Abstract
The epigenetic regulator TET2 is frequently mutated in hematological diseases. Mutations have been shown to arise in hematopoietic stem cells early in disease development and lead to altered DNA methylation landscapes and an increased risk of hematopoietic malignancy. Here, we show by genome-wide mapping of TET2 binding sites in different cell types that TET2 localizes to regions of open chromatin and cell-type–specific enhancers. We find that deletion of Tet2 in native hematopoiesis as well as fully transformed acute myeloid leukemia (AML) results in changes in transcription factor (TF) activity within these regions, and we provide evidence that loss of TET2 leads to attenuation of chromatin binding of members of the basic helix–loop–helix (bHLH) TF family. Together, these findings demonstrate that TET2 activity shapes the local chromatin environment at enhancers to facilitate TF binding and provides an example of how epigenetic dysregulation can affect gene expression patterns and drive disease development.
The tet methylcytosine dioxygenase (also known as ten-eleven translocation [TET]) enzymes (TET1-3) mediate active DNA demethylation of cytosines in CG dinucleotides. This occurs by processive TET-mediated oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC). The presence of 5hmC may lead to passive replication-dependent loss of DNA methylation, whereas 5fC and 5caC can be excised by thymine DNA glycosylase (TDG) and be replaced by unmodified cytosine via the base-excision repair (BER) pathway. Although targeting of TET1 to chromatin has been investigated (Williams et al. 2011; Wu et al. 2011), the mechanisms of TET2 recruitment to chromatin remain poorly understood (for review, see Rasmussen and Helin 2016).
Loss-of-function mutations of TET2 have been found in patients with a wide range of hematological diseases, including acute myeloid leukemia (AML) (Scourzic et al. 2015). More recently, high frequencies of TET2 mutations have also been observed in aging-associated clonal hematopoiesis (Genovese et al. 2014; Jaiswal et al. 2014; Xie et al. 2014) and in the poorly studied disorder clonal cytopenia of unknown significance (Kwok et al. 2015; Hansen et al. 2016). In previous studies, we and others identified a role of TET2 in protecting enhancer elements from aberrant DNA methylation (Hon et al. 2014; Lu et al. 2014; An et al. 2015; Rasmussen et al. 2015; Yamazaki et al. 2015). In addition, inhibition of TET proteins was shown to perturb chromatin architecture at enhancers in an embryonal carcinoma cell line undergoing neuronal differentiation (Mahé et al. 2017). Despite these results, direct TET2 binding at enhancers in hematopoietic cells has not been reported. In fact, previous studies mapping TET2 genome-wide occupancy in embryonic stem (ES) cells noted significant TET2 binding at CpG islands and promoters (Chen et al. 2013; Deplus et al. 2013; Peng et al. 2016) or at promoter-distal SALL4A binding sites located at enhancers (Xiong et al. 2016). These seemingly contradictory observations as well as the impact of aberrant DNA methylation at enhancer elements in hematopoietic cells remains to be resolved.
Gene expression is regulated by transcription factors (TFs) that bind DNA in a sequence-specific manner. Activation of a specific gene locus depends both on concentration of individual TFs as well as their ability to access the regulatory genomic elements that control gene expression. TF binding outside gene promoters is associated with low- or intermediate DNA methylation, enrichment of specific histone marks (e.g., monomethylation at histone H3 lysine 4 and acetylation of H3 lysine 27), as well as the presence of a nucleosome-depleted region (Stadler et al. 2011; Thurman et al. 2012). Although much work has focused on understanding the role of aberrant TF expression in leukemia, less is known about the role of the chromatin environment, and hence DNA methylation (Blattler and Farnham 2013), in modulating TF access to their cognate binding sites.
The occurrence of enhancer DNA hypermethylation and hematological malignancies upon TET2 mutation suggests that DNA methylation may pose a challenge for TF binding (Thurman et al. 2012). Although several TFs have been demonstrated to bind methylated DNA and induce DNA hypomethylation (e.g., CTCF and REST) (Lienert et al. 2011; Stadler et al. 2011), many TFs show an inherent binding preference in vitro for motifs with either methylated or unmethylated CpG sites (Hashimoto et al. 2014; Wang et al. 2017; Yin et al. 2017). As an illustration of this, global loss of DNA methylation in ES cells was shown to unmask previously inaccessible genomic binding sites for the TF NRF1 (Domcke et al. 2015). Moreover, DNA methylation and TET-mediated oxidation products potentially influence binding of epigenetic “readers” (e.g., Methyl-Binding Domain [MBD] proteins) (Song and Pfeifer 2016), histone variants and nucleosome remodeling enzymes (Conerly et al. 2010; Brunelle et al. 2015), and modify the physical properties of chromatin and the shape of DNA itself (Ngo et al. 2016).
Here, we generate and analyze genomic profiles from ES cells and several hematopoietic cell types to understand the mechanisms by which loss of TET2, and the resulting altered DNA methylation landscape, affects gene expression patterns and promotes the onset of hematological malignancies.
Results
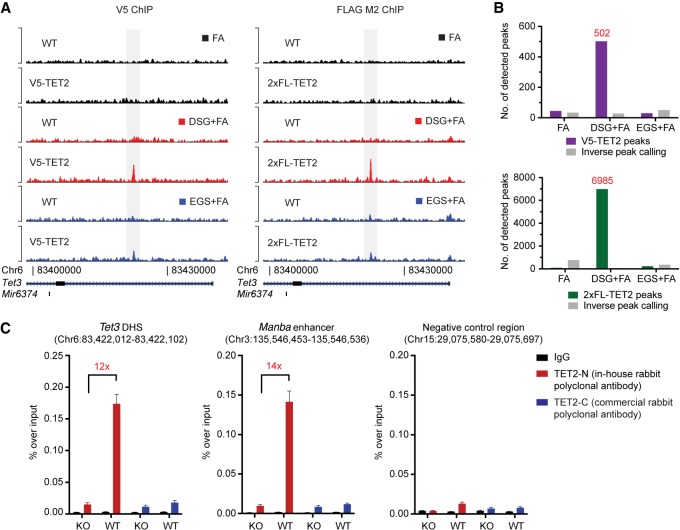
Depletion of TET2 in ES and hematopoietic cells results in widespread changes in the DNA methylation landscape (Ko et al. 2010; Asmar et al. 2013; Hon et al. 2014; An et al. 2015; Rasmussen et al. 2015; Yamazaki et al. 2015). However, it still remains unclear which regions are directly targeted by TET2 as opposed to being altered due to secondary effects of its depletion. Previous studies, in our laboratory and others, have been hampered by chromatin immunoprecipitation (ChIP) artifacts—recently coined as “Phantom peaks” (Jain et al. 2015)—and the lack of ChIP-grade antibodies against TET2. To circumvent this, we used CRISPR homology-directed repair to introduce ChIP-verified epitope tags into the endogenous Tet2 locus in murine ES cells. We identified, upon clonal expansion and characterization, two independent lines with in-frame integration of a V5-tag or a 2xFLAG (2xFL)-tag at the C terminus of Tet2 (Supplemental Fig. S1A,B). Analysis of ChIP coupled with high-throughput sequencing (ChIP-seq) using V5 or FLAG-specific antibodies revealed that the presence of the protein–protein crosslinker disuccinimidyl glutarate (DSG) in combination with formaldehyde (FA) greatly increased the signal-to-noise ratio of the ChIP signal (Fig. 1A) and increased the number of called peaks (Fig. 1B). In contrast, crosslinking conditions with formaldehyde alone, or the combination of the longer ethylene glycol bis(succinimidyl succinate) (EGS) crosslinker plus formaldehyde, were insufficient to capture these regions.
Figure 1.
TET2 chromatin immunoprecipitation and sequencing on wild-type and epitope-tagged cell lines. (A) Representative ChIP-seq tracks of either V5 (left) or 2xFL (right) ChIP experiments in ES cells using different crosslinking conditions. (FA) formaldehyde; (DSG) disuccinimidyl glutarate; (EGS) ethylene glycol bis(succinimidyl succinate). (B) Histograms showing the number of called peaks in the different crosslinking conditions in V5-TET2 (upper) or 2xFL-TET2 (lower)-expressing cells compared to parental cells without endogenously tagged TET2. The number of peaks enriched in parental cells are also shown (inverse peak calling). (C) ChIP-qPCR for TET2 binding in ES cells using antibodies raised against endogenous TET2. Two positive regions (an intronic DNase I hypersensitivity site in Tet3 and an enhancer region in the Manba gene) as well as a negative control region are shown. Data are presented as mean enrichment over input with error bars depicting technical triplicates. The average fold enrichment over Tet2 knockout control cells is indicated.
