The abundance of 100 proteins were measured in plasma of patients with invasive cervical cancer and in population controls using the proximity extension assay (PEA). A signature of 11 proteins was identified that distinguish cases and controls with a sensitivity of 0.96 at a specificity of 1.0. In a prospective replication cohort the panel achieved a sensitivity of 0.78 and a specificity 0.56 of separating samples collected at the time of diagnosis from samples collected prior to diagnosis.
Keywords: Cancer biomarker(s), Personalized medicine, Cervical cancer, Blood*, Clinical proteomics, Human Papillomavirus, Screening
Graphical Abstract
Highlights
HPV is being introduced as the primary test in cervical cancer screening programs.
New biomarkers are needed for co-testing of women HPV positive in screening.
Analysis of plasma from women with invasive cervical cancer identified a 11-marker panel.
This signature shows high sensitivity and specificity to identify women with cancer.
Abstract
Human papillomavirus (HPV) is recommended as the primary test in cervical cancer screening, with co-testing by cytology for HPV-positive women to identify cervical lesions. Cytology has low sensitivity and there is a need to identify biomarkers that could identify dysplasia that are likely to progress to cancer. We searched for plasma proteins that could identify women with cervical cancer using the multiplex proximity extension assay (PEA). The abundance of 100 proteins were measured in plasma collected at the time of diagnosis of patients with invasive cervical cancer and in population controls using the Olink Multiplex panels CVD II, INF I, and ONC II. Eighty proteins showed increased levels in cases compared with controls. We identified a signature of 11 proteins (PTX3, ITGB1BP2, AXIN1, STAMPB, SRC, SIRT2, 4E-BP1, PAPPA, HB-EGF, NEMO and IL27) that distinguished cases and controls with a sensitivity of 0.96 at a specificity of 1.0. This signature was evaluated in a prospective replication cohort with samples collected before, at or after diagnosis and achieved a sensitivity of 0.78 and a specificity 0.56 separating samples collected at the time of diagnosis of invasive cancer from samples collected prior to diagnosis. No difference in abundance was seen between samples collected prior to diagnosis or after treatment as compared with population controls, indicating that this protein signature is mainly informative close to time of diagnosis. Further studies are needed to determine the optimal window in time prior to diagnosis for these biomarker candidates.
Cervical cancer is the third most common cancer in women, with 485,000 cases worldwide in 2013. In many developing countries, cervical cancer is the most common cancer-related cause of death in women (1). Cervical cancer is caused by persistent infection with oncogenic forms of Human papilloma virus (HPV)1. Among the over 150 distinct types of HPV, a set of 12–16 are classified as high-risk types for cervical cancer, with HPV16 being the most oncogenic HPV type (2–4). In countries with an organized screening program, cervical cytology (Pap-smear) is the most common diagnostic test. Due to the low sensitivity of cytology (5–8), co-testing of HPV together with cytology has been introduced in the United States (9). The World Health Organization (WHO) has recommended the use HPV test as the primary screening test and that HPV infected women are followed up with colposcopy. The higher sensitivity of HPV as the primary screening test has resulted in an increase in the number of women with CIN2+ lesions identified (10). HPV testing has a higher sensitivity than cytology, but a single HPV-test has lower specificity due to the high prevalence of transient HPV infections. According to the present strategy, women that are HPV-positive and have normal cytology in the reflex-test are returned to the screening program. Some of these women will have a persistent HPV infection and consequently are at risk of developing lesions before the next screening occasion. To increase the specificity of screening, and distinguish between transient and persistent HPV infections, the HPV-test can be repeated 4–6 months after the primary screening test. This has been shown to reduce the number of HPV infections requiring management by about 40% (11, 12). An alternative way is to combine the HPV test with additional biomarkers indicating the persistence of infection or identifying early signs of cervical dysplasia (2, 3). Among the biomarkers proposed for co-testing of cytology, cyclin-dependent kinase inhibitor protein (p16INK4a) has been shown to be expressed at high levels in dysplastic epithelium. Dual staining of p16INK4a and KiU67 has been used to increase the diagnostic sensitivity of Pap-smear cytology (13). Several other markers have been suggested for early detection of cervical cancer in HPV positive women, but none of these is presently being used in clinical routine.
To identify novel protein biomarkers, efficient protein screening methods are needed. A variety of gel-based (2DE) and gel-free methods (LC-MS) have been used in cervical cancer cell culture systems and tissue samples (14, 15). In biomarker discovery studies, large sample sizes are needed to achieve statistical power and MS or gel-based methods have limited throughput. An alternative is to use targeted protein methods, such as the proximity extension assay (PEA) (16). This method focuses on a pre-selection of proteins and can be highly multiplexed to enable analysis of 92 proteins in 90 samples in a single experiment requiring as little as 1 μl of plasma for 92 proteins. PEA was recently used to identify protein biomarkers for insulin resistance and type-2 diabetes, by examining 92 proteins in over 1300 individuals (17), and in studies of 441 proteins in 1000 individuals from a population-based cohort (18). The aim of the present study was to search for plasma proteins that can be used to identify women at risk of developing cervical cancer using PEA.
