N-linked protein glycosylation (Pgl) in Campylobacter jejuni is required for chicken colonization and human virulence, yet its biological role remains unknown. pgl gene deletion resulted in a significant rearrangement of the C. jejuni proteome that leads to alterations in crucial phenotypes including stress response, nutrient uptake, electron transport and chemotaxis, and is essential for full activity of the Nap nitrate reductase. N-glycosylation therefore contributes to multiple “virulence” phenotypes in C. jejuni.
Keywords: N-Glycosylation, Glycoprotein Pathways*, Bacteria, Virulence, Microbiology, Bacterial pathogenesis, Stress response
Graphical Abstract
Highlights
Protein N-glycosylation is essential for nitrate reductase (Nap) activity in C. jejuni.
Removal of N-glycosylation results in a metabolic switch from Asp to Pro uptake.
N-glycosylation is required for optimal chemotaxis towards several substrates.
Loss of N-glycosylation reduces survival following temperature and osmotic shock.
Abstract
Campylobacter jejuni is a major gastrointestinal pathogen generally acquired via consumption of poorly prepared poultry. N-linked protein glycosylation encoded by the pgl gene cluster targets >80 membrane proteins and is required for both nonsymptomatic chicken colonization and full human virulence. Despite this, the biological functions of N-glycosylation remain unknown. We examined the effects of pgl gene deletion on the C. jejuni proteome using label-based liquid chromatography/tandem mass spectrometry (LC-MS/MS) and validation using data independent acquisition (DIA-SWATH-MS). We quantified 1359 proteins corresponding to ∼84% of the C. jejuni NCTC 11168 genome, and 1080 of these were validated by DIA-SWATH-MS. Deletion of the pglB oligosaccharyltransferase (ΔpglB) resulted in a significant change in abundance of 185 proteins, 137 of which were restored to their wild-type levels by reintroduction of pglB (Δaaz.batpglB::ΔpglB). Deletion of pglB was associated with significantly reduced abundances of pgl targets and increased stress-related proteins, including ClpB, GroEL, GroES, GrpE and DnaK. pglB mutants demonstrated reduced survival following temperature (4 °C and 46 °C) and osmotic (150 mm NaCl) shock and altered biofilm phenotypes compared with wild-type C. jejuni. Targeted metabolomics established that pgl negative C. jejuni switched from aspartate (Asp) to proline (Pro) uptake and accumulated intracellular succinate related to proteome changes including elevated PutP/PutA (proline transport and utilization), and reduced DctA/DcuB (aspartate import and succinate export, respectively). ΔpglB chemotaxis to some substrates (Asp, glutamate, succinate and α-ketoglutarate) was reduced and associated with altered abundance of transducer-like (Tlp) proteins. Glycosylation negative C. jejuni were depleted of all respiration-associated proteins that allow the use of alternative electron acceptors under low oxygen. We demonstrate for the first time that N-glycosylation is required for a specific enzyme activity (Nap nitrate reductase) that is associated with reduced abundance of the NapAB glycoproteins. These data indicate a multifactorial role for N-glycosylation in C. jejuni physiology.
Campylobacter jejuni is a Gram-negative, microaerophilic, helical, and motile bacterium that is a major cause of human gastroenteritis. More than 400 million people are infected annually worldwide (1–2). Infection typically occurs via contaminated water or consumption of under-cooked or inadequately prepared poultry for human consumption because avian species, such as chickens, are widely considered as asymptomatic hosts, although this paradigm has recently been questioned (3–4). Human symptoms include mild, non-inflammatory diarrhea, but may also be characterized by abdominal cramps, bloody diarrhea, vomiting and inflammation. Although the disease is mostly self-limiting, potential sequelae include Guillain-Barré Syndrome and Miller Fisher Syndrome, which are related immune-mediated disorders of the peripheral nervous system (5–6).
C. jejuni survives in the environment and then avian host before human ingestion, which requires a diverse metabolic capacity and tolerance to a range of environments, including different temperatures. C. jejuni grows preferentially between 37–42 °C but can survive at extremes (such as 4 °C; enabling viable bacteria to contaminate >75% of supermarket chicken (7)). C. jejuni lacks pathways for using carbohydrates as carbon sources and was considered asaccharolytic until the discovery of strains containing an l-fucose permease (FucP; (8)). The primary nutrient sources are therefore organic and amino acids (reviewed in (9–11)). C. jejuni preferentially uses (in order) the amino acids serine (Ser), aspartate (Asp), glutamate (Glu) and proline (Pro) (12–13). Glu and Asp uptake are partly mediated by the Peb locus (14–15) and C. jejuni contains several transducer-like protein (Tlp)1 chemoreceptors that sense Asp and other nutrients (16–17). The C4-dicarboxylate transporters, including DcuA/DcuB and DctA, are also involved in Asp uptake, as well as transport of fumarate and succinate (11). DcuB is responsible for succinate export under very low oxygen conditions (11). Pro appears to be the least preferred amino acid and is thought to be consumed only when other substrates are exhausted (18). C. jejuni also catabolizes other organic acids including lactate (transported by the LctP permease (19)), pyruvate, acetate, α-ketoglutarate (α-KG; transported by the KgtP permease) and malate (18). Finally, C. jejuni can respire at very low oxygen availabilities by using alternative electron acceptors such as nitrate, nitrite, fumarate, trimethylamine-N-oxide (TMAO) or dimethyl sulfoxide (DMSO) (20), for which several reductases have been identified (Nap, nitrate; Nrf, nitrite; Frd, fumarate; and Cj0264c/Cj0265c, TMAO/DMSO). Both electron donors, including NADH and formate dehydrogenases (21), and electron acceptors are required for optimum host colonization (22).
A unique molecular feature of C. jejuni is the ability to modify proteins via N- and O-linked glycosylation (23–24). The N-linked glycosylation system is encoded by the 10 gene pgl cluster, which is responsible for the biosynthesis and attachment of a heptasaccharide glycan (GalNAc-α1,4-GalNAc-α1,4-[Glcβ1,3]-GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-Bac-β1; Bac is bacillosamine [2,4-diacetamido-2,4,6 trideoxyglucopyranose]) to proteins at the consensus sequon Asp/Glu-X-Asn-X-Ser/Thr (X ≠ Pro), where Asn is the attachment site (25). Biosynthesis begins in the cytoplasm where the enzymes PglFED use nucleotide-activated monosaccharide precursors (UDP-GlcNAc) to synthesize bacillosamine (Bac; (26)), which is coupled to an inner membrane-bound lipid anchor (undecaprenylphosphate; Und-P) by PglC. Subsequent steps include the sequential addition of 5 N-acetylgalactosamine (GalNAc) residues (the first by PglA and subsequently by PglHJ) and a single branching hexose (glucose; Glc; attached by PglI). Once completed, the PglK flippase “flips” the Und-P-bound heptasaccharide into the periplasm where it is added to target proteins in the periplasm by the PglB oligosaccharyltransferase. PglB is assumed to transfer the glycan to solvent-exposed or disordered regions on already folded substrates (27), however more recent work implies that glycosylation in some cases may occur before folding is complete, and hence influence final tertiary structure (28). The protein N-glycan can be further modified by the addition of a terminal phosphoethanolamine (pEtN) moiety that is catalyzed by the EptC pEtN transferase, which also modifies lipid A and the flagellar protein FlgG (29–31). PglB also liberates the glycan from Und-P as a “free oligosaccharide” (fOS) that is involved in osmotic balance (32).
Our group and others have shown that the Pgl system can target more than 130 sequons from ∼80 membrane-associated proteins (25, 29, 33–35), including periplasmic proteins, lipoproteins, inner membrane proteins and at least one protein that is thought to be surface-exposed (the lipoprotein JlpA (36)). Deletion of genes from the pgl cluster, particularly pglB, result in C. jejuni that are attenuated for the ability to colonize chickens and impaired in adherence to, and invasion of, human epithelial cells (37–38). Despite this, the function of N-glycosylation in C. jejuni remains poorly understood. Determining the role played by the Pgl system is additionally complicated because many of the identified glycoproteins have no known function. Site-directed mutagenesis has been performed for a handful of sites, generally with no discernible effect on C. jejuni pathogenesis or specific protein functions (39); although a single study has shown glycosylation of Cj0011c is required for full competence, which may reflect incorrect protein localization without glycan attachment (40). Wider studies have shown a requirement for glycosylation in binding host lectins (41), and a potential role in protection from host-derived proteases (42), although the proteins for which this is required have not been identified.
In this study, we examined the global C. jejuni proteome response to the loss of the pglB oligosaccharyltransferase using label-based liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) and proteome-wide validation using data independent analysis (DIA-SWATH-MS). Loss of pglB resulted in alterations that included elevated stress response proteins, and decreased abundance of previously characterized glycoproteins. Changes to nutrient transporters were validated by monitoring the intracellular levels of their substrates, which showed a shift from Asp to Pro in the absence of glycosylation. Changes in C. jejuni metabolism also aligned with altered chemotaxis to corresponding carbon sources. Finally, we show that loss of glycosylation severely impacts low oxygen respiration and provide the first example of an enzyme function requiring glycosylation, as pgl-negative mutants are defective in nitrate reductase activity. This study confirms glycosylation is required for essential C. jejuni phenotypes and contributes to resistance against cell stress, nutrient sensing and transport, and reductase activity.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
For quantitative proteomics by tandem mass tag [TMT] labeling and DIA-SWATH-MS, 3 biological replicates of each C. jejuni NCTC 11168 strain (wild-type [WT], ΔpglB and ΔpglB::pglB) were processed in tandem. The 2 technical approaches were treated as a “discovery” set (label-based) and a “validation” set (DIA-SWATH-MS) and were performed on separate triplicate sample sets and processed independently of each other. Before TMT labeling, equal aliquots from each biological replicate were split to form an internal technical replicate. For targeted metabolomics experiments, 3 biological replicates were prepared in triplicate extractions. Mass spectrometry data have been deposited to the ProteomeXchange Consortium via PRIDE with the data set identifier PXD011646.
Bacterial Strains and Growth Conditions
C. jejuni strains (NCTC 11168 (WT), ΔpglB and ΔpglB::pglB, and JHH1 (WT), JHH1ΔpglB and JHH1 ΔpglFED and their growth were as previously described (29). ΔpglB strains and JHH1ΔpglFED were grown in the presence of kanamycin (Km; 30 μg/ml) and ΔpglB::pglB was grown in medium with added chloramphenicol (Cm; 10 μg/ml) for selection, and 25 μg/ml trimethoprim (all antibiotics Sigma, St. Louis, MO) for all strains when grown in MH medium. Cells were grown from glycerol stocks in Mueller-Hinton (MH) Broth (Oxoid, Basingstoke, UK) for 48 h at 37 °C under microaerophilic conditions (5% O2, 10% CO2, 85% N2). Cells were sub-cultured into fresh MH broth at an initial OD600 of 0.1 and grown until late exponential phase before being harvested and lyophilized before further use. For growth in the presence of nitrate, medium was supplemented with 5% (v/v) fetal bovine serum (FBS), with antibiotics as required, and pre-incubated under microaerophilic conditions for 24 h. Medium was then supplemented with 20 mm sodium nitrate before inoculation from a starter culture, as above.
Western Blotting
Western blotting was performed as described previously (36). Proteins were transferred from SDS-PAGE gels to polyvinylidene difluoride (PVDF) membrane for 1 h at 400 mA (4°C). PVDF membranes were blocked overnight at 4°C in 5% bovine serum albumin (BSA) in TBS-T (20 mm Tris, 150 mm NaCl, 0.1% Tween-20, pH 7.4). Membranes were probed with a 1:1000 JlpA-specific rabbit anti-serum overnight at 4°C. Proteins were detected using a 1:1000 dilution of goat-anti-rabbit immunoglobulin, followed by incubation with SupersignalTM West Pico Chemiluminescent substrate according to the manufacturer's instructions (Thermo Scientific, Waltham, MA) and visualized using a ChemiDocTM MP Imaging System (Bio-Rad, Hercules, CA).
Quantitative PCR (qPCR) of Select C. jejuni Genes
Cells were harvested and immediately placed in RNAprotect Bacteria Reagent (Qiagen, Hilden, Germany) as per the manufacturer's instructions. Cells were lysed by 4 rounds of bead-beating (0.1 mm beads, 5 m/s, 25 s) with 1 min rest periods on ice. Cell debris was removed by centrifugation for 3 min at 4000 × g, 4 °C. RNA was extracted using an RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions, including the additional DNase I and Proteinase-K treatment steps to optimize RNA quality. RNA was checked for purity using the 260/280 and 260/230 optical wavelength measurements on a NanoDrop 2000 (Thermo Scientific). RNA was stored at −80 °C. Primers (supplemental Table S1) were designed and optimized using Primer3Plus (43). One hundred nanograms of RNA was used as a template, mixed with 250 nm of forward and reverse primers, and made up to 20 μl with the constituents of the Kapa SYBR® FAST One-Step kit (Roche, Basel, Switzerland). Reactions were combined on ice in a MicroAmp™ Optical 96-well reaction plate (Applied Biosystems, Foster City, CA) and placed in a LightCycler® 480 (Roche) for thermal cycling and quantification. One-step qPCR was performed as per the manufacturer's instructions, with the following modifications: reverse transcription was increased to 7 min, and extension was increased to 5 s. All reactions were performed in technical duplicate. Quantification of gene expression was performed using the 2ΔΔCT method, on a total of n = 6 biological replicates with 16S used as the housekeeper gene.
