A method to study immunoglobulin (Ig) isotype- and site-specific N-glycosylation using nLC-MS/MS with stepped-energy higher energy collisional dissociation (HCD) has been established. This method empowers characterization of N-glycosylation microheterogeneity from human serum-derived IgG1, IgG4, IgA1, IgA2 and IgM, including sequence and glycosylation-site variants. This multi-isotype approach is a crucial step toward developing a platform to define disease-specific N-glycan signatures for different isotypes and thus help tune antibodes to induce protection.
Keywords: Antibodies*, N-Glycosylation, Glycoproteomics, Serum/Plasma*, Immunology*, Immunoglobulin G1 (IgG1), Immunoglobulin G4 (IgG4), Immunoglobulin M (IgM), Immunoglobulins A1 and A2 (IgA1 and IgA2), sequence variant
Graphical Abstract
Highlights
nLC-MS/MS method to analyze immunoglobulin (Ig) N-glycopeptides from human serum.
Multi-isotype, site-specific characterization of immunoglobulin N-glycosylation.
IgA2 sequence and glycosylation-site variant analyses.
Platform to define disease-specific N-glycan signatures for different Ig isotypes.
Abstract
Antibodies are critical glycoproteins that bridge the innate and adaptive immune systems to provide protection against infection. The isotype/subclass of the antibody, the co-translational N-glycosylation on the CH2 domain, and the remodeling of the N-linked glycans during passage through the ER and Golgi are the known variables within the Fc domain that program antibody effector function. Through investigations of monoclonal therapeutics, it has been observed that addition or removal of specific monosaccharide residues from antibody N-glycans can influence the potency of antibodies, highlighting the importance of thoroughly characterizing antibody N-glycosylation. Although IgGs usually have a single N-glycosylation site and are well studied, other antibody isotypes, e.g. IgA and IgM, that are the first responders in certain diseases, have two to five sites/monomer of antibody, and little is known about their N-glycosylation. Here we employ a nLC-MS/MS method using stepped-energy higher energy collisional dissociation to characterize the N-glycan repertoire and site occupancy of circulating serum antibodies. We simultaneously determined the site-specific N-linked glycan repertoire for IgG1, IgG4, IgA1, IgA2, and IgM in individual healthy donors. Compared with IgG1, IgG4 displayed a higher relative abundance of G1S1F and a lower relative abundance of G1FB. IgA1 and IgA2 displayed mostly biantennary N-glycans. IgA2 variants with the either serine (S93) or proline (P93) were detected. In digests of the sera from a subset of donors, we detected an unmodified peptide containing a proline residue at position 93; this substitution would strongly disfavor N-glycosylation at N92. IgM sites N46, N209, and N272 displayed mostly complex glycans, whereas sites N279 and N439 displayed higher relative abundances of high-mannose glycoforms. This multi-isotype approach is a crucial step toward developing a platform to define disease-specific N-glycan signatures for different isotypes to help tune antibodies to induce protection. Data are available via ProteomeXchange with identifier PXD010911.
Although the major known protective role of each antibody depends on its Fab domain, which is responsible for highly selective antigen binding and neutralization, antibodies can, through their constant domains, mediate multiple antibody-dependent (AD) functions, including cellular cytotoxicity (ADCC)1, cellular phagocytosis (ADCP), neutrophil phagocytosis (ADNP), and complement deposition (ADCD) (1). Antibody-mediated effector functions are critical for defensive response to HIV (2–4), Ebola (5), tuberculosis (6) and other infectious and autoimmune diseases. The antibody Fc CH2 domain binds to Fc-receptors (FcRs) on immune cells, inducing distinct responses that are dependent on the type of cell that has been activated. The immunoglobulin G (IgG) antibody Fc can modulate an immune response via (1) class switching (i.e. the selection of a specific subclass or isotype, each with different affinities for FcRs), or (2) the processing of the N-glycan that is co-translationally added to asparagine residue at position 180 on IgG (N180) and at analogous positions on the other subclass members; this modification is responsible for tuning the affinity of the antibody Fc to FcRs (Fig. 1A). Therefore, characterizing the CH2 domain N-glycosylation has great potential to illuminate how these glycan-mediated functions are regulated.
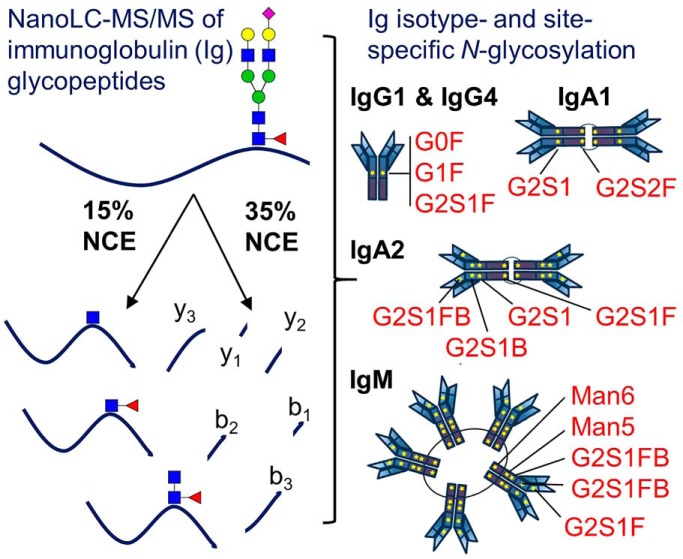
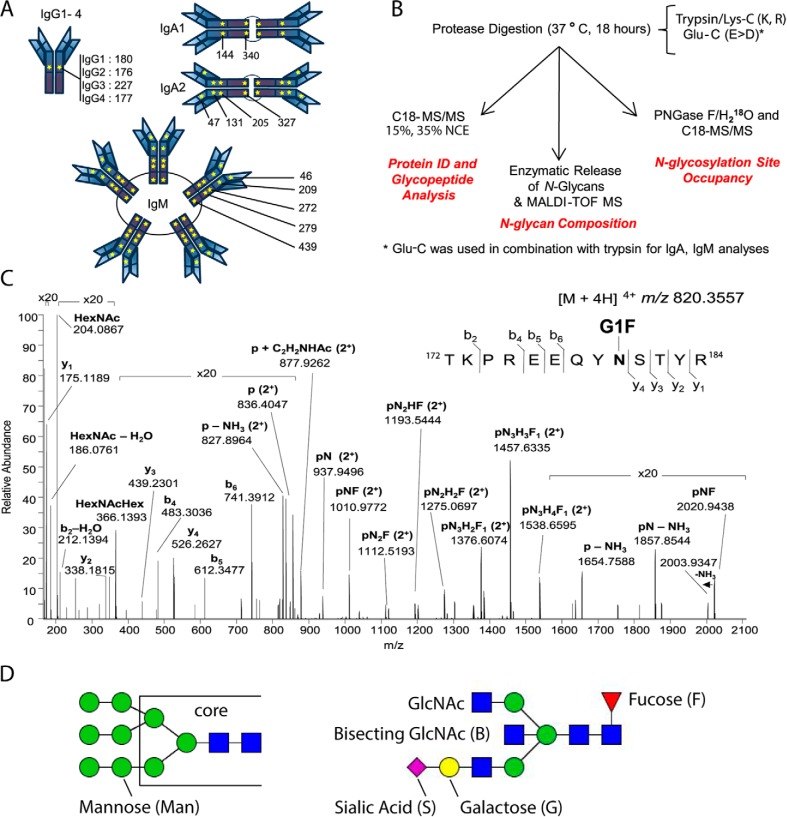
Fig. 1.
N-Glycosylation Sequons in the Immunoglobulin Fc Region Are Present in All Antibody Isotypes and Subclasses. A, Schematic of immunoglobulin N-glycosylation sequons. Stars represent the locations of N-glycosylation sequons within the Fc regions of IgG1–4, IgA1–2 and IgM. The location of the asparagine (N) within each sequon is numbered from the N terminus of the heavy chain constant region, and thus relates to the constant chain only. B, Schematic of the analytical workflow used in this study. nLC-MS/MS that employed C18 as the solid phase; stepped collision energy (15%, 35%) was used to observe fragments arising from cleavage of carbohydrate glycosidic linkages and peptide backbone fragments, respectively. Enzymatic release of N-glycans in H218O was achieved via the use of PNGase F. The PNGase release was also used to introduce 18O into formerly glycosylated asparagine residues, to estimate glycosylation site occupancy. C, Higher-energy collisional dissociation (HCD) spectrum of IgG1 N-glycopeptide 172TKPREEQYNSTYR184 + HexNAc4Hex4Fuc1 ([M + 4H]4+ m/z 820.3551), observed with 0.63 ppm error. Peptide b- and y-ions arising from breakage of the peptide bond and fragments arising from cleavage of glycosidic linkages, are labeled. D, Nomenclature for high mannose (left) and complex (right) N-linked glycans. High mannose and complex N-linked glycans share a common tri-mannosyl-chitobiose core (core). High mannose glycans are named by listing the total number of mannose (Man) residues. Man9, with two N-acetylglucosamine (GlcNAc) residues and nine mannose residues, is shown (left). A complex glycan consists minimally of the tri-mannosyl-chitobiose core with a single GlcNAc linked to each branch, and these residues are not listed. Monosaccharides beyond this basic structure are listed. G1S1FB (shown, right) is extended on one branch with galactose (G) and sialic acid (S), modified at the core with a fucose (F), and bisected with an N-acetylglucosamine (B).
Four IgG subclasses, IgG1–4, are present in human serum. All IgGs are composed of two heavy chains and two light chains, and each chain contains both a variable and a constant domain. IgG1, the most abundant subclass in serum, accounts for ∼90% of total serum IgG and is present in healthy individuals at a concentration of ∼10 mg/ml in plasma and serum (7). Among immunoglobulins, the structure-function relationship of IgG glycans is the best characterized, and intensive study of IgG1 N-glycan function, including contributions from the monoclonal therapeutics industry, has established that IgG1 N-glycans have specific structure-dependent functions (8–12). For example, through the development of Rituximab™, it has been established that the presence of core-fucosylated N-glycans interferes with FCGR3 binding, and this decreases the ability of fucosylated IgGs to drive ADCC (13–15). On the contrary, the presence of bisecting N-acetylglucosamine (GlcNAc) increases ADCC (14, 16–18). In addition, the presence of terminal sialic acid (NeuAc) has been demonstrated to be anti-inflammatory in multiple autoimmune diseases (19–21), and accumulation of agalactosylated (G0) IgG is used as a biomarker for autoimmune and infectious diseases (12, 22–24). Because N-glycans tune affinity for FcRs, N-glycan heterogeneity on the four IgG subclasses leads to the generation of a large variety of unique antibodies that may potentially act as “barcodes” to trigger distinct effector functions; however, little is understood about how this mechanism works.
Increased understanding of IgG glycosylation (25, 26) and the availability of monoclonal antibodies (9, 27, 28) have also advanced the field of therapeutics. Much less is known about the glycan repertoire of other antibody isotypes. Among these, immunoglobulin A (IgA) subclasses IgA1 and IgA2 are the most abundant antibodies; these are found as dimers at mucosal sites. Although the IgA1 and IgA2 constant domains are structurally like those of the IgGs (with each containing three Ig domains), their functions and the number of potential N-linked glycosylation sites vary. IgA1 has two N-linked sites on the Fc domain, N144 (enumerated starting from the N-terminal residue of the heavy constant chain) on the CH2 domain and N340 on the tail end of the CH3 domain (Fig. 1A) (29). The hinge region of IgA1 is also O-glycosylated (30). IgA2 possesses the two sites present on IgA1 and has two additional N-linked sites: N47 on the CH1 domain and N131 on CH2 (Fig. 1A), in addition to extensive O-linked glycosylation in its hinge region. In contrast, the heavy chain of secreted immunoglobulin M (IgM), like immunoglobulin E (IgE), has an additional Ig domain for a total of four Ig domains (31), whereas the IgG and IgA each have 3 Ig domains in the heavy chain region. The extra Ig domain is crucial for C1q binding and is not found on other antibody isotypes. The IgM antibody is found in serum as a pentamer (Fig. 1A). Each monomer of IgM has five N-linked glycosylation sites; these are spread across all domains of the heavy chain (N46, N209, N272, N279, and N439), meaning that there are 25 possible N-linked sites on the pentameric molecules (Fig. 1A). IgM is one of the first antibodies produced in response to infection and is important for clearance of viruses, bacteria, and parasites and capture of apoptotic cells (32). Although IgM N-glycans have been the subject of several studies, little is known about how IgM N-glycosylation impacts its function.
