Abstract
Background
In the current context of research on HIV reservoirs, offering new insights into the persistence of HIV DNA in infected cells, which prevents viral eradication, may aid in identifying cure strategies. This study aimed to describe the establishment of stable integrated forms among total HIV DNA during primary infection (PHI) and their dynamics during the natural history of infection.
Methods
Total and integrated HIV DNA were quantified in blood from 74 PHI patients and 97 recent seroconverters (<12 months following infection, “progression cohort”). The evolution of both markers over six years was modelled (mixed-effect linear models). Their predictive values for disease progression were studied (Cox models).
Findings
For most patients during PHI, stable integrated forms were a minority among total HIV DNA (median: 12%) and became predominant thereafter (median at AIDS stage: 100%). Both total and integrated HIV DNA increased over a six-year period. Patients from the progression cohort who reached clinical AIDS during follow-up (n = 34) exhibited higher total and integrated HIV DNA levels at seroconversion and a higher percentage of integrated forms than did slower progressors (n = 63) (median: 100% vs 44%). The integrated HIV DNA load was strongly associated with the risk of developing AIDS (aRR = 2.63, p = 0.002).
Interpretation
The profile of “rapid” or “slower” progression in the natural history of HIV infection appears to be determined early in the course of HIV infection. The strong predominance of unstable unintegrated forms in PHI may explain the great benefit of this early treatment, which induces a sharp decrease in total HIV DNA.
Fund
French National Agency for Research on AIDS and Viral Hepatitis.
Keywords: Total HIV DNA, Integrated HIV DNA, Reservoirs, Natural history, Primary HIV infection, Acquired immunodeficiency syndrome, Kinetics
Research in context.
Evidence before this study
HIV reservoirs represent a major barrier to eradicating HIV, making them one of the current priorities in HIV research. Understanding the pathogenic mechanisms underlying the persistence of HIV in reservoirs is therefore a crucial step. The level of total HIV DNA, a marker reflecting the size of the viral reservoir that is composed of both the persistent integrated forms and the more labile unintegrated ones, peaks during primary infection. During this time, short-lived cells contribute more than long-lived cells to the HIV reservoir. In clinical practice, the timing of the initiation of treatment is linked to the timing of the diagnosis. Previous studies revealed that the earlier the initiation of treatment, the more marked is the decrease in the total HIV DNA load, although a clear explanation for this observation is lacking. During treatment, the total HIV DNA is mostly composed of the integrated forms, but little is known about the dynamics of the different forms of HIV DNA during the natural history of HIV infection. Using Medline/PubMed, we identified the available evidence by searching with the keywords “total HIV DNA”, “integrated HIV DNA”, “natural history”, “primary infection”, and “AIDS”.
Added value of this study
We quantified both the integrated and total HIV DNA in the peripheral blood mononuclear cells of 171 untreated individuals from two large cohorts; one cohort was composed of patients with primary infections (ANRS-PRIMO cohort), while the other was composed of patients who were followed longitudinally over six years from the time of seroconversion to the development of a chronic infection or clinical AIDS (historical ANRS-SEROCO cohort). We found that integrated forms constitute a minority of the total HIV DNA during the primary infection and that they subsequently substantially increase to become predominant. A high integrated HIV DNA load during the first year after infection is strongly predictive of a rapid evolution towards clinical AIDS. Taken together with the results from previous studies, our results suggest a reason for the observation that the HIV primary infection period is the critical period during which antiretroviral therapy should be initiated, since that period is when the HIV DNA burden is mostly composed of labile forms – thereby protecting immune cells from infection. Conversely, the proportion of stable integrated forms increases after seroconversion, making treatments less efficient to diminish the reservoir during the chronic stage.
Implications of all the available evidence
Concomitant analysis of total HIV DNA and integrated HIV DNA loads contributes to the understanding of HIV persistence and pathogenesis. It also explains why early treatment initiation is the most effective strategy, leading to a marked and sustained decrease in the HIV DNA burden. Studying both markers may help identify HIV-infected individuals who will be the best candidates for new therapeutic strategies targeting HIV, including curative strategies.
Alt-text: Unlabelled Box
1. Introduction
Combined antiretroviral therapy (cART) has constituted major progress in the treatment of HIV infection and current recommendations are to treat all HIV-infected persons, regardless of the stage of infection. Although cART can efficiently block HIV replication, it cannot completely eliminate the virus from its reservoirs. In particular, infected resting CD4+ T-cells containing latent integrated HIV proviruses are refractory to current cART and represent a major hurdle preventing viral eradication [1,2]. Understanding the pathogenesis of HIV reservoirs is critical for developing and evaluating new therapeutic strategies aimed at viral eradication or functional cure.
Integrated HIV DNA is the most stable and functional form of the viral genome. It plays a major role in the pathogenesis of HIV infection and HIV reservoirs, even if it includes both replication-competent and defective genomes [[3], [4], [5]]. Cells containing integrated HIV DNA can produce new infectious virions upon stimulation and activation [6,7]. Proviruses persist indefinitely, partly due to the homeostatic proliferation of memory T-cells. In contrast, unintegrated forms, which include linear and episomal HIV DNA with 1- or 2-LTRs, are considered more labile and are surrogate markers of viral replication [8,9].
Total HIV DNA levels in peripheral blood mononuclear cells (PBMCs) have been described during both the natural history of HIV infection and cART [10]. It remains partially unexplained why the decrease in this surrogate marker of the reservoir is greater and faster when treatment is initiated during primary HIV infection (PHI) than during the chronic stage [[11], [12], [13]]. As for integrated HIV DNA, it has been mostly studied in patients on cART [[13], [14], [15]]. Only a few studies have measured integrated HIV DNA in untreated patients, most of which include fewer than 20 patients [14,[16], [17], [18]]. Longitudinal data are even more scarce for both markers: only one study to our knowledge has a follow-up of patients during untreated infection (17 patients during the first year, among which 10 still followed after 2.8 years) [18]. Moreover, some techniques do not allow a comparison between the amounts of total and integrated HIV DNA because of differences in assay standardization [19] and the long-term dynamics of integrated forms among total HIV DNA have never been described. Besides, all previous results showed a great inter-individual variability of total and integrated HIV DNA [14,16,17,20,21], enhancing the need for studies on large cohorts.
