Abstract
This study aimed at evaluating the transcriptional profile of apoptosis-related genes after in vitro stimulation of peripheral blood mononuclear cells (PBMCs) derived from individuals with periodontitis (P) and healthy nonperiodontitis (NP) control subjects with P. gingivalis HmuY protein. PBMCs from the P and NP groups were stimulated with HmuY P. gingivalis protein, and the expression of genes related to apoptosis was assessed by custom real-time polymerase chain reaction array (Custom RT2 PCR Array). Compared with the NP group, the P group showed low relative levels of apoptosis-related gene expression, downregulated for FAS, FAS ligand, TNFSF10 (TRAIL), BAK1, CASP9, and APAF1 after P. gingivalis HmuY protein stimulation. Furthermore, the P group exhibited low levels of relative gene expression, downregulated for CASP7 when the cells were not stimulated. Our data suggest that P. gingivalis HmuY protein might participate differently in the modulation of the intrinsic and extrinsic apoptosis pathways.
1. Introduction
Periodontitis is a multifactorial disease, with significant participation of the host, environmental factors, and bacterial components. It is known that keystone pathogens, such as Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola, in the subgingival biofilm elicit a host inflammatory response, which can lead to periodontal breakdown. [1]. The microbial diversity of the oral cavity is immense, and the host response during periodontitis is complex, with components of the innate and adaptive immune system that lead to chronic inflammation and bone resorption [2].
Recent metagenomic and mechanistic studies are consistent with a new periodontal pathogenesis model that proposes that periodontal diseases are caused by a synergistic and dysbiotic microbial community, not by a selected group of bacteria known as “periodontopathogens”. Bacteria found in low abundance in the microbiota have an effect throughout the community and are critical components for the development of dysbiosis. They are known as “keystone” pathogens [3]. However, an increased abundance in these known pathogens is observed, related to the presence as well as the severity of the disease, indicating a microbial variation in the dysbiotic process [4].
Virulence factors of P. gingivalis, the main keystone pathogen in periodontitis, can determine a great immunogenicity to stimulate innate and adaptive immune host responses. Among them are capsule components, lipopolysaccharide (LPS), fimbriae, and outer membrane proteins, such as associated and secreted gingipains. Gingipains specific for arginine (HRgpA and RgpB) and lysine (Kgp) play an important role in the infection, supporting the onset and progression of periodontitis [5].
P. gingivalis can invade the periodontal tissues and use a panel of virulence factors to evade the innate immune and inflammatory responses [6]. It has been shown that this pathogen has the ability to invade epithelial cells [7], which can be an escape mechanism from host defenses, favoring the penetration of the microorganism into the bloodstream and thus acting systemically in the human body [8].
P. gingivalis ability to obtain nutrients and evade the host immune response in the microenvironment is directly related to its survival, proliferation, and infection. One of the essential nutrients for its development is iron. This component is most abundant in the host in the form of heme [9]. The heme capture can be performed by a P. gingivalis HmuY, which sequesters and delivers heme to the outer membrane receptor HmuR [10–12].
There is a syntrophy among the species of the oral biofilm, revealing the interspecies cooperation in the acquisition of nutrients [13]. This mechanism reveals how HmuY works together with proteases produced also by other bacterial species to acquire heme from hemoglobin and may represent mutualism between P. gingivalis and Prevotella intermedia cohabiting the periodontal pocket [14]. However, HmuY is recognized by highly specific antibodies, suggesting that this protein can serve as a specific antigen of this bacterium [15].
P. gingivalis HmuY also seems to act in the programmed cell death process. Human peripheral blood mononuclear cells (PBMCs) cultured with the protein appear to be unable to complete the process of apoptosis, resulting in death characterized by the release of inflammatory cell content in the microenvironment, such as late apoptosis and necrosis, which can prolong the tissue destruction process [16]. This protein induces high levels of Bcl-2 in the mononuclear cells of individuals with periodontitis, resulting in the inhibition of apoptosis and thereafter enabling increased survival of CD3+ T cells, which prolong the chronic inflammatory condition in periodontitis [17].
The modulation of cell death molecules and mechanisms evolved by periodontopathogens during the establishment of periodontitis has been demonstrated [18–21]. However, the apoptosis of mononuclear cells under bacterial challenge is unclear. The present study identifies specific P. gingivalis components that are involved in these events, which is the novelty of the study. In addition, the role of HmuY in triggering the extrinsic and intrinsic apoptosis pathways is not well understood. Thus, this study was carried out to evaluate the transcriptional profile of genes involved in apoptosis mechanisms in PBMC under P. gingivalis HmuY protein stimulation.