Encouraged by these results, we tested if untagged endogenous TET2 could be mapped to chromatin using these crosslinking conditions. ES cell chromatin was crosslinked with DSG + FA and immunoprecipitation was performed using an antibody raised against the N terminus of TET2 (TET2-N) or a commercially available antibody directed against the TET2 C terminus (TET2-C). We observed a strong and reproducible enrichment of TET2-bound regions using the TET2-N antibody, and this signal was absent in knockout cells or at a negative control region (Fig. 1C). Thus, the combination of specific antibodies and optimized workflows allows for the precise mapping of TET2-bound genomic regions.
TET2 binds regions of open chromatin with enhancer features in embryonic stem cells
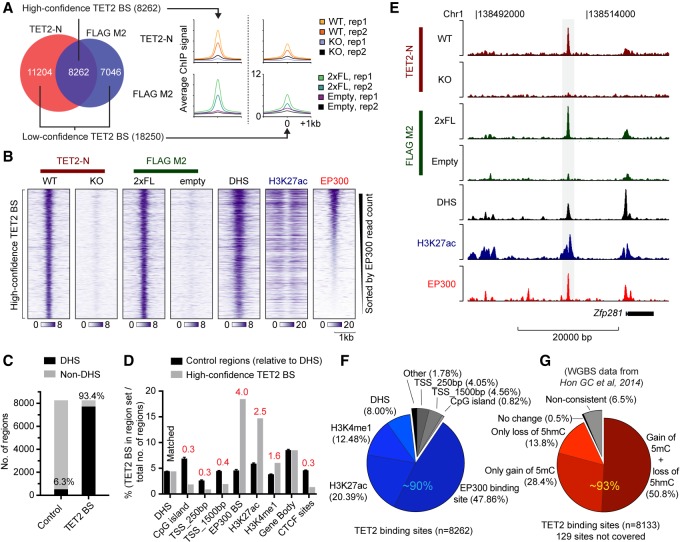
Next, we generated a high-quality ChIP-seq data set to determine genome-wide TET2 occupancy in ES cells. Quality control of biological duplicate experiments using antibodies to endogenous TET2 (TET2-N) or the FLAG-tagged TET2 expressed from the endogenous locus (FLAG M2) showed excellent correlation between replicate samples (Supplemental Fig. S1C–F). To avoid the false discovery of “phantom peaks,” TET2 binding sites were defined by enrichment over nonspecific ChIP-enriched regions identified in Tet2 knockout cells or parental cells without FLAG-tagged TET2. In total, this analysis revealed 26,512 TET2-bound regions of which approximately one-third, referred to as “high-confidence” TET2 binding sites, were identified with both antibodies and had a stronger ChIP-seq signal (Fig. 2A; Supplemental Fig. S1G,H). The vast majority (93.4%) of these 8262 sites were localized in regions of open chromatin defined by DNase I hypersensitivity (DHS) (Fig. 2B,C).
Figure 2.
TET2 binds regions of open chromatin with enhancer features in ES cells. (A) Venn diagram showing overlap of called peaks in TET2-N or FLAG M2 ChIP-seq experiments (left) as well as average ChIP signal from replicate samples (right). High- and low-confidence TET2 binding sites are defined, respectively, as regions showing evidence of TET2 binding in both peak sets (high) or only supported by a called positive region in one peak set (low). (B) Heat maps of ChIP-seq signal from wild-type TET2 or TET2 with two copies of a FLAG tag (2xFL). Tracks of H3K27ac and EP300 enrichment as well as regions of DHS in ES cells are also shown. The vertical axis contains all high-confidence TET2 binding sites defined in A, sorted by decreasing EP300 read counts. The horizontal axis is centered on TET2 peaks. (C) Histogram showing overlap of high-confidence TET2 binding sites with regions of DHS in ES cells. Random matched control regions (Control) were generated with same size, orientation, and distance relative to gene bodies. (D) Histogram showing overlap as C but with random matched control regions relative to DHS. The percentage of CpG islands, proximal (±250 bp) or distal (±1500 bp) transcription start sites (TSS), EP300 binding sites, H3K27ac, and H3K4me1 enriched domains, Gene body, and CTCF binding sites, overlapping TET2 high-confidence binding sites are shown. Data indicate the number of TET2 BS overlapping a given region set divided by the total number of loci in the region set (e.g., 175,237 DHS in ES cells) and fold enrichment over matched control are stated (red). (E) Representative ChIP-seq tracks showing a TET2-bound region upstream of the Zfp281 gene in ES cells. (F) Pie chart showing the distribution of high-confidence TET2 binding sites with respect to the indicated genomic regions. Each TET2 binding site is counted only once and excluded when moving clockwise from proximal promoters (e.g., 8% of TET2 high-confidence BS overlap with DHS but does not overlap with any of the preceding genomic elements). Blue hues indicate the fraction of TET2-bound regions with regulatory potential at promoter-distal sites (∼90% of all TET2-bound regions). (G) Pie chart as in F showing average change in DNA methylation (5mC and 5hmC) upon TET2 loss in ES cells (whole-genome bisulfite sequencing data [WGBS] from Hon et al. 2014) in high-confidence TET2 binding sites and flanking regions (±250 bp). Only CpG sites covered by more than 10 reads were included in the analysis. Each TET2 binding site is counted only once and excluded when moving clockwise. Red hues indicate the fraction (∼93%) of TET2-bound regions showing DNA methylation changes consistent with loss of TET2 catalytic activity (gain or 5mC or loss of 5hmC, or both). Regions in B–F enriched for DHS, EP300, H3K27ac, H3K4me1, and CTCF sites in ES cells were experimentally determined in the ENCODE Project (The ENCODE Project Consortium 2011).
To understand whether TET2 is enriched in specific regions of open chromatin in the genome, we compared TET2 binding sites to sets of random control regions (×5) matched for size, orientation, and distance to DHS sites. This showed a strong enrichment of TET2 high-confidence binding sites at DHS with enhancer features such as EP300 binding (fourfold), H3K27ac (2.5-fold), and H3K4me1 (1.6-fold), whereas TET2 binding at promoters, CpG islands, and CTCF binding sites was depleted (∼0.3-fold each) (Fig 2D). In agreement with this, we found that TET2 binding sites were located in CpG-sparse regions as compared to CpG islands (Supplemental Fig. S1I). This binding pattern is furthermore illustrated by a representative TET2-bound enhancer region 10 kb upstream of the master ES cell regulator Zfp281 (Fig. 2E). In total, up to 90% of TET2 binding sites are associated with features of distal regulatory elements (EP300, H3K27ac, H3K4me1, or DHS), and nearly half of TET2-bound regions overlap with promoter-distal EP300 binding sites (Fig. 2F). Chromatin occupancy of EP300 is a hallmark of active enhancers, and a biochemical interaction between EP300 and TET2 has recently been reported (Heintzman et al. 2007; Zhang et al. 2017). Thus, our data are in agreement with a model in which TET2 can be directly recruited to a subset of its chromatin targets via a direct interaction with EP300 (Zhang et al. 2017).
A previous study examined the genome-wide changes in 5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC) that occur upon Tet2 knockout in ES cells (Hon et al. 2014). We used this data set to identify epigenetic changes around (±250 bp) high-confidence TET2 binding sites. Genetic knockout of Tet2 is likely to result in gain of 5mC (due to absence of DNA demethylation), loss of 5hmC (stable product of methylcytosine oxidation by TET2), or both, at sites of TET2 recruitment. We did not observe a linear correlation between TET2 binding and changes in DNA methylation in Tet2 knockout (Supplemental Fig S1J,K). However, we found DNA methylation changes consistent with loss of TET2 catalytic activity (hyper-5mC, hypo-5hmC, or both) at the majority of high-confidence TET2 binding sites (∼93%) (Fig. 2G), thereby confirming that TET2 modifies the DNA methylation state. The analysis was repeated with the full set of 26,512 sites with evidence of TET2 binding (Supplemental Fig. S2A). Within this larger and less stringent set of regions, we observed a modest enrichment of TET2 binding at CpG islands and gene promoters (Supplemental Fig. S2B). However, in contrast to a pronounced DNA hypermethylation at sites cobound by TET2 and EP300, DNA methylation did not change at these CpG islands and promoters in Tet2-deficient ES cells (Supplemental Fig. S2C,D). This is consistent with the previously identified role of TET2 in maintaining a low level of DNA methylation at distal regulatory elements, whereas CpG islands and promoters are protected from DNA hypermethylation by additional mechanisms (Rasmussen and Helin 2016). It should be noted that most differentially methylated regions reported in Tet2 knockout ES cells (over 60,000 hyper-DMRs and 130,000 hypo-DMRs) (Hon et al. 2014) did not show detectable binding of TET2 in our data set (Supplemental Fig. S2E). This suggests that TET2 functions in the absence of robust and persistent binding at these sites and that some of these methylation changes (especially hypo-DMRs) can occur as a consequence of secondary events to TET2 depletion, or as a result of cell passaging (misincorporation during DNA replication) (Rasmussen et al. 2015). Taken together, our data show that TET2 binds to regions of open chromatin with enhancer features that undergo TET2-dependent DNA demethylation.