To better understand the establishment and maintenance of the HIV reservoir in the blood, the objectives of this work were i) to describe the contribution of integrated forms to total HIV DNA in untreated patients at different stages from PHI to chronic infection and ii) to evaluate the predictive value of these biomarkers at the time of seroconversion with respect to evolution towards clinical AIDS.
2. Materials and methods
2.1. Study population
Patients with a known or estimated date of infection from two ANRS French cohorts were selected. The ANRS-PRIMO cohort was approved by the Ile-de-France-3 Ethics Committee (CPP-1157). All patients from both cohorts gave written informed consent.
The ongoing ANRS-PRIMO CO6 cohort enrols patients presenting with PHI, as previously described [12,22]. All patients are treatment-naïve at inclusion. Patients enrolled between June 2015 and April 2016 with available frozen PBMCs samples from the time of their inclusion were selected for this study.
In the ANRS-SEROCO CO2 cohort, initiated in 1988, recently diagnosed HIV-infected individuals who were free of AIDS-related diseases at inclusion were enrolled [23]. Follow-up was scheduled every six months until 2009. Patients were selected within this cohort according to the following criteria: enrolment within 12 months following infection, no efficient cART received before inclusion and during the study until 1996, and having at least two frozen PBMCs samples available.
In both cohorts, the infection date was defined as the date of the incomplete Western blot minus one month, or the date of a primary symptomatic infection minus 15 days, or the midpoint between a negative and a positive HIV antibody test.
2.2. Quantification of total HIV DNA and integrated HIV DNA and determination of unintegrated HIV DNA
Total HIV DNA and integrated HIV DNA were quantified in all PBMCs samples. Total DNA was extracted with the QIAamp DNA Blood Mini Kit (Qiagen, France) according to the manufacturer's instructions.
First, total HIV DNA was quantified by real-time PCR with the Generic HIV DNA CELL kit (Biocentric, France), employing the 8E5 line as the standard [3]. Based on a previously reported technique [24], we developed a nested Alu-LTR PCR assay to quantify the integrated HIV DNA from the same extracts as total HIV DNA, using the HelaR7Neo cell-line as the standard with improved sensitivity and reproducibility (Supplementary Text 1). The quantification threshold varied depending on the number of available cells tested (threshold range [5–97] copies/106 PBMCs (0.70–1.97 log10)). The equivalence of HIV DNA copies between HeLaR7Neo and 8E5 standards was verified, allowing the determination of the percentage of integrated forms among total HIV DNA. Unintegrated HIV DNA levels were calculated by the difference in the number of copies between total and integrated HIV DNA.
2.3. Statistical analyses
Statistical analyses were performed using Stata software (v14.2, StataCorp, USA) and Prism software (v7, GraphPad, USA). Continuous variables were compared between groups by using Wilcoxon tests and qualitative variables with Fisher's exact tests. Spearman's correlation coefficient was employed to quantify the correlations between continuous variables. Cox models were used to determine the predictive values of HIV DNA levels for clinical AIDS onset. Mixed-effect linear models (MELMs) were employed to estimate changes in total HIV DNA and integrated HIV DNA over time, considering the value at enrolment as time t = 0 for each patient. Logarithm10 transformation of HIV RNA and HIV DNA levels was performed to fulfil the model assumptions. Integrated HIV DNA levels were set to half of the threshold when undetectable. To evaluate the robustness of our results, additional analyses were performed, setting the values of undetectable integrated HIV DNA levels to the threshold value, to zero, or to a range of randomized values between 1 and the threshold. All these analyses produced results similar to those obtained when the values of undetectable integrated HIV DNA levels were set to half the threshold value. P values<0.05 were considered significant.
3. Results
3.1. Patient characteristics
Seventy-four patients who presented during PHI (ANRS-PRIMO cohort [22]) were included, with a median [range] estimated time since infection of 1.2 [0.4–2.4] months (Fiebig II-III, n = 12; Fiebig IV, n = 7; Fiebig V, n = 35; Fiebig VI, n = 20). Ninety-seven recent seroconverters from the historical ANRS-SEROCO cohort were included (median estimated time between infection and enrolment: 6.2 [1.3–11.9] months) (called later “progression cohort”) [23]. The characteristics of the 171 study patients are presented in Table 1. As expected, the PHI patients had higher HIV RNA loads (p < 0.001) (see also Supplementary Fig. 1) and lower CD4+ T-cell counts than did those from the progression cohort (p = 0.036).
Table 1.
Baseline demographic and clinical characteristics of the patients.