EXPERIMENTAL PROCEDURES
Samples
The discovery cohort included plasma samples from healthy population controls (n = 50, denoted KA) from the Northern Sweden Population Health Study (NSPHS) (19) and as cases samples from women diagnosed with invasive cervical cancer from the Uppsala Biobank and the Uppsala Cancer Cohort (U-CAN) (n = 90, denoted UC) (Table 1). The replication cohort was based on samples from the Umeå Biobank and included plasma samples from women diagnosed with invasive cervical cancer (n = 90, denoted UM) and samples from the prospective Västerbotten Intervention Program (VIP) and from women participating in the mammography screening (MA) in Västerbotten, Sweden (20). All women included from VIP/MA developed invasive cervical cancer during the sample collection period. A set of these women (n = 16) were part of the UM-case cohort. Since the women were part of a prospective population-based study, the samples were collected at different time points before and after the date of diagnosis. In total, samples were obtained from 530 women included in VIP/MA, with between 1–12 samples from each woman, and a mean of 2.8 samples per woman, from before the date of diagnosis or after diagnosis (i.e. after treatment). The sample collection period ranged from 6166 days before to 7022 days after date of diagnosis. The plasma samples from the VIP and MA cohorts were collected between 1988 and 2014. All plasma samples in the two case cohorts were collected after the date of diagnosis but prior to the initiation of treatment. All women diagnosed with invasive cervical cancer were treated according to the standard clinical routine, including either chemotherapy or a combination of surgery and chemotherapy. Exclusion criteria for participation were pregnancy or self-reported previous cancer diagnosis. Plasma samples were collected in ethylenediaminetetraacetic acid (EDTA), separated and frozen within one hour after sampling and stored at −70 °C. In the VIP cohort, almost all samples were collected after overnight fasting. The sample aliquots analyzed had not previously been thawed.
Table I. Baseline information (age and time in freezer) for the two cohorts of women with invasive cervical cancer (Umeå Biobank, UM, Uppsala biobank, UC), and population controls (KA) used in the analysis, and the samples from the prospective study from UM.
| Discovery cohort |
Replication cohort |
|||
|---|---|---|---|---|
| Population controls (KA)** | Cancer cohort (UC) | Prospective | Cancer cohort (UM) | |
| Number of samples | 50 | 90 | 440 | 90 |
| Age at sample collection | ||||
| Minimum | 20 | 22.6 | 29.8 | 25.3 |
| Maximum | 86 | 86.4 | 73.8 | 96.9 |
| Mean | 58.4 | 46.5 | 52.3 | 60.8 |
| Standard deviation | 17.6 | 15.2 | 9.0 | 18.1 |
| Sample storage time in freezer* | ||||
| Minimum | 12 | 4.2 | 4 | 7 |
| Maximum | 12 | 8.9 | 30 | 13.7 |
| Mean | 12 | 6.4 | 17.7 | 10.3 |
| Standard deviation | 0 | 1.3 | 5.5 | 1.9 |
Notes: * Sample storage time in freezer until January 1, 2001.
** All samples were collected at one time point (2016) and stored for 8 years at −80 °C.
Ethical Considerations
The study was approved by the Regional Ethics Board in Uppsala, Sweden (Regionala Etikprövningsnämnden, Uppsala, Dnr 2016/145) and the Regional Ethics Board in Umeå, Sweden (Dnr 2013-314-32 M and 2012-229-31M).
Proximity Extension Assay
The abundance of 264 unique protein in plasma were analyzed in the discovery cohort using the Olink Multiplex CVD II, ONC II and INF I panels, and by the Olink Multiplex INF I panel in the replication cohort. All samples were then quantified by real-time PCR using the Fluidigm BioMark™ HD real-time PCR platform as described earlier (16, 21). Briefly, for each protein a dedicated pair of oligonucleotide-labeled antibodies bind to the targeted protein and if the two oligonucleotides are in proximity, a PCR target sequence is formed by a proximity-dependent DNA polymerization event and the resulting sequence is subsequently detected and quantified using real-time PCR. Each proximity extension assay (PEA) measurement has a specified lower detection limit (LOD) calculated based on negative controls that are included in each run and measurements below this limit were removed from further analysis. The Olink Multiplex data were reported in NPX (normalized protein expression levels), which are Ct values normalized by the subtraction of values for extension control, as well as an interplate control. The scale is then shifted using a run-time specific correction factor (normal background level). The final readout, NPX, is given on a log2-scale. Assay characteristics including detection limits and measurements of assay performance and validation for each protein are available at the manufacturer's webpage (http://www.olink.com). The analyses were performed using 1 μl of plasma for each panel of 92 proteins. To avoid batch effects, samples were randomized across plates. Each plate included inter-plate controls that were used to adjust for any plate differences. All samples in the discovery cohort were run at the same time and were randomized across plates. In addition to the full PEA panels, samples from the replication cohort were characterized using focused PEA panels which include up to 24 assays (including incubation, extension and detection controls) that are measured simultaneously in 192 samples as described earlier (21). The protein abundance levels on the focused PEA-panels from samples from the VIP- and MA-cohorts have been described earlier (22) and here that data was complemented with samples collected close to the date of diagnosis and samples from the UM-cohort. All samples in the replication cohort were run at the same time and were randomized across plates. The discovery cohort and the replication cohort were not run at the same time.
GWAS Lookup Analysis
All cis-SNPs (250 kbp downstream, 50 kbp upstream) of the genes encoding the 100 analyzed proteins were extracted published GWAS data (23).