Phenotypic Assays
Polymyxin B sensitivity was determined using Polymyxin B Etest® strips (Biomérieux, France). 1 ml of cultures grown as described above were spread on MH agar plates and an Etest® strip imbedded into the center. Plates were incubated for 48 h at 37 °C under microaerophillic conditions. Motility was assessed as per (29) using semi-solid MH medium with 0.4% agar. Plates were inoculated with 1 μl of an overnight biphasic culture (OD600 ∼0.5–0.6). Plates were incubated for 24 h at 37 °C under microaerophilic conditions and motility measured by diameter of bacterial spread.
Human Epithelial Caco-2 Cell Adherence and Invasion
Caco-2 cells were grown in 24 well plates for 24 h in DMEM + 10% fetal calf serum (FCS) at 37 °C with 10% CO2. Bacteria were grown on MH plates with 25 μg/ml trimethoprim and appropriate antibiotics for selection at 37 °C for 36 h under microaerophilic conditions. Bacteria were scraped from the plates and collected by centrifugation (3000 × g for 10 min) and added to the Caco-2 cells at a multiplicity of infection (MOI) of 100. After 4 h incubation (37 °C, 10% CO2), extracellular bacteria were removed by two rounds of washing in PBS. For adherence and invasion, cells were lysed with 0.1% Triton X-100 in PBS (15 min, 20 °C) and serial dilutions of the lysates plated on MH agar with selection antibiotics. CFU were enumerated after 48 h under microaerophilic conditions. To determine the proportion of invaded bacteria, cells were washed and then incubated in DMEM with 250 μg/ml gentamycin for 2 h before lysis and bacterial enumeration. Data provided are from a minimum of n = 6 replicates.
Stress Assays
Strains were cultured as above. For temperature stress, cells were collected and resuspended in fresh medium at an OD600 of 1.0 in 1 ml aliquots (n = 5 replicates) before exposure to 4 °C, 20 °C, 30 °C, 37 °C, 42 °C, and 46 °C for 30 mins; for oxidative stress, exposure to 5 mm H2O2 for 30 mins. For osmotic stress, cells were collected as above and adjusted to fresh MH medium supplemented with 150 mm NaCl and incubated for 2 h at 37 °C. Cultures were serially diluted and spread onto MH agar plates, which were incubated for 48 h at 37 °C under microaerophillic conditions before CFU enumeration.
Biofilm Formation
Overnight cultures were diluted to an OD600 of 0.1 in 24 well, flat-bottom cell culture plates containing fresh MH medium or 100% chicken exudate (44). Plates were incubated for 48 h under microaerophilic conditions with/without shaking at 120 rpm. Nonadherent, nonbiofilm cells were removed, and wells washed twice with 1 ml of sterile PBS. 1.2 ml of fresh MH broth supplemented with 0.01% (w/v) 2,3,5-triphenyltetrazolium chloride (TTC) was added to each well before further incubation at 37 °C in micraerophilic conditions for 72 h. TTC solution was removed and the wells air dried. Bound dye was dissolved using 20% acetone/80% ethanol and the A500 of the solution measured.
Chemotaxis Assays
Tube-based chemotaxis assays were performed as per (45) with minor modification. Briefly, cells were reconstituted in 250 μl 0.4% (w/v) PBS-agar to an OD600 of 1 and transferred to the bottom of a 2 ml tube and allowed to set. Plugs were overlaid with an additional 750 μl 0.4% (w/v) PBS-agar, which was also allowed to solidify. A sterile Whatman paper disk soaked in a saturated solution of each “chemoattractant” was laid on the surface. Tubes were incubated for 72 h at 37 °C under microaerophilic conditions, following which 200 μl 0.01% TTC in PBS was added. Following an additional incubation overnight, chemotactic behavior was assessed by the presence of 1,3,5-triphenylformazide (indicative of cellular respiration) and scored based on the intensity and position of the dye (+++, intense at surface; ++ significant migration; + some diffuse color; −, no reaction).
Targeted Metabolomics by LC-MS/MS
Lyophilized cells (n = 9 samples from 3 biological replicates) were reconstituted in ultrapure water to a concentration of 40 μg/ml and lysed by 6 rounds of 30 s bead beating. Metabolites were extracted by addition of chilled 1.8:2:2 water/methanol/chloroform. Samples were centrifuged for 20 min at 12,000 × g, 4 °C to induce phase separation, and the upper phase removed and dried. Samples were stored at −30 °C until required. LC-MS/MS was performed as per (46). Dried metabolites were reconstituted in ultrapure water and loaded onto either a Luna Phenyl-Hexyl column (50 mm x 1 mm × 5 μm particle size) (Phenomenex, Torrance, CA) or a Synergi Polar-RP (reversed phase), 80 Å column (50 mm × 1 mm × 4 μm particle size) (Phenomenex) using a Nexera UHPLC system (Shimadzu, Kyoto, Japan). For the Luna Phenyl-Hexyl column, samples were loaded in 100% buffer A (0.1% acetic acid) at 100 μl/min for 1 min, following which metabolites were eluted by adjusting the mobile phase composition to 100% buffer B (100% acetonitrile [MeCN]) over a 4.5 min linear gradient. For the Synergi Polar-RP column, samples were loaded in 95% buffer B (95% MeCN) at 100 μl/min for 3 min, following which metabolites were eluted by adjusting the mobile phase composition to 100% buffer A (5 mm ammonium acetate, 5% MeCN) over a 3.9 min linear gradient. Metabolites were eluted into a Q-TRAP 6500 mass spectrometer (SCIEX, Framingham, MA) operated in targeted MRM mode. Metabolites and their MS parameters (parent, product ion m/z [transitions], retention time, collision energy and declustering potential) are shown in supplemental Table S2. Data files were imported into Skyline (v. 4.1.0.18169), where peak areas were manually integrated. Statistical analysis was performed in Metaboanalyst (v.4.0) (47).
Peptide Sample Preparation for Quantitative Proteomics
Lyophilized cells were reconstituted in 100 mm HEPES, pH 7.5 and lysed by 6 rounds of 30 s bead beating. Proteins were precipitated by 1.8:2:2 water/methanol/chloroform. Protein pellets were washed twice with methanol, then solubilized in 8 m guanidine-HCl, 100 mm HEPES, pH 7.6. Free thiols were reduced with 10 mm dithiothreitol (DTT) and then alkylated with 20 mm iodoacetamide (IAA) for 1 h each. Proteins were diluted in 5 volumes of 100 mm triethylammonium bicarbonate (TEAB; pH 7.8) and digested with sequencing grade modified trypsin (Promega, Madison WI), which was added to make a protein/protease ratio of 30:1, overnight at 37 °C. Peptides were desalted by solid phase extraction using hydrophilic-lipophilic balance (HLB) cartridges (Waters, Bedford, MA) according to the manufacturer's instructions. After this step, samples could be lyophilized for subsequent DIA-SWATH-MS analysis (see below). For label-based proteomics, samples were labeled with TMT (Thermo Scientific) according to the manufacturer's instructions. Excess label was quenched with addition of hydroxylamine hydrochloride, following which labeled samples were pooled and desalted using HLB cartridges, as described above.
Peptide Separation by Offline Hydrophilic Interaction Liquid Chromatography (HILIC)
TMT labeled samples and a pooled, unlabeled sample used to generate a SWATH library, were fractionated offline by HILIC before LC-MS/MS. Samples were reconstituted in 90% MeCN, 0.1% trifluoroacetic acid (TFA) and loaded onto a 20 cm × 320 μm column packed in-house with 3 μm TSK-amide 80 HILIC particles (TOSOH Bioscience, King of Prussia, PA) in 100% buffer B (90% MeCN, 0.1% TFA) at 10 μl/min for 10 min using an Agilent 1200 LC system (Agilent Technologies, Santa Clara, CA). Peptides were eluted by adjusting the mobile phase to 70% buffer A (0.1% TFA) over a 40 min linear gradient at 6 μl/min and collected in 1 min fractions. Fractions were pooled where required and lyophilized before LC-MS/MS.
Quantitative Proteomics by LC-MS/MS
TMT labeled peptide fractions were reconstituted in 3% MeCN, 1% formic acid (FA) and loaded onto a 30 cm × 75 μm inner diameter (i.d.) pulled column packed in-house with 1.9 μm ReproSil-Pur 120 C18-AQ material (Dr. Maisch, Ammerbuch, Germany) in 95% buffer A (1% FA) at 350 nL/min using an EasyLC 1200 LC system (Thermo). Peptides were eluted by altering the mobile phase to 35% buffer B (80% MeCN, 1% FA) over a 90 min linear gradient directly into a Q Exactive™ HF Hybrid Quadrupole Mass Spectrometer (Thermo Scientific). The Q Exactive HF was configured to perform one full scan MS experiment (scan range 300 to 1650 m/z, resolution of 60,000, AGC of 3e6 and a maximum IT of 50 msec) with the top 15 precursors in fulfilment of the selection criteria (charge state 2–4, minimum intensity greater than 9.2e4, dynamic exclusion window of 40 s) selected for MS/MS (scan range 200 to 2000 m/z with a fixed first mass of 110 m/z, resolution of 15,000, automatic gain control [AGC] of 1e6, maximum IT of 50 msec, isolation window 1.4 m/z, normalized collision energy [NCE] set as 29).
Processing of Mass Spectrometry Files
Data files from TMT experiments were processed in Proteome Discoverer (v. 2.2) and searched against the UniProt C. jejuni NCTC 11168 database (UP000000799; organism ID 192222; release May 24, 2018 last modification; 1623 proteins (48)) with the SequestHT algorithm. Search parameters were set to enzyme full-trypsin digest with a maximum 2 missed cleavages and static modifications, carbamidomethyl (C); variable modifications, oxidation (M), TMT-6plex (peptide N-term, K), and using precursor and fragment ion tolerances of 20 ppm. Peptide level false discovery rate (FDR) was determined using Percolator (v. 2.08.01). Rank 1 peptide spectral matches (PSMs) corresponding to a 1% FDR were then exported, and reporter intensities normalized internally to total reporter ion signals across all channels. Peptides with ambiguous protein assignments were removed, as were peptides containing known N-glycosylation sites (loss of PglB will bias the quantitative data for that protein given the likely large increase in its non-glycosylated form in the mutant). All peptide identifications are contained within supplemental Data 11168 pgl_Peptide. Reporter signals for remaining PSMs were summed to find total reporter signals for identified proteins. For proteins with a minimum of 2 unique identified peptides, these values were imported into Perseus (v. 1.6.1.1) for statistical analysis. Missing values were imputed based on normal distributions, then summed reporter intensities were averaged with internal technical replicates to yield sample specific reporter intensities. One sample t-tests were performed on relevant log2 transformed reporter ratios using the 3 biological replicates via a Benjamini-Hochberg procedure and using an FDR cut-off = 0.05. A fold change corresponding to a 99% confidence interval of internal wild-type/wild-type comparisons across the 3 biological replicates was used to further narrow the list of proteins found to be significantly deviating from wild-type distributions.
Validation by Targeted DIA-SWATH-MS
Unlabeled digests and peptide fractions were reconstituted in 3% MeCN, 1% FA and loaded onto a 40 cm × 75 μm pulled column packed in-house with 3 μm ReproSil-Pur 120 C18-AQ material maintained at a column temperature of 45 °C at 200 nL/min in 98% buffer A (1% FA) using an Ekspert 425 nanoLC system (SCIEX). Peptides were eluted by altering the mobile phase composition to 28% buffer B (80% MeCN, 1% FA) over a 146 min linear gradient directly into a TripleTOF® 6600 mass spectrometer (SCIEX). For samples used for spectral library generation, the mass spectrometer was configured to perform one full scan MS experiment (scan range 350 to 1500 m/z, accumulation time 50 msec) with a maximum of 80 precursors in fulfilment of the selection criteria (charge state 2–5, intensity greater than 400, dynamic exclusion window of 20 s) selected for MS/MS in high sensitivity mode (scan range 100–1500 m/z, dynamic collision energy [DCE] set as true, mass tolerance of 20 ppm). For DIA-SWATH-MS experiments, the mass spectrometer was configured to perform one full scan MS experiment (scan range 350 to 1500 m/z, accumulation time 50 msec) followed by 80 variable SWATH windows over the mass range 350 to 1500 m/z (accumulation time 25 msec). Window size was determined using the SWATH variable window calculator tool (SCIEX) from summed total ion chromatograms (TIC) of library-generated samples. SWATH library data files were processed in Protein Pilot (v. 5.0) and searched against the UniProt C. jejuni NCTC 11168 proteome (as above) using the Paragon algorithm. Search parameters were sample set as identification; maximum 2 missed cleavages; Cys alkylation, IAA; digestion, trypsin; instrument, TripleTOF 6600; search effort as thorough ID, and using a detected protein threshold of 0.05. The group file was then imported into PeakView (v. 2.2.0.11391) using a total protein number corresponding to a global protein level FDR of 1%. SWATH files were processed against the resulting library using a peptide confidence threshold of 95%, 1% FDR and an extracted ion chromatogram (XIC) width of 20 ppm. Peptides containing known N-glycosites were removed and peptide areas were summed to yield a total protein area that was used for relative quantitation. All heat maps shown for visualization of proteomics data were generated in Morpheus (v. 1; https://software.broadinstitute.org/morpheus).