| Characteristics | PHI cohort a | Progression cohorta | p Valueb | Among the progression cohort |
||
|---|---|---|---|---|---|---|
| Rapid progressors | Slower progressors | p Valueb | ||||
| n | 74 | 97 | 34 | 63 | ||
| Male, n (%) | 69 (93.2%) |
79 (81.4%) |
0.040 | 29 (36.7%) |
50 (63.3%) |
0.589 |
| Caucasians, n (%) | 61 (82.4%) |
95 (97.9%) |
<0.001 | 34 (100.0%) |
61 (96.8%) |
0.540 |
| Median age at inclusion (IQR), years | 36 (29–45) |
30 (26–36) |
<0.001 | 31 (26–37) |
30 (26–36) |
0.530 |
| Sexual route of infection, n (%) | 72 (97.3%) |
88 (90.7%) |
0.117 | 32 (94.1%) |
56 (88.9%) |
0.487 |
| Median time between seroconversion and first sample (IQR), months | 1.0 (0.8–1.3) |
6.2 (4.2–8.5) |
<0.001 | 7.1 (4.4–9.5) |
6.0 (4.2–8.3) |
0.415 |
| Median time of follow-up (IQR), years | na | na | na | 4.5 (3.5–6.0) |
5.8 (4.6–6.3) |
0.008 |
| Median baseline CD4+ cell count (IQR), cells/mm3 | 449 (317–613) |
531 (390–666) |
0.036 | 481 (343–608) |
544 (429–706) |
0.124 |
| Median baseline CD8+ cell count (IQR), cells/mm3 | 761 (505–1253) |
846 (639–1157) |
0.177 | 927 (632–1157) |
832 (650–1144) |
0.972 |
| Median baseline CD4+/CD8+ cell ratio | 0.60 (0.37–0.91) |
0.57 (0.44–0.87) |
0.999 | 0.52 (0.41–0.77) |
0.65 (0.45–0.88) |
0.308 |
| Median plasma HIV RNA load (IQR), log copies/mL | 6.20 (5.30–6.98) |
4.16 (3.59–4.52) |
<0.001 | 4.37 (3.96–4.67) |
3.98 (3.47–4.39) |
0.002 |
| Median total HIV DNA load (IQR), log copies/106 PBMCs | 3.59 (3.29–4.03) |
3.10 (2.73–3.46) |
<0.001 | 3.22 (3.01–3.59) |
2.96 (2.63–3.38) |
0.008 |
| Median integrated HIV DNA load (IQR), log copies/106 PBMCs | 2.15 (0.95–3.16) |
2.90 (2.17–3.40) |
0.003 | 3.32 (2.97–3.66) |
2.53 (1.52–3.07) |
<0.001 |
na: non-applicable.
PHI refers to patients in the primary HIV infection stage (<3 months since the estimated infection date, ANRS-PRIMO cohort), while patients from the progression cohort were in their first year of infection (ANRS-SEROCO cohort). Rapid and slower progressors groups are defined according to their subsequent progression to clinical AIDS.
Obtained from Fisher's exact or Wilcoxon rank-sum tests.
Among the progression cohort, the following two groups could be defined regarding subsequent evolution during follow-up: individuals who developed AIDS (rapid progressors, n = 34) and those who had not developed AIDS by 1996 (slower progressors, n = 63) (Table 1). Notably the follow-up time was significantly longer for the slower progressors group. Hence, AIDS occurrence in rapid progressors was not due to a longer follow-up time.
3.2. Low percentage of integrated forms in PHI, followed by an increase in the first year post infection
We quantified the amounts of total HIV DNA and integrated HIV DNA in the samples taken at inclusion from all 171 patients. The results were combined to describe the profiles of both markers over the first year following infection (Fig. 1a). Loess curves showed a low level of integrated HIV DNA during PHI, which reached higher levels later in the first year following infection, while the level of total HIV DNA was high during PHI and decreased thereafter. Both markers reached a steadier state after 90 days post infection. The percentage of integrated HIV DNA among total HIV DNA was low for patients in the first 90 days after infection (median: 12%), while it became much higher between 3 and 12 months after infection (median: 65%). Accordingly, even though this percentage varied from 0.1 to 100% regardless of the delay between infection and enrolment, the number of patients with a high percentage of integrated HIV DNA increased with increasing time since infection (Fig. 1b).
Fig. 1.
Total HIV DNA and integrated HIV DNA loads in the first year following infection.
(a) Distribution with Loess regression of total HIV DNA (filled circles and solid line) and integrated HIV DNA (open circles and dotted line) according to the estimated time since infection (171 patients, one time point per patient at their inclusion in the ANRS-PRIMO cohort (PHI) or the ANRS-SEROCO cohort (progression cohort, divided into rapid and slower progressors)).
(b) Repartition of integrated among total HIV DNA percentages according to the time since infection. These percentages were divided into four categories defined by the quartile values obtained for the 171 patients (light blue to purple, with Q1 = 2%, Q2 = 28%, Q3 = 85%). Patients were classified into five groups defined by the time since infection (in days). The bar chart represents the percentage of patients belonging to each category of integrated/total HIV DNA percentage. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
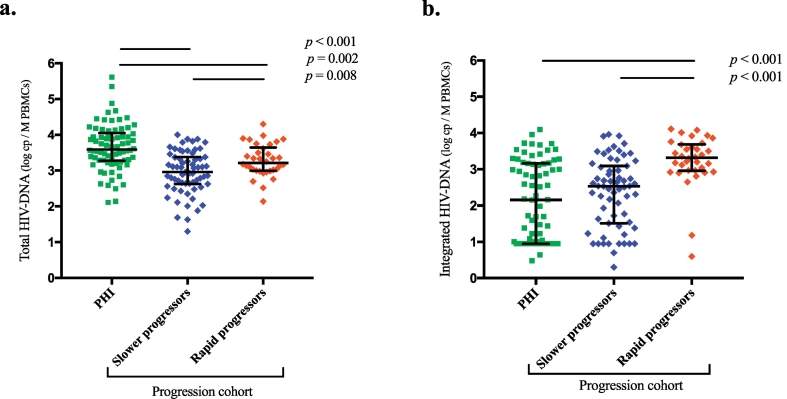
The median total HIV DNA level in PHI patients was 3.59 log10 copies/106 PBMCs [range: 2.11–5.61 log10], which was significantly higher than that in the 97 progression cohort patients (3.22 [2.14–4.30] log10 (p = 0.008) and 2.96 [1.30–4.00] log10 (p < 0.001) for rapid and slower progressors, respectively) (Table 1, Fig. 2a). Conversely, the median integrated HIV DNA levels in PHI patients (2.15 [0.48–4.10] log10 copies/106 PBMCs) and slower progressors (2.53 [0.30–3.96] log10) did not significantly differ from each other (p = 0.314) but were significantly lower than those in rapid progressors (3.32 [0.60–4.11] log10) (p < 0.001) (Table 1, Fig. 2b). Among patients from the progression cohort, both total HIV DNA and integrated HIV DNA levels were significantly higher in rapid progressors than in slower progressors (Table 1). Notably, the median percentage of integrated among total HIV DNA was 100% for rapid progressors, while it was 44% for slower progressors, indicating that integrated HIV DNA already represented the major form of HIV DNA in the rapid progressors group.