Cell-type–specific binding pattern of TET2 in myeloid hematopoietic versus ES cells
Next, we determined the genome-wide binding sites of TET2 in hematopoietic cells. To this end, we used myeloid cells immortalized by AML1-ETO with the potential for inducible deletion of Tet2 (Tet2fl/fl;AE;Rosa26+/CreERT2) (Rasmussen et al. 2015). Analysis of biological duplicate ChIP experiments using the TET2-N antibody (ChIP using a FLAG-tagged version of TET2 was not performed because this would require generating a novel FLAG-tagged Tet2 mouse line) identified 19,706 regions significantly enriched over knockout control. In total, only 7.4% of these regions were shared between ES cells and myeloid cells (Fig. 3A; Supplemental Fig. S2F,G).
Figure 3.
Cell-type–specific binding pattern of TET2 in myeloid hematopoietic cells. (A) Venn diagram showing overlap of TET2 binding sites in ES cells and myeloid hematopoietic cells. Binding sites are defined from biological replicate samples by enrichment over nonspecific ChIP-enriched regions in Tet2 knockout cells using the TET2-N antibody. (B) Pie chart showing distribution of TET2-bound regions in myeloid hematopoietic cells. Each TET2 binding site is counted only once and excluded when moving clockwise from proximal promoters (e.g., 10.1% of TET2 BS overlap with DHS but does not overlap with any of the preceding genomic elements). Blue hues indicate fraction (∼44%) of TET2-bound regions with regulatory potential at promoter-distal sites based on H3K27 acetylation and H3K4 methylation in myeloid cells as well as DHS in the related mouse erythroleukemia (MEL) cell line. (C) Representative ChIP-seq tracks showing TET2-bound enhancer regions upstream of the Gata2 gene in myeloid hematopoietic cells. (D) List of top enriched logos and their associated DNA-binding TFs identified in promoter-distal (−1.5 kb/+500 bp from TSS) TET2-bound regions in ES cells. (E) As in D, but for myeloid hematopoietic cells. Regions in B and C enriched for H3K27ac, H3K4me3, and H3K4me1 were experimentally determined in Rasmussen et al. (2015), and DHS in MEL cells was downloaded from the ENCODE Project (The ENCODE Project Consortium 2011).
We then overlapped the TET2 myeloid binding sites with various genomic regions. Although binding of TET2 could be detected in a subset of promoters and CpG islands, the majority of binding sites was promoter-distal (67.4%), and nearly half (44.4%) were associated with enhancer features (Fig. 3B). An illustrative example of this binding pattern is shown at enhancers upstream of the hematopoietic master regulator Gata2 (Fig. 3C). Since promoter DNA methylation patterns are largely unaffected by depletion of TET2 (Rasmussen et al. 2015), we decided to focus our analysis on promoter-distal TET2 binding sites. Identification of enriched Gene Ontology (GO) terms by Genomic Regions Enrichment of Annotations Tool (GREAT) (McLean et al. 2010) revealed a striking separation by cell type. For example, the GO terms “stem cell maintenance” and “blastocyst formation” were enriched for TET2 binding sites in ES cells, whereas “immune system process” was highly significant for sites in myeloid cells (Supplemental Fig. S2H,I). Next, we performed motif enrichment analysis of known DNA-binding TFs. The top enriched motifs belonged to master TFs of the respective cell types such as SOX2, KLF4, ESRRG, POU5F1, and NANOG in ES cells, and ERG, RUNX1, CEBPA, and GATA1 in myeloid cells (Fig. 3D,E). Together, these results indicate that TET2 is recruited to chromatin in a highly cell-type–specific manner and that TET2 binding at promoter-distal regulatory elements colocalizes with a wide range of predicted TF binding sites.
Differential analysis of chromatin accessibility reveals widespread changes in TF activity upon TET2 loss
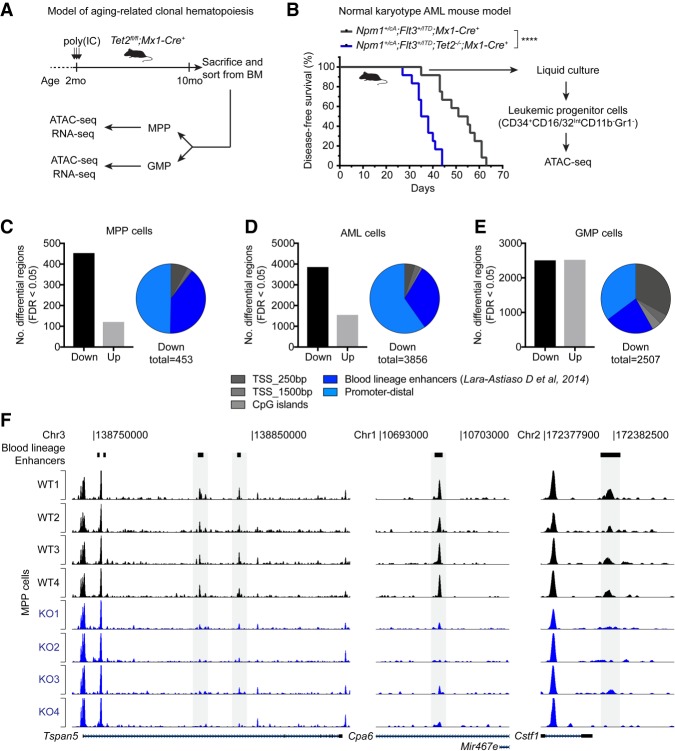
Analysis of chromatin accessibility in gene regulatory regions can be used as a genome-wide and locus-specific measure of nonhistone protein binding to DNA (Hesselberth et al. 2009). It therefore offers an opportunity to systematically assess whether particular sets of TFs (as defined by chromatin accessibility changes at their DNA-binding motifs) are affected by loss of TET2 and potentially drive the genomic and phenotypic changes observed in TET2-mutated hematopoietic cells. Therefore, we decided to perform genomic profiling by Assay for Transposase Accessible Chromatin and sequencing (ATAC-seq) in mouse ES cells as well as three different Tet2 knockout hematopoietic cell types—Multipotent progenitors (MPPs), Granulocyte-Monocyte progenitors (GMPs), and acute myeloid leukemia (AML) cells (Fig. 4A,B; Supplemental Fig. S3A,B).
Figure 4.
Differential analysis of chromatin accessibility reveals widespread changes in TF activity upon TET2 loss. (A) Diagram showing experimental setup to analyze native hematopoiesis in aged Tet2-deficient animals (see also Supplemental Fig. S3A). It has been demonstrated that native hematopoiesis is predominantly sustained by multipotent progenitors (MPPs) throughout the lifetime of the mouse (Sun et al. 2014; Busch et al. 2015). We therefore decided to perform genomic profiling on wild-type and Tet2-deficient MPPs as well as downstream myeloid-lineage progenitors (Granulocyte-Monocyte Progenitors [GMPs]). (B) Diagram showing experimental setup to analyze AML cells with and without deletion of Tet2. Genetically engineered mouse models (Lee et al. 2007; Quivoron et al. 2011; Vassiliou et al. 2011) carrying the indicated alleles were crossed and monitored for disease development. The Npm1+/cA;Flt3+/ITD genotype is sufficient to induce lethal AML (Mupo et al. 2013); however, ablation of Tet2 accelerates the onset of disease. Leaky Cre activity induces AML formation in the absence of pIpC-mediated induction of Mx1-Cre recombinase (Velasco-Hernandez et al. 2016). ATAC-seq analysis was performed on in vitro-grown leukemic progenitor cells isolated by FACS (see also Supplemental Fig. S3B). (****) P < 0.0001 (Log-rank [Mantel-Cox] test) in Kaplan-Meier plot. (C–E) Histogram showing the number of regions with significantly altered chromatin accessibility (FDR < 0.05) identified by diffBind analysis of wild-type and Tet2 knockout MPP cells (n = 4) (C), AML cells (n = 3) (D), and GMP cells (n = 4) (E). Pie charts indicate distribution of regions with decreased chromatin accessibility (down peaks) in Tet2 knockout cells with respect to promoters, CpG islands, and blood lineage enhancer regions (Lara-Astiaso et al. 2014). (F) Tracks of ATAC-seq signal in MPP cells from individual wild-type and Tet2 knockout animals (n = 4) at three representative genomic loci. Peaks that overlap a blood lineage enhancer with significantly decreased ATAC-seq reads in Tet2 knockout animals are highlighted in gray.
