Abstract
Here we use a chromosome-level genome assembly of a prairie rattlesnake (Crotalus viridis), together with Hi-C, RNA-seq, and whole-genome resequencing data, to study key features of genome biology and evolution in reptiles. We identify the rattlesnake Z Chromosome, including the recombining pseudoautosomal region, and find evidence for partial dosage compensation driven by an evolutionary accumulation of a female-biased up-regulation mechanism. Comparative analyses with other amniotes provide new insight into the origins, structure, and function of reptile microchromosomes, which we demonstrate have markedly different structure and function compared to macrochromosomes. Snake microchromosomes are also enriched for venom genes, which we show have evolved through multiple tandem duplication events in multiple gene families. By overlaying chromatin structure information and gene expression data, we find evidence for venom gene-specific chromatin contact domains and identify how chromatin structure guides precise expression of multiple venom gene families. Further, we find evidence for venom gland-specific transcription factor activity and characterize a complement of mechanisms underlying venom production and regulation. Our findings reveal novel and fundamental features of reptile genome biology, provide insight into the regulation of snake venom, and broadly highlight the biological insight enabled by chromosome-level genome assemblies.
Squamate reptiles have become important models for a broad range of research, including studies on genome structure (Alföldi et al. 2011), coevolution (Geffeney et al. 2002), development (Cohn and Tickle 1999), and regenerative biology (Secor and Diamond 1998). Among squamates, snakes represent an enriched system for studying a number of extreme or unique biological features. For example, snakes are an emerging model system for studying sex chromosome evolution, given their lack of apparent global dosage compensation (Vicoso et al. 2013), independent origins of ZW and XY sex determination systems (Gamble et al. 2017), and wide range of differentiation between sex chromosomes among lineages (Matsubara et al. 2006). Snakes also possess microchromosomes, which have been shown in birds to have intriguing and unique genome biology (Hillier et al. 2004; Backström et al. 2010) but are virtually uncharacterized in reptiles. Snake venom systems are the most intensely studied feature of snake biology due to their medical relevance (Mackessy 2010) and also because they provide a unique opportunity to study the evolution of a complex phenotype that required gene duplication, shifts in gene function and regulation, and numerous structural and physiological adaptations for venom storage and delivery. Although numerous studies have characterized the composition and activity of snake venoms, progress in understanding the genomic context for venom evolution and precise cellular and regulatory mechanisms underlying venom expression has been severely limited by the fragmentary nature of existing snake genome assemblies (Bradnam et al. 2013; Castoe et al. 2013; Vonk et al. 2013; Yin et al. 2016).
Here we leverage a chromosome-level assembly of the genome of the prairie rattlesnake (Crotalus viridis), assembled using a combination of second-generation sequencing and Hi-C scaffolding (Lieberman-Aiden et al. 2009), to study key questions about reptile and snake genome biology that have been previously difficult to address due to the fragmentary genome assemblies available for reptile species. We trace patterns of chromosome-level synteny and composition across amniotes, specifically exploring synteny between reptile and avian genomes and testing hypotheses about the evolution of GC-isochore structure in reptiles. We further characterize genome-wide chromatin contacts using Hi-C data to demonstrate differences between classes of chromosomes and distinctions from patterns observed in mammalian data sets. Rattlesnakes have highly differentiated ZW sex chromosomes (Matsubara et al. 2006), and we use our genome and additional resequenced genomes to identify the Z Chromosome, the pseudoautosomal region of Z and W Chromosomes, and an evolutionary stratum in the process of degeneration. We further studied patterns of partial dosage compensation and used inferred ancestral genome-wide expression levels to characterize the evolution of dosage compensation in snakes. Lastly, we use a combination of Hi-C chromatin contact data from the rattlesnake venom gland, RNA-seq data from diverse tissues, and the chromosomal locations of snake venom gene families to identify mechanisms of venom gene regulation in the venom gland.
Results
Genome assembly and annotation
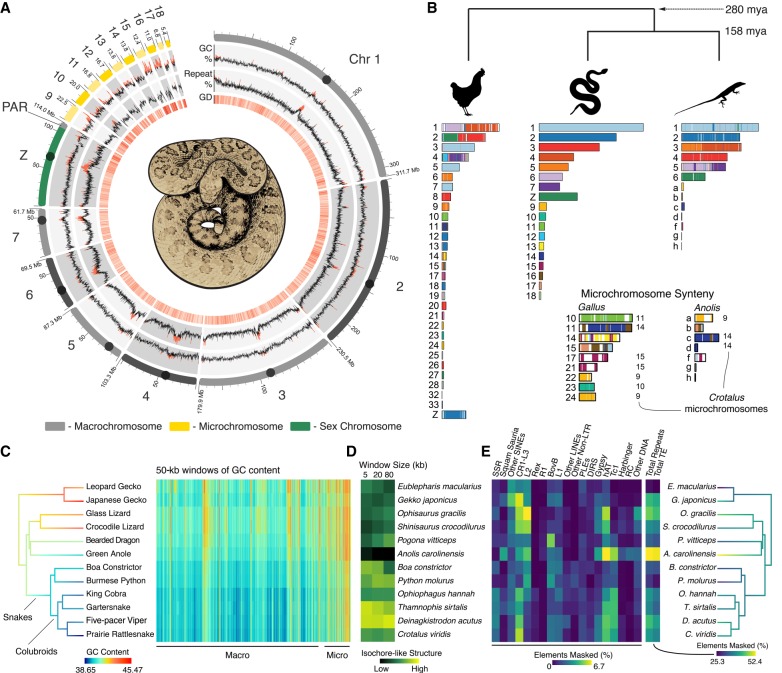
We sequenced and assembled a rattlesnake reference genome from a male prairie rattlesnake (Crotalus viridis viridis) that was sequenced at 1658-fold physical coverage using multiple approaches, including the Dovetail Genomics HiRise sequencing and assembly method (Putnam et al. 2016) that combines Chicago (Putnam et al. 2016; Rice et al. 2017) and Hi-C (Lieberman-Aiden et al. 2009) data, yielding a final scaffold length of 1.34 Gbp (Supplemental Fig. S1; Supplemental Tables S1, S2). Our annotation, which incorporated data from 24 RNA-seq libraries (Supplemental Table S3), included 17,352 protein-coding genes and an annotated repeat element content of 39.49% (Supplemental Tables S4, S5). Macrochromosomes were matched to scaffolds based on scaffold size and known chromosome-specific markers (Supplemental Table S6; Matsubara et al. 2006). Of six chromosomal markers from Matsubara et al. (2006) that did not map to predicted chromosomes in our rattlesnake assembly, we were able to corroborate the accuracy of our assembly for five using multiple lines of evidence, including cross-species synteny with Anolis and local Hi-C contact frequencies (Supplemental Methods; Supplemental Table S7; Supplemental Fig. S2). We also identified the rattlesnake Z Chromosome using multiple lines of evidence, which we discuss below. In our preliminary assembly, microchromosomes were overassembled into a single large scaffold, which we manually split based on multiple lines of evidence (see below and Supplemental Methods). The refined assembly had microchromosome scaffolds with lengths matching the predicted sizes of rattlesnake chromosomes (Baker et al. 1972). Our chromosome-level scaffolds include assembled telomeric and centromeric regions, with centromeres containing an abundant 164-bp monomer (Supplemental Fig. S3).