2. Materials and Methods
2.1. Participants
The present study was approved by the Feira de Santana State University Institutional Review Board (No. 0203.0.059.000-11). All volunteers in this study signed the free and informed consent form. A total of 8 individuals with severe periodontitis (P group) and 8 nonperiodontitis controls (NP group) were recruited between 2013 and 2014 from the College of Dentistry at the Feira de Santana State University, Bahia, Brazil. Volunteers with diabetes, pregnancy, smoking habit, any autoimmune disease, cardiovascular disease, prior periodontal treatment, and anti-inflammatories and antibiotics used within two and six months before inclusion in this study, respectively, were the exclusion criteria.
2.2. Disease Classification
A dentist (P.C.C.F.) was trained in the calibration process for periodontal examination (kappa interexaminer agreement value = 0.932) using a Williams periodontal probe (Hu-Friedy, Chicago, IL, USA). The following clinical parameters were evaluated: probing depth (PD), clinical attachment level (CAL), and bleeding on probing (BOP).
Participants were diagnosed as having periodontitis (P group) if they had at least four teeth with at least one site with a probing depth greater than or equal to 4 mm, clinical attachment loss of 3 mm or more, and bleeding on probing at the same site [22]. All individuals with periodontitis had a diagnosis of severe form of the disease, since they had at least 2 interproximal sites with clinical attachment loss greater than or equal to 6 mm (not affecting the same tooth) and at least 1 interproximal site with probing depth greater than or equal to 5 mm [23].
Those participants who did not meet these criteria were considered not to have periodontitis (NP group).
2.3. Antigen
The P. gingivalis HmuY protein (NCBI ID number: CAM 31898) was overexpressed, purified, and lyophilized as described previously [24] and subsequently dissolved in PBS to a final concentration of 2.5 μg/mL.
2.4. Blood Collection and Cell Culture
Peripheral venous blood (20 mL) was collected from the volunteers in heparinized tubes. Peripheral blood mononuclear cells (PBMCs) were obtained by density centrifugation in accordance with the manufacturer's guidelines (SepCell, StemCell Technologies Inc., USA) and then cultured in flat-bottom 24-well plates (106 cells/well) in RPMI (Roswell Park Memorial Institute) medium (LGCBio, São Paulo, SP, Brazil), containing 10% fetal calf serum (inactivated by heat) and 1% antibiotic/antimycotic drug (R&D Systems, Minneapolis, MN, USA). Cells (106 per well) were incubated with 5 μg/mL of pokeweed mitogen (PWM), 2.5 μg/mL of P. gingivalis HmuY protein, or in the absence of antigens (nonstimulated cells), under 5% CO2 in humid conditions for 48 h at 37°C.
2.5. Gene Expression Analysis
RNA was extracted from the cells using a miRNeasy Mini Kit (SABiosciences Corp.) in accordance with the manufacturer's instructions. cDNA was synthesized using 500 ng of the total RNA and a RT2 First Strand Kit (SABiosciences Corp.).
The Custom Human RT2 Profiler PCR Array: CAPH12794 (SABiosciences Corp.), designed for this study, was used for the sample analysis. Altogether, 45 different genes were simultaneously amplified in the sample using 384-well plates. To verify the presence of a single amplicon, a melting curve was performed.
PCR arrays were carried out using Applied Biosystems QuantStudio 12K Flex Real-Time PCR System 384-well block (Applied Biosystems). 10 μL of a mixture composed of 2× SABiosciences RT2 qPCR Master Mix and cDNA was applied into each well. The Ct values were calculated for each gene using the Applied Biosystems software. Baseline and threshold measurements were determined according to the manufacturer's guidelines (SABiosciences, Qiagen).
2.6. Statistical Analysis
The software supplied by Qiagen (https://www.qiagen.com/br/shop/genes-and-pathways/data-analysis-center-overview-page) identified glucuronidase beta (GUSB) as the most stably expressed housekeeping gene for data normalization. The fold change in gene expression (compared to positive control – PBMC – P group) was calculated using the ΔΔCt method. A more than twofold change in gene expression (compared to positive control – PBMC – P group) was considered as the up- or downregulation of a specific gene expression.
The RT2 Profiler PCR Array Data Analysis software does not perform any statistical analysis beyond the calculation of p values using Student's t-test based on 2^(-ΔCT) values for each gene P group compared to the NP group. The Microarray Quality Control (MAQC) published results indicating that a ranked list of genes based on fold change and such a p value calculation was sufficient to demonstrate reproducible results across multiple microarray and PCR Arrays including the RT2 Profiler PCR Arrays [25].
3. Results
The comparison between the periodontitis (P) and nonperiodontitis (NP) groups demonstrated that there were no statistically significant differences in the sex (p = 0.96) or age (p = 0.57) (Table 1). On the other hand, periodontal status was worse in the periodontitis individuals, who showed a higher percentage of teeth with bleeding on probing (p = 0.03), probing depth ≥ 4 mm (p = 0.001), and clinical attachment loss ≥ 3 mm (p = 0.001).
