cell, attenuating the translation of most mRNAs and, with it, global protein synthesis. However, a small subset of mRNAs with specialized features in their 5′ untranslated region are translated more efficiently in a regime of limited eIF2. This phenomenon, originally described in yeast (Abastado et al. 1991; Dever et al. 1992), is conserved in animal cells (Harding et al. 2000a; Lu et al. 2004; Vattem and Wek 2004). Some of the mRNAs implicated in this divergent response encode potent transcription factors that couple eIF2α phosphorylation to a gene expression program. As eIF2α is phosphorylated under diverse conditions, this program integrates otherwise unrelated stress signals giving rise to an integrated stress response (ISR) (Harding et al. 2003). The ISR's target genes affect the capacity to synthesize and secrete proteins, as well as important cell fate decisions (Pakos-Zebrucka et al. 2016).
Figure 5.
The integrated stress response (ISR). Translational control over client protein load is imposed by the endoplasmic reticulum (ER) stress-mediated PERK activation and phosphorylation of eIF2 on its α subunit. This figure highlights the inhibition the guanine nucleotide exchange factor (GEF), eIF2B, by phosphorylated eIF2 and the effects on messenger RNA (mRNA) translation of the resultant decline in availability of eIF2•GTP•tRNAiMet ternary complexes. Also cartooned are the phosphatase complexes that dephosphorylate eIF2 to terminate signaling in the ISR. Targeting this phosphatase complexes for pharmacological inhibition has emerged as a potentially useful way to combat certain diseases of protein misfolding, by enhancing ISR activity.
In theory, it should be possible to separate PERK's early and direct role in protein-folding homeostasis from its effects on ISR target genes by enforced expression of ATF4, CHOP, and other ISR-activating transcription factors. However, mimicking the dosage and temporal profile of a physiological ISR, required for such genetic rescue, is impractical. Therefore, at present, we cannot gauge the relative contribution of these two linked aspects of PERK action to its role in preserving the function of secretory cells.
Dephosphorylation of eIF2(αP)
PERK-mediated translational repression, as a short-term adaptation to the threat of protein misfolding in the ER, is counteracted by eIF2(αP) dephosphorylation. In mammals, this process is known to be directed by two structurally related regulatory subunits of a PP1-containing holophosphatase. PPP1R15A/GADD34 is expressed at low levels but is rapidly upregulated by the ISR, closing a negative feedback loop that contributes to the restoration of protein synthesis (Novoa et al. 2001, 2003; Brush et al. 2003), whereas PPP1R15B/CReP is constitutively present (Jousse et al. 2003).
Although combined deficiency of both regulatory subunits severely compromises cells, lack of the inducible PPP1R15A is well tolerated (Harding et al. 2009). Furthermore, cells and tissues of mice lacking PPP1R15A exhibit a measure of resistance to certain circumstances associated with higher levels of ER stress (Marciniak et al. 2004; Lin et al. 2008; D'Antonio et al. 2013). These findings speak to a failure of homeostasis, whereby the enhanced ability of the PPP1R15A-posessing (wild-type) cells to reverse the consequences of PERK activation, places them at a disadvantage compared to the PPP1R15A mutant cells, when both are exposed to the same challenge. It is tempting to interpret the consequences of eliminating PPP1R15A solely in light of its effect on protein synthesis and thus on proteostasis (Marciniak et al. 2004; Han et al. 2013); however, here too we must be reminded that the consequences of altered levels of eIF2(αP) are played out both in terms of its short-term effect on unfolded protein load, and also in terms of gene expression.