Synteny and chromosomal composition
The rattlesnake microchromosomes contain higher and more variable GC content than do the macrochromosomes and have particularly high gene density (Welch's two-sample t-test on 100-kb windows, P-value < 0.00001) and reduced repeat element content compared to macrochromosomes (Welch's two-sample t-test, P < 0.00001) (Fig. 1A). These patterns are similar to those in the chicken (Supplemental Fig. S4). Rattlesnake chromosomes are highly syntenic with those from Anolis, except for fusion/separation of Anolis Chromosome 3 into rattlesnake Chromosomes 4 and 5 (Fig. 1B; Supplemental Fig. S5). Our synteny inferences also confirm that Anolis Chromosome 6 is homologous to the sex chromosomes of rattlesnakes (Srikulnath et al. 2009).
Figure 1.
Structure and content of the rattlesnake genome. (A) Regional variation in GC content, genomic repeat content, and gene density (for 100-kb windows) are shown on to-scale chromosomes, with centromere locations represented by circles; values above the genome-wide median are red. GD is gene density, or the number of genes per 100-kb window; higher density shown in darker red. (B) Synteny between rattlesnake, chicken, and anole genomes. Colors on chicken and anole chromosomes correspond with homologous rattlesnake sequence. Numbers to the right of chromosomes in the microchromosome inset represent rattlesnake microchromosomes syntenic with a given chicken or anole chromosomes for >80% of their length. Divergence times are shown in millions of years (mya). (C) Patterns of GC content from genome alignment of 12 squamate species, with tree branches colored according to genomic GC content. The heat map to the right depicts GC content in 50-kb windows of aligned sequence, with macro- and microchromosome regions labeled below. (D) Genomic GC isochore structure measured by the standard deviation in GC content among 5-, 20-, and 80-kb windows. (E) Genomic repeat content among 12 squamate species, with tree branches colored by total genomic repeat content.
Despite conservation of squamate microchromosome homology, patterns of chicken-squamate homology suggest that there were major shifts between macro- and microchromosome locations for large syntenic regions early in amniote evolution. We find evidence for multiple macrochromosomal shifts in synteny between the chicken and squamate reptiles, some of which appear quite complex. For example, the chicken Chromosomes 1 and 2 show synteny patterns that are scattered across multiple squamate macrochromosomes, including the rattlesnake Z Chromosome. Furthermore, only about half of chicken microchromosomes are syntenic with squamate microchromosomes (Fig. 1B), while the rest of chicken microchromosomes share synteny with squamate macrochromosomes. Despite independent origins of some avian and squamate microchromosomes, there are broad similarities among squamate and avian microchromosomes (e.g., GC content variation, gene density) (Supplemental Fig. S4). Further, the presence of microchromosomes in most extant diapsids (Olmo 2005; Organ et al. 2008), the ancestral diapsid genome (O'Connor et al. 2018), amphibians (Voss et al. 2011), and fish (Braasch et al. 2015) broadly suggest that the majority of vertebrate evolution has been shaped by the distinctive but poorly understood biology of microchromosomes.
GC-isochore and repeat element evolution
Squamate reptiles have become important models for studying the evolution of genomic GC content and isochore structure, due to the loss of GC isochores in Anolis yet the apparent re-emergence of isochore structure in snakes (Fujita et al. 2011; Castoe et al. 2013). Comparisons of orthologous genomic regions across 12 squamates demonstrates that there have been two major transitions in genomic GC content, including a reduction in GC content from lizards to snakes and a further reduction in GC content within the colubroid snake lineage that includes the rattlesnake and cobra (Fig. 1C). This suggests that higher genome-wide GC content was likely the ancestral squamate condition and that snakes have evolved increased nucleotide composition variation through an increase in genomic AT content, rather than a buildup of GC-rich isochores. Based on studies of mammals (Duret and Galtier 2009), GC isochore structure is thought to be driven mainly by GC-biased gene conversion that results in GC-biased allele substitution in some genomic regions. The negative relationship between genomic GC content (Fig. 1C) and GC isochore structure (Fig. 1D; Supplemental Table S8) across squamate evolution indicates that this explanation may not apply to the apparent trends in snakes. Instead, GC content variation in snakes appears to be driven by AT-biased processes, including AT-biased substitution that was suggested by previous comparisons of lizard and snake genomes (Castoe et al. 2013). Similar to the patterns observed in GC content variation, genomic repeat element content has also undergone a major shift in colubroid snakes, which show substantial increases in transposable elements overall and specific increases in hAT and Tc1 DNA elements, CR1-L3 LINEs, and simple sequence repeats (SSRs) (Fig. 1E; Supplemental Fig. S6). It remains an open question, however, if shifts in GC content and genomic repeat landscapes are related in colubroid snakes (see also Pasquesi et al. 2018).
