Abstract
Deficiency in polycystin 1 triggers specific changes in energy metabolism. To determine whether defects in other human cystoproteins have similar effects, we studied extracellular acidification and glucose metabolism in human embryonic kidney (HEK-293) cell lines with polycystic kidney and hepatic disease 1 (PKHD1) and polycystic kidney disease (PKD) 2 (PKD2) truncating defects along multiple sites of truncating mutations found in patients with autosomal recessive and dominant PKDs. While neither the PKHD1 or PKD2 gene mutations nor their position enhanced cell proliferation rate in our cell line models, truncating mutations in these genes progressively increased overall extracellular acidification over time (P < 0.001 for PKHD1 and PKD2 mutations). PKHD1 mutations increased nonglycolytic acidification rate (1.19 vs. 1.03, P = 0.002), consistent with an increase in tricarboxylic acid cycle activity or breakdown of intracellular glycogen. In addition, they increased basal and ATP-linked oxygen consumption rates [7.59 vs. 5.42 (P = 0.015) and 4.55 vs. 2.98 (P = 0.004)]. The PKHD1 and PKD2 mutations also altered mitochondrial morphology, resembling the effects of polycystin 1 deficiency. Together, these data suggest that defects in major PKD genes trigger changes in mitochondrial energy metabolism. After validation in in vivo models, these initial observations would indicate potential benefits of targeting energy metabolism in the treatment of PKDs.
Keywords: Cas9 nucleases, CRISPR, ECAR, gene editing, gene targeting, OCR
INTRODUCTION
Polycystic kidney diseases (PKDs) are the most common inherited causes of end-stage renal disease (ESRD). Autosomal dominant PKD [ADPKD; Mendelian Inheritance in Man (MIM) nos. 173900 and 613095] leads to ESRD in most patients in their 50s. However, autosomal recessive PKD (ARPKD; MIM no. 263200) is a major cause of ESRD in children. Currently, most PKD patients can only be offered supportive care, which, unfortunately, has a limited impact on the onset of ESRD (21).
ADPKD is caused by mutations in PKD1 and PKD2 genes (3a, 14, 25, 26). PKD1 encodes polycystin 1 (PC-1), and PKD2 encodes polycystin 2 (PC-2). Among the most prominent consequences of PC-1 deficiency are changes in energy metabolism (13), including lower fatty acid oxidation (12), and, probably, increased glycolysis (18). Increased glycolysis in PC-1-deficient cells (18) was not observed by all investigators (12, 29) but is likely involved, since inhibition of glycolysis with 2-deoxyglucose (2-DG) ameliorated cystic kidney disease progression in two Pkd1-deficient mouse models (18). Translation of this therapeutic strategy to ADPKD patients is in progress (23). Similar to the effects of 2-DG, renal cystic disease progression in Pkd1-deficient mouse models was inhibited by commonly used hypoglycemic drugs [e.g., metformin and pioglitazone (15, 22)], and this therapeutic strategy is currently being explored in clinical trials (ClinicalTrials.gov NCT02656017 and NCT02697617).
Inhibition of glycolysis also altered renal cystic disease progression in other models (e.g., Han-SPRD Cy rat, Ift8-deficient mouse, and Pkhd1pck rat) (3, 17, 19). These data suggest that cellular bioenergetics are functionally relevant to additional forms of renal cystic disease. Therefore, we speculated that, similar to the effect of PC-1 deficiency, important changes in bioenergetics may also be triggered by defects in the other two major human PKD proteins, PC-2 and fibrocystin/polyductin, a protein encoded by the principal ARPKD gene polycystic kidney and hepatic disease 1 (PKHD1) (16, 28). As our initial step to address this question, we studied the in vitro effects of PKHD1 and PKD2 gene disruption along sites of common truncating mutations found in patients with ADPKD and ARPKD (Table 1). To assess the effects on bioenergetics, we evaluated the extracellular acidification rate (ECAR), as well as the mitochondrial respiration rate [reflected by oxygen consumption rate (OCR)] and cell proliferation.
Table 1.
Relevant PKHD1 and PKD2 truncating mutations
| Mutation (cDNA) | Number of Affected Families | Mutation (protein) | Positions of Engineered Protein Truncations in HEK-293 Cells |
|---|---|---|---|
| PKHD1* | |||
| c.126delT | 2 | F42fs | 36 |
| c.711_714del | 4 | M238fs | 221 |
| c.3761_3762delCCinsG | 26 | A1254fs | 1256 |
| c.9689delA | 24 | D3230fs | 3226 |
| PKD2** | |||
| c.958C>T | 14 | R320X | 321 |
| c.2224C>T | 6 | R742X | 746 |
| c.2407C>T | 8 | R803X | 804 |
| c.2419C>T | 13 | R807X | 805 and 808 |
PKHD1, polycystic kidney and hepatic disease 1 gene; PKD2, polycystic kidney disease 2 gene; HEK-293, human embryonic kidney cell line.
Based on the Mutation Database Autosomal Recessive Polycystic Kidney Disease.
Based on the Autosomal Dominant Polycystic Kidney Disease Database.
Our study design included six individual clones of cells with engineered protein truncations near sites of four different known PKHD1 truncating mutations and compared them with eight randomly selected wild-type (WT) clones with similar growth characteristics but without the engineered mutations. Our studies of clones with PKD 2 (PKD2) truncating mutations mimicked our approach for evaluation of PKHD1 truncating mutation effects.
To isolate cells with the desired engineered mutations, we chose to propagate these cells from a single clone. We found that this was technically challenging in the case of cell lines that express the typical characteristics of renal epithelial cells. However, we were able to clone human embryonic kidney (HEK-293) cells; therefore, we performed our studies in this cell line model, which has been extensively utilized in drug development as well as characterization of human ADPKD and ARPKD cystoproteins (4, 9, 31).
MATERIALS AND METHODS
Generation of cell line clones with PKHD1 and PKD2 truncating mutations.
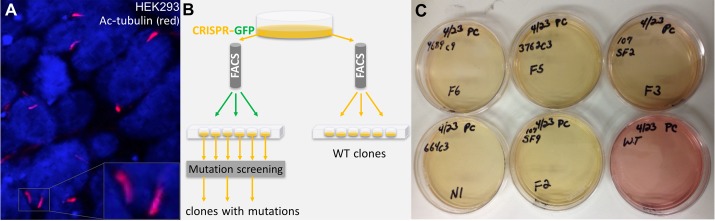
The PKHD1 truncating mutations were selected from the Mutation Database Autosomal Recessive Polycystic Kidney Disease (http://www.humgen.rwth-aachen.de) (1, 2), and the PKD2 truncating mutations were selected from the Autosomal Dominant Polycystic Kidney Disease Database (http://pkdb.mayo.edu) (5) (Table 1). Using CRISPR/Cas9 technology, we targeted sites of these truncating mutations in PKHD1 and PKD2 genes. Specifically, lyophilized synthetic oligonucleotides (Table 2) were reconstituted into stock solutions, and complementary oligonucleotide pairs were annealed in a thermocycler. The annealed oligonucleotides were ligated to the BbsI-linearized plasmid vector pX458 [plasmid pSpCas9(BB)-2A-GFP (PX458) 48138, Addgene, Cambridge MA] at 16°C for 12–15 h. The ligation reaction was used to transform Escherichia coli DH5a cells, plated on a Luria broth-ampicillin plate, and incubated overnight at 37°C. Multiple colonies from each bacterial plate were marked and selected for colony PCR to test for successful ligation of the annealed oligonucleotides into the plasmid vector. After the desired sequence of these plasmids was confirmed by Sanger sequencing, these plasmids were used for transfection of HEK-293 cells (American Type Culture Collection, Manassas, VA), the only clonable cell line that we were able to identify that survived an individual cell-derived clone propagation and did not lose the ability to express primary cilia (Fig. 1A).
Table 2.
Oligonucleotides used for PKHD1 and PKD2 targeting
| Mutation (cDNA) | CRISPR gRNA Sequence |
|---|---|
| PKHD1 | |
| c.126delT | 3′-GTAGCCTTGCAGGGGGAACGTGG-5′ |
| c.711_714del | 3′-CATGTGGAAGGCGACTACATCGG-5′ |
| c.1854delA | 5′-TGAAGGACACAATCATCTTCAGG-3′ |
| c.3761_3762delCCinsG | 5′-TGAAGGACACAATCATCTTCAGG-3′ |
| c.9689delA | 5′-GCATCGGGTATCTGGGGGGCTGG-3′ |
| PKD2 | |
| c.958C>T | 5′-ACTCGGAGTTGCCGTATTCGTGG-3′ |
| c.2224C>T | 3′-GAACTTCGACAAGATCTCAAAGG-5′ |
| c.2407C>T | 5′-CAGAGTCATCCAGGCTTCGAGGG-3′ |
| c.2419C>T | 5′-CAGAGTCATCCAGGCTTCGAGGG-3′ |
Protospacer adjacent motifs are underlined. PKHD1, polycystic kidney and hepatic disease 1 gene; PKD2, polycystic kidney disease 2 gene; gRNA, guide RNA.
