Abstract
Background
c-Src is a driver oncogene well-known for tumorigenic signaling, but little for metabolic function. Previous reports about c-Src regulation of glucose metabolism prompted us to investigate its function in other nutrient modulation, particularly in lipid metabolism.
Methods
Oil-red O staining, cell growth assay, and tumor volume measurement were performed to determine lipid amount and growth inhibitory effect of treatments in lung cancer cells and xenograft model. Gene expression was evaluated by immunoblotting and relative RT-PCR. Transcriptional activity of peroxisome proliferator-activated receptor gamma (PPARγ) was assessed by luciferase assay. Reactive oxygen species (ROS) was measured using ROS sensing dye. Oxygen consumption rate was evaluated by Seahorse XF Mito Stress Test. Clinical relevance of candidate proteins was examined using patient samples and public database analysis.
Findings
Inhibition of Src induced lipolysis and increased intracellular ROS. Src inhibition derepressed PPARγ transcriptional activity leading to induced expression of lipolytic gene fatty acid binding protein (FABP) 4 which accompanies reduced lipid droplets and decreased tumor growth. The reverse correlation of Src and FABP4 was confirmed in pair-matched lung cancer patient samples, and further analysis using public datasets revealed upregulation of lipolytic genes is associated with better prognosis of cancer patients.
Interpretation
This study provides an insight of how oncogenic factor Src concurrently regulates both cellular signaling pathways and metabolic plasticity to drive cancer progression.
Fund
National Research Foundation of Korea and Korea Health Industry Development Institute.
Keywords: PPARγ, Src, FABP4, Lipid, Lung cancer
Research in context.
Evidence before this study
Reprogramming lipid metabolism associated with redox balance control has been contributed to cancer development. As a driver oncogene, c-Src is well-studied in cancer signaling pathways. However, little is known for its involvement in cancer lipid metabolism.
Added value of this study
This study investigated biological function, downstream regulator and clinical relevance of oncogene Src regulating lipid metabolism in lung cancer. We showed that inhibition of Src can induce lipolysis by derepressing PPARγ activity which upregulates the expression of FABP4, a PPARγ's target gene. As lipid droplets are known as ROS scavenger, lipolysis induced by Src suppression consistently increased ROS accumulation in an FABP4 dependent manner. Furthermore, the negative correlation between Src and FABP4 was confirmed in pair-matched patient samples as well as from analyzing public database. This study also proposed lipolytic regulators as potential prognostic biomarkers which can be translated into the clinics.
Implications of all the available evidence
These findings expands our understandings of how oncogene Src concurrently regulates cellular signaling pathways as well as metabolic plasticity to drive cancer progression. Moreover, this study proposed a good prognosis of lipolysis which indicates that targeting lipid accumulation may be reasonable for lung cancer treatment.
Alt-text: Unlabelled Box
1. Introduction
Cancer metabolism, as one of key cancer hallmarks, has featured metabolic rewiring or adaptation of cancer cells in utilizing nutrients to overcome diverse pathophysiological stresses including oncogenic signaling, nutrient availability, and oxygen concentration [1]. Nutrient biochemistry has been extensively studied in cancer metabolism for fundamental nutrients including glucose, amino acids, and lipid [2]. Calorie restriction or nutrient starvation has become believed to benefit cancer patients for anti-cancer drug treatment [3]. Hence, targeting metabolic alteration would be considered as a novel therapeutic approach in cancer, in particular, with acquired drug resistance. These days, understanding cancer metabolism has become more detailed and complicated by linking to various oncogenic signaling pathways involving c-Myc, hypoxia inducible factor 1α (HIF1α), and K-ras [4]. The intimidate connection of biochemistry and molecular oncology in nutrient utilization would not only help better understanding of cancer biology, but also become a milestone for metabolic therapeutics to overcome anti-cancer drug resistance [1]. c-Src, an oncogenic non-receptor tyrosine kinase, potentiates diverse intracellular signaling cascades promoting cancer progression [5]. Recent studies for Src involvement in cancer metabolism reported critical roles of Src in glucose metabolism where it contributes to the Warburg effect or aerobic glycolysis by inactivating pyruvate dehydrogenase complex, and by phosphorylating hexokinase 1 and 2 leading to increased glycolysis [6,7]. These recent works would suggest the more extended potential of oncogenic Src functioning in other nutrient modulations (e.g. lipid, amino acid) in cancer.
Recently, several studies have reported that intratumoral lipid droplets contribute to cancer maintenance, aggressiveness, and drug resistance. For instance, intracellular lipid droplets become resource for ATP generation in glioblastoma [8], contribute to chemoresistance in colorectal cancer [9], and play a role as an antioxidant to protect tumor cells from oxidative stress in breast cancer [10]. Inter- or Intracellular lipid mobilization involves multiple mechanisms by which fatty acid binding protein (FABP) gene family is involved in modulating lipid fluxes and trafficking [11]. FABP4 knock-out mice showed decreased lipolysis [12], suggesting intracellular lipolytic function of FABP4 protein. Even with the manifest role of metabolic regulation in normal physiology, FABP4 function in cancer is less clear and even become controversial. While overexpressed FABP4 showed tumor suppressive function leading to apoptosis in prostate cancer, FABP4 upregulation in ovarian tumor metastasized into the omental area further promotes ovarian cancer metastasis into that area by transporting fatty acid from the surrounding adipocyte to ovarian tumor [13,14]. Similarly, the prognostic potential of FABP4 expression is controversially reported in lung cancer, which remains to be elucidated [15,16]. As an upstream factor of FABP4 expression [17], peroxisome proliferator-activated receptor gamma (PPARγ) belonging to the nuclear receptor superfamily is a master regulator in lipid metabolism by controlling networks of gene expression for lipid accumulation, lipolysis and white-to-brown transition in white adipocyte [[18], [19], [20]] which suggests leading role of PPARγ in lipid metabolism. Our recent study showed PPARγ as a tumor suppressor for lipid metabolic function in lung cancer where PPARγ-mediated fatty acid synthesis decreases intracellular nicotinamide adenine dinucleotide phosphate (NADPH) level, resulting in ROS-mediated cell growth suppression in lung cancer [21].
In the present study, we showed functional inhibition of oncogenic Src decreases lipid droplets by upregulating PPARγ-mediated FABP4 expression, which accompanies increased intracellular ROS. In addition, the higher expression of lipolysis genes, FABP4 and lipoprotein lipase (LPL) in tumor showed the better prognosis of lung and renal cancer patients. Taken together, this study provides a novel understanding of Src function in lipid metabolic reprogramming to promote tumorigenesis, and thus an insight of cancer therapeutics into targeting lipid metabolism in oncogene Src-driven tumors.
2. Materials and methods
2.1. Cell culture and reagents
Lung, renal cancer cell lines, and HEK293 cells were cultured in RPMI 1640 or DMEM medium supplemented with 5% or 10% fetal bovine serum (FBS), 50 U/mL penicillin, and 50 U/mL streptomycin at 37 °C with 5% CO2. Purchased are various chemicals including pioglitazone (Cat# sc-204848) from Santa Cruz, SU6656 (Cat# sc-203286A) from Santa Cruz or SU6656 (Cat# S7774) from Selleckchem, PP2 (Cat# 1767-1) from BioVision, HTS01037 (Cat# 10699-10) from Cayman Chemical and Stattic (Cat# S7947), Thiazolyl Blue Tetrazolium Blue (Cat# M2128) or Oil-red O (Cat# O1391) from Sigma-Aldrich. Included are cell lines for relevant experiments in this study (Table S1).
2.2. Plasmids
Expression vectors include pCDNA-BLRP tagged wtPPARγ and pCDNA vector as described previously [21,22], wtSrc-GFP [[23], [24], [25]] kindly provided by M. Frame (The Beatson Institute for Cancer Research, Glasgow, Scotland) and Yoav I. Henis (Tel Aviv University, Tel Aviv, Israel), pCDNA c-Abl Δ1-81 [26] from Yosef Shaul (Weizmann Institute of Science, Rehovot, Israel), pLL-EGFR-vIII from Jong Bae Park (National Cancer Center, Goyang, Korea), pCMV-Stat3 and pCMV control from Ki Woo Kim (Yonsei University, Seoul, Korea). Yes-EGFP and pcFlag-Fyn-wt were donated by Bernardo Mainou (unpublished) (Addgene plasmid #110497) and Lars Rönnstrand [27] (Addgene plasmid #74509), respectively. Various mutant constructs including constitutive active SrcY527F-GFP, kinase-dead SrcK295 M-GFP, SrcR175A-GFP with inactivated SH3 domain, SrcW118A-GFP with inactivated SH2 domain, and phospho-dead mutant PPARγY78F were generated using Pfu Plus 5× PCR Master Mix from Elpis Biotech (Cat# EBT1403) following the site-directed mutagenesis method as in literature [28]. Refer to supplementary table S2 for primer sequences.