Table 1.
Clinical findings of patients with periodontitis (P) and nonperiodontitis (NP) subjects.
| NP | P | p value | |
|---|---|---|---|
| Number of men/women | 9/17 | 7/13 | 0.958 |
| Age (years) (mean ± SD) | 37.92 ± 10.8 | 41.95 ± 9.7 | 0.565 |
| % Bleeding on probing (mean ± SD) | 8.45 ± 10.7 | 37.97 ± 16.9 | 0.028 |
| % Probing depth ≥ 4 (mean ± SD) | 1.23 ± 1.84 | 14.93 ± 8.91 | 0.001 |
| % Clinical attachment loss ≥ 3 (mean ± SD) | 19.60 ± 15.53 | 57.04 ± 17.25 | 0.001 |
SD = standard deviation, p = significance level (p ≤ 0.05).
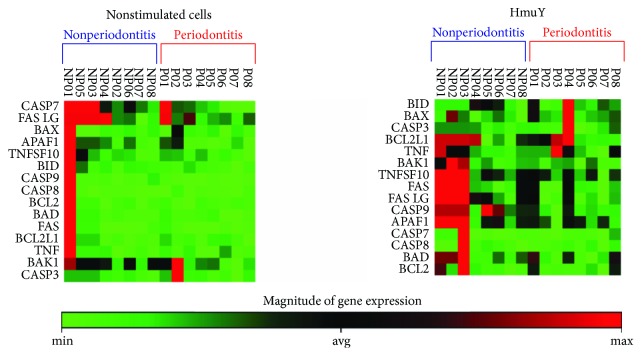
3.1. Expression Profile of Apoptosis Gene Targets: Heat Map
The analysis of the mRNA expression pattern indicated differences between the P and NP groups both in cells without stimulation (nonstimulated cells) and under P. gingivalis HmuY protein stimulus (HmuY). The variability in the pattern of global gene expression between the P and NP samples indicated heterogeneity, particularly in those stimulated with HmuY protein. Under both conditions of culturing, stimulation with HmuY (HmuY) and without antigenic stimulation (nonstimulated cells), PBMC of individuals of the NP group showed a higher number of genes upregulated, and on the other hand, individuals with periodontitis showed a downregulated gene expression profile. Furthermore, differences were more evident in the NP and P groups when stimulated with P. gingivalis HmuY protein (HmuY), as can be observed in Figure 1.
Figure 1.
Gene cluster identification involved in apoptosis mechanisms by PBMC from individuals with periodontitis (P) and nonperiodontitis (NP) under P. gingivalis HmuY protein (HmuY) stimulus and in the absence of stimulus (nonstimulated cells). The green-black-red color gradient represents relative levels of gene expression, indicating under-even-over regulation, respectively.
In nonstimulated cells, the expression of CASP7 and FASLG was upregulated in a higher number of individuals of the NP group (3 and 4 individuals, respectively). Patient NP1, without the diagnosis of periodontitis, showed upregulation in most of the genes examined. In the P group, the upregulation of CASP7, FAS, BAK, and CASP3 occurred in one case for each gene and in two cases for BCL2L1. Most of the genes evaluated were downregulated in the cells cultured without stimulation in this group.
BCL2L1 was upregulated in four out of eight individuals without periodontitis after HmuY stimulation, while only one individual showed upregulation when the cells were cultured without stimulus. Similarly, three individuals showed upregulation of TNFSF10, FAS, FASLG, CASP9, and APAF1 under HmuY challenge. In the P group, after stimulation with HmuY protein, the individual who showed upregulation in BID, BAX, CASP3, and BCL2L1 also showed indetermination in TNF, TNFSF10, FAS, FASLG, CASP9, and APAF1, differently from the cells grown without stimulus, which showed downregulation in the case of all these genes. The other individual who showed upregulation in BCL2L1 also demonstrated upregulation in TNF. The pattern of regulation changed from downregulation to indetermination in most of the cases.