Fig. 1.
CRISPR/Cas9-based targeting of human embryonic kidney (HEK-293) cells. A: representative immunofluorescent micrograph shows HEK-293 cells with primary cilia [acetylated (Ac) tubulin: red]; nuclei are stained with DAPI (blue). B: CRISPR/Cas9 targeting of truncating mutations in polycystic kidney and hepatic disease 1 (PKHD1) and polycystic kidney disease 2 (PKD2) genes of the HEK-293 cells. After transfection with CRISPR/Cas9 plasmids (see materials and methods), clones were sorted by green fluorescent protein (GFP) to obtain single-cell clones. Once confluent, these cells were used for genomic DNA isolation and PCR amplification of PKHD1 or PKD2 target sequences to define clones with the sequence modifications at target PKHD1 and PKD2 sites. FACS, fluorescein-activated cell sorting. C: color of culture medium pH indicator (phenol red) changes more rapidly in clones of HEK-293 cells that carry homozygous PKHD1 truncating mutations (yellow medium) than in wild-type (WT) cells (red-pink medium). A similar, although less prominent, change was observed in clones with PKD2 truncating mutations.
Transfection complex mixtures for each plasmid were prepared by addition of 2 µg of plasmid DNA into 100 µl of DMEM by vortex and 8 µl of LipoD293 (SignaGen Laboratories, Rockville, MD) into 100 µl of DMEM by vortex, gentle mixing, and incubation at room temperature for 15 min. HEK-293 cells were trypsinized and counted to obtain individual pellets containing 1.2 ×106 cells. Pellets were resuspended in 200 µl of specific transfection complex and incubated at 37°C for 15 min. Thereafter, 2 ml of complete culture medium were added, and the cell suspension was placed into a single well of a six-well plate. Cultures were maintained for 12–18 h, and then complete medium was replaced. The presence of green fluorescent protein (GFP)-positive cells was determined 24–48 h posttransfection. Cells were sorted by GFP to obtain single-cell clones 3–5 days posttransfection.
As the initial step for cell sorting and subsequent cloning, cells were trypsinized, counted, and diluted to ~0.5 × 106/ml for sorting. Sorting was performed using a cell sorter (FACSAria, BD Biosciences, Franklin Lakes, NJ) (Fig. 1B). To isolate single-cell clones, one cell per well was placed into a 96-well plate. WT clones were sorted without fluorescent markers, while all CRISPR/Cas9-edited clones were sorted on the basis of positive GFP determination. Postsorting clones were maintained under normal culture conditions and observed for colony formation. Once wells were near confluence, cells were trypsinized and transferred to six-well plates. Confluent wells in the six-well plates were trypsinized; half were used for continued culture, and half were used for genomic DNA isolation and downstream PCR amplification of PKHD1 or PKD2 target sequences.
We generated 145 clones from the PKHD1 targeting (40 from nonhomologous end-joining transfections and 105 from homologous-driven repair) and 116 clones from the PKD2 targeting (all from homologous-driven repair). Among these clones, we identified those with truncating mutations near sites of known human truncating mutations (Table 1). We used these clones as a model to study the effects of truncating mutations across PKHD1 and PKD2 genes.
Cell culture.
The HEK-293 cells were cultured in DMEM (Invitrogen/Thermo Fisher Scientific, Carlsbad, CA) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone, Logan, UT) and 1× penicillin-streptomycin (Pen-Strep, Gibco/Thermo Fisher Scientific, Waltham, MA). Cells were routinely split using 0.25% trypsin-EDTA (Invitrogen). Cultures were maintained in a humidified incubator at 37°C and 5% CO2. For all assays, six individual replicates of each mutant and WT clones were plated in random order to avoid positional bias.
Proliferation assay.
Cell proliferation assays were performed by incubation of cells with 0.5 mg/ml thiazolyl blue tetrazolium bromide (MTT; Sigma, St. Louis, MO). Cells were placed in 96-well plates at 2 × 104 cells/well. At 48 h postplating, the culture medium was removed, and 100 µl of MTT-containing medium were added per well. Cells were incubated for 90 min; then MTT solution was removed, and cells were washed twice with PBS. DMSO (100 µl) was added to each well, and absorbance at 570 nm was measured. Cell proliferation was also analyzed using the bromodeoxyuridine (BrdU) cell proliferation ELISA kit (Abcam, Cambridge, UK) at 24 and 48 h after addition of BrdU to the cell cultures. Absorbance measurements for the MTT and BrdU assays were performed using a Synergy HT plate reader (BioTek, Winooski, VT).
pH assays.
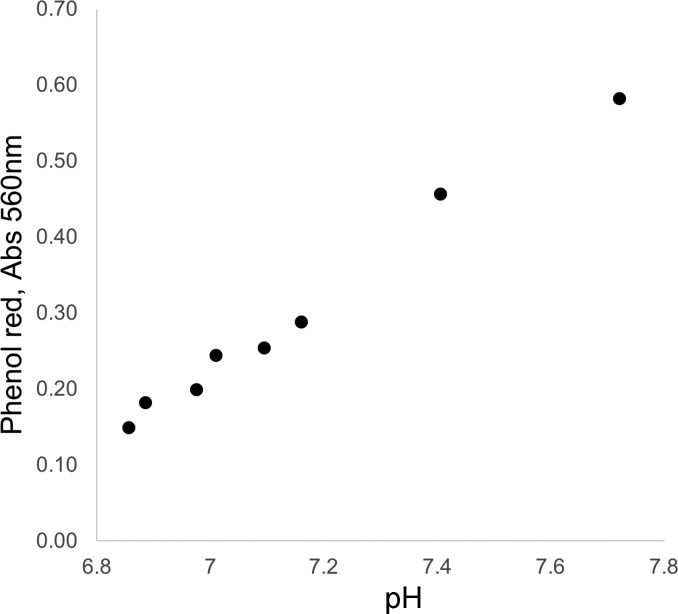
Cells were trypsinized, counted, and plated, 6 wells per clone, in 96-well plates at 1 × 104 or 2 × 104 cells/well on day 0. The cells were maintained in a humidified incubator at 37°C and 5% CO2. Each well contained 200 µl of complete DMEM (Invitrogen) containing 39.85 µM phenolsulfonphthalein (phenol red), a pH indicator widely used in research and industry. Starting on day 3 (D3) postplating, absorbance at 560 nm (Abs560, the absorbance maximum for phenol red) for each well was measured daily through day 8 (D8) using a BioTek Synergy HT plate reader. All data are reported as Abs560; the formula pH = (Abs560 + 3.328)/0.507 can be used for conversion of the Abs560 data to pH (also see the Abs560 vs. pH plot in Fig. 2).
Fig. 2.
Standard curve plot: phenol red absorbance at 560 nm (Abs560) vs. pH. Based on a linear-curve fit, pH can be calculated as follows: pH = (Abs560 + 3.328)/0.507.
Measurement of glycolytic function effects on ECAR and OCR.
Glycolytic metabolism of the HEK-293 cell clones was determined using an extracellular flux analyzer (model XFe96, Agilent Technologies, Santa Clara, CA) and the glycolytic stress test, as described by Kramer et al. (7). The extracellular flux analyzer simultaneously measures real-time ECAR, an indicator of glycolysis, and OCR, an indicator or mitochondrial function.
Specifically, the mutant and WT HEK-293 clones were grown in XFe96 plates for 24 h in DMEM to form uniform monolayers (2 × 104 cells/well). The culture medium was replaced by the XF assay medium without glucose and pyruvate 1 h before the glycolytic stress test. Initially, the ECAR and OCR components were measured in basal XF medium, and then 5 mM glucose, 1 µg/ml oligomycin (an inhibitor of ATP synthase), and 100 mM 2-DG (an inhibitor of hexokinase) were injected sequentially, with two measurements after each injection. The duration of the assay was <90 min, and cell viability remained >90% over the course of the assay. At the end of the assay period, the cells were gently washed using PBS (200 µl), lysed using radioimmunoprecipitation assay lysis buffer, and assayed for protein by the DC (Lowry) protein assay. All OCR- and ECAR-related measurements were normalized to the protein content in each well. The ECAR-to-OCR ratio was determined following oligomycin injection. Also, we used ECAR values after addition of glucose as an indicator of basal glycolytic rate, ECAR after subsequent addition of oligomycin as an indicator of maximal glycolytic capacity, the difference between maximal glycolysis capacity and basal glycolytic rate as glycolytic reserve, and the baseline ECAR after addition of 2-DG as an indicator of nonglycolytic acidification. The two most significant proton sources that contribute to nonglycolytic acidification, the tricarboxylic acid (TCA) cycle and breakdown of intracellular glycogen, were not evaluated in this study. In addition, the difference between basal OCR and OCR after addition of oligomycin was used as an indicator of ATP-linked OCR, addition of FCCP was used to determine maximal OCR, the difference between maximal and basal OCR was used as an indicator of OCR reserve capacity, and OCR after addition of antimycin A was used as an indicator of nonmitochondrial OCR (6).
Mass spectrometry analyses of TCA (Krebs) cycle metabolites.