Reductase Assays
Nitrate and nitrite reductase activities were assayed as described (20) and performed on a minimum of n = 5 replicates. Cells were harvested after 24 h and washed with 25 ml 10 mm Tris-HCl (pH 7.5) and intact cells were resuspended in 1 ml of 10 mm Tris-HCl (pH 7.5). Benzyl viologen-linked assays were carried out in a 1 ml assay volume containing 10 mm Tris-HCl (pH 7.5), 0.1 mm benzyl viologen, 20 mm sodium dithionite and 5 mm electron acceptor (sodium nitrate/sodium nitrite). Cells were added to the buffer-plus-viologen mixture and sparged with nitrogen gas for 10 min. 20 mm sodium dithionite was added to the assay mixture until A578 stabilized. The assay was initiated by addition of an anaerobic solution of 5 mm sodium nitrate/nitrite. After 5 min, the reaction was stopped and the A578 determined for each sample and displayed as a function of total protein as determined by QubitTM Protein Assay Kit (Invitrogen, Carlsbad, CA).
RESULTS
Proteomics Reveals Multiple Alterations Associated with Loss of Protein N-glycosylation
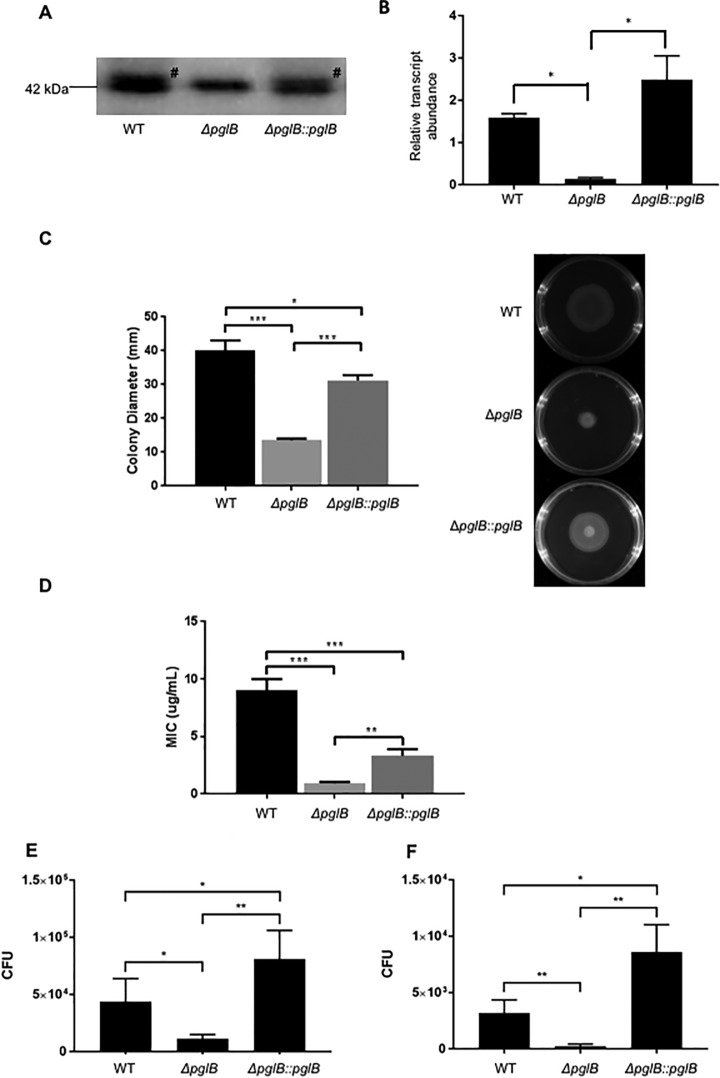
To better understand the role of N-glycosylation in C. jejuni, we compared wild-type (WT) strain NCTC 11168 with a pglB deletion mutant (ΔpglB) and a pglB restored mutant (ΔpglB::pglB). We first confirmed that pglB deletion resulted in the loss of N-glycosylation by probing C. jejuni proteins with anti-JlpA anti-serum. JlpA is a singly glycosylated lipoprotein in C. jejuni NCTC 11168, although it is doubly glycosylated in other strains (36). Deletion of pglB resulted in loss of the JlpA glycoform, which was restored by complementation (Fig. 1A). Quantitative PCR (qPCR) further confirmed that expression of pglB was lost in ΔpglB and restored levels were not significantly different to those observed for WT (Fig. 1B). We also confirmed that loss of pglB resulted in a motility defect (Fig. 1C) and attenuation of resistance to the antimicrobial peptide polymyxin B (Fig. 1D), as previously described (29). Finally, we observed a significant reduction in C. jejuni NCTC 11168 human epithelial Caco-2 cell adherence and invasion in ΔpglB that was restored in ΔpglB::pglB (Fig. 1E–1F).
Fig. 1.
N-glycosylation influences multiple C. jejuni phenotypes. A, Western blotting using anti-JlpA serum showing deletion of pglB results in loss of N-glycosylated JlpA. # indicates position of glycosylated band (addition of 1406 Da); B, qPCR confirms deletion (ΔpglB) and restoration (ΔpglB::pglB) of pglB gene expression (* p < 0.05); C, Decreased motility associated with pglB deletion; (left) colony diameter in mm taken from (right) migration on semi-solid agar (*** p < 0.001, * p < 0.05); D, Polymyxin B resistance is attenuated in ΔpglB (*** p < 0.001; ** p < 0.01); E, Adherence to and F, invasion of Caco-2 cells is reduced in C. jejuni ΔpglB (** p < 0.005; * p < 0.05).
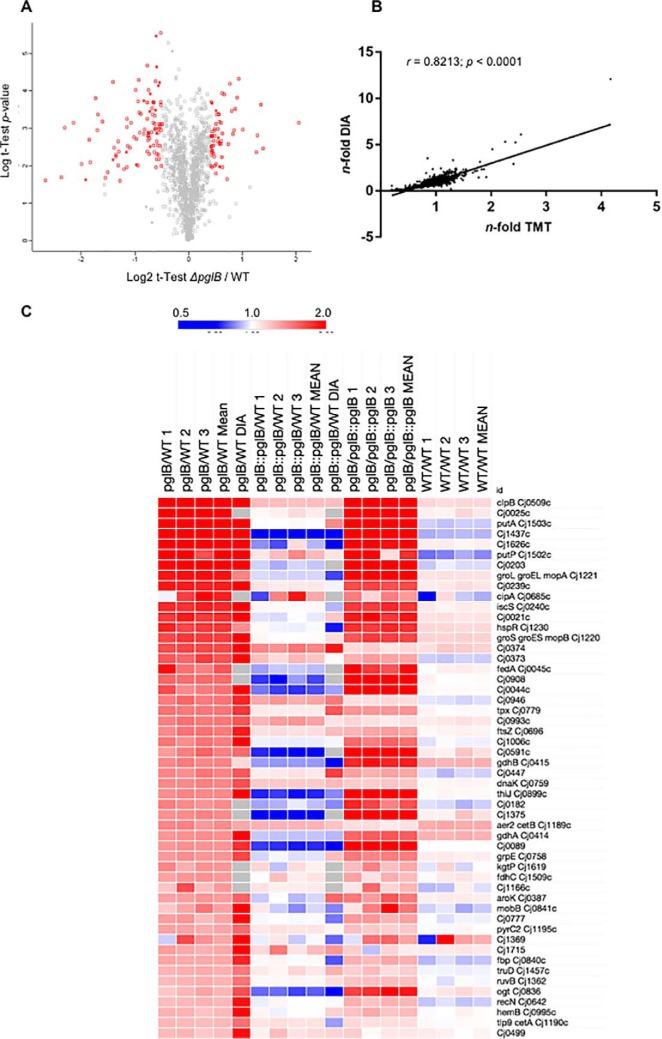
To better understand molecular phenotypes associated with N-linked glycosylation in C. jejuni, we conducted label-based (TMT labeling) quantitative proteomics of WT, ΔpglB and ΔpglB::pglB post-trypsin digest and offline HILIC fractionation followed by RPLC-MS/MS. This approach enabled the identification of 1359 proteins (≥ 2 peptides per protein at an FDR of 1%) that were quantified in all of 3 biological replicates and that collectively represent ∼84% of the predicted C. jejuni NCTC 11168 proteome (total 1623 predicted genes/proteins; Fig. 2A and supplemental Data S1). We also analyzed additional biological replicates using DIA-SWATH-MS to act as a validation cohort on our quantitative discovery proteomics data set. DIA-SWATH-MS enabled the relative quantitation of 1080 proteins (∼67% of the C. jejuni NCTC 11168 predicted proteome; supplemental Data S2). Both the ΔpglB versus WT and ΔpglB::pglB versus WT DIA-SWATH-MS data sets correlated well with the original label-based TMT data (Pearson correlation r = 0.8213 and r = 0.7232, respectively [both at p < 0.0001]; Fig. 2B and supplemental Fig. S1). For both data sets, Log2 data were converted to n-fold change for subsequent data handling. We defined significant fold change for further analysis as those proteins demonstrating a >± 1.4-fold change between ΔpglB and WT in the label-based data (with additional included proteins being those with a >± 1.3-fold change and a >± 1.5-fold change in the DIA-SWATH-MS data set; all at p < 0.05) and that were significantly returned toward their WT abundance in the complemented ΔpglB::pglB strain. Any proteins showing a conflicting abundance change (defined here as significantly up- or downregulated in the label-based or DIA-SWATH-MS data sets, and with the opposite significant change in the other data set) were not considered for further analysis. We also assessed the biological variance across replicates by determining the relative abundance of all 1359 proteins between 3 WT proteome replicates. This analysis found only 8 proteins with significantly altered mean relative abundance representing a false quantitation rate of ∼0.006% across the data set (supplemental Data S2). The complete quantitative proteome data sets are shown in heat map format in supplemental Fig. S2. Relative quantitation highlighted 185 proteins at significantly altered abundance in ΔpglB compared with WT (64 at elevated abundance and 121 at reduced abundance); however, of these 48 were not restored significantly to WT levels in ΔpglB::pglB. Removal of these from the final data set left 137 significantly altered proteins on deletion of pglB relative to the WT, of which 51 were present at increased abundance and 86 were present at reduced abundance (Fig. 2C–2D). DIA-SWATH-MS validated 111 of these changes (81.0% of the total changing data set; 42 and 69 proteins for the increased and decreased data sets, respectively) and there were no proteins showing a conflicting abundance change across the two technical approaches. We next took the complete cohort of proteins that were identified as either elevated or reduced in abundance in ΔpglB and restored significantly toward WT levels by complementation of pglB and employed STRING db to identify functional relationships.
Fig. 2.