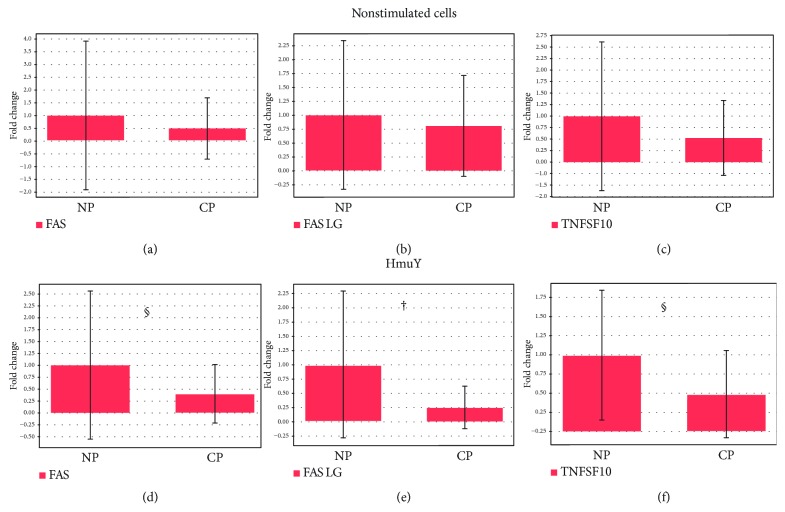
3.2. Apoptosis-Related Gene Expression in the Extrinsic Pathway
PBMC from the P group showed downregulation and lower levels for FAS ligand compared to those from the NP group (p = 0.01), as observed in Figure 2, under stimulation with P. gingivalis HmuY protein. Under the same conditions, the downregulation of FAS and TNFSF10 in the P group, but not statistically significant (p = 0.09; p = 0.06, respectively) (Figures 2(d) and 2(f)), was observed. When PBMCs were cultured without stimulus, no statistically significant difference was found for FAS (p = 0.31), FAS ligand (p = 0.43), or TNFSF10 (TRAIL) (p = 0.22) expression between the P and NP groups (Figures 2(a)–2(c)).
Figure 2.
Apoptosis-related extrinsic pathway gene expression in the PBMC of the (a-c) P and NP groups without stimuli and under (d-f) P. gingivalis HmuY protein stimulation. (a, d) FAS (TNF receptor superfamily, member 6); (b, e) FAS LG (TNF superfamily, member 6 ligand); (c, f) TNFSF10 (tumor necrosis factor ligand superfamily-10). †p = 0.01 and §p < 0.1.
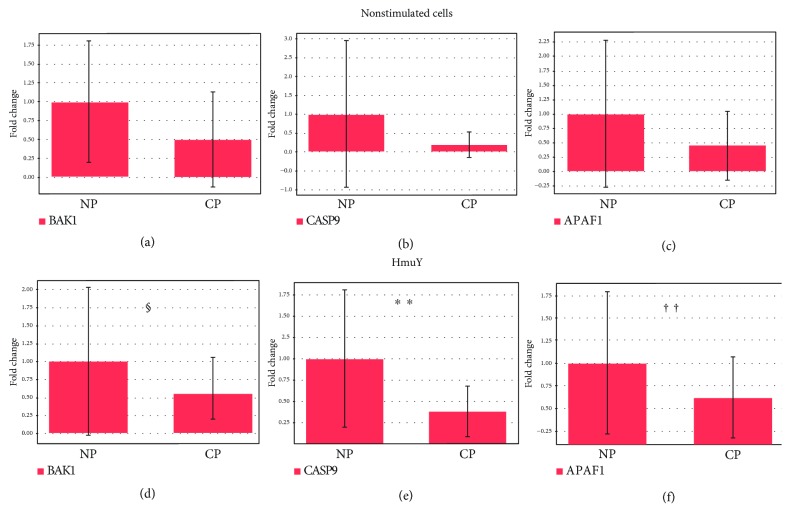
3.3. Apoptosis-Related Gene Expression in the Intrinsic Pathway
The downregulation and lower levels of CASP9 (p = 0.003) and APAF1 (p = 0.048) in PBMC in the P group compared to PBMC from the NP group were observed, when the cells were cultured under stimulation of P. gingivalis HmuY protein (Figures 3(e) and 3(f)). Downregulation was found in the P group for the expression of BAK1, compared to the NP group, but this difference is not statistically significant (p = 0.085) (Figure 3(a)). There was no statistically significant difference between the P and NP groups in the BAK1 (p = 0.35), CASP9 (p = 0.25), and APAF1 (p = 0.6) expression for unstimulated PBMC (nonstimulated cells) (Figures 3(d)–3(f)).
Figure 3.
Apoptosis-related intrinsic pathway gene expression in the PBMC of the (a-c) P and NP groups without stimuli and under (d-f) P. gingivalis HmuY protein stimulation. (a, d) BAK1 (BCL2-antagonist/killer 1); (b, e) CASP9 (caspase 9); (c, f) APAF1 (apoptotic peptidase activating factor-1). ∗∗p = 0.01, ††p = 0.05, and §p < 0.1.