Studies of mutated and WT HEK-293 cell lines were performed with ~106-cell aliquots. Specifically, we analyzed triplicates of four individual mutated clones and one triplicate of pooled WT HEK-293 clones (without PKD gene mutation). After a brief 1× ice-cold PBS wash, cells were extracted using 1.0 ml of HPLC-grade ice-cold methanol, scraped on ice, and placed on an orbital rotor for 30 min at 4°C. Thereafter, lysates were transferred and centrifuged at 1,000 g at 4°C for 20 min. The supernatant was stored at −80°C for subsequent analyses.
Standards were generated as a master mix of all compounds at 100 μg/ml in H2O and serially diluted to 10 times the final concentrations (0.05–10 μg/ml, 9 standards). Standards were further diluted to 1× in methanol to a total volume of 1 ml and dried under a gentle stream of N2. For cell extracts, 1 ml of each was transferred to a glass tube and dried under a gentle stream of N2. Standards and samples were resuspended in 50 µl of 5% acetic acid and vortexed for 15 s. Amplifex keto reagent (50 µl; SCIEX, Concord, ON, Canada) was added to each sample and allowed to react for 1 h at room temperature. Standards and samples were then dried under a gentle stream of N2 and resuspended in 1 ml and 200 µl of 0.1% formic acid, respectively.
Samples were analyzed by liquid chromatography (LC)-multiple reaction ion-monitoring mass spectrometry (MS). LC was performed using the LC20AC HPLC system (Shimadzu, Columbia, MD) with a Synergi Hydro-RP 4-µm 80-Å 250 × 2-mm ID column (Phenomenex, Torrance, CA). Mobile phases were as follows: 0.1% formic acid (phase A) and methanol-0.1% formic acid (phase B). Compounds were eluted using a 5–40% linear gradient of phase B from 1 to 7 min, a column wash of 40–100% phase B from 7 to 10 min, and reequilibration at 5% phase B from 10.5 to 15 min. Column eluant was passed into an electrospray ionization interface of an API 4000 triple-quadrupole MS (SCIEX). The following mass transitions were monitored in the positive ion mode: m/z 261/118 for α-ketoglutarate, m/z 247/144 for oxaloacetate, and m/z 204/144, 204/118, and 204/60 for pyruvate. In the negative mode, the following transitions are monitored: m/z 115/71 for fumarate, m/z 89/43 for lactate, m/z 117/73 for succinate, m/z 133/115 for malate, m/z 173/85 for cis-aconitate, m/z 191/87 for citrate, m/z 191/73 for isocitrate, m/z 147/129 for 2-hydroxyglutarate, m/z 146/102 for glutamate, m/z 145/42 for glutamine, and m/z 132/88 for aspartate. The 16 transitions were each monitored for 35 ms, with a total cycle time of 560 ms. MS parameters were as follows: collisionally activated dissociation (CAD) 4, curtain gas (CUR) 15, ion source gas 1 (GS1) 60, ion source gas 2 (GS2) 30, temperature of ion source (TEM) 600, and ionization voltage (IS) −3,500 V for negative-polarity mode and 4,500 V for positive-polarity mode. MultiQuant software (version 3.0.1, SCIEX) was used to compare peak areas of metabolites in the sample extracts with those in the known standards to calculate metabolite concentrations.
Characterization of mitochondrial morphology.
We used transmission electron microscopy and two clones with the highest culture medium acidification rate among WT clones (WT clones 2 and 3; see Fig. 6B, left and Fig. 9B, left), two clones with PKHD1 truncating mutations (PKHD1 c.106delA and c.662dupA; see Fig. 6B, clones 221 and 36), and two clones with PKHD2 truncating mutations (PKD2 c.2415_2420delCCTCG and c.2422_2423insA; see Fig. 9B, clones 805 and 808) to characterize the effects of PKHD1 and PKD2 mutations on mitochondrial morphology. Confluent cells were fixed with a modified Karnovsky’s fixative (2.5% electron microscopy-grade glutaraldehyde in 0.1 M phosphate buffer), rinsed several times with PBS, postfixed in 1% osmium tetroxide in cacodylate buffer for 1 h, and rinsed with PBS for 1 h. Then the samples were dehydrated through a series of graded ethyl alcohols from 50 to 100% (50%, 70%, and 95% for 10 min each, followed by 3 changes of 100% for 15 min). After dehydration, the infiltration process included two changes of 100% propylene oxide for 15 min each and was followed by a 50:50 mixture of propylene oxide and the embedding resin (Embed 812, Electron Microscopy Sciences, Fort Washington, PA) for 12–18 h. The samples were transferred to fresh 100% embedding medium twice for ≥1 h each time. The tissue was then embedded in a fresh change of 100% embedding medium and incubated for 12–18 h in the oven at 60°C for polymerization. Thereafter, a diamond histology knife and an ultramicrotome were used to cut the blocks into thick (1- to 2-μm) sections, which were stained with Toluidine blue and used as a reference to trim blocks for thin sectioning. The appropriate blocks were then cut into thin sections using a diamond knife (Diatome, Electron Microscopy Sciences) at 70–100 nm (silver to pale gold using color interference). These sections were mounted on copper grids and stained with 2% aqueous uranyl acetate and lead citrate (Leica). Ultrathin sections were examined on a Tecnai Spirit 120-kV transmission electron microscope (FEI, Hillsboro, OR), and digital micrographs were captured with a charge-coupled-device camera (Advanced Microscopy Techniques, Woburn, MA).
Fig. 6.
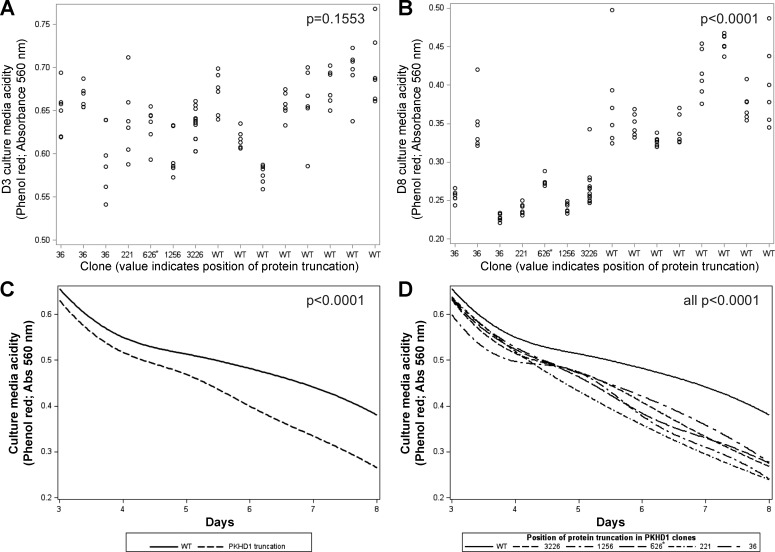
Truncating mutations in the polycystic kidney and hepatic disease 1 (PKHD1) gene increase cell culture medium acidification. A: at day 3 (D3), PKHD1 gene mutations had no significant effect on cell culture medium acidity, reflected by absorbance at 560 nm (Abs560, maximum for phenol red; P = 0.1553). B: by day 8 (D8), the effect of PKHD1 mutation on increase in cell culture medium acidity, reflected by Abs560, was significant (P < 0.0001). C: increase in culture medium acidity was greater overall in cell clones with PKHD1 truncating mutations over time (reflected by decrease in Abs560, P < 0.0001) when 6 values from D3 to D8 were used for trajectory analyses. D: each of the PKHD1 truncating mutations showed a greater increase in acidity over time than WT (all P < 0.0001). The 626* label designates the PKHD1 p.K626_M627del clone that was not included in these statistical analyses; however, when it was included, results of the above analyses were nearly identical.
Fig. 9.
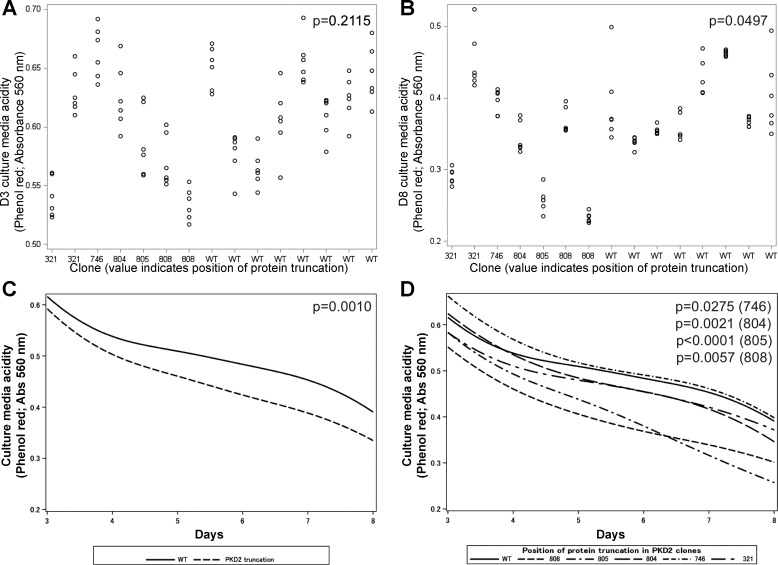
Truncating mutations in the polycystic kidney disease 2 (PKD2) gene increase cell culture medium acidification. A: at day 3 (D3), PKD2 gene mutations had no significant effect on cell culture medium acidity, reflected by absorbance at 560 (Abs560, maximum for phenol red; P = 0.2115). B: by day 8 (D8), the effect of PKHD1 mutation on increase in acidity of cell culture medium, reflected by Abs560, was significant (P = 0.0497). C: over time, increase in culture medium acidity was greater overall in cell clones with PKD2 truncating mutations (reflected by decrease in Abs560; P = 0.0010) when 6 values from D3 to D8 were used for trajectory analyses. D: with the exception of p.I321Y*, each of the PKD2 truncating mutations showed a significantly greater increase in acidity over time than wild-type (WT).