Analysis of regions with differential chromatin accessibility in MPP and AML cells revealed that the majority of differential regions (∼75%) were less accessible in Tet2-deficient cells. In addition, 88% of peaks with loss of DNA accessibility in MPP cells were associated with increased DNA methylation in hematopoietic stem cells with ablation of TET activity (Supplemental Fig. S3C; An et al. 2015). Finally, a considerable fraction of these peaks in both MPP and AML cells (30%–40%) overlapped with previously annotated enhancers in blood lineages (Fig. 4C,D,F; Lara-Astiaso et al. 2014). This suggests that increased DNA methylation plays a direct role in restricting DNA binding within these regions. In contrast, differentially accessible regions in GMPs were balanced between higher and lower chromatin accessibility (Fig. 4E). To gain further insight into this, we overlapped ATAC-seq differential regions in GMPs with TET2 ChIP data obtained from the GMP-like myeloid hematopoietic cells. We found significantly higher normalized TET2 read counts in regions with reduced accessibility upon Tet2 knockout compared to those with increased accessibility (Supplemental Fig. S3D). This suggests that TET2 is likely to have a direct role to maintain open chromatin (and TF binding), whereas the increases in chromatin accessibility at other regions may be due to accumulated epigenetic dysregulation or associated secondary events in response to TET2 loss (e.g., imbalance of differentiation-associated cytokines and growth factors).
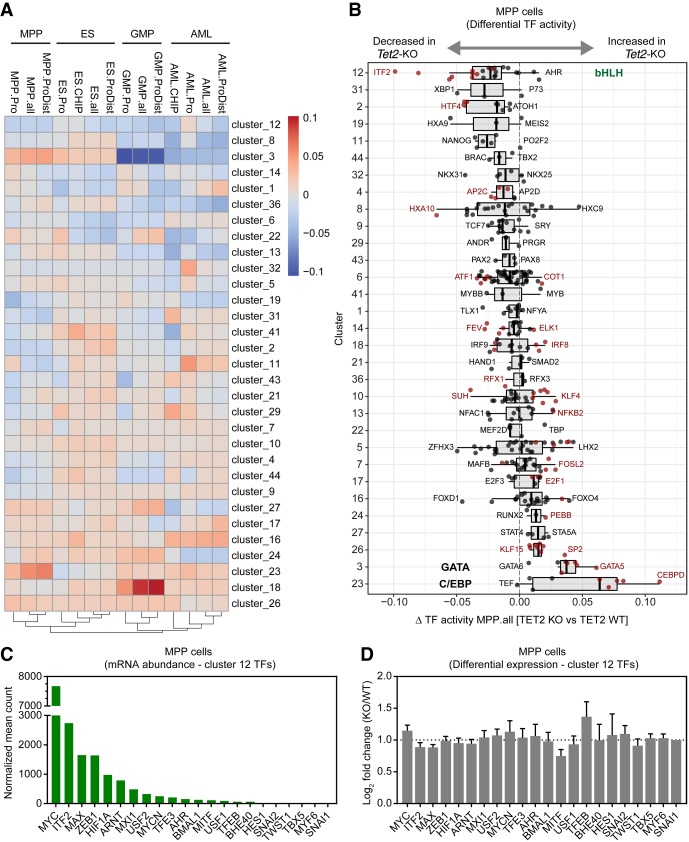
To identify TF binding events that are affected by TET2 loss in native chromatin, we developed a novel computational method, diffTF (Berest et al. 2018), to assess “TF activity” on a genome-wide scale using profiles of chromatin accessibility by ATAC-seq. In this method, we compare the accessibility changes at putative binding sites within ATAC-seq peaks for each TF and compare this distribution to the background distribution of accessibility fold changes for all other TFs. If the putative binding sites of a TF are overall less open in the Tet2 mutant cells, we define this TF to be less “active” in Tet2 knockout cells and vice versa. Thus “TF activity” is here defined as the TF being associated with increased chromatin accessibility at its target sites. We predicted differential TF activity using a comprehensive collection of DNA-binding motifs for mouse TFs (Kulakovskiy et al. 2016). However, although this analysis identified numerous TFs with altered activity in one or several cell types, the high similarity of DNA-binding motifs in groups of related TFs (e.g., GATA family) makes it challenging to pinpoint an observed effect to a single group member. Therefore, we clustered TFs based on similarity of DNA-binding motifs (Supplemental Table S1) and compared the overall differential activity of clusters in wild-type and Tet2 knockout (Fig. 5A,B).
Figure 5.
Summary of diffTF analysis and ATAC-seq profiles of multiple cell types with loss of TET2. (A) The heatmap represents the summary of diffTF analyses between Tet2 knockout and wild type across multiple cell types. The color scale of the heatmap corresponds to the Z-scores of the weighted mean difference values (TF activity) obtained from diffTF. Negative values indicate that the TF cluster has lower activity in Tet2 knockout samples compared to the wild type, and vice versa for positive values. Only TF clusters with three or more TFs are shown. For each cell type, the analysis was run on different sets of peaks: promoter regions only (−1.5 kb/+500 bp from TSS; labeled celltype.Pro) and putative enhancer regions (+100 kb/−100 kb from TSS excluding promoter regions; celltype.ProDist), in addition to the full set of peaks (celltype.all). For the cell types for which we mapped TET2 binding sites by ChIP-seq (ES cells and AML cells), diffTF was also run using ATAC-seq peaks intersected with TET2 ChIP in the corresponding cell type (celltype.CHIP). TET2-bound regions in AML cells were determined in bulk AML cells (containing both the leukemic precursor population that was used for ATAC-seq analysis as well as more differentiated cells) (Supplemental Fig. S3B). (B) Box plot showing weighted mean difference values obtained from diffTF analyses for each TF cluster in MPP cells comparing Tet2 knockout versus wild type. Individual TFs within a cluster are shown (black dots), and TFs passing a significance threshold (P-value < 0.1) are highlighted (red dots). The predominant TF identity of selected clusters (12, 3, and 23) are marked. (C) Histogram showing mean normalized counts for Cluster 12 TFs obtained from RNA-seq data generated from wild-type and Tet2 knockout MPP cells (n = 4). The TFs within Cluster 12 are ranked based on mean normalized abundance. (D) DESeq2 differential expression values of Cluster 12 TFs ranked as in C. No statistically significant changes were observed (FDR < 0.05).
Loss of TET2 resulted in widespread changes in TF activity in all observed cell types. In GMP cells, we detected a strong signature of aberrant lineage differentiation characterized by increased activity of the IRF family of TFs (Cluster 18) and decreased activity of GATA family members (Cluster 3). Conversely, MPP cells showed a pronounced increase in chromatin accessibility in predicted binding sites of the GATA and CCAAT/Enhancer binding protein family (Clusters 3 and 23) and AML, GMP, and MPP cells were characterized by a loss of activity of members of the HOX protein family (Cluster 8). Finally, Tet2 knockout ES cells exhibited a prominent decrease in activity of a large TF cluster containing steroid hormone receptors including estrogen receptor alpha (Cluster 6). Several recent studies are consistent with a direct role of DNA methylation and TET2 catalytic function to facilitate activity and/or expression of members of these TF clusters: (1) CEBPB has been reported to have a preference for binding to motifs with a methylated cytosine, thus potentially implicating TET2 as a factor that restricts C/EBP family TF binding to chromatin (Mann et al. 2013; Zhu et al. 2016); (2) consistent with loss of HOX gene activity in AML cells, TET2 catalytic activity has been implicated in maintenance of expression of the HOXA cluster in various cell types (Bocker et al. 2012); and (3) TET2 has recently been shown to mediate recruitment of estrogen receptor alpha by demethylation of estrogen responsive enhancers in the breast cancer cell line MCF-7 (Wang et al. 2018). Together, these results suggest that loss of TET2 catalytic activity, and the resulting alterations in DNA methylation and hydroxymethylation, has pleiotropic effects on chromatin accessibility and TF binding, most likely related to cell-type–specific chromatin structure and TF expression patterns.