Statistical Analysis
All calculations and illustrations were carried out in R (R core team) (24) (version 3.2.3). Here, individual protein levels (NPX) were normalized by plate and sampling round using the MDimNormn-package (25). Significance levels for each measured protein between cases and controls in the discovery cohort were calculated using the two-sided rank-based Spearman test (Wilcoxon). In the analysis of the discovery cohort, the protein NPX-values were normalized for the age of the individual at time of collection of the plasma sample. Individual proteins were then normalized to have mean equal to zero and unit variation using the observations in the control group (KA). The parameters obtained from the control group were then used to normalize the cases (UM) in the same way. In the analysis of the replication cohort, the protein NPX-values were adjusted for storage-time and chronological age at sample collection as described earlier (22). Samples acquired earlier than 3 years (1,095.75 days) from diagnosis in the replication control group (VIP+MA, n = 127) where normalized to have mean equal to zero and unit variation using the observations. The remaining samples in the VIP, MA and UM cohorts where then normalized using the same parameters. Proteins with all measurements below detection limit in either the discovery or replication data were excluded from further analysis. In total, 16 such proteins were removed (IL-2, IL-22-RA1, IL-13, IL-33, IFN-gamma, IL-2RB, IL-1-alpha, TSLP, PD-L1, IL-24, ARTN, TNF, IL-20, IL-4, LIF, and NRTN). For the remaining proteins, samples with measurements below the detection limit were replaced by the lowest value present after normalization. Only proteins available from both the discovery and replication cohort was used in the final analysis. Two of the remaining proteins were included on multiple panels (VEGF-A and HGF were present in both the ONC2 and INF2 panels) and only the measurements from the INF2 panel were included in the final data set consisting of 100 unique proteins. Power analysis was carried out using the “pwr”-package (26) in R and for the discovery cohort we have 0.95 power at 0.05 significant level to detect differences of 0.64 units in the normalized data. For the comparison between the samples taken at time of diagnosis and the samples used for normalization in the replication data, we are powered to detect differences of 0.48 units.
Model Building
Univariate linear models associating time to diagnosis and protein value were built using the “glm” function in R. The fraction of variance explained in the protein values by time to diagnosis was calculated using the Cox-Snell method as implemented by the “RsqGLM”-function from the R-package modEvA (version 1.3.2) (27). A multivariate Naïve Bayes model was built using the “caret”-package (28) (version 6.0–80) in R using 50% of the discovery cohort and all available proteins. The validation set was chosen to contain the same frequency of cases and controls as the whole set. The model was then trained using a cross-validation approach with 5 folds on the training data only. A second model was built using the same data for a subset of the proteins. These proteins were selected by first running feature selection with the “rfe”-function from the “caret” package, allowing selections of 5 to 25, 35, 45, 50 … or all proteins. The feature selection returned 11 proteins which were then used to build a second model using the same parameters and data as above. The trained models were then applied to the remaining 50% of the discovery data and to the prospective cohort. Finally, samples from the prospective cohort were split into bins of samples based on their diagnose-offset in days, in order to evaluate the potential of the protein markers in screening. In the binned-analysis, the first bin (longest time before diagnosis) was used as controls and each consecutive bin as cases. ROC-curves and performance measurements for all predictions were generated using the pROC R-package (29).
RESULTS
Univariate Analysis of Protein Biomarkers
After quality control and normalization (see Methods for details) 100 unique proteins remained and were available in both the discovery and replication cohorts. Two proteins (VEGF-A and HGF) were measured in duplicates and were found to show strong correlations with significant correlation coefficients (Spearman's rho) ranging from 0.53 to 0.93 with p values below 6.4 × 10−41.
Power-analysis in the discovery data show that our study is powered to detect differences of at least 0.64 units in the normalized data (see Methods for details). Univariate testing in the discovery cohort (t test, adjusted for multiple hypothesis testing using Bonferroni adjustment) resulted in 48 significant differences. We reasoned that useful biomarkers should have reliable measurements in the controls and higher values in the cases than the controls. Consequently, we removed 20 proteins that showed significantly lower levels in the cases as compared with controls from further analysis. Out of the remaining 28 proteins, 13 (46.4%) also showed a significant difference in the replication cohort between samples collected at the time of diagnosis and those collected at least 3 years before diagnosis (Fig. 1A, supplemental Table S2). The biomarker with the lowest p values in the replication data (supplemental Table S2) was PTX3 (p = 5.97 × 10−17, Fig. 1B). Among the 28 proteins, all but one protein (CD244, supplemental Table S2), showed a higher abundance value in the cases than the controls both in the discovery and the replication cohorts. Also, CD244 was found to have higher values in the cases in the discovery cohort (+0.78 units, p = 1.4 × 10−4), but lower values in the cases in the replication cohort (−0.59 units, p = 3.7 × 10−5).
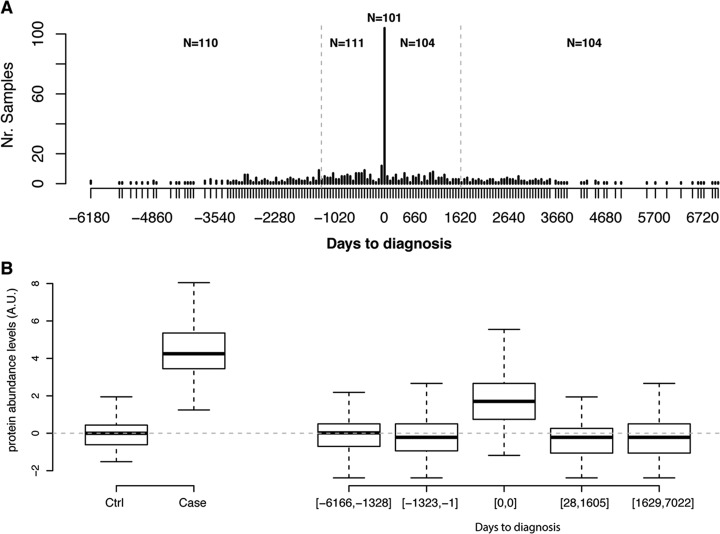
Fig. 1.
Replication of univariate analyses. A, Distribution of days to diagnosis for samples in the replication cohort. The gray dotted vertical lines indicate the cut-offs used in the binned analysis. B, Distribution of normalized protein abundance levels in the cohorts used for PTX3. The top and the bottom of the box represents the 25th and 75th percentile and the band inside the box the median value. The whiskers are calculated as 1.5× the interquartile range. Cohorts ordered as in (A). The dotted gray line indicates the mean of the groups used to determine normalization coefficients, e.g. the control samples from the discovery cohort and the samples from the replication cohort collected at least 3 years before diagnosis (See Methods for details).