Earlier studies on antibody N-glycans have determined that glycosylation varies by sex, age (33), and geographic location and is antigen- and pathogen-specific (22, 34, 35). For example, HIV-specific antibodies targeting either the viral envelope or matrix proteins display glycosylation profiles distinct from those that target influenza (34). Further, treatment with an experimental vaccine induces an antigen-specific profile that differs from bulk antibody N-glycosylation (34). Mounting evidence points to glycosylation as tunable by signals that arise during vaccine and B cell priming, e.g. patients treated with either a protein or vector-based antigen display distinct glycosylation profiles (34, 36). Similarly, accumulation of agalactosylated, inflammatory IgG is observed in people with active tuberculosis, compared with those with latent infections (6). Therefore, it is clearly important to understand changes in antibody N-glycosylation during disease onset, because such changes can serve as biomarkers that may be exploited to improve the effectiveness of further vaccination (37). Antibody glycosylation is not static or predetermined but programmed and potentially altered throughout the course of an infection. Previous studies have shown that IgM N-glycans are functionally important and that abolishing antibody N-glycosylation impacts their secretion and function (38). However, there has to date been minimal focus on characterization of site-specific N-glycosylation on either IgM or IgA derived from human serum. Further, although N-glycan function has been explored in monoclonal antibodies, no clear relationship between glycan structure(s) and function(s) in polyclonal antibodies has yet been established, especially for IgA and IgM, the human serum antibodies with multiple N-glycosylation sites. Thus, our knowledge of their roles will benefit from a site-specific understanding of N-glycan heterogeneity and this information can direct future vaccine development (37).
Glycoproteomic-based methods for the analysis of immunoglobulins, primarily IgG derived both from serum and monoclonal sources, have contributed greatly to the understanding of the impact of N-glycan heterogeneity on the activity of these important molecules (25, 39–43). Recently, collision-induced dissociation (CID) with elevated dissociation energy (44) and higher-energy collisional dissociation (HCD) with stepped collision energy (45, 46) have shown great promise for glycopeptide analysis. Using the latter approach, a glycopeptide precursor ion is fragmented using two or more collision energies, and the product ions from these independent fragmentation events are subsequently combined and detected to generate a single product ion spectrum. This is advantageous because fragmentation of glycopeptides with low collision energy favors cleavage of glycosidic linkages, whereas fragmentation with higher collision energy favors cleavage of the peptide backbone; detection of both sets of glycopeptide product ions in a single step facilitates glycopeptide identification with high confidence. We have applied this promising approach here, to expand our understanding of site-specific immunoglobulin N-glycosylation.
EXPERIMENTAL PROCEDURES
Reagents
Peptide N-glycosidase F (PNGase F) was purchased from New England Biolabs (NEB, Ipswich, MA). CaptureSelect Affinity Matrix against human IgG1, IgG4, IgA and IgM, GlycanAssure APTS (8-aminopyrene-1,3,6-trisulfonic acid) kits, and MS grade trypsin/Lys-C (tosyl phenylalanine chloromethyl ketone treated) and were purchased from Thermo-Fisher Scientific (Waltham, MA). Endoproteinase Glu C was purchased from Roche (Mannheim, Germany). Pierce C18 Tips (100 μl), dithiothreitol (DTT), ammonium bicarbonate (NH4HCO3), iodoacetamide and Amicon Ultra filters for protein purification were purchased from Sigma Aldrich (St. Louis, MO). Small (10 μl) C18 ZipTips were purchased from Millipore (Billerica, MA). Heavy water (H218O, 99%) was purchased from Cambridge Isotope Laboratories (Andover, MA). Acid Citrate Dextrose (ACD) tubes were obtained from BD Vacutainer (Franklin Lakes, NJ).
Sample Selection
Human serum was obtained from Research Blood Components (Boston, MA), following American Association of Blood Banks guidelines. An IRB-approved consent form was collected from each donor, giving permission to use their blood for research purposes. Blood from healthy males and females between the ages 18 and 65 was collected in ACD-A tubes (BD Medical Vacutainer #364606; Becton Dickinson) and centrifuged. Following centrifugation, serum samples were transferred to 50 ml conical tubes, packaged and shipped at room temperature on the day of collection. The product was not stored or frozen before shipment.
Enrichment of Immunoglobulins
Antibodies (IgG1, IgG3, IgG4, IgA1, IgA2, and IgM) from 10 ml of donor sera prepared as described above were enriched using isotype-specific CaptureSelect Affinity Matrix resin (Thermo-Fisher). IgG2 was not isolated because of the lack of an IgG2-specific resin. Although IgG3 was isolated from this set of donor sera our results for IgG3 will be reported in a separate publication. In brief, 10 ml serum was dialyzed and filtered and added to 500 μl of resuspended and washed resin (based on 2.5–8 mg/ml binding capacity) and incubated for 1 h with end-to-end mixing at room temperature. The enriched antibody was eluted using 0.1 m acetic acid and immediately neutralized with 1 m Tris, pH 7.5. The neutralized eluate was then concentrated, and buffer exchanged into phosphate buffered saline (PBS) using an Amicon concentrator tube (50-kDa cutoff). Enrichment and purity were confirmed by Luminex analysis (Millipore-Sigma) according to the manufacturer's instructions and the concentration of each sample was assessed by human anti-Ig ELISA (Thermo-Fisher).
Proteolysis of Immunoglobulins
Between 2–20 μg of each serum-derived immunoglobulin sample (IgG1, IgG4, IgA1, IgA2, and IgM) were suspended in 50 mm NH4HCO3, pH 7.8, reduced with 5 mm DTT, and alkylated with 15 mm iodoacetamide. IgG1 and IgG4 samples were incubated with trypsin at a ratio of 1:50 of enzyme to total protein at 37 °C for 18 h. IgA1, IgA2, and IgM samples were incubated with endoproteinase GluC at a ratio of 1:20 of enzyme to total protein at 37 °C for 18 h, followed by incubation with trypsin/Lys-C at a ratio of 1:50 of enzyme to total protein, for 18 h. Samples were dried to remove volatile buffers, and peptides were purified using C18 tips (100 μl), according to the manufacturer's protocol.
Nano LC-MS/MS Glycopeptide Analysis of Immunoglobulin Proteolysis Products
For nano-liquid chromatography-tandem mass spectrometry (nLC-MS/MS) analysis, immunoglobulin-derived peptide/glycopeptide samples were analyzed on a Q Exactive HF Hybrid Quadrupole-Orbitrap™ mass spectrometer (Thermo Scientific) equipped with a nanoACQUITY UPLC system (Waters, Milford, MA) interfaced through a Triversa Nanomate (Advion, Ithaca, NY) electrospray ionization source. For chromatographic separation, a nanoACQUITY UPLC Symmetry C18 Trap Column (100 Å, 5 μm, 180 μm × 20 mm, Waters) column was used for trapping, and an ACQUITY UPLC Peptide BEH C18 nanoACQUITY column (130 Å, 1.7 μm, 150 μm × 100 mm, Waters) was used for separation. The peptide trapping step was performed at 4 μl/min for 4 min with 1% acetonitrile and 0.1% formic acid (Solvent A). Following the trapping step, peptides were separated on the analytical column according to the following conditions: 0–1 min: 2% B, 1–3 min: 2–5% B, 3–43 min: 5–40% B (solvent A: 1% acetonitrile and 0.1% formic acid in water; solvent B: 99% acetonitrile, 1% water and 0.1% formic acid). MS scans were acquired with the following settings: 60,000 resolution @ m/z 400, scan range m/z 370–2000, 1 μscan/MS, automatic gain control (AGC) target 1 × 106, and a maximum injection time of 100 ms. MS2 scans were acquired with the following settings: 15,000 resolution at m/z 400, 2 μscan/MS, AGC target of 1 × 106, maximum injection time of 200 ms, isolation window of 2.0 m/z, isolation offset of 0.4 m/z, stepped normalized collision energy (SNCE) of 15 and 35%, exclusion of charge states 1 and >6, and dynamic exclusion for 8 s. Profile data were recorded for MS and MS2 scans. To calculate the total areas of the peaks corresponding to the [M + nH]n+ selected peptides, ion chromatograms of all detected charge states were extracted, and the areas were summed.
Site-Occupancy Determination
Each of the peptide/glycopeptide mixtures obtained by treatment of an immunoglobulin sample with protease(s) was split into two equal amounts and dried in a centrifugal evaporator. First, 2 μl GlycoBuffer 2 (NEB, 50 mm sodium phosphate, pH 7.5) was aliquoted and dried under vacuum; next, 20 μl of H218O (99% 18O, Cambridge Isotopes) was added. After mixing, each of the buffer solutions containing H218O was transferred to one of the tubes containing a dried aliquot of immunoglobulin peptides. Next, 1 μl (500 units) of PNGase F was added to one tube of peptides in each pair of aliquots; the second tube was not treated with the glycosidase and served as the control. The contents were gently mixed by vortexing, and the tubes were placed on the Thermomixer at 37 °C for 16 h. The treated samples were dried under vacuum and stored at −20 °C; before analysis they were dissolved in 10 μl of 1% acetonitrile and 99% water containing 0.1% formic acid and desalted via C18 ZipTip™ according to the manufacturer's protocol. Peptides/glycopeptides were eluted from ZipTips™ into 50% acetonitrile/50% water containing 0.1% formic acid, dried under vacuum, and stored at −20 °C.
Nano LC-MS/MS Analysis of Immunoglobulin Protease Digests for Site Occupancy Determination
After PNGase F/H218O treatment, the peptide samples were analyzed on a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer (Thermo Scientific) equipped with a nanoACQUITY UPLC system (Waters) and a Triversa Nanomate (Advion). For chromatographic separation, a nanoACQUITY UPLC Symmetry C18 Trap Column (100 Å, 5 μm, 180 μm × 20 mm, Waters) column was used for trapping, and an ACQUITY UPLC Peptide BEH C18 nanoACQUITY Column (130 Å, 1.7 μm, 150 μm × 100 mm, Waters) column was used for separation. The peptide trapping and separation were performed as described above for peptide/glycopeptide analysis. MS scans were acquired with the following settings: 70,000 resolution @ m/z 400, scan range m/z 370–1880, 1 μscan/MS, AGC target 1 × 106, and a maximum injection time of 100 ms. MS2 scans were acquired with the following settings: 17,500 resolution at m/z 400, AGC target of 5 × 105, maximum injection time of 60 ms, isolation window of 2.0 m/z, isolation offset of 0.4 m/z, a normalized collision energy (NCE) of 27%, exclusion of charge states 1 and >8, underfill ratio of 1.2%, and dynamic exclusion for 8 s. Profile data were recorded for MS and MS2 scans.
MS/MS Data Interpretation
To confirm protein identity and determine amino acid sequences, nLC-MS/MS data files were converted to mgf and mzML formats with the ProteoWizard MSConvert (47) tool with peak picking enabled, followed by analysis via Mascot Daemon v2.4.0 with the Reviewed UniProtKB/Swiss-Prot Homo sapiens protein sequence database (last modified March 13, 2018). For all data sets, cleavage rules were applied for each specific protease (trypsin/Lys-C: K, R, P1′≠ P; or endoproteinase Glu-C-trypsin: K, R, D, E) and peptides with up to two missed cleavages were considered. The following peptide modifications were considered: methionine oxidation (variable), deamidation (variable), deamidation with 18O (variable), and carbamidomethylation (fixed). To assign glycopeptide tandem mass spectra, nLC-MS/MS data were processed using Byonic v2.13.17 (Protein Metrics, San Carlos, CA), with a custom protein sequence database consisting of IgG1, IgG4, IgA1, IgA2, or IgM sequences. Cleavage rules were consistent with those used in the corresponding Mascot searches (trypsin or Glu-C-trypsin). The following peptide modifications were considered: methionine oxidation (variable), carbamidomethylation (fixed), and a Byonic N-glycan database consisting of 182 human N-glycans (with ≤1 fucose). Glycopeptide matches with scores ≥100 or ≥3 diagnostic ions were accepted after manual confirmation. Extracted ion chromatograms (EICs) of carbohydrate oxonium ions at m/z 204.087 (HexNAc) and m/z 366.140 (HexNAc1Hex1) were generated and assigned glycopeptides and retention times were compared with ensure that all major glycopeptides could be accommodated within the chosen limits. The analyses performed here are enough to establish glycan composition and limited topological information. EICs of glycopeptide precursors were generated manually for the purpose of determining relative abundances using Xcalibur v2.2 (Thermo-Fisher) using a 0.02 Da window (0.01 Da on either side of the theoretical precursor m/z). For custom searches of IgA2 variant sequences and generation of sequence coverage plots, PEAKS Studio v8.5 (Bioinformatics Solutions Inc., Waterloo, ON, Canada) was employed. IgA2 variant IgA2m(1) (J00221), IgA2m(2) (M60192) and IgA2n (S71043) sequences from UniProt and IMGT (brackets) were considered. The same amino acid modifications were considered in PEAKS searches as with Mascot searches. Error tolerances of 5 ppm, and 2 missed cleavages, were allowed.
