Abstract
Background
Osteoblasts participating in the inflammation regulation gradually obtain concerns. However, its role in joint inflammation of rheumatoid arthritis (RA) is largely unknown. Here, we investigated the role of osteoblastic pleckstrin homology domain-containing family O member 1 (PLEKHO1), a negative regulator of osteogenic lineage activity, in regulating joint inflammation in RA.
Methods
The level of osteoblastic PLEKHO1 in RA patients and collagen-induced arthritis (CIA) mice was examined. The role of osteoblastic PLEKHO1 in joint inflammation was evaluated by a CIA model and a K/BxN serum-transfer arthritis (STA) model which were induced in osteoblast-specific Plekho1 conditional knockout mice and mice expressing high Plekho1 exclusively in osteoblasts, respectively. The effect of osteoblastic PLEKHO1 inhibition was explored in a CIA mice model and a non-human primate arthritis model. The mechanism of osteoblastic PLEKHO1 in regulating joint inflammation were performed by a series of in vitro studies.
Results
PLEKHO1 was highly expressed in osteoblasts from RA patients and CIA mice. Osteoblastic Plekho1 deletion ameliorated joint inflammation, whereas overexpressing Plekho1 only within osteoblasts exacerbated local inflammation in CIA mice and STA mice. PLEKHO1 was required for TRAF2-mediated RIP1 ubiquitination to activate NF-κB for inducing inflammatory cytokines production in osteoblasts. Moreover, osteoblastic PLEKHO1 inhibition diminished joint inflammation and promoted bone formation in CIA mice and non-human primate arthritis model.
Conclusions
These data strongly suggest that the highly expressed PLEKHO1 in osteoblasts contributes to joint inflammation in RA. Targeting osteoblastic PLEKHO1 may exert dual therapeutic action of alleviating joint inflammation and promoting bone formation in RA.
Keywords: Osteoblast, PLEKHO1, Rheumatoid arthritis, Inflammation, Bone formation
Research in context.
Evidence before this study
Osteoblasts are well-known cell types to modulate bone formation. Recent evidence suggested that osteoblasts could produce proinflammatory molecules in response to several challenges. PLEKHO1 is a scaffold protein mediating the interactions with various proteins in multiple signaling pathways, including the osteogenic bone morphogenetic protein signaling pathway and the inflammatory related pathways. Our previous studies indicated that targeting PLEKHO1 in osteoblasts could promote bone formation in osteoporotic rodents.
Added value of this study
Currently, little is known whether osteoblasts contribute to the severity of the local inflammatory process in rheumatoid arthritis (RA), and what molecule event is involved in this progression. Our studies found that PLEKHO1 was highly expressed in osteoblasts of articular specimens from RA patients, and these highly expressed PLEKHO1 in osteoblast mediated joint inflammation in RA. Moreover, we elucidated that PLEKHO1 was required for TRAF2-mediated RIP1 ubiquitination to activate NF-κB for inducing inflammatory cytokines production in osteoblasts. Early inhibition of osteoblastic PLEKHO1 could diminish joint inflammation and promote bone formation in collagen-induced arthritis animal models.
Implications of all the available evidence
Our studies uncovered a novel role of osteoblastic PLEKHO1 in regulating joint inflammation in RA. Targeting osteoblastic PLEKHO1 could serve as a new pharmacological target with dual action of inflammation inhibition and bone formation augmentation in RA.
Alt-text: Unlabelled Box
1. Introduction
Rheumatoid arthritis (RA) is characterized by chronic inflammation and progressive bone destruction [1,2]. Osteoblasts are well-known cell types to modulate bone formation. Interestingly, increasing line of evidence suggests that osteoblasts could not only produce proinflammatory molecules in response to bacterial challenge but also contribute to inflammation through the recruitment of leukocytes to the sites of infection during osteomyelitis [3,4]. However, whether osteoblasts contribute to local joint inflammation in RA progression is still unknown.
Pleckstrin homology domain-containing family O member 1 (PLEKHO1, also known as casein kinase-2 interacting protein-1 (CKIP-1)) is a scaffold protein mediating the interactions with various proteins in multiple signaling pathways, including the osteogenic bone morphogenetic protein (BMP) signaling pathway and the inflammatory phosphoinositide 3-kinase (PI3K)-Akt pathways [5,6]. Accordingly, the Plekho1 systemic knock out mice showed the higher bone mass than their wide-type controls, indicating that PLEKHO1 may play a role in regulating bone formation [7]. In our previous studies, we further demonstrated that loss of PLEKHO1 in osteoblasts alleviated the age-related bone formation reduction, and osteoblast-targeted Plekho1 siRNA inhibition could promote bone formation in aging and osteoporotic rodents [8,9]. On the other hand, several recent studies have reported that PLEKHO1 was also involved in cytokine signaling in response to inflammation [10,11]. However, it remains unclear whether PLEKHO1 contributes to the local joint inflammation and reduction of bone formation in RA pathogenesis.
In this study, we found that PLEKHO1 was highly expressed in osteoblasts of joint bone specimens from RA patients and collagen-induced arthritis (CIA) mice. By genetic approach, we found that deletion of osteoblastic Plekho1 resulted in remarkable amelioration of local inflammation in CIA mice and K/BxN serum-transfer arthritis (STA) mice, whereas genetic overexpression of Plekho1 only in osteoblasts of CIA mice and STA mice dramatically exacerbated joint inflammation. More importantly, we elucidated that PLEKHO1 was required for TRAF2-mediated RIP1 ubiquitination to activate NF-κB for inducing inflammatory cytokines production in osteoblasts. Finally, we also found that early inhibition of osteoblastic PLEKHO1 diminished joint inflammation and promoted bone formation in CIA mice and the non-human primate arthritis model.
Together, this study uncovers a novel role of osteoblastic PLEKHO1 in joint inflammation regulation during RA. Targeting osteoblastic PLEKHO1 could serve as a new pharmacological target with dual action of inflammation inhibition and bone formation augmentation in RA.
2. Methods
2.1. Human joint bone tissue preparation
Tibial plateau samples from knee joints of subjects with RA (fulfilled the 1987 revised American College of Rheumatology criteria) were obtained at knee joint replacement surgery and provided by Shanghai Guanghua Hospital (China). Subjects with malignancy, diabetes or other severe diseases in the previous 5 years were excluded from our study. For non-RA control, we selected age- and gender-matched subjects who were diagnosed as severe trauma (TM) and underwent knee joint replacement surgery in Shanghai Guanghua Hospital and Shenzhen 8th People Hospital (China). Half of the bone samples were fixed with 4% buffered formalin before decalcification, the other half were stored in liquid nitrogen until use. All the clinical procedures were approved by the Committees of Clinical Ethics in the Shanghai Guanghua Hospital and Shenzhen 8th People Hospital. All subjects gave informed consent prior to surgery.
2.2. Cells
Murine osteoblastic cell line, MC3T3E1, was purchased from the Cell Resource Center, Peking Union Medical College (China) and cultured in α-MEM (Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS, Thermo Fisher Scientific) at 37°C in a humidified 5% CO2 incubator. Human osteoblast–like cell line, MG-63, was purchased from ATCC and cultured in EMEM (Thermo Fisher Scientific) containing 10% FBS at 37°C in a humidified 5% CO2 incubator. Human embryonic kidney HEK293T cell was purchased from ATCC and cultured in DMEM (Thermo Fisher Scientific) containing 10% FBS at 37°C in a humidified 5% CO2 incubator. Primary osteoblast precursor cells were isolated from the calvarial bone of newborn C57BL/6 mice (1- to 2-day-old) through enzymatic digestion with α-MEM containing 0.1% collagenase (Life technologies, Grand Island, NY, USA) and 0.2% dispase II (Life Technologies, Grand Island, NY, USA). The isolated osteoblast precursor cells were promoted with osteogenic α-MEM medium with 10% FBS, 1% penicillin-streptomycin, 5mM β-glycerol phosphate (Sigma-Aldrich, St. Louis, MO, USA), 0.1mg/ml ascorbic acid (Sigma-Aldrich, St. Louis, MO, USA) and 10nM dexamethasone (Sigma-Aldrich, St. Louis, MO, USA) for 9 days and culture medium was replaced every 2–3 days [12]. Primary fibroblast-like synoviocytes (FLS) were isolated from the ankle joints of C57BL/6 mice by collagenase digestion [13]. Isolated FLS were grown in DMEM high glucose, GlutaMAX (Life Technologies, Grand Island, NY, USA) supplemented with 10% FBS and 1% penicillin-streptomycin. As no bone marrow-derived lineage cells were detected after passage 3, cultured FLS were used between passages 4–8. CD4+T lymphocytes were isolated from the spleen single-cell suspension of C57BL/6 mice by specific magnetic beads-based negative-selection (Miltenyi Biotec, Bergisch Gladbach, Germany). Neutrophils were isolated from peripheral blood of C57BL/6 mice by specific magnetic beads sorting (Miltenyi Biotec, Bergisch Gladbach, Germany) [14]. Before use, osteoblasts were confirmed to express osteocalcin, synovial fibroblasts were confirmed to express vascular cell adhesion molecule 1 and not F4/80 or CD45, CD4+T lymphocytes were confirmed to express CD3 and CD4, and neutrophils were confirmed to express Gr-1 and CD11b.
2.3. Co-culture and migration experiment
For osteoblasts-FLS co-culture: FLS were grown in co-culture with osteoblasts from Plekho1osx−/− mice, Plekho1osx Tg mice or WT littermates. Co-culture experiments were conducted in 6-well transwell coculture plates (Millipore, Billerica, MA, USA). The primary osteoblasts (5×105/well) were placed in the lower well and FLS (5×105/well) in the inserts. TNF-α was added for 24h. After experiment, the inserts were removed from the transwell coculture plates. FLS in the inserts were collected and firstly detected the osteocalcin mRNA expression by quantitative real time PCR to exclude the possible contamination of osteoblasts in the same co-culture system. Then, the mRNA levels of IL-1β and IL-6 in FLS were detected by quantitative real time PCR. For osteoblasts-CD4+T cells co-culture: Isolated CD4+T lymphocytes were grown in co-culture with osteoblasts from Plekho1osx−/− mice, Plekho1osx Tg mice or WT littermates. Coculture experiments were conducted in 6-well transwell coculture plates (Millipore, Billerica, MA, USA). The primary osteoblasts (5×105/well) were placed in the lower well and CD4+T lymphocytes (1×106/well) in the inserts. TNF-α was added for 24h. After experiment, CD4+T lymphocytes were collected and firstly detected the osteocalcin mRNA expression by quantitative real time PCR to exclude the possible contamination of osteoblasts. Then, the mRNA levels of IL-1β and IL-6 in CD4+T lymphocytes were detected by quantitative real time PCR. For osteoblasts-neutrophils co-culture: Isolated neutrophils were plated onto 3.0μm pore polycarbonate membrane transwell inserts, which were placed into the osteoblasts culture wells in the presence of TNF-α. Neutrophils were allowed to migrate through the membrane for 2 h. Migrated cells attached to the bottom of the well were stained by chloroacetate esterase and the positive cells were counted to determine the changes in chemotactic activity of osteoblasts.
2.4. siRNA inhibition
Plekho1 siRNA and non-sense siRNA (random siRNA) were purchased from GenePharma (Shanghai, China). Transfection was performed with Lipofectamine™ 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, murine osteoblastic cell line MC3T3E1 were seeded in a 6-well plate. Then, 4μl of 20 μM Plekho1 siRNA and 2.5μl of Lipofectamine™ 2000 were diluted in 0.25ml of Opti-MEM separately for each well. After 5 min of incubation at room temperature, the diluted Plekho1 siRNA and Lipofectamine™ 2000 were mixed gently and allowed to stand for 20 min at room temperature. Then, cells were washed by PBS. And 1.5 ml MEM without serum and antibiotics was added to each well. Followed by adding 0.5 ml of Opti-MEM containing the Lipofectamine™ 2000-siRNA complex and incubated for 6 h at 37 °C. Non-sense siRNA transfected by Lipofectamine™ 2000 were used as negative control. Then, the Lipofectamine™ 2000-siRNA complex was removed and replaced with fresh complete MEM. Next, the cells were stimulated by 10 ng/ml TNF-α for another 45 min. The supernatant and the cell lysis were collected for ELISA and western blot analysis, respectively.
2.5. Plasmid construction and transfection
HEK293T cells were cultured and used for transfection. Plasmids of different epitope-tagged human PLEKHO1 and TRAF2, including both full-length proteins and truncated mutants, as well as human RIP1 were constructed by PCR, followed by subcloning into expression vector pcDNA3.1 [[15], [16], [17], [18]]. Plasmid of HA-tagged human ubiquitin-K63 was from Addgene. Transfection was performed using Lipofectamine 2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer's instructions [7].
2.6. Immunoprecipitation and immunoblotting
At 36 h after the transfection, HEK293T were lysed in HEPES lysis buffer (20 mM HEPES pH 7.2, 50 mM NaCl, 0.5% Triton X-100, 1mM NaF, 1mM dithiothreitol) supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitors (10mM NaF and 1mM Na3VO4). Immunoprecipitations were performed using anti-Flag or anti-Myc antibody and protein A/G-agarose (Santa Cruz Biotechnology) at 4 °C. PLEKHO1 and TRAF2, including full-length proteins and truncated mutants, in lysates or immunoprecipitates were examined by anti-Myc (Cell Signaling Technology) and anti-Flag (Sigma-Aldrich, St. Louis, MO, USA) primary antibodies, respectively, and the appropriate secondary antibodies in immunoblotting, followed by detection with SuperSignal chemiluminescence kit (Thermo Fisher Scientific) [6,7,19].
2.7. In vivo ubiquitylation assay
HEK293T cells were transfected with Myc-RIP1, HA-Ub-K63, Flag-PLEKHO1 or Flag-ΔLZ and Flag-TRAF2 or Flag-ΔTRAF C. At 36 h after the transfection, cells were lysed in HEPES lysis buffer and then incubated with anti-Myc antibody (Sigma-Aldrich, St. Louis, MO, USA) for 3 h and protein A/G-agarose beads for a further 8 h at 4 °C. After three washes, ubiquitination of RIP1 was detected by immunoblotting with an anti-HA antibody (Cell Signaling Technology) [7,20].