2.3. siRNA transfection
Calu6 cells were transfected with negative control siRNA (Bioneer, Cat #SN-1002) or SMARTpool siGENOME SRC siRNA 50 μM (Dharmacon, Cat# M-003175-03-0005) and/or SMARTpool siGENOME FABP4 siRNA 50 μM (Dharmacon, Cat# M-008853-00-0005). All siRNA transfections were performed with lipofectamin 3000 (Invitrogen, Cat# L3000015) according to the manufacturer's recommendation. All siRNA target sequence are listed in table S3.
2.4. Immunoblot and immunoprecipitation assays
Cells or homogenized tumor tissues were lysed using RIPA buffer followed by immunoblot assay as previously reported [21]. Primary antibodies include β-actin (Abcam Cat# ab6276, RRID:AB_2223210), 4HNE (Abcam Cat# ab46545, RRID:AB_722490), GAPDH (Santa Cruz Biotechnology Cat# sc-25778, RRID:AB_10167668), FYN (BD Cat# 610164, RRID:AB_397565), phospho-Stat3 (Y705) (Cell Signaling Technology Cat# 9131, RRID:AB_331586), Stat3 (Cell Signaling Technology Cat# 9139, RRID:AB_331757), pSrc (Y416) (Cell Signaling Technology Cat# 6943, RRID:AB_10013641), Src (Cell Signaling Technology Cat# 2108, RRID:AB_331137), PPARγ (Cell Signaling Technology Cat# 2435, RRID:AB_2166051), FABP4 (Cell Signaling Technology Cat# 3544, RRID:AB_2278257), GFP (Cell Signaling Technology Cat# 2955, RRID:AB_1196614), Cyclin A2 (Cell Signaling Technology Cat# 4656, RRID:AB_2071958), PARP (Cell Signaling Technology Cat# 9542, RRID:AB_2160739), pAMPK (Cell Signaling Technology Cat# 2535, RRID:AB_331250), AMPK (Cell Signaling Technology Cat# 5832, RRID:AB_10624867), Cleaved Caspase 9 (Cell Signaling Technology Cat# 9501, RRID:AB_331424), and Cleaved Caspase 3 (Cell Signaling Technology Cat# 9664, RRID:AB_2070042). Secondary antibodies include HRP conjugated anti-mouse IgG (Abcam Cat# ab6728, RRID:AB_955440), and anti-rabbit IgG (Innovative Research Cat# G-21234, RRID:AB_1500696). For immunoprecipitation assay, HEK293 cells were transfected with 300 ng of biotin-protein ligase (BirA), 1000 ng of BLRP-tagged wtPPARγ or pCDNA control, 1000 ng of wtSrc-GFP, kinase dead SrcK295 M-GFP or pEGFP control followed by 5 μM of SU6656 treatment for 24 h. As previously reported [21], immunoprecipitation was carried out using streptavidin resin beads (Thermo) at 4 °C overnight and washed four times in phosphate-buffered saline with 0∙5% NP40. The immunoprecipitates were resolved by SDS-PAGE and immunoblotted using antibodies including PPARγ (Cell Signaling Technology Cat# 2435, RRID:AB_2166051) and GFP (Cell Signaling Technology Cat# 2955, RRID:AB_1196614). For quantification, the signal intensity of the blot was measured using ImageJ.
2.5. Relative RT-PCR analysis
Total RNAs were extracted using TRIzol reagent (Invitrogen) and followed by reverse-transcription to cDNA using ReverTra Ace® qPCR Master Mix (Toyobo) with the treatment of DNase as the manufacturer's instruction. Relative RT-PCR assay was conducted using ABI Prism 7900 HT Sequence Detection System (Applied Biosystems). Triplicates of each PCR reaction were performed using SYBR green real-time PCR master mixes (Life Technologies). Delta-delta Ct method was used to analyze data using 18S rRNA as an internal reference and the change in expression of gene of interest was normalized to untreated control sample [29]. In all RT-PCR, the y-axis represents fold change in gene expression. Refer to supplementary Table S4 for primer sequences.
2.6. Oil-red O (ORO) staining
Cells were seeded on coverslips and treated with compounds as indicated in each figure. Lipid staining was performed as described previously using 0∙5% ORO solution [21]. Lipid droplets in the pictures were quantified using ImageJ software as in the literature [30].
2.7. Luciferase assay
Luciferase assay was performed using HEK293 cells transfected with 200 ng of renilla luciferase as a control reporter vector, 600 ng of TK-PPRE3x-Luc plasmid expressing luciferase gene under the control of PPAR response element and thymidine kinase promoter, and 600 ng of plasmids of interest or control as indicated. Note that luciferase assays were carried out in parallel with immunoblot assays. Renilla or firefly luciferase activity was measured using Dual Luciferase Reporter Assay System (Promega). Relative light unit (RLU) represents PPARγ transcriptional activity.
2.8. Cell growth assay
To carry out cell proliferation assay, ten thousands of cells were seeded in commercially available culture media. After treatment, cells were stained with 0∙4% methylene blue in 50% methanol. For MTT assay, 5 mg/mL MTT solution was prepared by dissolving Thiazolyl Blue Tetrazolium Blue in PBS. Cell viability was evaluated by incubating with MTT solution for two hours, and followed by dissolving blue formazan crystal in DMSO and measuring optical density at 570 nm.
2.9. Intracellular ROS measurement
Intracellular ROS level was measured using CM-H2DCF-DA (2′-7′dichlorofluorescin diacetate) (Molecular Probes) as previously described [31]. For ROS measurement, cells were washed twice with Krebs-Ringer Bicarbonate (KRB) buffer and incubated in 1 or 2∙5 μM of CM-H2DCF-DA solution for H1993 or Calu6 cells, respectively, for 15 min at 37 °C. After additional washings with the KRB buffer, ten different regions of cell images were taken using inverted microscope (IX81, Olympus) equipped with an array laser Nipkow spinning disk (CSU10, Yokogawa Electric Corporation). Fluorescence intensity was quantified using MetaMorph software (Molecular Devices).
2.10. Mitochondrial oxygen consumption rate
Both Calu6 and H1993 cells were plated at 2500 cells/well in a 96-well microplate. Cells were treated at the following day with drug or vehicle for three days. The analysis procedure for mitochondrial stress test was followed as the manufacturer instruction (Agilent). Oxygen consumption rate was measured basally and following drug injection including 2 μM oligomycin, 0∙5 μM FCCP and 0∙5 μM each rotenone/antimycin A. The reads were normalized by protein amounts measured using Pierce BCA Protein Assay Kit (Thermo).
2.11. Gene expression analysis
A microarray dataset (GSE4824) of lung cancer cell lines from GEO database was analyzed for lipid metabolic genes using Matrix 1.29 (Matrix Service Company) [32,33]. For expression analysis of Src, FABP4 and LPL genes, the cohort Provisional TCGA data for lung adenocarcinoma and renal cell carcinoma were obtained from cbioportal.com [34,35]. Pearson correlation analysis between the genes was carried out in patients who have upregulation of the genes indicated by z-score > 2.
2.12. Patient samples
Fresh-frozen tissue samples were obtained from tissue bank in department of pathology (Wonju Severance Christian Hospital). Five pairs of lung tissues include lung adenocarcinoma and the corresponding normal tissues from the same patients, which are histologically confirmed. The sample acquirement was approved under the Committee of Institutional Review Board (Approval number: CR318314).
2.13. Survival analysis
For prognostic analysis, datasets were obtained from TCGA and GEO databases. The datasets include clinical information and gene expression of the cohort Provisional TCGA database, GSE8894, and GSE11117 for lung cancer patients or GDC TCGA kidney clear cell carcinoma for renal cancer patients [[34], [35], [36], [37], [38]]. Kaplan-Meier plot shows patient survival on FABP4 or LPL expression and log-rank test shows statistical significance.