Basic helix–loop–helix (bHLH) TFs are adversely affected by loss of TET2 in hematopoietic cells
To understand whether the observed TF activity changes could potentially explain the altered hematopoietic differentiation program observed in Tet2 knockout hematopoietic cells, we focused on a cluster of basic helix–loop–helix (bHLH) TFs (Cluster 12), whose activity was consistently decreased upon TET2 loss in all cell types and this decrease was observed at sites of TET2 binding (e.g., see “AML.ChIP” and “ES.ChIP”) (Fig. 5A,B). Cluster 12 consists of 22 TFs that preferentially bind enhancer-box (E-box) elements (5′-CANNTG-3′) in the genome. E-box motifs often contain a central CpG site, and 5mC, as well as the TET-mediated oxidation products 5hmC and 5fC, in this motif have been shown to impair in vitro DNA binding (Wang et al. 2017; Yin et al. 2017). Analysis of mRNA expression of Cluster 12 TFs in MPP cells revealed that the most abundantly expressed TFs were the MYC/MAX, MAX/MXI1, and HIF1A/ARNT heterodimers, as well as the E proteins ITF2 and ZEB1. Therefore, it is likely that the observed changes in chromatin accessibility originate from altered binding activity of these TFs as other Cluster 12 TF members are lowly expressed or absent (Fig. 5C). None of the abundant TFs were found to be differentially expressed in Tet2 knockout MPP cells, thus suggesting that loss of TET2 results in decreased TF binding activity rather than inducing changes in TF expression (Fig. 5D).
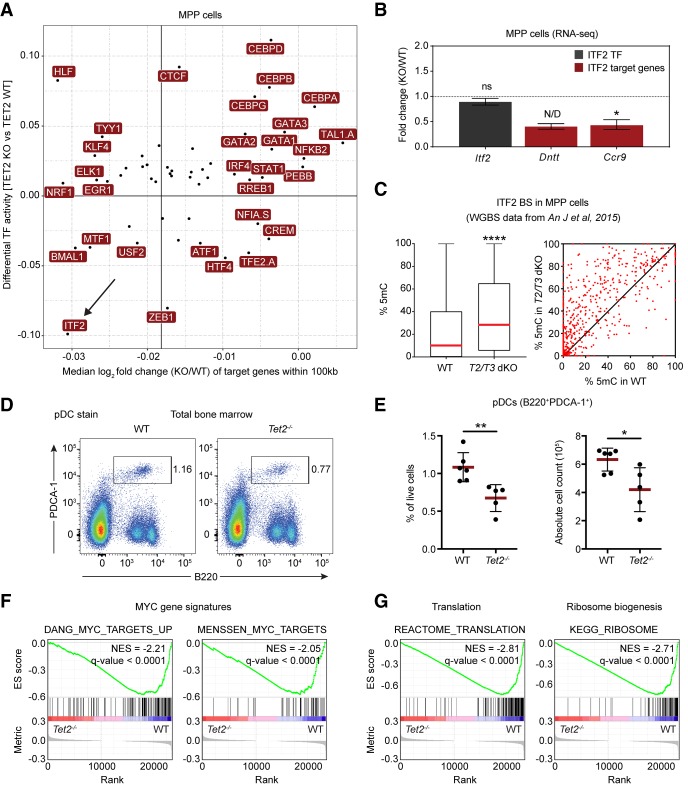
We reasoned that a potential loss of TF binding may result in decreased expression of TF target genes. To analyze the effect of TF binding on gene expression, we generated expression profiles from wild-type and Tet2 knockout MPP cells and determined the expression of genes within 100 kb of the predicted TF binding sites. This analysis revealed that loss of TET2 leads to decreased expression of genes with predicted ITF2 binding sites (Fig. 6A). In support of this, we also observed a decrease in expression of the validated ITF2 target genes, Ccr9 and Dntt, in Tet2 knockout MPPs (Fig. 6B), and analysis of DNA methylation changes in predicted ITF2 binding sites revealed an overall increase in 5mC levels in hematopoietic stem cells upon ablation of TET activity (Fig. 6C).
Figure 6.
Basic helix–loop–helix (bHLH) TFs are adversely affected by loss of TET2 in hematopoietic cells. (A) Correlation of changes in TF activity and target gene expression. The scatter plot shows a positive correlation (Pearson's r = 0.4; P-value < 0.005) between differential TF activity, as defined by diffTF, and the median log2 fold change of putative target genes (TF binding site within ±100 kb of the promoter region) of the same TF. The accentuated vertical line represents the median expression of all target genes used in the analysis. (B) DESeq2 expression values from RNA-seq in MPPs showing expression of Itf2 (gray) and the two validated ITF2 target genes Dntt and Ccr9 (red). Data are presented as mean ± SEM (n = 4). (ns) not significant; (N/D) not calculated due to outlier sample; (*) P < 0.05 (Q-value). (C) Box (left) and scatter plot (right) showing average 5mC changes at predicted ITF2 binding sites in wild-type and Tet2/Tet3 double knockout (T2/T3 dKO) LSK (LineagenegSca1+cKit+) cells (An et al. 2015). Box plot shows median (red line) and 25th and 75th quantile, and the scatter plot shows 5mC changes for individual ITF2 sites. Only CpG sites covered by more than 10 reads were included in the analysis. (****) P < 0.0001 (paired two-tailed Student's t-test). (D) Representative FACS plots showing the population of plasmacytoid dendritic cells (pDC) in bone marrow of aged wild-type and Tet2-deficient animals. (E) Scatter dot plots showing the percentage (left) and absolute cell count (right) of pDCs in the bone marrow of aged wild-type and Tet2-deficient animals. Line and error bars represent mean ± SD (n = 5–6). (*) P < 0.05; (**) P < 0.01 (unpaired two-tailed Student's t-test). (F) Enrichment plots from GSEA showing decreased expression of MYC target genes in Tet2-deficient MPP cells (Menssen and Hermeking 2002; Zeller et al. 2003). (G) Enrichment plots from GSEA showing decreased expression in Tet2-deficient MPPs of genes involved in ribosome biogenesis and translation (see Supplemental Table S2 for full list of gene signatures).
Mice with hematopoietic-specific knockout of a single allele of Itf2 display defects in differentiation of plasmacytoid dendritic cells (pDCs) (Cisse et al. 2008). Therefore, we reasoned that a change in ITF2 binding activity should recapitulate this. Phenotypic analysis of aged Tet2 knockout mice revealed a partial impairment of pDC development that is highly similar to previously published observations in Itf2 heterozygous knockout bone marrow (Fig 6D,E; Cisse et al. 2008). Overall, these results suggest that the knockout of Tet2 phenocopies the haploinsufficiency of Itf2 in hematopoietic stem cells by decreasing the binding activity of ITF2 rather than changing its expression. This would derail normal pDC differentiation in the bone marrow. Frequent mutations of TET2 have been found in patients with Blastic plasmacytoid dendritic cell neoplasm (BPDCN), a rare myeloid neoplasm characterized by proliferation of aberrant pDC precursor cells (Scourzic et al. 2015). Thus, TET2 may prevent BPDCN by keeping the binding sites of ITF2 accessible and thus maintaining the expression of ITF2-regulated genes.
Within Cluster 12, MYC was by far the most abundantly expressed TF in hematopoietic stem cells. It has previously been reported that DNA methylation inhibits MYCN chromatin binding in Tet1-3 triple knockout ES cells (Yin et al. 2017), and a recent study has uncovered that TET2 is recruited to MYC binding sites by SNIP1 in cancer cell lines (Chen et al. 2018). However, analysis of chromatin accessibility by diffTF at MYC binding sites (predicted individually from all available MYC DNA-binding motifs) did not show a significant difference in wild-type and Tet2 knockout, possibly due to a complex MYC binding pattern at E-boxes throughout the genome. To further analyze if MYC activity could be affected by TET2, we performed gene set enrichment analysis (GSEA) on RNA-seq data from wild-type and Tet2 knockout MPP cells. This analysis revealed a pronounced down-regulation of MYC target gene signatures (Fig. 6F; Supplemental Table S2; Menssen and Hermeking 2002; Zeller et al. 2003). Furthermore, we also found a general down-regulation of gene signatures of translation, ribosome biogenesis and metabolism, which are processes known to be controlled by MYC (Fig. 6G; Supplemental Table S2). Thus, transcriptome analysis suggests that MYC activity is indeed adversely affected in Tet2 knockout MPP cells, likely through altered DNA methylation at MYC target sites and decreased MYC TF chromatin binding. In support of this, conditional knockout of Myc has been shown to result in hematopoietic stem cell expansion (Wilson et al. 2004) similar to what has been observed in aged Tet2 knockout animals (Moran-Crusio et al. 2011; Quivoron et al. 2011). Together, these results suggest that impairment of MYC activity may confer a competitive advantage to Tet2 knockout hematopoietic stem cells, and that impairment of other bHLH TFs, such as ITF2, renders terminal differentiation inefficient.