Capillary Electrophoresis (CE)
Between 10–20 μg of previously isolated IgG1 and IgG4 underwent two enrichment steps to ensure purity and IgGs alone were captured using Protein G beads (NEB). The Fab region was then separated from the Fc using IdeZ Protease according to the manufacturer's directions (NEB). The remaining protocol was performed using a GlycanAssure kit (Thermo-Fisher). Briefly, the supernatant (Fc portion) was collected and treated with PNGase F for one hour at 50 °C to release the glycans from the Fc domain. Glycans were then purified using GlycanAssure™ magnetic beads that bind free glycans. Glycan binding to beads and precipitation of contaminating proteins were performed using 100% acetonitrile. Bead-bound glycans were washed and eluted off the beads according to the manufacturer's directions. Eluted glycans were labeled with APTS for two hours at 50 °C. Unreacted dye was removed from the glycans using GlycanAssure beads. Bead-bound glycans were washed three times and were eluted in HPLC-grade water. The eluted glycans were analyzed by capillary electrophoresis (3500xL, ABI Sequencer, Thermo-Fisher) using the GlycanAssure™ software (Thermo-Fisher).
N-glycan Release, Permethylation, and nLC-MS/MS Analysis
N-glycans were released from 10 μg of each sample with 500 units of PNGase F in 50 mm ammonium bicarbonate buffer overnight at 37 °C. Samples were passed through SepPak C18 (1 cc) cartridges (Waters), and the flow-through was dried. Samples were resuspended in DMSO, then subjected to permethylation with periodic addition of methyl iodide (Sigma-Aldrich) according to an established protocol (48, 49). Following permethylation, glycans were dried, then dissolved in 10% acetonitrile/90% water with 0.1% formic acid, then desalted using a C18 ZipTip™. Glycans were eluted into 10 μl of 60% acetonitrile/40% water containing 0.1% formic acid, and 1/10th of each sample was spotted onto a steel target plate with 0.5 μl of 1 mm sodium acetate, 0.5 μl 2,5-dihydroxybenzoic acid (DHB)(20 mg/ml), and dried under vacuum. MALDI-TOF MS analysis was performed on an UltrafleXtreme TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA) using 10% laser power and summing 500 shots/spectrum. Following MALDI-TOF MS analysis, the remaining sample was dried, then dissolved in 4 μl of 10% water/90% acetonitrile with 0.1% formic acid, and 3 μl of each sample were injected for nLC-MS/MS analysis using a 6550 Q-TOF MS with 1200 series nanoflow pumps and a ChipCube-ESI source fitted with a custom HPLC-Chip with a 360 nL Polaris™ C18-A 5 μm trapping column and a 150 mm × 75 μm Polaris™ C18-A 3 μm analytical column (all from Agilent Corp., Santa Clara, CA). After an injection of the sample onto the trapping column, the column was washed at a flow rate of 1.5 μl/min for 4 min using 10% acetonitrile and 0.1% formic acid, and the sample mixture was separated on the analytical column using a gradient from 10% to 65% acetonitrile with 0.1% formic acid at a flow rate of 0.2 μl/min. The mass spectrometer was operated in positive mode using the high-resolution, extended dynamic range (2 GHz) setting. MS and MS2 spectra were recorded from m/z 300–3000. The ion source gas temperature was set to 225 °C, and the flow was set at 11 liters/min, with a capillary voltage of 1900 V. Precursors ≥1000 counts and charge states ≥2 were selected for fragmentation, and alternating collision energy according to the equation y = mx + b, with the first collision energy set based on the following parameters: y being the collision energy, slope m = 0.9, x representing the charge state, and the offset b = 2.0; for the second collision event, slope m = 2.0 and the offset b = 5.0. Spectra were collected in centroid mode.
Experimental Design and Statistical Rationale
Blood was obtained from four healthy individuals; using CaptureSelect Resin (Thermo), antibodies were isolated from the serum fraction of each blood sample. Antibody purity was assessed using Luminex assays targeting each isotype and subclass (Millipore Sigma). For bulk antibody glycopeptide analyses, we selected HIV-negative donors. Each antibody isotype and subclass (IgG1, IgG4, IgA1, IgA2, and IgM) was isolated from the same donor (n = 4), and each glycopeptide preparation was analyzed by nLC-MS/MS in triplicate, to verify that the glycopeptide abundances remained consistent through each run and to account for any variation because of spray instability, etc. Each individual donor is considered a biological replicate in this case (n = 4); hence, every antibody isotype was analyzed as an n = 12 (4 biological replicates, 3 technical replicates). We compared N-glycans between antibody isotypes from the same donors; IgG1 was selected as a control because the N-glycosylation of IgG1 has been extensively characterized. The reports of experimental design and the results from the analyses are in accordance with MIRAGE guidelines (50). To monitor nLC-MS/MS performance, we used a commercial source of human alpha-1-acid glycoprotein (AGP), performed proteolysis of AGP with trypsin (with a 1:20 ratio of trypsin to protein), and injected 100 ng of AGP peptides per run to monitor instrument performance throughout the set of experiments. All purified antibodies were run unblinded but glycopeptide assignments were verified using Mascot. Because IgG1 and IgG4 contain glycopeptides that differ by only a single amino acid, the statistical significance between the IgG1 and IgG4 glycoform distributions at this site was determined, to measure the impact of sequence (subtype) differences on N-glycan distribution. The Holm-Sidak method was used, with alpha = 0.05 using Prism GraphPad 7. Each glycan type was analyzed individually, without assuming a consistent standard deviation.
RESULTS
We designed a nLC-MS/MS method to characterize site-specific antibody Fc N-glycosylation across antibody isotypes and subclasses and applied this method to the analysis of immunoglobulins from human serum (Fig. 1B). Our goal in establishing this method is to enable the comparison of antibody N-glycosylation across many patient-derived serum samples from healthy and disease states and to enable the study of how glycans impact antibody function. At the outset, immunoglobulins (IgG1, IgG4, IgA1, IgA2, and IgM) were purified from the sera of four healthy donors, subjected to proteolysis, and analyzed in triplicate via nLC-MS/MS using HCD fragmentation with stepped normalized collision energy (15%, 35%) to facilitate the observation of fragments arising from cleavage of (1) glycosidic bonds that yield information about the glycan composition and topology, and (2) the peptide backbone that yield amino acid sequence information. We show site-specific bulk antibody N-glycosylation from small (∼100 ng) amounts of IgG1, IgG4, IgA1, IgA2, and IgM purified from healthy human serum using nLC-MS/MS without glycopeptide enrichment (Fig. 1B, 1C). We also characterized IgG3 N-glycosylation from the same donors but will report these results in a separate publication.
Site Specific N-Glycosylation of Immunoglobulin G (IgG) Subclasses
To establish that our method effectively surveyed immunoglobulin glycoforms, we first analyzed IgG1 N-glycosylation, as it has previously been characterized by multiple research groups (8–12). Purified IgG1 was subjected to proteolysis with trypsin, and 100 ng of each peptide sample was analyzed (in triplicate) via nLC-MS/MS using reversed phase (C18) separation and stepped collision energy. To verify the identity and assess the purity of each sample, Mascot analyses were performed on all the resulting data files. The protein search results were scrutinized to confirm that IgG1 was the top hit, and that there were no unique sequences belonging to other immunoglobulin isotypes and subclasses (see supplementary information). Minor components in the samples included IgG2, IgG3, alpha-1-acid glycoprotein, and immunoglobulin light chains. Because all assignments and quantification were performed at the glycopeptide level, the extraneous proteins did not contribute to the nLC-MS results reported for any of the immunoglobulin heavy chains. To examine the glycoform microheterogeneity at the single IgG1 N-glycosylation site, we used Byonic to assign glycopeptide tandem mass spectra, and manually verified the search results. Based on these analyses, we identified 20 IgG1 N-glycoforms, all sharing the same amino acid sequence that corresponds to the IgG1 Fc tryptic peptide containing N180 (enumerated starting from the N-terminal residue of the constant heavy chain). Glycopeptide tandem mass spectra were also examined for clues about N-glycan topology. For example, the positive-ion MS2 spectra of glycopeptides with core-fucosylated N-linked glycans have a characteristic set of fragment ions consisting of the intact peptide with (1) HexNAc1 (+203.0794 u), (2) HexNAc1Fuc1 (+349.1373 u), and (3) HexNAc2Fuc1 (+552.2167 u) that are derived from the fucosylated chitobiose core; these are all singly or multiply protonated and are annotated as pN, pNF, and pN2F, respectively, on the plotted mass spectra (Fig. 1C). This topological information is consistent with the known presence of core-fucosylation on IgG1 N-glycans. Precursor mass accuracy (better than 2 ppm), MS2 fragment ions, and retention time (within 30 s) were used as criteria to accept glycopeptide assignments, and EICs were generated for the m/z value corresponding to each glycopeptide.
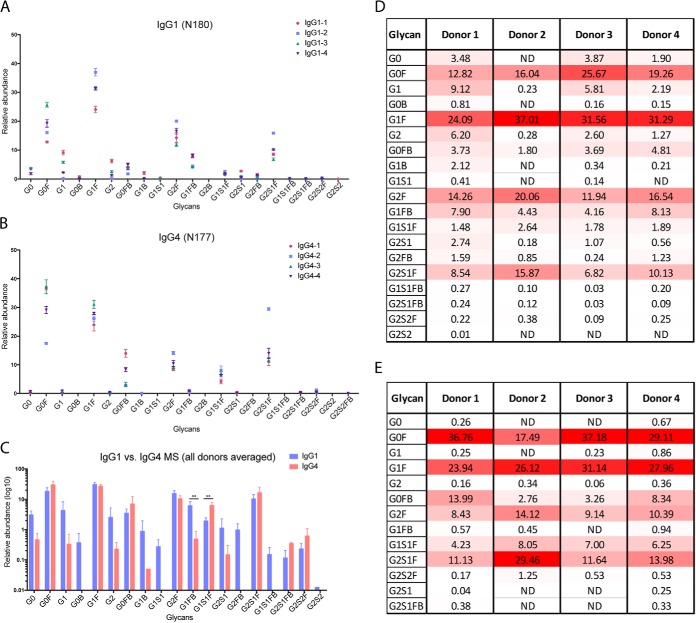
Consistent with previous findings (26), our results show that bulk IgG1 in healthy donor serum is highly fucosylated; the five most abundant glycoforms are core fucosylated (Fig. 2A). Some glycoforms contain a bisecting GlcNAc residue. In addition to observing extensive glycan heterogeneity on the single IgG1 Fc site within individual samples, we also detected donor-to-donor variability within bulk antibodies from healthy individuals. To rule out the possibility that the observed variability was method related, IgG1 samples from each donor were analyzed in triplicate. However, although donor-dependent variability clearly exists, the overall trends in glycoform abundance are consistent between donors, and the most abundant IgG1 glycoforms were the same among the four healthy donors (Fig. 2A, 2C, 2D). Next, we performed an orthogonal nLC-MS/MS analysis of released, permethylated N-glycans from IgG1 to verify the glycoform distributions and add additional structural details to the glycan assignments. The released and permethylated N-glycans from purified IgG1 were also analyzed by MALDI-TOF MS and capillary electrophoresis (CE) to verify that our glycopeptide nLC-MS/MS methods accurately represent the distribution of sialylated glycoforms (see supplementary information). Permethylated glycan analyses agreed with our glycopeptide results and confirmed that the most abundant glycoforms were fucosylated but not sialylated, although low abundance sialylated glycoforms were also detected with both methods. Because the released glycans may include some contributions from co-precipitated glycoproteins, the results for the MALDI-TOF MS, nLC-MS/MS and CE analyses can differ slightly from the glycopeptide results. Given that the different subclasses of IgGs are functionally distinct, variability in Fc glycosylation among healthy individuals may account for at least part of the functional differences observed. As noted earlier, we did not separately analyze IgG2 N-glycans, because of the low abundance of IgG2 and the difficulty of purifying it.
Fig. 2.