2.8. Animals
Osteoblast-specific Plekho1 knockout mice (Plekho1osx−/− mice), Plekho1 knockout mice (Plekho1−/− mice) and osteoblast-specific Plekho1 overexpressing (Plekho1osx Tg) mice were prepared according to our previous reports [21]. We generated the Plekho1osx−/− mice based on the Cre-loxP strategy. Briefly, we created the heterozygous mice carrying the mutant allele with LoxP sites harboring exon 3 to exon 6 of Plekho1 gene (Plekho1fl/−), which were then crossed with Osx-Cre mice (Beijing Biocytogen Co., Ltd., China) to generate the Plekho1osx−/− mice. To generate Plekho1−/− mice, Plekho1fl/+ mice were crossed with CMV-Cre mice (Beijing Biocytogen Co., Ltd., China). To generate Plekho1osx Tg mice, we created a mouse strain carrying the ROSA26-PCAG-STOPfl-Plekho1-eGFP allele, and crossed them with the Osx-Cre mice to generate the Plekho1osx Tg mice that overexpressing PLEKHO1 in osteoblasts. DBA/1 mice and C57BL/6J mice were from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The human TNF transgenic (hTNFtg) mice (strain Tg197; genetic background, C57BL/6) were obtained from Dr. G. Kollias (Institute of Immunology, Biomedical Sciences Research Center “Alexander Fleming,” Vari, Greece). KRN T cell receptor (TCR) transgenic mice (B10.BR genetic background) were obtained from Dr. D. Mathis and Dr. C. Benoist (Harvard Medical School, Boston, Massachusetts, USA) and maintained on a C57BL/6 background. NOD/Lt mice were purchased from HFK Bioscience Co.,Ltd. (Beijing, China) and were used for breeding of K/BxN arthritic mice. Male Plekho1osx−/− mice were bred with female hTNFtg mice to obtain Plekho1osx−/−/hTNFtg mice. Male Plekho1−/− mice were bred with female Plekho1osx Tg mice to generate offspring (Plekho1−/−/Plekho1osx Tg mice) that express high PLEKHO1 exclusively in osteoblasts. All mice data were generated from age- and sex-matched littermates. Female cynomolgus monkeys (Macaca fascicularis), 3–4 years old, were purchased from PharmaLegacy Laboratories Co., Ltd. (Shanghai, China). All the animals were maintained under standard animal housing conditions (12-h light, 12-h dark cycles and free access to food and water). All procedures on mice were approved by the Ethical Animal Care and Use Committee in China Academy of Chinese Medical Sciences (EARIA2014310) and Hong Kong Baptist University (HASC/12-13/0045). All procedures on monkeys were approved by the Ethical Animal Care and Use Committee in Hong Kong Baptist University (HASC/15-16/0134).
2.9. Collagen-induced arthritis (CIA) mice model
The induction of CIA was performed as previously described with slight modification [22]. Briefly, type II collagen (bovine type II collagen for DBA/1 mice, chicken type II collagen for C57BL/6J mice) (Sigma-Aldrich, St. Louis, MO, USA) was dissolved at a concentration of 2 mg/ml in 0.05 M acetic acid and emulsified with complete Freund's adjuvant (CFA) (Sigma-Aldrich, St. Louis, MO, USA). To induce CIA in C57BL/6J mice, 100 mg of desiccated killed Mycobacterium tuberculosis H37Ra (BD Biosciences) was suspended in 30 ml of CFA [23]. On day 0, the CIA mice were immunized with 0.1 ml of collagen by intradermal injection at the base of the tail. On day 21, mice were given a booster dose of collagen through the same route.
2.10. K/BxN serum-transfer arthritis (STA) mice model
Pro-arthritic serum was isolated from K/BxN mice (generated by crossing KRN TCR transgenic mice with NOD/Lt mice) as previously described [24]. K/BxN arthritis was induced by the intraperitoneal injection of diluted K/BxN serum (75 μl serum with 75 μl endotoxin-free PBS) in C57BL/6J mice on day 0 and 2 [25].
2.11. Collagen-induced arthritis (CIA) non-human primate model
The induction of CIA in cynomolgus monkeys was performed as previously described with slight modification [26]. Briefly, bovine type II collagen (4 mg/ml) was emulsified in an equal volume of CFA. Ten monkeys were immunized with 1 ml of the cold emulsion by intradermal injections divided over 20 sites: 19 sites on the back and 1 site at the base of the tail, and after a three-week interval, boosted with the same immunizing procedure. During the study, body weight was recorded once a week. Onset of CIA was monitored by arthritic score, once a monkey fulfilled the criteria of 5% of the maximum arthritic score, it was assigned to the respective treatment group.
2.12. Treatment
For mice siRNA treatment experiment: DBA/1 mice were induced by type II collagen. After CIA was successfully established (day 28 after primary immunization, a mouse with a score of one or above was regarded as arthritic), parts of the mice were sacrificed before treatment as CIA baseline (CIA-BL). The remaining CIA mice were divided into (AspSerSer)6-liposome-Plekho1 siRNA group, (AspSerSer)6-liposome-non sense siRNA group, (AspSerSer)6-liposome group, and vehicle control group. The mice in each group received six periodic intravenous injections of (AspSerSer)6-liposome-Plekho1 siRNA, (AspSerSer)6-liposome-non sense siRNA, (AspSerSer)6-liposome, and PBS, respectively, with a siRNA dose of 5.89 mg/kg at an interval of one week.
For non-human primate siRNA treatment experiment: After CIA was successfully established, one monkey was sacrificed before siRNA treatment as CIA baseline (CIA-BL). The remaining CIA monkeys were divided into three groups: (AspSerSer)6-liposome-Plekho1 siRNA group, (AspSerSer)6-liposome-non sense siRNA group, and vehicle control group. The monkeys in each group received five periodic intravenous injections of (AspSerSer)6-liposome-Plekho1 siRNA, (AspSerSer)6-liposome-non sense siRNA and PBS, respectively, with a siRNA dose of 1 mg/kg at an interval of one week.
2.13. Evaluation of arthritis severity
Clinical severity of arthritis in mice was evaluated according to the following visual scoring system [22]: 0 = no swelling or erythema; 1 = Erythema and mild swelling confined to the tarsals or ankle joint; 2 = Erythema and mild swelling extending from the ankle to the tarsals; 3 = Erythema and moderate swelling extending from the ankle to metatarsal joints; 4 = Erythema and severe swelling encompass the ankle, foot and digits, or ankylosis of the limb. Each limb was assigned a score of 0 to 4, with a maximum possible score of 16 for each mouse.
The severity of arthritis in monkeys was assessed by observing the degree of swelling and rigidity at the metacarpophalangeal (MCP), proximal interphalangeal (PIP), and distal interphalangeal (DIP) joints of both the hands and feet, as well as the wrists, ankle, elbow, and knee (total 64 joints) weekly until the end of experiment, as reported in previous literature with slight modifications [27]. Each joint was assessed in accordance with the evaluation criteria shown as follows: 0 = normal; 1 = swelling not visible but can be determined by touch; 2 = swelling slightly visible and can be confirmed by touch; 3 = swelling clearly visible and/or noticeable joint deformity. Each joint is examined as per the evaluation criteria. The arthritis score for each animal was designated as the sum of individual joint score, with maximum score of 192.
2.14. Evaluation of joint swelling
Joint swelling at PIP joints of each cynomolgus monkey was measured by vernier caliper (Mitutoyo, Neuss, Germany) every week and calculated using the following formula: PIP swelling(mm2) = horizontal diameter × vertical diameter × 3.14 × 0.25.
2.15. Radiographic examination
Radiographs of the animals were obtained using a Portable X-ray SP-VET-4.0 unit (Sedecal, Madrid, Spain). In the anesthetized condition, the hands and feet of the monkeys were radiographed in the dorso-palmar position. Radiology projections of the arthritic hands and feet were performed weekly to assess the extent of joint destruction, bone erosion and joint space narrowing. The digital radiographs images of the hands and feet were acquired and the PIP joints were graded according to the modified Larsen's method with minor modifications [28]. The scoring criteria were as follows: 1 = Minor deformity in joint cartilage layers, and/or subchondral bone regions; 2 = Severe deformity in joint cartilage layers and subchondral bone regions, small amount of osteophytes present. The joint cavity fuzzy, but visible; 3 = Severe Grade 2 changes + Large amount of osteophytes present. The joint cavity was indistinguishable or invisible; 4 = Severe Grade 3 changes + the joint cavity undetectable. Bones appeared sclerotic or ankyolosing with major joint disfiguration. The scoring was done by two blinded professional investigators respectively. The radiologic score of PIP joints (4 × 4) for each animal ranged from 0 to 64.
2.16. Routine blood tests and blood biochemistry
During the course of the non-human primate studies, blood samples (2 ml) were collected in EDTA treated tubes at various time points and processed appropriately to run assessments for hematology, coagulation, serum chemistry. Hematology and coagulation measurements were made on whole blood and analyzed with automated hematology analyzer (Sysmex XT-2000iV™, UK) and Amax Destiny Plus™ analyzer (Trinity Biotech), respectively. Serum chemistry was determined with biochemical analyzer Hitachi-7180 (Hitachi, Tokyo, Japan).
2.17. ELISA
Ankle joints of mice were pulverized using a mortar and pestle filled with liquid nitrogen. Tissue was transferred to 15 ml tubes, placed on dry ice and resuspended in 1 ml PBS/200 mg of tissue and homogenized using a tissue homogeniser. Joint homogenates were centrifuged for 10 min at 500g at 4°C. Supernatants were transferred to 1.5 ml tubes, centrifuged at 15,000g for 5 min and collected for analysis. IL-1β and IL-6 quantifications were determined by ELISA according the instructions of the manufacturer (eBioscience, San Diego, CA, US), data was multiplied by a dilution factor for conversion from pg/ml to pg/g [29].
2.18. Total RNA isolation, reverse transcription and quantitative real-time PCR
Total RNA was isolated from the cultured cells and isolated cells using the RNeasy® Mini Kit (QIAGEN, Dusseldorf, Germany). The concentration of total RNA was determined using a spectrophotometer. cDNA was synthesized from 0.5 μg of total RNA using a commercial first-strand cDNA synthesis kit (QIAGEN, Dusseldorf, Germany). Real time PCR reactions were performed using SYBR Green in a 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, California, USA). The employed primer sequences in the study were listed in Supplementary Table 1. The amplification conditions were as follows: 50 °C for 2 min, 95 °C for 10 min, 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. The fluorescence signal emitted was collected by ABI PRISM® 7900HT Sequence Detection System and the signal was converted into numerical values by SDS 2.1 software (Applied Biosystems). The mRNA expression level of the target gene was first calculated from the Relative Standard Curve Method by the SDS 2.1 software. The threshold cycle (CT) value, which represents the relative expression of each target gene, was determined from the corresponding curve. Then, the relative expression of mRNA was determined by dividing the target amount by endogenous control amount to obtain a normalized target value. The relative mRNA expression was calculated using the 2-△△Ct method as follow: △△Ct = (Ct target gene – Ct reference gene) experimental group - (Ct target gene – Ct reference gene) WT-OB group) [8].
2.19. Western blot analysis
The bone specimens and cells were snap-frozen in liquid nitrogen and mechanically homogenized in lysis buffer. Protein fractions were collected by centrifugation at 15,000g at 4 °C for 10 min and then subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% BSA and incubated with specific antibodies overnight. A horseradish peroxidase–labeled secondary antibody (Abcam) was added and visualized using an enhanced chemiluminescence kit (Pierce). Antibodies to the following proteins were used: GAPDH (Sigma), PLEKHO1 (Santa Cruz), IKKβ (Cell Signaling Technology), p-IKKα/β (Cell Signaling Technology), p65 (Cell Signaling Technology) and p-p65 (Abcam). The relative amounts of the transferred proteins were quantified by scanning the auto-radiographic films with a gel densitometer (Bio-Rad, USA) and normalized to the corresponding GAPDH level [30].
2.20. Laser capture micro-dissection (LCM)
For LCM, the hind paw of mice and the hand of monkeys were decalcified in 10% ethylenediaminetetraacetic acid (EDTA) and embedded in OCT. Then, the series frozen sections (5 μm) were prepared in a cryostat (CM3050; Leica Microsystems, Wetzlar, Germany) at −20 °C. Adjacent sections were mounted on either glass slides or polyethylene membrane–equipped slides (P.A.L.M., Bernried, Germany). The sections mounted on glass slides were performed immunostaining to identify the osteocalcin-positive cells. Briefly, the cryosections were air dried at room temperature, fixed in ice-cold acetone for 10 min, permeabilized in 0.1% Triton X-100 at room temperature for 20 min, and blocked in 5% donkey serum in PBS. The sections were incubated overnight at 4 °C with rabbit polyclonal anti-osteocalcin antibody (Santa Cruz Biotechnology, Inc.). After three washes in PBS, the sections were incubated with Alexa Fluor 488-conjugated donkey anti-rabbit IgG (Invitrogen). Finally, the sections were mounted with medium containing (4′,6-diamidinole-2-phenolindole) DAPI (Vector laboratories) and examined under a fluorescence microscope to identify the osteocalcin-positive staining cells. The adjacent sections mounted on membrane-coated slides were stained with neutral red for 1 min at room temperature. After brief rinsing in water, the sections were air-dried. Osteocalcin-positive staining cells in adjacent sections were isolated by microdissection with an upgraded laser pressure catapulting microdissection system (P.A.L.M.) using a pulsed 355 nm diode laser in the Leica LMD 7000 Laser Microdissection System. About 100–200 identified cells were collected in reaction tube containing 5 μl lysis buffer for RNA extraction and subsequent real time PCR analysis following above mentioned protocols [30].
2.21. Immunofluorescence staining
The bone specimens were fixed with 4% buffered formalin and embedded with O.C.T. after decalcification with 10% EDTA. The frozen sections (5 μm thickness) were cut in a freezing cryostat at −20 °C. The sections were air dried at room temperature, fixed in ice-cold acetone for 10 min, permeablilized with 0.1% Triton X-100 at room temperature for 20 min, and blocked in 5% donkey serum in PBS. The sections were then incubated overnight at 4 °C with the mixture of goat polyclonal osteocalcin antibody (Santa Cruz Biotechnology, Inc.), and rabbit polyclonal PLEKHO1 antibody (Santa Cruz Biotechnology, Inc.). Following three washes in PBS, the sections were incubated with the mixture of Alexa Fluor 488-conjugated donkey anti-goat antibody, and Alexa Fluor 568-conjugated donkey anti-rabbit antibody (Invitrogen) for 1 h. Negative control experiments were done by omitting the primary antibodies. The sections were mounted with the medium containing DAPI and then examined under a fluorescence microscope (DM6000B, Leica image analysis system) [30,31] .