2.14. Xenograft experiment
Animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Yonsei University Wonju College of Medicine (Approval number: YWC-170907-3). To establish the mouse xenograft model, five millions of Calu6 cells were subcutaneously injected into the right flank region of 4-week old female Balb/c nude mice. When tumors grew enough to be tangible, mice were randomly divided into two groups (Vehicle group n = 5, SU6656 group n = 7). Mice were intraperitoneally treated with SU6656 (20 mg/kg) every other day for 23 days. Tumor volume and body weight were measured every two days, and tumor weight was measured after sacrificing mice at the end of the experiment. Tumor volumes were determined with a digital caliper and calculated using the formula ½ × (width2 × length). The animal experiments were compliant with the animal care guideline of Yonsei University Wonju College of Medicine.
2.15. Statistics
All graphing and statistical analysis including two-tailed Student's t-test, ANOVA, Pearson correlation coefficient and log-rank test were performed using GraphPad Prism version 6.0. Data were presented as mean ± SEM (n ≥ 3).
3. Results
3.1. Oncogenic Src is involved in lipid droplet accumulation in cancer cells
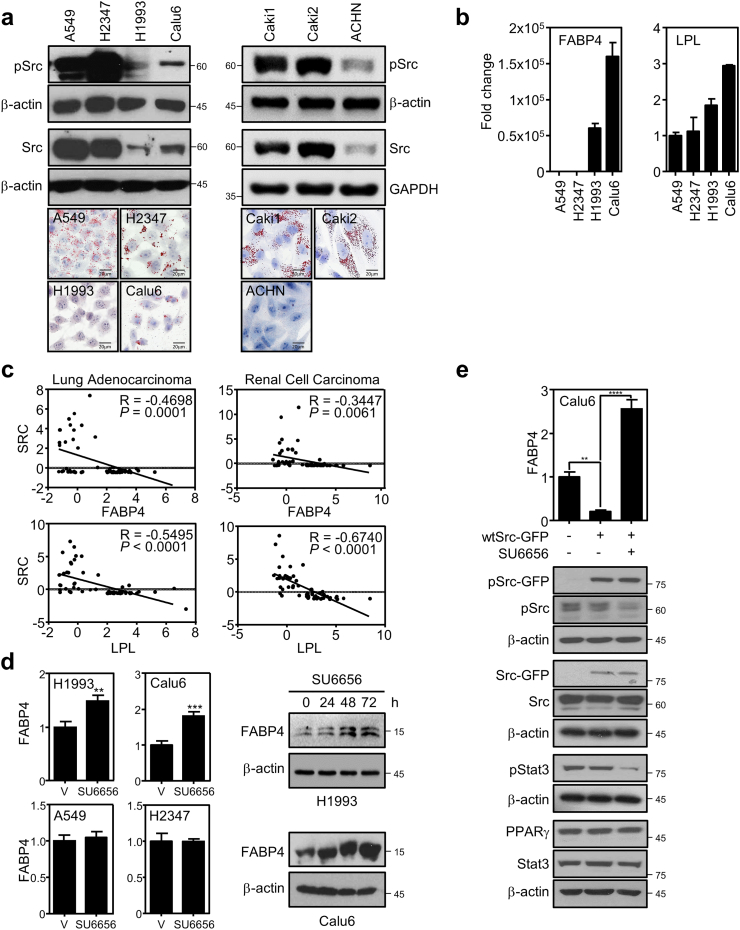
Since recent studies proposed oncogenic Src regulation of glucose metabolism, we asked if the Src is also involved in regulating cancer lipid metabolism [6,7]. To answer for this question, we first investigated how Src expression is correlated to intracellular lipid amount in several cancer cell lines. For comparative study, we selected a panel of cancer cell lines including non-small cell lung cancer (NSCLC) and renal cell carcinoma (RCC). The NSCLC cells include A549 and H2347 cells with high expression of Src and H1993 and Calu6 cells with low Src expression. The RCC cells include Caki1, Caki2 cells for high Src expression and ACHN cells for low Src expression. To see the intracellular lipid contents in the panel, we carried out Oil-red O (ORO) staining that interestingly revealed the more lipid storage in the high Src expression group compared to the low expression group (Fig. 1a). From the correlation of Src to lipid accumulation, we further wanted to identify downstream factors of Src to see the underlying mechanism by analyzing lipid metabolic genes in the four lung cancer cell lines using the microarray datasets available in public (Fig. S1a). Interestingly, we found that FABP4 and LPL genes in the microarray dataset were highly upregulated in Calu6 and H1993 cells, which is confirmed using the RT-PCR assay (Fig. S1a and 1b). Using The Cancer Genome Atlas (TCGA) data obtained from the Provisional dataset of cbioportal [34,35], further biostatistics analysis showed significant reverse Pearson correlation between Src and FABP4 or LPL in both lung cancer and RCC, which is consistent with the result from in vitro NSCLC cell lines in Fig. 1a (Fig. 1c). However, RCC cells unexpectedly showed very low expression of FABP4 and LPL genes in cancer cell line encyclopedia (CCLE) database [39] (Fig. S1b). Thus, lung cancer cell lines have been used for further mechanistic studies. To see the functional hierarchy between Src and the lipolytic genes, we pharmacologically inhibited kinase activity of Src by treating specific inhibitor SU6656 [40]. Interestingly, we found that SU6656 treatment increased FABP4 expression in the cell lines with low Src expression but not in the high Src expressing group while LPL expression was not affected or marginally induced by the Src inhibitor (Fig. 1d and S1c). Consistent with this, knockdown of Src significantly increased FABP4 expression while exogenous overexpression of Src significantly suppressed FABP4 expression or marginally suppressed LPL expression which was rescued by SU6656 treatment in Calu6 cells (Fig. S1d, 1e, and S1e). Collectively, our data show that Src negatively regulates lipolysis gene expression, which may result in intracellular lipid accumulation in cancer.
Fig. 1.