Comparison of Site-Specific IgG1 and IgG4 Fc N-Glycosylation using nLC-MS/MS. A, IgG1 Fc N-glycopeptide relative abundance, based on nLC-MS/MS analysis of IgG1 tryptic peptides. Immunoglobulin subclasses were isolated from a set of healthy donors (n = 4) and each sample was analyzed in triplicate. Glycopeptide assignments were made based on precursor m/z, peptide backbone fragment ions, carbohydrate fragment ions, and retention time. B, IgG4 Fc N-glycopeptide relative abundance, assigned as in (A). C, Comparison of IgG1 and IgG4 Fc N-glycopeptide relative abundance, based on glycoform. (Blue = IgG1, Red = IgG4) Differences in bisecting, fucosylated glycoforms and sialylated, fucosylated glycoforms are noted. ** indicates statistical significance (p < 0.001). D, Heat map of IgG1 glycoforms. E, Heat map of IgG4 glycoforms. The heat maps shows the N-glycosylation site, putative N-glycan, and average relative abundance (%) for each glycoform for donors 1–4. The heat map is scaled according to relative abundance for each donor sample, with the highest abundance glycoform in each sample colored red, the lowest colored white, and intermediate glycoforms colored on a linear intensity scale between the two extremes.
Next, we analyzed IgG4 glycoforms using the nLC-MS/MS method described above and confirmed the assignments of the IgG4 peptides that contain the occupied glycosylation site N177 (enumerated starting from the N-terminal residue of the heavy constant chain) by verifying the protein identity and the sequence of the IgG4 glycopeptide (for the tandem MS data, see supplemental Fig. S1). The IgG1 and IgG4 subclasses share the most abundant glycans on their Fc regions (Fig. 2B, 2E). However, for IgG4, we detected a smaller range of IgG4 glycoforms than we had observed for IgG1, despite our analyzing equal quantities of IgG1 and IgG4 and our determination that both sites are fully occupied (see below). Nevertheless, we did detect a greater number of IgG4 glycoforms than were reported for a previous effort that also used nLC-MS/MS (26). Because both IgG1 and IgG4 samples were derived from the same donors, it is clear that the observed differences in glycosylation are Ig-subclass-specific. Interestingly, we found relative abundances of G1 and G2 glycoforms trending toward a decrease in all four IgG4 samples, as compared with IgG1. Despite donor-to-donor variability, trends in glycoform abundances were constant across the IgG4 samples, with the same high abundance glycoforms shared among all donors (Fig. 2C). To determine the differences between the distributions of IgG1 and IgG4 glycoforms, we averaged the sets of glycans observed for all four donors and confirmed that IgG4 had notably fewer glycoforms than IgG1. The observed differences tended to occur primarily in low abundance components, including those that contain bisecting GlcNAc (B) and NeuAc (S), such as G0B, G1S1, G2FB, G1S1FB and G2S2. Indeed, these glycoforms were not detected in IgG4 (Fig. 2D). In addition, there were significant differences between the high abundance glycoforms that were present in the two subclasses. For example, we detected higher levels of the fucosylated, bisecting N-glycoform G1FB in IgG1 compared with IgG4, and higher levels of the sialylated, fucosylated glycoform G1S1F in IgG4 (Fig. 2C).
To (1) confirm the amino acid sequence of each glycopeptide and (2) estimate the site occupancy of each N-glycosylation site, we treated all samples with PNGase F in the presence of H218O, resulting in the removal of N-linked glycans from the occupied Asn sites, accompanied by hydrolysis of the formerly occupied Asn residues to Asp residues, with incorporation of 18O into these formerly N-glycosylated sites. We found only very slight variations in the peptide sequences. We confirmed that IgG1 and IgG4 Fc N-glycosylation sites are highly occupied (>99.7%) (Table I). Our results emphasize the need to explore N-glycosylation profiles in tandem with antibody function, as the intriguing differences documented here are likely to impact antibody function. In this regard, as noted above, we also analyzed IgG3 N-glycosylation from the same donor samples, but we plan to report the results of the IgG3 analyses separately, to deal more completely with the biological implications of glycoform variations in this protein.
Table I. Estimated site occupancy of IgG1 and IgG4 Fc N-glycosylation sites from serum of healthy donors.
| IgG1 | Unmodified peptide area | Labeled D18O peptide area | % Occupied | IgG4 | Unmodified peptide area | Labeled D18O peptide area | % Occupied |
|---|---|---|---|---|---|---|---|
| Donor 1A | 3.59E+05 | 2.21E+08 | 99.84% | Donor 1A | 1.31E+06 | 9.07E+08 | 99.86% |
| Donor 1B | 2.84E+05 | 2.13E+08 | 99.87% | Donor 1B | 1.31E+06 | 8.76E+08 | 99.85% |
| Donor 1C | 2.90E+05 | 2.10E+08 | 99.86% | Donor 1C | 1.00E+06 | 8.29E+08 | 99.88% |
| Donor 2A | 2.55E+05 | 1.52E+08 | 99.83% | Donor 2A | 4.92E+06 | 2.35E+09 | 99.79% |
| Donor 2B | 2.79E+05 | 1.43E+08 | 99.80% | Donor 2B | 3.66E+06 | 2.47E+09 | 99.85% |
| Donor 2C | 2.24E+05 | 1.42E+08 | 99.84% | Donor 2C | 4.99E+06 | 2.38E+09 | 99.79% |
| Donor 3A | 1.03E+05 | 5.70E+07 | 99.82% | Donor 3A | 1.33E+06 | 1.06E+09 | 99.87% |
| Donor 3B | 7.66E+04 | 5.45E+07 | 99.86% | Donor 3B | 1.31E+06 | 9.84E+08 | 99.87% |
| Donor 3C | 7.20E+04 | 5.50E+07 | 99.87% | Donor 3C | 2.60E+06 | 1.06E+09 | 99.75% |
| Donor 4A | 8.96E+04 | 8.10E+07 | 99.89% | Donor 4A | 7.37E+05 | 8.02E+08 | 99.91% |
| Donor 4B | 7.95E+04 | 7.87E+07 | 99.90% | Donor 4B | 5.88E+05 | 7.26E+08 | 99.92% |
| Donor 4C | 7.41E+04 | 6.94E+07 | 99.89% | Donor 4C | 2.48E+05 | 5.94E+08 | 99.96% |
For each analysis, extracted ion chromatograms (EICs) of the unmodified peptide and the formerly glycosylated peptide labeled with 18O were generated, and the area under the curve was calculated. To estimate site occupancy, labeled (18O) peptide area was divided by the summed area of the labeled and unmodified peptide. Results of triplicate analyses are shown.
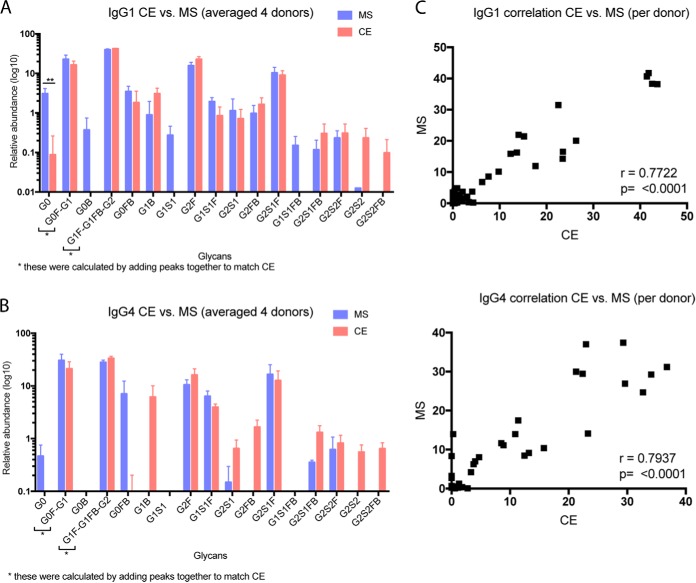
IgG glycans have been studied extensively via both CE and MS. To perform another validation of the nLC-MS/MS based method, we released Fc-specific glycans from purified IgG1 and IgG4 and analyzed the released glycans via CE. As with our mass spectrometry-based IgG analysis above, we did not include CE analysis of IgG2 because we do not have a method to purify adequate amounts of this very low abundance antibody. Our CE results for both IgG1 and IgG4 were comparable to those obtained using our nLC-MS/MS method, showing very minimal differences in the glycan profiles (Fig. 3). In both methods, the trends in abundances of glycans remained the same. However, we found that there are some glycoforms, including G0B, G1S1, and G1S1FB, which could only be detected via MS-based methods, and other glycoforms, especially several low-abundance, disialylated N-glycans, that we detected only in IgG1, and only with the CE method (Fig. 3A). As was the case for IgG1, use of CE resulted in detection of twelve glycoforms for IgG4 (Fig. 3B), whereas our nLC-MS/MS method only found ten. For both IgG1 and IgG4, this difference occurred only with respect to glycans of very low abundance that make up less than 1% of the total profile. The abundances of the assigned glycoforms from the nLC-MS/MS and CE-based methods correlated strongly within each subclass, with correlation values of 0.7722 (p = <0.0001) and 0.7937 (p = <0.0001) for IgG1 and IgG4, respectively (Fig. 3C). Our nLC-MS/MS showed lower levels (or no signal) for the low abundance disialylated species (Fig. 3A, 3B), either because of lower-efficiency detection of the target glycans or, more likely, the presence of co-precipitated glycoproteins in the CE-analyzed samples. We emphasize that the y axis of Fig 3B is on a log scale, which may have the effect of exaggerating the significance of minor components.
Fig. 3.
Comparison of IgG1 and IgG4 Glycoforms using Capillary Electrophoresis of Released N-Glycans. A, Relative abundance (log10) of released IgG1 N-glycans determined via capillary electrophoresis (CE) versus glycopeptide nLC-MS/MS (n = 4, serum from healthy donors analyzed in triplicate). Note that CE is unable to separate certain glycoforms, so to compare this method to MS-based methods, the relevant multiple glycoforms distinguished in MS analyses were summed. B, Relative abundance (log10) of released IgG4 N-glycans determined via capillary CE versus glycopeptide nLC-MS/MS (n = 4, serum from healthy donors analyzed in triplicate). C, Correlation of CE versus MS analyses for IgG1 glycoforms. D, Correlation of CE versus MS analyses for IgG4 glycoforms.
Despite the minor differences in the results obtained using the two methods, our nLC-MS/MS-based glycopeptide method provided enough knowledge of glycoform heterogeneity, enabled assignment of glycoforms not detected by CE, and required a practical amount of starting material. Therefore, we concluded that we could extend this approach to undertake the analysis of the N-glycosylation of the multimeric antibodies IgA1, IgA2, and IgM.
As we have discussed above, several techniques have been applied to assess N-glycosylation heterogeneity in immunoglobulins, including the analysis of released N-glycans using CE or MS, and glycopeptide analysis via nLC-MS/MS. Among these, glycopeptide MS/MS has major advantages: (1) glycopeptide fragmentation facilitates determination of the site of glycan attachment, and, as a corollary to this first point, (2) the relationship between the glycan and protein is maintained. This is especially important in samples that may contain multiple glycoprotein impurities even after antibody pulldown, and in glycoproteins such as IgA and IgM, that have multiple N-glycosylation sites. In this context, site-specific glycosylation analysis can be achieved across multiple glycosylation sites within the same protein, and across multiple proteins. Our key goal was to develop a method to assign site-specific Fc glycosylation in antibodies within the same donor sample, as Fab and Fc glycans are functionally different. Strategies that rely on the release of N-glycans from intact proteins fail to meet this criterion, but nLC-MS/MS analyses of glycopeptides enable us to distinguish Fab from Fc glycans.
Site Specific N-Glycosylation of Immunoglobulin A (IgA) Subclasses
Methods that rely on the release of N-linked glycans are poorly suited for the site-specific analysis of IgA1, IgA2, and IgM, because of the presence of multiple N-glycosylation sites on each of these molecules. Secretory IgA1 has two N-glycosylation sequons: one in the CH2 domain that interacts with its cognate receptor, and the other on the tail domain whose function is unknown. An additional challenge arises from the fact that IgA1 and IgA2 share a region of partial sequence identity within the CH2 domain, and this region contains an N-glycosylation sequon.