2.22. MicroCT analysis
MicroCT analyses were performed using a high-speed μCT (viva CT 40, Scanco Medical, Switzerland) at energy of 70 kVp and intensity of 114 μA with high resolution and voxel size of 10.5 μm (mouse) or 19 μm (monkey), respectively. In mouse studies, the hind limbs were scanned and the proximal tibia was selected for analysis. In non-human primate studies, the fore-hand was scanned and the PIP joint was selected for analysis. 3D reconstruction analysis was performed with SCANCO software (SCANCO Medical, Switzerland). Standard image analysis procedures were used to determine the trabecular and the cortical parameters. The following parameters were determined: bone mineral density (BMD), trabecular fraction (BV/TV), trabecular number (Tb.N) and trabecular thickness (Tb.Th) [32].
2.23. Histology evaluation
The hind paw of mice and the hand of monkeys were fixed in 4% formaldehyde, decalcified with EDTA and embedded in paraffin. The serial sections of the paw and hand were examined for inflammation using hematoxylin and eosin staining. Inflammation was scored according to the following criteria: 0 = normal; 1 = minimal infiltration of inflammatory cells in perijoint area; 2 = mild infiltration; 3 = moderate infiltration; and 4 = marked infiltration [33]. Scoring was executed blindly by two investigators and mean values were calculated.
2.24. Bone histomorphometric analysis
To assess bone formation, the animals were administered with xylenol orange (90 mg/kg) and calcein green (10 mg/kg) for labeling bone formation surface on day 12 and day 2 before sacrifice by intraperitoneal injection, respectively. The hind paw of mice and the hand of monkeys were fixed in 70% ethanol. The samples were trimmed from surrounding soft tissues, dehydrated in a series of ethanol solutions (70%, 80%, 90%, 95% and 100%), and embedded in methyl methacrylate. Non-decalcified histological sections (10 μm thick) were made using a diamond saw microtome. The sections for light microscopic observations were performed by the modified Goldner's trichrome, while unstained sections were prepared for fluorescent microscope observations. In mice study, the calcaneus area within the hind paw was chosen for histomorphometric analysis. In non-human primate study, the PIP joint was selected for analysis. All the measurements were performed using the professional image software (ImageJ, NIH, USA and BIOQUANT OSTEO, Version 13.2.6, Nashville, TN, USA). The following bone histomorphometric parameters for bone remodeling activity were analyzed, including mineral apposition rate (MAR), bone formation rate/bone surface (BFR/BS), osteoblast surface (Ob.S/BS), osteoblast number (Ob.N/B.Pm), osteoclast surface (Oc.S/BS) and osteoclast number (Oc.N/B·Pm) [8].
2.25. Statistical analysis
For statistical analysis, data were expressed as the mean ± standard deviation. In general, statistical differences among groups were analyzed by one-way analysis of variance (ANOVA) with a Tukey's multiple comparisons test to determine group differences in the study parameters. Especially, the dynamic detection data were compared by means of two-way ANOVA with subsequent Bonferroni posttests. All statistical analyses were performed using Graphpad Prism Software, 6.05 (Graph Pad Software Inc., San Diego, CA). P values <.0.05 are considered significant.
3. Results
3.1. Osteoblastic PLEKHO1 is upregulated in RA patients and CIA mice
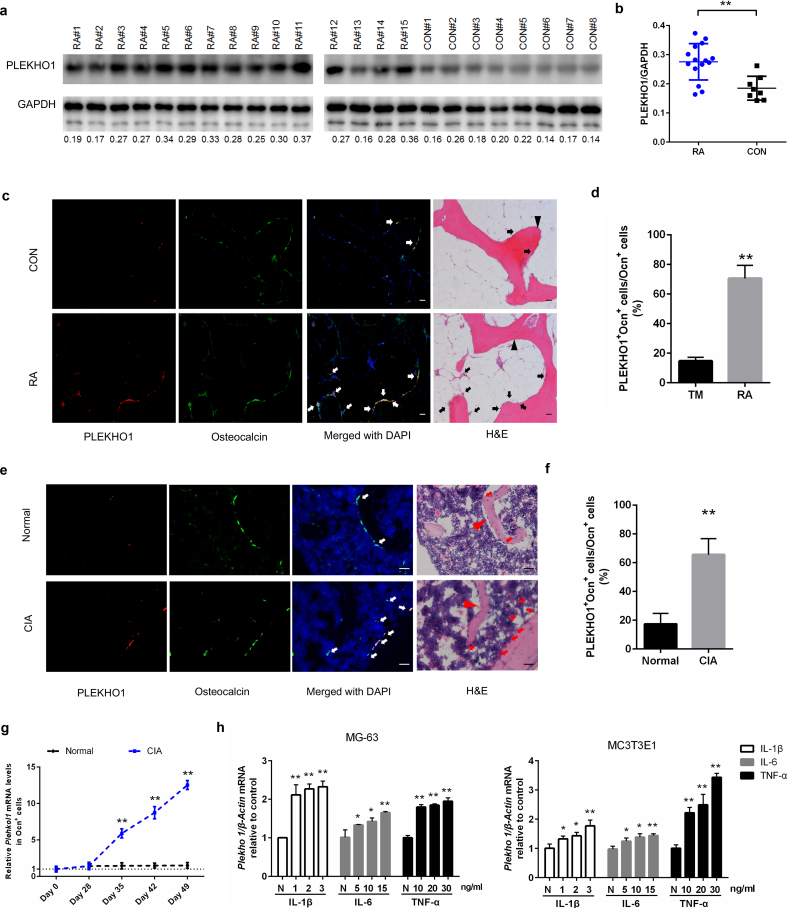
To assess whether PLEKHO1 has a role in RA pathology, we firstly investigated the expression of PLEKHO1 in knee joint bone tissues from RA patients and control individuals with severe trauma (TM). We observed the high levels of PLEKHO1 protein in knee joint bone tissues from RA patients when compared to TM controls (Fig. 1a, b). Immunofluorescence staining further showed more instances of PLEKHO1 colocalization with osteocalcin positive (Ocn+) cells in knee joint bone tissues from RA patients as compared to the TM controls (Fig. 1c). Quantitatively, the percentage of cells co-expressing PLEKHO1 and Ocn in Ocn+ cells of bone tissues from knee joints was also remarkably higher in RA patients as compared to TM controls (Fig. 1d).
Fig. 1.
Highly expressed PLEKHO1 in osteoblasts of RA patients and CIA mice. (a,b) Comparison of PLEKHO1 levels in bone tissues from knee joint between RA and TM patients underwent knee joint replacement surgery by western blot. a, Representative electrophoretic bands of the samples from fifteen RA patients and eight TM patients are shown. The numbers below the bands represent a semiquantitative value. b, The semiquantitative data of the protein levels. (c,d) Comparison of PLEKHO1 expression (red) within osteocalcin positive (Ocn+) osteoblasts (green) in bone tissues from knee joint between RA and TM patients by immunofluorescence analysis. c, Representative fluorescent micrographs of the samples from five RA patients and five TM patients are shown. Merged images with DAPI staining showed co-staining of PLEKHO1 and Ocn+ osteoblasts (arrows, yellow). H&E staining of the same sections is shown, and black arrowheads point to bone-formation surfaces, enriched by those cells with pink, which is a merged colour of green (osteoblast marker) and blue (DAPI staining for nuclei) in the immunofluorescence staining. Scale bars, 20 μm. d, Comparison of the percentage of cells co-expressing Ocn and PLEKHO1 in Ocn+ cells from bone tissues of knee joint between RA patients and TM patients. (e,f) Comparison of PLEKHO1 expression (red) within Ocn+ osteoblasts (green) in bone tissues of ankle joint from hind paws of CIA and normal mice on day 49 after primary immunization by immunofluorescence analysis. e, Representative fluorescent micrographs of the bone samples from ten CIA mice and ten normal mice are shown. Merged images with DAPI staining showed co-staining of PLEKHO1 and Ocn+ osteoblasts (arrows, yellow). H&E staining of the same sections is shown, and red arrowheads point to bone-formation surfaces. Scale bars: 20 μm. f, Comparison of the percentage of cells co-expressing Ocn and PLEKHO1 in Ocn+ cells from bone tissues of ankle joint from hind paws between CIA mice and normal mice on day 49 after primary immunization. (g) Time course changes of Plekho1 mRNA level in Ocn+ osteoblasts from CIA and normal mice by laser-capture microdissection (LCM) combined with qPCR. (h) Levels of Plekho1 mRNA induced by proinflammatory cytokines (IL-1β, IL-6 and TNF-α) in human osteoblast–like cell line MG-63 (left) and mouse osteoblast–like cell line MC3T3E1 (right) by real time PCR analysis. N: no cytokines, served as control. Note: RA, rheumatoid arthritis; TM, severe trauma. All data are the mean ± s.d. *P < 0.05, **P < 0.01. Two-way ANOVA with Bonferroni posttests was performed. Comparisons between two groups were performed using a Student's t-test. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To further investigate the dynamic changes of osteoblastic PLEKHO1 in RA progression, we examined the time course changes in PLEKHO1 expression within osteoblasts in CIA mice, a classic animal model of RA. In line with the above findings in RA patients' bone specimens, we also found the higher percentage of cells co-expressing PLEKHO1 and Ocn in Ocn+ cells at the ankle joint in hind paws of CIA mice on day 49 after primary immunization compared to those normal controls (Fig. 1e, f). Then, we collected Ocn+ cells of ankle joints from CIA group by laser capture micro-dissection (LCM) at different time point after collagen immunization. Further real time PCR analysis indicated that the Plekho1 mRNA level in Ocn+ cells at ankle joint from CIA group was increased over time from day 28 and significantly higher than that in control group on day 35, 42 and 49, respectively (Fig. 1g).
The above data suggested that the PLEKHO1 was upregulated in osteoblasts of joint bone tissues within the local inflammatory environment of RA. Consistently, we further found that the levels of Plekho1 mRNA were markedly upregulated in both human osteoblast–like cell line MG-63 and mouse osteoblast–like cell line MC3T3E1 after stimulated with the recombinant proinflammatory cytokines IL-1β, IL-6 and TNF-α, respectively (Fig. 1h).
3.2. Osteoblastic Plekho1 deletion attenuates joint inflammation in inflammatory arthritis mice
To test whether deletion of osteoblastic Plekho1 would attenuate joint inflammation during inflammatory arthritis, we generated the heterozygous mice carrying the mutant allele with LoxP sites harboring exon 3 to exon 6 of Plekho1 gene (Plekho1fl/−). These mice were then crossed with Osx-Cre mice to generate the osteoblast-specific Plekho1 conditional knockout (Plekho1osx−/−) mice (Supplementary Fig. 1a-b). Real time PCR analysis showed that the Plekho1 mRNA level was hardly detected in Ocn+ cells (osteoblasts) from Plekho1osx−/− mice when compared to control littermates (hereafter WT mice, including Plekho1fl/fl mice, Osx+/−;Plekho1fl/− mice, and Osx+/− mice), whereas no significant difference in Plekho1 mRNA level within Ocn− cells (non-osteoblasts) was found between Plekho1osx−/− mice and WT mice (Supplementary Fig. 1c).
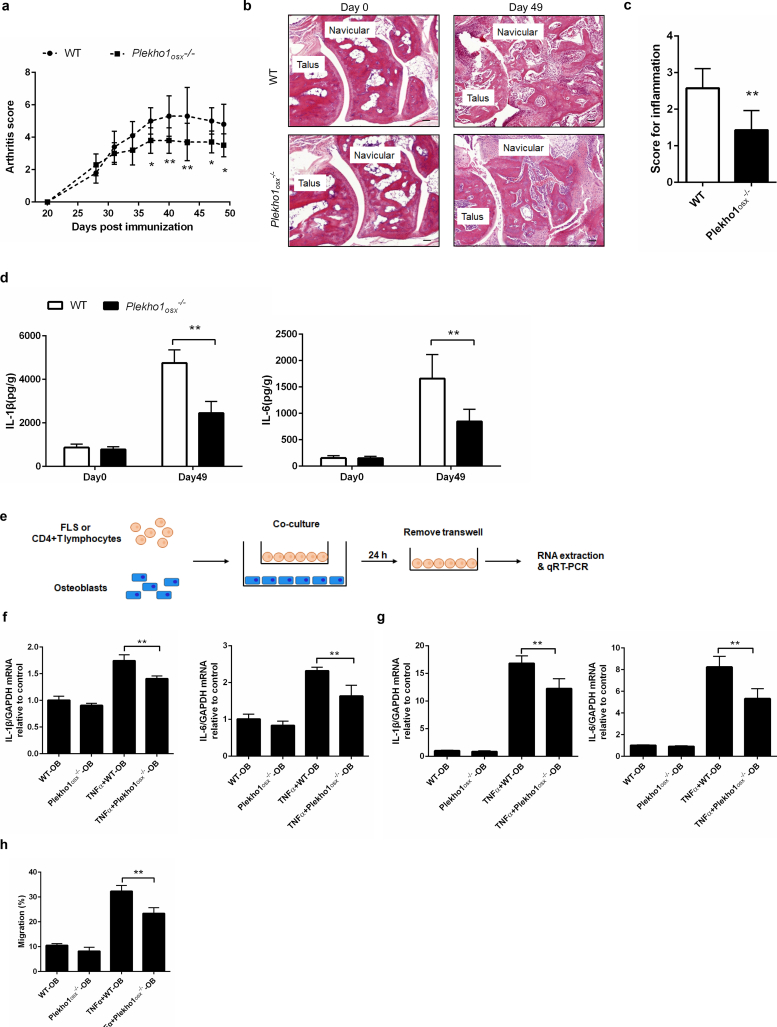
Subsequently, we examined the joint inflammation changes of Plekho1osx−/− mice after induction with type II chicken collagen. We found that Plekho1osx−/−-CIA mice showed obvious attenuation in joint inflammation. The arthritis score in Plekho1osx−/−-CIA mice was significantly lower than that in WT-CIA mice from day 37 to day 49 after first immunization (Fig. 2a). Histological evaluation showed that the score for inflammation in ankle joint from the hind paws was significantly lower in Plekho1osx−/−-CIA mice than that in WT-CIA mice on day 49 after primary immunization (Fig. 2b, c). Moreover, ELISA assay showed that the IL-1β and IL-6 levels in ankle joint of hind paws from Plekho1osx−/−-CIA mice were also remarkably lower on day 49 when compared with the WT-CIA mice (Fig. 2d). As previous studies regarded that the joint inflammation progression was mainly caused by the activated fibroblast-like synoviocytes (FLS) and immune cells including infiltrating lymphocytes and neutrophils [34], we then detected the cross-talk between osteoblasts and FLS, CD4 + T lymphocytes as well as neutrophils by employing an in vitro co-culture system which could experimentally simulate the temporo-spatial complexity of the in vivo cross-talk between osteoblasts and inflammatory cells in the joint. The mature osteoblasts differentiated from the primary osteoblast precursor cells isolated from the calvarial bone of newborn Plekho1osx−/− mice and WT littermates in osteogenic medium were confirmed by the expression of osteocalcin. The FLS, CD4 + T lymphocytes and neutrophils were obtained from WT mice by collagenase digestion and specific magnetic beads-based selection, respectively. Thereafter, the FLS, CD4 + T lymphocytes or neutrophils were co-cultured with Plekho1osx−/− or WT osteoblasts, respectively (Fig. 2e). After co-culture, the cells were collected for real time PCR analysis. The results showed that under TNF-α stimulation, the mRNA levels of IL-1β and IL-6 in the CD4 + T lymphocytes and FLS were remarkably decreased after co-culture with osteoblasts from Plekho1osx−/− mice when compared with the WT control group (Fig. 2f, g). The migration of neutrophils was also inhibited after co-culture with osteoblasts from Plekho1osx−/− mice (Fig. 2h). These results hinted an important role of osteoblastic PLEKHO1 in modulating local inflammatory response in RA.