Sex chromosome evolution
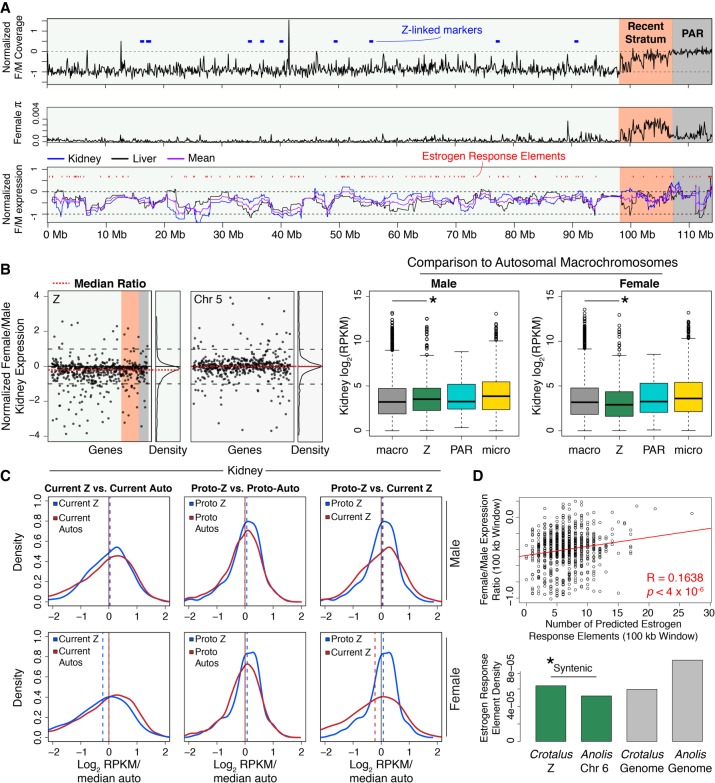
Snake sex chromosomes have evolved multiple times, apparently from different autosomal chromosomes (Gamble et al. 2017), and colubroid Z/W Chromosomes are homologous with Anolis Chromosome 6 (Srikulnath et al. 2009; Vicoso et al. 2013). We identified a single 114-Mb scaffold as the rattlesnake Z Chromosome that contains known Z-linked genes (Supplemental Table S6; Matsubara et al. 2006) and demonstrates roughly half female (ZW) versus male (ZZ) mapped genomic read coverage based on additional male and female samples we sequenced (Fig. 2A; Supplemental Table S9; Supplemental Fig. S7). We also identified the recombining pseudoautosomal region (PAR) of the Z Chromosome as the distal 7.2-Mb region that shows equal male-female genomic read coverage (Fig. 2A). The PAR is GC-rich relative to the genomic background and the remaining Z Chromosome (42.9%) (Supplemental Fig. S8), similar to the PAR of the collard flycatcher (Smeds et al. 2014), suggesting that common processes may drive increased PAR GC content in independently evolved snake and avian sex chromosomes. The rattlesnake PAR also exhibits distinctive patterns of repeat element content (Supplemental Fig. S9) and a higher density of genes than the remaining Z Chromosome (Fisher's exact test, P = 4.46 × 10−7) (Supplemental Fig. S10). Adjacent to the PAR, we identified an evolutionary stratum (“Recent Stratum”) that shows near-autosomal female genomic read coverage (Fig. 2A, top panel). We hypothesize that recombination was most recently suppressed in this region and that substantial homology is retained between Z and W Chromosomes. Consistent with this hypothesis, we observe elevated nucleotide diversity (π) across this region specifically in females (Fig. 2A; Supplemental Fig. S11), likely due to reads mapping to divergent Z- and W-linked gametologs. These results suggest that a number of W-linked gametologs have either been retained during Z/W divergence or are still in the process of degeneration, as has been suggested for birds (Bellott et al. 2017). To further understand the evolutionary origins of the Recent Stratum, we compared mappings of female and male resequencing data for the prairie rattlesnake with those from the pygmy rattlesnake (Sistrurus catenatus) and five-pacer viper (Deinagkistrodon acutus). Both species exhibit similar patterns of intermediate female normalized coverage across the Recent Stratum (Supplemental Fig. S7), suggesting that this evolutionary stratum evolved prior to the divergence between the prairie rattlesnake and five-pacer viper greater than 30 million years ago (Zheng and Wiens 2016). Collectively, the features of the Recent Stratum suggest that recombination suppression and degeneration are ongoing processes in pit vipers, despite the already high differentiation between Z and W Chromosomes (Matsubara et al. 2006).
Figure 2.
The Z Chromosome of the rattlesnake and the evolution of snake dosage compensation. (A) Normalized (log2) female/male genomic read coverage, female π, and windowed (30-gene) log2 normalized female/male gene expression. Known Z-linked markers (Matsubara et al. 2006) shown as blue blocks. In expression plot, red marks represent predicted estrogen response elements (EREs). On each plot, the pseudoautosomal region (PAR) and Recent Stratum are highlighted in gray and orange, respectively. (B) Normalized (log2) female/male kidney gene expression per gene (black dots) across the Z shown next to expression on Chromosome 5, a similarly sized autosome (left panels). The red dashed lines are the median ratios, and relative density is shown to the right of each panel. Gene expression (log2 RPKM) distributions for male and female across macrochromosomes, Z Chromosome, the PAR, and microchromosomes (center and right panels). Asterisks depict significant differences between autosomal and Z Chromosome expression. (C) Density plots of current and inferred ancestral patterns of gene expression (log2 RPKM) in male and female kidney, respectively. Dashed lines represent the median of each distribution. (D) EREs drive partial dosage compensation. The correlation (red line) between predicted EREs and female/male gene expression ratios in 100-kb windows (top panel) is shown with evidence for accumulation of EREs on the rattlesnake Z (bottom panel). Each bar shows the density of EREs found in specific chromosomes (rattlesnake Z and Anolis 6 shown in green) and genome-wide (gray bars). The asterisk depicts a significantly greater density of EREs on the rattlesnake Z than Anolis Chromosome 6.