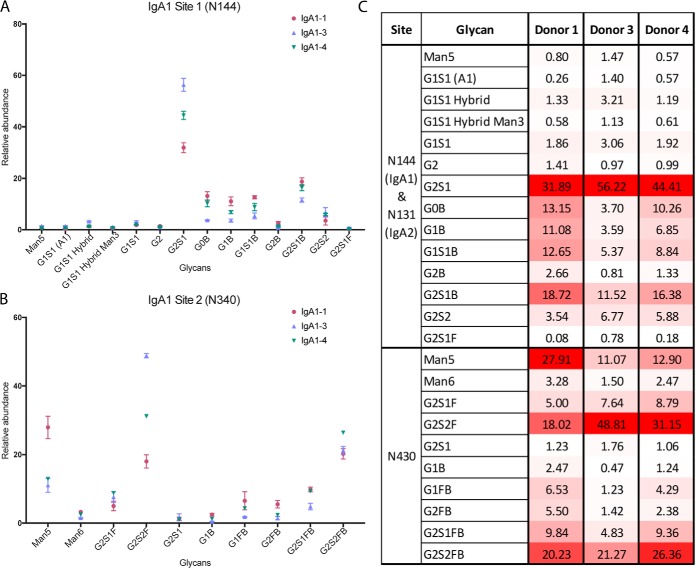
To address these challenges and to obtain glycopeptides of a convenient size for tandem MS analysis, IgA1 and IgA2 were separated, treated with Glu-C and trypsin and analyzed via nLC-MS/MS. We subsequently confirmed that our separation was successful by identifying sequences unique to each of the subclasses via Mascot analysis (see supplementary information). Separation of IgA1 and IgA2 enabled us to examine how glycan heterogeneity at the shared N-glycosylation sequon differs between the subclasses. We determined that IgA1 glycopeptides from the IgA1 CH2 site at N144 are dominated by N-glycans with free terminal galactose, with the exception of one glycoform (G0B) (Fig. 4A, and supplemental Fig. S2). We found that four of the six most abundant glycoforms are monosialylated. Most interestingly, we detected no fucosylation and very little disialylation, and it therefore became clear that the glycan repertoire of IgA1 is distinct from that of IgG1. For example, in IgG1 and IgG4, we determined that G2S1 is scarce; however, we found that it was the most abundant glycoform in IgA1. Additionally, at this site, we also determined the presence of N-glycans with compositions consistent with bisecting GlcNAc and a low level of minimally processed high mannose N-glycans. As previously discussed, IgA1 and IgA2 share sequence identity at this site; we determined that the glycoform distributions of the two isoforms were the same (Fig. 5B). Given that this site is within the CH2 domain, which is known to interact with the Fc receptor, IgA1 and IgA2 may have similar potential to interact with the receptor. We determined that the glycans on IgA1 site 2 (N340) were larger than those on the first site and consisted mostly of biantennary (disialylated and digalactosylated) forms (Fig. 4B–4C, and supplemental Fig. S3). Perhaps the most interesting difference we found between the two sites was that almost all complex N-glycans on this tailpiece were core fucosylated (Fig. 4C). Like the CH2 domain site, this site also had some high-mannose type glycans, suggesting that these regions are less accessible to processing by glycosyltransferases than is the single site in IgG1 which contains only complex N-glycans. We also estimated the site occupancy of IgA1 glycosylation sites after treatment with PNGase F in the presence of H218O and determined that both IgA1 sites were >99.2% occupied (Table II).
Fig. 4.
IgA1 Glycoforms at Sites N144 and N340. A, IgA1 site N144 N-glycopeptide relative abundance, based on nLC-MS/MS analysis of IgA1 site N144 trypsin/Glu-C peptides. Immunoglobulin subclasses were isolated from a set of healthy donors (n = 4) and each sample was analyzed in triplicate. Glycopeptide assignments were made based on precursor m/z, peptide backbone fragment ions, carbohydrate fragment ions, and retention time. B, IgA1 site N340 N-glycopeptide relative abundances, based on nLC-MS/MS analysis of IgA1 site N340 trypsin/Glu-C peptides. C, Heat map of IgA1 glycoforms. The heat map shows the N-glycosylation site, putative N-glycan, and average relative abundance (%) for each glycoform for donors 1, 3, and 4. Donor 2 was excluded, as an insufficient amount of IgA1 was isolated for glycopeptide detection. The heat map is scaled according to relative abundance for each donor sample, with the highest abundance glycoform at each site and within each sample colored red, the lowest colored white, and intermediate glycoforms colored on a linear intensity scale between the two extremes.
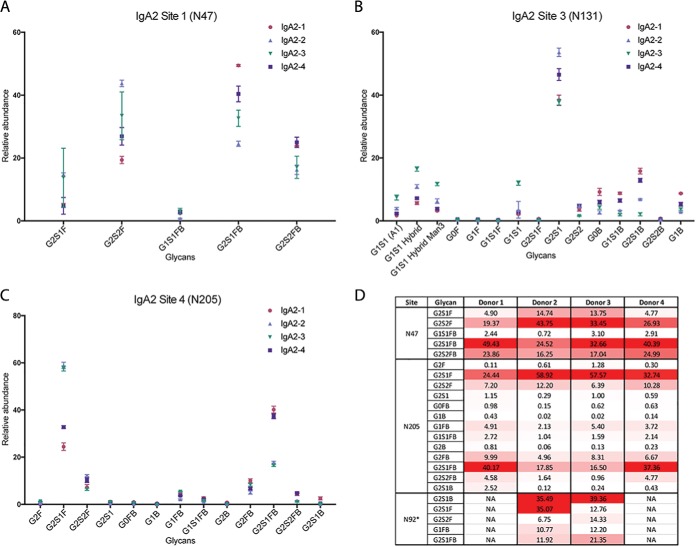
Fig. 5.
IgA2 Glycoforms at Sites N47, N131, and N205. A, IgA2 site N47 N-glycopeptide relative abundance. B, IgA2 site N131 N-glycopeptide relative abundance. C, IgA2 site N205 N-glycopeptide relative abundance. For both sites, abundances and standard deviation are shown for IgA2 from four healthy individuals, each run in triplicate. D, Heat map of IgA2 glycoforms. The heat map shows the N-glycosylation site, putative N-glycan, and average relative abundance (%) for each glycoform for donors 1–4. The heat map is scaled according to relative abundance for each donor sample, with the highest abundance glycoform at each site and within each sample colored red, the lowest colored white, and intermediate glycoforms colored on a linear intensity scale between the two extremes.
Table II. Estimated site occupancy of IgA1 N-glycosylation sites N144 and N340.
| IgA1-N144 | Unmodified peptide area | Labeled D18O peptide area | % Occupied | IgA1-N340 | Unmodified peptide area | Labeled D18O peptide area | % Occupied |
|---|---|---|---|---|---|---|---|
| Donor 1A | ND | 4.46E+08 | HIGH | Donor 1A | ND | 1.08E+07 | HIGH |
| Donor 1B | 4.11E+06 | 4.84E+08 | 99.16% | Donor 1B | ND | 2.09E+07 | HIGH |
| Donor 1C | 4.00E+06 | 4.75E+08 | 99.17% | Donor 1C | ND | 2.26E+07 | HIGH |
| Donor 2A | ND | 1.15E+09 | HIGH | Donor 2A | ND | 4.12E+07 | HIGH |
| Donor 2B | ND | 1.18E+09 | HIGH | Donor 2B | ND | 4.39E+07 | HIGH |
| Donor 2C | ND | 1.18E+09 | HIGH | Donor 2C | ND | 4.49E+07 | HIGH |
| Donor 3A | 2.12E+06 | 4.22E+08 | 99.50% | Donor 3A | ND | 2.43E+06 | HIGH |
| Donor 3B | 2.58E+06 | 4.23E+08 | 99.39% | Donor 3B | ND | 2.33E+06 | HIGH |
| Donor 3C | 2.63E+06 | 3.91E+08 | 99.33% | Donor 3C | ND | 2.59E+06 | HIGH |
| Donor 4A | 5.85E+06 | 7.33E+08 | 99.21% | Donor 4A | ND | 2.45E+07 | HIGH |
| Donor 4B | 5.23E+06 | 6.99E+08 | 99.26% | Donor 4B | ND | 2.81E+07 | HIGH |
| Donor 4C | 5.72E+06 | 6.72E+08 | 99.16% | Donor 4C | ND | 2.88E+07 | HIGH |
Results of triplicate analyses are shown.
Next, we investigated the N-glycosylation of IgA2, which has 4–5 N-glycosylation sites, as the number of N-glycosylation sequons present varies between IgA2 allotypes (29, 51). IgA2 is ∼10-fold less abundant than IgA1. We purified IgA1 and IgA2 separately, analyzed equal amounts of IgA1 and IgA2, and verified the success of our purification by identifying sequences unique to each isoform (see Supplementary Information), and we were able to observe glycopeptides spanning all the N-glycosylation sites, except for the last site, N327. IgA2 has three known allotypes, and two of these allotypes, IgA2m(2) and IgA2n, share an additional N-glycosylation sequon on the CH1 domain that is absent in the IgA2m(1) allotype (52). IgA2m(1), the third known allotype, contains a proline residue at position 93 and hence N92 is not within a N-glycosylation sequon (NXS/T, X≠P). IgA2m(2) and IgA2n have serine at position 93 and may be glycosylated at N92. We detected both the proline- and serine-containing peptides in donor samples (see supplemental Fig. S4–S5); two out of four samples used in this study had the IgA2m(1) allotype, with a proline at position 93 and a total of four N-glycosylation sequons (51). We also documented glycosylation at IgA2 site N92 in the donor samples with the IgA2m(2) allotype, both by detection of the formerly-glycosylated, 18O-labeled peptide (supplemental Fig. S6) and by direct observation of glycopeptides with complex N-glycans at this site (supplemental Fig. S7). We found that all N-glycans on IgA2 site N47 are highly fucosylated, digalactosylated, and sialylated (Fig. 5A, 5D, and supplemental Fig. S8). The observed glycan compositions indicate that there are similar ratios of N-glycans with and without bisecting GlcNAc. In total, we detected a limited number of glycans at this site. Notably, IgA2 Site N47 was the only site that we found glycopeptide evidence of N-glycosylation where were unable to estimate site occupancy. After treatment with PNGase F in the presence of H218O, we detected a precursor m/z that matched the theoretical m/z, charge state and predicted retention time predicted for this deglycosylated, labeled peptide. However, despite repeated attempts to target and fragment this ion, we were unable to obtain a well-defined MS2 spectrum, probably because of the extremely low abundance of the precursor ion. It is possible that some of this peptide was lost during the peptide cleanup step that we performed after PNGase F treatment and before nLC-MS/MS analysis.
IgA2 site N131 is the CH2 domain glycosylation site that shares sequence identity with IgA1. As previously discussed, the glycoforms at this site are the same in the two subclasses (Fig. 5B, and supplemental Fig. S2). At site N205, G2S1F and G2S1FB are the dominant N-glycoforms (like IgA2 Site N47), and several lower abundance N-glycans are also present (Fig. 5C, and supplemental Fig. S9). There is a fifth N-glycosylation sequon (“site 5”) at N327, but we did not detect any glycopeptides at this site, nor could we detect any 18O-labeled peptide at this site after treatment with PNGase F in the presence of H218O. However, studies performed on IgA2 derived from saliva and human milk have found evidence of N-glycosylation at IgA2 site N327 (53–55). Based on our estimates of N-glycosylation site occupancy, IgA2 sites N131 and N205 are highly occupied (Table III). Although IgA1 and IgA2 function(s) are not understood as well as those of the IgG subclasses, there is speculation that the functions of these subclasses differ from one another. Consistent with this idea, we have found that there are significant differences in the N-glycosylation of IgA1 and IgA2.
Table III. Estimated site occupancy of IgA2 N-glycosylation sites N131 and N205.
| IgA2-N131 | Unmodified peptide area | Labeled D18O peptide area | % Occupied | IgA2-N205 | Unmodified peptide area | Labeled D18O peptide area | % Occupied |
|---|---|---|---|---|---|---|---|
| Donor 1A | 2.54E+06 | 1.38E+09 | 99.82% | Donor 1A | ND | 9.87E+08 | HIGH |
| Donor 1B | 3.02E+06 | 1.29E+09 | 99.77% | Donor 1B | ND | 9.41E+08 | HIGH |
| Donor 1C | 2.57E+06 | 1.21E+09 | 99.79% | Donor 1C | ND | 9.90E+08 | HIGH |
| Donor 2A | ND | 6.25E+09 | HIGH | Donor 2A | ND | 4.03E+09 | HIGH |
| Donor 2B | ND | 6.18E+09 | HIGH | Donor 2B | ND | 3.76E+09 | HIGH |
| Donor 2C | ND | 5.74E+09 | HIGH | Donor 2C | ND | 3.84E+09 | HIGH |
| Donor 3A | 1.11E+06 | 5.21E+08 | 99.79% | Donor 3A | ND | 8.33E+08 | HIGH |
| Donor 3B | 1.07E+06 | 5.23E+08 | 99.80% | Donor 3B | ND | 7.96E+08 | HIGH |
| Donor 3C | 8.35E+05 | 4.53E+08 | 99.82% | Donor 3C | ND | 8.39E+08 | HIGH |
| Donor 4A | 7.25E+06 | 2.11E+09 | 99.66% | Donor 4A | ND | 1.72E+09 | HIGH |
| Donor 4B | 3.22E+06 | 2.20E+09 | 99.85% | Donor 4B | ND | 1.87E+08 | HIGH |
| Donor 4C | 2.88E+06 | 1.97E+09 | 99.85% | Donor 4C | ND | 1.73E+09 | HIGH |
Results of triplicate analyses are shown.