Fig. 2.
Baseline HIV DNA levels during primary infection and recent seroconversion, according to further progression to clinical AIDS.
Scatter dot plots display (a) total HIV DNA and (b) integrated HIV DNA loads for patients in the primary infection phase (PHI, ANRS-PRIMO cohort, <3 months since the estimated time of infection) and for the rapid progressors and slower progressors groups among the progression cohort (ANRS-SEROCO cohort, <1 year since infection). The bars represent median and interquartile range values.
Overall total HIV DNA levels were significantly correlated with the integrated HIV DNA (Supplementary Fig. 2) and plasma HIV RNA levels (Supplementary Fig. 3). Interestingly, for PHI patients, the HIV RNA load showed the strongest correlation with the unintegrated HIV DNA level, a marker of ongoing viral replication, while for patients from the progression cohort at the inclusion time (both rapid and slower progressors), the HIV RNA load was more strongly correlated with the integrated HIV DNA level than with the level of unintegrated forms (Supplementary Fig. 4–5).
3.3. Increase in total HIV DNA and integrated HIV DNA loads during years of untreated infection
Among the 97 HIV-infected individuals from the progression cohort, the median time between the first and the last sample was 4.5 years (range: [1.5–7.7]) or 5.8 years [2.9–8.1] for rapid progressors or slower progressors, respectively. HIV DNA kinetics were studied based on a total of 340 samples, with a median of four frozen cell samples per patient (range: [2–6]). Total HIV DNA levels significantly increased over time (p < 0.001), showing a + 0.627 log10 (+5371 copies/106 PBMCs) increase in rapid progressors and a + 0.574 log10 (+2508 copies/106 PBMCs) increase in slower progressors over six years (Fig. 3a); the increases were similar in the two groups (p = 0.740). The integrated HIV DNA level also significantly increased over the same six-year period, exhibiting similar increases (p = 0.300) in rapid progressors (+0.538 log10 (+5122 copies/106 PBMCs), p = 0.008) and slower progressors (+0.796 log10 (+1779 copies/106 PBMCs), p < 0.001) (Fig. 3b). Over time, slower progressors maintained lower levels of both total HIV DNA and integrated HIV DNA than rapid progressors, while the percentage of integrated forms tended to increase only in slower progressors (p = 0.056). During the same six-year period, CD4+ T-cell counts decreased in both groups (p < 0.001), with a steeper decline in rapid progressors than in slower progressors (−427 and − 222 CD4+ T-cells/mm3respectively, p = 0.002) (Supplementary Fig. 6).
Fig. 3.
Increase in total HIV DNA and integrated HIV DNA over six years of untreated infection.
(a) Mixed-effect linear models (MELMs) describing the evolution of total HIV DNA over six years for rapid progressors (n = 34, 111 samples) and slower progressors (n = 63, 229 samples).
(b) MELMs describing the evolution of integrated HIV DNA over six years for rapid progressors (107 samples) and slower progressors (207 samples).
Samples obtained after six years of follow-up were not considered for inclusion in the MELMs to have enough results at each time point.
Twenty-four patients among the rapid progressors had an available sample from the 12 months before clinical AIDS occurrence or at the AIDS stage. The median total HIV DNA and integrated HIV DNA loads were 3.64 log10 (IQR [3.42–3.98]) and 3.69 log10 [3.31–3.97], respectively. The median percentage of integrated forms was 100% (IQR [82.3–100]).
3.4. High predictive value of the integrated HIV DNA load for evolution towards clinical AIDS
Given the observed differences in the viral parameters at inclusion between the rapid and slower progressors groups, we evaluated their predictive value for evolution towards AIDS. The CD4+ T-cell counts at inclusion were similar between the two groups (p = 0.124) (Table 1) and were not included in the models. Univariate Cox models showed that a one-log increase in the amount of total HIV DNA, integrated HIV DNA, or HIV RNA at the baseline was significantly associated with a 2.48- to 3.12-fold increase in the risk of developing AIDS (Table 2). Since the amounts of total HIV DNA and integrated HIV DNA were strongly linked, two separate multivariate Cox models were applied, with adjustment for sex, age and HIV RNA level at enrolment. In the model including total HIV DNA, a higher HIV RNA load was associated with an increased risk of progression to AIDS (adjusted risk ratio for a one-log positive difference (aRR) = 2.02, p = 0.044) (Table 2). In the second multivariate model, only the integrated HIV DNA load was strongly associated with the risk of progression (aRR = 2.63, p = 0.002) (Table 2).
Table 2.
Factors associated with the evolution towards clinical AIDS according to baseline levels of total HIV DNA and integrated HIV DNA in univariate and multivariate analyses.
| Univariate analysis |
Multivariate analysis including total HIV DNA |
Multivariate analysis including integrated HIV DNA |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RR 95% CI | p Value | aRR 95% CI | p Value | aRR 95% CI | p Value | |||||
| Sex | Male | 1 | 1 | 1 | ||||||
| Female | 0.50 [0.19–1.32] |
0.161 | 1.00 [0.34–2.99] |
0.995 | 1.09 [0.38–3.14] |
0.870 | ||||
| Age at inclusion a | 1.02 [0.98–1.06] |
0.249 | 1.01 [0.97–1.05] |
0.737 | 0.70 [0.97–1.05] |
0.699 | ||||
| log10 HIV RNA (copies/mL)b | 2.48 [1.45–4.23] |
0.001 | 2.02 [1.02–4.00] |
0.044 | 1.42 [0.74–2.73] |
0.290 | ||||
| log10 total HIV DNA (copies/106 PBMCs)b | 2.55 [1.24–5.22] |
0.011 | 1.47 [0.61–3.54] |
0.392 | ||||||
| log10 integrated HIV DNA (copies/106 PBMCs)b | 3.12 [1.78–5.46] |
0.000 | 2.63 [1.41–4.91] |
0.002 | ||||||
For a one-year positive difference.