Discussion
Although DNA methylation landscapes in TET2 mutant cells have been extensively characterized (Ko et al. 2010; Asmar et al. 2013; Hon et al. 2014; Rasmussen et al. 2015; Yamazaki et al. 2015), much less is known about the functional consequences that ultimately drive hematopoietic stem cell expansion and malignancy. Here, we resolve the inconsistency between regions reported to be bound by TET2 (Chen et al. 2013; Deplus et al. 2013; Peng et al. 2016) and those affected by TET2 catalytic function. We show that TET2 is predominantly recruited to promoter-distal regions of open chromatin, including enhancers. Therefore, it is likely that it is epigenetic perturbation of these elements that results in dysregulated hematopoiesis of TET2-mutated stem cells, possibly through pleiotropic effects on many genes that together derail homeostasis and differentiation. A recent study has reported a direct biochemical interaction between the histone acetyltransferase EP300 and TET2 (Zhang et al. 2017). In ES cells, we find that approximately half of high-confidence TET2 binding sites colocalize with promoter-distal EP300-enriched regions (Fig. 2B,F), and nearly half of all EP300-enriched regions show evidence of TET2 binding (Supplemental Fig. S2B). Taken together these results may suggest that EP300 is involved in recruiting TET2 to chromatin through direct protein–protein interactions. However, a direct validation of this and a comprehensive analysis of the role of additional potential recruiters, such as WT1 (Rampal et al. 2014; Wang et al. 2015), remain an area of active investigation.
To dissect the impact of TET2 loss in hematopoiesis, we investigated genome-wide changes in enhancer function using chromatin accessibility as a measure of activity in a native chromatin context. We report widespread changes, of which a majority of differentially open regions in all tested cell types, except GMP, were less accessible upon ablation of TET2. This is consistent with a recent study showing that most DNA methylation-sensitive regulatory elements (∼86%) are inhibited by DNA methylation (Lea et al. 2018). To perform an unbiased analysis of TF signatures that were globally enriched or depleted, we developed a novel computational tool, diffTF, to assess differential TF binding from ATAC-seq profiles (Berest et al. 2018). We could detect signatures of aberrant activity of several clusters of hematopoietic TFs such as IRF, C/EBP, GATA, and HOX protein families in different hematopoietic cell types. In addition to these changes, we also found a strong and consistent loss of activity for a cluster of bHLH TFs (including MYC and ITF2) that preferentially bind enhancer-box (E-box) elements in the genome. To examine the molecular and phenotypic consequences of this, we uncovered that (1) MYC and ITF2 target genes were down-regulated in Tet2 knockout hematopoietic stem cells; (2) ITF2 binding sites were DNA hypermethylated upon ablation of TET activity; and (3) Tet2 knockout mice show hallmarks of Myc-deficient and Itf2-deficient hematopoiesis. Thus, our results support a model by which TET2 recruitment and catalytic functions are necessary to shape the chromatin structure for permissive TF binding.
It should be noted that although inferring differential TF occupancy from ATAC-seq data provides a global view of the transcriptional network of a cell, it does have limitations: The use is dependent on collections of TF binding motifs of high quality (not applicable to TFs with poorly defined or unknown DNA-binding motifs) as well as the inherent impact of binding of specific TFs on nucleosome density. For instance, it is possible that additional TF binding events that were not identified in this study may also require TET2 activity; however, such TFs may not show strong global differences or are not detected from TF motif analysis of chromatin accessibility alone. Vice versa, due to the high similarity of DNA-binding motifs within certain TF families, the role of a TF identified by this approach should subsequently be characterized to confirm the identity as well as the biological significance.
Frequent mutations in the DNA methylation machinery have been found in patients suffering from hematological diseases as well as solid cancers (Rasmussen and Helin 2016). Yet, whether DNA methylation is causally involved in shaping gene expression patterns, rather than passively mirroring transcriptional states, is still a matter of debate (Schübeler 2015). Here, we present data in support of a role of TET2-mediated cytosine modifications at enhancers to facilitate TF binding and fidelity of target gene expression in hematopoietic cells. Rather than being an obligate activator, TET2 functions to reinforce binding of some TFs and contributes in this manner to enhancer activity and gene expression. Analysis of chromatin maturation after replication fork passage has indicated that open chromatin at cell-type–specific enhancers are only slowly reestablished through competition of TFs with newly deposited nucleosomes (Ramachandran and Henikoff 2016). Thus, cells that naturally divide rapidly and must undergo coordinated cell state transitions (e.g., hematopoietic stem and progenitor cells) may be particularly sensitive to epigenetic disturbances of TF binding kinetics. Comprehensive analysis of enhancer function in TET2-mutated hematopoiesis will foster a greater understanding of the role of epigenetic dysregulation in disease and may lead to discoveries of potential clinical significance.
Methods
ES cell lines
Mouse ES cell lines were derived from blastocysts harvested from Tet2fl/fl animals (Moran-Crusio et al. 2011). To generate a clean Tet2 knockout ES cell line (to use as ChIP control), cells were transiently transfected with a Cre recombinase-expressing plasmid and subcloned to identify a constitutive Tet2 knockout ES cell line. Endogenous tagging of TET2 was performed using CRISPR homology-directed repair with a single-stranded DNA oligonucleotide repair template. Briefly, a sgRNA targeting the insertion site was cloned into pSpCas9(BB)-2A-GFP (PX458) (Addgene #48138). This plasmid was cotransfected in ES cells with oligonucleotides encoding either two copies of Flag (DYKDDDDKDYKDDDDK) or a single copy of V5 tag (GKPIPNPLLGLDST) as well as homology arms (60 bp each). Transfected cells were single-cell sorted, and the resulting clones were screened for stable expression of epitope-tagged TET2 by Western blot. All mouse ES cell lines were cultured in feeder-free, gelatinized plates in “Serum/2i/LIF” conditions: Dulbecco's Modified Eagle Medium (DMEM) supplemented with 15% fetal bovine serum, 2 mM Glutamax (Gibco), 0.1 mM 2-Mercaptoethanol (Gibco), 1× nonessential amino acids (Gibco), 1× Pen/Strep (Gibco), 3 µM GSK3 inhibitor (CHIR99021), 1 µM MEK1 inhibitor (PD0325901), and leukemia inhibitory factor (LIF).
Mice
For analysis of aging-related Tet2-deficient hematopoiesis, cohorts of age-matched litter mates (8-wk old) of Tet2fl/fl or Tet2fl/fl;Mx1-Cre+ mice (Quivoron et al. 2011) were injected three times intraperitoneally with 250 µg polyinosinic-polycytidylic acid (PolyI:C LMW, InvivoGen) at experimental days 0, 2, and 4. The mice were subsequently allowed to age and sacrificed at 10 mo of age. To generate a Tet2 knockout AML model, the following genetically modified mouse lines Npm1cA-Flox (Vassiliou et al. 2011), Flt3-ITD (Lee et al. 2007), and Tet2fl/fl;Mx1-Cre+ (Quivoron et al. 2011) were intercrossed and Npm1+/cA-Flox;Fl3+/ITD;Mx1-Cre+ or Npm1+/cA; Fl3+/ITD;Tet2fl/fl;Mx1-Cre+ mice were monitored for disease development. AML cells were harvested and 2.5 × 104 c-Kit+Gr1−Mac1− AML splenocytes transplanted into sublethally irradiated (650Rad) Ly5.1 recipient animals by tail vein injection. Ly5.1 mice were maintained on medicated water (Ciprofloxacin 100 µg/mL) for 3 wk following the irradiation procedure. To establish in vitro culture of AML cells, c-Kit+Gr1−Mac1− splenocytes harvested from moribund mice were purified by FACS and cultured in suspension in nontissue culture treated plasticware in StemPro-34 SFM media (Thermo Fisher Scientific) supplemented with 2 mM GlutaMAX (Gibco), 1× Pen/Strep (Gibco), 0.1 mM 2-mercaptoethanol (Sigma-Aldrich), as well as the cytokines SCF (50 ng/mL), IL3 (10 ng/mL), and IL6 (10 ng/mL) (Peprotech). All animal work was carried out in compliance with ethical regulation under license by the Danish regulatory authority.