Src negatively regulates lipolysis. (a) Correlation of endogenous Src expression to lipid droplets in a panel of lung cancer (left) and renal cell carcinoma cell lines (right). Together with basal protein expression of pSrc and Src, lipid contents were assayed using Oil-red O staining in the panel. (b) Expression of lipolytic genes in lung cancer cells. The mRNA expression of FABP4 and LPL was shown using the RT-PCR assay. (c) Reverse correlation between Src and FABP or LPL. The analysis showed negative Pearson's correlation coefficient for the expression pattern between Src and FABP4 as well as Src and LPL. For biostatistics analysis, data were obtained for lung adenocarcinoma or RCC patients from the cbioportal database. (d-e) Pharmacological inhibition of Src induces FABP4 expression. (d) Expression of FABP4 mRNA (left) and protein (right) was assayed upon treatment of Src inhibitor SU6656 compound in the lung cancer cell lines. Cells were treated with 5 μM of SU6656 for 24, 48, and 72 h and assayed for FABP4 protein expression or 24 h for mRNA expression. (e) Oncogenic Src was exogenously overexpressed in Calu6 cells and followed by FABP4 expression upon SU6656 treatment. Cells were transfected with pEGFP control or wtSrc-GFP followed by SU6656 treatment for 24 h. Data represent mean ± SEM of triplicates. Similar results were observed in at least two independent experiments. Asterisks refer to **P ≤ 0.01; ***P ≤ 0∙001; ****P ≤ 0∙001 (Student's t-test (d) and one-way ANOVA, Tukey's post test (e)). In all RT-PCR data, y-axis represents fold change in gene expression. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Src inhibition induces PPARγ-mediated FABP4 expression
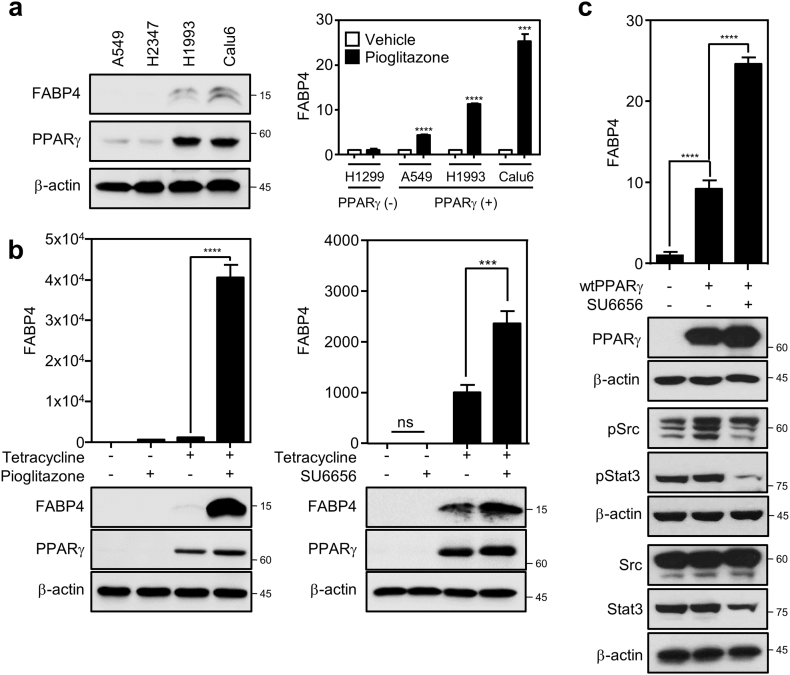
As knowing that Src is involved in cancer lipid metabolism, we next wondered the underlying mechanism by which Src suppresses lipolytic gene expression to control intracellular lipid metabolism in cancer. Since previously reported that FABP4 is a direct target gene of PPARγ in lipid metabolism [17], we investigated if PPARγ is involved in the Src regulation of FABP4 expression and intracellular lipid droplet in cancer. We first found positive correlation of basal PPARγ and FABP4 expression in the NSCLC panel and further PPARγ activation by pioglitazone strongly induced FABP4 expression in a PPARγ activation-dependent manner, confirming that FABP4 is a direct target of the receptor in cancer (Fig. 2a and S2a). Interestingly, note that LPL, also known as a PPARγ target gene in the literature [41], is not strongly induced by PPARγ activation in cancer cells (Fig. S2b). To further confirm the PPARγ specific regulation of FABP4 expression, we next took advantage of two independent gain-of-functional systems, an inducible cell line for PPARγ expression and a PPARγ-negative cell line with exogenously overexpressed PPARγ. Regarding the inducible system, we previously developed a human bronchial epithelial cell line (HBEC-C1-PPARγ) which inducibly expresses PPARγ mRNA upon tetracycline treatment [22]. We found that FABP4 expression is dramatically induced, but not or marginally for LPL expression, on PPARγ activation by pioglitazone treatment (Fig. 2b left and S2c). Furthermore, FABP4 expression was also markedly induced by the Src specific inhibitor treatment only under tetracycline-induced overexpression of PPARγ, indicating that Src-mediated FABP4 suppression is dependent on PPARγ (Fig. 2b right). Note that pStat3 was decreased showing the specific inhibition of Src activity under SU6656 treatment in HBEC cells (Fig. S2d). In addition, this pharmacological result from the HBEC cells was confirmed using H1299 cells with exogenous overexpression of PPARγ. Consistently, the overexpression of PPARγ in the PPARγ-negative H1299 cells showed significant induction of FABP4 mRNA expression, which was further upregulated under SU6656 treatment (Fig. 2c). Taken together, this suggests that Src inhibition releases PPARγ activity leading to induction of FABP4 expression in cancer.
Fig. 2.
Src suppression increases PPARγ-mediated FABP4 expression. (a) Induced expression of FABP4 mRNA upon PPARγ activation. Basal expression of FABP4 and PPARγ was determined using immunoblot assay in A549, H2347, H1993 and Calu6 cells (left). Using RT-PCR assay, FABP4 mRNA induction was evaluated in PPARγ-negative cells (H1299) and PPARγ-positive cells (A549, H1993, Calu6) treated with DMSO or 10 μM of pioglitazone for 24 h (right). (b) FABP4 expression upon PPARγ activation or Src inhibition in HBEC-C1-PPARγ cells. HBEC-C1-PPARγ cells were treated with tetracycline overnight, followed by 10 μM of pioglitazone or 5 μM of SU6656 treatment for 24 h. QPCR (upper) and immunoblot (lower) assays were performed to determine FABP4 expression. (c) FABP4 expression in H1299 cells with PPARγ overexpression. H1299 cells were transfected with empty vector or PPARγ and followed by 5 μM of SU6656 treatment for 24 h. Data represent mean ± SEM of triplicates. Similar results were observed in at least two independent experiments. Asterisks refer to ***P ≤ 0∙001; ****P ≤ 0∙001 (Student's t-test (a) and one-way ANOVA, Tukey's post test (b,c)). In all RT-PCR data, y-axis represents fold change in gene expression.
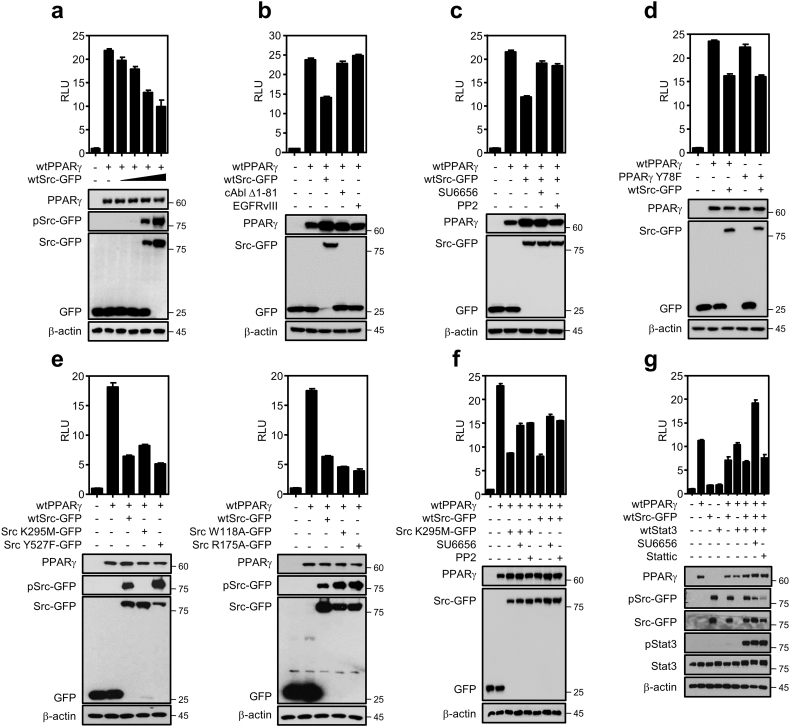
3.3. Kinase activity of Src is dispensable for suppressing PPARγ activity
Having demonstrated that pharmacological inhibition of Src releases PPARγ transcriptional activity accompanying lipolytic gene induction, we further wanted to understand the more detailed regulatory mechanism of Src on the receptor activity. Using luciferase assay for PPARγ transactivation activity, we first confirmed that Src inhibits transcriptional activity of PPARγ in a dose-dependent manner of Src expression (Fig. 3a). Note that luciferase construct PPRE3x-luc contains PPARγ response element as previously reported [21]. To see whether the PPARγ inhibition is Src specific or not, included are other tyrosine kinases, cAbl kinase (Δ1–81) and epidermal growth factor receptor variant III (EGFRvIII) that are constitutive active forms of tyrosine kinases as negative controls [26,42] or other Src family kinases including Fyn and Yes [5]. Note that transfection efficiency of cAbl kinase and EGFRvIII is represented as mRNA expression in Fig. S3a. The inhibition of PPARγ-activated luciferase activity was mediated only by Src but not by the other kinases including Fyn and Yes, and consistently treatment of Src specific inhibitors, SU6656 and PP2, rescued the PPARγ transcriptional activity (Fig. 3b, S3b, and 3c). This suggests that Src is a specific kinase regulating PPARγ transcriptional activity. Based on our previous report that Src phosphorylates at tyrosine 78 of PPARγ, we next examined if the Src suppression is mediated by posttranslational modification of the nuclear receptor PPARγ [43]. To answer for this question, a PPARγ mutant construct replaced tyrosine with phenylalanine at residue 78 was tested using luciferase assay. We showed the mutant PPARγ construct is transcriptionally active and, surprisingly enough, found the Src is still able to inhibit the mutant PPARγ activity (Fig. 3d). This suggests the tyrosine 78 phosphorylation is not required for Src suppression of the PPARγ transcriptional activity. Along with this, we further evaluated the PPARγ transcriptional activity using multiple Src kinase mutants [23]. This includes various mutant Src constructs for being constitutive active (Y527F), kinase-dead (K295M), or SH2 domain (W118A) and SH3 domain (R175A) which are defective for protein-protein interaction. Strikingly, both kinase-dead (K295 M) and constitutive active (Y527F) forms of Src are still able to suppress PPARγ transcriptional activity to the similar extent of inhibition as wildtype Src (Fig. 3e). Moreover, the suppression of PPARγ activity by both types of Src is rescued by Src inhibitors, indicating additional inhibitory role of Src inhibitors beyond simply suppressing the Src kinase activity (Fig. 3f). It is important to note that kinase-dead mutant of Src showed similar binding ability to PPARγ as wildtype Src does (Fig. S3c). This indicates Src suppression of PPARγ activity may also occur through direct interaction between Src and PPARγ. In addition, as a well-known downstream factor of Src, Stat3 was previously reported to interact with PPARγ [44]. To see if Stat3 is an intermediator for Src suppression of PPARγ, we executed the similar transactivation assay including Stat3 co-transfection with PPARγ in the presence or absence of Stat3 specific inhibitor Stattic. Interestingly, exogenous overexpression of Stat3 showed neither activation nor inhibition of PPARγ transactivation. Further, Stattic treatment showed no rescue of PPARγ transcriptional activity in the presence of Src, indicating that Src suppression of PPARγ activity is independent of Stat3 (Fig. 3g). Taken together, our data suggests that Src suppresses FABP4 expression by inhibiting PPARγ transcriptional activity in a kinase activity-independent manner, which may be regulated through protein interaction with PPARγ.