Discovery of Multivariate Protein Biomarker Signatures
Using all 80 protein biomarkers (supplemental Table S1) with higher values in cases compared with controls in the discovery cohort, we proceeded with creating a multivariate model predicting cervical cancer from controls. To this end, the discovery cohort was split into a training (50%) and a validation (50%) proportion and a Naïve Bayes classifier was developed using the training proportion only. The final model, based on all 80 proteins, achieved close to perfect classification in the training proportion (Fig. 2A), with a sensitivity of 0.98 (95% CI 0.93–1.0) at a specificity of 1.0 (95% CI 1.0–1.0). The output from the model is a probability of having cancer, and the threshold for calling cancer was set at 0.75. When the same model and threshold was applied to the validation proportion in the discovery cohort, similar results were obtained (sensitivity = 0.96, 95% CI 0.89–1.0, specificity = 1.0, 95% 1.0–1.0, Fig. 2B). Because the underlying data was normalized to have zero mean and unit variance in both cohorts, the model was assumed to be immediately applicable to the replication cohort.
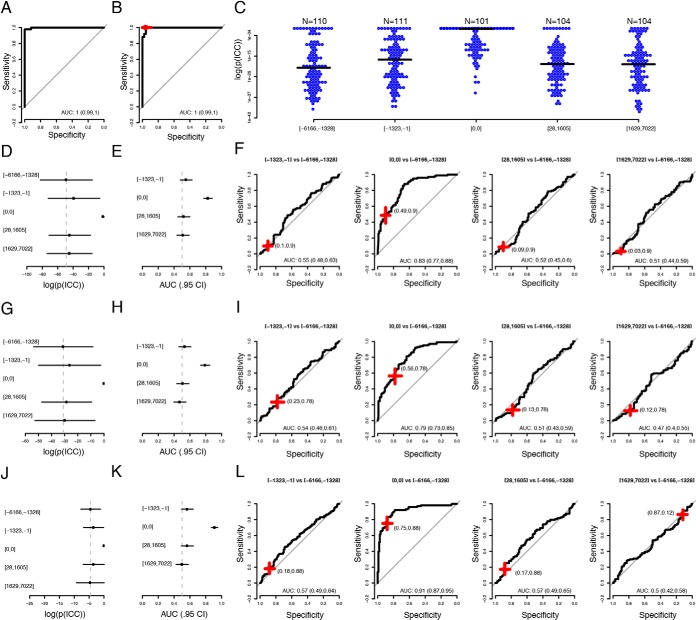
Fig. 2.
Multivariate analyses. A, Aggregated ROC-curve for the test-samples in the cross-validation training of the model using the full discovery data with 80 proteins. AUC given with 95% confidence interval. B, Performance of the trained model in the withheld discovery validation set. The red-cross represent the performance at the cut-off determined using the training data. C, Distribution of model-output (log(p)) when applied to the replication cohort in bins. The black line represents median value in each bin. D, Median (dot) and median absolute deviation (lines) of probabilities (model output) in the same bins as in (C). The dotted vertical line indicates the median in the bin with samples collected 6166 to 1328 days before diagnosis. E, AUCs for discriminations of the samples collected 6166 to 1328 days before diagnosis and the subsequent bins. Horizontal black lines indicate 95% confidence interval. The dotted vertical line indicates and AUC = 0.5. F, ROC-curves in the same bins as in (E). The red crosses indicate performance at the cut-off determined in the training proportion of the discovery data. The width and breadth of the cross represent 95% confidence interval of the sensitivity and specificity. G–I, Same as (D–F) but for the 11-protein signature. J–L, As (G–I) but for the 11-protein signature retrained as in (A–B) but with samples used for normalization in the replication cohort as controls and the samples taken at diagnosis in the replication cohort as cases.
We then proceeded to split the replication data into five bins with samples ordered by diagnose-offset in days, starting with the samples collected the longest time prior to diagnosis. The samples collected at the time of diagnosis were all kept in a separate bin and the samples collected before or after diagnosis were split into equal sized groups. Each bin contained between 101 and 111 samples. The model was then applied to all bins. The probability of having cancer was found to be significantly higher (Wilcoxon-test, Bonferroni adjusted, q-values <1.87 × 10−15) in the group with samples collected at diagnosis, compared with any other group (Fig. 2C, 2D). No other nominally significant (Wilcoxon-test) higher probability was found in any other comparison. In order to evaluate the ability to use the protein model as a screening method, we used the bin with samples collected the longest time before cancer diagnosis (1328 to 6166 days) as controls and compared this group with each consecutive bin as cases. In this analysis, the bin containing the samples collected at time of diagnosis resembles the set-up in the discovery cohort and achieved an AUC of 0.83 (95% CI 0.77–0.88) (Fig. 1E). At the threshold defined in the training proportion of the discovery cohort, this corresponds to a sensitivity of 0.90 (95% CI 0.84–0.95) at a specificity of 0.49 (95% CI 0.39–0.58). For the other groups, with samples collected 1323 to 1 days before diagnosis, 28 to 1605 or 1629 to 7022 days after diagnosis, the AUCs ranged from 0.51 to 0.55 (Fig. 2F).
We then proceeded with a feature-selection step in the model generation (see Methods for details) and this generated a signature consisting of 11 proteins (PTX3, ITGB1BP2, AXIN1, STAMPB, SRC, SIRT2, 4E-BP1, PAPP-A, HB-EGF, NEMO and IL-27). This signature achieved perfect classification in the training proportion of the discovery data at a probability threshold of 0.73. Similar performance as for the full protein model was achieved in the validation proportion of the discovery data (sensitivity 0.96, 95% CI 0.89–1, specificity 1.0, 95% CI 1.0–1.0) and in the replication data (Fig. 2G, 2H). In the bin containing samples collected at diagnosis an AUC of 0.79 (95% CI 0.73–0.85) was obtained with a sensitivity of 0.78 (95% CI 0.70–0.85) and a specificity 0.56 (95% CI 0.47–0.65) at the threshold defined in the analysis of the discovery data. Again, no discrimination was seen in the groups between samples collected before or after diagnosis (Fig. 2I).