Chromatin immunoprecipitation and sequencing
ChIP-seq experiments were carried out using a two-step crosslinking procedure with 2 mM disuccinimidyl glutarate (DSG) and 1% formaldehyde (FA). The genomic coordinates of all region sets derived from TET2 ChIP analysis in various cell types can be found in Supplemental Table S3. See Supplemental Methods for a full description of experimental protocol, processing, and analysis of ChIP data sets.
ATAC-seq library generation and analysis
ATAC-seq libraries were generated and sequenced as described previously (Buenrostro et al. 2013; Lara-Astiaso et al. 2014). See Supplemental Methods for a full description of experimental protocol and data preprocessing steps.
diffTF analysis
In order to define potential difference in TF activity based on the chromatin accessibility (ATAC-seq) data, we developed a computational pipeline called diffTF (deposited at https://git.embl.de/grp-zaugg/diffTF; documented at https://difftf.readthedocs.io) (Berest et al. 2018). This method takes as input predicted TF binding sites defined by genome-wide PWM scanner (PWMscan) (Ambrosini et al. 2018) overlapped with the open regions from ATAC-seq. For the PWM database source, we used HOCOMOCO v10 database (Kulakovskiy et al. 2016), in which we replaced CDX4 and EVX1 motifs (that were retracted from this database) with PWM models from the recent methyl-SELEX study (Yin et al. 2017). We then extended TF binding sites by 100 bp in each direction and calculated a fold change of chromatin accessibility between two conditions. Finally, we removed potential CG-content bias from the data and used an analytical approach for significance assessment (Supplemental Methods). The final weighted mean difference value for each TF corresponds to the genome-wide difference in accessibility between two conditions. We used the matrix-clustering tool from the RSAT suite (Castro-Mondragon et al. 2017) on the combined PWM data set with the following parameters: -lth Ncor 0.4 -lth cor 0.6 and observed 88 clusters of TFs.
External data sets
The following experimentally determined data sets were downloaded from the ENCyclopedia Of DNA Elements (ENCODE; https://www.encodeproject.org/): DNase-seq “ENCFF001YOK” and “ENCFF00PUJ”, EP300 binding sites “ENCFF001YAD”, H3K27 acetylation “ENCFF001XWY”, H3K4 monomethylation “ENCFF001XWY”, and CTCF binding sites “ENCFF001YAC”. Active and poised enhancers in myeloid hematopoietic cells were obtained from the NCBI Gene Expression Omnibus (GEO) under accession number GSE59591. DNA methylation data from wild-type and Tet2 knockout cells were obtained from GEO: GSE48519 and GSE72630.
Data access
The ChIP-seq, ATAC-seq, and RNA-seq data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE115972. The source code for the diffTF software used in this study is available as Supplemental Code and at https://git.embl.de/grp-zaugg/diffTF.
Supplementary Material
Acknowledgments
We thank members of the Helin laboratory for advice and discussion. K.D.R. was supported by a postdoctoral fellowship from the Danish Medical Research Council (FSS 1333-00120B), K.N. was supported by Program for Advancing Strategic International Networks to Accelerate the Circulation of Talented Researchers, JSPS (S2704), and L.S.-C. was supported by a Marie Sklodowska-Curie individual fellowship (Horizon 2020 Framework Programme, grant agreement no. H2020-MSCA-IF-2017-796341). The work in the Helin laboratory was supported by grants to K.H. from The European Research Council (294666_DNAMET), the Danish Cancer Society, the Danish National Research Foundation (DNRF82), and through a center grant from the Novo Nordisk Foundation (NNF17CC0027852). This work was also supported by the National Institutes of Health (NIH, P30 CA008748).
Author contributions: K.D.R. designed the study, performed the majority of experiments, analyzed data, and wrote the first draft of the manuscript. I.B. and J.B.Z. provided genomic data analysis and interpretation (ATAC-seq, RNA-seq, ChIP-seq) and designed the diffTF software. S.K., K.N., L.S.-C., and M.T.P. performed experiments. G.S.V. and J.C. provided essential reagents and intellectual input. K.H. and J.B.Z. designed and supervised the study, analyzed data, secured funding, and wrote the manuscript with K.D.R.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239277.118.
References
- Ambrosini G, Groux R, Bucher P. 2018. PWMScan: a fast tool for scanning entire genomes with a position-specific weight matrix. Bioinformatics 34: 2483–2484. 10.1093/bioinformatics/bty127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, González-Avalos E, Chawla A, Jeong M, López-Moyado IF, Li W, Goodell MA, Chavez L, Ko M, Rao A. 2015. Acute loss of TET function results in aggressive myeloid cancer in mice. Nat Commun 6: 10071 10.1038/ncomms10071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmar F, Punj V, Christensen J, Pedersen MT, Pedersen A, Nielsen AB, Hother C, Ralfkiaer U, Brown P, Ralfkiaer E, et al. 2013. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica 98: 1912–1920. 10.3324/haematol.2013.088740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berest I, Arnold C, Reyes-Palomares A, Palla G, Rasmussen KD, Helin K, Zaugg J. 2018. Quantification of differential transcription factor activity and multiomic-based classification into activators and repressors: diffTF. bioRxiv 10.1101/368498 [DOI] [PubMed]
- Blattler A, Farnham PJ. 2013. Cross-talk between site-specific transcription factors and DNA methylation states. J Biol Chem 288: 34287–34294. 10.1074/jbc.R113.512517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocker MT, Tuorto F, Raddatz G, Musch T, Yang FC, Xu M, Lyko F, Breiling A. 2012. Hydroxylation of 5-methylcytosine by TET2 maintains the active state of the mammalian HOXA cluster. Nat Commun 3: 818 10.1038/ncomms1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunelle M, Nordell Markovits A, Rodrigue S, Lupien M, Jacques PÉ, Gévry N. 2015. The histone variant H2A.Z is an important regulator of enhancer activity. Nucleic Acids Res 43: 9742–9756. 10.1093/nar/gkv825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch K, Klapproth K, Barile M, Flossdorf M, Holland-Letz T, Schlenner SM, Reth M, Höfer T, Rodewald HR. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature 518: 542–546. 10.1038/nature14242 [DOI] [PubMed] [Google Scholar]
- Castro-Mondragon JA, Jaeger S, Thieffry D, Thomas-Chollier M, van Helden J. 2017. RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res 45: e119 10.1093/nar/gkx314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Chen Y, Bian C, Fujiki R, Yu X. 2013. TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493: 561–564. 10.1038/nature11742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, Lin HP, Zhou WJ, He CX, Zhang ZY, Cheng ZL, Song JB, Liu P, Chen XY, Xia YK, et al. 2018. SNIP1 recruits TET2 to regulate c-MYC target genes and cellular DNA damage response. Cell Rep 25: 1485–1500.e4. 10.1016/j.celrep.2018.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier C, den Hollander NS, Kant SG, et al. 2008. Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135: 37–48. 10.1016/j.cell.2008.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conerly ML, Teves SS, Diolaiti D, Ulrich M, Eisenman RN, Henikoff S. 2010. Changes in H2A.Z occupancy and DNA methylation during B-cell lymphomagenesis. Genome Res 20: 1383–1390. 10.1101/gr.106542.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplus R, Delatte B, Schwinn MK, Defrance M, Méndez J, Murphy N, Dawson MA, Volkmar M, Putmans P, Calonne E, et al. 2013. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J 32: 645–655. 10.1038/emboj.2012.357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, Schübeler D. 2015. Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528: 575–579. 10.1038/nature16462 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2011. A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol 9: e1001046 10.1371/journal.pbio.1001046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese G, Kähler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. 2014. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 371: 2477–2487. 10.1056/NEJMoa1409405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JW, Westman MK, Sjö LD, Saft L, Kristensen LS, Ørskov AD, Treppendahl M, Andersen MK, Grønbæk K. 2016. Mutations in idiopathic cytopenia of undetermined significance assist diagnostics and correlate to dysplastic changes. Am J Hematol 91: 1234–1238. 10.1002/ajh.24554 [DOI] [PubMed] [Google Scholar]