Fig. 2.
Genetic deletion of osteoblastic Plekho1 leads to amelioration of joint inflammation in CIA mice. (a) Time course changes in arthritis score from Plekho1osx−/− mice and WT mice (including Plekho1fl/fl mice, Osx+/−;Plekho1fl/− mice, and Osx+/− mice) after induction with type II chicken collagen. (b) Representative histological images in the ankle joint of hind paws from Plekho1osx−/−-CIA mice and WT-CIA mice on day 49 after primary immunization. Scale bars, 50 μm. (c) Histological inflammation score in the ankle joint of hind paws from Plekho1osx−/−-CIA mice and WT-CIA mice on day 49 after primary immunization. (d) Time course changes of IL-1β and IL-6 levels in the ankle joint of hind paws from Plekho1osx−/−-CIA mice and WT-CIA mice by ELISA examination. (e) A schematic diagram illustrating the design of the co-culture experiments. The CD4 + T lymphocytes or fibroblast-like synoviocytes (FLS) derived from WT mice were co-cultured with the osteoblasts derived from Plekho1osx−/− mice or WT mice, with or without the stimulation of TNF-α. (f) IL-1β and IL-6 mRNA levels in CD4 + T lymphocytes after co-culture with osteoblasts from WT mice and Plekho1osx−/− mice. (g) IL-1β and IL-6 mRNA levels in FLS after co-culture with osteoblasts from WT mice and Plekho1osx−/− mice. (h) Migration of neutrophils after co-culture with osteoblasts from WT mice and Plekho1osx−/− mice. All data are the mean ± s.d. n = 10 per group. *P < 0.05, **P < 0.01. For a, Two-way ANOVA with subsequent Bonferroni posttests was performed. For c & d, a Student's t-test was performed. For f-h, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed.
Because osteoblastic PLEKHO1 also exerts an inhibitory role in the osteoblastic bone formation during aging-related bone loss and osteoporosis [8,9], we further examined the bone phenotypes of Plekho1osx−/− -CIA mice. MicroCT analysis showed a better organized architecture and a higher bone mass in Plekho1osx−/−-CIA mice compared to WT-CIA mice on day 49 after primary immunization, both in cortical bone and trabecular bone (Supplementary Fig. 2a). The values of bone mineral density (BMD), trabecular fraction (BV/TV), trabecular thickness (Tb.Th) and trabecular number (Tb.N) of the tibial bones decreased mildly from baseline (day 0) in Plekho1osx−/−-CIA mice but rapidly in WT-CIA mice (Supplementary Fig. 2b). Dynamic bone histomorphometric analysis showed a larger width between the xylenol and the calcein labeling bands in Plekho1osx−/−-CIA mice compared to WT-CIA mice (Supplementary Fig. 2c).The values of mineral apposition rate (MAR), bone formation rate (BFR/BS), osteoblasts surface (Ob.S/BS) and osteoblast number (Ob.N/B·Pm) were also decreased slower in Plekho1osx−/−-CIA mice than those in WT-CIA mice after primary immunization. The values of osteoclasts surface (Oc.S/BS) and osteoclast number (Oc.N/B·Pm) showed a slower increase in Plekho1osx−/−-CIA mice than those in WT-CIA mice after primary immunization (Supplementary Fig. 2d). These results demonstrated that osteoblastic PLEKHO1 could negatively regulate bone formation in CIA mice as well.
In addition, to further evaluate the role of osteoblastic PLEKHO1 in local inflammation regulation during RA, we induced Plekho1osx−/− mice with K/BxN serum, which could induce the acute arthritis that was more sensitive in mice with C57BL/6 background. Of note, we again observed less joint inflammation in Plekho1osx−/−-STA mice, including the lower arthritis score since day 8 and lower histological inflammation score in ankle joint of hind paws on day 12 compared to the control mice (Supplementary Fig. 3a-c). Moreover, ELISA assay showed that the IL-1β and IL-6 levels in ankle joint of hind paws from Plekho1osx−/−-STA mice were also remarkably lower on day 12 when compared with the WT-STA mice (Supplementary Fig. 3d).
3.3. Overexpressing Plekho1 only in osteoblasts exacerbates joint inflammation in inflammatory arthritis mice
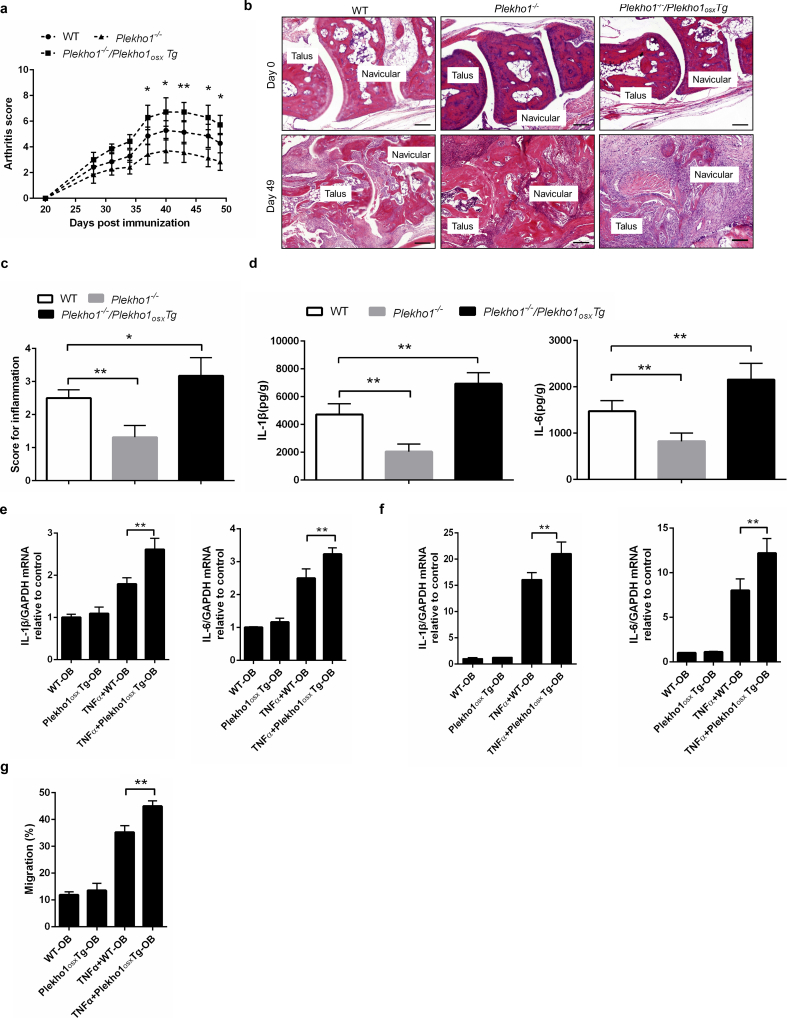
To exclude the interference of PLEKHO1 in other cells, we created another mouse strain carrying the ROSA26-PCAG-STOPfl-Plekho1-eGFP allele, and crossed them with the Osx-Cre mice to generate the osteoblast-specific Plekho1 overexpressing (Plekho1osx Tg) mice that overexpressing PLEKHO1 in osteoblasts. Then, we crossed Plekho1−/− mice with Plekho1osx Tg mice to obtain mice that express high PLEKHO1 exclusively in osteoblasts (Plekho1−/−/Plekho1osx Tg mice) (Supplementary Fig. 4a, 4b). Real time PCR analysis showed that the expression of Plekho1 mRNA was remarkably higher in Ocn+ cells (osteoblasts) but hardly detected in Ocn− cells (non-osteoblasts) in Plekho1−/−/Plekho1osx Tg mice when compared to control littermates (hereafter WT mice, including Osx+/−;Plekho1fl/− mice and Osx+/−;Plekho1fl/fl mice). In contrast, the expression of Plekho1 mRNA was hardly detected in both Ocn+ cells and Ocn− cells in Plekho1−/− mice (Supplementary Fig. 4c). After CIA induction, we found that the exclusively high PLEKHO1 expression in osteoblasts exacerbated the disease severity and joint inflammation progression in Plekho1−/−/Plekho1osx Tg-CIA mice, including the higher arthritis score since day 35 after primary immunization and the higher pathological inflammation score of ankle joint from the hind paws as compared to WT-CIA mice on day 49 after primary immunization (Fig. 3a-c). ELISA assay showed that the IL-1β and IL-6 protein levels in ankle joint of hind paws from Plekho1−/−/Plekho1osx Tg-CIA mice were also obviously higher when compared to WT-CIA mice on day 49 after primary immunization (Fig. 3d). In addition, we isolated primary osteoblast precursor cells from calvarial bone of newborn Plekho1osx Tg mice and WT littermates, these precursor cells were induced maturation for co-culture with CD4 + T lymphocytes, FLS and neutrophils of WT mice under TNF-α stimulation, respectively. We found that the IL-1β and IL-6 mRNA levels in CD4 + T lymphocytes and FLS were significantly higher after co-culture with osteoblasts from Plekho1osx Tg mice when compared with the WT control group (Fig. 3e, f). The migration of neutrophils was also enhanced after co-culture with osteoblasts from Plekho1osx Tg mice (Fig. 3g).
Fig. 3.
Overexpressed Plekho1 in osteoblasts exacerbates inflammation in Plekho1−/−/Plekho1osxTg mice with CIA. (a) Disease progression assessed by arthritic score in WT-CIA, Plekho1−/−-CIA and Plekho1−/−/Plekho1osxTg-CIA mice. *P < 0.05, **P < 0.01,compared to WT-CIA group. (b) Representative histological images in the ankle joint of hind paws from WT-CIA, Plekho1−/−-CIA and Plekho1−/−/Plekho1osxTg-CIA mice. Scale bars, 50 μm. (c) Histological inflammation score in the ankle joint of hind paws from WT-CIA, Plekho1−/−-CIA and Plekho1−/−/Plekho1osxTg-CIA mice. (d) IL-1β and IL-6 levels in the ankle joint of hind paws from WT-CIA, Plekho1−/−-CIA and Plekho1−/−/Plekho1osxTg-CIA mice on day 49 after primary immunization by ELISA examination. (e) IL-1β and IL-6 mRNA levels in CD4 + T lymphocytes after co-culture with osteoblasts from WT mice and Plekho1osxTg mice. (f) IL-1β and IL-6 mRNA levels in fibroblast-like synoviocytes after co-culture with osteoblasts from WT mice and Plekho1osxTg mice. (g) Migration of neutrophils after co-culture with osteoblasts from WT mice and Plekho1osxTg mice. All data are the mean ± s.d. n = 9 per group. *P < 0.05, **P < 0.01. For a, a two-way ANOVA with subsequent Bonferroni posttests was performed. For c-g, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed.
We also induced Plekho1−/−/Plekho1osx Tg mice with K/BxN serum. Consistently, we again observed severer joint inflammation in Plekho1−/−/Plekho1osx Tg-STA mice, including the faster increase in arthritis score and the higher histological inflammation score in the ankle joint of hind paws compared to the control littermates (Supplementary Fig. 5a-c). IL-1β and IL-6 protein levels in ankle joint of hind paws from Plekho1−/−/Plekho1osx Tg-STA mice were significantly higher on day 12 when compared with the WT-STA mice (Supplementary Fig. 5d). These results further demonstrated the important role of osteoblastic PLEKHO1 in modulating joint inflammation in RA.
3.4. PLEKHO1 is required for TRAF2-mediated RIP1 ubiquitination to activate NF-κB for inducing inflammatory cytokines production in osteoblasts
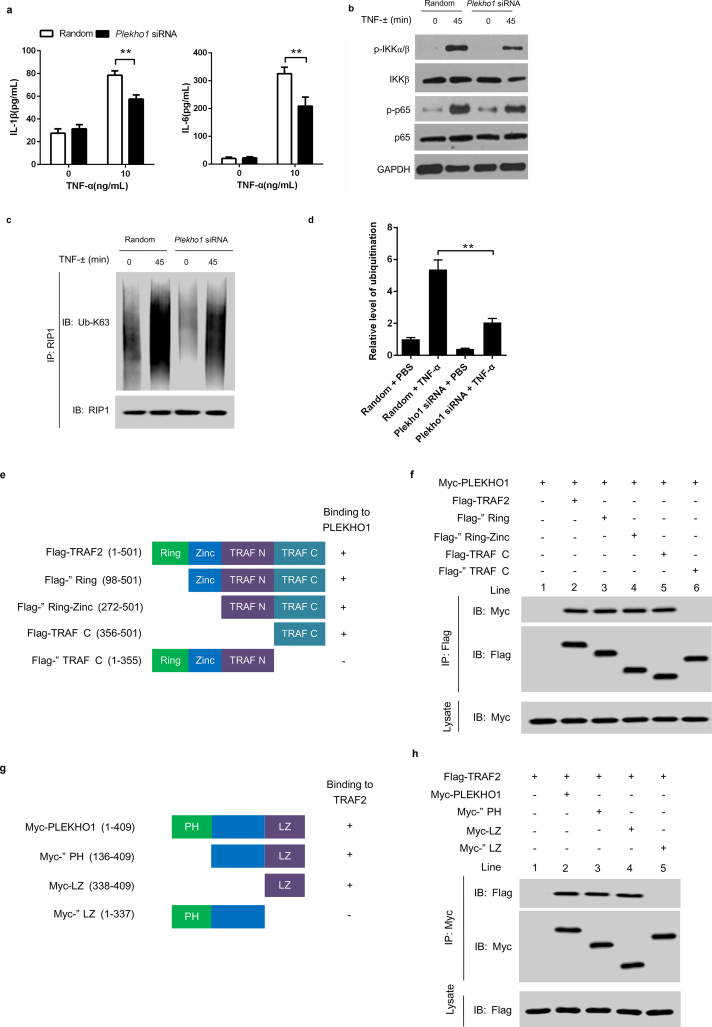
Then, to detect whether PLEKHO1 could affect TNF-α-stimulated osteoblasts releasing inflammatory effectors so as to modulate inflammation, we used a mouse osteoblast–like cell line MC3T3E1. We found that after Plekho1 silencing by Plekho1 siRNA, the IL-1β and IL-6 production in the supernatant of TNF-α-stimulated MC3T3E1 cells were remarkably lower than the random siRNA group (Fig. 4a). To further confirm it in vivo, we crossed hTNFtg mice, which could spontaneously develop arthritis at the age of four week due to the overexpressed human TNF transgene, with Plekho1osx−/− mice to obtain Plekho1osx−/−/hTNFtg mice. Notably, Plekho1osx−/−/hTNFtg mice showed a distinct alleviation of disease severity. The arthritis score in Plekho1osx−/−/hTNFtg mice was significantly lower than that in hTNFtg mice from 7 to 10 weeks after birth (Supplementary Fig. 6a). Histological analysis also indicated that the joint inflammation was significantly ameliorated in Plekho1osx−/−/hTNFtg mice compared to hTNFtg mice, which was reflected by lower histological inflammation score at week 10 (Supplementary Fig. 6b, 6c). These results implied that the osteoblastic PLEKHO1 on joint inflammation regulation might be related with TNF-α involved signal mechanism. As previous studies have demonstrated that NF-κB signal pathway played an important role in TNF-α induced inflammatory cytokines production [35], we therefore detected the change of related proteins in this signal pathway after specifically silencing Plekho1 by Plekho1 siRNA. We found that IKKα/β and p65 phosphorylation as well as RIP1 ubiquitination were inhibited under TNF-α stimulation in Plekho1-silenced MC3T3E1 cells, although no significantly change on production of IKKβ and p65 (Fig. 4b-d).