The performance of the 11-protein signature in the replication cohort was found to be good (AUC = 0.79, Fig. 2I), but not as good as in the validation part of the discovery cohort (AUC = 1.0, Fig. 2B). We reasoned that since the PEA provide a readout of protein abundance levels on a relative scale and this scale is not necessarily the same between runs, the normalization used employed might not be enough to allow the same model with the same cut-off to be applied in a second cohort. To investigate this, we re-trained the 11-protein signature in the replication cohort using the samples used to define the normalization parameters (Methods for details) as controls and the samples collected at the time of diagnosis as cases (Fig. 2J–2L). In the same comparisons as above, the model then achieved an AUC of 0.91 separating samples collected at the time of diagnosis from the samples collected 6166 to 1328 days before diagnosis. This approach did not, however, significantly improve the performance in the other time-intervals used in the prospective cohort, with all 95% confidence intervals including an AUC of 0.5 (Fig. 2K, 2L).
Genetic Variability in the Genes Encoding Candidate Biomarkers
To determine the contribution of genetic variability in the genes encoding the proteins in the multivariate signatures, we used the data from our previously published GWAS for cervical cancer (23). Using a window of 250 kb upstream of each gene (to include the promotor region) and 50 kb downstream of each gene, a total of 7355 SNPs was found in the 80 genes and 637 within the 11 genes. Of these, 916 of the SNP in the 80 genes and 24 SNPs in the 11 genes showed a nominally significant (p < 0.05) difference between cases and controls in the GWAS data. Adjusting for multiple hypothesis testing, 137 SNPs of the 916 remained significant, while none of the 24 SNPs in the 11 genes did. All 137 SNPs were retrieved because of their location in cis with either TNFB, MIC-A/B or RAGE and were located within a 1.2Mb region on chr6. This region sits within the major histocompatibility complex loci (30), which is associated with a high number of human autoimmune and infectious diseases. The lowest p value among these 137 SNPs was found for rs2516448 (p < 4.87 × 10−13) located 7.4kb downstream of MIC-A. This SNP was however not among the SNPs detected in a GWAS for protein abundance levels in MIC-A previously published (18). The lowest p value among the 24 nominally significant SNPs within the genes encoding the 11 proteins was found for rs1405 (p = 0.003), located in the first intron of the gene encoding PAPP-A. Taken together, the difference in abundance levels between cases and controls does not appear to be affected to any significant degree by genetic variability related to cervical cancer found in the genes encoding these proteins.
Biomarker Levels Before Diagnosis and After Treatment
The multivariate models generated above did not show any significant predictive power before or after time of diagnosis, as compared with the samples collected 1328 to 6166 days before diagnosis. In order to evaluate single proteins as early indicators of invasive cervical cancer, we estimated the variance explained in the protein values by the time-to-diagnosis. This was done by building linear models with protein values as response and time to diagnosis, either before or after, as variable. Among all 80 proteins, the explained variance in the protein abundance levels ranged from 3.76 × 10−6 to 6.27%. Two models remained significant after multiple hypothesis testing (p < 0.05/2/80 = 3.13 × 10−4), for CCL and FR-alpha, both decreasing before diagnosis with coefficients ranging from −1.87 × 10−4 to −1.69 × 10−4 (supplemental Table S3). For the proteins in the 11-biomaker signature, the explained variance in the protein abundance levels ranged from 1.76 × 10−4 to 2.6%. None of the models generated remained significant after multiple hypothesis correction.
Finally, we investigated if there was any difference among the proteins when observing the samples in the same bins as used in the multivariate modeling, by comparing the samples taken immediately before or after diagnosis with samples collected the longest time before diagnosis (supplemental Table S4). We restricted this analysis to the 28 proteins that were found to have a significant increase in abundance level at time of diagnosis as compared with controls in the discovery cohort. Among these proteins, none had even a nominally significant difference between samples collected 1328 to 6166 days before diagnosis and samples collected 28 to 1605 days after diagnosis. As reported above, 13 of the 28 proteins had significantly increased abundance levels compared with the group collected at time of diagnosis. Lastly, we compared the samples collected 1 to 1323 days before diagnosis with the samples collected 1328 to 6166 days before diagnosis. One protein, FR-alpha, was found to have nominally different values but this difference did not remain significant after adjustment for multiple hypothesis testing.
DISCUSSION
We used PEA to study 100 unique proteins and identified a biomarker signature with 11 proteins showing a significant difference in plasma levels between samples collected at the time of diagnosis of invasive cervical cancer and population controls. The proteins identified distinguished between cervical cancer cases and population controls with a high sensitivity and specificity.
Five of the 11 proteins in our biomarker signature, namely Pentraxin-related protein (PTX3), Proto-oncogene tyrosine kinase Src (SRC), Eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), Interleukin-27 (IL-27) and SIR2-like protein 2 (SIRT2), have previously been associated with cervical cancer.