Site Specific N-Glycosylation of Immunoglobulin M (IgM)
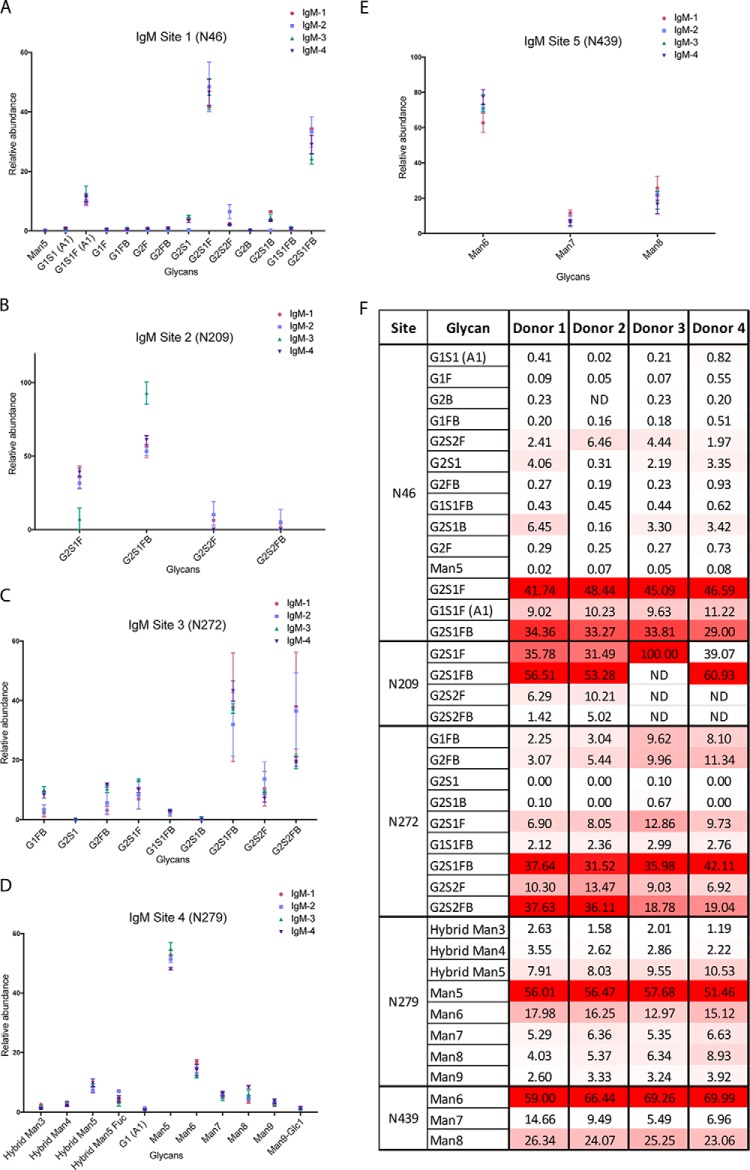
The IgM heavy chain has five N-linked glycosylation sites, located at N46, N209, N272, N279, and N439 (all enumerated starting from the N terminus of the heavy constant chain); it is known that the first three sites have high levels of complex glycans, whereas high-mannose N-glycans predominate on the last two sites (56–59). IgM samples from our set of donors (n = 4) were treated with trypsin and endoproteinase Glu-C, and these proteolytic digests were analyzed directly via nLC-MS/MS, in triplicate, to determine how site-specific N-glycosylation varied among the five heavy chain sites in the serum IgM of healthy individuals. At site N46, two of the three most abundant glycans were sialylated and fucosylated, but did not contain a bisecting N-acetylglucosamine (Fig. 6A, 6F, supplemental Fig. S10). This is in contrast to the N-glycans at site N209, where the most abundant glycoform has a composition consistent with that of a bisected N-glycan (Fig. 6B, supplemental Fig. S11), and site N272, where two of the top three most abundant glycans also have compositions consistent with those of bisected N-glycans (Fig. 6C, supplemental Fig. S12). At site N279, high-mannose glycans were among the most abundant forms observed, and at site N439, high mannose glycans were detected exclusively (Fig. 6D, 6E; supplemental Figs. S13–14). IgM heavy chain sites 1–4 are highly occupied (Table IV), whereas site 5 demonstrated much lower site occupancy. Indeed, our estimates suggest that only about one third of IgM molecules in serum are have N-glycan occupancy at site 5. This differs from the findings of ES Moh et al. (59), who analyzed glycans from pentameric and hexameric recombinant IgM, and found that the occupancy of IgM site 5 was ∼60%. This observed discrepancy may arise from the fact that their study was performed on recombinant IgM, whereas our findings are drawn from IgM isolated from human sera. Indeed, this highlights the need for our sensitive, high-throughput nLC-MS/MS approach, which can be applied to the analysis of patient-derived immunoglobulins. Of note, we detected IgM HC glycopeptides of variable lengths that contained N439 but differed by the presence/absence of the C-terminal tyrosine residue. Although this may be an experimental artifact because of nonspecific proteolysis, it is also possible that this arises from variable termination of the protein translation or in vivo C-terminal truncation. We also released IgM glycans using PNGase F and analyzed permethylated glycans via nLC-MS/MS to assess the glycan composition of bulk IgM glycans (see supplemental information). The glycan compositions observed via nLC-MS/MS are consistent with our glycopeptide analyses; the released glycans included high mannose and bisected N-linked glycan structures.
Fig. 6.
IgM Glycoforms at Sites N46, N209, N272, N279, and N439. A, IgM site N46 N-glycopeptide relative abundance. B, IgM site N209 N-glycopeptide relative abundance. C, IgM site N272 N-glycopeptide relative abundance. D, IgM site N279 N-glycopeptide relative abundance. E, IgM site N439 N-glycopeptide relative abundance. For all sites, abundances and standard deviation are shown for IgM from four healthy individuals, each run in triplicate. F, Heat map of IgM glycoforms. The heat map shows the N-glycosylation site, putative N-glycan, and average relative abundance (%) for each glycoform for donors 1–4. The heat map is scaled according to relative abundance for each donor sample, with the highest abundance glycoform at each site and within each sample colored red, the lowest colored white, and intermediate glycoforms colored on a linear intensity scale between the two extremes.
Table IV. Estimated site occupancy of IgM N-glycosylation sites N46, N209, N272, N279 and N439.
| Unmodified peptide area | Labeled D18O peptide area | % Occupied | Unmodified peptide area | Labeled D18O peptide area | % Occupied | ||
|---|---|---|---|---|---|---|---|
| IgM-N46 | IgM-N209 | ||||||
| Donor 1A | ND | 2.38E+08 | HIGH | Donor 1A | ND | 5.11E+07 | HIGH |
| Donor 1B | ND | 2.41E+08 | HIGH | Donor 1B | ND | 5.52E+07 | HIGH |
| Donor 1C | ND | 2.49E+08 | HIGH | Donor 1C | ND | 6.10E+07 | HIGH |
| Donor 2A | ND | 3.10E+08 | HIGH | Donor 2A | ND | 4.76E+07 | HIGH |
| Donor 2B | ND | 2.96E+08 | HIGH | Donor 2B | ND | 5.35E+07 | HIGH |
| Donor 2C | ND | 3.05E+08 | HIGH | Donor 2C | ND | 6.36E+07 | HIGH |
| Donor 3A | ND | 1.18E+08 | HIGH | Donor 3A | ND | 1.79E+07 | HIGH |
| Donor 3B | ND | 1.15E+08 | HIGH | Donor 3B | ND | 1.80E+07 | HIGH |
| Donor 3C | ND | 1.14E+08 | HIGH | Donor 3C | ND | 3.30E+07 | HIGH |
| Donor 4A | ND | 1.10E+08 | HIGH | Donor 4A | ND | ND | - |
| Donor 4B | ND | 9.49E+07 | HIGH | Donor 4B | ND | ND | - |
| Donor 4C | ND | 9.91E+07 | HIGH | Donor 4C | ND | 1.16E+05 | HIGH |
| IgM-N272 | IgM-N279 | ||||||
| Donor 1A | ND | 9.43E+06 | HIGH | Donor 1A | ND | 3.42E+09 | HIGH |
| Donor 1B | ND | 8.63E+06 | HIGH | Donor 1B | ND | 3.26E+09 | HIGH |
| Donor 1C | ND | 7.26E+06 | HIGH | Donor 1C | ND | 3.22E+09 | HIGH |
| Donor 2A | ND | 6.78E+06 | HIGH | Donor 2A | ND | 4.42E+09 | HIGH |
| Donor 2B | ND | 7.08E+06 | HIGH | Donor 2B | ND | 4.49E+09 | HIGH |
| Donor 2C | ND | 7.75E+06 | HIGH | Donor 2C | ND | 4.62E+09 | HIGH |
| Donor 3A | ND | 8.21E+06 | HIGH | Donor 3A | ND | 1.85E+09 | HIGH |
| Donor 3B | ND | 8.20E+06 | HIGH | Donor 3B | ND | 1.75E+09 | HIGH |
| Donor 3C | ND | 8.09E+06 | HIGH | Donor 3C | ND | 1.65E+09 | HIGH |
| Donor 4A | ND | 2.83E+06 | HIGH | Donor 4A | ND | 1.64E+09 | HIGH |
| Donor 4B | ND | 1.86E+06 | HIGH | Donor 4B | ND | 1.51E+09 | HIGH |
| Donor 4C | ND | 1.37E+06 | HIGH | Donor 4C | ND | 1.32E+09 | HIGH |
| IgM-N439 | |||||||
| Donor 1A | 1.85E+07 | 9.29E+06 | 33.39% | ||||
| Donor 1B | 1.65E+07 | 8.16E+06 | 33.09% | ||||
| Donor 1C | 1.70E+07 | 7.73E+06 | 31.21% | ||||
| Donor 2A | 2.81E+07 | 1.43E+07 | 33.70% | ||||
| Donor 2B | 2.68E+07 | 1.52E+07 | 36.13% | ||||
| Donor 2C | 2.82E+07 | 1.47E+07 | 34.31% | ||||
| Donor 3A | 3.36E+06 | 1.73E+06 | 34.06% | ||||
| Donor 3B | 2.95E+06 | 1.55E+06 | 34.46% | ||||
| Donor 3C | 3.40E+06 | 1.77E+06 | 34.26% | ||||
| Donor 4A | 7.37E+06 | 6.01E+06 | 44.91% | ||||
| Donor 4B | 7.50E+06 | 4.76E+06 | 38.84% | ||||
| Donor 4C | 6.34E+06 | 4.54E+06 | 41.73% |
Results of triplicate analyses are shown.
DISCUSSION
Over several decades, evidence has accumulated that immunoglobulin Fc N-glycosylation influences antibody:Fc receptor binding, with profound implications for the subsequent inflammatory and immune responses (8–14, 22–24, 60). Moreover, the N-glycosylation of immunoglobulins varies based on the exposure of individuals to specific pathogens (22, 35, 36), and geographical location (33), and this variation provides strong evidence that glycosylation is not random or stochastic but programmed in response to environmental stimuli. Therefore, the potential exists to design vaccines that maximize antibody function by teaching cells how to favor production of specific glycosylated forms. If this can be achieved, it will empower vaccination strategies for HIV and many other infectious diseases. However, achieving this vision requires a deeper understanding of how antibody N-glycosylation impacts function, which in turn requires a robust method to analyze antibody N-glycosylation.
N-glycosylation is a heterogeneous protein modification, and development of an understanding of which antibody glycoforms promote favorable immune responses is essential to the development of more effective vaccines. Although the role of IgG1 N-glycosylation in relation to antibody function is well documented, less is known about the N-glycosylation of IgG2, IgG3, IgG4, IgA1, IgA2, and IgM. Several groups have developed methods for surveying immunoglobulin Fc N-glycosylation (25, 26, 51, 56–59, 61–63). However, these methods frequently focus on a single antibody isotype, and, in several cases, the reports were based on the analysis of recombinant protein rather than antibodies isolated from human donors (51, 59, 62). We sought here to develop a complete nLC-MS/MS method for site-specific N-glycosylation analysis of the IgG, IgA and IgM isotypes and subclasses from the same set of human serum samples. Site-specific analysis of N-glycosylation, via the analysis of glycopeptides, is a particularly important feature of this effort, as IgA1, IgA2, and IgM each contain multiple N-glycosylation sites, and determination of the N-glycoform distribution associated with each site is critical to developing an understanding of the role of N-glycosylation in immunoglobulin function.