To analyze mitochondrial morphology, we evaluated the following established mitochondrial shape descriptors: surface area (mitochondrial size, μm2); perimeter (μm); aspect ratio [computed as (major mitochondria axis)/(minor mitochondria axis), reflecting length-to-width ratio); form factor [computed as (perimeter2)/(4π·surface area), reflecting complexity and branching aspect of mitochondria]; and circularity [computed as 4π(surface area/perimeter2)], which together with roundness [computed as 4(surface area)/(π·major axis2)] represents two-dimensional indexes of sphericity (values of 1 indicate perfect spheroids). These analyses were obtained blindly using ImageJ (version 1.48, National Institutes of Health, Bethesda, MD), and the analyses were replicated independently by two individuals proficient in the use of this software for quantitative image data analyses.
Statistical analyses.
Statistical evaluations were performed with the SAS 9.3 statistical software package (SAS Institute, Cary, NC). For each outcome, we first determined the proportion of total variability accounted for by differences between clones (vs. between replicates within clones) using the intraclass correlation coefficient. As expected, these analyses indicated high similarity among replicates within each clone (i.e., most variability was due to between-clone differences), violating the assumption of independence of standard statistical tests. Thus, hierarchical linear models were used to appropriately account for the nested structure of the data (30), with replicates modeled as nested within clones. The effects of truncation were tested with two analyses, one comparing WT with all truncations combined (an overall truncation effect) and the other testing differences among all individual truncations. In the latter, truncation was modeled as a categorical variable with five levels for PKHD1 (4 different truncations and WT) and six levels for PKD2 (5 different truncations and WT). A significant omnibus test was followed by specific contrasts comparing each truncation with WT. In addition to these models of individual outcomes, we also evaluated trajectories of culture medium acidity between D3 and D8 using a three-level hierarchical model (days nested within replicates, which were nested within clones) (20). These models simultaneously tested truncation effects on baseline (D3) acidity and its change over time (linear slope; estimates reflect daily change between D3 and D8). For mitochondrial morphology phenotypes, the intraclass correlation coefficients were low, indicating independence of replicates within each clone. Therefore, these phenotypes were analyzed using general linear models.
RESULTS
HEK-293 clones with PKHD1 and PKD2 mutations.
After confirming that HEK-293 cells express primary cilia (Fig. 1A), we used this cell line to generate multiple clones carrying four different truncations of PKHD1 at four sites corresponding to common ARPKD truncating mutations (Table 1, Fig. 3). For subsequent analyses, we used those clones that were homozygous for the generated genetic defect, with the exception of a clone with two different truncating mutations (p.L3226Yfs*6 and p.L3226Cfs*6) at position 3226. In the case of PKD2, we engineered clones with five different truncations across four mutation sites (Table 1, Fig. 4). Untransfected HEK-293 cells were cloned similarly to HEK-293 cells carrying the PKHD1 and PKD2 mutations (Fig. 1B). Eight of these randomly selected clones with growth characteristics comparable to the PKHD1 and PKD2 mutant clones were used as WT controls.
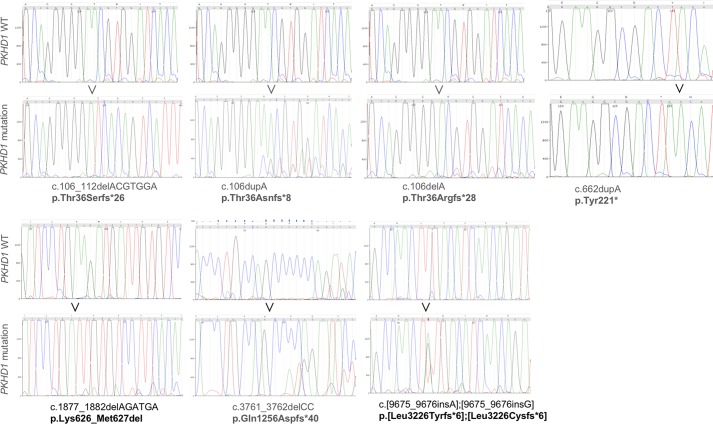
Fig. 3.
Targeted regions of the polycystic kidney and hepatic disease 1 (PKHD1) gene in individual clonal human embryonic kidney (HEK-293) cell lines. Individual panels show sequences of targeted PKHD1 regions from wild-type (WT) and mutant cell lines; labels show mutation description in the PKHD1 cDNA and resulting protein defects.
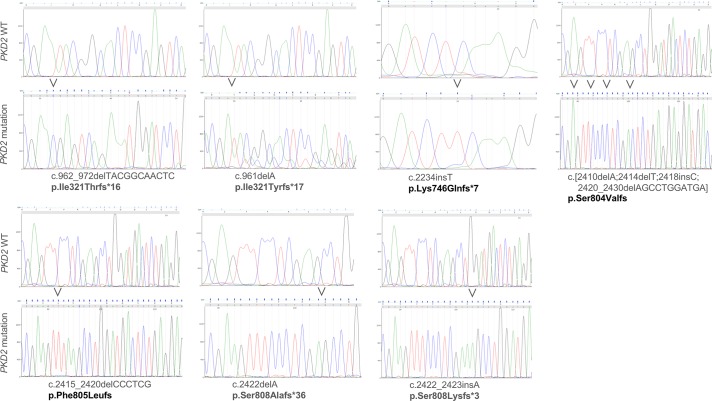
Fig. 4.
Targeted regions of the polycystic kidney disease 2 (PKD2) gene in individual clonal human embryonic kidney (HEK-293) cell lines. Individual panels show sequences of targeted PKD2 regions from wild-type (WT) and mutant cell lines; labels show mutation description in the PKHD1 cDNA and resulting protein defects. In the case of p.Ser804Valfs and p.Phe805Leufs, frameshifts extend beyond the end of targeted exons.
Truncating mutations in PKHD1 or PKD2 genes do not alter proliferation of HEK-293 cells.
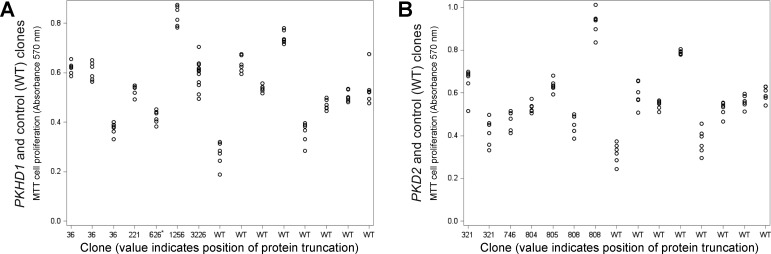
The variability in MTT proliferation assay data between individual clones was high (93%) compared with variability within each clone (7%; Fig. 5A). The PKHD1 gene mutations had no overall effect on outcomes of the 48-h MTT proliferation assay (vs. WT: b = 0.09, P = 0.2499).
Fig. 5.
Truncating mutations in polycystic kidney and hepatic disease 1 (PKHD1) or polycystic kidney disease 2 (PKD2) genes have no major effect on human embryonic kidney (HEK-293) cell proliferation. A: PKHD1 gene truncations had no overall effect on proliferation rate estimated by thiazolyl blue tetrazolium bromide (MTT) proliferation assay [vs. wild-type (WT): b = 0.09, P = 0.2499]. The 626* label designates the PKHD1 p.K626_M627del clone that was not included in these statistical analyses; however, when it was included, results were nearly identical. B: PKD2 gene mutations had no overall effect on MTT-based proliferation rates (vs. WT: b = 0.02, P = 0.5452).
For PKD2 gene mutations, as expected, the basal MTT-dependent signal varied between individual clones, consistent with differences in the metabolic pathways leading to MTT reduction (Fig. 5B). Similar to PKHD1 results, the PKD2 gene mutations had no overall effect on MTT-based proliferation rates (vs. WT: b = 0.02, P = 0.5452). Also, proliferation estimated by MTT did not differ between WT and specific PKD2 truncation positions [F(5,80) = 0.60, P = 0.7013].