Fig. 3.
Src suppression of PPARγ transcriptional activity in a kinase activity-independent manner. PPARγ transcriptional activity was assessed using luciferase assay in which both PPARγ and TK-PPRE3x-luc constructs were co-transfected into HEK293 cells with various forms of kinase constructs including Src, EGFR, and c-Abl as indicated in each figure. (a) Src repression of PPARγ transcriptional activity in a dose-dependent manner of wtSrc-GFP construct. Note that the amount of PPARγ is 0 or 600 ng, and wtSrc-GFP is increased as 0, 6, 60, 300, or 600 ng, respectively. (b) Src specific-suppression of PPARγ transcriptional activity. Luciferase assay was carried out for PPARγ transcriptional activity in the presence of several oncogenic tyrosine kinases including cAbl (Δ1-81), EGFRvIII and wtSrc-GFP. (c) Rescue of PPARγ activity by Src-specific inhibitors. Included are Src specific inhibitors, SU6656 and PP2, targeting kinase activity. (d-e) Src inhibition of PPARγ transcriptional activity independent of PPARγY78 phosphorylation (d) and also independent of Src kinase activity (e). Luciferase assay was performed in various conditions including PPARγY78F construct in the presence of wtSrc-GFP (d) or wt-PPARγ construct in the presence of different mutant forms of Src (e). Note that included are kinase-dead Src K295M-GFP, constitutive active Src Y527F-GFP constructs, or SrcW118A and SrcR175A Src mutants for SH2 or SH3 domain, respectively. (f) Both wildtype Src and kinase-dead Src suppress PPARγ activity. Using luciferase assay, PPARγ transactivation was assayed with wildtype or kinase-dead Src construct upon treatment of two specific inhibitors. (g) Src suppression of PPARγ transactivation is independent of Stat3 activity. PPARγ transactivation was assayed with Stat3 co-transfection in the presence or absence of 2 μM Stattic treatment targeting Stat3 for assessing PPARγ transcriptional activity. Note that all transfections in HEK293 cells were confirmed using immunoblot assay in each figure. RLU indicates relative light unit. Similar results were observed in at least two independent experiments.
Both in vitro and in vivo models confirm Src regulation of lipid droplets in an FABP4-dependent manner.
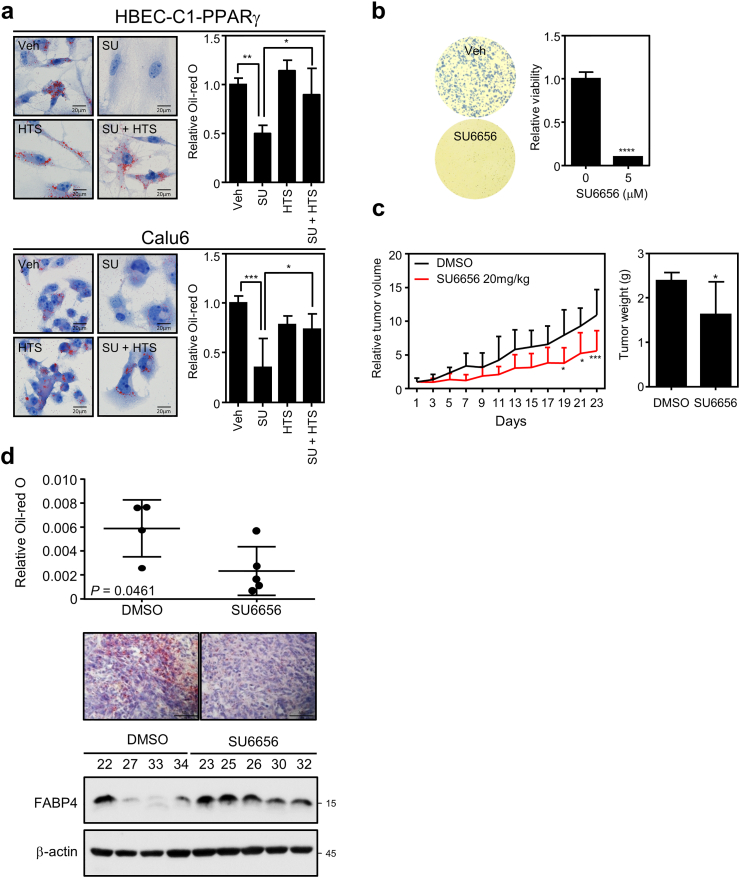
Identification of the molecular mechanism by which Src suppresses PPARγ-FABP4 axis further pushed us to verify the lipid accumulation upon FABP4 inhibition. The treatment of Src inhibitor SU6656 reproducibly reduced lipid droplets, which is rescued by FABP4 specific inhibitor HTS01037 in both Calu6 and HBEC-C1-PPARγ lung cell lines (Fig. 4a). Consistent with pharmacological approach, Src knockdown also reduced lipid droplets which was rescued by FABP4 inhibitor treatment or FABP4 knockdown (Fig. S4a and S4b). We next established in vivo xenograft tumor model using Calu6 cells to confirm the in vitro results. The xenografted athymic nude mice were intraperitoneally administered with 20 mg/kg SU6656 or vehicle every other day for 23 days. Consistent with the anti-proliferative effect of SU6656 in Calu6 cells (Fig. 4b), the in vivo treatment of SU6656 showed decreased tumor growth without any change of body weight (Fig. 4c, S4c and S4d). Moreover, SU6656 treatment significantly decreased intratumoral lipid accumulation, which consistently accompanied the upregulated FABP4 expression in the group with drug treatment (Fig. 4d). Taken together, this data support that inhibition of Src induces FABP4 expression to decrease lipid accumulation, which may contribute to suppress cancer progression.
Fig. 4.