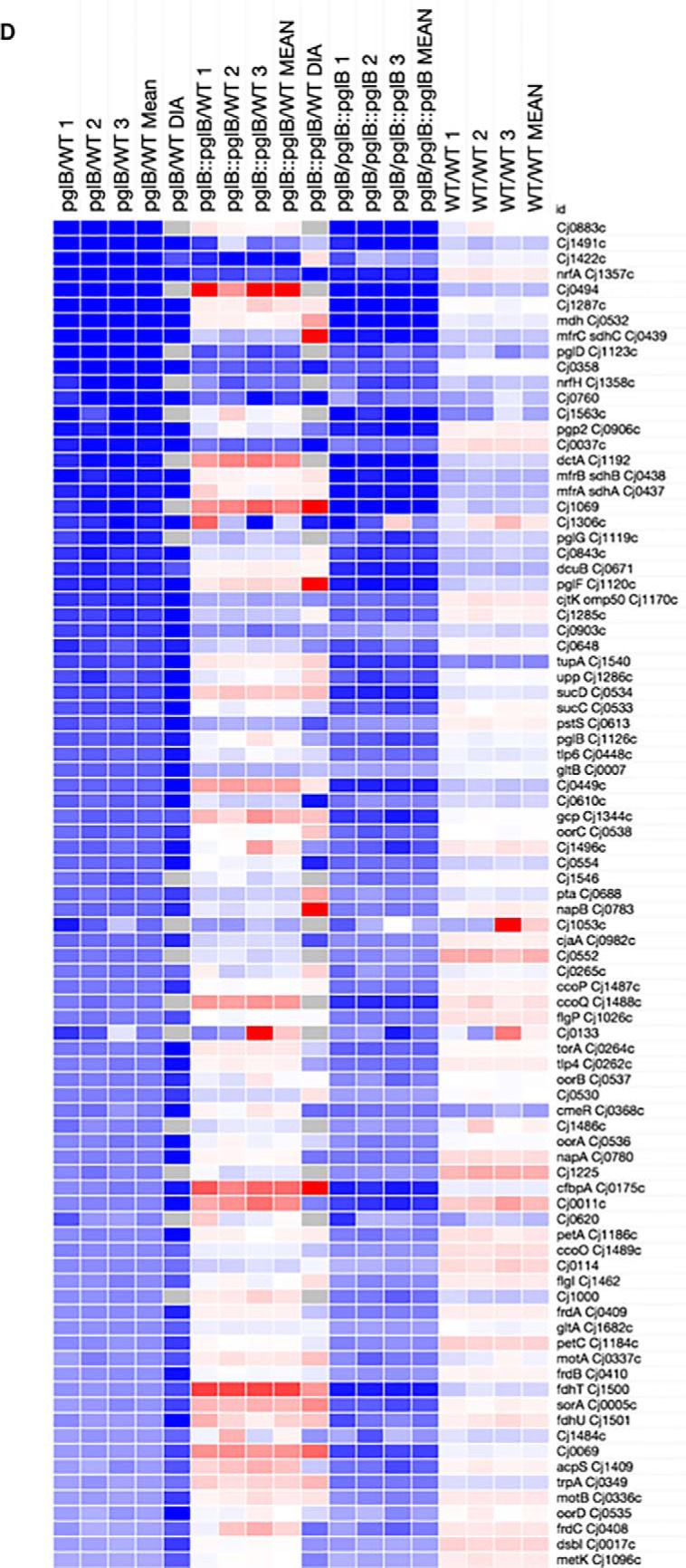
Differentially abundant proteins in C. jejuni NCTC 11168 ΔpglB compared with WT identified by quantitative proteomics. A, Volcano plot for ΔpglB compared with WT; x axis represents averaged Log2(ΔpglB/WT), y axis represents log10(p value). Significantly differentially abundant proteins are highlighted in red (p < 0.05); B, Correlation plot based on n-fold changes observed in the discovery set (n-fold TMT) and validation set (n-fold DIA) between ΔpglB and WT, Pearson correlation r = 0.8213 at a p < 0.0001 determined from 1067 aligned proteins. Heat maps showing proteins present at significantly increased (C) and decreased (D) abundance in ΔpglB compared with WT. Data from each label-based replicate and comparison, and mean n-fold change, were mapped for all comparisons. For inclusion, regulated proteins needed to return toward WT levels in ΔpglB::pglB and be significantly different between ΔpglB and ΔpglB::pglB. DIA-SWATH-MS validation is shown (“DIA”) and no proteins with an “opposite” change between discovery and validation sets was included. Values are gray where the protein was not identified by DIA-SWATH-MS.
Stress-related Proteins Are Significantly Induced in the Absence of Protein N-glycosylation
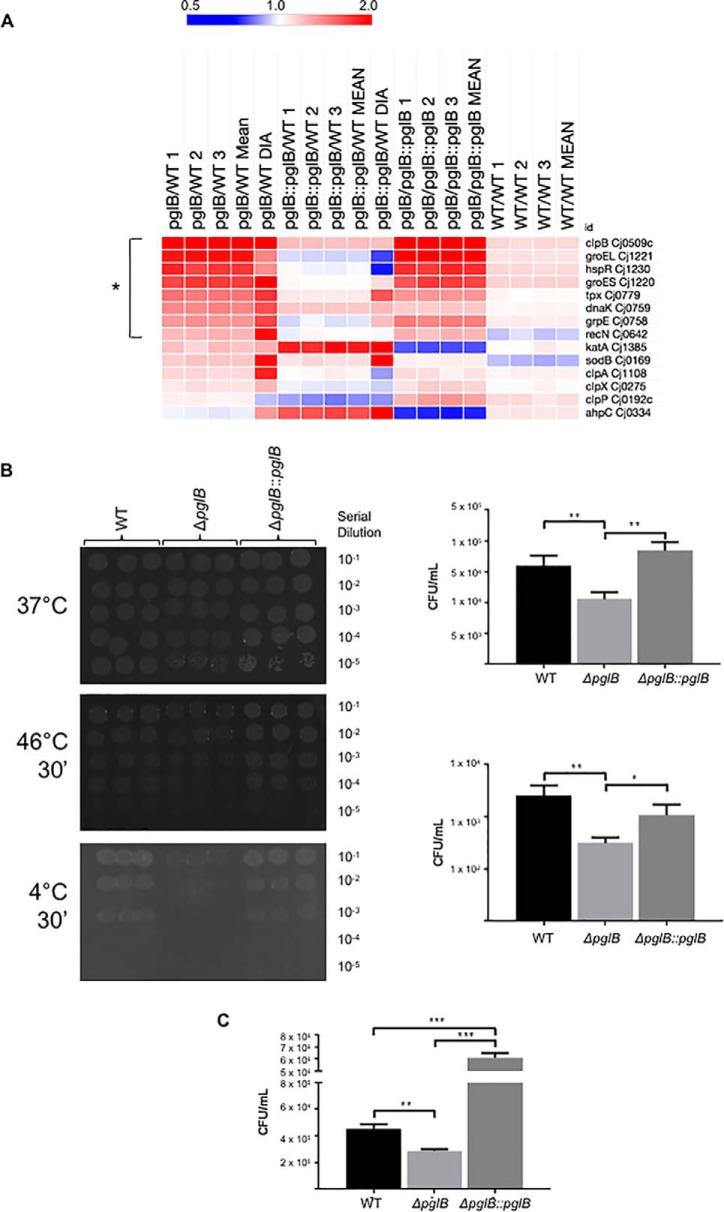
The proteins with significantly increased abundance in ΔpglB (Fig. 2C) were clustered with STRING db (supplemental Fig. S3A). The major induced functional group in the ΔpglB mutant was associated with cell stress (Fig. 3A). These proteins included ClpB (mean 4.2-fold induction in the label-based data, and 12.1-fold in DIA-SWATH-MS), GroEL (1.92- and 1.49-fold), GroES (1.75- and 2.20-fold), DnaK (1.48- and 1.69-fold), GrpE (1.42- and 1.72-fold) and the heat shock transcriptional regulator HspR (1.78- and 1.45-fold), indicating that loss of N-glycosylation may lead to stress effects and/or protein unfolding (Fig. 3A). We also observed increased abundance of the thiol peroxidase Tpx (1.54- and 1.77-fold). We next examined the complete data set to determine the response of other stress related proteins, particularly those involved in antioxidant defense. We observed non-significantly elevated levels of catalase (KatA) and superoxide dismutase (SodB) in ΔpglB, however restoration of pglB lead to a further, significant induction of KatA and increased abundance of AhpC, while not altering the ΔpglB-associated abundance of SodB (Fig. 3A). We next examined whether loss of pglB results in changes to cell stress resistance. We tested pgl-negative C. jejuni for their ability to recover from temperature (30 min at 4 °C or 46 °C followed by overnight growth at 37 °C), osmotic (2 h in medium supplemented with 150 mm NaCl) and hydrogen peroxide (30 min in medium supplemented with 5 mm H2O2) shock. We observed no differences in recovery from 30 min temperature shock at 20 °C–44 °C (data not shown), however ΔpglB C. jejuni recovered very poorly after temperature shocks at 4 °C and 46 °C compared with WT and ΔpglB::pglB (Fig. 3B). Consistent with previous work showing growth defects of glycosylation negative C. jejuni under increased osmotic stress (NaCl and sucrose) that may be related to fOS (32), we also observed poor recovery of ΔpglB following salt shock (Fig. 3C). Surprisingly, we found both ΔpglB and ΔpglB::pglB were enhanced for H2O2 stress resistance relative to WT, with the pglB complemented strain also showing significantly higher recovery than ΔpglB (supplemental Fig. S4). These results are however consistent with our proteomics data for antioxidant proteins (Fig. 3A) in ΔpglB::pglB. Biofilm formation is a mechanism C. jejuni may employ for protection against environmental stress and has previously been associated with motility and chemotaxis. We tested these C. jejuni strains using 4 models of biofilm growth; in MH medium (with and without shaking) and in 100% chicken exudate (“juice” (44); with and without shaking). In both growth models, ΔpglB demonstrated significantly enhanced biofilm formation in the absence of shaking (supplemental Fig. S5). In MH medium with shaking however, both ΔpglB and ΔpglB::pglB were almost completely devoid of biofilm-forming ability. Adding exudate restored both mutants to WT levels, with no significant difference between any strains (supplemental Fig. S5). Collectively, these data indicate that glycosylation negative C. jejuni display an active stress response during basal growth that likely inhibits their ability to further induce stress-related proteins on sudden imposition of environmental stress.
Fig. 3.
C. jejuni ΔpglB is associated with increased levels of stress-related proteins and is attenuated for stress resistance. A, Heat map of differentially regulated (bracketed *) and other common non-regulated stress-related proteins; B, Survival of ΔpglB is compromised by temperature shocks at 4 °C and 46 °C; (left) plate-based enumeration of bacteria following shock at (top) 37 °C (no change), (middle) 46 °C, and (bottom) 4 °C; (right) CFU/ml plotted from plate assays for (top) 46 °C and (bottom) 4 °C (** p < 0.01, * p < 0.05); C, Survival of ΔpglB is compromised by osmotic shock in 150 mm NaCl (*** p < 0.001, ** p < 0.01).
Proteins Involved in Nutrient Transport and Chemosensing Are Altered in N-glycosylation Negative C. jejuni
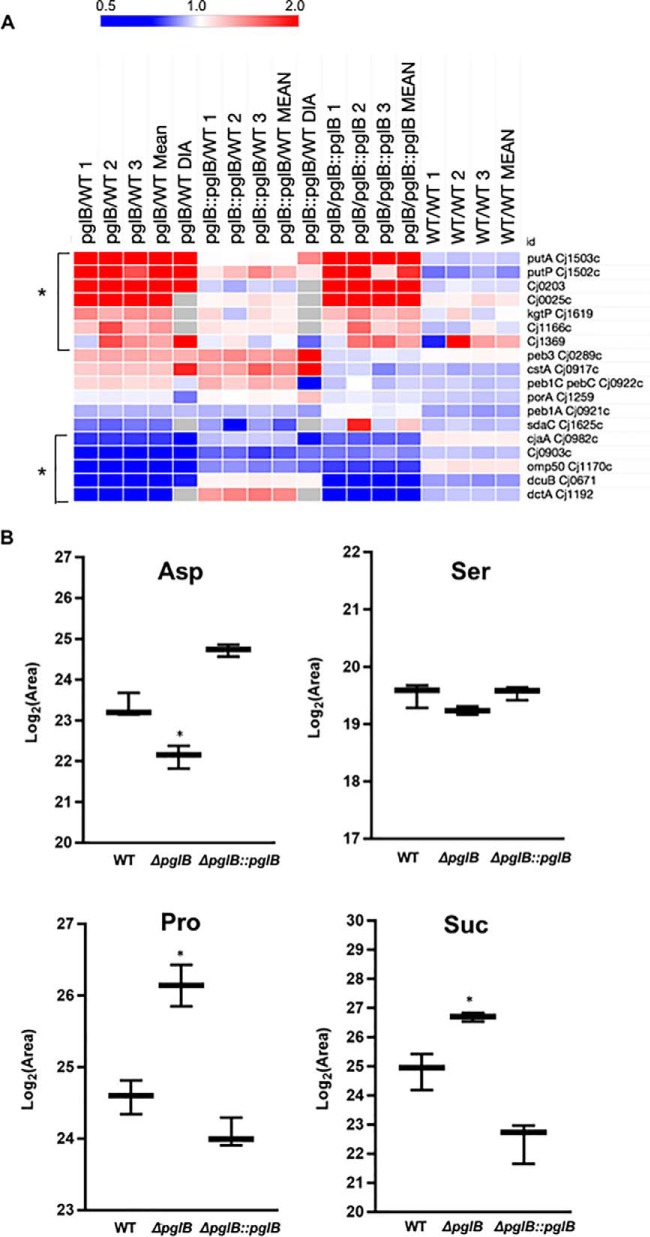
Cluster analysis of the up- and downregulated protein sets showed that proteins involved in nutrient acquisition were differentially abundant in the pglB negative background (Fig. 4A and supplemental Fig. S3A–S3B). We observed significantly increased abundances of the Pro uptake and utilization proteins PutP (2.26- and 5.32-fold in label-based and DIA-SWATH-MS proteomics, respectively) and PutA (2.54- and 6.09-fold), the putative citrate transporter Cj0203 (2.02- and 4.51-fold), a putative C4-dicarboxylate transporter (Cj0025; 2.64-fold), the α-KG permease KgtP (1.41-fold), a putative integral membrane Ser/Thr exporter (Cj1166c; 1.40-fold) and a putative permease (Cj1369; 1.35- and 2.10-fold). There were also concurrent significant decreases in the low oxygen Asp, fumarate and succinate transport proteins, DctA (0.39-fold) and DcuB (0.45- and 0.56-fold), a putative Na+/Ala symporter (Cj0903c; 0.51- and 0.27-fold) and CjaA (Cj0982c; a putative amino acid transport protein that can bind Cys). Other characterized amino acid transporters were not restored to WT levels by reintroduction of pglB or were not significantly altered in abundance (e.g. serine transporter SdaC [Cj1625c], Peb3, Peb1C and Peb1A; Fig. 4A). To determine whether these proteome alterations were reflected at the phenotypic level, we employed targeted metabolomics using LC-MS/MS to assay the intracellular levels of known C. jejuni carbon sources, as well as metabolism-related substrates and products (supplemental Data S3). Metabolomics confirmed that ΔpglB mutants were significantly depleted in intracellular Asp (0.43-fold of WT, p < 0.05; Fig. 4B) and enriched in Pro (2.95-fold higher in ΔpglB, p < 0.05; Fig. 4B) compared with WT C. jejuni. Restoration of pglB returned both Asp and Pro to WT levels. Pro is additionally converted to either Glu or 1-pyrroline-5-carboxylate by PutA, and a small increase in intracellular Glu (1.39-fold, n.s.; supplemental Fig. S6A) was observed in ΔpglB. Intracellular Ala was decreased in ΔpglB (0.55-fold compared with WT) consistent with protein abundance and the proposed but not proven function of Cj0903c (supplemental Fig. S6A). We observed no significant change in Ser (Fig. 4B), which is generally considered the preferred substrate after Asp in C. jejuni. All other amino acids were not significantly altered (supplemental Fig. S6A).