The outcomes of BrdU analyses resembled those based on the MTT assay. The variability in BrdU proliferation assay data between individual clones was high (39%) compared with the variability within each clone (61%). The PKHD1 gene mutations had no overall effect on the outcomes of the BrdU proliferation data (vs. WT: b = 0.04, P = 0.2947 for 24 h and b = −0.12, P = 0.14 for 48 h). The outcomes of BrdU analyses for PKD2 gene mutations also showed no overall effect on BrdU-based proliferation rates at 24 h (vs. WT: b = −0.04, P = 0.2865). At 48 h, PKD2 mutations were associated with lower BrdU proliferation (vs. WT: b = −0.19, P = 0.0203). The BrdU proliferation estimates did not reach statistical significance for comparisons between WT and specific PKD2 truncation positions [F(5,32) = 2.49, P = 0.0513].
Together, the MTT and BrdU assay data demonstrate that neither truncating mutations in PKHD1 nor truncating mutations in PKD2 were associated with increased proliferation of HEK-293 cells.
Truncating mutations in the PKHD1 gene increase cell culture medium acidification.
We studied medium acidification daily between D3 and D8 after aliquots of 104 cells were plated in six replicates from each clone (Table 1). Each of these studies was replicated using 2 × 104 plated cells with similar outcomes.
At D3, the variability in acidity was moderate between (60%), as well as within (40%), individual clones (Fig. 6A). The PKHD1 gene mutations had no significant effect on cell culture medium acidity at D3, reflected by Abs560 (vs. WT: b = −0.030, P = 0.1553). Similarly, there was no effect of PKHD1 truncation position on medium acidity at D3 [F(4,76) = 0.83, P = 0.5110].
At D8, variability in acidity was greater between (86%) than within (14%) individual clones (Fig. 6B). By D8, the PKHD1 gene mutations significantly increased cell culture medium acidity, reflected by Abs560 (vs. WT: b = −0.117, P < 0.0001). The effect of specific PKHD1 truncating mutations on medium acidification was also significant [F(4,76) = 8.23, P < 0.0001], with each truncation showing greater acidity than WT (b = −0.102 to −0.141, all P < 0.0010). Adjustment of D8 Abs560 data for the D3 baseline yielded identical results (b = −0.091 to −0.130, all P < 0.0017). These results point to the equivalent effect of different PKHD1 truncations on cell culture medium acidification in this model.
When the Abs560 data for each of the 6 days between D3 and D8 were used for trajectory analyses, the trajectories of clones with PKHD1 gene mutations did not differ from WT at D3 (b = −0.017, P = 0.2807) but showed a greater increase in cell culture medium acidity over time (i.e., steeper decline in Abs560 values) (per day: b = −0.020, P < 0.0001; Fig. 6C); Abs560 and pH are in a near-linear relationship in the studied range (Fig. 2). Likewise, specific PKHD1 truncating mutations did not differ from WT at D3 (b = −0.011 to −0.036, all P > 0.2331), but each showed a greater increase in acidity over time (per day: b = −0.018 to −0.029, all P < 0.0001; Fig. 6D). Replication of all the above analyses with 2 × 104 plated cells yielded similar results (data not shown).
Truncating mutations in the PKHD1 gene increase ECAR and OCR.
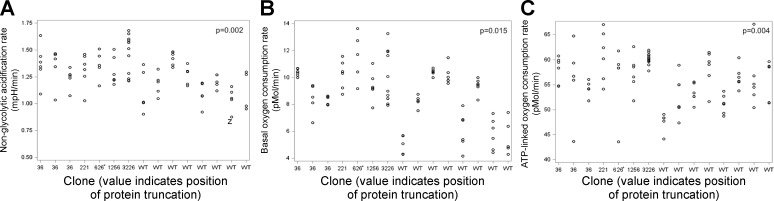
The variability in ECAR was moderately high between individual clones (75%) compared with ECAR variability within each clone (25%). Overall, PKHD1 gene mutations increased ECAR (vs. WT: b = 4.825, P = 0.0293). However, there was no effect of individual PKHD1 truncating mutations [F(4,71) = 1.67, P = 0.1668], suggesting that the differences between each truncation and WT were relatively small. The PKHD1 mutations increased the nonglycolytic acidification rate (1.19 vs. 1.03, P = 0.002), as indicated by ECAR after addition of 2-DG (Fig. 7A). A possible explanation for the PKHD1 mutation-induced increase in ECAR is increased glycolytic flux and TCA cycle activity [consistent with the metabolomics data (see below) and the increased cell culture medium acidification] or breakdown of intracellular glycogen.
Fig. 7.
Truncating mutations in the polycystic kidney and hepatic disease 1 (PKHD1) gene increase extracellular acidification rate (ECAR) and oxygen consumption rate (OCR). A: PKHD1 truncating mutations increased nonglycolytic acidification rate in human embryonic kidney (HEK-293) cells (P = 0.002). B: PKHD1 truncating mutations increased basal OCR (P = 0.015). C: PKHD1 truncating mutations increased ATP-linked OCR (P = 0.004).
Variability in OCR, an indicator of mitochondrial respiration, was also high between individual clones (80%) compared with OCR variability within clones (20%). Together, there was a trend for the PKHD1 gene mutations to increase OCR (vs. WT: b = 31.866, P = 0.0648). However, there was no effect of individual PKHD1 truncating mutations [F(4,71) = 1.21, P = 0.3129], indicating that the differences between each truncation and WT were relatively small. The PKHD1 mutations increased basal OCR (7.59 vs. 5.42, P = 0.015; Fig. 7B) and ATP-linked OCR (the difference between the basal OCR and OCR after addition of oligomycin; 4.55 vs. 2.98, P = 0.004; Fig. 7C). Neither maximal OCR, OCR reserve capacity, nor nonmitochondrial OCR was significantly altered by the PKHD1 mutations.
In contrast to OCR and ECAR alone, the variability in the OCR-to-ECAR ratio was low between individual clones (3%) but very high within clones (97%). The PKHD1 gene mutations did not affect the OCR-to-ECAR ratio (vs. WT: b = −0.3988, P = 0.2948). There was also no effect of individual PKHD1 truncating mutations [F(4,71) = 0.53, P = 0.7174]. (While ECAR is a robust indicator of glycolysis in most cell types when highly aerobic cells are stressed, CO2 production from the mitochondria can contribute to ECAR, and the change in the OCR-to-ECAR ratio is a marker of susceptibility to this effect.)
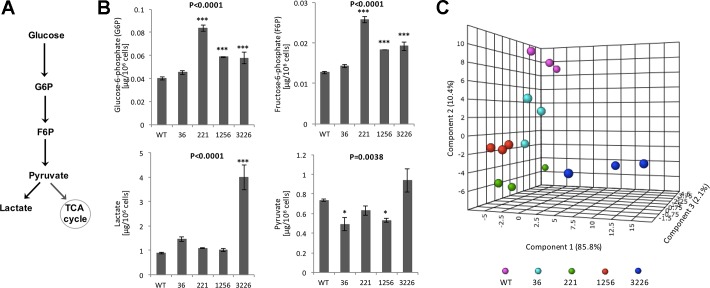
Together, the ECAR and OCR data point to “stressed,” more energetic metabolism in HEK-293 cell clones that carry PKHD1 truncating mutations. This concept was confirmed by the MS-based assessment of glycolysis and TCA (Krebs) cycle metabolites. Compared with a pool of WT clones, PKHD1 gene mutations increased levels of glucose 6-phosphate [F(4,10) = 38.96, P < 0.0001], fructose 6-phospate [F(4,10) = 66.72, P < 0.0001], and lactate [F(4,10) = 32.16, P < 0.0001]; pyruvate levels were decreased overall [F(4,10) = 7.92, P = 0.0038] (Fig. 8). The effect of the PKHD1 gene mutations on TCA cycle metabolites was negligible (data not shown); perhaps this effect is specific to the HEK-293 cell line.
Fig. 8.
Truncating mutations in the polycystic kidney and hepatic disease 1 (PKHD1) gene increase glycolysis. A: metabolic pathway of glycolysis converts glucose to pyruvate via a series of intermediate metabolites. Its initial components are glucose 6-phosphate (G6P) and fructose 6-phosphate (F6P); pyruvate and lactate are the end products of glycolysis. TCA, tricarboxylic acid cycle. B: liquid chromatography-multiple reaction ion-monitoring mass spectrometry was used to determine key metabolites of the glycolytic pathway. Compared with a pool of wild-type (WT) human embryonic kidney (HEK-293) clones, PKHD1 gene mutations increased levels of G6P [F(4,10) = 38.96, P < 0.0001], F6P [F(4,10) = 66.72, P < 0.0001], and lactate [F(4,10) = 32.16, P < 0.0001]; overall, pyruvate levels were decreased [F(4,10) = 7.92, P = 0.0038]. C: principal component loading plot of components 1, 2, and 3 extracted from factor analysis of the individual glycolytic components. The cluster representing biological replicates of pooled wild-type (WT) HEK-293 clones is distinctly separated from clusters of HEK-293 cells with PKHD1 truncating mutations.
Amino acid deletion at the site of PKHD1 missense mutations alters extracellular acidification and glucose metabolism similar to truncating mutations.