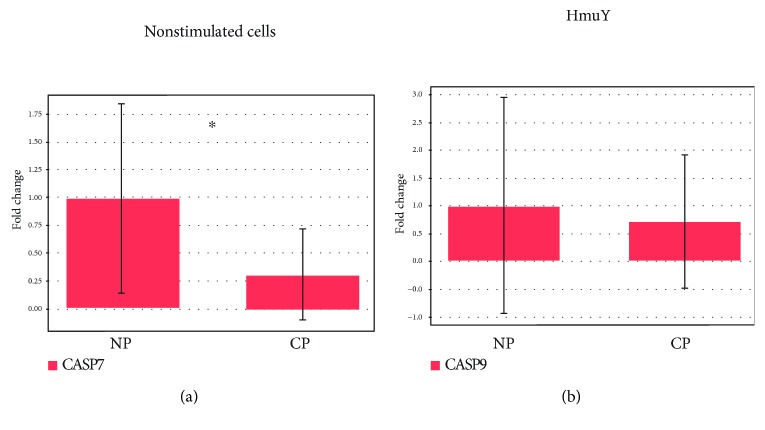
3.4. Caspase 7 Gene Expression
PBMC from the P and NP groups stimulated with P. gingivalis HmuY protein (HmuY) did not demonstrate a statistically significant difference for caspase 7 (p = 0.40) (Figure 4(a)). However, there was downregulation and lower levels for caspase 7 in unstimulated cells from the P group in comparison to the NP group (p = 0.05) (Figure 4(b)).
Figure 4.
Caspase 7 expression in the PBMC of the (a) P and NP groups without stimulation (nonstimulated cells) and under (b) P. gingivalis HmuY protein stimulation. Caspase 7 (apoptosis-related cysteine peptidase). ∗p = 0.05.
4. Discussion
The most common outcome in inflammatory/infectious conditions is cell death by necrosis [26]. However, previous studies have demonstrated both induction and inhibition of apoptosis mechanisms in different cells types through the challenge of oral microorganisms and/or their components [21, 27–31]. It was demonstrated that the upregulation of apoptosis-related proteins occurred in gingival epithelial cells challenged with Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola [32]. In addition, P. gingivalis products seem to be able to induce the expression of genes related to apoptosis in osteogenic bone marrow stromal cells [33] and THP-1 cells [34]. The potential of P. gingivalis HmuY protein to interfere with cell death mechanisms in PBMC has been shown previously [16, 17].
In this study, the involvement of genes encoding apoptosis-related proteins that participate in the intrinsic and extrinsic pathways of cell death that were expressed differently in the PBMC of the P and NP groups was demonstrated. In the extrinsic pathway, HmuY was able to downregulate the expression of FASL in individuals with periodontitis. In the intrinsic pathway, P. gingivalis HmuY protein downregulated the caspase 9 and APAF1 expression in cells from diseased individuals. Thus, it seems that P. gingivalis uses this protein to suppress apoptosis of the defense host cells through both pathways, corroborating previous studies that proposed a role of delaying apoptosis [16] and increasing the production of BCL-2 [17] by HmuY.
Several studies have demonstrated many factors to suppress or inhibit apoptosis, showing the dependence of genes which encode pro- and antiapoptotic proteins in the balance of this process [35, 36]. In the presence of a bacterial challenge, it is possible to observe DNA damage associated with apoptosis and expression of p53, BCL-2 [37], FAS, FASL, and active caspase 3 in gingival tissues [38]. P. gingivalis can induce apoptosis in human gingival epithelial cells through the increase of FASL expression and the increase of the gene transcription mediated by NFκB [39]. The oligomerization of APAF1, induced by its binding to cytochrome c, forms an apoptosome, a known structure that recruits and activates a caspase initiator, caspase 9, which, in turn, cleaves and activates caspase effector caspase 3 and caspase 7, leading to apoptosis [40].
Regarding FAS, TNFSF10 (extrinsic pathway), and BAK1 (intrinsic pathway), consistently with the results shown above, the downregulation of mRNA expression was observed in cells from individuals with periodontitis under HmuY stimulus. However, the differences found between this group and individuals without the disease showed borderline p values. Therefore, these results must be confirmed in studies using a larger sample size to improve the statistical power. These findings showed that P. gingivalis HmuY protein may impair or retard the process of apoptosis by downregulation of genes that initiate the extrinsic and intrinsic signaling pathways in periodontitis. It is known that in the extrinsic pathway, apoptosis can be induced by cell surface receptors, such as FAS, TNFR1, and TNFSF10 [41–43], while intrinsic apoptotic stimuli can activate BAX and BAK and, consequently, cause mitochondrial outer membrane permeabilization [40].
In a previous study, using P. gingivalis lipopolysaccharide, the upregulation of mRNA for IL-1β and FAS ligand was identified in a mouse periodontitis model [44]. Furthermore, the expression of mRNA for FAS and FAS-L in human gingival epithelial cells (CEGH) was upregulated by heat-killed P. gingivalis, and programmed cell death was induced [39].
In the assessment of caspase 7, downregulation in the mRNA expression in cells from periodontitis individuals cultured without stimulus in comparison to cells from control individuals has been demonstrated. This difference was not observed when the cells were cultured in the presence of P. gingivalis HmuY protein. Caspase 7 has been found in inflammatory conditions induced by bacterial infections to be involved as an effector molecule in apoptosis responsible for inducing cell detachment and ROS production in a redundant way with caspase 3 [45, 46].