Either way, the benefit of attenuated PPP1R15A activity to certain mouse models of human diseases associated with ER stress has engendered interest in the details of eIF2(αP) dephosphorylation and in the prospects of targeting the process for inhibition. The crystal structure of the complex between the carboxyl terminus of the PPP1R15 regulatory subunits and catalytic PP1 subunit explains their tight binding, but not the selectivity for eIF2(αP) (Chen et al. 2015; Choy et al. 2015). Further observations indicate that substrate specificity is imparted by G-actin joining the PPP1R15–PP1 complex as a third component (Chambers et al. 2015; Chen et al. 2015). However, a recent study reported on dramatic acceleration of eIF2(αP) dephosphorylation by either regulatory subunit (in the absence of G-actin) and on the selective susceptibility of the PPP1R15A–PP1 complex to inhibition by the drug Guanabenz and its close derivative, Sephin1 (Carrara et al. 2017). The latter biochemical observations have been greeted enthusiastically, as they suggest the possibility of tuning ER proteostasis by targeting the rate of eIF2(αP) dephosphorylation (Das et al. 2015). Unfortunately, the proposed inhibitory effect of Guanabenz/Sephin1 on eIF2(αP) dephosphorylation in vitro or in cultured cells could not be confirmed in subsequent studies (Crespillo-Casado et al. 2017, 2018). Thus, the promise of targeting the PPP1R15 holophosphatase to restore proteostasis by affecting the early adaptation to ER stress likely has yet to be realized.
CONCLUDING REMARKS
This review has focused on early events in the UPR that do not require new macromolecule synthesis. However, as they act upstream of the extensively studied changes in gene expression and cellular phenotypes brought about by physiological and pathological alterations in protein-folding homeostasis, they stand to influence the organismal response and the implications of the UPR in a variety of disease conditions (Karagöz et al. 2018).
These early events seem especially important when considering the prospects of pharmacologic modulation of the UPR. The aforementioned attempts to target eIF2(αP) dephosphorylation are an interesting case in point, as it seems likely that further progress will depend on assays that reproduce the relevant enzymatic reactions in vitro. But it is also possible to consider how small molecules that shift the equilibrium between LD monomers and dimers might affect signaling in the IRE1 or PERK branches of the UPR. Such small molecules may be discovered by direct biophysical assays that exploit the recent in vitro models for IRE1 activity. Thus, the question of how the UPR is activated and therefore the validity of the in vitro assays that aim to recapitulate relevant aspects of that process may assume practical significance, beyond the curiosity of scholars.
ACKNOWLEDGMENTS
We thank Niko Amin-Wetzel for comments on the manuscript. This work was supported by a Wellcome Trust Principal Research Fellowship to D.R. (Wellcome 200848/Z/16/Z) and a Wellcome Trust Strategic Award to the Cambridge Institute for Medical Research (Wellcome 100140).
Footnotes
Editors: Richard I. Morimoto, F. Ulrich Hartl, and Jeffery W. Kelly
Additional Perspectives on Protein Homeostasis available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abastado JP, Miller PF, Jackson BM, Hinnebusch AG. 1991. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol Cell Biol 11: 486–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abravaya K, Myers MP, Murphy SP, Morimoto RI. 1992. The human heat shock protein hsp70 interacts with HSF, the transcription factor that regulates heat shock gene expression. Genes Dev 6: 1153–1164. [DOI] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nunez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. 2016. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167: 1867–1882, e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin-Wetzel N, Saunders RA, Kamphuis MJ, Rato C, Preissler S, Harding HP, Ron D. 2017. A J-protein co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell 171: 1625–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, et al. 2012. Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem 55: 7193–7207. [DOI] [PubMed] [Google Scholar]
- Bakunts A, Orsi A, Vitale M, Cattaneo A, Lari F, Tade L, Sitia R, Raimondi A, Bachi A, van Anken E. 2017. Ratiometric sensing of BiP-client versus BiP levels by the unfolded protein response determines its signaling amplitude. eLife 6: e27518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke J, Feige MJ, Hendershot LM. 2015. BiP and its nucleotide exchange factors Grp170 and Sil1: Mechanisms of action and biological functions. J Mol Biol 427: 1589–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotti A, Zhang Y, Hendershot L, Harding H, Ron D. 2000. Dynamic interaction of BiP and the ER stress transducers in the unfolded protein response. Nat Cell Biol 2: 326–332. [DOI] [PubMed] [Google Scholar]