Patterns of gene expression between heterogametic and homogametic sexes in organisms with differentiated sex chromosomes are of broad interest because of the diversity of mechanisms that can result in dosage compensation (Graves 2016). To investigate dosage compensation in the rattlesnake, we compared female and male RNA-seq data from kidney and liver tissues across the rattlesnake Z Chromosome (Supplemental Table S9). We find evidence from both tissues for lower overall expression in the female (Fig. 2B, left panel; Supplemental Fig. S12), consistent with previous conclusions that female colubroids lack complete dosage compensation (Vicoso et al. 2013) but also that this ratio is higher than expected if there were no dosage compensation (i.e., log2 female/male expression > −1, Wilcoxon signed-rank tests, P-values < 2.2 × 10−16). We also find that chromosome-wide gene expression is higher on the Z than on autosomes for males (Mann–Whitney U tests, P-values < 0.0002), yet lower on the Z than on autosomes for females (P-values < 0.02) (Fig. 2B). Consistent with this, the Z is also enriched for male-biased genes and depauperate in female-biased genes, relative to autosomes (Fisher's exact tests, P-values < 2 × 10−5) (Supplemental Fig. S13).
To understand how patterns of gene expression on the Z have evolved, we compared current Z gene expression in our rattlesnake samples to inferred ancestral (i.e., proto-Z) expression, based on expression levels in autosomal orthologs of the rattlesnake Z genes in the anole and chicken (following Julien et al. 2012; Marin et al. 2017). We find that current male Z expression has not changed from the inferred male proto-Z expression level (Fig. 2C; Supplemental Fig. S12) but that current female Z expression is lower than ancestral female expression (Mann–Whitney U tests, P-values < 0.005). This finding suggests that female Z expression diminished after the establishment of sex chromosomes in the rattlesnake. Combined with evidence that current male Z expression is higher than autosomes, these findings raise the question of whether ancestral expression levels predisposed the proto-Z (e.g., Anolis Chromosome 6) to become the rattlesnake Z. We addressed this by comparing inferred ancestral Z and autosomal expression (Fig. 2C) and find that the ancestor of the rattlesnake Z shows higher expression in both sexes than ancestral autosomes (Mann–Whitney U tests, P-values < 0.02). These findings suggest that, due to the enrichment of male-specific function and the overall elevated level of expression, characteristics of the rattlesnake Z ancestor may have favored its transition from autosome to sex chromosome.
No mechanisms underlying partial dosage of genes or regions have been identified in snakes. The ratio of female/male gene expression is regionally variable across the rattlesnake Z, suggesting partial dosage compensation driven by regional or gene-specific mechanisms (Fig. 2A, bottom panel). We hypothesized that an inherently female-biased regulatory mechanism, estrogen response elements (EREs), might explain dosage-compensated regions and tested for a relationship between the ratio of female/male expression and the number of predicted EREs in 100-kb windows of the Z Chromosome. There is a positive relationship between ERE density and female/male expression for rattlesnakes on the rattlesnake Z (Fig. 2D), yet we do not find this relationship for the analogous comparison of Anolis female/male expression and ERE density on Anolis Chromosome 6 (Supplemental Fig. S14). We also find that that the rattlesnake Z Chromosome has a much higher density of EREs than Anolis Chromosome 6 (two-sample Z test, P-value < 2.2 × 10−16) (Fig. 2D) and is enriched for EREs compared to the genomic background (Fisher's exact test, P-value < 2.2 × 10−16), despite a much higher density of EREs in the Anolis genome overall. To further understand if ERE accumulation is a general feature of snake Z Chromosome evolution, we also analyzed Z Chromosome and autosomal sequences of the five-pacer viper and find consistent evidence for ERE enrichment on the Z Chromosome compared to Anolis Chromosome 6 (Fisher's exact test, P-value = 0.00016) (Supplemental Fig. S15). Our results illustrate that the evolution of the pit viper Z Chromosome has involved regional accumulation of EREs, which may be an important mechanism underlying regional dosage compensation.
Hi-C exposes unique microchromosome biology
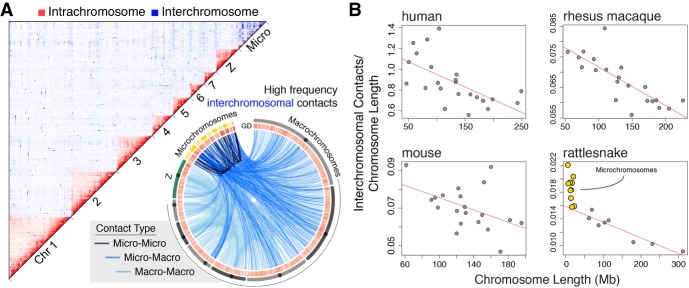
Our analyses of the first chromatin contact data for a nonmammalian vertebrate (Fig. 3A) demonstrate broad similarities in chromatin structure across vertebrate macrochromosomes, yet unique features of snake microchromosomes. We find that patterns of intra- and inter-chromosomal chromatin contacts across rattlesnake macrochromosomes are consistent with patterns observed in mammals, such that when interchromosomal contact frequencies are normalized by chromosome length, they show a consistent negative linear relationship across species (Fig. 3B). Rattlesnake microchromosomes deviate significantly from this macrochromosomal pattern and share disproportionately high frequencies of contacts with other chromosomes, including other microchromosomes (Fig. 3A,B). Indeed, the initial overassembly (Supplemental Fig. S16) of microchromosomes into a single scaffold was likely driven by these unexpected high contact frequencies among microchromosomes, which significantly exceed assumptions used for assembly that are based on mammalian macrochromosomes (t-test, P < 0.000001). These findings highlight the uniqueness of microchromosome interactions within the nucleus of the rattlesnake venom gland and beg the question of whether distinctive chromatin contacts are a consistent feature of microchromosomes in other amniotes.
Figure 3.