Fig. 4.
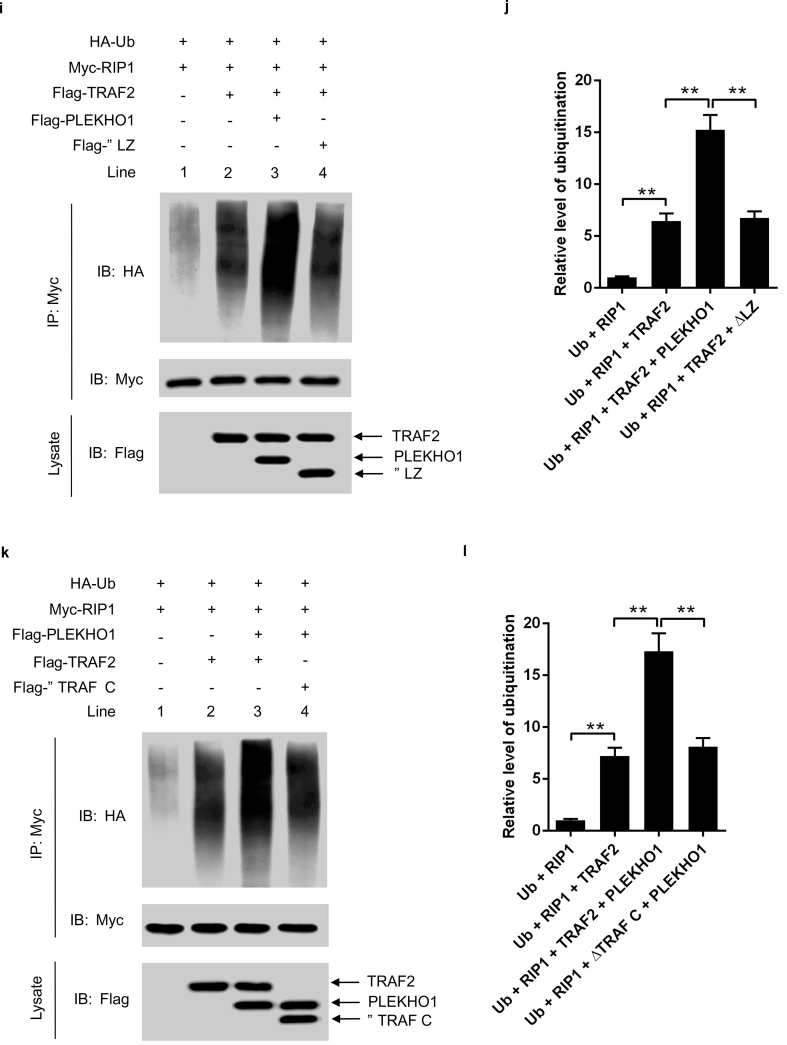
PLEKHO1 interacts with TRAF2 to promote the TRAF2-mediated ubiquitination of RIP1. (a) Levels of inflammatory cytokines (IL-1β, IL-6) produced by MC3T3E1 cells transfected with Plekho1 siRNA or random siRNA and subjected to TNF-α (10 ng/ml). (b) MC3T3E1 cells transfected with Plekho1 siRNA or random siRNA were stimulated with TNF-α (10 ng/ml) for 45 min, and harvested for detection of IKKα/β and p65 phosphorylation. (c) MC3T3E1 cells transfected with Plekho1 siRNA or random siRNA were stimulated with TNF-α (10 ng/ml) for 45 min, and harvested for detection of RIP1 ubiquitination. (d) Quantification of relative level of RIP1 ubiquitination. (e, f) Immunoprecipitation using anti-Flag antibody to examine the PLEKHO1-TRAF2 interaction and mapping of the PLEKHO1-interacting domain in TRAF2. e, The Flag-tagged full-length TRAF2 (Flag-TRAF2) and various truncated mutants (Flag-ΔRing, Flag-ΔRing-Zinc, Flag-TRAF N and Flag-ΔTRAF C) were shown with amino acid numbers. f, HEK293T cells were transfected with Myc-PLEKHO1 and Flag-TRAF2 or truncated mutants as indicated. Full-length TRAF2 or truncated mutants was immunoprecipitated by anti-Flag antibody. PLEKHO1 in immunoprecipitates or whole-cell lysate was detected by anti-Myc antibody. (g, h) Immunoprecipitation using anti-Myc antibody to examine the PLEKHO1-TRAF2 interaction and mapping of the TRAF2-interacting domain in PLEKHO1. g, The Myc-tagged full-length PLEKHO1 (Myc-PLEKHO1) and various truncated mutants (Myc-ΔPH, Myc-LZ, and Myc-ΔLZ) were shown with amino acid numbers. h, HEK293T cells were transfected with Flag-TRAF2 and Myc-PLEKHO1 or truncated mutants as indicated. Full-length PLEKHO1 or truncated mutants was immunoprecipitated by anti-Myc antibody. TRAF2 in immunoprecipitates or whole-cell lysate was detected by anti-Flag antibody. (i) In vivo ubiquitination assay of RIP1 in HEK293T cells transfected with Myc-RIP1, HA-Ubiquitin (HA-Ub) and Flag-TRAF2, along with Flag-PLEKHO1 or Flag-ΔLZ. Ubiquitination of RIP1 was detected in Myc immunoprecipitates. (j) Quantification of relative level of RIP1 ubiquitination. (k) In vivo ubiquitination assay of RIP1 in HEK293T cells transfected with Myc-RIP1, HA-Ub and Flag-PLEKHO1, along with Flag-TRAF2 or Flag-ΔTRAF C. Ubiquitination of RIP1 was detected in Myc immunoprecipitates. (l) Quantification of relative level of RIP1 ubiquitination. Note: IP, Immunoprecipitation; IB, immunoblotting. Data are representative of three independent experiments. **P < 0.01. Comparisons between two groups were performed using a Student's t-test.
Previous studies demonstrated that TNF-α up-regulated the expression of inflammatory mediators through TRAF2-mediated NF-κB activation in MC3T3E1 cells [36]. In addition, PLEKHO1 could interact with the member of TRAF family to regulate the downstream signal [6], we therefore hypothesized that PLEKHO1 might interact with TRAF2 to promote the activation of NF-κB signal pathways. TRAF2 contains a ring finger domain, a zinc finger domain, a TRAF N domain and a TRAF C domain [17,37,38]. To test whether TRAF2 could interact with PLEKHO1 and which domain in TRAF2 was essential for their interaction, we constructed Myc-tagged full-length PLEKHO1 (Myc-PLEKHO1), Flag-tagged full-length TRAF2 (Flag-TRAF2) and various TRAF2 truncated mutants including Flag-ΔRing, Flag-ΔRing-Zinc, Flag-TRAF N and Flag-ΔTRAF C (Fig. 4e). HEK293T cells were transfected with Myc-PLEKHO1 and Flag-TRAF2 or truncated mutants. After immunoprecipitation using anti-Flag antibody, we detected that TRAF2 could interact with PLEKHO1 (Fig. 4f, Lane 2). However, deletion of TRAF C domain (ΔTRAF C) abolished the interaction between TRAF2 and PLEKHO1 (Fig. 4f, Lane 6), whereas other truncated mutants (ΔRing, ΔRing-Zinc and TRAF C) maintained the binding with PLEKHO1 as that of full-length TRAF2 (Fig. 4f, Lane 3–5). In addition, PLEKHO1 is composed of an N-terminal pleckstrin homology (pH)-containing domain and a C-terminal leucine zipper (LZ)-containing domain [7,16]. We also constructed PLEKHO1 truncated mutants (Myc-ΔPH, Myc-LZ and Myc-ΔLZ) to identify the key domain in PLEKHO1 that interacted with TRAF2 (Fig. 4g). Data from immunoprecipitation showed that the PLEKHO1 mutant with deletion of LZ-containing domain (ΔLZ) could not interact with TRAF2 (Fig. 4h, Lane 5), but other PLEKHO1 mutants could still bound to TRAF2 (Fig. 4h, Lane 2–4).
As TRAF2 could ubiquitinate RIP1, which is essential for NF-κB activation [39], we then determined whether PLEKHO1 affected TRAF2-mediated RIP1 ubiquitination. We performed in vivo ubiquitination assay in HEK293T cells transfected with Myc-RIP1, HA-Ubiquitin (HA-Ub), Flag-PLEKHO1 or Flag-ΔLZ and Flag-TRAF2 or Flag-ΔTRAF C. Our results demonstrated that PLEKHO1 promoted the TRAF2-mediated ubiquitination of RIP1 (lane 3 in Fig. 4i, lane 3 in Fig. 4k, Fig. 4j, Fig. 4l). Nevertheless, PLEKHO1 mutant with deletion of c-terminal LZ-containing domain had no obvious promotive effects on TRAF2-mediated ubiquitination of RIP1 (lane 4 in Fig. 4i, Fig. 4j). Disruption of TRAF2-PLEKHO1 interaction by deletion of ΔTRAF C in TRAF2 also diminished the promotive effect of PLEKHO1 on TRAF2-mediated RIP1 ubiquitination (lane 4 in Fig. 4k, Fig. 4l). All these results indicated that PLEKHO1 could interact with TRAF2 to promote the TRAF2-mediated ubiquitination of RIP1.
3.5. Osteoblastic PLEKHO1 inhibition alleviates joint inflammation and bone formation reduction in CIA mice
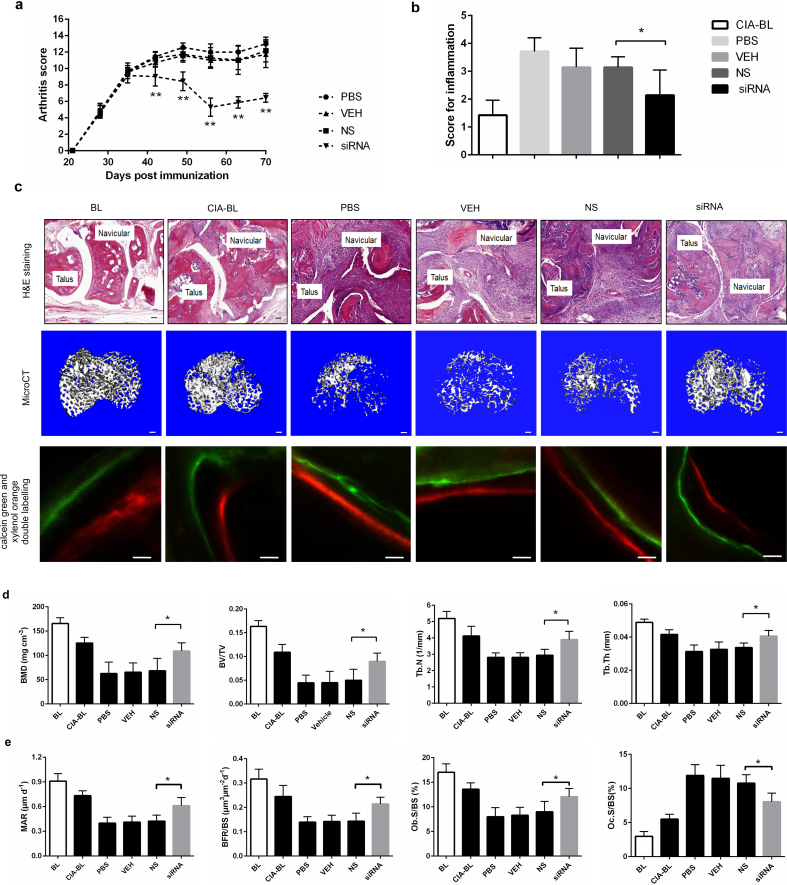
Next, to test whether early inhibition of osteoblastic PLEKHO1 could alleviate joint inflammation and promote bone formation in mice with established arthritis, we used a cross-species Plekho1 siRNA and a nucleic acid delivery system targeting osteoblasts, which were both established by our group [5,8]. This cross-species Plekho1 siRNA could target the four species (human, monkey, rat and mouse). In vivo studies demonstrated that periodic intravenous injections of this cross-species Plekho1 siRNA promoted bone formation and reversed bone loss in the osteoporotic mice. On the other hand, to facilitate delivering this cross-species Plekho1 siRNA to osteoblasts, we have developed a targeted delivery system – (AspSerSer)6-liposome. We confirmed that this targeted delivery system could facilitate Plekho1 siRNA specifically achieve high gene knockdown efficiency in osteoblasts. Therefore, we then performed six consecutive administration of this cross-species Plekho1 siRNA encapsulated by the osteoblast-targeting delivery system ((AspSerSer)6-liposome-Plekho1 siRNA, hereafter siRNA) in DBA/1 mice at an interval of one week from day 28 after primary immunization with bovine type II collagen, because arthritis was clinically evident at this time point (Supplementary Fig. 7a). We firstly confirmed that the level of Plekho1 mRNA was significantly downregulated in Ocn+ cells of siRNA-treated CIA mice after six-week treatment (on day 70 after primary immunization) (Supplementary Fig. 7b). Thereafter, we found that the arthritis score was significantly lower in siRNA-treated CIA mice when compared to mice treated with (AspSerSer)6-liposome-non sense siRNA (hereafter NS), (AspSerSer)6-liposome (hereafter VEH) or PBS (Fig. 5a). Histological evaluation showed that the score of joint inflammation in siRNA-treated CIA mice was also significantly lower when compared to three other control groups after treatment (Fig. 5b, c). ELISA assay showed that the IL-1β and IL-6 levels in ankle joint of hind paws from siRNA-treated CIA mice were also significantly lower when compared with the three other control groups (Supplementary Fig. 7c). Subsequently, microCT analysis showed better organized microarchitecture and a higher bone mass in tibial bone of siRNA-treated CIA mice compared to mice treated with NS, VEH or PBS after treatment (Fig. 5c). The values of BMD, BV/TV, Tb.N and Tb.Th of the tibial bones had remarkable restoration in siRNA-treated CIA mice, whereas such restoration was not observed in CIA mice treated with NS, VEH or PBS (Fig. 5d). Similarly, we found a larger width between the xylenol and calcein labeling bands in the mice treated with siRNA compared to the mice treated with NS, VEH or PBS. The values of MAR, BFR/BS and Ob.S/BS were significantly higher, whereas the value of Oc.S/BS were significantly lower in siRNA group when compared with three other control groups after treatment (Fig. 5c, Fig. 5e).