To our knowledge, this study represents the first examination of site-specific immunoglobulin N-glycosylation spanning multiple isotypes and subclasses from individuals within a single set of human serum samples. It was achieved without glycopeptide enrichment; this feature makes it more amenable to high-throughput use and avoids the potential for bias during enrichment. Site-specific glycosylation analysis is well suited for the analysis of immunoglobulins purified from biological samples, as it enables differentiation between glycans derived from the target protein versus those from co-isolated proteins. To emphasize this point, we remind the reader that, after purification, protein identity was verified by performing a protein sequence database search on each nLC-MS/MS data file, and the results demonstrated that, despite our best attempts to purify each immunoglobulin, nontarget proteins were still present. Nevertheless, because the N-glycans were still attached to definitive peptides, nLC-MS/MS enabled us to focus specifically on glycosylation of target proteins by ensuring that the glycopeptides of interest were derived from the target glycoprotein. This is a huge advantage over approaches that employ N-glycan release as the first step, followed by analysis (such as CE, MALDI-TOF MS, or LC-MS/MS) of native or derivatized glycan pools. Indeed, these alternative approaches likely err in reporting the abundance of certain glycoforms because of the presence of glycans released from non-target proteins. Similarly, this method has an advantage over CE of chromophore-tagged N-glycans, as demonstrated by the nLC-MS/MS separation and detection of multiple IgG1 glycoforms that co-eluted during CE (Fig. 2, 3). Of course, the method also distinguishes Fc from Fab glycans, which have different functional outcomes, specifically for IgAs and IgMs where no available enzyme successfully cleaves Fab from the Fc domain. Nevertheless, the reader should note that, compared with nLC-MS/MS, the CE results indicated higher levels of the low abundance disialyated species (Fig. 3A, 3B), and it has not yet been determined whether this is because of CE's higher-efficiency detection of the target glycans or the presence of co-precipitated glycoproteins in the CE-analyzed samples.
Site-specific N-glycosylation analysis can enable a greater understanding of how Fc glycosylation impacts function. General trends observed in this study included the detection of differences in the abundances of specific glycoforms in IgG1 versus IgG4, in which we found higher levels of the fucosylated, bisected N-glycan G1FB in IgG1, and lower levels of the sialylated and fucosylated glycoform G1S1F. In the multimeric immunoglobulins IgA1, IgA2 and IgM, we were able to discern clear differences in glycoform types and abundances between glycosylation sites, which can serve as the basis for functional studies. For example, there are higher levels of complex, fucosylated N-linked glycans on IgA1 site N340 compared with site N144. In IgA2, a similar pattern emerged, in which sites N47 and N205 contained mostly fucosylated N-glycans, whereas site N131 (which shares a common sequence with IgA1 site N144) has a profile very similar to the equivalent IgA1 site, with higher relative abundances of non-fucosylated glycoforms. We also detected multiple allotypes of IgA2, one with a unique N-glycosylation sequon. Glycoproteomic methods are particularly well suited to understanding changes in glycosylation between sequence variants, as both the glycan and sequence information are preserved. Finally, in IgM we found that sites N46, N209, and N272 have mostly complex glycoforms, whereas sites N279 and N439 display primarily high-mannose N-linked glycans, highlighting the complex mechanism by which glycosylation machinery works, in addition to antibody-specific glycosylation.
The development of this nLC-MS/MS workflow using stepped normalized collision energy enables production of (glyco)peptide backbone fragments and glycan fragments that aid in the interpretation of peptide sequence and glycan topology that are essential for site-specific characterization of immunoglobulin N-glycosylation. The results presented here show differences in immunoglobulin N-glycosylation between well studied and less accessible isotypes and subclasses, and we were able to gauge donor-to-donor variability, and therefore distinguish changes in Ig isotype, subclass glycosylation independent of donor-to-donor variability. It is essential to understand immunoglobulin N-glycosylation from human serum, as this gives us the most relevant readout of the glycosylation.
The motivation for this work is to elucidate the glycoform patterns of circulating immunoglobulins and facilitate studies of N-glycan structure-function relationships in immunoglobulin subclasses, with the goal that antibody N-linked glycosylation can be controlled via vaccination to produce responsive, functional (favorably glycosylated) immunoglobulins. The data presented herein demonstrate that nLC-MS/MS has the analytical capability to define site-specific glycoform distributions of individual Ig classes derived from human serum.
DATA AVAILABILITY
Peptide, glycopeptide and formerly-N-glycosylated peptide mass spectrometry data have been deposited to the ProteomeXchange Consortium (64) via the PRIDE (65) partner repository with the dataset identifier (project ID) PXD010911 and (URL) 10.6019/PXD010911.
Supplementary Material
Footnotes
* This research is supported by NIH grants F32 CA196157 (to KBC), NIH grant 2R37AI080289 (to GA) and P41 GM104603, S10 OD010724 and S10 OD021651 (to CEC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental Figures.
This article contains supplemental Figures.
1 The abbreviations used are:
- ADCC
- antibody-dependent cellular cytotoxicity
- ACD
- acid citrate dextrose solution
- AGC
- automatic gain control
- ADCP
- antibody-dependent cellular phagocytosis
- ADNP
- antibody-dependent neutrophil phagocytosis
- ADCD
- antibody-dependent and complement deposition
- APTS
- 8-aminopyrene-1,3,6-trisulfonic acid
- CE
- Capillary electrophoresis
- DHB
- 2,5-dihydroxybenzoic acid
- DTT
- dithiothreitol
- EIC
- extracted ion chromatogram
- ER
- endoplasmic reticulum
- Fc
- fragment crystallizable
- FcR
- fc-receptor
- IgA
- immunoglobulin A
- IgE
- immunoglobulin E
- IgG
- immunoglobulin G
- IgM
- immunoglobulin M
- NCE
- normalized collision energy
- nLC-MS/MS
- nano-liquid chromatography–tandem mass spectrometry
- PNGase F
- peptide N-glycosidase F.
REFERENCES
- 1. Nimmerjahn F., and Ravetch J. V. (2008) Fcgamma receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad R., Sindhu S. T., Toma E., Morisset R., Vincelette J., Menezes J., and Ahmad A. (2001) Evidence for a correlation between antibody-dependent cellular cytotoxicity-mediating anti-HIV-1 antibodies and prognostic predictors of HIV infection. J. Clin. Immunol. 21, 227–233 [DOI] [PubMed] [Google Scholar]
- 3. Baum L. L. (2010) Role of humoral immunity in host defense against HIV. Curr. HIV/AIDS Rep. 7, 11–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bournazos S., Klein F., Pietzsch J., Seaman M. S., Nussenzweig M. C., and Ravetch J. V. (2014) Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmaljohn A., and Lewis G. K. (2016) Cell-targeting antibodies in immunity to Ebola. Pathog. Dis. 74, ftw021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lu L. L., Chung A. W., Rosebrock T. R., Ghebremichael M., Yu W. H., Grace P. S., Schoen M. K., Tafesse F., Martin C., Leung V., Mahan A. E., Sips M., Kumar M. P., Tedesco J., Robinson H., Tkachenko E., Draghi M., Freedberg K. J., Streeck H., Suscovich T. J., Lauffenburger D. A., Restrepo B. I., Day C., Fortune S. M., and Alter G. (2016) A functional role for antibodies in tuberculosis. Cell 167, 433–443.e414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Plebani A., Ugazio A. G., Avanzini M. A., Massimi P., Zonta L., Monafo V., and Burgio G. R. (1989) Serum IgG subclass concentrations in healthy subjects at different age: age normal percentile charts. Eur. J. Pediatr. 149, 164–167 [DOI] [PubMed] [Google Scholar]
- 8. de Jong S. E., Selman M. H., Adegnika A. A., Amoah A. S., van Riet E., Kruize Y. C., Raynes J. G., Rodriguez A., Boakye D., von Mutius E., Knulst A. C., Genuneit J., Cooper P. J., Hokke C. H., Wuhrer M., and Yazdanbakhsh M. (2016) IgG1 Fc N-glycan galactosylation as a biomarker for immune activation. Sci. Rep. 6, 28207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forthal D. N., Gach J. S., Landucci G., Jez J., Strasser R., Kunert R., and Steinkellner H. (2010) Fc-glycosylation influences Fcγ receptor binding and cell-mediated anti-HIV activity of monoclonal antibody 2G12. J. Immunol. 185, 6876–6882 [DOI] [PubMed] [Google Scholar]
- 10. Kaneko Y., Nimmerjahn F., and Ravetch J. V. (2006) Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. Science 313, 670–673 [DOI] [PubMed] [Google Scholar]
- 11. Moore J. S., Wu X., Kulhavy R., Tomana M., Novak J., Moldoveanu Z., Brown R., Goepfert P. A., and Mestecky J. (2005) Increased levels of galactose-deficient IgG in sera of HIV-1-infected individuals. AIDS 19, 381–389 [DOI] [PubMed] [Google Scholar]
- 12. Parekh R. B., Dwek R. A., Sutton B. J., Fernandes D. L., Leung A., Stanworth D., Rademacher T. W., Mizuochi T., Taniguchi T., and Matsuta K. (1985) Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 316, 452–457 [DOI] [PubMed] [Google Scholar]
- 13. Okazaki A., Shoji-Hosaka E., Nakamura K., Wakitani M., Uchida K., Kakita S., Tsumoto K., Kumagai I., and Shitara K. (2004) Fucose depletion from human IgG1 oligosaccharide enhances binding enthalpy and association rate between IgG1 and FcgammaRIIIa. J. Mol. Biol. 336, 1239–1249 [DOI] [PubMed] [Google Scholar]
- 14. Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., and Shitara K. (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 [DOI] [PubMed] [Google Scholar]
- 15. Yamane-Ohnuki N., Kinoshita S., Inoue-Urakubo M., Kusunoki M., Iida S., Nakano R., Wakitani M., Niwa R., Sakurada M., Uchida K., Shitara K., and Satoh M. (2004) Establishment of FUT8 knockout Chinese hamster ovary cells: an ideal host cell line for producing completely defucosylated antibodies with enhanced antibody-dependent cellular cytotoxicity. Biotechnol. Bioeng. 87, 614–622 [DOI] [PubMed] [Google Scholar]
- 16. Davies J., Jiang L., Pan L. Z., LaBarre M. J., Anderson D., and Reff M. (2001) Expression of GnTIII in a recombinant anti-CD20 CHO production cell line: Expression of antibodies with altered glycoforms leads to an increase in ADCC through higher affinity for FC gamma RIII. Biotechnol. Bioeng. 74, 288–294 [PubMed] [Google Scholar]
- 17. Kurimoto A., Kitazume S., Kizuka Y., Nakajima K., Oka R., Fujinawa R., Korekane H., Yamaguchi Y., Wada Y., and Taniguchi N. (2014) The absence of core fucose up-regulates GnT-III and Wnt target genes: a possible mechanism for an adaptive response in terms of glycan function. J. Biol. Chem. 289, 11704–11714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nishima W., Miyashita N., Yamaguchi Y., Sugita Y., and Re S. (2012) Effect of bisecting GlcNAc and core fucosylation on conformational properties of biantennary complex-type N-glycans in solution. J. Phys. Chem. B 116, 8504–8512 [DOI] [PubMed] [Google Scholar]
- 19. Anthony R. M., Wermeling F., Karlsson M. C., and Ravetch J. V. (2008) Identification of a receptor required for the anti-inflammatory activity of IVIG. Proc. Natl. Acad. Sci. U.S.A. 105, 19571–19578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nimmerjahn F., and Ravetch J. V. (2007) The antiinflammatory activity of IgG: the intravenous IgG paradox. J. Exp. Med. 204, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pagan J. D., Kitaoka M., and Anthony R. M. (2018) Engineered sialylation of pathogenic antibodies in vivo attenuates autoimmune disease. Cell 172, 564–577.e513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ackerman M. E., Crispin M., Yu X., Baruah K., Boesch A. W., Harvey D. J., Dugast A. S., Heizen E. L., Ercan A., Choi I., Streeck H., Nigrovic P. A., Bailey-Kellogg C., Scanlan C., and Alter G. (2013) Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J. Clin. Invest. 123, 2183–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Parekh R. B., Roitt I. M., Isenberg D. A., Dwek R. A., Ansell B. M., and Rademacher T. W. (1988) Galactosylation of IgG associated oligosaccharides: reduction in patients with adult and juvenile onset rheumatoid arthritis and relation to disease activity. Lancet 1, 966–969 [DOI] [PubMed] [Google Scholar]