Src regulation of tumor growth and lipid droplet is FABP4-dependent in vitro and in vivo. (a) Lipid staining (left) and quantification (right) in HBEC-C1-PPARγ (upper) and Calu6 (lower) cells. Cells were treated with 5 μM of SU6656 (SU), 20 μM of HTS01037 (HTS) or both for 3 days. (b) In vitro cell growth assay upon Src inhibitor treatment. Cell proliferation (left) and MTT (right) assays showed inhibition of cell proliferation upon SU6656 treatment for 7 days in Calu6. (c-d) In vivo analysis of the xenograft tumors by inhibiting oncogenic Src. Five millions of Calu6 cells were injected into the flank region of athymic nude mice. When tumors were tangible, mice were intraperitoneally administered with vehicle (n = 5) or SU6656 20 mg/kg (n = 7) for 23 days every other day. (c) Tumor growth suppression upon SU6656 treatment. Both tumor volume (left) and tumor weight (right) were measured every other day or at the end of the experiment, respectively. Tumor growth were represented as mean relative tumor size ± SEM. Statistical analysis was determined using 2-way ANOVA. (d) Intratumoral lipid amount and FABP4 protein expression. Intratumoral lipid content (upper) or FABP4 expression (lower) were assayed in the residual tumor tissues at the end of in vivo experiment. Note that a pair of representative figures was shown for lipid staining. Values are mean ± SEM. Statistical significance was assessed using Student's t-test. Asterisks refer to *P ≤ 0∙05; **P ≤ 0∙01; ***P ≤ 0∙001; ****P ≤ 0∙0001 (One-way ANOVA, Tukey's post test (a), Student's t-test (b, c (right) and d) and two-way ANOVA, Sidak's post test (c (left))).
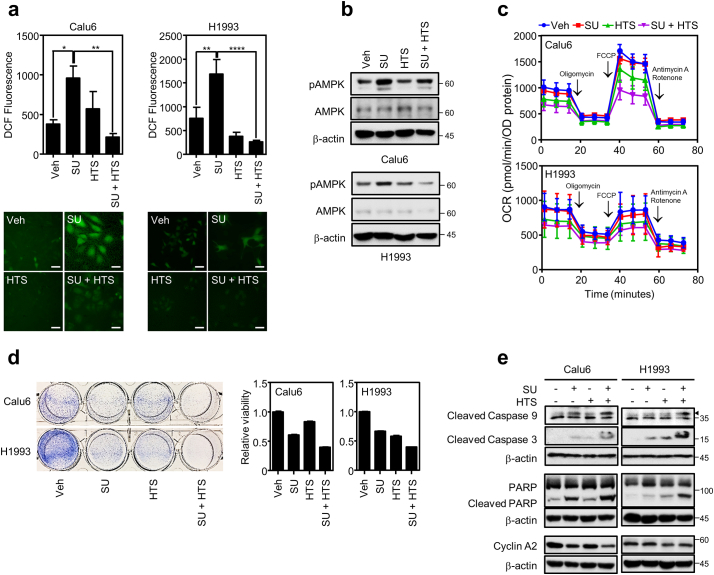
3.4. Src suppression induces FABP4-mediated intracellular ROS generation
Here, we wanted to see how Src inhibition-mediated lipolysis contributes to lung cancer growth inhibition. To this point, as a previous study reported that lipid droplets play an antioxidant role contributing to cancer cell survival [10], we investigated if lipid droplet reduction by Src suppression might affect intracellular ROS level. Using CM-H2DCF-DA as a ROS sensing dye, we measured intracellular ROS level with Src and/or FABP4 inhibition. As shown in Fig. 5a, Src suppression by SU6656 treatment showed significantly increased intracellular ROS level, which was markedly blunted by FABP4 inhibitor HTS01037, suggesting that FABP4 activity is associated with the increased ROS. Consistent with the pharmacological inhibition, Src knockdown also induced ROS accumulation which is blunted by FABP4 knockdown (Fig. S5a). Regarding biological consequences relevant to the intracellular ROS accumulation, we further examined lipid peroxidation and AMPK activation under treatment of Src and/or FABP4 inhibitor [45,46]. The Src inhibitor increased pAMPK which FABP4 inhibitor treatment turned back to the normal level, while lipid peroxidation remains unchanged (Fig. 5b and S5b). Since FABP4 facilitates lipolysis and β-oxidation [47], we next wondered if the Src inhibition-mediated lipolysis also led to increased β-oxidation of fatty acid. Seahorse assay measuring mitochondrial respiration showed no significant difference for oxygen consumption rate (OCR) between SU6656 and vehicle treatments, whereas HTS01037 treatment decreased basal level of intracellular OCR in Calu6 and H1993 lung cancer cell lines (Fig. 5c), which suggest that lipolysis may not provide fatty acid for β-oxidation-mediated ATP production. Along with the ROS generation, we also examined cell growth response under treatment of both inhibitors. The Src inhibitor alone induced both apoptosis and cell cycle arrest in Calu6 and induced apoptosis in H1993. However, the treatment of the FABP4 specific inhibitor showed no rescue of the SU6656-mediated cell growth suppression and even additive growth suppression with Src inhibition. (Fig. 5d and e), which might be due to the suppression of basal OCR dominating the lipolysis inhibition effect. Taken together, our data suggest that Src suppression induces FABP4 to decrease lipid droplets which subsequently induce endogenous ROS level.
Fig. 5.
Intracellular ROS generation upon lipolysis regulation. (a) Measurement of intracellular ROS. Cells were treated with 5 μM of SU6656 (SU) in the presence or absence of 20 μM of FABP4 inhibitor HTS01037 (HTS) for 3 days and followed by incubation with 1 μM of CM-H2DCF-DA for H1993 or 2∙5 μM of CM-H2DCF-DA for Calu6 cells. Pictures were taken using confocal microscope followed by quantitative analysis using MetaMorph 6.3 software (Molecular Devices). Scale bar = 20 μm (b) AMPK phosphorylation upon inhibitor treatment. Cells treated with SU or HTS for 3 days were assayed for AMPK and pAMPK expression in H1993 and Calu6 cells. (c) Oxygen consumption rates in lung cancer cell lines with inhibitor treatment. Calu6 and H1993 cells were treated with SU, HTS or both for 3 days. The OCRs were measured as in materials and methods. (d) Cell growth assay with inhibitor treatment. Cell proliferation and MTT assays show cell viability under SU, HTS, or both treatment for 3 days. (e) Protein expression involved in regulation of cell cycle or apoptosis in Calu6 and H1993 upon indicated treatment for 24 h. Values are mean ± SEM. Results represent data from at least two independent experiments. Asterisks refer to *P ≤ 0∙05; **P ≤ 0∙01; ****P ≤ 0∙0001 (One-way ANOVA, Tukey's post test).
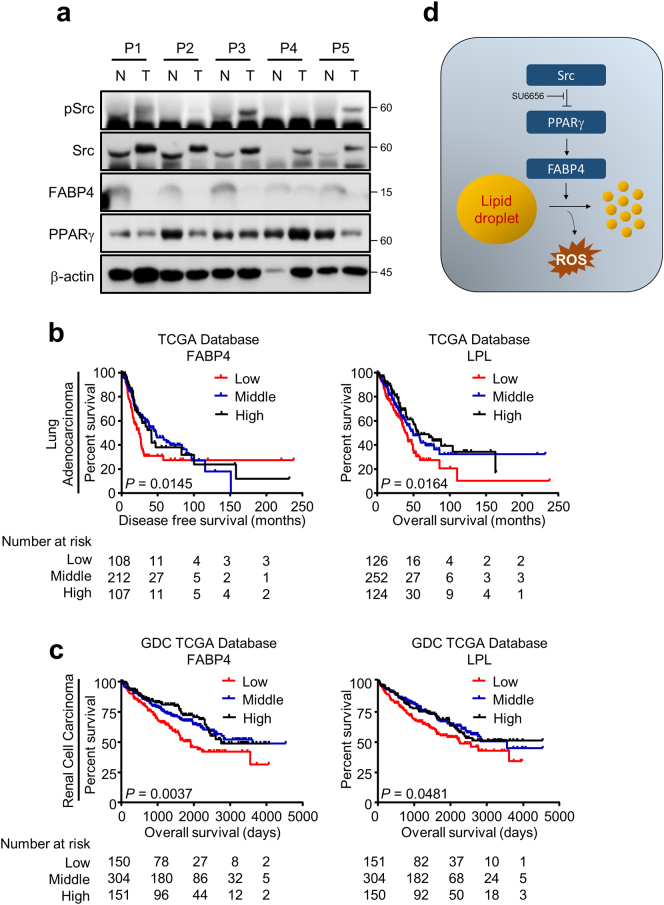
3.5. Increased expression of lipolytic genes represents the better prognosis of lung and renal cancer patients
In this study, we have shown that PPARγ derepression by Src inhibition induces FABP4 expression followed by lipid droplet breakdown, which potentially contributes to decreased tumor progression by increasing endogenous ROS level. Hereby, we wanted to know if the biological finding provides any translational interpretation in the clinical setting. To that end, five pair-matched normal and tumor tissues were obtained from lung cancer patients under IRB approval. We found Src expression is higher in the tumors compared to the corresponding normal tissues while basal PPARγ expression is consistently opposite to the Src expression pattern, and more interestingly, the FABP4 expression showed the reverse correlation to the Src expression in the panel (Fig. 6a and table S5). This is consistent with the previous analysis using the public datasets shown in Fig. 1c. Furthermore, using multiple datasets from TCGA and GEO databases, we next sought to determine the prognostic value of lipolysis gene FABP4 and LPL. Interestingly, our data showed that the higher FABP4 and LPL expression is significantly associated with the better survival of both disease free and overall survival in lung cancer as well as RCC (Fig. 6b, c and S6), suggesting tumor suppressive role of FABP4 and LPL in lung cancer and RCC. Based on our study, here the schematic model was proposed in Fig. 6d. Taken together, the biological finding for the oncogenic Src function in lipolytic gene regulation and thus cancer lipid metabolism would be therapeutically translated into the clinics.