Fig. 5.
Osteoblastic PLEKHO1 inhibition leads to amelioration of articular inflammation and promotion of bone formation in CIA mice. (a) Time course changes in arthritis score from CIA mice treated with PBS, VEH, NS, and siRNA, respectively. (b) Histological inflammation score in the hind paws from the CIA mice after 6-week treatment in the respective group. (c) Representative histological images (Scale bars, 50 μm), microCT images (proximal tibial bones, Scale bars, 100 μm), calcein green and xylenol orange double labelling (Scale bars, 10 μm) from the hind paws of mice in each group at day 70 after primary immunization. (d) The values of microCT parameters (BMD, BV/TV, Tb.N and Tb.Th) of the proximal tibial bones from the CIA mice after treatment in the respective group. (e) The values of bone histomorphometric parameters (MAR, BFR/BS, Ob.S/BS and Oc.S/BS) in the hind paws from the CIA mice after treatment in the respective group. All data are the mean ± s.d. *P < 0.05, **P < 0.01. n = 7 per group. For a, a two-way ANOVA with subsequent Bonferroni posttests was performed. For b,d,e, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed. Note: BL, baseline; CIA-BL, collagen-induced arthritis baseline; VEH, (AspSerSer)6-liposome; NS, (AspSerSer)6-liposome -NS siRNA; siRNA, (AspSerSer)6- liposome -Plekho1 siRNA. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Plekho1 siRNA delivered to osteoblasts improves arthritis symptoms and promotes bone formation in a collagen-induced non-human primate arthritis model
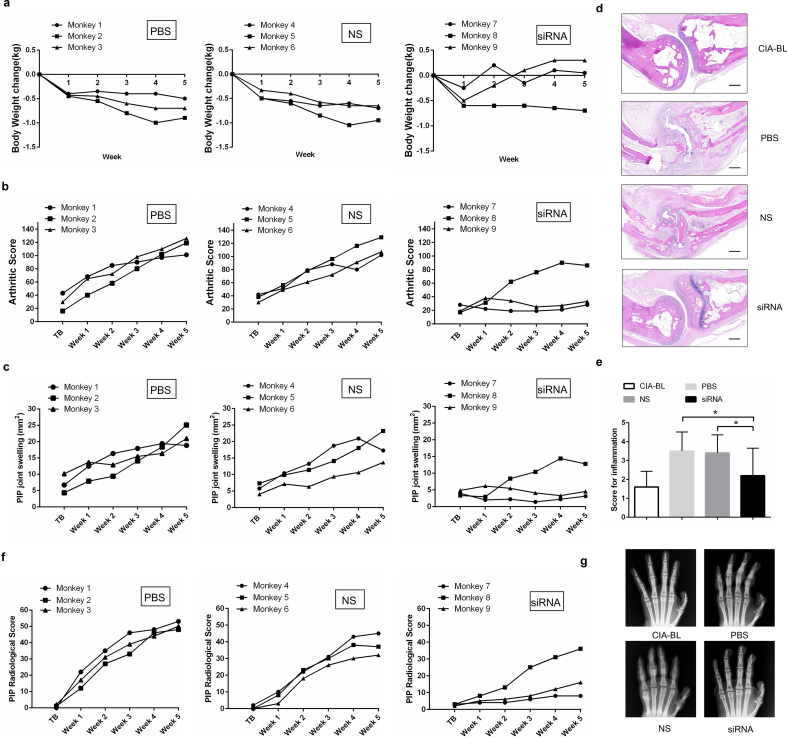
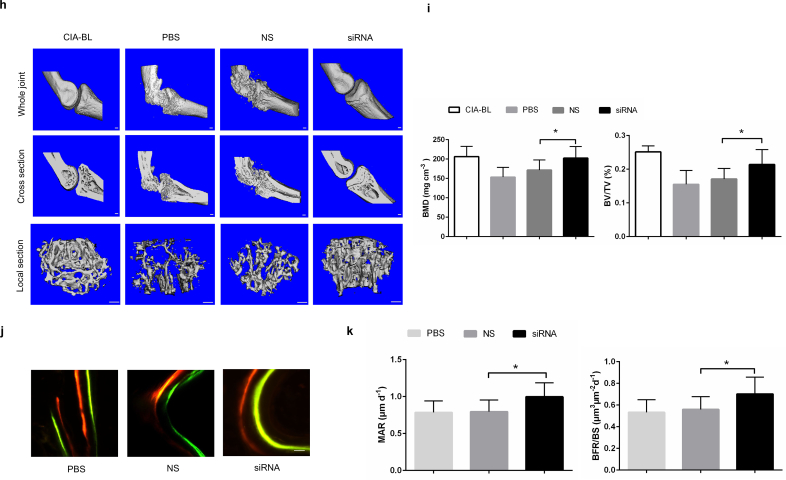
To facilitate our findings to clinical translation, we carried out Plekho1 siRNA inhibition experiments in cynomolgus monkeys (Supplementary Fig. 8a). We firstly confirmed that the Plekho1 mRNA expression in osteoblasts was significantly inhibited till 5 weeks after treatment with siRNA (Supplementary Fig. 8b). Thereafter, we found that the body weight, an important indicator for disease progression, in NS and PBS-treated monkeys were gradually decreased during 5-week treatment, whereas the body weight in siRNA-treated monkeys were continuously increased except one monkey (Monkey 8) (Fig. 6a). We also found that the arthritis score and the proximal interphalangeal point (PIP) joint swelling were continuously increased in NS and PBS-treated monkeys, whereas these two parameters in siRNA-treated monkeys maintained at a low level during 5-week treatment except Monkey 8 (Fig. 6b, c). We performed histological evaluation and found that the joint inflammation in siRNA-treated monkeys was significantly attenuated than that of NS and PBS-treated monkeys after treatment (Fig. 6d, e). We then used X ray to assess the bony structure of the PIP joints and found that the siRNA treatment could alleviate the continuous increase of the PIP radiological score. After treatment, the average PIP radiological score in PBS, NS and siRNA-treated monkeys were 43.33 ± 5.51, 43.67 ± 7.09, 20 ± 14.42, respectively (Fig. 6f, g). Consistent with the X ray findings, microCT reconstruction images also showed the better organized bony and joint structure in siRNA-treated monkeys compared to NS and PBS-treated monkeys after treatment (Fig. 6h). The values of BMD and BV/TV were significantly higher in siRNA-treated monkeys compared to NS and PBS-treated monkeys after treatment (Fig. 6i). Furthermore, we found a larger width between the two labeling bands in the siRNA-treated monkeys compared to NS and PBS-treated monkeys after treatment (Fig. 6j). The values of bone formation parameters MAR and BFR/BS were also significantly higher in siRNA-treated monkeys compared to NS and PBS-treated monkeys after treatment (Fig. 6k). In addition, we found that siRNA treatment did not cause any visible differences in appearance, behavior or sign of organ damage when examined by necropsy. To further evaluate its safety, we performed routine blood, blood coagulation and blood biochemistry tests, and found no statistically significant differences in these parameters among the three groups during 5-week treatment (Supplementary Fig. 8c-e).
Fig. 6.
RNA interference-based silencing of Plekho1 with osteoblast-selective delivery alleviates articular inflammation and inhibits bone erosion in non-human primate arthritis model induced by type II bovine collagen. (a) Body weight monitoring over time of each cynomolgus monkey after treatment. (b) Analysis of arthritis score over time of each cynomolgus monkey after treatment. (c) PIP joints swelling monitoring over time of each cynomolgus monkey after treatment. (d) Representative histological change of PIP joints from the hand of cynomolgus monkey in each group (four PIPs in each cynomolgus monkey, three cynomolgus monkeys in each group). Scale bar, 500 μm. (e) Comparison of histological inflammation score in each group. (f) Radiological score monitoring over time by X ray examination of the hand in each cynomolgus monkey. (g) Representative X ray image of the hand in each group (three cynomolgus monkeys in each group). (h) Representative 3D microarchitecture of the PIP in each group (four PIPs in each cynomolgus monkey, three cynomolgus monkeys in each group), obtained by microCT examination. Scale bars, 1.0 mm. (i) Analysis of the three-dimensional architecture parameters (BMD and BV/TV) for monkeys in each group. (j) Bone formation was examined by sequential labels with fluorescent dye in nondecalcified bone sections from cynomolgus monkeys. Representative fluorescent micrographs of PIP showed the xylenol (red) and calcein (green) labels in each group (four PIPs in each cynomolgus monkey, three cynomolgus monkeys in each group). Scale bar, 10 μm. (k) Analysis of bone histomorphometric parameters (MAR and BFR/BS) from cynomolgus monkeys in each group. All data are the mean ± s.d. *P < 0.05. A one-way ANOVA with subsequent Tukey's multiple comparisons test was performed. Note: CIA-BL, collagen-induced arthritis baseline; NS, (AspSerSer)6-liposome -NS siRNA; siRNA, (AspSerSer)6- liposome -Plekho1 siRNA; TB, treatment begins; PIP, proximal interphalangeal. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
This study uncovered a new role of osteoblastic PLEKHO1 in regulating joint inflammation in RA pathogenesis. We further demonstrated that the osteoblastic PLEKHO1 inhibition could not only augment bone formation but also inhibit joint inflammation in RA.
Previous views suggest that the joint inflammation in RA is mainly mediated by the activated inflammatory cells such as FLS and immune cells including infiltrated T lymphocytes and leukocytes [40]. However, it remains unclear whether other cell populations also contribute to regulate the local joint inflammation. Albeit osteoblasts are among the dominant cell types in the joint, the current perspectives restrict their role in the modulation of bone formation in RA. Interestingly, our study showed that osteoblasts could directly contribute to regulate the local joint inflammation during RA development. The inflammation environment of RA could induce the expression of PLEKHO1 within osteoblasts, as evidenced by the aberrant high levels of PLEKHO1 in osteoblasts of the joint bone samples from both RA patients and CIA mice as well as the increased PLEKHO1 expression in both human and mouse osteoblast-like cells stimulated by various inflammatory cytokines. More surprisingly, we found that genetic deletion of Plekho1 gene in osteoblasts markedly attenuated the local joint inflammation in inflammatory arthritic mouse models, as supported by the low arthritis scores and the histological inflammation scores as well as downregulated inflammatory cytokines (IL-1β and IL-6) in joint samples from the Plekho1osx−/− mice induced by type II collagen or K/BxN serum. Moreover, conditional overexpression of osteoblastic Plekho1 in the Plekho1 systemic knockout mice (Plekho1−/−/Plekho1osx Tg) significantly exacerbated the joint inflammation in CIA model and STA model. All these findings indicate that the osteoblasts with high PLEKHO1 expression contribute to the joint inflammation in RA. Despite the regulatory role on BMP signaling, PLEKHO1 has been identified to regulate the cytokine signaling response to inflammation [10,11]. A recent study showed that overexpression of PLEKHO1 triggered the activation of pro-inflammatory pathways in the HEK293 model system [41]. Consistently, we found lower levels of inflammatory cytokines (IL-1β and IL-6) in CD4 + T cells and FLS as well as decreased migration of neutrophils after co-culture with the Plekho1-depleted osteoblasts derived from the Plekho1osx−/− mice when compared to those co-cultured with Plekho1-intact osteoblasts. Furthermore, the above changes in the inflammatory cells were reversed after co-culture with the Plekho1-overexpressed osteoblasts derived from the Plekho1osx Tg mice. All these results suggest that the elevated osteoblastic PLEKHO1 could regulate the inflammatory cells that were commonly activated in the local inflammatory environment in RA joint. Cellular interactions are believed to be of importance in the pathogenesis of RA. These interactions are facilitated by the cytokines released from the activated cells that then, through both autocrine and paracrine mechanisms, induce the production of other proinflammatory cytokines, which together contribute to the pathogenesis of the disease [42]. FLS, CD4 + T lymphocytes and neutrophils could be activated by cytokine-driven stimuli, including IL-1β, IL-6, IL-18 and so on [43]. Moreover, in our study, we further found that silence of osteoblastic Plekho1 could remarkably decrease the production of IL-1β and IL-6. We thought that PLEKHO1 affecting the secretion of inflammatory cytokines in osteoblasts might be one of the ways in which osteoblasts affected the activities of FLS, CD4 + T lymphocytes and neutrophils.
TNF-α plays a pivotal role in regulating the inflammatory response in RA through inducing proinflammatory cytokines release, activating inflammatory cells and promoting leukocytes accumulation [42]. Recently, emerging evidence indicated that osteoblasts could respond to the stimulations of TNF-α by secretion of several inflammatory effectors [44,45]. Our studies showed that silence of osteoblastic Plekho1 could remarkably decrease the production of the important proinflammatory cytokines (IL-1β and IL-6). Moreover, we observed remarkable alleviation of local inflammation in human TNF-α transgenic mice with osteoblastic deficiency of Plekho1, implying that osteoblastic PLEKHO1 on joint inflammation regulation might be related with TNF-α involved signal mechanism. Therefore, to further investigate the molecular mechanism of osteoblastic PLEKHO1 modulating the joint inflammation in RA, we performed a series of in vitro studies. We found that the phosphorylation of IKKα/β and p65 as well as the ubiquitination of RIP1 in NF-κB signal pathway were remarkably inhibited in TNF-α-stimulated osteoblasts after Plekho1 silence. Moreover, we confirmed that PLEKHO1 could bind to TRAF2 and promote TRAF2-mediated RIP1 ubiquitination in TNF-α-stimulated osteoblasts, and the domains for the interaction were LZ-containing domain of PLEKHO1 and C domain of TRAF2 respectively. In contrast to the previous studies which demonstrated that lipid mediator sphingosine-1-phosphate specifically bound to TRAF2 and increased TRAF2-catalyzed ubiquitination of RIP1 [20], our study found a new cofactor “PLEKHO1” required for TRAF2-mediated RIP1 ubiquitination to activate NF-kB for inducing inflammatory cytokines production (Supplementary Fig. 9).