The Pentraxin-related protein (PTX3) plays a key role in resistance to microbes and regulation of inflammation and has been implicated in cancer development (31). Increased PTX3 expression has been associated with differentiation of cervical tumors and a PTX3-knockdown has been shown to suppress the metastatic potential of cervical cancer cells. Therefore, PTX3 is believed to contribute to tumorigenesis of human cervical cancer cells and has been discussed as a possible biomarker for cervical cancer (32). In our analysis, the cases had 1.7× higher NPX-values as compared with the controls, corresponding to more than twice as high protein levels in plasma. In cervical cancer, SRC activity is deregulated by HPV E6/E7 onco-proteins (33). Expression of SRC has been shown to increase gradually from normal cervical epithelium to cervical cancer in situ and cervical squamous cell carcinoma, and expression levels correlate with survival of cervical cancer patients. We found that patients with invasive cervical cancer had on average 1.8 higher NPX-values than the population controls. High SRC expression has also been reported to be a negative prognostic marker for recurrence-free survival of cervical squamous cell carcinoma, indicating that SRC could be a valuable biomarker in the follow up phase (34). 4E-BP1 is a translational repressor and higher levels of this protein has been associated with poorer survival after surgical treatment of cervical cancer (35). IL-27 has been shown to be highly expressed in cervical cancer cell lines (36). In our discovery cohort, cases had on average 1.7 times higher IL27 NPX values than the controls. Finally, expression of SIRT2, both on a transcript and protein level, has been shown to be up-regulated in cervical cancer as compared with normal tissue (37). In our study the cases in the discovery cohort had on average 3.7 times higher NPX values than the population controls. The previous study (37) did not find higher levels of SIRT2 during progression from preneoplasia to squamous cell carcinoma, which is relevant from a screening prospective. Similarly, our 11-biomarker signature did not perform as well as for samples collected at time of diagnosis. In general, the direction of difference in abundance of these proteins between our cases and controls is consistent with earlier reports. A further set of four proteins (PAPP-A, ITGB1BP2, AXIN1 and NEMO) have previously been associated with other types of cancer, but, as to the best of our knowledge, this is the first study that shows a direct association with cervical cancer. Pappalysin-1 (PAPP-A) is a metalloproteinase, and studies in mice identify PAPP-A as a pregnancy-dependent oncogene, with extended lactation being protective against PAPP-A-mediated carcinogenesis (38).
The locus containing the gene ITGB1BP2 (also known as Melusin), encoding the Integrin beta 1 binding protein 2 protein, has been shown to harbor genetic duplications in cervical cancers, but the expression does not appear to be up-regulated (39). AXIN1 (Axis inhibition protein 1) has positive and negative regulatory function in the Wnt-beta-catenin signaling pathway, with a specific function in regulation of apoptosis in human melanoma (40). NEMO (NF-κB essential modulator) is a regulatory subunit of the NF-κB classical activation pathway, which is associated with tumor promotion and can be activated by viral infections or through signaling by pro-inflammatory cytokines (41). Interesting, the NF-κB pathway has been shown to be downregulated by HPV leading to persistent infections but activated again in high grade intraepithelial neoplasia and cervical cancers (42).
Using samples from a prospective population-based study, we examined the changes in abundance relative to the time of diagnosis. In order to achieve groups of comparable size, samples from 0 to 3.6 years prior to diagnosis and samples from 0 to 4.5 years after diagnosis, were compared with samples taken earlier than 3.6 years before diagnosis, and to samples collected at time of diagnosis. No individual protein differences between these groups remained statistically significant after Bonferroni correction. In addition, although the multivariate models showed good discrimination between samples collected at diagnosis and the controls, the models were not able to discriminate samples taken before or after diagnosis from the samples used as controls. The results indicate that the protein abundance differences seen in the case-control comparison reflect changes that occurred closer to diagnosis than the samples that were available from the prospective cohort. This may imply that in order to identify women at risk of developing cervical cancer, there is a need to have a shorter screening interval than the 3-years presently used in the organized screening of cervical cancer in Sweden.
Our current study is limited by the number of samples that were available with collection dates close to the time of diagnosis. This low sample-rate reduces our power to detect changes in the groups before diagnosis and this also means that is difficult to pin-point the underlying biological processes that drives the changes in biomarker levels. We are also restricted in our modeling by possible intra-assay variation between the discovery and replication cohorts. Our 11-biomarker model achieved and AUC of 0.79 in the replication cohort compared with 1.0 in the discovery cohort. This could however be increased to an AUC of 0.91 when retraining the coefficients of the model on the values in the replication cohort. This suggests that the 11-biomarker model can distinguish cases from controls, but that the normalization procedure employed here matching samples taken long before diagnosis in the time-series with the controls from the discovery cohort is insufficient to capture all of this variation.
In summary, we have identified several protein biomarker candidates for invasive cervical cancer, both individually and multivariate signatures. To determine the clinical utility of these biomarker there is a need to examine samples from women with different precancerous stages (CIN1, CIN2, CIN3 and Invasive cervical cancer) as well as to study series of samples collected closer to the date of diagnosis than available in the prospective cohort we studied, to determine the window in time when the protein panel is informative of future cancer development.
Supplementary Material
Footnotes
* This study was supported by the Swedish Cancer Foundation, The Swedish Foundation for Strategic Research and The Swedish Research Council.
 This article contains supplemental Tables. The study was co-funded by OLINK Proteomics AB.
This article contains supplemental Tables. The study was co-funded by OLINK Proteomics AB.
1 The abbreviations used are:
- HPV
- human papilloma virus
- PEA
- proximity extension assay
- U-CAN
- Uppsala cancer cohort
- VIP
- vasterbotten intervention program
- NPX
- normalized protein expression levels.