- 24. Rademacher T. W., Parekh R. B., Dwek R. A., Isenberg D., Rook G., Axford J. S., and Roitt I. (1988) The role of IgG glycoforms in the pathogenesis of rheumatoid arthritis. Springer Semin. Immunopathol. 10, 231–249 [DOI] [PubMed] [Google Scholar]
- 25. Huffman J. E., Pucic-Bakovic M., Klaric L., Hennig R., Selman M. H., Vuckovic F., Novokmet M., Kristic J., Borowiak M., Muth T., Polasek O., Razdorov G., Gornik O., Plomp R., Theodoratou E., Wright A. F., Rudan I., Hayward C., Campbell H., Deelder A. M., Reichl U., Aulchenko Y. S., Rapp E., Wuhrer M., and Lauc G. (2014) Comparative performance of four methods for high-throughput glycosylation analysis of immunoglobulin G in genetic and epidemiological research. Mol. Cell Proteomics 13, 1598–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wuhrer M., Stam J. C., van de Geijn F. E., Koeleman C. A., Verrips C. T., Dolhain R. J., Hokke C. H., and Deelder A. M. (2007) Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 7, 4070–4081 [DOI] [PubMed] [Google Scholar]
- 27. Reichert J. M., Rosensweig C. J., Faden L. B., and Dewitz M. C. (2005) Monoclonal antibody successes in the clinic. Nat. Biotechnol. 23, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 28. Sondermann P., and Szymkowski D. E. (2016) Harnessing Fc receptor biology in the design of therapeutic antibodies. Curr. Opin. Immunol. 40, 78–87 [DOI] [PubMed] [Google Scholar]
- 29. Goritzer K., Maresch D., Altmann F., Obinger C., and Strasser R. (2017) Exploring site-specific N-glycosylation of HEK293 and plant-produced human IgA isotypes. J. Proteome Res. 16, 2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wada Y., Dell A., Haslam S. M., Tissot B., Canis K., Azadi P., Backstrom M., Costello C. E., Hansson G. C., Hiki Y., Ishihara M., Ito H., Kakehi K., Karlsson N., Hayes C. E., Kato K., Kawasaki N., Khoo K. H., Kobayashi K., Kolarich D., Kondo A., Lebrilla C., Nakano M., Narimatsu H., Novak J., Novotny M. V., Ohno E., Packer N. H., Palaima E., Renfrow M. B., Tajiri M., Thomsson K. A., Yagi H., Yu S. Y., and Taniguchi N. (2010) Comparison of methods for profiling O-glycosylation: Human Proteome Organisation Human Disease Glycomics/Proteome Initiative multi-institutional study of IgA1. Mol. Cell Proteomics 9, 719–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Czajkowsky D. M., and Shao Z. (2009) The human IgM pentamer is a mushroom-shaped molecule with a flexural bias. Proc. Natl. Acad. Sci. U.S.A. 106, 14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ehrenstein M. R., and Notley C. A. (2010) The importance of natural IgM: scavenger, protector and regulator. Nat. Rev. Immunol. 10, 778–786 [DOI] [PubMed] [Google Scholar]
- 33. Pucić M., Knezević A., Vidic J., Adamczyk B., Novokmet M., Polasek O., Gornik O., Supraha-Goreta S., Wormald M. R., Redzić I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., Wuhrer M., Rudd P. M., Josić D., and Lauc G. (2011) High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell Proteomics 10, M111.010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mahan A. E., Jennewein M. F., Suscovich T., Dionne K., Tedesco J., Chung A. W., Streeck H., Pau M., Schuitemaker H., Francis D., Fast P., Laufer D., Walker B. D., Baden L., Barouch D. H., and Alter G. (2016) Antigen-specific antibody glycosylation is regulated via vaccination. PLoS Pathog. 12, e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ackerman M. E., Mikhailova A., Brown E. P., Dowell K. G., Walker B. D., Bailey-Kellogg C., Suscovich T. J., and Alter G. (2016) Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PLoS Pathog. 12, e1005315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chung A. W., Ghebremichael M., Robinson H., Brown E., Choi I., Lane S., Dugast A. S., Schoen M. K., Rolland M., Suscovich T. J., Mahan A. E., Liao L., Streeck H., Andrews C., Rerks-Ngarm S., Nitayaphan S., de Souza M. S., Kaewkungwal J., Pitisuttithum P., Francis D., Michael N. L., Kim J. H., Bailey-Kellogg C., Ackerman M. E., and Alter G. (2014) Polyfunctional Fc-effector profiles mediated by IgG subclass selection distinguish RV144 and VAX003 vaccines. Sci. Transl. Med. 6, 228ra238. [DOI] [PubMed] [Google Scholar]
- 37. Mehta N., and Alter G. (2017) Opportunities to exploit antibody glycosylation in vaccination. Future Virol. 12, 325–328 [Google Scholar]
- 38. Sitia R., Rubartelli A., and Hämmerling U. (1984) The role of glycosylation in secretion and membrane expression of immunoglobulins M and A. Mol. Immunol. 21, 709–719 [DOI] [PubMed] [Google Scholar]
- 39. Reusch D., Haberger M., Falck D., Peter B., Maier B., Gassner J., Hook M., Wagner K., Bonnington L., Bulau P., and Wuhrer M. (2015) Comparison of methods for the analysis of therapeutic immunoglobulin G Fc-glycosylation profiles-Part 2: Mass spectrometric methods. MAbs 7, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Trbojevic-Akmacic I., Vilaj M., and Lauc G. (2016) High-throughput analysis of immunoglobulin G glycosylation. Expert Rev. Proteomics 13, 523–534 [DOI] [PubMed] [Google Scholar]
- 41. Falck D., Jansen B. C., de Haan N., and Wuhrer M. (2017) High-throughput analysis of IgG Fc glycopeptides by LC-MS. Methods Mol. Biol. 1503, 31–47 [DOI] [PubMed] [Google Scholar]
- 42. O'Flaherty R., Trbojevic-Akmacic I., Greville G., Rudd P. M., and Lauc G. (2018) The sweet spot for biologics: recent advances in characterization of biotherapeutic glycoproteins. Expert Rev. Proteomics 15, 13–29 [DOI] [PubMed] [Google Scholar]
- 43. Wada Y., Azadi P., Costello C. E., Dell A., Dwek R. A., Geyer H., Geyer R., Kakehi K., Karlsson N. G., Kato K., Kawasaki N., Khoo K. H., Kim S., Kondo A., Lattova E., Mechref Y., Miyoshi E., Nakamura K., Narimatsu H., Novotny M. V., Packer N. H., Perreault H., Peter-Katalinic J., Pohlentz G., Reinhold V. N., Rudd P. M., Suzuki A., and Taniguchi N. (2007) Comparison of the methods for profiling glycoprotein glycans–HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology 17, 411–422 [DOI] [PubMed] [Google Scholar]
- 44. Khatri K., Staples G. O., Leymarie N., Leon D. R., Turiak L., Huang Y., Yip S., Hu H., Heckendorf C. F., and Zaia J. (2014) Confident assignment of site-specific glycosylation in complex glycoproteins in a single step. J. Proteome Res. 13, 4347–4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang H., Yang C., and Sun T. (2018) Characterization of glycopeptides using a stepped higher-energy C-trap dissociation approach on a hybrid quadrupole orbitrap. Rapid Commun. Mass Spectrom 32, 1353–1362 [DOI] [PubMed] [Google Scholar]
- 46. Bollineni R. C., Koehler C. J., Gislefoss R. E., Anonsen J. H., and Thiede B. (2018) Large-scale intact glycopeptide identification by Mascot database search. Sci. Rep. 8, 2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., and Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ciucanu I. K., F. (1984) A Simple and Rapid Method for the Permethylation of Carbohydrates. Carbohydr. Res. 131, 209–217 [Google Scholar]
- 49. Ciucanu I., and Costello C. E. (2003) Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J. Am. Chem. Soc. 125, 16213–16219 [DOI] [PubMed] [Google Scholar]
- 50. Struwe W. B., Agravat S., Aoki-Kinoshita K. F., Campbell M. P., Costello C. E., Dell A., Ten F., Haslam S. M., Karlsson N. G., Khoo K. H., Kolarich D., Liu Y., McBride R., Novotny M. V., Packer N. H., Paulson J. C., Rapp E., Ranzinger R., Rudd P. M., Smith D. F., Tiemeyer M., Wells L., York W. S., Zaia J., and Kettner C. (2016) The minimum information required for a glycomics experiment (MIRAGE) project: sample preparation guidelines for reliable reporting of glycomics datasets. Glycobiology 26, 907–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yoo E. M., Yu L. J., Wims L. A., Goldberg D., and Morrison S. L. (2010) Differences in N-glycan structures found on recombinant IgA1 and IgA2 produced in murine myeloma and CHO cell lines. MAbs 2, 320–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chintalacharuvu K. R., Raines M., and Morrison S. L. (1994) Divergence of human alpha-chain constant region gene sequences. A novel recombinant alpha 2 gene. J. Immunol. 152, 5299–5304 [PubMed] [Google Scholar]
- 53. Ramachandran P., Boontheung P., Xie Y., Sondej M., Wong D. T., and Loo J. A. (2006) Identification of N-linked glycoproteins in human saliva by glycoprotein capture and mass spectrometry. J. Proteome Res. 5, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 54. Picariello G., Ferranti P., Mamone G., Roepstorff P., and Addeo F. (2008) Identification of N-linked glycoproteins in human milk by hydrophilic interaction liquid chromatography and mass spectrometry. Proteomics 8, 3833–3847 [DOI] [PubMed] [Google Scholar]
- 55. Jia W., Lu Z., Fu Y., Wang H. P., Wang L. H., Chi H., Yuan Z. F., Zheng Z. B., Song L. N., Han H. H., Liang Y. M., Wang J. L., Cai Y., Zhang Y. K., Deng Y. L., Ying W. T., He S. M., and Qian X. H. (2009) A strategy for precise and large scale identification of core fucosylated glycoproteins. Mol. Cell Proteomics 8, 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chapman A., and Kornfeld R. (1979) Structure of the high mannose oligosaccharides of a human IgM myeloma protein. II. The minor oligosaccharides of high mannose glycopeptide. J. Biol. Chem. 254, 824–828 [PubMed] [Google Scholar]
- 57. Chapman A., and Kornfeld R. (1979) Structure of the high mannose oligosaccharides of a human IgM myeloma protein. I. The major oligosaccharides of the two high mannose glycopeptides. J. Biol. Chem. 254, 816–823 [PubMed] [Google Scholar]
- 58. Arnold J. N., Wormald M. R., Suter D. M., Radcliffe C. M., Harvey D. J., Dwek R. A., Rudd P. M., and Sim R. B. (2005) Human serum IgM glycosylation: identification of glycoforms that can bind to mannan-binding lectin. J. Biol. Chem. 280, 29080–29087 [DOI] [PubMed] [Google Scholar]
- 59. Moh E. S., Lin C. H., Thaysen-Andersen M., and Packer N. H. (2016) Site-specific N-glycosylation of recombinant pentameric and hexameric human IgM. J. Am. Soc. Mass. Spectrom 27, 1143–1155 [DOI] [PubMed] [Google Scholar]
- 60. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., and Dwek R. A. (2001) Glycosylation and the immune system. Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 61. Wormald M. R., Rudd P. M., Harvey D. J., Chang S. C., Scragg I. G., and Dwek R. A. (1997) Variations in oligosaccharide-protein interactions in immunoglobulin G determine the site-specific glycosylation profiles and modulate the dynamic motion of the Fc oligosaccharides. Biochemistry 36, 1370–1380 [DOI] [PubMed] [Google Scholar]
- 62. Mattu T. S., Pleass R. J., Willis A. C., Kilian M., Wormald M. R., Lellouch A. C., Rudd P. M., Woof J. M., and Dwek R. A. (1998) The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcalpha receptor interactions. J. Biol. Chem. 273, 2260–2272 [DOI] [PubMed] [Google Scholar]
- 63. Royle L., Roos A., Harvey D. J., Wormald M. R., van Gijlswijk-Janssen D., Redwan el R. M., Wilson I. A., Daha M. R., Dwek R. A., and Rudd P. M. (2003) Secretory IgA N- and O-glycans provide a link between the innate and adaptive immune systems. J. Biol. Chem. 278, 20140–20153 [DOI] [PubMed] [Google Scholar]
- 64. Deutsch E. W., Csordas A., Sun Z., Jarnuczak A., Perez-Riverol Y., Ternent T., Campbell D. S., Bernal-Llinares M., Okuda S., Kawano S., Moritz R. L., Carver J. J., Wang M., Ishihama Y., Bandeira N., Hermjakob H., and Vizcaino J. A. (2017) The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 45, D1100–D1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vizcaino J. A., Csordas A., del-Toro N., Dianes J. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Peptide, glycopeptide and formerly-N-glycosylated peptide mass spectrometry data have been deposited to the ProteomeXchange Consortium (64) via the PRIDE (65) partner repository with the dataset identifier (project ID) PXD010911 and (URL) 10.6019/PXD010911.