In addition to the clones with PKHD1 truncating mutations, we also generated a PKHD1 p.K626_M627del clone (c.1877_1882delAGATGA; Fig. 2) with a two-amino acid deletion at the site of human missense mutations p.K626R and p.M627K (1, 2). Since this clone contains a nontruncating mutation, we discuss it separately from the truncating mutations. Nonetheless, we studied it together with truncating mutations and noted that its extracellular acidification and glucose metabolism characteristics followed the same pattern as clones with PKHD1 truncating mutations (Figs. 5–7). The outcomes of statistical analyses with and without this p.K626-M627del clone were nearly identical.
Truncating mutations in the PKD2 gene increase cell culture medium acidification.
The study design for evaluation of PKD2 truncation effects on cell culture acidification mimicked the above-described approach used for characterization of PKHD1 truncation effects.
At D3, the variability in acidity was greater among (76%) than within (24%) individual clones (Fig. 9A). The PKD2 gene mutations had no significant overall effect on cell culture medium acidity at D3, reflected by Abs560 (vs. WT: b = −0.012, P = 0.2115). However, there was a significant effect of specific PKD2 truncation positions on medium acidity at D3 [F(5,80) = 2.44, P = 0.0176]. Examination of specific clones showed that only the truncation at protein position 808 differed from WT, showing greater acidity (b = −0.064, P = 0.0047).
At D8, the majority of variability in acidity was between (85%), rather than within (15%), individual clones (Fig. 9B). The PKD2 gene mutations had an overall effect on cell culture medium acidity, reflected by Abs560 (vs. WT: b = −0.028, P = 0.0497). There was also a significant effect of specific PKD2 truncation positions on medium acidity at D8 [F(5,80) = 2.49, P = 0.0379]. Examination of specific clones showed that truncation at protein positions 805 and 808 differed from WT, showing greater acidity (b = −0.1325, P = 0.0084 and b = −0.895, P = 0.0159). Adjustment of D8 Abs560 data for the D3 baseline yielded identical results for the 805 clone.
When the Abs560 data for each of the 6 days between D3 and D8 were used for trajectory analyses, the trajectories of clones with PKD2 gene mutations did not differ from WT at D3 (b = −0.0295, P = 0.1128) but showed a greater increase in cell culture medium acidity over time (i.e., a steeper decline in Abs560 values; per day: b = −0.0074, P = 0.0010; Fig. 9C). Analysis of the effects of specific PKD2 truncating mutations showed differences at D3 [F(5,384) = 3.41, P = 0.0050] and in slope over time [F(5,384) = 12.34, P < 0.0001]. Specific contrasts revealed that, at D3, only truncation at position 808 differed from WT, showing greater acidity (b = −0.077, P = 0.0005). Across D3–D8, the increase in acidity over time was greater in clones with four of the five truncations than WT: 746 (b = −0.007, P = 0.0275), 804 (b = −0.010, P = 0.0021), 805 (b = −0.023, P < 0.0001), and 808 (b = −0.007, P = 0.0057) (Fig. 9D). Replication with 2 × 104 cells yielded no overall truncation effects on baseline or slope of acidity (P = 0.1792 and P = 0.4035). For analyses of specific mutations, acidity was greater in truncation 808 than WT (b = −0.095, P = 0.0014) and the increase in acidity was steeper over time in truncation 805 than WT (b = −0.080, P = 0.0224).
These data also point to the value of studying trajectories (vs. single end points). In the case of PKD2 mutations, their effect on culture medium acidification was not significant at D3 or just passed the significance threshold of P < 0.05 at D8 (Fig. 9, A and B). However, the significance of the PKD2 mutation effects substantially increased when the effects of mutations on individual clones were obtained at multiple time points during the D3–D8 interval that were analyzed as a trajectory (Fig. 9, A and B).
Effect of truncating mutations in the PKD2 gene on ECAR and OCR in HEK-293 cells did not reach statistical significance.
The variability in ECAR was moderately high between individual clones (71%) compared with ECAR variability within each clone (29%). There were no differences in ECAR overall between PKD2 gene mutations and WT (b = 1.140, P = 0.1864) or for specific truncating mutations [F(5,74) = 1.64, P = 0.1591]. The studied PKD2 mutations did not significantly alter glycolysis rate, glycolytic capacity, glycolytic reserve, or nonglycolytic acidification rate.
The variability in OCR was also high between individual clones (83%) compared with OCR variability within clones (17%). There were no differences between PKD2 gene mutations and WT in OCR overall (b = 11.194, P = 0.0848) or for specific truncating mutations [F(5,74) = 1.60, P = 0.1700].
In contrast to OCR and ECAR alone, variability in the OCR-to-ECAR ratio was similar between (56%) and within (44%) individual clones. The PKD2 gene mutations did not affect the OCR-to-ECAR ratio (vs. WT: b = −0.132, P = 0.5262). There was also no effect of individual PKD2 truncating mutations [F(5,74) = 1.08, P = 0.3773]. The studied PKD2 mutations did not significantly alter basal OCR, ATP-linked OCR, maximal OCR, OCR reserve capacity, or nonmitochondrial OCR.
Truncating mutations in PKHD1 or PKD2 genes alter mitochondrial morphology in HEK-293 cells.
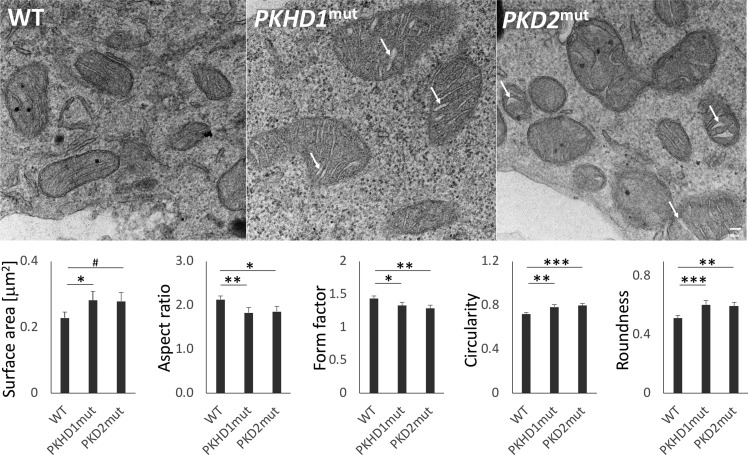
The effect of PKHD1 and PKD2 truncating mutations on mitochondrial morphology was assessed using transmission electron microscopy in two clones with the highest culture medium acidification rate among clones in each studied group (WT clones and clones with truncating mutations in PKHD1 and PKD2). Mitochondria from clones with truncating mutations in PKHD1 and PKD2 expressed a more rounded phenotype, with dilated cristae, than typically thin tubular cristae of WT mitochondria (Fig. 10).
Fig. 10.
Truncating mutations in polycystic kidney and hepatic disease 1 (PKHD1) and polycystic kidney disease 2 (PKD2) genes alter mitochondrial morphology. Top: representative transmission electron microscopy images of mitochondria from wild-type (WT) human embryonic kidney (HEK-293) cell line clones (n = 80 mitochondria) show thin tubular cristate. In contrast, mitochondria from PKHD1-deficient (PKHD1mut, n = 71) and PKD2-deficient (PKD2mut, n = 65) HEK-293 cell line clones express a more rounded swollen-like phenotype with cristae abnormalities including cristae dilatation (arrows). Scale bar = 100 nm. Bottom: aspect ratio, form factor, circularity, and roundness calculations were based on values obtained in μm. #P < 0.1, *P < 0.05, **P < 0.01, ***P < 0.001.
Specifically, the PKHD1 mutations were associated with increased size (reflected in surface area; vs. WT: b = 0.0536, P = 0.0452), decreased aspect ratio (b = −0.3004, P = 0.0087) and form factor (b = −0.1093, P = 0.0307), and increased circularity (b = 0.0657, P = 0.0026) and roundness (b = 0.0914, P = 0.0009). Similarly, the PKD2 mutations were associated with decreased aspect ratio (vs. WT: b = −0.2747, P = 0.0190) and form factor (b = −0.1508, P = 0.0038) and increased circularity (b = 0.0780, P = 0.0005) and roundness (b = 0.0817, P = 0.0037). The trend toward increased mitochondria size in clones with PKD2 mutations did not reach statistical significance (b = 0.0507, P = 0.0648). Together, these data support a model in which PKHD1 and PKD2 genes regulate mitochondrial morphology.
DISCUSSION
Increased epithelial cell proliferation is one of the hallmark phenotypes observed in kidneys from PKD patients and multiple PKD models (reviewed in Refs. 27, 32). Therefore, it is surprising that our current study did not demonstrate a significant effect of PKHD1 or PKD2 truncating mutations on cell proliferation. Our data suggest that the degree of metabolic changes exceeds the effects of these mutations on cell proliferation. Alternatively, it is possible that PKHD1 or PKD2 mutations alone have a minimal direct effect on proliferation of otherwise unaltered cells and that the previously observed enhanced proliferation of epithelial cells in PKD kidneys is triggered by other processes or cells, such as those involved in kidney development, function, and repair (e.g., paracrine regulation, cell-matrix interaction, and responses to injury or changes in flow). The latter hypothesis is supported by focal distribution of frequent cell clusters of proliferation and apoptosis in epithelial cysts (8). Finally, it is possible that PKHD1 or PKD2 mutations can stimulate proliferation; however, this effect is negligible in cells with a highly proliferative phenotype, such as HEK-293 cells (vs. relatively slowly proliferating epithelial cells in tubules of normal kidneys).