Thereby, the negative modulation through the inhibition of the apoptotic process can enable P. gingivalis to infect host cells and provide an escape mechanism from the host immune system. These findings suggest that this keystone pathogen can reduce or inhibit PBMC apoptosis through downregulation of mRNA genes which are involved in the intrinsic and extrinsic pathways of early proapoptotic signaling in programmed cell death under P. gingivalis HmuY protein stimulation in individuals with periodontitis. This process could lead to continuous production of proinflammatory cytokines and chemokines in the microenvironment of the inflamed periodontal tissue, improving survival and chemotaxis of cells involved in immune defense mechanisms. It can result in a chronic inflammatory state of destruction of periodontal tissues and possibly in the establishment of a dysbiotic process in affected individuals.
We are aware that a limitation of the present study is the relatively small sample size, which suggests the need for further studies with larger numbers of participants to establish the role of the P. gingivalis HmuY protein in inflammatory and molecular mechanisms of cell death, providing new insights into the immunopathogenesis of periodontitis. In addition, the experiments were carried out using PBMCs and cannot be extrapolated to the microenvironment of the periodontal lesion, in which resident cells play a major role in the pathogenesis of the disease in vivo.
In conclusion, P. gingivalis HmuY protein might contribute to the survival of PBMC in periodontitis by regulating the transcriptional profile of genes involved in apoptosis, such as FASL, caspase 9, APAF1, FAS, TNFSF10 (TRAIL), and BAK1. It can lead to the perpetuation and aggravation of the inflammatory condition of the periodontal structures through the release of more inflammatory mediators which contribute to the breakdown of tissues.
Acknowledgments
This study was supported by the Research Support Foundation of the State of Bahia, Brazil (grant no. APP019/2011), the National Science Center, Poland (NCN; grant no. 2015/17/B/NZ6/01969), and a student fellowship from the Ministry of Education, Brazil. The authors thank the Laboratory of Immunology and Molecular Biology and Laboratory Allergy and Acarology at UFBA for allowing the use of the real-time PCR system.
Data Availability
The data underlying the findings of this study are available in the online repository: https://repositorio.ufba.br/ri/bitstream/ri/21723/1/TESE%20PAULO%20CIRINO.pdf [47].
Conflicts of Interest
The authors have declared no competing interests.
References
- 1.Deo V., Bhongade M. L. Pathogenesis of periodontitis: role of cytokines in host response. Dentistry Today. 2010;29(9):60–62. [PubMed] [Google Scholar]
- 2.Huang N., Gibson F. C. Immuno-pathogenesis of periodontal disease: current and emerging paradigms. Current Oral Health Reports. 2014;1(2):124–132. doi: 10.1007/s40496-014-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajishengallis G., Lamont R. J. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. European Journal of Immunology. 2014;44(2):328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutin S., Hagenfeld D., Zimmermann H., et al. Clustering of subgingival microbiota reveals microbial disease ecotypes associated with clinical stages of periodontitis in a cross-sectional study. Frontiers in Microbiology. 2017;8:p. 340. doi: 10.3389/fmicb.2017.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zenobia C., Hajishengallis G. Porphyromonas gingivalis virulence factors involved in subversion of leukocytes and microbial dysbiosis. Virulence. 2015;6(3):236–243. doi: 10.1080/21505594.2014.999567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bostanci N., Belibasakis G. N. Porphyromonas gingivalis: an invasive and evasive opportunistic oral pathogen. FEMS Microbiology Letters. 2012;333(1):1–9. doi: 10.1111/j.1574-6968.2012.02579.x. [DOI] [PubMed] [Google Scholar]
- 7.Njoroge T., Genco R. J., Sojar H. T., Hamada N., Genco C. A. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infection and Immunity. 1997;65(5):1980–1984. doi: 10.1128/iai.65.5.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walter C., Zahlten J., Schmeck B., et al. Porphyromonas gingivalis strain-dependent activation of human endothelial cells. Infection and Immunity. 2004;72(10):5910–5918. doi: 10.1128/IAI.72.10.5910-5918.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olczak T., Simpson W., Liu X., Genco C. A. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiology Reviews. 2005;29(1):119–144. doi: 10.1016/j.femsre.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Olczak T., Siudeja K., Olczak M. Purification and initial characterization of a novel Porphyromonas gingivalis HmuY protein expressed in Escherichia coli and insect cells. Protein Expression and Purification. 2006;49(2):299–306. doi: 10.1016/j.pep.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 11.Olczak T., Sroka A., Potempa J., Olczak M. Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Archives of Microbiology. 2008;189(3):197–210. doi: 10.1007/s00203-007-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wójtowicz H., Guevara T., Tallant C., et al. Unique structure and stability of HmuY, a novel heme-binding protein of Porphyromonas gingivalis. PLoS Pathogens. 2009;5(5, article e1000419) doi: 10.1371/journal.ppat.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hajishengallis G., Lamont R. J. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends in Microbiology. 2016;24(6):477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne D. P., Potempa J., Olczak T., Smalley J. W. Evidence of mutualism between two periodontal pathogens: co-operative haem acquisition by the HmuY haemophore of Porphyromonas gingivalis and the cysteine protease interpain a (InpA) of Prevotella intermedia. Molecular Oral Microbiology. 2013;28(3):219–229. doi: 10.1111/omi.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smiga M., Bielecki M., Olczak M., Smalley J. W., Olczak T. Anti-HmuY antibodies specifically recognize Porphyromonas gingivalis HmuY protein but not homologous proteins in other periodontopathogens. PLoS One. 2015;10(2, article e0117508) doi: 10.1371/journal.pone.0117508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trindade S. C., Olczak T., Gomes-Filho I. S., et al. Porphyromonas gingivalis antigens differently participate in the proliferation and cell death of human PBMC. Archives of Oral Biology. 2012;57(3):314–320. doi: 10.1016/j.archoralbio.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Carvalho-Filho P. C., Trindade S. C., Olczak T., et al. Porphyromonas gingivalis HmuY stimulates expression of Bcl-2 and Fas by human CD3+T cells. BMC Microbiology. 2013;13(1):p. 206. doi: 10.1186/1471-2180-13-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pradeep A. R., Suke D. K., Prasad M. V. R., et al. Expression of key executioner of apoptosis caspase-3 in periodontal health and disease. Journal of Investigative and Clinical Dentistry. 2016;7(2):174–179. doi: 10.1111/jicd.12134. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y., Hou J., Peng L., et al. The pro-apoptotic and pro-inflammatory effects of calprotectin on human periodontal ligament cells. PLoS One. 2014;9(10, article e110421) doi: 10.1371/journal.pone.0110421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ao M., Miyauchi M., Inubushi T., et al. Infection with Porphyromonas gingivalis exacerbates endothelial injury in obese mice. PLoS One. 2014;9(10, article e110519) doi: 10.1371/journal.pone.0110519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q., Pan C., Teng D., et al. Porphyromonas gingivalis modulates Pseudomonas aeruginosa-induced apoptosis of respiratory epithelial cells through the STAT3 signaling pathway. Microbes and Infection. 2014;16(1):17–27. doi: 10.1016/j.micinf.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Gomes-Filho I. S., Cruz S. S., Rezende E. J. C., et al. Exposure measurement in the association between periodontal disease and prematurity/low birth weight. Journal of Clinical Periodontology. 2007;34(11):957–963. doi: 10.1111/j.1600-051X.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- 23.Page R. C., Eke P. I. Case definitions for use in population-based surveillance of periodontitis. Journal of Periodontology. 2007;78(7s):1387–1399. doi: 10.1902/jop.2007.060264. [DOI] [PubMed] [Google Scholar]
- 24.Olczak T., Wojtowicz H., Ciuraszkiewicz J., Olczak M. Species specificity, surface exposure, protein expression, immunogenicity, and participation in biofilm formation of Porphyromonas gingivalis HmuY. BMC Microbiology. 2010;10(1):p. 134. doi: 10.1186/1471-2180-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi L., Campbell G., Jones W. D., et al. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nature Biotechnology. 2010;28(8):827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green D. R., Llambi F. Cell death signaling. Cold Spring Harbor Perspectives in Biology. 2015;7(12) doi: 10.1101/cshperspect.a006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Q., Sztukowska M., Ojo A., Scott D. A., Wang H., Lamont R. J. FOXO responses to Porphyromonas gingivalis in epithelial cells. Cellular Microbiology. 2015;17(11):1605–1617. doi: 10.1111/cmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimoto T., Fujita T., Kajiya M., et al. Involvement of smad2 and Erk/Akt cascade in TGF-β1-induced apoptosis in human gingival epithelial cells. Cytokine. 2015;75(1):165–173. doi: 10.1016/j.cyto.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 29.Zhao P., Liu J., Pan C., Pan Y. NLRP3 inflammasome is required for apoptosis of Aggregatibacter actinomycetemcomitans-infected human osteoblastic MG63 cells. Acta Histochemica. 2014;116(7):1119–1124. doi: 10.1016/j.acthis.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Dittmann C., Doueiri S., Kluge R., Dommisch H., Gaber T., Pischon N. Porphyromonas gingivalis suppresses differentiation and increases apoptosis of osteoblasts from New Zealand obese mice. Journal of Periodontology. 2015;86(9):1095–1102. doi: 10.1902/jop.2015.150032. [DOI] [PubMed] [Google Scholar]