For a one-log positive difference
4. Discussion
The presence of long-lived infected cells that are not targeted by cART currently prevents viral eradication and cure. Understanding HIV pathogenesis by generating new insights into the dynamics of the HIV DNA components during infection is critical for conceiving of therapeutic strategies targeting these reservoirs.
This was the first large study to quantify both total HIV DNA and integrated HIV DNA, which is the main persistent form of HIV [25,26], in samples collected during PHI, chronic infection, and at various time points until the AIDS stage, which was made possible by two large French cohorts. The ANRS-SEROCO cohort is one of the rare large historical cohorts that included patients in the 1990s who, at that time, remained largely untreated during their follow-up and who had available longitudinal frozen cell samples.
One of the objectives of this study was to explore the link between the amount of stable proviruses and the risk of HIV disease progression, with the hypothesis that unstable unintegrated forms may have a lesser impact on long-term evolution. From the time of recent seroconversion, the amount of total HIV DNA was higher in rapidly progressing patients than in slower progressors, which agreed with its known predictive value for HIV disease progression [10,27]. Here, we report for the first time the predictive value of the amount of integrated HIV DNA, which appeared to be even more strongly predictive of the risk of developing clinical AIDS. Integrated HIV DNA is probably mostly responsible for the predictive value of total HIV DNA observed in previous studies, since this stable form of HIV persistence drives the course of infection [2,5]. Integrated HIV DNA is indeed the major source of the viral replication and the HIV RNA level reflects the proportion of cells containing this HIV stable form. Yet, the pathogenic impact of integrated HIV DNA is not only through the viral replication since its predictive value is independent of the HIV RNA level. Another potential means to drive the pathogenesis could be the production of transcripts and antigens from defective integrated genomes, which do not result in HIV RNA viremia, but contribute to the immune activation, as previously described [28]. The concomitant study of total HIV DNA and integrated HIV DNA showed that the percentage of integrated forms is highly informative. Indeed, this percentage was already high in rapid progressors at a time point within 12 months from seroconversion and remained high during disease progression, confirming that early events determine the profile of a “rapid progressor” or “slower progressor”. Total HIV DNA being strongly predictive of progression towards AIDS in PHI [29], the few PHI patients with a high proportion of integrated forms might be at a particularly high risk of becoming rapid progressors.
These data are in line with the very low levels of integrated HIV DNA observed in the unique group of elite suppressors compared to patients under efficient cART with similar total HIV DNA levels [30]. The low levels of integrated HIV DNA may be the consequence of more efficient specific cytotoxic T-lymphocyte (CTL) responses. CTLs could have preferentially destroyed cells harbouring integrated viral DNA, in which the viral transcription and translation into proteins is more efficient, eventually leading to the maintenance of lower proportions of integrated forms [31]. Rapid progressors may have less efficient CTL responses and therefore maintain higher levels of integrated HIV DNA.
Studies examining total HIV DNA have shown that the establishment of reservoirs is a very early event in PHI [32]. Although integrated HIV DNA is included in total HIV DNA, concomitant study of these two markers showed that they exhibit very different kinetics in the first year of infection, where the first three months after infection stand apart from the following months and years. Integrated HIV DNA levels were found to be low for most patients during PHI, while total HIV DNA levels were particularly high at that time, which was consistent with previous data showing a peak during this stage [29,32]. We observed a slight short decrease in integrated HIV DNA levels after the first weeks of infection (Fiebig III-IV), which is in agreement with the results previously reported for 19 untreated PHI patients from whom samples were collected during earlier stages (17 at Fiebig I-II stages), which showed a peak of integrated HIV DNA in week 2 and a decrease until week 6 after enrolment [18]. Ananworanich's work presented some results describing the evolution of total HIV DNA and integrated HIV DNA for ten patients during 144 weeks [18]. However, only the median results were presented for these two biomarkers and the proportions of stable forms among total HIV DNA and their evolution over time were not detailed for the different individuals. Thanks to a far greater number of patients and longer follow-up time, we showed here a more extensive increase of the proportion of integrated forms than in this previous work. With techniques allowing the comparison between the levels of total HIV DNA and integrated HIV DNA, we show the benefit of studying the percentage of integrated forms, their heterogeneity between patients and their impact on the outcome of untreated HIV infection. Although several PHI patients had undetectable integrated HIV DNA levels, the technique used here has a low quantification threshold and we are confident these results reflect truly low levels of integrated forms. The longer follow-up time also allowed us to highlight the progressive increase of total HIV DNA, which was not significant in previous shorter studies.
Taking all these results into account, we propose a model for the evolution of HIV DNA forms in blood during the natural history of HIV infection (Fig. 4a). During PHI, the labile linear and episomal HIV DNA forms are particularly abundant and mostly constitute evidence of active replication in recently infected activated cells [17]. This observation might be explained by the simultaneous cytokine storm and high immune activation detected at this stage. Later during the first year following infection, while the infection is evolving towards a steady state, integrated HIV DNA becomes the major component of the total HIV DNA in most patients. Both HIV DNA markers continue to increase over the subsequent years of follow-up (Fig. 4a). The expansion of HIV reservoirs can be linked to several factors: the infection of new cells due to continuous viral replication, the persistence of long-lived infected cells, and the proliferation of infected cells [[33], [34], [35]]. The last two factors might explain the increasing contribution of integrated HIV DNA forms, which are transmitted from a mother cell to all daughter cells, while unintegrated forms are diluted during cell division.
Fig. 4.
Proposed models for the dynamics of the different HIV DNA forms in blood reservoirs.
HIV DNA loads are represented in log10 copies/106 PBMC. Note the breaks in the time axes.