Fig. 4.
C. jejuni ΔpglB is associated with changes in nutrient acquisition proteins that correlate with abundance of intracellular nutrients. A, Heat map of differentially regulated (bracketed *) and other known nutrient transporters and utilization proteins; B, Intracellular abundance of Asp, Ser, Pro and succinate (Suc) (* p < 0.05). The complete data set of 34 metabolites is shown in supplemental Fig. S6.
We also assayed organic acids that can be imported by C. jejuni. We observed significantly elevated intracellular levels of malate (2.57-fold relative to WT; supplemental Fig. S6B) in ΔpglB. This was consistent with reduced abundances of both malate oxidoreductase (Cj1287c; 0.28- and 0.06-fold of WT in ΔpglB as determined by label-based quantitative proteomics and DIA-SWATH-MS, respectively) and malate dehydrogenase (Mdh; 0.29- and 0.22-fold of WT) (Fig. 2D). Intracellular levels of citrate and pyruvate were slightly elevated (1.38- and 1.66-fold, respectively) but not above the chosen significant fold-change threshold, whereas lactate reached this threshold (1.92-fold increase in ΔpglB compared with WT) but with p > 0.05. Significant changes were observed in intracellular fumarate (2.25-fold; supplemental Fig. S6B) and succinate (3.39-fold; Fig. 4B), and these were consistent with reduced levels of DctA and DcuB, as well as major abundance differences observed for C. jejuni respiration/electron transport proteins, including methylmenaquinol:fumarate reductase (MfrABC; formerly annotated as “succinate dehydrogenase,” SdhABC; (49–50)), fumarate reductase (FrdABC; with both fumarate reductase and succinate dehydrogenase activity (51)) and succinyl-CoA ligase (SucCD) (Fig. 2D).
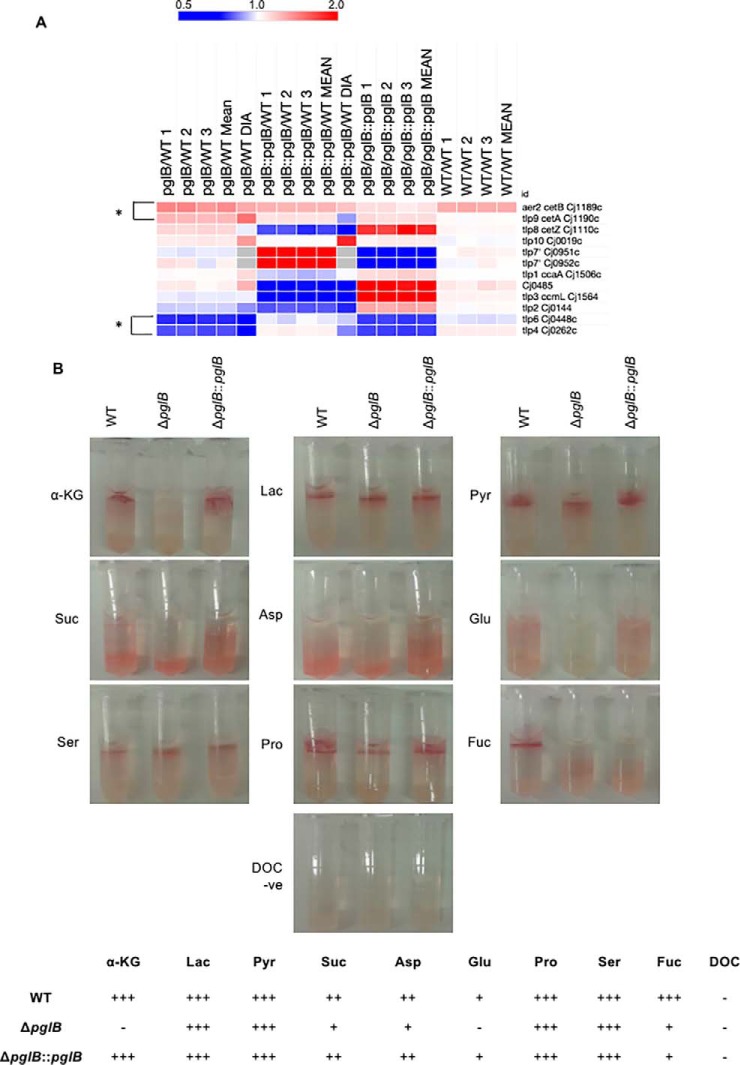
Given the above results, we next wished to determine whether loss of N-linked glycosylation influenced chemotaxis toward C. jejuni substrates. Our proteomics data reflected significant abundance changes in 4 Tlp chemoreceptor-like proteins (Fig. 5A). CetA (Tlp9) and CetB were increased in abundance (1.28- and 1.56-fold; and 1.46- and 1.34-fold, respectively), whereas Tlp4 and Tlp6 were decreased in abundance (0.65- and 0.38-fold; and 0.59- and 0.39-fold, respectively) in ΔpglB compared with WT. The remainder of the Tlp family were not significantly altered, however, Tlp3/CcmL and Tlp7 (Cj0951c/Cj0952c) were significantly up- and downregulated, respectively, in ΔpglB compared with ΔpglB::pglB, indicating they were significantly altered in the complemented strain alone relative to WT (Fig. 5A). We also included Cj0485 as this protein has recently been shown to mediate chemoattraction toward l-fucose (45). Like Tlp3, Cj0485 was significantly more abundant in ΔpglB than in ΔpglB::pglB, mainly as a result of significantly reduced abundance in the complemented strain relative to WT. We next conducted chemotaxis assays on C. jejuni substrates using a modified disk-based/TTC staining method (45). Deletion of pglB did not influence chemotactic response to lactate, pyruvate, Pro or Ser, however we observed a complete loss of chemotaxis to α-KG and Glu, as well as reduced migration toward Asp and succinate (Fig. 5B). A similar reduced chemotactic response was observed for fucose, however, unlike the other chemoattractants, chemotactic response to fucose was not restored in ΔpglB::pglB. The known chemoattractants for C. jejuni Tlps are provided in supplemental Fig. S7. Chemoattractants for Tlp4 are not well characterized, although Tlp6 is known to facilitate migration toward several substrates including Asp, Glu, succinate and TCA cycle intermediates (52).
Fig. 5.
C. jejuni ΔpglB is associated with changes in transducer-like proteins (Tlps) that correlate with altered chemotaxis toward C. jejuni chemoattractants. A, Heat map of differentially regulated (bracketed *) and all other known C. jejuni NCTC11168 Tlps; B, Representative chemoattraction assays for 9 substrates and sodium deoxycholate (DOC, negative control; n = 6 replicates) (upper); and scoring for each chemoattractant (lower) across the strains.
The pgl Pathway and Known Glycoproteins Are Influenced by Loss of pglB
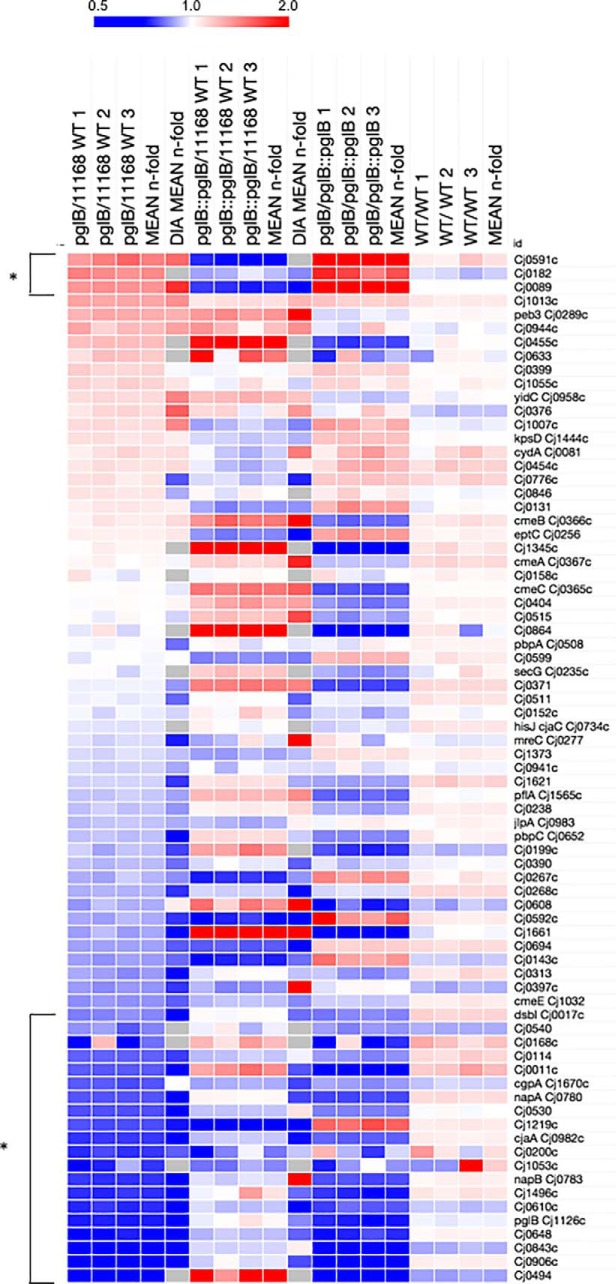
Our data have shown that N-linked glycosylation is required for multiple C. jejuni phenotypes. We now examined how the pgl cluster and known glycoprotein abundances were influenced by removal of glycosylation. First, we observed that deletion of pglB leads to commensurate decreased abundance of PglD (0.31-fold), PglG (0.44-fold) and PglF (0.45- and 0.26-fold [label-based and DIA-SWATH-MS, respectively]) (Fig. 2D). PglA (0.30- and 0.10-fold) and PglC (0.51- and 0.29-fold) were also reduced in abundance but not sufficiently restored by reintroduction of pglB to be included in the final data set. PglIHJK were not altered in the pglB background. It is well-known that PglF (the first step in the biosynthesis of Bac) is the rate-limiting step for the Pgl system (26) and here was the only Pgl protein completely restored to WT levels in ΔpglB::pglB. Western blotting with anti-JlpA anti-sera confirmed that glycosylation was fully restored in the pglB complemented strain (Fig. 1A), despite some pathway members remaining at lower abundance than observed in WT. We next examined the set of proteins with altered abundance to determine whether known N-glycoproteins were enriched. There are currently 81 known N-glycoproteins and these comprise 5.0% of the complete predicted C. jejuni proteome (n = 1623). In these experiments, we identified 75 of these proteins from 1359 in the quantitative set (5.5%; Fig. 6). Three glycoproteins were contained within the set of 51 upregulated proteins in ΔpglB (5.9%; Cj0591c, Cj0182 and Cj0089) and finally, 12 glycoproteins were within the significantly downregulated protein set in this background (14.0%), confirming a 2.5-fold enrichment against the total proteome. Further, an additional 8 proteins were significantly downregulated but were only partially restored in ΔpglB::pglB and therefore did not reach significance, despite some attenuation of their ΔpglB levels. Examination of the response of the entire glycoprotein data set (Fig. 6) showed the bulk of these proteins were downregulated, outside of the n-fold threshold applied. The vast majority of C. jejuni glycoproteins are functionally unknown and hence a specific test set was difficult to obtain. We observed no change to the tripartite antibiotic efflux system CmeABC (all 3 members are glycosylated) despite reduced resistance to the antimicrobial peptide polymyxin B (Fig. 1D), nor any effects on JlpA (confirmed by Western blotting; Fig. 1A), Peb3, the EptC pEtN transferase or CjaC glycoproteins (Fig. 6).