- Blond-Elguindi S, Fourie AM, Sambrook JF, Gething MJ. 1993. Peptide-dependent stimulation of the ATPase activity of the molecular chaperone BiP is the result of conversion of oligomers to active monomers. J Biol Chem 268: 12730–12735. [PubMed] [Google Scholar]
- Brush MH, Weiser DC, Shenolikar S. 2003. Growth arrest and DNA damage-inducible protein GADD34 targets protein phosphatase 1α to the endoplasmic reticulum and promotes dephosphorylation of the α subunit of eukaryotic translation initiation factor 2. Mol Cell Biol 23: 1292–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunney TD, Cole AR, Broncel M, Esposito D, Tate EW, Katan M. 2014. Crystal structure of the human, FIC-domain containing protein HYPE and implications for its functions. Structure 22: 1831–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlino A, Toledo H, Skaleris D, DeLisio R, Weissbach H, Brot N. 1992. Interactions of liver Grp78 and Escherichia coli recombinant Grp78 with ATP: Multiple species and disaggregation. Proc Natl Acad Sci 89: 2081–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson L, Lazarides E. 1983. ADP-ribosylation of the Mr 83,000 stress-inducible and glucose-regulated protein in avian and mammalian cells: Modulation by heat shock and glucose starvation. Proc Natl Acad Sci 80: 4664–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Ali MM. 2015a. Crystal structures reveal transient PERK luminal domain tetramerization in endoplasmic reticulum stress signaling. EMBO J 34: 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Prischi F, Nowak PR, Kopp MC, Ali MM. 2015b. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. eLife 4: 03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrara M, Sigurdardottir A, Bertolotti A. 2017. Decoding the selectivity of eIF2α holophosphatases and PPP1R15A inhibitors. Nat Struct Mol Biol 24: 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey AK, Moehlman AT, Zhang J, Servage KA, Kramer H, Orth K. 2017. Fic-mediated deAMPylation is not dependent on homo-dimerization and rescues toxic AMPylation in flies. J Biol Chem 292: 21193–21204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavener DR, Gupta S, McGrath BC. 2010. PERK in β cell biology and insulin biogenesis. Trends Endocrinol Metab 21: 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Petrova K, Tomba G, Vendruscolo M, Ron D. 2012. ADP ribosylation adapts an ER chaperone response to short-term fluctuations in unfolded protein load. J Cell Biol 198: 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers JE, Dalton LE, Clarke HJ, Malzer E, Dominicus CS, Patel V, Moorhead G, Ron D, Marciniak SJ. 2015. Actin dynamics tune the integrated stress response by regulating eukaryotic initiation factor 2α dephosphorylation. eLife 4: 04872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Rato C, Yan Y, Crespillo-Casado A, Clarke HJ, Harding HP, Marciniak SJ, Read RJ, Ron D. 2015. G-actin provides substrate-specificity to eukaryotic initiation factor 2α holophosphatases. eLife 4: e04871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy MS, Yusoff P, Lee IC, Newton JC, Goh CW, Page R, Shenolikar S, Peti W. 2015. Structural and functional analysis of the GADD34:PP1 eIF2α phosphatase. Cell Rep 11: 1885–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JS, Walter P. 1996. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87: 391–404. [DOI] [PubMed] [Google Scholar]
- Cox JS, Chapman RE, Walter P. 1997. The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8: 1805–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. 2005. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc Natl Acad Sci 102: 18773–18784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespillo-Casado A, Chambers JE, Fischer PM, Marciniak SJ, Ron D. 2017. PPP1R15A-mediated dephosphorylation of eIF2α is unaffected by Sephin1 or Guanabenz. eLife 6: e26109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespillo-Casado A, Claes Z, Choy MS, Peti W, Bollen M, Ron D. 2018. A Sephin1-insensitive tripartite holophosphatase dephosphorylates translation initiation factor 2α. J Biol Chem 293: 7766–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Antonio M, Musner N, Scapin C, Ungaro D, Del Carro U, Ron D, Feltri ML, Wrabetz L. 2013. Resetting translational homeostasis restores myelination in Charcot–Marie–Tooth disease type 1B mice. J Exp Med 210: 821–838. [Google Scholar]