Genome-wide chromosomal contacts in the rattlesnake venom gland. (A) 2D heat map of intrachromosomal (red) and interchromosomal (blue) contacts among rattlesnake chromosomes (top). Locations of interchromosomal contacts (bottom), where light blue lines are contacts between macrochromosomes, medium blue lines are micro-to-macrochromosome contacts, and dark blue lines are contacts between microchromosomes. (B) Comparison of interchromosomal contacts normalized by chromosome length versus chromosome length for different species from Hi-C data sets. Red lines depict negative linear relationships for macrochromosomes.
Venom evolution and regulation
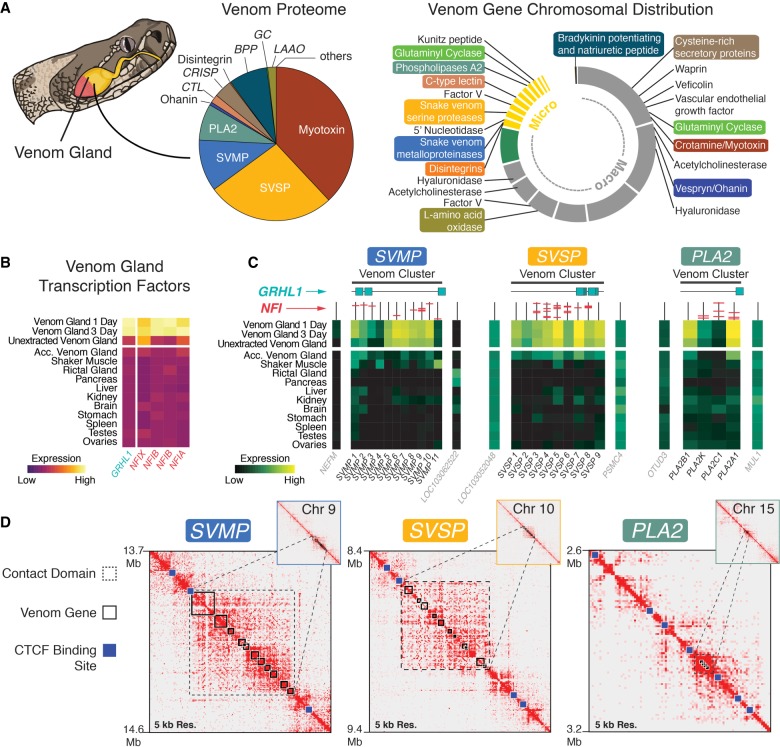
While numerous studies have characterized the diversity of venom composition among snake species (e.g., Mackessy 2008; Casewell et al. 2009, 2012; Rokyta et al. 2012), the chromosomal location of venom genes and mechanisms underlying the regulation of venom remain poorly understood. Our rattlesnake genome provides the genomic location and context for snake venom genes (Fig. 4A; Supplemental Tables S10, S11; Supplemental Fig. S17) and demonstrates that microchromosomes are enriched for these genes (i.e., 37% of all venom genes are found on microchromosomes which represent 10% of the genome; Fisher's exact test, P = 0.0017) (Fig. 4A). Moreover, microchromosome-linked venom gene families include three of the most abundant and well-characterized components of rattlesnake venom (snake venom metalloproteinases, SVMPs; snake venom serine proteinases, SVSPs; and type IIA phospholipases A2, PLA2s) (Fig. 4A)—each of these families is located on a different microchromosome (Fig. 4A; Supplemental Fig. S17). The other major component of prairie rattlesnake venom, myotoxin (crotamine), is located on Chromosome 1 (Fig. 4A). To identify patterns of venom gene family evolution, we conducted phylogenetic estimates of each of the microchromosome-linked families listed above (including nonvenom paralogs). We inferred that each venom family represents a distinct set of tandemly duplicated genes derived from a single ancestral homolog that gave rise to a monophyletic cluster of venom paralogs (Supplemental Figs. S18, S19). While this has been proposed previously (Ikeda et al. 2010; Vonk et al. 2013), the contiguity of our genome provides new definitive evidence that this duplicative mechanism explains the origin of multiple unlinked snake venom gene clusters.
Figure 4.
Genomic context for venom gene regulation and production. (A) Pie chart of the venom proteome with relative abundance of venom families (redrawn from Saviola et al. 2015). Chromosomal location of venom gene families (right); colored labels correspond to families from the proteome chart. (B) Gene expression across tissues of transcription factors (TFs) significantly up-regulated in the venom gland. (C) Heat maps of gene expression across tissues for venom genes in each of the three focal venom gene families and the genes immediately flanking (i.e., outside of) each venom cluster (labeled in gray). Vertical lines above each gene represent their promoters, with predicted NFI binding sites shown in red. Predicted GRHL1 binding sites in venom clusters are shown as turquoise squares. (D) Hi-C heat maps showing contact domains (black dashed boxes), for the SVMP, SVSP, and PLA2 venom genes (solid black boxes). Blue squares are predicted CTCF binding sites. Values to the left of heat maps are start and end coordinates (in Mb) of each region, visualized at 5-kb resolution.
The depletion of venom is followed by the rapid expression, synthesis, and storage of proteins in the venom gland lumen over the course of several days. To investigate the regulation of venom production, we compared gene expression between venom glands and body tissues and identified a set of 12 transcription factors (TFs) with significantly higher expression in the venom gland (Fig. 4B; Supplemental Table S12; Supplemental Fig. S20). Many of these TFs were linked to the secretory demands of the venom gland (e.g., the unfolded protein response of the endoplasmic reticulum: ATF6 and CREB3L2) or repair of the glandular epithelium (e.g., ELF5). While the potential involvement of these TFs in regulating venom production cannot be entirely ruled out, we did not find evidence of predicted binding sites that would suggest a role in directly regulating venom genes (Supplemental Table S12). Five transcription factors, however, stood out as candidates for regulating venom gene expression based on their known regulatory functions, links to established mechanisms of venom production, and the proximity of their predicted binding sites to venom genes (Fig. 4B).