Increased extracellular medium acidification rate in studied clones of HEK-293 cells with homozygous PKHD1 or PKD2 truncating mutations (vs. WT clones) is consistent with the previously reported faster medium acidification rate of PC-1-deficient Pkd1−/− mouse embryonic fibroblasts (18). In our current study, extracellular medium acidification rate was higher in all clones with PKHD1 truncating mutations (vs. WT clones) when medium acidity was evaluated at D8 or as a trajectory of daily values between D3 and D8. The PKD2 truncating mutations had similar, although less impressive, overall effects on the slope of decline in medium acidity in analyses based on D8 (adjusted for D3) data and a trajectory of daily values between D3 and D8.
The increase in ECAR and OCR observed in our current study in clones of cells with PKHD1 truncating mutations (vs. WT clones) was not observed in immortalized cell lines enriched in either proximal or distal tubules from two Pkd1 conditional mice (subsequently inactivated Pkd1 using lentiviral cre expression) or in a mouse collecting duct cell line with shRNA Pkd1 knockdown (12). Since we did not observe significant changes in ECAR and OCR in clones carrying PKD2 truncating mutations, it is possible that the ECAR- and OCR-enhancing effect is unique to the PKHD1 truncating mutations and that the PKD2 truncating mutations alter energetic metabolism differently [e.g., via previously described impaired fatty acid metabolism in Pkd1-deficient cells (12)]. Alternatively, it is possible that defects in all three PKD genes may alter ECAR and OCR; however, this effect was below the resolution of the assays and experimental design of the previously reported Pkd1 (12) and the current PKD2 studies.
The increased mitochondrial roundness and circularity, as well as mitochondrial cristae dilatation, in cells with PKHD1 and PKD2 truncating mutations suggest that these genes participate in the shaping of normal mitochondrial morphology. Similar mitochondrial changes were recently described in patients and mice with a mutated PKD1 gene (10). PKD1 encodes a mitochondrial matrix protein that regulates mitochondrial morphology and function (10). Together, these data suggest that a dysfunction of PKD1, PKD2, and PKHD1 genes disrupts the same mitochondria-regulating pathway that is required for proper renal tubule diameter regulation during development and regeneration after injury.
While most typical forms of ARPKD are caused by mutations in a single gene, PKHD1, this gene encodes multiple alternatively spliced isoforms predicted to form both membrane-bound and secreted variants (11). While it is conceivable that the position of PKHD1 truncating mutations influences the spectrum of the alternatively spliced isoforms, it was not known whether different truncating mutations uniquely alter energy metabolism. The current study demonstrated the lack of such an effect in the studied model. Similarly, we observed no effect of the position of PKD2 truncating mutations on extracellular medium acidification.
In summary, we observed an increase in extracellular acidification rate in clones of cells with homozygous PKHD1 and PKD2 truncating mutations in a human HEK-293 cell line model. Together with the previously reported faster acidification in mouse Pkd1-deficient cells, these data suggest that enhanced extracellular medium acidification is a common feature for defects in all three PKD genes. At least in the case of PKHD1 and PKD2, this effect was not altered by the site of truncating mutations. In the case of PKHD1 truncating mutations, we also observed an increase in both ECAR and OCR, pointing to a stress-dependent increase in energy utilization. The major strength of this study is the use of human-derived clones with multiple mutations, multiple WT controls, and multiple replicates for each of the studied genes. The major limitation of this study is the use of the HEK-293 cell-based in vitro model. We used this approach as an initial step toward more expensive and time-consuming preclinical in vivo investigations of the studied PKD gene defects. Validation of this initial observation in such studies would point to a potential benefit of targeting glucose or energy metabolism in ARPKD and ADPKD.
GRANTS
This work was supported in part by National Institutes of Health (NIH)-funded University of Alabama at Birmingham (UAB) Hepato/Renal Fibrocystic Disease Core Center Grant P30 DK-074038 and R01 DK-097423, Grant 1-I01-BX002298 from the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, and the Detraz Endowed Research Fund in Polycystic Kidney Disease (to M. Mrug). Additional support was provided by NIH-funded UAB-University of California San Diego O'Brien Center Grant 1P30 DK-079337. The mass spectrometer used for this study was obtained from a grant from the UAB Health Services Foundation General Endowment Fund.
DISCLOSURES
M. Mrug reports grants and consulting fees outside the submitted work from Otsuka Pharmaceuticals and Sanofi. None of the other authors has any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
P.C., S.M., J.M.P., A.K.C., R.A.K., P.D.B., V.M.D.-U., B.K.Y., and M.M. conceived and designed research; P.C., J.Z., B.K.C., L.S.W., and T.F.B. performed experiments; P.C., J.Z., S.M., B.K.C., L.S.W., S.B., V.M.D.-U., and M.M. analyzed data; P.C., J.Z., S.M., B.K.C., L.S.W., S.B., P.D.B., V.M.D.-U., B.K.Y., and M.M. interpreted results of experiments; P.C., S.M., A.K.C., L.S.W., and M.M. prepared figures; A.K.C., L.S.W., and M.M. drafted manuscript; P.C., S.M., B.K.C., L.S.W., S.B., V.M.D.-U., and M.M. edited and revised manuscript; P.C., J.Z., S.M., B.K.C., J.M.P., A.K.C., L.S.W., T.F.B., S.B., R.A.K., P.D.B., V.M.D.-U., B.K.Y., and M.M. approved final version of manuscript.
REFERENCES
- 1.Bergmann C, Senderek J, Küpper F, Schneider F, Dornia C, Windelen E, Eggermann T, Rudnik-Schöneborn S, Kirfel J, Furu L, Onuchic LF, Rossetti S, Harris PC, Somlo S, Guay-Woodford L, Germino GG, Moser M, Büttner R, Zerres K. PKHD1 mutations in autosomal recessive polycystic kidney disease (ARPKD). Hum Mutat 23: 453–463, 2004. doi: 10.1002/humu.20029. [DOI] [PubMed] [Google Scholar]
- 2.Bergmann C, Senderek J, Schneider F, Dornia C, Küpper F, Eggermann T, Rudnik-Schöneborn S, Kirfel J, Moser M, Büttner R, Zerres K. PKHD1 mutations in families requesting prenatal diagnosis for autosomal recessive polycystic kidney disease (ARPKD). Hum Mutat 23: 487–495, 2004. doi: 10.1002/humu.20019. [DOI] [PubMed] [Google Scholar]
- 3.Blazer-Yost BL, Haydon J, Eggleston-Gulyas T, Chen JH, Wang X, Gattone V, Torres VE. Pioglitazone attenuates cystic burden in the PCK rodent model of polycystic kidney disease. PPAR Res 2010: 274376, 2010. doi: 10.1155/2010/274376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Burn TC, Connors TD, Dackowski WR, Petry LR, Van Raay TJ, Millholland JM, Venet M, Miller G, Hakim RM, Landes GM, Kilnger KW, Qian F, Onuchic LF, Watnick T, Germino GG, Doggett NA; The American PKD1 Consortium . Analysis of the genomic sequence for the autosomal dominant polycystic kidney disease (PKD1) gene predicts the presence of a leucine-rich repeat. Hum Mol Genet 4: 575–582, 1995. doi: 10.1093/hmg/4.4.575. [DOI] [PubMed] [Google Scholar]
- 4.Cai Y, Maeda Y, Cedzich A, Torres VE, Wu G, Hayashi T, Mochizuki T, Park JH, Witzgall R, Somlo S. Identification and characterization of polycystin-2, the PKD2 gene product. J Biol Chem 274: 28557–28565, 1999. doi: 10.1074/jbc.274.40.28557. [DOI] [PubMed] [Google Scholar]