Fig. 6.
High FABP4 or LPL expression shows the better prognosis. (a) Expression of pSrc, Src, PPARγ and FABP4 in pair-matched patient tissues. Five pair-matched tissue samples were obtained from lung cancer patients upon IRB approval, and followed by measuring proteins of interest expression. (b–c) Kaplan-Meier plots for lung and renal cancer patient survival. Data were obtained from TCGA database for lung cancer and RCC or from GEO database (Fig. S6) for lung cancer. Disease free or overall survival was analyzed for groups with high (high, >75th percentile), in-between (middle, from 25th percentile to 75th percentile) or low expression (low, <25th percentile) of FABP4 or LPL in lung (c) and renal (c) cancer patients. Note that log-rank test was used for the statistical analysis. (d) Proposed model for Src regulation on lipid droplets.
4. Discussion
Cancer cells display diverse metabolic reprogramming including the Warburg effect for aerobic glycolysis and reductive carboxylation in citric acid cycle to utilize glutamine into intracellular lipid storage. This metabolic adaptation enables cancer cells to deal with bioenergetics for energy requirement, reducing power for intracellular ROS control, and biosynthesis of building blocks for cancer proliferation, which are essential prerequisites for cancer survival. Amongst major nutrient sources (e.g., glucose, fatty acid, protein), intracellular lipid has become a crucial source for energy demand, but also proposed for protecting cancer cells from cellular ROS stress [48]. Metabolic alteration in fast growing cancers indispensably accompanies high ROS generation which should be resolved for maintaining cancer progression. To handle this hazardous situation, cancer cells have extensively advanced various ROS scavenging systems balancing intracellular redox homeostasis [49]. Many of recent studies have proposed that the metabolic adaptability of cancer shows intimidate connection to the oncogenic alterations in the tumorigenic process [4]. Src is a well-known oncogene regulating key signal transduction pathways to promote tumor progression [5]. Recent studies reported Src-mediated metabolic rewiring of cancer cells in which Src inhibits pyruvate dehydrogenase and activates hexokinase 1 and 2, contributing to the Warburg effect or aerobic glycolysis [6,7]. Considering Src regulation of glucose utilization in cancer, one might reasonably think of Src involvement in metabolism of other intracellular nutrients including amino acids and lipid. Thus, we hereby hypothesized oncogenic Src regulates cancer lipid metabolism for cancer maintenance and progression. To that end, we executed several independent approaches using in vitro as well as in vivo systems including a panel of cancer cell lines and xenograft model. The preclinical results were further correlated to the patient outcome of survival using public databases. In summary, we found that Src inhibition induces lipolysis leading to cancer growth inhibition, which notably accompanies several significant biological and clinical consequences as follows: 1) Inhibition of oncogenic Src induces lipolysis. 2) Mechanistically, we identified Src-PPARγ-FABP4 axis in regulating intracellular lipid mobilization in cancer. Strikingly, the Src inhibition of PPARγ is independent of kinase activity function. 3) High expression of lipolysis genes, FABP4 or LPL, shows the better prognosis of lung and renal cancer patients.
From this study, it is of interest that a couple of issues would be raised to be discussed further. Firstly, we found the mode of Src action regulating PPARγ activity is independent of kinase activity, which may be controlled by the interaction of the oncogene to the receptor. We previously reported that Src phosphorylates PPARγ at tyrosine 78 residue to regulate inflammation and insulin sensitivity [43]. However but interestingly, the Src inhibition of PPARγ activity in this study turns out to be independent of tyrosine 78 phosphorylation of the receptor. This would support the notion for the potential interaction between oncogene Src and PPARγ. Further, it is also possible for some factors to be involved between Src and PPARγ which may explain the binary modes of Src action for regulating PPARγ transcriptional activity. Secondly, LPL expression was marginally responded to the PPARγ activation in this study while it is known as a PPARγ target in other study [41]. It would be of interest to identify other transcriptional factors modulating the LPL expression and thus regulating cancer lipid metabolism. Lastly, the Src inhibition-mediated cancer growth suppression was not rescued by the pharmacological inhibition of FABP4 and unexpectedly showed the more additive growth suppression when treated with the Src inhibitor SU6656 compound. Also, we interestingly found that the FABP4 inhibition markedly decreased the basal level of OCR, potentially leading to the overall reduction of the intracellular ATP level. Consequently, this might be compounding or dominating the potential rescue of the FABP4 inhibitor for the SU6656-mediated cell growth effect by inhibiting intracellular ROS generation.
Taken together, this study provides evidence on how the oncogene Src to modulate lipid droplets contributing to the intracellular ROS decrease, and an insight of the metabolic plasticity for cancer cells to cope with the cellular stress encountered during the oncogene-driven cancer progression. This finding extends our understanding of the oncogenic Src as a metabolic regulator beyond a simple signal transducer, which could be potentially developed into Src-targeted therapeutic strategies linking signal transduction and metabolic regulation in cancer.
Funding sources
This work was financially supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Medical Research Center Program (2017R1A5A2015369), and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI17C0039).
Declaration of interests
The authors report no declaration of interest.
Author contributions
Conceived the experiments and wrote the paper: Y.J., T.N.M.H. Performed the experiments: T.N.M.H., M.-K.K., and V.T.A.V. Analyzed the data: T.N.M.H., J.-W.C., J.H.C., H.-W.K., S.-K.C., K.-S.P., and Y.J.
Acknowledgements
We thank Dr. Ai N.H. Phan for her technical supports, Dr. Ji-Hee Kim for providing renal cell lines, and Dr. Luong D. Ly for his support on ROS measurement.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.015.