Though neither TFs nor transcriptional mechanisms regulating venom production have been precisely identified, there is evidence that following venom depletion, venom production is triggered by a1-adrenoceptors that activate the ERK signaling pathway (Kerchove et al. 2008). One of the venom-gland up-regulated TF genes was GRHL1, which is known to function in epidermal barrier formation and repair (Ting et al. 2005) and is regulated directly by ERK (Kim and McGinnis 2011). We also identified a set of four Nuclear Factor 1 (NFI) TFs, all of which share the same predicted binding site and are classified as RNA polymerase II core promoter binding TFs. NFI TFs are known to drive tissue-specific expression (Gronostajski 2000) and function in chromatin remodeling and transactivation (Fane et al. 2017). Predicted binding sites of GRHL1 tend to occur in close proximity to venom genes (average within 79 kb of a venom gene), and predicted NFI binding sites are present in the promoter regions of a large proportion (∼72%) of venom genes (Fig. 4C; Supplemental Tables S12, S13; Supplemental Fig. S19). We also found that genes flanking venom clusters (and lacking venom-specific expression) lacked NFI binding sites and were, on average, further (86 kb) away from the nearest GRHL1 binding site; binding sites for either set of TFs were not, however, statistically enriched in venom gene regions compared to the genomic background (Supplemental Table S13; Supplemental Methods). The up-regulation of GRHL1 and NFI TFs upon venom depletion and the presence of their predicted binding sites in venom gene clusters suggests these TFs may play a direct role in the regulation of venom, although the distribution of their binding sites does not entirely explain variation in venom gene expression (e.g., Fig. 4C), suggesting that other TFs and potentially other mechanisms also contribute to venom regulation.
Because our results indicated that the specificity of venom gene expression is not fully explained by venom-specific TF activity, we tested for evidence that venom is also regulated by specific chromatin structure and organization. We performed Hi-C sequencing of a 1-d post-extraction venom gland, which enabled us to capture chromatin contacts associated with venom production. Genomic regions containing venom clusters show a specific structure within discrete high-frequency chromatin contact regions, representing venom-specific topologically associated chromatin domains (TADs) (Fig. 4D; Supplemental Fig. S21; Dixon et al. 2016). These “venom TADs” are flanked by predicted binding sites of CTCF, which coordinates DNA looping and insulates transcriptional activity. Consistent with our chromatin data, we find that genes flanking venom TADs exhibit varied expression profiles across tissues, while genes within venom TADs show high venom gland specificity (Fig. 4C,D), indicating a strong insulating regulatory effect of TAD boundaries surrounding venom cluster regions. Collectively, these findings suggest that venom gene regulation is driven by synergistic interactions between tightly regulated chromatin structure and highly expressed TFs that are responsive to venom depletion.
Discussion
Our results provide new perspectives on the structure and function of amniote genomes, mechanisms and evolution of dosage compensation, and the biology and regulation of snake venom. These findings further demonstrate the potential for a new generation of well-assembled genomes to facilitate advances in our understanding of the diversity of genome biology across otherwise poorly characterized lineages of the tree of life. Much of what is known about reptile genome biology comes from studies of lizards and birds (e.g., Hillier et al. 2004; Warren et al. 2010; Alföldi et al. 2011), thus a primary motivation of this study was to use the highly contiguous rattlesnake genome to compare and contrast aspects of snake genome biology with those of other reptiles and amniotes. For example, studies of bird genomes have shown that avian microchromosomes are gene-rich and therefore functionally important. Despite the semi-independent origins of microchromosomes in squamate reptiles and birds, snake microchromosomes exhibit many of the same compositional patterns (i.e., gene density, GC and repeat content) as microchromosomes in birds (Fig. 1). Moreover, as the first species with microchromosomes to be examined using Hi-C, we find that rattlesnake microchromosomes exhibit fundamentally different patterns of chromatin contact, with proportionally higher inter-chromosomal contact frequencies than macrochromosomes in snakes or mammals (Fig. 3; Lieberman-Aiden et al. 2009; Rao et al. 2014; Darrow et al. 2016). This discovery highlights the unique structure and function of microchromosomes and raises the question of whether the uniqueness of snake microchromosome chromatin structure is a feature common to all amniote vertebrate microchromosomes. Future analyses using Hi-C or other data to compare microchromosome structure and nuclear contact patterns will be key to address the generality of links between microchromosome structure, organization, and function across vertebrate lineages.
A major goal of comparative genomics is to understand the patterns and mechanisms that lead to the observed variation in genome structure and function across species. Previous comparative analyses have demonstrated unique patterns of genome structure and content in squamate reptiles that are distinct from those observed in other major amniote lineages (i.e., birds and mammals) (Alföldi et al. 2011; Fujita et al. 2011; Pasquesi et al. 2018). Our comparative analyses of 12 squamate genomes provide new insight and context for understanding the evolution of unique genomic features of squamates (Fig. 1C–E). For example, our results indicate that snakes have re-evolved genomic GC-isochore structure while also evolving reduced overall genomic GC content. The confluence of these patterns raises the intriguing possibility that snake isochore structure has evolved not through an accumulation of GC content (i.e., GC-biased gene conversion), as observed in mammals and birds (Duret and Galtier 2009; Weber et al. 2014), but through the accumulation of AT content via AT-biased substitutions (Castoe et al. 2013) or other mechanisms. This observation in snakes, together with the extremely varied GC landscapes across squamates (Fig. 1C,D; Fujita et al. 2011; Castoe et al. 2013), raises a number of questions, including whether mechanisms outside of GC-biased gene conversion contribute to GC isochore structure in vertebrates and whether GC-biased gene conversion plays a major role in squamate genome evolution.
Due to the independent origins of distinct sex determination systems (Gamble et al. 2017) and variation in differentiation between sex chromosomes among lineages (Matsubara et al. 2006; Vicoso et al. 2013), snakes have become an important model system for investigating sex chromosome evolution. Through our analyses of the rattlesnake Z Chromosome, we identified the recombining pseudoautosomal region of the highly differentiated Z and W Chromosomes and an evolutionary stratum bearing the hallmarks of recombination suppression and degeneration on the W Chromosome. These findings indicate that even through the rattlesnake Z and W are highly differentiated, further differentiation and recombination suppression between the Z and W are ongoing (Fig. 2). Despite the independent origins of Z/W Chromosomes in rattlesnakes and birds, there are similarities in the patterns of GC-richness of the pseudoautosomal regions of sex chromosomes in both lineages, suggesting that common processes may drive increased pseudoautosomal region GC content across divergent amniote lineages.