- Das I, Krzyzosiak A, Schneider K, Wrabetz L, D'Antonio M, Barry N, Sigurdardottir A, Bertolotti A. 2015. Preventing proteostasis diseases by selective inhibition of a phosphatase regulatory subunit. Science 348: 239–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. 2000. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott–Rallison syndrome. Nat Genet 25: 406–409. [DOI] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. 1992. Phosphorylation of initiation factor 2α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596. [DOI] [PubMed] [Google Scholar]
- Dorner A, Wasley L, Kaufman R. 1992. Overexpression of GRP78 mitigates stress induction of glucose regulated proteins and blocks secretion of selective proteins in Chinese hamster ovary cells. EMBO J 11: 1563–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P, Goepfert A, Stanger FV, Harms A, Schmidt A, Schirmer T, Dehio C. 2012. Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482: 107–110. [DOI] [PubMed] [Google Scholar]
- Freiden PJ, Gaut JR, Hendershot LM. 1992. Interconversion of three differentially modified and assembled forms of BiP. EMBO J 11: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner BM, Walter P. 2011. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333: 1891–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham H, Woolery AR, Tracy C, Stenesen D, Kramer H, Orth K. 2014. Unfolded protein response-regulated Drosophila Fic (dFic) protein reversibly AMPylates BiP chaperone during endoplasmic reticulum homeostasis. J Biol Chem 289: 36059–36069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Back SH, Hur J, Lin YH, Gildersleeve R, Shan J, Yuan CL, Krokowski D, Wang S, Hatzoglou M, et al. 2013. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat Cell Biol 15: 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H, Zhang Y, Ron D. 1999. Translation and protein folding are coupled by an endoplasmic reticulum resident kinase. Nature 397: 271–274. [DOI] [PubMed] [Google Scholar]
- Harding H, Novoa I, Zhang Y, Zeng H, Wek RC, Schapira M, Ron D. 2000a. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6: 1099–1108. [DOI] [PubMed] [Google Scholar]
- Harding H, Zhang Y, Bertolotti A, Zeng H, Ron D. 2000b. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5: 897–904. [DOI] [PubMed] [Google Scholar]
- Harding H, Zeng H, Zhang Y, Jungreis R, Chung P, Plesken H, Sabatini D, Ron D. 2001. Diabetes mellitus and exocrine pancreatic dysfunction in Perk−/− mice reveals a role for translational control in survival of secretory cells. Mol Cell 7: 1153–1163. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. 2003. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 11: 619–633. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Scheuner D, Chen JJ, Kaufman RJ, Ron D. 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2α (eIF2α) dephosphorylation in mammalian development. Proc Natl Acad Sci 106: 1832–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding HP, Zyryanova AF, Ron D. 2012. Uncoupling proteostasis and development in vitro with a small molecule inhibitor of the pancreatic endoplasmic reticulum kinase, PERK. J Biol Chem 287: 44338–44344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot LM, Ting J, Lee AS. 1988. Identity of the immunoglobulin heavy-chain-binding protein with the 78,000-dalton glucose-regulated protein and the role of posttranslational modifications in its binding function. Mol Cell Biol 8: 4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. 2006. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 313: 104–107. [DOI] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol 186: 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Toyama T, Nakamura Y, Tamada K, Shimizu H, Ninagawa S, Okada T, Kamei Y, Ishikawa-Fujiwara T, Todo T, et al. 2017. UPR transducer BBF2H7 allows export of type II collagen in a cargo- and developmental stage-specific manner. J Cell Biol 216: 1761–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Okamoto H. 1980. Translational control of proinsulin synthesis by glucose. Nature 283: 100–102. [DOI] [PubMed] [Google Scholar]