In addition, we observed that the osteoblast-specific Plekho1-depleted CIA mice showed an apparent attenuation in bone formation reduction and osteoblast-targeted Plekho1 siRNA treatment could promote bone repair in CIA rodents, which could be explained by the above documented role of PLEKHO1 as a critical ubiquitylation modulator for regulating the BMP signaling [7]. Certainly, PLEKHO1's effect on local inflammation regulation might also be considered as one of the mechanisms for its bone formation modulation, as the common view regarded that the inflammation could impair the function of osteoblasts-mediated bone formation in RA [46].
Considering the role of osteoblastic PLEKHO1 in regulating joint inflammation and bone erosion repair during RA, inhibition of it may exert dual beneficial effect on RA treatment. Thus, to facilitate its clinical translation, we evaluated the effect of osteoblast-targeted PLEKHO1 inhibition by administrating Plekho1 siRNA encapsulated by our developed osteoblast-targeting delivery system in a CIA mouse model and a non-human primate arthritis model. We demonstrated that osteoblast-targeting Plekho1 siRNA treatment not only improved arthritis symptoms but also promoted bone formation in both animal models. Moreover, the siRNA treatment didn't cause any significant side effects. However, there are still some limitations in our study. For example, in non-human primate experiment, the number of animals in each group is not big, we couldn't get more convincing data. Even so, these results will still facilitate the clinical translation of RNA interference-based therapeutic strategy in RA, since currently reports about siRNA-based therapeutic approaches for rheumatic diseases are almost from rodent experiments [47].
In summary, our studies uncovered a novel role of osteoblastic PLEKHO1 in mediating the joint inflammation during RA development. Targeting osteoblastic PLEKHO1 may be a promising therapeutic strategy with dual action of inflammation inhibition and bone formation augmentation for RA.
The following are the supplementary data related to this article.
Establishment of the osteoblast-specific Plekho1 conditional knockout (Plekho1osx-/-) mice. (a) Schematic diagram for development strategy to generate Plekho1floxp/- mice. (b) Identification of Plekho1osx-/- mice by PCR analysis. Plekho1fl/fl (271bp), Osx Cre (445bp). (c) Real time-PCR analysis for Plekho1 mRNA in Ocn+ cells isolated from WT mice (including Plekho1fl/fl mice, Osx+/-;Plekho1fl/- mice and Osx+/- mice) and Plekho1osx-/- mice (n=8 subjects each)
Bone phenotype changes in Plekho1osx-/- mice with CIA. (a) Representative 3D microarchitecture of the proximal tibia from Plekho1osx−/−-CIA mice and WT-CIA mice, obtained by microCT examination. Left: Cortical bone, Scale bars, 1mm. Right: Trabecular bone, Scale bars, 100 μm. (b) Comparison of the microCT parameters (BMD, BV/TV, Tb.N and Tb.Th) between Plekho1osx−/−-CIA mice and WT-CIA mice. (c) Bone formation examined by sequential labeling with fluorescent dye in non-decalcified bone sections. Representative fluorescent micrographs in the hind paws from Plekho1osx−/−-CIA mice and WT-CIA mice showing xylenol (red) and calcein (green) labeling. Scale bar, 10 μm. (d) Comparison of the bone histomorphometry parameters (MAR, BFR/BS, Ob.S/BS, Ob.N/B.Pm, Oc.S/BS and Oc.N/B.Pm) between Plekho1osx−/−-CIA mice and WT-CIA mice. *P < 0.05, **P < 0.01. Comparisons between two groups were performed using a Student’s t test. Note: BMD, bone mineral density; BV/TV, relative bone volume; Tb.Th, trabecular thickness, Tb.N, trabecular number, MAR, mineral apposition rate; BFR/BS, bone formation rate/bone surface; Ob.S/BS: osteoblast surface; Ob.N/B.Pm: osteoblast number; Oc.S/BS: osteoclast surface; Oc.N/B.Pm: osteoclast number; tBMD, trabecular BMD; tBV/TV, trabecular; cBMD, cortical BMD, cBV/TV, cortical BV/TV.
Genetic deletion of osteoblastic Plekho1 leads to amelioration of joint inflammation in K/BxN serum-transfer arthritis (STA) mice. (a) Time course changes in arthritis score from Plekho1osx−/− mice and WT mice (including Plekho1fl/fl mice, Osx+/-;Plekho1fl/- mice, and Osx+/- mice) after induction with K/BxN serum. (b) Representative histological images in the ankle joint of hind paws from Plekho1osx−/−-STA mice and WT-STA mice on day 12 after primary immunization. Scale bars, 50 μm. (c) Histological inflammation score in the ankle joint of hind paws from Plekho1osx−/−-STA mice and WT-STA mice on day 12 after primary immunization. (d) IL-1β and IL-6 levels in the ankle joint of hind paws from Plekho1osx−/−-STA mice and WT-STA mice on day 12 after primary immunization by ELISA examination. All data are the mean ± s.d. n = 9 per group. *P < 0.05, **P < 0.01. Two-way ANOVA with subsequent Bonferroni posttests was performed. Comparisons between two groups were performed using a Student’s t test.
Establishment of Plekho1-/-/Plekho1osxTg mice. (a) Schematic diagram for development strategy to generate ROSA26-PCAG-STOPfl-Plekho1-eGFP knock-in mice. (b) Identification of Plekho1osxTg mice by PCR analysis. Plekho1-GTfl/fl (236bp), Osx Cre (445bp). (c) Real time-PCR analysis for Plekho1 mRNA in Ocn+ cells isolated from Plekho1-/-/Plekho1osxTg mice, Plekho1-/- mice and Plekho1osxTg mice. **P < 0.01, compared to WT mice (including Osx+/-;Plekho1fl/- mice and Osx+/-;Plekho1fl/fl mice).
Overexpressed Plekho1 in osteoblasts exacerbates inflammation in Plekho1-/-/Plekho1osxTg mice with STA. (a) Disease progression assessed by arthritic score in WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice. *P < 0.05, compared to WT mice. (b) Representative histological images in the ankle joint of hind paws from WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice. Scale bars, 50μm. (c) Histological inflammation score in the ankle joint of hind paws from WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice. (d) IL-1β and IL-6 levels in the ankle joint of hind paws from WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice on day 12 after primary immunization by ELISA examination. All data are the mean ± s.d. n = 9 per group. *P < 0.05, **P < 0.01. For a, a two-way ANOVA with subsequent Bonferroni posttests was performed. For c-d, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed.
Deficiency of Plekho1 in osteoblasts alleviates joint inflammation in Plekho1osx−/−/hTNFtg mice. (a) Disease progression assessed by joint swelling score in hTNFtg and Plekho1osx−/−/hTNFtg mice from week 3 to 10. (b) Representative histological images in the ankle joint of the hind paws from hTNFtg and Plekho1osx−/−/hTNFtg mice. Scale bars, 50 μm. (c) Histological inflammation score in the ankle joint of the hind paws of hTNFtg and Plekho1osx−/−/hTNFtg mice. All data are the mean ± s.d. *P < 0.05, **P < 0.01. n = 10 per group. Two-way ANOVA with subsequent Bonferroni posttests was performed. Comparisons between two groups were performed using a Student’s t test.
Data from CIA mice treated with osteoblast-targeting delivery system encapsulated Plekho1 siRNA. (a) A schematic diagram for illustrating the experimental design. (b) Real-time PCR analysis of Plekho1 mRNA levels in Ocn+ cells isolated by LCM in the articular bone from the hind paws collected from the groups of mice indicated at day 70 after primary immunization. (c) Levels of IL-1β and IL-6 in ankle joint from the hind paws of the CIA mice after treatment in the respective group. All data are the mean ± s.d. *P < 0.05. **P < 0.01. n = 7 per group. A one-way ANOVA with subsequent Tukey's multiple comparisons test was performed. Notes: EB, experiment begins; TB, treatment begins; BL, baseline; CIA-BL, collagen-induced arthritis baseline; VEH, (AspSerSer)6-liposome; NS, (AspSerSer)6-liposome-NS siRNA; siRNA, (AspSerSer)6-liposome-Plekho1 siRNA.
Data from non-human primate arthritis model treated with osteoblast-selective Plekho1 siRNA. (a) A schematic diagram for illustrating the experimental design. (b) Real-time PCR analysis of Plekho1 mRNA levels in osteoblasts isolated by LCM in the articular bone from the PIP of the hand collected from the groups of cynomolgus monkeys indicated. (c) Changes of routine blood tests among the three groups. (d) Changes of blood coagulation tests among the three groups. (e) Changes of blood biochemistry tests among the three groups. All data are the mean ± s.d. **P < 0.01. For b, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed. For c-e, a two-way ANOVA with subsequent Bonferroni's multiple comparisons test was performed. Note: BL, baseline; CIA-BL, collagen-induced arthritis baseline; NS, (AspSerSer)6-liposome -NS siRNA; siRNA, (AspSerSer)6- liposome -Plekho1 siRNA; EB, experiment begins; TB, treatment begins; PIP, proximal interphalangeal.
A model of the role of PLEKHO1 in TRAF2-mediated NF-κB activation initiated by TNF-α. Binding of TNF-α to the trimeric TNFR1 results in the recruitment of TRADD, which then recruits TRAF2 and RIP1. PLEKHO1 can interact with TRAF2 to promote TRAF2-mediated RIP1 ubiquitination, which acts as a scaffold to recruit and activate IKK complexes. IKK then phosphorylates IκBα leading to the activation of NF-κB signaling pathway
Primers used for real-time PCR
Author contributions
G.Z. and A.L. supervised the whole project. X.H., J.L., C.L., S.A.B., K.Z. and L.D. performed the major research and wrote the manuscript in equal contribution. B.G., D.L., C.L., Q.G. and D.F. provided the technical support. Y.B., H.F., L.X. and X.P. collected human bone specimens. C.X. and B.Z. provided their professional expertise.
Competing financial interests
The authors declare no competing financial interests.
Acknowledgments
We thank the technical staff from Institute for Advancing Translational Medicine in Bone & Joint Diseases, Hong Kong Baptist University and Institute of Basic Theory, China Academy of Chinese Medical Sciences for providing critical comments and technical support. This study was supported by the Hong Kong General Research Fund (HKBU479111, HKBU478312, HKBU 12114416, HKBU262913, HKBU261113, HKBU12122516), the Natural Science Foundation Council of China (81272045, 81470072, 81700780), the Research Grants Council & Natural Science Foundation Council of China (N_HKBU435/12), the Hong Kong Baptist University Strategic Development Fund (SDF15-0324-P02), and the National Key R&D Program of China (2018YFC1705205). The funding institutions had not any role in study design, data collection, data analysis, interpretation or writing of the report in this study.