Although previous studies have found evidence of a lack of global dosage compensation on the Z Chromosome in females (Vicoso et al. 2013; Yin et al. 2016), the evolution of gene expression and incomplete dosage compensation as the snake Z Chromosome evolved has not been studied. Our comparison of female and male Z Chromosome expression with inferred ancestral expression provides new evidence that, in comparison to the ancestral proto-Z autosome, male expression has remained largely constant, while female expression has become reduced after the establishment of the sex chromosomes (Fig. 2C). We also found that chromosome-wide gene expression on the proto-Z was higher in both sexes than on other autosomes, raising the possibility that autosomes with these expression characteristics may be more likely (e.g., predisposed) to become sex chromosomes. We further demonstrated high gene-specific or regional variation in dosage compensation in the rattlesnake and provide the first report that a female-biased transcriptional regulatory mechanism that modulates expression in other reptiles (Rice et al. 2017), estrogen response elements, does explain some of the variation in dosage compensation across the Z Chromosome. Specifically, we found that the density of estrogen response elements positively correlates with female gene expression across the Z Chromosome (Fig. 2D) and that these elements have accumulated on the Z Chromosome following its divergence from its autosomal homolog (Chromosome 6) in the anole lizard. Evidence for ERE accumulation on the Z Chromosome of the rattlesnake and the five-pacer viper further indicates that ERE accumulation occurred early in the evolution of the snake Z Chromosome and provides evidence for the potential role of EREs in dosage compensation in ZW systems.
Despite venom representing the most intensively studied feature of snake biology, previous fragmentary snake genome assemblies have provided limited genomic context for snake venom evolution and regulation. Leveraging the first chromosome-level genome assembly for a snake, our precise chromosomal localization of genome genes revealed that numerous important components of snake venom (Mackessy 2008) are located on snake microchromosomes (Fig. 4A), further underscoring the functional importance of snake microchromosomes. Our integrated analysis of venom systems provides new evidence for a role of GRHL1 in venom gene regulation, thereby linking a transcriptional regulatory mechanism to a previously known regulatory stimulus (ERK signaling) (Kim and McGinnis 2011) shown to trigger venom production (Kerchove et al. 2008). Analyses of Hi-C chromatin contact information from recently depleted venom glands provided new evidence for the tight regulation of chromatin in and around venom gene clusters, to the extent that venom genes occupy venom-specific topologically associated domains (venom TADs) bounded by CTCF binding sites, and genes within versus outside the boundaries of these venom TADs show distinct expression profiles (Fig. 4). Collectively, our results provide new evidence for the coordinated roles of chromatin organization and transcription factor activity in the process of venom gene regulation.
Methods
Genome assembly and annotation
Animal procedures were conducted with approved and registered IACUC protocols. Chicago and Hi-C libraries were constructed from genomic DNA from the liver and venom gland of a single male Crotalus viridis viridis, and assembly was performed using the Dovetail Genomics HiRise v2.1.3-59a1db48d61f assembler. A previous version of this assembler (Putnam et al. 2016) is available as an open-source distribution at https://github.com/DovetailGenomics/HiRise_July2015_GR; however, Dovetail Genomics has not made the HiRise version used on this assembly available as open source software at this time. Chicago and Hi-C data were used to improve an existing fragmentary assembly (CroVir2.0; NCBI accession SAMN07738522) (Supplemental Tables S1, S2), which was constructed using multiple short-read sequencing libraries in combination with long-insert mate-pair libraries (Supplemental Table S1). Information about input assembly breaks and Chicago assembly scaffold joins can be found in Supplemental Material 2. Genomic DNA for these libraries was extracted from snap-frozen liver tissue using standard phenol-chloroform-isoamyl DNA extraction methods. We generated 24 transcriptomic libraries from 16 different tissue types (Supplemental Table S3) to generate a de novo rattlesnake transcriptome, which we assembled using Trinity v.20140717 (Grabherr et al. 2011) with default settings. De novo transcriptome assembly resulted in 801,342 transcripts, including 677,921 Trinity-annotated genes, with an average length of 559 bp and N50 length of 718 bp.
We annotated repeat elements present in the improved genome assembly using libraries from complete squamate genomes (Supplemental Methods) constructed using RepeatModeler v.1.0.9 (Smit and Hubley 2015). De novo and homology-based predictions were then performed using RepeatMasker v.4.0.6 (Smit et al. 2015). We used MAKER v.2.31.8 (Cantarel et al. 2008) to annotate protein-coding genes using empirical evidence for gene prediction from our de novo transcriptome assembly detailed above and protein data sets of all annotated protein-coding genes for Anolis carolinensis (Alföldi et al. 2011), Python molurus bivittatus (Castoe et al. 2013), Thamnophis sirtalis (Perry et al. 2018), Ophiophagus Hannah (Vonk et al. 2013), and Deinagkistrodon acutus (Yin et al. 2016). Prior to running MAKER, we used BUSCO v. 2.0.1 (Simão et al. 2015) and the full C. viridis genome assembly to iteratively train AUGUSTUS v. 3.2.3 (Stanke and Morgenstern 2005) HMM models based on 3950 tetrapod vertebrate benchmarking universal single-copy orthologs (BUSCOs) (Supplemental Table S4). The resulting annotation consisted of 17,486 genes, and we ascribed gene IDs based on homology using reciprocal best-BLAST (with e-value thresholds of 1 × 10−5) and stringent one-way BLAST (with an e-value threshold of 1 × 10−8) searches against protein sequences from NCBI for Anolis, Python, and Thamnophis.
