We congratulate Wu et al.1 on their successful application of single-nucleus RNA sequencing to characterize renal cell types underrepresented by single-cell RNA sequencing. A particular strength of their workflow is the detection of cells such as podocytes, mesangial cells, and glomerular endothelial cells, which can be underrepresented by conventional single-cell sequencing. These cell types have previously been described as distinct clusters using single-cell sequencing at markedly higher total cell numbers, such as the recent landmark renal single-cell sequencing paper by Park et al.2
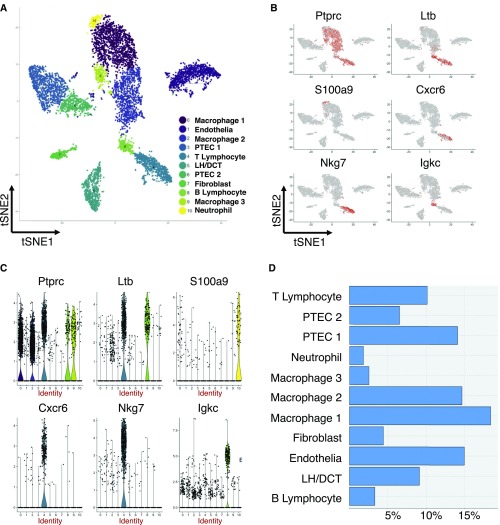
As a group characterizing renal injuries using a similar single-cell, high-throughput, droplet-based platform, we are struck by the differences in cell classification between those described by Wu et al. using single-nucleus sequencing and those seen in our own single-cell sequencing of similar kidneys after unilateral ureteric obstruction (UUO). In contrast to the authors, we detect multiple leukocytes in our samples and identify clusters of distinct subsets of mononuclear phagocytes (35%), neutrophils (1.8%), B lymphocytes (3.3%), and T lymphocytes (10%) from a library of 6213 transcriptomes (Figure 1). This correlates well with existing histologic studies and FACS analysis of digested UUO kidneys showing nearly 40% of renal cells are CD45-positive leukocytes3–5 and is in contrast to a single, small macrophage cluster found on the single-nucleus analysis. Several groups have reported the importance of both T and B lymphocytes in the pathogenesis of UUO, yet single-nuclear RNA sequencing seems not to identify such cells, with representative lymphocyte marker genes suggested by Park et al. instead expressed in sparse cells scattered across multiple clusters in the single-nucleus dataset.
Figure 1.
Single cell sequencing identifies multiple distinct leukocyte populations in the obstructed murine kidney. (A) t-SNE plot demonstrating distinct cluster formation, annotated by cell type. (B) Leukocyte gene expression projected across these t-SNE clusters. (C) Leukocyte gene expression plotted across clusters, natural logarithmic scale. (D) Cell frequency as percentage of total library size. LH/DCT, loop of Henle/distal convoluted tubule; PTEC, proximal tubular epithelial cell; t-SNE, t-Distributed Stochastic Neighbour Embedding.
It is probable that differing technologies may introduce a cell-specific bias into the library preparation. Single-nucleus sequencing appears to bring improved identification of glomerular cell types. Single-cell sequencing may be more biased toward leukocytes, to the detriment of those glomerular cells. The advantages of single-nucleus sequencing are important and include the reduction of stress-response gene signatures and the ability to sequence historical frozen samples. However, we wonder if current protocols lack the ability to characterize the full spectrum of leukocytes present within the kidney. We propose that complimentary roles exist for both single-cell and single-nucleus sequencing, and researchers may wish to consider the cell-type bias in any workflow, and their own cell of interest, when designing sequencing experiments.
Disclosures
None.
Acknowledgments
Work in the Ferenbach laboratory is funded by the Wellcome Trust and Kidney Research UK.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Wu H, Kirita Y, Donnelly EL, Humphreys BD: Advantages of single-nucleus over single-Cell RNA sequencing of adult kidney: Rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 30: 23–32, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Park J, Shrestha R, Qiu C, Kondo A, Huang S, Werth M, et al. : Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360: 758–763, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eis V, Luckow B, Vielhauer V, Siveke JT, Linde Y, Segerer S, et al. : Chemokine receptor CCR1 but not CCR5 mediates leukocyte recruitment and subsequent renal fibrosis after unilateral ureteral obstruction. J Am Soc Nephrol 15: 337–347, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Liu L, Kou P, Zeng Q, Pei G, Li Y, Liang H, et al. : CD4+ T Lymphocytes, especially Th2 cells, contribute to the progress of renal fibrosis. Am J Nephrol 36: 386–396, 2012 [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Zhu F, Wang X, Yao W, Wang M, Pei G, et al. : Continuous AMD3100 treatment worsens renal fibrosis through regulation of bone marrow derived pro-angiogenic cells homing and T-cell-related inflammation. PLoS One 11: e0149926, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]