- 5.Harris PC, Rossetti S. Molecular diagnostics for autosomal dominant polycystic kidney disease. Nat Rev Nephrol 6: 197–206, 2010. doi: 10.1038/nrneph.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill BG, Benavides GA, Lancaster JR Jr, Ballinger S, Dell’Italia L, Jianhua Z, Darley-Usmar VM. Integration of cellular bioenergetics with mitochondrial quality control and autophagy. Biol Chem 393: 1485–1512, 2012. doi: 10.1515/hsz-2012-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer PA, Ravi S, Chacko B, Johnson MS, Darley-Usmar VM. A review of the mitochondrial and glycolytic metabolism in human platelets and leukocytes: implications for their use as bioenergetic biomarkers. Redox Biol 2: 206–210, 2014. doi: 10.1016/j.redox.2013.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanoix J, D’Agati V, Szabolcs M, Trudel M. Dysregulation of cellular proliferation and apoptosis mediates human autosomal dominant polycystic kidney disease (ADPKD). Oncogene 13: 1153–1160, 1996. [PubMed] [Google Scholar]
- 9.Lantinga-van Leeuwen IS, Leonhard WN, Dauwerse H, Baelde HJ, van Oost BA, Breuning MH, Peters DJ. Common regulatory elements in the polycystic kidney disease 1 and 2 promoter regions. Eur J Hum Genet 13: 649–659, 2005. doi: 10.1038/sj.ejhg.5201392. [DOI] [PubMed] [Google Scholar]
- 10.Lin CC, Kurashige M, Liu Y, Terabayashi T, Ishimoto Y, Wang T, Choudhary V, Hobbs R, Liu LK, Lee PH, Outeda P, Zhou F, Restifo NP, Watnick T, Kawano H, Horie S, Prinz W, Xu H, Menezes LF, Germino GG. A cleavage product of polycystin-1 is a mitochondrial matrix protein that affects mitochondria morphology and function when heterologously expressed. Sci Rep 8: 2743, 2018. doi: 10.1038/s41598-018-20856-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menezes LF, Cai Y, Nagasawa Y, Silva AM, Watkins ML, Da Silva AM, Somlo S, Guay-Woodford LM, Germino GG, Onuchic LF. Polyductin, the PKHD1 gene product, comprises isoforms expressed in plasma membrane, primary cilium, and cytoplasm. Kidney Int 66: 1345–1355, 2004. doi: 10.1111/j.1523-1755.2004.00844.x. [DOI] [PubMed] [Google Scholar]
- 12.Menezes LF, Lin CC, Zhou F, Germino GG. Fatty acid oxidation is impaired in an orthologous mouse model of autosomal dominant polycystic kidney disease. EBioMedicine 5: 183–192, 2016. doi: 10.1016/j.ebiom.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Menezes LF, Zhou F, Patterson AD, Piontek KB, Krausz KW, Gonzalez FJ, Germino GG. Network analysis of a Pkd1-mouse model of autosomal dominant polycystic kidney disease identifies HNF4α as a disease modifier. PLoS Genet 8: e1003053, 2012. doi: 10.1371/journal.pgen.1003053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mochizuki T, Wu G, Hayashi T, Xenophontos SL, Veldhuisen B, Saris JJ, Reynolds DM, Cai Y, Gabow PA, Pierides A, Kimberling WJ, Breuning MH, Deltas CC, Peters DJ, Somlo S. PKD2, a gene for polycystic kidney disease that encodes an integral membrane protein. Science 272: 1339–1342, 1996. doi: 10.1126/science.272.5266.1339. [DOI] [PubMed] [Google Scholar]
- 15.Muto S, Aiba A, Saito Y, Nakao K, Nakamura K, Tomita K, Kitamura T, Kurabayashi M, Nagai R, Higashihara E, Harris PC, Katsuki M, Horie S. Pioglitazone improves the phenotype and molecular defects of a targeted Pkd1 mutant. Hum Mol Genet 11: 1731–1742, 2002. doi: 10.1093/hmg/11.15.1731. [DOI] [PubMed] [Google Scholar]
- 16.Onuchic L, Furu L, Nagasawa Y, Hou X, Eggermann T, Ren Z, Bergmann C, Senderek J, Esquivel E, Zeltner R, Rudnik-Schöneborn S, Mrug M, Sweeney W, Avner E, Zerres K, Guay-Woodford L, Somlo S, Germino G. PKHD1, the polycystic kidney and hepatic disease 1 gene, encodes a novel large protein containing multiple immunoglobulin-like plexin-transcription-factor domains and parallel β-helix 1 repeats. Am J Hum Genet 70: 1305–1317, 2002. doi: 10.1086/340448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Riwanto M, Kapoor S, Rodriguez D, Edenhofer I, Segerer S, Wüthrich RP. Inhibition of aerobic glycolysis attenuates disease progression in polycystic kidney disease. PLoS One 11: e0146654, 2016. doi: 10.1371/journal.pone.0146654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe I, Chiaravalli M, Mannella V, Ulisse V, Quilici G, Pema M, Song XW, Xu H, Mari S, Qian F, Pei Y, Musco G, Boletta A. Defective glucose metabolism in polycystic kidney disease identifies a new therapeutic strategy. Nat Med 19: 488–493, 2013. doi: 10.1038/nm.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sas KM, Yin H, Fitzgibbon WR, Baicu CF, Zile MR, Steele SL, Amria M, Saigusa T, Funk J, Bunni MA, Siegal GP, Siroky BJ, Bissler JJ, Bell PD. Hyperglycemia in the absence of cilia accelerates cystogenesis and induces renal damage. Am J Physiol Renal Physiol 309: F79–F87, 2015. doi: 10.1152/ajprenal.00652.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singer JD, Willett JB. Applied Longitudinal Data Analysis. New York: Oxford University Press, 2003. [Google Scholar]
- 21.Spithoven EM, Kramer A, Meijer E, Orskov B, Wanner C, Caskey F, Collart F, Finne P, Fogarty DG, Groothoff JW, Hoitsma A, Nogier MB, Postorino M, Ravani P, Zurriaga O, Jager KJ, Gansevoort RT; ERA-EDTA Registry; EuroCYST Consortium; WGIKD; EuroCYST Consortium; WGIKD . Analysis of data from the ERA-EDTA Registry indicates that conventional treatments for chronic kidney disease do not reduce the need for renal replacement therapy in autosomal dominant polycystic kidney disease. Kidney Int 86: 1244–1252, 2014. doi: 10.1038/ki.2014.120. [DOI] [PubMed] [Google Scholar]
- 22.Takiar V, Nishio S, Seo-Mayer P, King JD Jr, Li H, Zhang L, Karihaloo A, Hallows KR, Somlo S, Caplan MJ. Activating AMP-activated protein kinase (AMPK) slows renal cystogenesis. Proc Natl Acad Sci USA 108: 2462–2467, 2011. doi: 10.1073/pnas.1011498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Testa F, Busutti M, Chiaravalli M, Leonelli M, Capistrano M, Scolari F, Spotti D, Boletta A, Magistroni R. Design of a phase I clinical trial with 2-deoxy-d-glucose in autosomal dominant polycystic kidney disease (Abstract) J Am Soc Nephrol 27: 769A, 2016. [Google Scholar]
- 25.The European Polycystic Kidney Disease Consortium The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. Cell 77: 881–894, 1994. doi: 10.1016/0092-8674(94)90137-6. [DOI] [PubMed] [Google Scholar]
- 26.The International Polycystic Kidney Disease Consortium Polycystic kidney disease: the complete structure of the PKD1 gene and its protein. Cell 81: 289–298, 1995. doi: 10.1016/0092-8674(95)90339-9. [DOI] [PubMed] [Google Scholar]
- 27.Trudel M. c-Myc signalling in the genetic mechanism of polycystic kidney disease. In: Polycystic Kidney Disease, edited by Li X. Brisbane, Australia: Codon Publications, 2015. doi: 10.15586/codon.pkd.2015.ch10. [DOI] [PubMed] [Google Scholar]
- 28.Ward CJ, Hogan MC, Rossetti S, Walker D, Sneddon T, Wang X, Kubly V, Cunningham JM, Bacallao R, Ishibashi M, Milliner DS, Torres VE, Harris PC. The gene mutated in autosomal recessive polycystic kidney disease encodes a large, receptor-like protein. Nat Genet 30: 259–269, 2002. doi: 10.1038/ng833. [DOI] [PubMed] [Google Scholar]
- 29.Warner G, Hein KZ, Nin V, Edwards M, Chini CC, Hopp K, Harris PC, Torres VE, Chini EN. Food restriction ameliorates the development of polycystic kidney disease. J Am Soc Nephrol 27: 1437–1447, 2016. doi: 10.1681/ASN.2015020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woltman H, Feldstain A, MacKay JC, Rocchi M. An introduction to hierarchical linear modeling. Tutor Quant Methods Psychol 8: 52–69, 2012. doi: 10.20982/tqmp.08.1.p052. [DOI] [Google Scholar]
- 31.Zhang MZ, Mai W, Li C, Cho SY, Hao C, Moeckel G, Zhao R, Kim I, Wang J, Xiong H, Wang H, Sato Y, Wu Y, Nakanuma Y, Lilova M, Pei Y, Harris RC, Li S, Coffey RJ, Sun L, Wu D, Chen XZ, Breyer MD, Zhao ZJ, McKanna JA, Wu G. PKHD1 protein encoded by the gene for autosomal recessive polycystic kidney disease associates with basal bodies and primary cilia in renal epithelial cells. Proc Natl Acad Sci USA 101: 2311–2316, 2004. doi: 10.1073/pnas.0400073101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou JX and Li X. Apoptosis in polycystic kidney disease: from pathogenesis to treatment. In: Polycystic Kidney Disease, edited by Li X. Brisbane, Australia: Codon Publications, 2015. doi: 10.15586/codon.pkd.2015.ch9. [DOI] [PubMed] [Google Scholar]