(a) Model for HIV DNA dynamics during the natural history of infection. During early PHI, total HIV DNA levels rise rapidly to reach a peak. Integrated HIV DNA shows a concomitant but much lower peak; the majority of HIV DNA is composed of unintegrated forms at that time [18]. Both total HIV DNA and integrated HIV DNA display a slight decrease thereafter, before reaching a more stable state. During chronic infection, both HIV blood biomarkers progressively increase, and the proportion of integrated forms increases.
(b, c) Models of HIV DNA dynamics under treatment, depending on the timing of cART initiation. Efficient treatment initiation stops or significantly decreases viral replication and the infection of new cells [43]. Unintegrated HIV DNA forms, resulting from viral replication, are eliminated because of their lability or cell death, or they are diluted during cell division, without new production as a result of cART. In contrast, stable integrated HIV DNA can persist for a longer time. Thus, cART initiation at PHI (b), when the integrated HIV DNA level is low, induces a more pronounced and extended decrease of total HIV DNA than cART initiation during the chronic stage, when the predominance of stable integrated forms is already higher (c) [11,12]. Efficient treatment initiation stops the evolution of CD4+ T-cell subsets contributions to HIV reservoirs; they remain mainly composed of short-lived cells when cART is initiated during PHI [38,40], while long-lived cells are the major contributor to HIV DNA when cART is initiated during chronic infection [35,40]. This difference might explain the continued decrease in total HIV DNA after several years of treatment when cART is initiated early.
At the AIDS stage, integrated forms represented the vast majority of HIV DNA in almost all patients, which demonstrates that during this profound immunodeficiency, the presence of stable reservoirs is definitely unaffected by cART.
The models in Fig. 4, depicting the evolution of the different viral forms over time, propose an explanation for why very early treatment is highly beneficial. Such early treatment has been proven to be more effective in reducing total HIV DNA levels and restoring immune functions [11,12,36,37]. The rapid and steep initial decline of total HIV DNA (Fig. 4b) could be explained mainly by the elimination of the labile unintegrated forms. Early cART initiation can also limit the establishment of persistent integrated HIV DNA forms and prevent the progressive increase of blood reservoirs observed in the natural history of the infection. Moreover, previous studies have shown that HIV-infected blood cells during PHI are mostly short-lived T-cells [38], which is an additional factor contributing to reservoir instability during this stage. Treating HIV-infected individuals as soon as the PHI stage allows partial preservation of the most long-lived memory T-cells from infection and the maintenance of a predominant contribution of the short-lived ones [[36], [37], [38], [39], [40]]. The slower but continued decrease in total HIV DNA at later times following cART initiation might be explained by the death of these short-lived infected cells [11,12]. In contrast, in patients treated during the chronic stage (Fig. 4c), total HIV DNA decreases only shortly after cART initiation, due to the elimination of infected activated cells [41]. Integrated HIV DNA, which is highly predominant in latently infected cells, persists at high levels thereafter. Long-lived highly proliferative central-memory T-cells have been shown to be the main contributors to HIV blood reservoirs during the chronic stage and in patients treated since the chronic stage, contributing to more stable reservoirs [35,40]. Taken together, these results suggest that during PHI, the existing reservoirs, which are mainly composed of short-lived cells infected with unstable viral forms, are easily eliminated through the early initiation of cART. In contrast, in the chronic phase, the blood reservoirs are mainly composed of stable proviruses in long-lived quiescent cells and are unaffected by cART. This could explain the greater impact of treatment when initiated during PHI compared to the chronic phase, which is an additional reason for recommending the treatment of all HIV-infected individuals, including the initiation of treatment as soon as possible in those diagnosed during PHI.
We acknowledge that in the present study we quantified total and integrated HIV DNA on PBMC and not CD4+ T cells. Nevertheless, in a previous study on patients from the same ANRS-SEROCO cohort, we showed a high correlation of total HIV DNA levels expressed either by copies/106 PBMC, or copies/106 CD4+ T-cells or copies/mL of whole blood [42]. The predictive value level on disease progression was similar whatever the expression of the total HIV DNA load. Besides, we only investigated blood reservoirs. Despite the continuous circulation of lymphocytes between the blood and tissues, the dynamics of integrated HIV DNA and total HIV DNA in infected CD4+ T-cell subsets and their relative contributions could differ in lymphoid tissues because of different inflammation and cellular activation levels, which should therefore also be explored.
In conclusion, we used two large cohorts of untreated HIV-infected individuals to describe the evolution of total and integrated HIV DNA loads and their association with disease evolution. We showed that integrated forms constitute a minority of the HIV DNA present during PHI and progressively become predominant over time. However, patients with high levels of integrated HIV DNA shortly after infection are at a higher risk of rapid progression towards AIDS, indicating that the rapidity of progression is determined soon after infection. These results highlight that PHI is a key period, and they contribute to the understanding of why cART initiation during this time is most effective in decreasing the HIV DNA burden, thereby protecting immune cells from infection. Concomitant analysis of total HIV DNA and integrated HIV DNA loads contributes to the understanding of HIV persistence and pathogenesis. Studying both markers may help to identify HIV-infected individuals who will be the best candidates for new therapeutic strategies targeting HIV, including cure and/or remission strategies.
Acknowledgments
Acknowledgements
This work is dedicated to the memory of Jean-Florian Mettetal, Denis Bucquet, and Christiane Deveau. We thank all the patients and clinicians who participated in this study, Laurent Tran and Anne Persoz for data management and the laboratory technicians (EA 7327, Université Paris Descartes) for their technical assistance. We thank Michaela Müller-Trutwin and Asier Sáez-Cirión for helpful discussions.
Declaration of interests
VAF reports grants from ANRS, during the conduct of the study; other from Janssen Cilag (for travel and registration fees for participation in conferences), other from ViiV Healthcare (for travel and registration fees for participation in conferences) outside of the submitted work. LM reports grants from ANRS, during the conduct of the study. Other authors have nothing to disclose.