REFERENCES
- 1. Global Burden of Disease Cancer, C., et al. (2017) Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 3, 524–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sahasrabuddhe V. V., Luhn P., and Wentzensen N. (2011) Human papillomavirus and cervical cancer: biomarkers for improved prevention efforts. Future Microbiol. 6, 1083–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schiffman M., et al. (2011) Human papillomavirus testing in the prevention of cervical cancer. J. Natl. Cancer Inst. 103, 368–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsikouras P., Zervoudis S., Manav B., Tomara E., Iatrakis G., Romanidis C., Bothou A, and Galazios G. (2016) Cervical cancer: screening, diagnosis and staging. J BUON, 21, 320–325 [PubMed] [Google Scholar]
- 5. Agorastos T., Chatzistamatiou K., Katsamagkas T., Koliopoulos G., Daponte A., Constantinidis T., Constantinidis T.C. and HERMES study group. (2015) Primary screening for cervical cancer based on high-risk human papillomavirus (HPV) detection and HPV 16 and HPV 18 genotyping, in comparison to cytology. PLoS ONE 10, e0119755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castle P. E., Stoler M. H., Wright T. C. Jr, Sharma A., Wright T. L., and Behrens C. M. (2011) Performance of carcinogenic human papillomavirus (HPV) testing and HPV16 or HPV18 genotyping for cervical cancer screening of women aged 25 years and older: a subanalysis of the ATHENA study. Lancet Oncol. 12, 880–890 [DOI] [PubMed] [Google Scholar]
- 7. Chatzistamatiou K., Moysiadis T, Moschaki V, Panteleris N, and Agorastos T. (2016) Comparison of cytology, HPV DNA testing and HPV 16/18 genotyping alone or combined targeting to the more balanced methodology for cervical cancer screening. Gynecol. Oncol. 142, 120–127 [DOI] [PubMed] [Google Scholar]
- 8. Cobucci R., Maisonnette M., Macêdo E., Santos Filho F. C., Rodovalho P., Nóbrega M. M., and Goncalves A. (2016) Pap test accuracy and severity of squamous intraepithelial lesion. Indian J. Cancer 53, 74–76 [DOI] [PubMed] [Google Scholar]
- 9. Saslow D., Solomon D., Lawson H. W., Killackey M., Kulasingam S. L., Cain J. M., Garcia F. A., Moriarty A. T., Waxman A. G., Wilbur D. C., Wentzensen N., Downs L. S. Jr, Spitzer M., Moscicki A. B., Franco E. L, Stoler M. H., Schiffman M., Castle P. E., Myers E. R., Chelmow D., Herzig A., Kim J. J., Kinney W., Herschel W. L., and Waldman J. (2012) American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. J. Low Genit. Tract Dis. 16, 175–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ronco G., Dillner J., Elfström K. M., Tunesi S., Snijders P. J., Arbyn M., Kitchener H., Segnan N., Gilham C., Giorgi-Rossi P., Berkhof J., Peto J., Meijer C. J. and; International HPV screening working group. (2014) Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet 383, 524–32 [DOI] [PubMed] [Google Scholar]
- 11. Gustavsson I., Aarnio R., Berggrund M., Hedlund-Lindberg J., Strand A.S., Sanner K., Wikström I., Enroth S., Olovsson M., and Gyllensten U. (2018) Randomised study shows that repeated self-sampling and HPV test has more than two-fold higher detection rate of women with CIN2+ histology than Pap smear cytology. Br. J. Cancer 118, 896–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gyllensten U., Sanner K., Gustavsson I., Lindell M., Wikström I., and Wilander E. (2011) Short-time repeat high-risk HPV testing by self-sampling for screening of cervical cancer. Br. J. Cancer 105, 694–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. von Knebel Doeberitz M., Reuschenbach M., Schmidt D., and Bergeron C. (2012) Biomarkers for cervical cancer screening: the role of p16(INK4a) to highlight transforming HPV infections. Expert Rev. Proteomics 9, 149–163 [DOI] [PubMed] [Google Scholar]
- 14. Di Domenico F., De Marco F., and Perluigi M. (2013) Proteomics strategies to analyze HPV-transformed cells: relevance to cervical cancer. Expert Rev. Proteomics 10, 461–472 [DOI] [PubMed] [Google Scholar]
- 15. Kontostathi G., Zoidakis J., Anagnou N. P., Pappa K. I., Vlahou A., and Makridakis M. (2016) Proteomics approaches in cervical cancer: focus on the discovery of biomarkers for diagnosis and drug treatment monitoring. Expert Rev. Proteomics 13, 731–45 [DOI] [PubMed] [Google Scholar]
- 16. Assarsson E., Lundberg M., Holmquist G., Björkesten J., Thorsen S. B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G., Andersson A. C., Lindstedt P., Stenvang J., Gullberg M., and Fredriksson S. (2014) Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS ONE 9, e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nowak C., Sundström J., Gustafsson S., Giedraitis V., Lind L., Ingelsson E., and Fall T. (2016) Protein biomarkers for insulin resistance and type 2 diabetes risk in two large community cohorts. Diabetes 65, 276–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Enroth S., Maturi V., Berggrund M., Enroth S. B., Moustakas A., Johansson Å., and Gyllensten U. (2018) Systemic and specific effects of antihypertensive and lipid-lowering medication on plasma protein biomarkers for cardiovascular diseases. Sci. Rep. 8, 5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Igl W., Johansson A., and Gyllensten U. (2010) The Northern Swedish Population Health Study (NSPHS)–a paradigmatic study in a rural population combining community health and basic research. Rural Remote Health 10, 1363. [PubMed] [Google Scholar]
- 20. Hallmans G., Agren A., Johansson G., Johansson A., Stegmayr B., Jansson J. H., Lindahl B., Rolandsson O., Söderberg S., Nilsson M., Johansson I., and Weinehall L. (2003) Cardiovascular disease and diabetes in the Northern Sweden Health and Disease Study Cohort - evaluation of risk factors and their interactions. Scand. J. Public Health Suppl. 61, 18–24 [DOI] [PubMed] [Google Scholar]
- 21. Assarsson E., and Lundberg M. (2017) Development and validation of customized PEA biomarker panels with clinical utility. Advancing precision medicine: Current and future proteogenomic strategies for biomarker discovery and development. Science 32–36 [Google Scholar]
- 22. Enroth S., Hallmans G., Grankvist K., and Gyllensten U. (2016) Effects of long-term storage time and original sampling month on biobank plasma protein concentrations. EBioMedicine, 12, 309–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen D., Chen D., Juko-Pecirep I., Hammer J., Ivansson E., Enroth S., Gustavsson I, Feuk L., Magnusson P. K., McKay J. D., Wilander E., and Gyllensten U. (2013) Genome-wide association study of susceptibility loci for cervical cancer. J. Natl. Cancer Inst. 105, 624–633 [DOI] [PubMed] [Google Scholar]
- 24. R Develpment Core Team, R: A language and environment for statistical computing. 2015, Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 25. Hong M. G., Lee W., Nilsson P., Pawitan Y., and Schwenk J. M. (2016) Multidimensional normalization to minimize plate effects of suspension bead array data. J. Proteome Res. 15, 3473–3480 [DOI] [PubMed] [Google Scholar]
- 26. Champely S. pwr: Basic Functions for Power Analysis. 2018 [Google Scholar]
- 27. Barbosa A. M., Brown J. A., and Jimenez-Valverde A. (2016) Real R, Model Evaluation and Analysis. https://R-Forge.R-project.org/projects/modeva/.