- Hashimoto H, Olanrewaju YO, Zheng Y, Wilson GG, Zhang X, Cheng X. 2014. Wilms tumor protein recognizes 5-carboxylcytosine within a specific DNA sequence. Genes Dev 28: 2304–2313. 10.1101/gad.250746.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318. 10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. 2009. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods 6: 283–289. 10.1038/nmeth.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, Yen CA, Ye Z, Mao SQ, Wang BA, et al. 2014. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 56: 286–297. 10.1016/j.molcel.2014.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D, Baldi S, Zabel A, Straub T, Becker PB. 2015. Active promoters give rise to false positive ‘Phantom Peaks’ in ChIP-seq experiments. Nucleic Acids Res 43: 6959–6968. 10.1093/nar/gkv637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman P V, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. 2014. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 371: 2488–2498. 10.1056/NEJMoa1408617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. 2010. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 468: 839–843. 10.1038/nature09586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva A V, Kasianov AS, Ashoor H, Ba-alawi W, Bajic VB, Medvedeva YA, Kolpakov FA, et al. 2016. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res 44: D116–D125. 10.1093/nar/gkv1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok B, Hall JM, Witte JS, Xu Y, Reddy P, Lin K, Flamholz R, Dabbas B, Yung A, Al-Hafidh J, et al. 2015. MDS-associated somatic mutations and clonal hematopoiesis are common in idiopathic cytopenias of undetermined significance. Blood 126: 2355–2361. 10.1182/blood-2015-08-667063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, Zaretsky I, Jaitin DA, David E, Keren-Shaul H, Mildner A, Winter D, Jung S, et al. 2014. Immunogenetics. Chromatin state dynamics during blood formation. Science 345: 943–949. 10.1126/science.1256271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea AJ, Vockley CM, Johnston RA, Del Carpio CA, Barreiro LB, Reddy TE, Tung J. 2018. Genome-wide quantification of the effects of DNA methylation on human gene regulation. eLife 7: e37513 10.7554/eLife.37513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Tothova Z, Levine RL, Anderson K, Buza-Vidas N, Cullen DE, McDowell EP, Adelsperger J, Fröhling S, Huntly BJP, et al. 2007. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell 12: 367–380. 10.1016/j.ccr.2007.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. 2011. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet 43: 1091–1097. 10.1038/ng.946 [DOI] [PubMed] [Google Scholar]
- Lu F, Liu Y, Jiang L, Yamaguchi S, Zhang Y. 2014. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev 28: 2103–2119. 10.1101/gad.248005.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahé EA, Madigou T, Sérandour AA, Bizot M, Avner S, Chalmel F, Palierne G, Métivier R, Salbert G. 2017. Cytosine modifications modulate the chromatin architecture of transcriptional enhancers. Genome Res 27: 947–958. 10.1101/gr.211466.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann IK, Chatterjee R, Zhao J, He X, Weirauch MT, Hughes TR, Vinson C. 2013. CG methylated microarrays identify a novel methylated sequence bound by the CEBPB|ATF4 heterodimer that is active in vivo. Genome Res 23: 988–997. 10.1101/gr.146654.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hermeking H. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci 99: 6274–6279. 10.1073/pnas.082005599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, Figueroa ME, Vasanthakumar A, Patel J, Zhao X, et al. 2011. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 20: 11–24. 10.1016/j.ccr.2011.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mupo A, Celani L, Dovey O, Cooper JL, Grove C, Rad R, Sportoletti P, Falini B, Bradley A, Vassiliou GS. 2013. A powerful molecular synergy between mutant Nucleophosmin and Flt3-ITD drives acute myeloid leukemia in mice. Leukemia 27: 1917–1920. 10.1038/leu.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo TT, Yoo J, Dai Q, Zhang Q, He C, Aksimentiev A, Ha T. 2016. Effects of cytosine modifications on DNA flexibility and nucleosome mechanical stability. Nat Commun 7: 10813 10.1038/ncomms10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Li Y, Xi Y, Li W, Li J, Lv R, Zhang L, Zou Q, Dong S, Luo H, et al. 2016. MBD3L2 promotes Tet2 enzymatic activity for mediating 5-methylcytosine oxidation. J Cell Sci 129: 1059–1071. 10.1242/jcs.179044 [DOI] [PubMed] [Google Scholar]
- Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, Do Cruzeiro M, Delhommeau F, Arnulf B, Stern MH, et al. 2011. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 20: 25–38. 10.1016/j.ccr.2011.06.003 [DOI] [PubMed] [Google Scholar]
- Ramachandran S, Henikoff S. 2016. Transcriptional regulators compete with nucleosomes post-replication. Cell 165: 580–592. 10.1016/j.cell.2016.02.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, Li Y, Ahn J, Abdel-Wahab O, Shih A, et al. 2014. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 9: 1841–1855. 10.1016/j.celrep.2014.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Helin K. 2016. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev 30: 733–750. 10.1101/gad.276568.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen KD, Jia G, Johansen J V, Pedersen MT, Rapin N, Bagger FO, Porse BT, Bernard OA, Christensen J, Helin K. 2015. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 29: 910–922. 10.1101/gad.260174.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D. 2015. Function and information content of DNA methylation. Nature 517: 321–326. 10.1038/nature14192 [DOI] [PubMed] [Google Scholar]
- Scourzic L, Mouly E, Bernard OA. 2015. TET proteins and the control of cytosine demethylation in cancer. Genome Med 7: 9 10.1186/s13073-015-0134-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Pfeifer GP. 2016. Are there specific readers of oxidized 5-methylcytosine bases? Bioessays 38: 1038–1047. 10.1002/bies.201600126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, Wirbelauer C, Oakeley EJ, Gaidatzis D, Tiwari VK, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495. 10.1038/nature10716 [DOI] [PubMed] [Google Scholar]
- Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho YJ, Klein A, Hofmann O, Camargo FD. 2014. Clonal dynamics of native haematopoiesis. Nature 514: 322–327. 10.1038/nature13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489: 75–82. 10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassiliou GS, Cooper JL, Rad R, Li J, Rice S, Uren A, Rad L, Ellis P, Andrews R, Banerjee R, et al. 2011. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet 43: 470–475. 10.1038/ng.796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Hernandez T, Säwén P, Bryder D, Cammenga J. 2016. Potential pitfalls of the Mx1-Cre system: implications for experimental modeling of normal and malignant hematopoiesis. Stem Cell Rep 7: 11–18. 10.1016/j.stemcr.2016.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xiao M, Chen X, Chen L, Xu Y, Lv L, Wang P, Yang H, Ma S, Lin H, et al. 2015. WT1 recruits TET2 to regulate its target gene expression and suppress leukemia cell proliferation. Mol Cell 57: 662–673. 10.1016/j.molcel.2014.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Hashimoto H, Zhang X, Barwick BG, Lonial S, Boise LH, Vertino PM, Cheng X. 2017. MAX is an epigenetic sensor of 5-carboxylcytosine and is altered in multiple myeloma. Nucleic Acids Res 45: 2396–2407. 10.1093/nar/gkw1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ozark PA, Smith ER, Zhao Z, Marshall SA, Rendleman EJ, Piunti A, Ryan C, Whelan AL, Helmin KA, et al. 2018. TET2 coactivates gene expression through demethylation of enhancers. Sci Adv 4: eaau6986 10.1126/sciadv.aau6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen J V, Cloos PAC, Rappsilber J, Helin K. 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348. 10.1038/nature10066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche AC, Knabenhans C, Macdonald HR, Trumpp A. 2004. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev 18: 2747–2763. 10.1101/gad.313104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D'Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y. 2011. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393. 10.1038/nature09934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. 2014. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 20: 1472–1478. 10.1038/nm.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Zhang Z, Chen J, Huang H, Xu Y, Ding X, Zheng Y, Nishinakamura R, Xu GL, Wang H, et al. 2016. Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Mol Cell 64: 913–925. 10.1016/j.molcel.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, He R, Taby R, Vasanthakumar A, Macrae T, et al. 2015. TET2 mutations affect non-CpG island DNA methylation at enhancers and transcription factor–binding sites in chronic myelomonocytic leukemia. Cancer Res 75: 2833–2843. 10.1158/0008-5472.CAN-14-0739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F, et al. 2017. Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356: eaaj2239 10.1126/science.aaj2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4: R69 10.1186/gb-2003-4-10-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, Luo J, Yen RC, Li Y, Baylin SB. 2017. Acetylation enhances TET2 function in protecting against abnormal DNA methylation during oxidative stress. Mol Cell 65: 323–335. 10.1016/j.molcel.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Wang G, Qian J. 2016. Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet 17: 551–565. 10.1038/nrg.2016.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.