References
- 1.Walsh N.C. Rheumatic diseases: the effects of inflammation on bone. Immunol Rev. 2005;208:228–251. doi: 10.1111/j.0105-2896.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 2.Muller-Ladner U. Mechanisms of disease: the molecular and cellular basis of joint destruction in rheumatoid arthritis. Nat Clin Pract Rheumatol. 2005;1(2):102–110. doi: 10.1038/ncprheum0047. [DOI] [PubMed] [Google Scholar]
- 3.Chauhan V.S., Marriott I. Differential roles for NOD2 in osteoblast inflammatory immune responses to bacterial pathogens of bone tissue. J Med Microbiol. 2010;59(7):755–762. doi: 10.1099/jmm.0.015859-0. [DOI] [PubMed] [Google Scholar]
- 4.Dapunt U. The osteoblast as an inflammatory cell: production of cytokines in response to bacteria and components of bacterial biofilms. BMC Musculoskelet Disord. 2016;17 doi: 10.1186/s12891-016-1091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo B. Therapeutic RNA interference targeting CKIP-1 with a cross-species sequence to stimulate bone formation. Bone. 2014;59:76–88. doi: 10.1016/j.bone.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L. CKIP-1 regulates macrophage proliferation by inhibiting TRAF6-mediated Akt activation. Cell Res. 2014;24(6):742–761. doi: 10.1038/cr.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu K. Targeting WW domains linker of HECT-type ubiquitin ligase Smurf1 for activation by CKIP-1. Nat Cell Biol. 2008;10(8):994–1002. doi: 10.1038/ncb1760. [DOI] [PubMed] [Google Scholar]
- 8.Zhang G. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat Med. 2012;18(2):307–314. doi: 10.1038/nm.2617. [DOI] [PubMed] [Google Scholar]
- 9.Liu J. Increased PLEKHO1 within osteoblasts suppresses Smad-dependent BMP signaling to inhibit bone formation during aging. Aging Cell. 2017;16(2):360–376. doi: 10.1111/acel.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L.Q. The PH domain containing protein CKIP-1 binds to IFP35 and Nmi and is involved in cytokine signaling. Cell Signal. 2007;19(5):932–944. doi: 10.1016/j.cellsig.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Sakamoto T. CKIP-1 is an intrinsic negative regulator of T-cell activation through an interaction with CARMA1. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou X. Multiple functions of Osterix are required for bone growth and homeostasis in postnatal mice. Proc Natl Acad Sci U S A. 2010;107(29):12919–12924. doi: 10.1073/pnas.0912855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardy R.S. Characterisation of fibroblast-like synoviocytes from a murine model of joint inflammation. Arthritis Res Ther. 2013;15(1) doi: 10.1186/ar4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berger M. Neutrophils express distinct RNA receptors in a non-canonical way. J Biol Chem. 2012;287(23):19409–19417. doi: 10.1074/jbc.M112.353557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang L. CKIP-1 recruits nuclear ATM partially to the plasma membrane through interaction with ATM. Cell Signal. 2006;18(9):1386–1395. doi: 10.1016/j.cellsig.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y. CKIP-1 couples Smurf1 ubiquitin ligase with Rpt6 subunit of proteasome to promote substrate degradation. EMBO Rep. 2012;13(11):1004–1011. doi: 10.1038/embor.2012.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoeflich K.P. Mediation of TNF receptor-associated factor effector functions by apoptosis signal-regulating kinase-1 (ASK1) Oncogene. 1999;18(42):5814–5820. doi: 10.1038/sj.onc.1202975. [DOI] [PubMed] [Google Scholar]
- 18.Takeuchi M., Rothe M., Goeddel D.V. Anatomy of TRAF2. Distinct domains for nuclear factor-kappaB activation and association with tumor necrosis factor signaling proteins. J Biol Chem. 1996;271(33):19935–19942. doi: 10.1074/jbc.271.33.19935. [DOI] [PubMed] [Google Scholar]
- 19.Sondarva G. TRAF2-MLK3 interaction is essential for TNF-alpha-induced MLK3 activation. Cell Res. 2010;20(1):89–98. doi: 10.1038/cr.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez S.E. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 2010;465(7301):1084–1088. doi: 10.1038/nature09128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu J. Increased PLEKHO1 within osteoblasts suppresses Smad-dependent BMP signaling to inhibit bone formation during aging. Aging Cell. 2017;16(2):360–376. doi: 10.1111/acel.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brand D.D., Latham K.A., Rosloniec E.F. Collagen-induced arthritis. Nat Protoc. 2007;2(5):1269–1275. doi: 10.1038/nprot.2007.173. [DOI] [PubMed] [Google Scholar]
- 23.Inglis J.J. Collagen-induced arthritis in C57BL/6 mice is associated with a robust and sustained T-cell response to type II collagen. Arthritis Res Ther. 2007;9(5) doi: 10.1186/ar2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D.M. Mast cells: a cellular link between autoantibodies and inflammatory arthritis. Science. 2002;297(5587):1689–1692. doi: 10.1126/science.1073176. [DOI] [PubMed] [Google Scholar]
- 25.Johnsen A.K. Genome-wide and species-wide dissection of the genetics of arthritis severity in heterogeneous stock mice. Arthritis Rheum. 2011;63(9):2630–2640. doi: 10.1002/art.30425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato A. Early effects of tocilizumab on bone and bone marrow lesions in a collagen-induced arthritis monkey model. Exp Mol Pathol. 2008;84(3):262–270. doi: 10.1016/j.yexmp.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Horai N. Muscle wasting associated with pathologic change is a risk factor for the exacerbation of joint swelling in collagen-induced arthritis in cynomolgus monkeys. BMC Musculoskelet Disord. 2013;14 doi: 10.1186/1471-2474-14-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rau R., Herborn G. Development and evaluation of a modified version of Larsen's scoring method to assess radiologic changes in rheumatoid arthritis. Zeitschrift Fur Rheumatologie. 1997;56(3):127–135. doi: 10.1007/s003930050028. [DOI] [PubMed] [Google Scholar]
- 29.Rioja I. Joint cytokine quantification in two rodent arthritis models: kinetics of expression, correlation of mRNA and protein levels and response to prednisolone treatment. Clin Exp Immunol. 2004;137(1):65–73. doi: 10.1111/j.1365-2249.2004.02499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G. Continuous occurrence of both insufficient neovascularization and elevated vascular permeability in rabbit proximal femur during inadequate repair of steroid-associated osteonecrotic lesions. Arthritis Rheum. 2009;60(10):2966–2977. doi: 10.1002/art.24847. [DOI] [PubMed] [Google Scholar]
- 31.Walsh N.C. Osteoblast function is compromised at sites of focal bone erosion in inflammatory arthritis. J Bone Miner Res. 2009;24(9):1572–1585. doi: 10.1359/jbmr.090320. [DOI] [PubMed] [Google Scholar]
- 32.Poliachik S.L. Transient muscle paralysis disrupts bone homeostasis by rapid degradation of bone morphology. Bone. 2010;46(1):18–23. doi: 10.1016/j.bone.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guma M. Caspase 1-independent activation of Interleukin-1 beta in neutrophil-predominant inflammation. Arthritis Rheum. 2009;60(12):3642–3650. doi: 10.1002/art.24959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choy E. Understanding the dynamics: pathways involved in the pathogenesis of rheumatoid arthritis. Rheumatology (Oxford) 2012;51(Suppl. 5):v3–11. doi: 10.1093/rheumatology/kes113. [DOI] [PubMed] [Google Scholar]
- 35.Hayden M.S., Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 36.Tsai C.L. TNF-alpha induces matrix metalloproteinase-9-dependent soluble intercellular adhesion molecule-1 release via TRAF2-mediated MAPKs and NF-kappaB activation in osteoblast-like MC3T3-E1 cells. J Biomed Sci. 2014;21:12. doi: 10.1186/1423-0127-21-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aggarwal B.B. Signalling pathways of the TNF superfamily: a double-edged sword. Nat Rev Immunol. 2003;3(9):745–756. doi: 10.1038/nri1184. [DOI] [PubMed] [Google Scholar]
- 38.Zheng C., Yin Q., Wu H. Structural studies of NF-kappaB signaling. Cell Res. 2011;21(1):183–195. doi: 10.1038/cr.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z.J. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol. 2005;7(8):758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McInnes I.B., Schett G. MECHANISMS OF DISEASE the pathogenesis of rheumatoid arthritis. New England J Med. 2011;365(23):2205–2219. doi: 10.1056/NEJMra1004965. [DOI] [PubMed] [Google Scholar]
- 41.Juhasz K. Casein kinase 2-interacting protein-1, an inflammatory signaling molecule interferes with TNF reverse signaling in human model cells. Immunol Lett. 2013;152(1):55–64. doi: 10.1016/j.imlet.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Brennan F.M., McInnes I.B. Evidence that cytokines play a role in rheumatoid arthritis. J Clin Invest. 2008;118(11):3537–3545. doi: 10.1172/JCI36389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McInnes I.B., Schett G. Cytokines in the pathogenesis of rheumatoid arthritis. Nat Rev Immunol. 2007;7(6):429–442. doi: 10.1038/nri2094. [DOI] [PubMed] [Google Scholar]
- 44.Shen F. Cytokines link osteoblasts and inflammation: microarray analysis of interleukin-17- and TNF-alpha-induced genes in bone cells. J Leukoc Biol. 2005;77(3):388–399. doi: 10.1189/jlb.0904490. [DOI] [PubMed] [Google Scholar]
- 45.Confalone E., D'Alessio G., Furia A. IL-6 induction by TNFalpha and IL-1beta in an osteoblast-like cell line. Int J Biomed Sci. 2010;6(2):135–140. [PMC free article] [PubMed] [Google Scholar]
- 46.Baum R., Gravallese E.M. Impact of inflammation on the osteoblast in rheumatic diseases. Curr Osteoporos Rep. 2014;12(1):9–16. doi: 10.1007/s11914-013-0183-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apparailly F., Jorgensen C. siRNA-based therapeutic approaches for rheumatic diseases. Nat Rev Rheumatol. 2013;9(1):56–62. doi: 10.1038/nrrheum.2012.176. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Establishment of the osteoblast-specific Plekho1 conditional knockout (Plekho1osx-/-) mice. (a) Schematic diagram for development strategy to generate Plekho1floxp/- mice. (b) Identification of Plekho1osx-/- mice by PCR analysis. Plekho1fl/fl (271bp), Osx Cre (445bp). (c) Real time-PCR analysis for Plekho1 mRNA in Ocn+ cells isolated from WT mice (including Plekho1fl/fl mice, Osx+/-;Plekho1fl/- mice and Osx+/- mice) and Plekho1osx-/- mice (n=8 subjects each)
Bone phenotype changes in Plekho1osx-/- mice with CIA. (a) Representative 3D microarchitecture of the proximal tibia from Plekho1osx−/−-CIA mice and WT-CIA mice, obtained by microCT examination. Left: Cortical bone, Scale bars, 1mm. Right: Trabecular bone, Scale bars, 100 μm. (b) Comparison of the microCT parameters (BMD, BV/TV, Tb.N and Tb.Th) between Plekho1osx−/−-CIA mice and WT-CIA mice. (c) Bone formation examined by sequential labeling with fluorescent dye in non-decalcified bone sections. Representative fluorescent micrographs in the hind paws from Plekho1osx−/−-CIA mice and WT-CIA mice showing xylenol (red) and calcein (green) labeling. Scale bar, 10 μm. (d) Comparison of the bone histomorphometry parameters (MAR, BFR/BS, Ob.S/BS, Ob.N/B.Pm, Oc.S/BS and Oc.N/B.Pm) between Plekho1osx−/−-CIA mice and WT-CIA mice. *P < 0.05, **P < 0.01. Comparisons between two groups were performed using a Student’s t test. Note: BMD, bone mineral density; BV/TV, relative bone volume; Tb.Th, trabecular thickness, Tb.N, trabecular number, MAR, mineral apposition rate; BFR/BS, bone formation rate/bone surface; Ob.S/BS: osteoblast surface; Ob.N/B.Pm: osteoblast number; Oc.S/BS: osteoclast surface; Oc.N/B.Pm: osteoclast number; tBMD, trabecular BMD; tBV/TV, trabecular; cBMD, cortical BMD, cBV/TV, cortical BV/TV.
Genetic deletion of osteoblastic Plekho1 leads to amelioration of joint inflammation in K/BxN serum-transfer arthritis (STA) mice. (a) Time course changes in arthritis score from Plekho1osx−/− mice and WT mice (including Plekho1fl/fl mice, Osx+/-;Plekho1fl/- mice, and Osx+/- mice) after induction with K/BxN serum. (b) Representative histological images in the ankle joint of hind paws from Plekho1osx−/−-STA mice and WT-STA mice on day 12 after primary immunization. Scale bars, 50 μm. (c) Histological inflammation score in the ankle joint of hind paws from Plekho1osx−/−-STA mice and WT-STA mice on day 12 after primary immunization. (d) IL-1β and IL-6 levels in the ankle joint of hind paws from Plekho1osx−/−-STA mice and WT-STA mice on day 12 after primary immunization by ELISA examination. All data are the mean ± s.d. n = 9 per group. *P < 0.05, **P < 0.01. Two-way ANOVA with subsequent Bonferroni posttests was performed. Comparisons between two groups were performed using a Student’s t test.
Establishment of Plekho1-/-/Plekho1osxTg mice. (a) Schematic diagram for development strategy to generate ROSA26-PCAG-STOPfl-Plekho1-eGFP knock-in mice. (b) Identification of Plekho1osxTg mice by PCR analysis. Plekho1-GTfl/fl (236bp), Osx Cre (445bp). (c) Real time-PCR analysis for Plekho1 mRNA in Ocn+ cells isolated from Plekho1-/-/Plekho1osxTg mice, Plekho1-/- mice and Plekho1osxTg mice. **P < 0.01, compared to WT mice (including Osx+/-;Plekho1fl/- mice and Osx+/-;Plekho1fl/fl mice).
Overexpressed Plekho1 in osteoblasts exacerbates inflammation in Plekho1-/-/Plekho1osxTg mice with STA. (a) Disease progression assessed by arthritic score in WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice. *P < 0.05, compared to WT mice. (b) Representative histological images in the ankle joint of hind paws from WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice. Scale bars, 50μm. (c) Histological inflammation score in the ankle joint of hind paws from WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice. (d) IL-1β and IL-6 levels in the ankle joint of hind paws from WT-STA, Plekho1-/--STA and Plekho1-/-/Plekho1osxTg-STA mice on day 12 after primary immunization by ELISA examination. All data are the mean ± s.d. n = 9 per group. *P < 0.05, **P < 0.01. For a, a two-way ANOVA with subsequent Bonferroni posttests was performed. For c-d, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed.
Deficiency of Plekho1 in osteoblasts alleviates joint inflammation in Plekho1osx−/−/hTNFtg mice. (a) Disease progression assessed by joint swelling score in hTNFtg and Plekho1osx−/−/hTNFtg mice from week 3 to 10. (b) Representative histological images in the ankle joint of the hind paws from hTNFtg and Plekho1osx−/−/hTNFtg mice. Scale bars, 50 μm. (c) Histological inflammation score in the ankle joint of the hind paws of hTNFtg and Plekho1osx−/−/hTNFtg mice. All data are the mean ± s.d. *P < 0.05, **P < 0.01. n = 10 per group. Two-way ANOVA with subsequent Bonferroni posttests was performed. Comparisons between two groups were performed using a Student’s t test.
Data from CIA mice treated with osteoblast-targeting delivery system encapsulated Plekho1 siRNA. (a) A schematic diagram for illustrating the experimental design. (b) Real-time PCR analysis of Plekho1 mRNA levels in Ocn+ cells isolated by LCM in the articular bone from the hind paws collected from the groups of mice indicated at day 70 after primary immunization. (c) Levels of IL-1β and IL-6 in ankle joint from the hind paws of the CIA mice after treatment in the respective group. All data are the mean ± s.d. *P < 0.05. **P < 0.01. n = 7 per group. A one-way ANOVA with subsequent Tukey's multiple comparisons test was performed. Notes: EB, experiment begins; TB, treatment begins; BL, baseline; CIA-BL, collagen-induced arthritis baseline; VEH, (AspSerSer)6-liposome; NS, (AspSerSer)6-liposome-NS siRNA; siRNA, (AspSerSer)6-liposome-Plekho1 siRNA.
Data from non-human primate arthritis model treated with osteoblast-selective Plekho1 siRNA. (a) A schematic diagram for illustrating the experimental design. (b) Real-time PCR analysis of Plekho1 mRNA levels in osteoblasts isolated by LCM in the articular bone from the PIP of the hand collected from the groups of cynomolgus monkeys indicated. (c) Changes of routine blood tests among the three groups. (d) Changes of blood coagulation tests among the three groups. (e) Changes of blood biochemistry tests among the three groups. All data are the mean ± s.d. **P < 0.01. For b, a one-way ANOVA with subsequent Tukey's multiple comparisons test was performed. For c-e, a two-way ANOVA with subsequent Bonferroni's multiple comparisons test was performed. Note: BL, baseline; CIA-BL, collagen-induced arthritis baseline; NS, (AspSerSer)6-liposome -NS siRNA; siRNA, (AspSerSer)6- liposome -Plekho1 siRNA; EB, experiment begins; TB, treatment begins; PIP, proximal interphalangeal.
A model of the role of PLEKHO1 in TRAF2-mediated NF-κB activation initiated by TNF-α. Binding of TNF-α to the trimeric TNFR1 results in the recruitment of TRADD, which then recruits TRAF2 and RIP1. PLEKHO1 can interact with TRAF2 to promote TRAF2-mediated RIP1 ubiquitination, which acts as a scaffold to recruit and activate IKK complexes. IKK then phosphorylates IκBα leading to the activation of NF-κB signaling pathway
Primers used for real-time PCR