Funding sources
The ANRS-PRIMO and SEROCO cohorts are sponsored by the ANRS (French National Agency for Research on AIDS and Viral Hepatitis). This work was funded by the ANRS. The funders of the study had no role in study design, data collection, data analysis, data interpretation, writing the report, or decision to submit for publication.
Authors' contributions
VAF, CR, LM, and FB designed the study. PT, TL, FB, CR, LM, and VAF interpreted the results. PT and TL produced the virological data. FB and AE gathered the data from the cohorts. CG, MB, and JPV included patients and collected the clinical data. PT, TL, FB, CR, LM, and VAF analysed the data. PT, TL, FB, OD, JPV, CG, CR, LM, and VAF participated in drafting and/or revising the manuscript. VAF was responsible for virological analyses. LM was responsible for statistical analyses and models. AM, MG, and OD provided technical support. VAF supervised the study. All authors critically reviewed and approved the manuscript. PT and TL, as well as LM and VAF, contributed equally to this work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.02.016.
Appendix A. Supplementary data
Supplementary material
References
- 1.Churchill M.J., Deeks S.G., Margolis D.M., Siliciano R.F., Swanstrom R. HIV reservoirs: what, where and how to target them. Nat Rev Microbiol. 2016;14(1):55–60. doi: 10.1038/nrmicro.2015.5. [DOI] [PubMed] [Google Scholar]
- 2.Murray J.M., Zaunders J.J., McBride K.L. HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during antiretroviral therapy. J Virol. 2014;88(6):3516–3526. doi: 10.1128/JVI.03331-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avettand-Fenoel V., Chaix M.L., Blanche S. LTR real-time PCR for HIV-1 DNA quantitation in blood cells for early diagnosis in infants born to seropositive mothers treated in HAART area (ANRS CO 01) J Med Virol. 2009;81(2):217–223. doi: 10.1002/jmv.21390. [DOI] [PubMed] [Google Scholar]
- 4.Bruner K.M., Murray A.J., Pollack R.A. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho Y.C., Shan L., Hosmane N.N. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plantin J., Massanella M., Chomont N. Inducible HIV RNA transcription assays to measure HIV persistence: pros and cons of a compromise. Retrovirology. 2018;15(1):9. doi: 10.1186/s12977-017-0385-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Descours B., Avettand-Fenoel V., Blanc C. Immune responses driven by protective human leukocyte antigen alleles from long-term nonprogressors are associated with low HIV reservoir in central memory CD4 T cells. Clin Infect Dis. 2012;54(10):1495–1503. doi: 10.1093/cid/cis188. [DOI] [PubMed] [Google Scholar]
- 8.Sharkey M., Triques K., Kuritzkes D.R., Stevenson M. In vivo evidence for instability of episomal human immunodeficiency virus type 1 cDNA. J Virol. 2005;79(8):5203–5210. doi: 10.1128/JVI.79.8.5203-5210.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mexas A.M., Graf E.H., Pace M.J. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. AIDS. 2012;26(18):2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avettand-Fenoel V., Hocqueloux L., Ghosn J. Total HIV-1 DNA, a marker of viral reservoir dynamics with clinical implications. Clin Microbiol Rev. 2016;29(4):859–880. doi: 10.1128/CMR.00015-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hocqueloux L., Avettand-Fenoel V., Jacquot S. Long-term antiretroviral therapy initiated during primary HIV-1 infection is key to achieving both low HIV reservoirs and normal T cell counts. J Antimicrob Chemother. 2013;68(5):1169–1178. doi: 10.1093/jac/dks533. [DOI] [PubMed] [Google Scholar]
- 12.Laanani M., Ghosn J., Essat A. Impact of the timing of initiation of antiretroviral therapy during primary HIV-1 infection on the decay of cell-associated HIV-DNA. Clin Infect Dis. 2015;60(11):1715–1721. doi: 10.1093/cid/civ171. [DOI] [PubMed] [Google Scholar]
- 13.Koelsch K.K., Boesecke C., McBride K. Impact of treatment with raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir. AIDS. 2011;25(17):2069–2078. doi: 10.1097/QAD.0b013e32834b9658. [DOI] [PubMed] [Google Scholar]
- 14.Murray J.M., McBride K., Boesecke C. Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing transmission afterwards. AIDS. 2012;26(5):543–550. doi: 10.1097/QAD.0b013e328350fb3c. [DOI] [PubMed] [Google Scholar]
- 15.Kiselinova M., De Spiegelaere W., Buzon M.J., Malatinkova E., Lichterfeld M., Vandekerckhove L. Integrated and total HIV-1 DNA Predict ex vivo viral outgrowth. PLoS Pathog. 2016;12(3) doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinzone M.R., Graf E., Lynch L. Monitoring integration over time supports a role for cytotoxic T lymphocytes and ongoing replication as determinants of reservoir size. J Virol. 2016;90(23):10436–10445. doi: 10.1128/JVI.00242-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu W., Jiao Y., Lei R. Rapid turnover of 2-LTR HIV-1 DNA during early stage of highly active antiretroviral therapy. PLoS One. 2011;6(6) doi: 10.1371/journal.pone.0021081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ananworanich J., Chomont N., Eller L.A. HIV DNA set point is rapidly established in acute HIV infection and dramatically reduced by early ART. EBioMedicine. 2016;11:68–72. doi: 10.1016/j.ebiom.2016.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eriksson S., Graf E., Dahl V. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2) doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cortes F.H., Passaes C.P., Bello G. HIV controllers with different viral load cutoff levels have distinct virologic and immunologic profiles. J Acquir Immune Defic Syndr. 2015;68(4):377–385. doi: 10.1097/QAI.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vandergeeten C., Fromentin R., Merlini E. Cross-clade ultrasensitive PCR-based assays to measure HIV persistence in large cohort studies. J Virol. 2014;88(21):12385–12396. doi: 10.1128/JVI.00609-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghosn J., Deveau C., Chaix M.L. Despite being highly diverse, immunovirological status strongly correlates with clinical symptoms during primary HIV-1 infection: a cross-sectional study based on 674 patients enrolled in the ANRS CO 06 PRIMO cohort. J Antimicrob Chemother. 2010;65(4):741–748. doi: 10.1093/jac/dkq035. [DOI] [PubMed] [Google Scholar]