- 28. Kuhn M., Wing J., Weston S., Williams A., Keefer C., Engelhardt A., Cooper T., Mayer Z., Kenkel B., R Core Team, Benesty M., Lescarbeau R., Ziem A., Scrucca L., Tang Y., Candan C., and Hunt T.. caret: Classification and Regression Training. 2018 [Google Scholar]
- 29. Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J. C., and Müller M. (2011) pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Matzaraki V., Kumar V., Wijmenga C., and Zhernakova A. (2017) The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 18, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Giacomini A., Ghedini G.C., Presta M., and Ronca R. (2018) Long pentraxin 3: A novel multifaceted player in cancer. Biochim. Biophys. Acta Rev. Cancer 1869, 53–63 [DOI] [PubMed] [Google Scholar]
- 32. Ying T. H., Lee C. H., Chiou H. L., Yang S. F., Lin C. L., Hung C. H., Tsai J. P., and Hsieh Y. H. (2016) Knockdown of Pentraxin 3 suppresses tumorigenicity and metastasis of human cervical cancer cells. Sci. Rep. 6, 29385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Al Moustafa A. E., Yasmeen A., and Alachkar A. (2011) Src inhibitors are promising therapy molecules for human cervical carcinomas. Med. Hypothese, 77, 812–814 [DOI] [PubMed] [Google Scholar]
- 34. Hou T., Xiao J., Zhang H., Gu H., Feng Y., and Li J. (2013) Phosphorylated c-Src is a novel predictor for recurrence in cervical squamous cell cancer patients. Int. J. Clin. Exp. Pathol. 6, 1121–1127 [PMC free article] [PubMed] [Google Scholar]
- 35. Benavente S., Vergés R., Hermosilla E., Fumanal V., Casanova N., García A., Ramón Y Cajal S., and Giralt J. (2009) Overexpression of phosphorylated 4E-BP1 predicts for tumor recurrence and reduced survival in cervical carcinoma treated with postoperative radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 75, 1316–1322 [DOI] [PubMed] [Google Scholar]
- 36. Zhang B., Xie F., Dong C. L., Gu C. J., Cheng J., Wang Y., Xu X. Z., Pu H., Wu Y. B., Qi X. W., Li D. J., Yu J. J., and Li M. Q. (2017) The cross talk between cervical carcinoma cells and vascular endothelial cells mediated by IL-27 restrains angiogenesis. Am. J. Reprod. Immunol. 78, e12706. [DOI] [PubMed] [Google Scholar]
- 37. Singh S., Kumar P. U., Thakur S., Kiran S., Sen B., Sharma S., Rao V. V., Poongothai A. R., and Ramakrishna G. (2015) Expression/localization patterns of sirtuins (SIRT1, SIRT2, and SIRT7) during progression of cervical cancer and effects of sirtuin inhibitors on growth of cervical cancer cells. Tumour Biol. 36, 6159–6171 [DOI] [PubMed] [Google Scholar]
- 38. Takabatake Y., Oxvig C., Nagi C., Adelson K., Jaffer S., Schmidt H., Keely P. J., Eliceiri K. W., Mandeli J., and Germain D. (2016) Lactation opposes pappalysin-1-driven pregnancy-associated breast cancer. EMBO Mol. Med. 8, 388–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Narayan G., Bourdon V, Chaganti S, Arias-Pulido H, Nandula S. V., Rao P. H., Gissmann L., Dürst M., Schneider A., Pothuri B., Mansukhani M., Basso K., Chaganti R. S., and Murty V. V. (2007) Gene dosage alterations revealed by cDNA microarray analysis in cervical cancer: identification of candidate amplified and overexpressed genes. Genes Chromosomes Cancer 46, 373–384 [DOI] [PubMed] [Google Scholar]
- 40. Biechele T. L., Kulikauskas R. M., Toroni R. A., Lucero O. M., Swift R. D., James R. G., Robin N. C., Dawson D. W., Moon R. T., and Chien A. J. (2012) Wnt/beta-catenin signaling and AXIN1 regulate apoptosis triggered by inhibition of the mutant kinase BRAFV600E in human melanoma. Sci. Signal. 5, ra3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Karin M., and Greten F. R. (2005) NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 5, 749–759 [DOI] [PubMed] [Google Scholar]
- 42. Tilborghs S., Corthouts J., Verhoeven Y., Arias D., Rolfo C., Trinh X. B., and van Dam P. A. (2017) The role of Nuclear Factor-kappa B signaling in human cervical cancer. Crit Rev Oncol. Hematol. 120, 141–150 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.