- Jousse C, Oyadomari S, Novoa I, Lu P, Zhang Y, Harding HP, Ron D. 2003. Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J Cell Biol 163: 767–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz GE, Acosta-Alvear D, Nguyen HT, Lee CP, Chu F, Walter P. 2017. An unfolded protein-induced conformational switch activates mammalian IRE1. eLife 6: e30700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Karagöz GE, Acosta-Alvear D, Walter P. 2018. The unfolded protein response: Detecting and responding to fluctuations in the protein folding capacity of the endoplasmic reticulum. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. 2014. A draft map of the human proteome. Nature 509: 575–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Kimata YI, Shimizu Y, Abe H, Farcasanu IC, Takeuchi M, Rose MD, Kohno K. 2003. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol Biol Cell 14: 2559–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Oikawa D, Shimizu Y, Ishiwata-Kimata Y, Kohno K. 2004. A role for BiP as an adjustor for the endoplasmic reticulum stress-sensing protein Ire1. J Cell Biol 167: 445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Ishiwata-Kimata Y, Ito T, Hirata A, Suzuki T, Oikawa D, Takeuchi M, Kohno K. 2007. Two regulatory steps of ER-stress sensor Ire1 involving its cluster formation and interaction with unfolded proteins. J Cell Biol 179: 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke E, Goswami D, Southworth D, Griffin PR, Agard DA. 2014. Glucocorticoid receptor function regulated by coordinated action of the Hsp90 and Hsp70 chaperone cycles. Cell 157: 1685–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozutsumi Y, Segal M, Normington K, Gething MJ, Sambrook J. 1988. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332: 462–464. [DOI] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324. [DOI] [PubMed] [Google Scholar]
- Laitusis AL, Brostrom MA, Brostrom CO. 1999. The dynamic role of GRP78/BiP in the coordination of mRNA translation with protein processing. J Biol Chem 274: 486–493. [DOI] [PubMed] [Google Scholar]
- Lau E, Cao Q, Ng DC, Bleakley BJ, Dincer TU, Bot BM, Wang D, Liem DA, Lam MP, Ge J, et al. 2016. A large dataset of protein dynamics in the mammalian heart proteome. Sci Data 3: 160015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford BE, Jacobs DF. 1986. Translational control of ADP-ribosylation in eukaryotic cells. Eur J Biochem 161: 661–667. [DOI] [PubMed] [Google Scholar]
- Lin W, Kunkler PE, Harding HP, Ron D, Kraig RP, Popko B. 2008. Enhanced integrated stress response promotes myelinating oligodendrocyte survival in response to interferon-γ. Am J Pathol 173: 1508–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CY, Schroder M, Kaufman RJ. 2000. Ligand-independent dimerization activates the stress-response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem 275: 24881–24885. [DOI] [PubMed] [Google Scholar]
- Liu CY, Xu Z, Kaufman RJ. 2003. Structure and intermolecular interactions of the luminal dimerization domain of human IRE1α. J Biol Chem 278: 17680–17687. [DOI] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. 2004. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol 167: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai TC, Munakata T, Tran DM, Takagi H, Kimata Y. 2018. A chimeric mutant analysis in yeast cells suggests BiP independent regulation of the mammalian endoplasmic reticulum-stress sensor IRE1α. Biosci Biotechnol Biochem 82: 1527–1530. [DOI] [PubMed] [Google Scholar]
- Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, Jungreis R, Nagata K, Harding HP, Ron D. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18: 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack KE, Misselwitz B, Plath K, Rapoport TA. 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-α factor across the ER membrane. Cell 97: 553–564. [DOI] [PubMed] [Google Scholar]
- Misselwitz B, Staeck O, Matlack KE, Rapoport TA. 1999. Interaction of BiP with the J-domain of the Sec63p component of the endoplasmic reticulum protein translocation complex. J Biol Chem 274: 20110–20115. [DOI] [PubMed] [Google Scholar]
- Moore K, Hollien J. 2015. Ire1-mediated decay in mammalian cells relies on mRNA sequence, structure, and translational status. Mol Biol Cell 26: 2873–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zeng H, Harding H, Ron D. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2a. J Cell Biol 153: 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novoa I, Zhang Y, Zeng H, Jungreis R, Harding HP, Ron D. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J 22: 1180–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa D, Kimata Y, Kohno K, Iwawaki T. 2009. Activation of mammalian IRE1α upon ER stress depends on dissociation of BiP rather than on direct interaction with unfolded proteins. Exp Cell Res 315: 2496–2504. [DOI] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. 2016. The integrated stress response. EMBO Rep 17: 1374–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panniers R, Henshaw EC. 1983. A GDP/GTP exchange factor essential for eukaryotic initiation factor 2 cycling in Ehrlich ascites tumor cells and its regulation by eukaryotic initiation factor 2 phosphorylation. J Biol Chem 258: 7928–7934. [PubMed] [Google Scholar]
- Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, El-Samad H, Walter P. 2010. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb R, Zhang ZR, Appathurai S, Mariappan M. 2015. A functional link between the co-translational protein translocation pathway and the UPR. eLife 4: e07426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Chambers JE, Crespillo-Casado A, Avezov E, Miranda E, Perez J, Hendershot LM, Harding HP, Ron D. 2015a. Physiological modulation of BiP activity by trans-protomer engagement of the interdomain linker. eLife 4: e08961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rato C, Chen R, Antrobus R, Ding S, Fearnley IM, Ron D. 2015b. AMPylation matches BiP activity to client protein load in the endoplasmic reticulum. eLife 4: e12621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rato C, Perera LA, Saudek V, Ron D. 2017a. FICD acts bifunctionally to AMPylate and de-AMPylate the endoplasmic reticulum chaperone BiP. Nat Struct Mol Biol 24: 23–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Rohland L, Yan Y, Chen R, Read RJ, Ron D. 2017b. AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation. eLife 6: e29428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. 2010. Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci 107: 14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid DW, Chen Q, Tay AS, Shenolikar S, Nicchitta CV. 2014. The unfolded protein response triggers selective mRNA release from the endoplasmic reticulum. Cell 158: 1362–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412: 300–307. [DOI] [PubMed] [Google Scholar]
- Rodriguez F, Arsene-Ploetze F, Rist W, Rudiger S, Schneider-Mergener J, Mayer MP, Bukau B. 2008. Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones. Mol Cell 32: 347–358. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Chen AJ, Nakayasu ES, Lazar CS, Zbornik EA, Worby CA, Koller A, Mattoo S. 2015. A novel link between Fic (filamentation induced by cAMP)-mediated adenylylation/AMPylation and the unfolded protein response. J Biol Chem 290: 8482–8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Nakai A, Sokawa Y, Hirayoshi K, Nagata K. 1993. Modulation of the phosphorylation of glucose-regulated protein, GRP78, by transformation and inhibition of glycosylation. Exp Cell Res 205: 76–83. [DOI] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. 1998a. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev 12: 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Vattem KM, Sood R, An J, Liang J, Stramm L, Wek RC. 1998b. Identification and characterization of pancreatic eukaryotic initiation factor 2α-subunit kinase, PEK, involved in translational control. Mol Cell Biol 18: 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram A, Plumb R, Appathurai S, Mariappan M. 2017. The Sec61 translocon limits IRE1α signaling during the unfolded protein response. eLife 6: e27187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd-Corlett A, Jones E, Seghers C, Gething MJ. 2007. Lobe IB of the ATPase domain of Kar2p/BiP interacts with Ire1p to negatively regulate the unfolded protein response in Saccharomyces cerevisiae. J Mol Biol 367: 770–787. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Ogura T, Tatsuta T, Bukau B. 1998. Levels of DnaK and DnaJ provide tight control of heat shock gene expression and protein repair in Escherichia coli. Mol Microbiol 30: 567–581. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258. [DOI] [PubMed] [Google Scholar]
- Vattem KM, Wek RC. 2004. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci 101: 11269–11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Li J, Tao J, Sha B. 2018. The luminal domain of the ER stress sensor protein PERK binds misfolded proteins and thereby triggers PERK oligomerization. J Biol Chem 293: 4110–4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol 10: a32870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieteska L, Shahidi S, Zhuravleva A. 2017. Allosteric fine-tuning of the conformational equilibrium poises the chaperone BiP for post-translational regulation. eLife 6: e29430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Bocking T, Wolf M, Grigorieff N, Kirchhausen T, Harrison SC. 2010. Structure of clathrin coat with bound Hsc70 and auxilin: Mechanism of Hsc70-facilitated disassembly. EMBO J 29: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. 2002. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol 22: 3864–3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu CY, Back SH, Clark RL, Peisach D, Xu Z, Kaufman RJ. 2006. The crystal structure of human IRE1 luminal domain reveals a conserved dimerization interface required for activation of the unfolded protein response. Proc Natl Acad Sci 103: 14343–14348. [DOI] [PMC free article] [PubMed] [Google Scholar]