Appendix A. Supplementary data
Supplementary material
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Tennant D.A., Duran R.V., Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat Rev Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 3.O'Flanagan C.H., Smith L.A., McDonell S.B., Hursting S.D. When less may be more: calorie restriction and response to cancer therapy. BMC Med. 2017;15:106. doi: 10.1186/s12916-017-0873-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagarajan A., Malvi P., Wajapeyee N. Oncogene-directed alterations in cancer cell metabolism. Trends Cancer. 2016;2:365–377. doi: 10.1016/j.trecan.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang S., Yu D. Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends Pharmacol Sci. 2012;33:122–128. doi: 10.1016/j.tips.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y., Cai Q., Shenoy A.K. Src drives the Warburg effect and therapy resistance by inactivating pyruvate dehydrogenase through tyrosine-289 phosphorylation. Oncotarget. 2016;7:25113–25124. doi: 10.18632/oncotarget.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Wang S., Jiang B. c-Src phosphorylation and activation of hexokinase promotes tumorigenesis and metastasis. Nat Commun. 2017;8:13732. doi: 10.1038/ncomms13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bensaad K., Favaro E., Lewis C.A. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9:349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 9.Cotte A.K., Aires V., Fredon M. Lysophosphatidylcholine acyltransferase 2-mediated lipid droplet production supports colorectal cancer chemoresistance. Nat Commun. 2018;9:322. doi: 10.1038/s41467-017-02732-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarc E., Kump A., Malavasic P., TO Eichmann, Zimmermann R., Petan T. Lipid droplets induced by secreted phospholipase A2 and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. Biochim Biophys Acta. 2018;1863:247–265. doi: 10.1016/j.bbalip.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 11.Hotamisligil G.S., Bernlohr D.A. Metabolic functions of FABPs--mechanisms and therapeutic implications. Nat Rev Endocrinol. 2015;11:592–605. doi: 10.1038/nrendo.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheja L., Makowski L., Uysal K.T. Altered insulin secretion associated with reduced lipolytic efficiency in aP2−/− mice. Diabetes. 1999;48:1987–1994. doi: 10.2337/diabetes.48.10.1987. [DOI] [PubMed] [Google Scholar]
- 13.De Santis M.L., Hammamieh R., Das R., Jett M. Adipocyte-fatty acid binding protein induces apoptosis in DU145 prostate cancer cells. J Exp Ther Oncol. 2004;4:91–100. [PubMed] [Google Scholar]
- 14.Nieman K.M., Kenny H.A., Penicka C.V. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17:1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z., Shen Q., Xie H. Elevated expression of FABP3 and FABP4 cooperatively correlates with poor prognosis in non-small cell lung cancer (NSCLC) Oncotarget. 2016;7:46253–46262. doi: 10.18632/oncotarget.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu Y.L., Hung J.Y., Lee Y.L. Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget. 2017;8:104831–104854. doi: 10.18632/oncotarget.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tontonoz P., Hu E., Graves R.A., Budavari A.I., Spiegelman B.M. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 18.Schadinger S.E., Bucher N.L., Schreiber B.M., Farmer S.R. PPARgamma2 regulates lipogenesis and lipid accumulation in steatotic hepatocytes. Am J Physiol Endocrinol Metab. 2005;288:E1195–E1205. doi: 10.1152/ajpendo.00513.2004. [DOI] [PubMed] [Google Scholar]
- 19.Festuccia W.T., Laplante M., Berthiaume M., Gelinas Y., Deshaies Y. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia. 2006;49:2427–2436. doi: 10.1007/s00125-006-0336-y. [DOI] [PubMed] [Google Scholar]
- 20.Ohno H., Shinoda K., Spiegelman B.M., Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phan A.N.H., Vo V.T.A., Hua T.N.M. PPARgamma sumoylation-mediated lipid accumulation in lung cancer. Oncotarget. 2017;8:82491–82505. doi: 10.18632/oncotarget.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J., Sato M., Choi J.W. Nuclear receptor expression and function in human lung cancer pathogenesis. PLoS One. 2015;10 doi: 10.1371/journal.pone.0134842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shvartsman D.E., Donaldson J.C., Diaz B., Gutman O., Martin G.S., Henis Y.I. Src kinase activity and SH2 domain regulate the dynamics of Src association with lipid and protein targets. J Cell Biol. 2007;178:675–686. doi: 10.1083/jcb.200701133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timpson P., Jones G.E., Frame M.C., Brunton V.G. Coordination of cell polarization and migration by the Rho family GTPases requires Src tyrosine kinase activity. Curr Biol. 2001;11:1836–1846. doi: 10.1016/s0960-9822(01)00583-8. [DOI] [PubMed] [Google Scholar]
- 25.Sandilands E., Cans C., Fincham V.J. RhoB and actin polymerization coordinate Src activation with endosome-mediated delivery to the membrane. Dev Cell. 2004;7:855–869. doi: 10.1016/j.devcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 26.Keshet R., Bryansker Kraitshtein Z., Shanzer M., Adler J., Reuven N., Shaul Y. c-Abl tyrosine kinase promotes adipocyte differentiation by targeting PPAR-gamma 2. Proc Natl Acad Sci U S A. 2014;111:16365–16370. doi: 10.1073/pnas.1411086111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chougule R.A., Kazi J.U., Ronnstrand L. FYN expression potentiates FLT3-ITD induced STAT5 signaling in acute myeloid leukemia. Oncotarget. 2016;7:9964–9974. doi: 10.18632/oncotarget.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H., Naismith J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. doi: 10.1186/1472-6750-8-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Mehlem A., Hagberg C.E., Muhl L., Eriksson U., Falkevall A. Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease. Nat Protoc. 2013;8:1149–1154. doi: 10.1038/nprot.2013.055. [DOI] [PubMed] [Google Scholar]
- 31.Xu S., Nam S.M., Kim J.H. Palmitate induces ER calcium depletion and apoptosis in mouse podocytes subsequent to mitochondrial oxidative stress. Cell Death Dis. 2015;6:e1976. doi: 10.1038/cddis.2015.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou B.B., Peyton M., He B. Targeting ADAM-mediated ligand cleavage to inhibit HER3 and EGFR pathways in non-small cell lung cancer. Cancer Cell. 2006;10:39–50. doi: 10.1016/j.ccr.2006.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong Y., Xie Y., Lee W. Research resource: diagnostic and therapeutic potential of nuclear receptor expression in lung cancer. Mol Endocrinol. 2012;26:1443–1454. doi: 10.1210/me.2011-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami E., Gao J., Dogrusoz U. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao J., Aksoy B.A., Dogrusoz U. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitra R., Lee J., Jo J. Prediction of postoperative recurrence-free survival in non-small cell lung cancer by using an internationally validated gene expression model. Clin Cancer Res. 2011;17:2934–2946. doi: 10.1158/1078-0432.CCR-10-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang P., Cheng C.L., Chang Y.H. Molecular gene signature and prognosis of non-small cell lung cancer. Oncotarget. 2016;7:51898–51907. doi: 10.18632/oncotarget.10622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grossman R.L., Heath A.P., Ferretti V. Toward a shared vision for cancer genomic data. N Engl J Med. 2016;375:1109–1112. doi: 10.1056/NEJMp1607591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barretina J., Caponigro G., Stransky N. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blake R.A., Broome M.A., Liu X. SU6656, a selective src family kinase inhibitor, used to probe growth factor signaling. Mol Cell Biol. 2000;20:9018–9027. doi: 10.1128/mcb.20.23.9018-9027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamauchi T., Kamon J., Waki H. The mechanisms by which both heterozygous peroxisome proliferator-activated receptor gamma (PPARgamma) deficiency and PPARgamma agonist improve insulin resistance. J Biol Chem. 2001;276:41245–41254. doi: 10.1074/jbc.M103241200. [DOI] [PubMed] [Google Scholar]
- 42.Zeineldin R., Ning Y., Hudson L.G. The constitutive activity of epidermal growth factor receptor vIII leads to activation and differential trafficking of wild-type epidermal growth factor receptor and erbB2. J Histochem Cytochem. 2010;58:529–541. doi: 10.1369/jhc.2010.955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choi S., Jung J.E., Yang Y.R. Novel phosphorylation of PPARgamma ameliorates obesity-induced adipose tissue inflammation and improves insulin sensitivity. Cell Signal. 2015;27:2488–2495. doi: 10.1016/j.cellsig.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 44.Ju K.D., Lim J.W., Kim H. Peroxisome proliferator-activated receptor-gamma inhibits the activation of STAT3 in cerulein-stimulated pancreatic acinar cells. J Cancer Prev. 2017;22:189–194. doi: 10.15430/JCP.2017.22.3.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayala A., Munoz M.F., Arguelles S. Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev. 2014;2014 doi: 10.1155/2014/360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cardaci S., Filomeni G., Ciriolo M.R. Redox implications of AMPK-mediated signal transduction beyond energetic clues. J Cell Sci. 2012;125:2115–2125. doi: 10.1242/jcs.095216. [DOI] [PubMed] [Google Scholar]
- 47.Furuhashi M., Saitoh S., Shimamoto K., Miura T. Fatty acid-binding protein 4 (FABP4): pathophysiological insights and potent clinical biomarker of metabolic and cardiovascular diseases. Clin Med Insights Cardiol. 2014;8:23–33. doi: 10.4137/CMC.S17067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tirinato L., Pagliari F., Limongi T. An overview of lipid droplets in cancer and cancer stem cells. Stem Cells Int. 2017;2017 doi: 10.1155/2017/1656053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marengo B., Nitti M., Furfaro A.L. Redox homeostasis and cellular antioxidant systems: crucial players in cancer growth and therapy. Oxid Med Cell Longev. 2016;2016:6235641. doi: 10.1155/2016/6235641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material