- 23.Hubert J.B., Burgard M., Dussaix E. Natural history of serum HIV-1 RNA levels in 330 patients with a known date of infection. The SEROCO Study Group. AIDS. 2000;14(2):123–131. doi: 10.1097/00002030-200001280-00007. [DOI] [PubMed] [Google Scholar]
- 24.Brussel A., Delelis O., Sonigo P. Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA. Methods Mol Biol. 2005;304:139–154. doi: 10.1385/1-59259-907-9:139. [DOI] [PubMed] [Google Scholar]
- 25.O'Doherty U., Swiggard W.J., Jeyakumar D., McGain D., Malim M.H. A sensitive, quantitative assay for human immunodeficiency virus type 1 integration. J Virol. 2002;76(21):10942–10950. doi: 10.1128/JVI.76.21.10942-10950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinzone M.R., Di Rosa M., Cacopardo B., Nunnari G. HIV RNA suppression and immune restoration: can we do better? Clin Dev Immunol. 2012;2012:515962. doi: 10.1155/2012/515962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouzioux C., Hubert J.B., Burgard M. Early levels of HIV-1 DNA in peripheral blood mononuclear cells are predictive of disease progression independently of HIV-1 RNA levels and CD4+ T cell counts. J Infect Dis. 2005;192(1):46–55. doi: 10.1086/430610. [Epub 2005 May 31] [DOI] [PubMed] [Google Scholar]
- 28.Imamichi H., Dewar R.L., Adelsberger J.W. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proc Natl Acad Sci U S A. 2016;113(31):8783–8788. doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goujard C., Bonarek M., Meyer L. CD4 cell count and HIV DNA level are independent predictors of disease progression after primary HIV type 1 infection in untreated patients. Clin Infect Dis. 2006;42(5):709–715. doi: 10.1086/500213. [DOI] [PubMed] [Google Scholar]
- 30.Graf E.H., Mexas A.M., Yu J.J. Elite suppressors harbor low levels of integrated HIV DNA and high levels of 2-LTR circular HIV DNA compared to HIV+ patients on and off HAART. PLoS Pathog. 2011;7(2) doi: 10.1371/journal.ppat.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasternak A.O., Lukashov V.V., Berkhout B. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology. 2013;10:41. doi: 10.1186/1742-4690-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ananworanich J., Sacdalan C.P., Pinyakorn S. Virological and immunological characteristics of HIV-infected individuals at the earliest stage of infection. J Virus Erad. 2016;2:43–48. doi: 10.1016/S2055-6640(20)30688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacchus-Souffan C.F.M., Abdel-Mohsen M., Hoh R., Ahn H., Hecht F., Martin J. 9th International AIDS Society Conference; Paris, France: 2017. Early initiation of long-term ART may favor interventions to eradicate HIV: analysis of cell turnover and persistent HIV reservoirrs in distinct CD4+ T cells in suppressed HIV disease. [Google Scholar]
- 34.Pasternak A.O., Jurriaans S., Bakker M., Berkhout B., Lukashov V.V. Steady increase in cellular HIV-1 load during the asymptomatic phase of untreated infection despite stable plasma viremia. AIDS. 2010;24(11):1641–1649. doi: 10.1097/QAD.0b013e32833b3171. [DOI] [PubMed] [Google Scholar]
- 35.Chomont N., El-Far M., Ancuta P. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li J.Z., Gandhi R.T. The sooner, the better: more evidence that early antiretroviral therapy lowers viral reservoirs in HIV-infected infants. J Infect Dis. 2014;210(10):1519–1522. doi: 10.1093/infdis/jiu298. [DOI] [PubMed] [Google Scholar]
- 37.Ananworanich J., Vandergeeten C., Chomchey N. 20th Conference on Retroviruses and Opportunistic Infections (CROI) Atlanta, GA, 3-6 March 2013. CROI; 2013. Early ART intervention restricts the seeding of the HIV reservoir in long-lived central memory CD4 T cells. [Abst. 47] [Google Scholar]
- 38.Cheret A., Bacchus-Souffan C., Avettand-Fenoel V. Combined ART started during acute HIV infection protects central memory CD4+ T cells and can induce remission. J Antimicrob Chemother. 2015;70(7):2108–2120. doi: 10.1093/jac/dkv084. [DOI] [PubMed] [Google Scholar]
- 39.Bacchus C., Cheret A., Avettand-Fenoel V. A single HIV-1 cluster and a skewed immune homeostasis drive the early spread of HIV among resting CD4+ cell subsets within one month post-infection. PLoS One. 2013;8(5) doi: 10.1371/journal.pone.0064219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gantner P., Barnig C., Partisani M. Distribution and reduction magnitude of HIV-DNA burden in CD4+ T cell subsets depend on art initiation timing. AIDS. 2018;32(7):921–926. doi: 10.1097/QAD.0000000000001770. [DOI] [PubMed] [Google Scholar]
- 41.Murray J.M., Zaunders J., Emery S. HIV dynamics linked to memory CD4+ T cell homeostasis. PLoS One. 2017;12(10) doi: 10.1371/journal.pone.0186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avettand-Fenoel V., Boufassa F., Galimand J., Meyer L., Rouzioux C., Group ASCS HIV-1 DNA for the measurement of the HIV reservoir is predictive of disease progression in seroconverters whatever the mode of result expression is. J Clin Virol. 2008;42(4):399–404. doi: 10.1016/j.jcv.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Cardozo E.F., Andrade A., Mellors J.W., Kuritzkes D.R., Perelson A.S., Ribeiro R.M. Treatment with integrase inhibitor suggests a new interpretation of HIV RNA decay curves that reveals a subset of cells with slow integration. PLoS Pathog. 2017;13(7) doi: 10.1371/journal.ppat.1006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material