Fig. 6.
C. jejuni ΔpglB is associated with decreased abundances of known glycoproteins. Heat map of all identified and previously observed glycoproteins (n = 75 glycoproteins observed of 81 known); (bracketed *) present at significantly altered abundance.
N-glycosylation Is Required for Optimal Function of C. jejuni Nitrate Reductase (NapA/NapB)
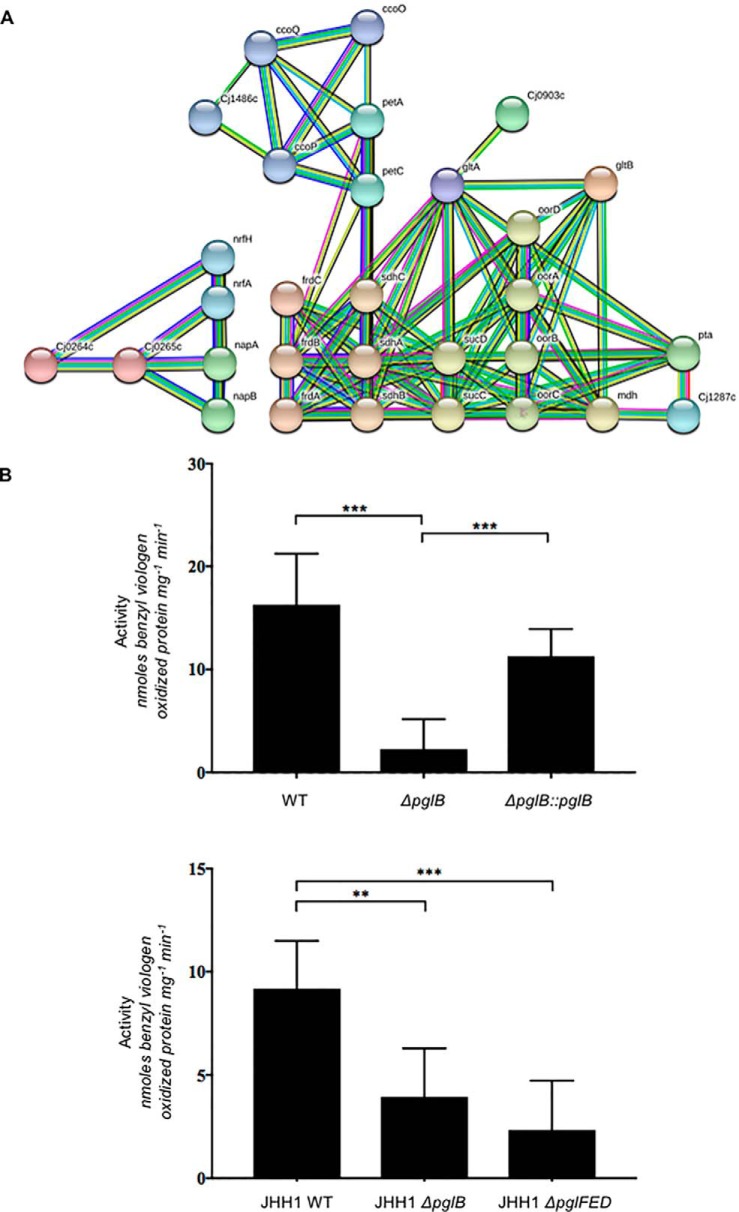
The major functional group associated with the downregulated protein set was respiration and the electron transport chain, particularly those reductases required as alternate electron acceptors under low oxygen conditions, as all subunits of the nitrate (NapA/NapB), nitrite (NrfA/NrfH) and TMAO/DMSO (Cj0264c/Cj0265c) reductases, as well as FrdABC, were significantly downregulated in glycosylation negative C. jejuni (Fig. 2D and Fig. 7A). Importantly, of these proteins, both NapA and NapB, which constitute the nitrate reductase complex, are the only glycoproteins of known function in the downregulated glycoprotein data set (Fig. 6). We therefore tested C. jejuni NCTC 11168 WT, ΔpglB and ΔpglB::pglB for their nitrate reductase activity using the benzyl viologen reductase assay with 5 mm sodium nitrate as the electron acceptor. ΔpglB showed an ∼7-fold reduction in activity, which was restored to near WT levels in ΔpglB::pglB (Fig. 7B). Addition of 20 mm sodium nitrate to growth medium also reduced the growth of ΔpglB at the end of exponential phase compared with WT and ΔpglB::pglB (supplemental Fig. S8A). Given that we also observed significantly reduced abundances of NrfA (0.23- and 0.05-fold of WT in ΔpglB)/NrfH (0.35-fold, label-based proteomics only) that comprise the nitrite reductase, we performed the above reductase assay with 5 mm sodium nitrite. We observed no change in enzyme activity for NrfA (supplemental Fig. S8B). To determine whether decreased nitrate reductase function was the result of effects on gene expression, we performed qPCR on the napA and napB genes. These data show that napA and napB expression are unaltered in ΔpglB compared with WT (supplemental Fig. S8C). We finally wished to confirm that the NapA/NapB phenotype was not specific to C. jejuni NCTC 11168, and second, that it was not specific to pglB oligosaccharyltransferase deletions. We therefore performed nitrate reductase assays on the clinical JHH1 strain (53) in which we deleted both the pglB gene (JHH1ΔpglB) and the first 3 genes in the pgl pathway to make a biosynthetic mutant (JHH1ΔpglFED). Although JHH1 WT showed slightly lower activity than NCTC 11168, JHH1ΔpglB was still significantly attenuated compared with this WT (∼2.5-fold lower). Additionally, JHH1ΔpglFED also showed a significant loss of nitrate reductase activity (∼3.5-fold lower compared with JHH1 WT). These data confirm that N-glycosylation is required for wild-type nitrate reductase activity in C. jejuni, consistent with the reduction in abundance of the NapA/NapB proteins as detected by quantitative proteomics. This reduced protein abundance is independent of gene expression.
Fig. 7.
C. jejuni ΔpglB is associated with decreased abundances of electron transport chain proteins, particularly electron accepting reductases. A, STRING db cluster analysis of proteins present at reduced abundance shows connectivity between proteins, including alternative electron acceptors used under low oxygen conditions (NapA/NapB [nitrate reductase], NrfA/NrfH [nitrite reductase], Cj0264c/Cj0265c [TMAO/DMSO reductase]); B, Nitrate reductase assay (upper) C. jejuni NCTC 11168 WT, ΔpglB, ΔpglB::pglB; (lower) C. jejuni JHH1 WT, JHH1ΔpglB, JHH1ΔpglFED (*** p < 0.001, * p < 0.01).
DISCUSSION
Protein N-glycosylation is required for C. jejuni chicken colonization and human gastrointestinal epithelial cell adherence and invasion. Therefore, as one of very few confirmed virulence determinants, along with motility and chemotaxis, the Pgl system represents a potential anti-microbial target (54). Better understanding of the phenotypes associated with N-glycosylation is crucial in determining the contribution of this pathway to pathogenesis. Our approach was to employ quantitative proteomics to direct subsequent phenotypic investigations. To ensure the data were robust, only proteins quantified in 3 biological replicates (label-based quantitative proteomics acting as a “test” discovery set) were included in our ensuing analyses, and we also included a “validation” cohort (DIA-SWATH-MS) that enabled independent alignment of 1080/1359 (79.5%) proteins from additional biological replicates. Cluster-based analysis of the up- and downregulated proteins in a pglB oligosaccharyltransferase-deficient C. jejuni mutant (with protein levels returned significantly toward their WT levels in a pglB-restored mutant, ΔpglB::pglB) indicated 5 major functional groups associated with the Pgl system; stress response (elevated), nutrient acquisition and utilization (members in both elevated and diminished groups), known N-glycoproteins (predominantly diminished), nutrient sensing (members in both groups), and most significantly, respiration (all diminished; n = 34/86 [39.5% of the downregulated protein set]).
Nitrate reductase (Nap) is the first example of an enzyme function that relies on N-glycosylation in this organism, and potentially one of the first examples across all forms of life. The loss of Nap activity associated with deletion of glycosylation raises the question of how the modification could influence function. Expression of the C. jejuni 90 kDa catalytic subunit napA in E. coli produces functional enzyme (55), suggesting that glycosylation of NapA is not required for activity outside of C. jejuni and that complex formation with NapB is not an absolute requirement for function. Indeed, previous studies have shown that amino acids essential for salt bridge formation between NapA/NapB, particularly Glu76, are not present in either C. jejuni or E. coli NapA (56). Hence, NapA/NapB have been shown to interact only weakly in E. coli (56) and the interaction is not essential for catalysis. NapA is a molybdoprotein that requires molybdenum (Mo) from the co-factor, Mo guanosine dinucleoside, which itself is synthesized by MobA/MobB in C. jejuni. MobB was significantly upregulated in ΔpglB (1.37- and 2.29-fold for label-based and DIA-SWATH-MS, respectively), suggesting enough Mo is present for cofactor biosynthesis. Mo transport is encoded by the ModABC transport system, which is required for NapA activity and repressed when enough Mo is present (57). Both ModA and ModC were decreased in abundance in ΔpglB, however these changes were not restored by pglB complementation, further confirming Mo availability does not generate the ΔpglB Nap phenotype.
We also speculated that loss of Nap activity might be associated with a change in the menaquinone (MK) pool that acts as the sole electron carrier in the C. jejuni periplasm. MK is synthesized by an atypical futalosine pathway (58) encoded by at least 4 genes (mqnACD and Cj0117), of which only MqnA (Cj1285c) was present at reduced abundance in ΔpglB. Our data show a decrease of specific electron donors to the MK pool, most notably MfrABC (formerly designated SdhABC (49–50)), whereas the nuo operon-encoded NADH dehydrogenase (Cj1566–1579) and formate dehydrogenase (Cj1508–1511) remained unaltered (supplemental Data S2). These results suggest both enough MK and electrons are available for respiration. Despite this, almost all C. jejuni electron acceptors, including those that are oxygen-dependent (predominantly a cb-type cytochrome c oxidase [Cj1487–1490; ccoNOQP] which acts via a proton-translocating cytochrome bc complex [encoded by petABC] and a periplasmic c-type cytochrome [Cj1153]), fumarate reductase FrdABC (which also acts as a succinate dehydrogenase electron donor (51)) and low oxygen alternative electron acceptors (Nap, Nfr, and Cj0264c/Cj0265c) are present at significantly reduced abundances in N-glycosylation negative bacteria. Electron acceptors such as Nap are crucial in maintaining proton motive force (PMF) in the periplasm and for balancing H+ accumulation in the cytoplasm, in the presence of functional ATP synthase (59). Our data show no change in any ATP synthase subunit (Suppl. Data S2), suggesting a model in which H+ is driven into the cytoplasm causing intracellular acidosis and osmotic imbalance, and a depletion of H+ and thus, PMF, in the periplasm. Indeed, the ΔpglB data overlap significantly with the groups of proteins differentially regulated in studies of C. jejuni hyperosmotic and acid stress (60–61). This scenario also correlates with the changes we observed in intracellular stress response proteins, despite glycosylation occurring solely in the periplasm. Although it remains speculation, it is noteworthy that flagellar motility is fundamentally powered by PMF through the flagellar motor proteins MotA/MotB (62), which were both (along with the motor supporting protein, FlgP; Fig. 2D), significantly downregulated in ΔpglB compared with WT, and restored in ΔpglB::pglB. motA/motB mutants are defective for motility (63), therefore reduced abundances of these proteins, and a loss of PMF required for their activity, may combine to generate the motility defect seen in ΔpglB C. jejuni. Finally, nitrate consumed under physiological conditions by Nap provides nitrite for the nitrite reductase NrfA. Our data showed significantly reduced abundance of NrfA and NrfH, which is thought to transfer electrons from MK to NrfA, in ΔpglB. We therefore anticipated reduced NrfA activity however, supply of exogenous nitrite enabled nitrite reductase activity. This suggests the lower abundance of these proteins in our data relate to expression and the lack of available substrate. Conversely however, it is also plausible that reduced Nap activity is related to loss of PMF and accumulation of acidifying H+, because acidified nitrite is known to kill C. jejuni, including as part of the host innate immune response (64).