- 31.Bostanci N., Thurnheer T., Aduse-Opoku J., Curtis M. A., Zinkernagel A. S., Belibasakis G. N. Porphyromonas gingivalis regulates TREM-1 in human polymorphonuclear neutrophils via its gingipains. PLoS One. 2013;8(10, article e75784) doi: 10.1371/journal.pone.0075784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bostanci N., Bao K., Wahlander A., Grossmann J., Thurnheer T., Belibasakis G. N. Secretome of gingival epithelium in response to subgingival biofilms. Molecular oral Microbiology. 2015;30(4):323–335. doi: 10.1111/omi.12096. [DOI] [PubMed] [Google Scholar]
- 33.Reddi D., Belibasakis G. N. Transcriptional profiling of bone marrow stromal cells in response to Porphyromonas gingivalis secreted products. PLoS One. 2012;7(8, article e43899) doi: 10.1371/journal.pone.0043899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carayol N., Chen J., Yang F., et al. A dominant function of IKK/NF-κB signaling in global lipopolysaccharide-induced gene expression. The Journal of Biological Chemistry. 2006;281(41):31142–31151. doi: 10.1074/jbc.M603417200. [DOI] [PubMed] [Google Scholar]
- 35.Bugueno I. M., Khelif Y., Seelam N., et al. Porphyromonas gingivalis differentially modulates cell death profile in Ox-LDL and TNF-α pre-treated endothelial cells. PLoS One. 2016;11(4, article e0154590) doi: 10.1371/journal.pone.0154590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu X., Wang Y., Lin J., et al. Lipopolysaccharides-induced suppression of innate-like B cell apoptosis is enhanced by CpG oligodeoxynucleotide and requires toll-like receptors 2 and 4. PLoS One. 2016;11(11, article e0165862) doi: 10.1371/journal.pone.0165862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tonetti M. S., Cortellini D., Lang N. P. In situ detection of apoptosis at sites of chronic bacterially induced inflammation in human gingiva. Infection and Immunity. 1998;66(11):5190–5195. doi: 10.1128/iai.66.11.5190-5195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gamonal J., Bascones A., Acevedo A., Blanco E., Silva A. Apoptosis in chronic adult periodontitis analyzed by in situ DNA breaks, electron microscopy, and immunohistochemistry. Journal of Periodontology. 2001;72(4):517–525. doi: 10.1902/jop.2001.72.4.517. [DOI] [PubMed] [Google Scholar]
- 39.Brozovic S., Sahoo R., Barve S., et al. Porphyromonas gingivalis enhances FasL expression via up-regulation of NFκB-mediated gene transcription and induces apoptotic cell death in human gingival epithelial cells. Microbiology. 2006;152(3):797–806. doi: 10.1099/mic.0.28472-0. [DOI] [PubMed] [Google Scholar]
- 40.Tait S. W. G., Green D. R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nature Reviews Molecular Cell Biology. 2010;11(9):621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 41.Giogha C., Lung T. W. F., Pearson J. S., Hartland E. L. Inhibition of death receptor signaling by bacterial gut pathogens. Cytokine & Growth Factor Reviews. 2014;25(2):235–243. doi: 10.1016/j.cytogfr.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Amarante-Mendes G. P., Griffith T. S. Therapeutic applications of TRAIL receptor agonists in cancer and beyond. Pharmacology & Therapeutics. 2015;155:117–131. doi: 10.1016/j.pharmthera.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunetti G., Oranger A., Mori G., et al. TRAIL effect on osteoclast formation in physiological and pathological conditions. Frontiers in Bioscience. 2011;E3(3):1154–1161. doi: 10.2741/e318. [DOI] [PubMed] [Google Scholar]
- 44.Firth J. D., Ekuni D., Irie K., Tomofuji T., Morita M., Putnins E. E. Lipopolysaccharide induces a stromal-epithelial signalling axis in a rat model of chronic periodontitis. Journal of Clinical Periodontology. 2013;40(1):8–17. doi: 10.1111/jcpe.12023. [DOI] [PubMed] [Google Scholar]
- 45.Brentnall M., Rodriguez-Menocal L., De Guevara R. L., Cepero E., Boise L. H. Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell Biology. 2013;14(1):p. 32. doi: 10.1186/1471-2121-14-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamkanfi M., Kanneganti T. D. Caspase-7: a protease involved in apoptosis and inflammation. The International Journal of Biochemistry & Cell Biology. 2010;42(1):21–24. doi: 10.1016/j.biocel.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho-Filho P. C. Cytokine production and mRNA expression in peripheral blood mononuclear cells under stimuli of Porphyromonas gingivalis rHmuY in chronic periodontitis. [Ph.D. thesis] Salvador: Federal University of Bahia, Available from: UFBA Institutional repository; 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying the findings of this study are available in the online repository: https://repositorio.ufba.br/ri/bitstream/ri/21723/1/TESE%20PAULO%20CIRINO.pdf [47].