Loss of nitrate reductase function may also explain N-glycosylation-associated phenotypes. The gut is oxygen-poor and hence, alternative electron acceptors, such as nitrate that is enriched in the gastrointestinal tract, are crucial in enabling the establishment of infection (65) and ΔnapA C. jejuni are attenuated for chicken colonization (22). Additionally, ΔnapA C. jejuni are deficient in biofilm formation under microaerobic conditions, while displaying enhanced biofilm formation under anaerobic conditions (66), a model-specific response like that seen in biofilm growth for ΔpglB. Secondly, nitrate accumulation has previously been shown to inhibit the succinate transporter DcuB from exporting succinate under restricted oxygen conditions (11). We observed elevated intracellular succinate consistent with both the observed reduction in DcuB abundance and, in the presence of decreased FrdABC, a commensurate decrease in the succinyl-CoA ligase (SucCD) component of the TCA cycle. Deletion of succinate dehydrogenase activity encoded by FrdABC leads to an inability to use Glu and Pro, as well as a colonization deficit in chickens (51). Accumulation of intracellular succinate has been observed as a correlating factor in catabolite repression and associated with the transcription factors RacRS, Cj1000 and CsrA (67). Neither RacRS or CsrA were differentially regulated in our data set, whereas Cj1000 was significantly downregulated, suggesting intracellular succinate accumulation was unrelated to this phenomenon. DctA also transports succinate, as well as Asp, into the cell (11). Given that downregulation of DctA was associated with the observed reduction in intracellular Asp, we believe that nitrate accumulation leading to inhibition of DcuB and loss of succinate export is the most likely explanation for this metabolic phenotype in pglB-deficient C. jejuni.
Loss of chemotaxis in ΔpglB C. jejuni toward α-KG, Glu, fucose, succinate and Asp was detected. Fucose chemotaxis is mediated by Cj0485 (45), which was heavily downregulated in the pglB complemented C. jejuni but only partially downregulated in ΔpglB. This is reflected in the chemotaxis assay where loss of migration toward fucose was observed for both strains relative to the WT. Reduced chemoattraction toward Glu, Asp, and succinate was likely mediated by reduced ΔpglB abundance of Tlp6, which mediates chemotactic response to these nutrients, as well as other organic acids (52). Loss of chemotaxis toward α-KG was perhaps surprising, given the elevated abundance of the KgtP α-KG transporter, and metabolomics identified no change in intracellular α-KG in pgl-deficient C. jejuni, suggesting these two molecular changes balance each other at the phenotypic level. Reduced chemotaxis toward Asp and succinate may be a response to both the intracellular accumulation of succinate, and the reduced abundance of the Asp transporter DctA. Given the complexity of Asp transport in C. jejuni however (with Asp associated with Peb1A, DctA, DcuA and DcuB (11, 14–15)), and that multiple Tlp respond to Asp, this phenotype remains to be fully understood.
Some phenotypes we observed are potentially more difficult to associate with glycosylation. For example, the apparent switch from Asp to Pro uptake, and intracellular increases in Pro, may be associated with osmotic balance as elevated cellular Pro has previously been shown as a mechanism in osmotolerance (68–69). Second, it may reflect reduced DctA abundance leading to decreased intracellular Asp and hence a requirement for alternative nutrients, although we observed no change to intracellular Ser, nor levels of the Ser transporter SdaC, suggesting Ser was not limited under our chosen conditions. The underlying relationship between increased Pro and N-glycosylation remains to be determined. It is interesting to note however, that both PutP and DctA contain N-linked glycosylation sequons (PutP, 397DKNSS401; DctA, 387EANAT391) contained outside predicted transmembrane spanning regions, and in predicted tryptic peptides well beyond the scope of reliable glycopeptide analysis (PutP, Lys within the sequon would be sterically hindered by glycosylation leaving a large peptide of 48 amino acids [5126.75 Da even without glycosylation]; DctA, 49 amino acid predicted peptide [4664.56 Da without glycosylation]. If such sites are glycosylated it will require the use of non-trypsin proteases to capture them for subsequent MS analysis.
Given the significant over-representation of known glycoproteins within the set of proteins identified at decreased abundance in pglB negative C. jejuni, we speculate that N-glycosylation may play a global role in protection against degradative processes, most specifically protecting against periplasmic proteases. A role in protein protection has been seen previously on exposure of C. jejuni to the host (42). To explore this in the context of nitrate reductase activity, we confirmed that loss of NapA and NapB protein abundance was unrelated to gene expression, as no change was observed for napA/napB transcript levels. Our Nap data, together with our observation of an enrichment of known glycoproteins within the downregulated proteomic data set, provide further evidence that a major function of glycosylation is to protect exposed loop regions of already folded substrates that are often targets for proteases. Despite this evidence however, some glycoproteins do appear stable in the pgl-negative background, suggesting that glycosylation is only protective in some structural contexts. Identification of more “resistant” sites, protein structural modeling and membrane topology predictions may aid in determining those sites more amenable to stability without glycosylation.
Glycosylation negative C. jejuni display a loss of electron transport proteins that is likely to cause a cellular proton imbalance leading to hyperosmotic stress and acidosis. The proteomic phenotype therefore involves cytoplasmic stress response proteins that are involved in protection against molecular damage caused by these and other unfavorable environments, despite glycosylation itself occurring in the periplasm. Subsequently, pgl-deficient C. jejuni are more sensitive to additional stress effects, such as those seen here for shocks at extreme temperatures, or further osmotic shock. These results are consistent with those attributed previously to the role of fOS in maintaining osmotic balance (32), which is another mechanism likely to be involved in phenotypes observed here. Our data have implications beyond the molecular. Survival on supermarket chicken meat at refrigeration temperatures is a major food chain issue in the poultry-human nexus (70). Our discovery that glycosylation negative C. jejuni are poorly able to survive a relatively short exposure to these low temperatures provides further evidence that novel antimicrobials that inhibit this pathway, even without bactericidal activity, could dramatically reduce food product contamination for human consumption. In conclusion, we have shown that glycosylation contributes to several C. jejuni phenotypes including nutrient uptake and utilization, chemosensing, electron transport, biofilm formation, motility and cell stress. Finally, we have shown for the first time, that a C. jejuni enzyme function depends on N-glycosylation for wild-type activity.
DATA AVAILABILITY
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via PRIDE with the data set identifier PXD011646.
Supplementary Material
Acknowledgments
We thank Dr. Ben Crossett, David Maltby and Angela Connolly for technical advice and support, and Desmond Li, Alexander Rookyard, and Zeynep Sumer-Bayraktar for assistance. This work was facilitated by access to Sydney Mass Spectrometry, a Core Research Facility of the University of Sydney.
Footnotes
* This work was supported in part by a National Health and Medical Research Council (NHMRC) Project Grant (APP1106878; to S.J.C.). J.A.C and L.M. are supported by Australian Postgraduate Awards. W.P.K. is supported by a University of Sydney/Faculty of Science Postgraduate Award in Microbial Vaccinology.
 This article contains supplemental Figures and Tables.
This article contains supplemental Figures and Tables.
1 The abbreviations used are:
- Tlp
- transducer-like protein
- TMAO
- trimethylamine-N-oxide
- DMSO
- dimethyl sulfoxide
- fOS
- free oligosaccharide
- FBS
- fetal bovine serum
- PVOF
- polyvinylidene difluoride.
REFERENCES
- 1. Ruiz-Palacios G. M. (2007) The health burden of Campylobacter infection and the impact of antimicrobial resistance: playing chicken. Clin. Infect. Dis. 44, 701–703 [DOI] [PubMed] [Google Scholar]
- 2. Kaakoush N. O., Castaño-Rodriguez N., Mitchell H. M., and Man S. M. (2015) Global epidemiology of Campylobacter infection. Clin. Microbiol. Revs. 28, 687–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silva J., Leite D., Fernandes M., Mena C., Gibbs P. A., and Teixeira P. (2011) Campylobacter spp. as a foodborne pathogen: a review. Front. Microbiol. 2, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Awad W. A., Hess C., and Hess M. (2018) Re-thinking chicken-Campylobacter jejuni interaction: a review. Avian Pathol. 47, 352–363 [DOI] [PubMed] [Google Scholar]
- 5. Nachamkin I., Allos B. M., and Ho T. (1998) Campylobacter species and Guillain-Barré Syndrome. Clin. Microbiol. Revs. 11, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poropatich K. O., Fischer-Walker C. L., and Black R. E. (2010) Quantifying the association between Campylobacter infection and Guillain-Barré Syndrome: a systematic review. J. Health Popul. Nutr. 28, 545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ransom G. M., Kaplan B., McNamara A. M., and Wachsmuth I. K. (2000) Campylobacter prevention and control: the USDA Food Safety and Inspection Service role and new food safety approaches, p. 511–528. In Nachamkin I. and Blaser M. J. (ed.), Campylobacter, 2nd ed ASM Press, Washington, D.C. [Google Scholar]
- 8. Stahl M., Friss L. M., Nothaft H., Liu X., Li J., Szymanski C. M., and Stintzi A. (2011) L-fucose utilization provides Campylobacter jejuni with a competitive advantage. Proc. Natl. Acad. Sci. U.S.A. 108, 7194–7199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stahl M., Butcher J., and Stintzi A. (2012) Nutrient acquisition and metabolism by Campylobacter jejuni. Front. Cell. Infect. Microbiol. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hofreuter D. (2014) Defining the metabolic requirements for the growth and colonization capacity of Campylobacter jejuni. Front. Cell Infect. Microbiol. 4, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wösten M. M., van de Lest C. H., van Dijk L., and van Putten J. P. (2017) Function and regulation of the C4-dicarboxylate transporters in Campylobacter jejuni. Front. Microbiol. 8, 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hofreuter Novik D., V, and Galan J. E. (2008) Metabolic diversity in Campylobacter jejuni enhances specific tissue colonization. Cell Host Microbe 4, 425–433 [DOI] [PubMed] [Google Scholar]
- 13. Guccione E., Leon-Kempis M. D., Pearson B. M., Hitchin E., Mulholland F., van Diemen P. M., Stevens M. P., and Kelly D. J. (2008) Amino acid-dependent growth of Campylobacter jejuni: key roles for aspartase (AspA) under microaerobic and oxygen-limited conditions and identification of AspB (Cj0762), essential for growth on glutamate. Mol. Microbiol. 69, 77–93 [DOI] [PubMed] [Google Scholar]
- 14. Pei Z. H., Burucoa C., Grignon B., Baqar S., Huang X. Z., Kopecko D. J., Bourgeois A. L., Fauchere J. L., and Blaser M. J. (1998) Mutation in the peb1A locus of Campylobacter jejuni reduces interactions with epithelial cells and intestinal colonization of mice. Infect. Immun. 66, 938–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Leon-Kempis M. D., Guccione E., Mulholland F., Williamson M. P., and Kelly D. J. (2006) The Campylobacter jejuni PEB1a adhesin is an aspartate/glutamate-binding protein of an ABC transporter essential for microaerobic growth on dicarboxylic amino acids. Mol. Microbiol. 60, 1262–1275 [DOI] [PubMed] [Google Scholar]
- 16. Chandrashekhar K., Kassem I. I., and Rajashekara G. (2017) Campylobacter jejuni transducer like proteins: chemotaxis and beyond. Gut Microbes 8, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rahman H., King R. M., Shewell L. K., Semchenko E. A., Hartley-Tassell L. E., Wilson J. C., Day C. J., and Korolik V. (2014) Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog. 10, e1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wright J. A., Grant A. J., Hurd D., Harrison M., Guccione E. J., Kelly D. J., and Maskell D. J. (2009) Metabolite and transcriptome analysis of Campylobacter jejuni in vitro growth reveals a stationary-phase physiological switch. Microbiology 155, 80–94 [DOI] [PubMed] [Google Scholar]
- 19. Thomas M. T., Shepherd M., Poole R. K., van Vliet A. H. M., Kelly D. J., and Pearson B. M. (2011) Two respiratory enzyme systems in Campylobacter jejuni NCTC 11168 contribute to growth on L-lactate. Environ. Microbiol. 13, 48–61 [DOI] [PubMed] [Google Scholar]
- 20. Sellars M. J., Hall S. J., and Kelly D. J. (2002) Growth of Campylobacter jejuni supported by respiration of fumarate, nitrate, nitrite, trimethylamine-N-oxide, or dimethyl sulfoxide requires oxygen. J. Bacteriol. 184, 4187–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weerakoon D. R., Borden N. J., Goodson C. M., Grimes J., and Olson J. W. (2009) The role of respiratory donor enzymes in Campylobacter jejuni host colonization and physiology. Microb. Pathog. 47, 8–15 [DOI] [PubMed] [Google Scholar]
- 22. Weingarten R. A., Grimes J. L., and Olson J. W. (2008) Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 74, 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Szymanski C. M., Yao R. J., Ewing C. P., Trust T. J., and Guerry P. (1999) Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32, 1022–1030 [DOI] [PubMed] [Google Scholar]
- 24. Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., North S. J., Panico M., Morris H. R., Dell A., Wren B. W., and Aebi M. (2002) N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science 298, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 25. Kowarik M., Young N. M., Numao S., Schulz B. L., Hug I., Callewaert N., Mills D. C., Watson D. C., Hernandez M., Kelly J. F., Wacker M., and Aebi M. (2006) Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 25, 1957–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morrison M. J., and Imperiali B. (2014) The renaissance of bacillosamine and its derivatives: pathway characterization and implications in pathogenicity. Biochemistry 53, 624–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kowarik M., Numao S., Feldman M. F., Schulz B. L., Callewaert N., Kiermaier E., Catrein I., and Aebi M. (2006) N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science 314, 1148–1150 [DOI] [PubMed] [Google Scholar]
- 28. Silverman J. M., and Imperiali B. (2016) Bacterial N-glycosylation efficiency is dependent on the structural context of target sequons. J. Biol. Chem. 291, 22001–22010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scott N. E., Nothaft H., Edwards A. V. G., Labbate M., Djordjevic S. P., Larsen M. R., Szymanski C. M., and Cordwell S. J. (2012) Modification of the Campylobacter jejuni N-linked glycan by EptC protein-mediated addition of phosphoethanolamine. J. Biol. Chem. 287, 29384–29396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cullen T. W., and Trent M. S. (2010) A link between the assembly of flagella and lipooligosaccharide of the Gram negative bacterium Campylobacter jejuni. Proc. Natl. Acad. Sci. U.S.A. 107, 5160–5165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cullen T. W., Madsen J. A., Ivanov P. L., Brodbelt J. S., and Trent M. S. (2012) Characterization of unique modification of flagellar rod protein FlgG by Campylobacter jejuni lipid A phosphoethanolamine transferase, linking bacterial locomotion and antimicrobial peptide resistance. J. Biol. Chem. 287, 3326–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nothaft H., Liu X., McNally D. J., Li J. J., and Szymanski C. M. (2009) Study of free oligosaccharides derived from the bacterial N-glycosylation pathway. Proc. Natl. Acad. Sci. U.S.A. 106, 15019–15024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., Michael F. S., Aberg E., and Szymanski C. M. (2002) Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J. Biol. Chem. 277, 42530–42539 [DOI] [PubMed] [Google Scholar]
- 34. Scott N. E., Parker B. L., Connolly A. M., Paulech J., Edwards A. V., Crossett B., Falconer L., Kolarich D., Djordjevic S. P., Højrup P., Packer N. H., Larsen M. R., and Cordwell S. J. (2011) Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-inked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics 10, M000031-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scott N. E., Marzook N. B., Cain J. A., Solis N., Thaysen-Andersen M., Djordjevic S. P., Packer N. H., Larsen M. R., and Cordwell S. J. (2014) Comparative proteomics and glycoproteomics reveal increased N-linked glycosylation and relaxed sequon specificity in Campylobacter jejuni NCTC11168 O. J. Proteome Res. 13, 5136–5150 [DOI] [PubMed] [Google Scholar]
- 36. Scott N. E., Bogema D. R., Connolly A. M., Falconer L., Djordjevic S. P., and Cordwell S. J. (2009) Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J. Proteome Res. 8, 4654–4664 [DOI] [PubMed] [Google Scholar]
- 37. Karlyshev A. V., Everest P., Linton D., Cawthraw S., Newell D. G., and Wren B. W. (2004) The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiol. 150, 1957–1964 [DOI] [PubMed] [Google Scholar]
- 38. Kelly J., Jarrell H., Millar L., Tessier L., Fiori L. M., Lau P. C., Allan B., and Szymanski C. M. (2006) Biosynthesis of the N-linked glycan in Campylobacter jejuni and addition onto protein through block transfer. J. Bacteriol. 188, 2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kakuda T., and DiRita V. J. (2006) Cj1496c encodes a Campylobacter jejuni glycoprotein that influences invasion of human epithelial cells and colonization of the chick gastrointestinal tract. Infect. Immun. 74, 4715–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larsen J. C., Szymanski C., and Guerry P. (2004) N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81–176. J. Bacteriol. 186, 6508–6514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Sorge N. M., Bleumink N. M., van Vliet S. J., Saeland E., van der Pol W. L., van Kooyk Y., and van Putten J. P. (2009) N-glycosylated proteins and distinct lipooligosaccharide glycoforms of Campylobacter jejuni target the human C-type lectin receptor MGL. Cell. Microbiol. 11, 1768–1781 [DOI] [PubMed] [Google Scholar]
- 42. Alemka A., Nothaft H., Zheng J., and Szymanski C. M. (2013) N-glycosylation of Campylobacter jejuni surface proteins promotes bacterial fitness. Infect. Immun. 81, 1674–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Untergasser A., Nijveen H., Rao X., Bisseling T., Geurts R., and Leunissen J. A. M. (2007) Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Brown H. L., Reuter M., Salt L. J., Cross K. L., Betts R. P., and van Vliet A. H. (2014) Chicken juice enhances surface attachment and biofilm formation of Campylobacter jejuni. Appl. Environ. Microbiol. 80, 7053–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dwivedi R., Nothaft H., Garber J., Kin L X, Stahl M., Flint A., van Vliet A. H. M., Stintzi A., and Szymanski C. M. (2016) L-fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol. Microbiol. 101, 575–589 [DOI] [PubMed] [Google Scholar]
- 46. Wei R., Li G. D., and Seymour A. B. (2010) High-throughput and multiplexed LC/MS/MRM method for targeted metabolomics. Anal. Chem. 82, 5527–5533. [DOI] [PubMed] [Google Scholar]
- 47. Chong J., Soufan O., Li C., Caraus I., Li S. Z., Bourque G., Wishart D. S., and Xia J. G. (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46, W486–W494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parkhill J., Wren B. W., Mungall K., Ketley J. M., Churcher C., Basham D., Chillingworth T., Davies R. M., Feltwell T., Holroyd S., Jagels K., Karlyshev A. V., Moule S., Pallen M. J., Penn C. W., Quail M. A., Rajandream M. A., Rutherford K. M., van Vliet A. H. M., Whitehead S., and Barrell B. G. (2000) The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403, 665–668 [DOI] [PubMed] [Google Scholar]
- 49. Guccione E., Hitchcock A., Hall S. J., Mulholland F., Shearer N., van Vliet A. H. M., and Kelly D. J. (2010) Reduction of fumarate, mesaconate and crotonate by Mfr, a novel oxygen-regulated periplasmic reductase in Campylobacter jejuni. Environ. Microbiol. 12, 576–591 [DOI] [PubMed] [Google Scholar]
- 50. Kassem I. I., Khatri M., Sanad Y. M., Wolboldt M., Saif Y. M., Olson J. W., and Rajashekara G. (2014) The impairment of methylmenaquinol:fumarate reductase affects hydrogen peroxide susceptibility and accumulation in Campylobacter jejuni. Microbiology Open 3, 168–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weingarten R. A., Taveirne M. E., and Olson J. W. (2009) The dual-functioning fumarate reductase is the sole succinate:quinone reductase in Campylobacter jejuni and is required for full host colonization. J. Bacteriol. 191, 5293–5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chandrashekhar K., Gangaiah D., Pina-Mimbela R., Kassem I. I., Jeon B. H., and Rajashekara G. (2015) Transducer like proteins of Campylobacter jejuni 81–176: role in chemotaxis and colonization of the chicken gastrointestinal tract. Front. Cell. Infect. Microbiol. 5, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cordwell S. J., Len A. C. L., Touma R. G., Scott N. E., Falconer L., Jones D., Connolly A., Crossett B., and Djordjevic S. P. (2008) Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8, 122–139 [DOI] [PubMed] [Google Scholar]
- 54. De Schutter J. W., Morrison J. P., Morrison M. J., Ciulli A., and Imperiali B. (2017) Targeting bacillosamine biosynthesis in bacterial pathogens: development of inhibitors to a bacterial amino-sugar acetyltransferase from Campylobacter jejuni. J. Med. Chem. 60, 2099–2118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mintmier B., McGarry J. M., Sparacino-Watkins C. E., Sallmen J., Fischer-Schrader K., Magalon A., McCormick J. R., Stolz J. F., Schwarz G., Bain D. J., and Basu P. (2018) Molecular cloning, expression and biochemical characterization of periplasmic nitrate reductase from Campylobacter jejuni. FEMS Microbiol. Lett. 365, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jepson B. J., Mohan S., Clarke T. A., Gates A. J., Cole J. A., Butler C. S., Butt J. N., Hemmings A. M., and Richardson D. J. (2007) Spectropotentiometric and structural analysis of the periplasmic nitrate reductase from Escherichia coli. J. Biol. Chem. 282, 6425–6437 [DOI] [PubMed] [Google Scholar]
- 57. Taveirne M. E., Sikes M. L., and Olson J. W. (2009) Molybdenum and tungsten in Campylobacter jejuni: their physiological role and identification of separate transporters regulated by a single ModE-like protein. Mol. Microbiol. 74, 758–771 [DOI] [PubMed] [Google Scholar]
- 58. Li X., Apel D., Gaynor E. C., and Tanner M. E. (2011) 5′-methylthioadenosine nucleosidase is implicated in playing a key role in a modified futalosinepathway for menaquinone biosynthesis in. Campylobacter jejuni. J. Biol. Chem. 286, 19392–19398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen M. T., and Lo C. J. (2016) Using biophysics to monitor the essential protonmotive force in bacteria. Adv. Exp. Med. Biol. 915, 69–79 [DOI] [PubMed] [Google Scholar]
- 60. Cameron A., Frirdich E., Huynh S., Parker C. T., and Gaynor E. C. (2012) Hyperosmotic stress response in Campylobacter jejuni. J. Bacteriol. 194, 6116–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Reid A. N., Pandey R., Palyada K., Whitworth L., Doukhanine E., and Stintzi A. (2008) Identification of Campylobacter jejuni genes contributing to acid adaptation by transcriptional profiling and genome-wide mutagenesis. Appl. Environ. Microbiol. 74, 1598–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Beeby M., Ribardo D. A., Brennan C. A., Ruby E. G., Jensen G. J., and Hendrixson D. R. (2016) Diverse high-torque bacterial flagellar motors assemble wide stator rings using a conserved protein scaffold. Proc. Natl. Acad. Sci. U.S.A. 113, E1917–E1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mertins S., Allan B. J., Townsend H. G., Köster W., and Potter A. A. (2013) Role of motAB in adherence and internalization in polarized Caco-2 cells and in cecal colonization of Campylobacter jejuni. Avian Dis. 57, 116–122 [DOI] [PubMed] [Google Scholar]
- 64. Iovine N. M., Pursnani S., Voldman A., Wasserman G., Blaser M. J., and Weinrauch Y. (2008) Reactive nitrogen species contribute to innate host defense against Campylobacter jejuni. Infect. Immun. 76, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Winter S. E., Winter M. G., Xavier M. N., Thiennimitr P., Poon V., Keestra A. M., Laughlin R. C., Gomez G., Wu J., Lawhon S. D., Popova I. E., Parikh S. J., Adams L. G., Tsolis R. M., Stewart V. J., and Bäumler A. J. (2013) Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kassem I. I., Khatri M., Esseili M. A., Sanad Y. M., Saif Y. M., Olson J. W., and Rajashekara G. (2012) Respiratory proteins contribute differentially to Campylobacter jejuni's survival and in vitro interaction with hosts' intestinal cells. BMC Microbiol. 12, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. van der Stel A. X., van de Lest C. H. A., Huynh S., Parker C. T., van Putten J. P. M., Wösten M. M. S. M. (2018) Catabolite repression in Campylobacter jejuni correlates with intracellular succinate levels. Environ. Microbiol. 20, 1374–1388 [DOI] [PubMed] [Google Scholar]
- 68. Hofreuter D., Mohr J., Wensel O., Rademacher S., Schreiber K., Schomburg D., Gao B., and Galán J. E. (2012) Contribution of amino acid catabolism to the tissue specific persistence of Campylobacter jejuni in a murine colonization model. PLoS ONE 7, e50699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Pérez-Arellano I., Carmona-Álvarez, Gallego F. J., and Cervera J. (2010) Molecular mechanisms modulating glutamate kinase activity. Identification of the proline feedback inhibitor binding site. J. Mol. Biol. 404, 890–901 [DOI] [PubMed] [Google Scholar]
- 70. Bhaduri S., and Cottrell B. (2004) Survival of cold-stressed Campylobacter jejuni on ground chicken and chicken skin during frozen storage. Appl. Environ. Microbiol. 70, 7103–7109 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry data have been deposited to the ProteomeXchange Consortium via PRIDE with the data set identifier PXD011646.