Hi-C sequencing and analysis
We dissected the venom glands from the genome animal 1 d and 3 d after venom was initially extracted in order to track a time-series of venom production. A subsample of the 1-d venom gland was sent to Dovetail Genomics, where DNA was extracted and replicate Hi-C sequencing libraries were prepared according to their protocol (see above). We also extracted total RNA from both 1-d and 3-d venom gland samples, along with tongue and pancreas tissue from the Hi-C genome animal. mRNA-seq libraries were generated and sequenced at Novogene on two separate lanes of the Illumina HiSeq 4000 platform using 150-bp paired-end reads (Supplemental Table S3).
Raw Illumina paired-end reads were mapped and processed using the Juicer pipeline (Durand et al. 2016) to produce Hi-C maps binned at multiple resolutions, as low as 5-kb resolution, and for the annotation of contact domains. All contact matrices used for further analysis were KR-normalized in Juicer. Topologically associated chromatin domains (TADs) were called using Juicer's Arrowhead algorithm for finding contact domains at various resolutions (5-kb, 10-kb, 25-kb, 50-kb, and 100-kb) with default settings (Durand et al. 2016). One hundred seventy-five TADs were identified at 5-kb resolution, 16 at 10-kb, 53 at 25-kb, 175 at 50-kb, and 126 at 100-kb. Additionally, TADs were annotated at 20-kb resolution using the HiCExplorer software (Ramírez et al. 2018). Raw reads were mapped and processed separately through HiCExplorer, and 1262 TADs were called at 20-kb resolution using the default settings with the P-value set to 0.05. We further identified TADs by eye at finer-scale (i.e., 5-kb) resolution.
We compared intra- and interchromosomal contact frequencies in the rattlesnake venom gland to the following mammalian Hi-C data sets: human lymphoblastoma cells (Rao et al. 2014) and human retinal epithelial cells, mouse kidney, and rhesus macaque tissue (Darrow et al. 2016).
Chromosome identification and synteny analysis
We determined the identity of chromosomes using a BLAST search of the chromosome-specific markers linked to snake chromosomes from Matsubara et al. (2006), downloaded from NCBI (accessions SAMN00177542 and SAMN00152474). We kept the best alignment per cDNA marker as its genomic location in the C. viridis genome, except when a marker hit two high-similarity matches on different chromosomes. The vast majority of markers linked to a specific macrochromosome (i.e., Chromosomes 1–7) (Supplemental Tables S6, S7) in Elaphe quadrivirgata mapped to a single genomic scaffold.
We identified a single 114-Mb scaffold corresponding to the Z Chromosome, as 10 out of 11 Z-linked markers mapped to this scaffold. To further vet this as the Z-linked region of the genome, we mapped reads from male and female C. viridis (Supplemental Table S8) to the genome using BWA (Li and Durbin 2009) with default settings, quantified coverage in 100-kb windows, and normalized windowed coverage by the median autosomal value per sex. The female exhibited roughly half the coverage of the male for much of the candidate Z Chromosome and nowhere else in the genome (Supplemental Fig. S7).
To explore broad-scale structural evolution across reptiles, we used the rattlesnake genome to perform in silico painting of the chicken (Gallus gallus version 5) and green anole (Anolis carolinensis version 2) genomes. Briefly, we divided the rattlesnake genome into 2.02 million potential 100-bp markers. For each of these markers, we used BLAST to record the single best hit in the target genome requiring an alignment length of at least 50 bp. This resulted in 41,644 potential markers in Gallus and 103,801 potential markers in Anolis. We then processed markers on each chromosome by requiring at least five consecutive markers supporting homology to the same rattlesnake chromosome. We consolidated each group of five consecutive potential markers as one confirmed marker. We also performed a traditional gene-based synteny analysis for comparison (Supplemental Methods; Supplemental Fig. S5), which yielded results consistent with our k-mer-based approach.
Sex chromosome analyses
The Z Chromosome was identified using the methods above, and the pseudoautosomal region (PAR) was identified based on an equal ratio of female:male genomic read coverage. The ‘Recent Stratum’ was identified using a comparison of female and male nucleotide diversity (π). To quantify gene expression on the rattlesnake Z Chromosome and across the genome, we prepared RNA-seq libraries from liver and kidney tissue from two males and females and sequenced them on an Illumina HiSeq using 100-bp paired-end reads (Supplemental Table S9). Per gene female-to-male (F/M) ratios of expression on the Z Chromosome were normalized by taking the log2 of the ratio of female and male Z expression values, each scaled first to the median expression level of autosomal genes in female and male, respectively. To explore regional variation in the current F/M gene expression ratio across the Z Chromosome, we performed a sliding window analysis of the log2 F/M expression ratio with a window size of 30 genes and a step size of one gene. Comparisons of current gene expression to inferred ancestral autosomal expression were performed using kidney and liver RNA-seq data from anole lizard and chicken males and females, following previously described methods (Julien et al. 2012; Marin et al. 2017). Additional details of these analyses are provided in the Supplemental Methods.
We predicted estrogen response elements (EREs; i.e., ESR1 binding sites) using the conserved ESR1 position weight matrix and binding site prediction using PoSSuM Search (Beckstette et al. 2006). We quantified the number of predicted EREs and the average current F/M gene expression ratio (see above) along the Z Chromosome in 100-kb windows and tested for a relationship between these variables using a Pearson's correlation coefficient. We also quantified the number of predicted EREs in the entire genome, as well as the entire Anolis genome. We then compared the density of EREs (i.e., number of EREs divided by total scaffold length) between the rattlesnake and Anolis genomes, and between the rattlesnake Z Chromosome and Anolis Chromosome 6, specifically. We tested for ERE enrichment on the Z Chromosome compared to Anolis Chromosome 6 using a Fisher's exact test.
Venom analyses
Venom homologs in the rattlesnake genome were identified and annotated using representatives from 38 known venom gene families (Supplemental Methods; Supplemental Table S10). Three venom gene families that are especially abundant, both in terms of presence in the venom proteome (Fig. 4A) and in copy number, in the venom of C. viridis are phospholipases A2, snake venom metalloproteinases, and snake venom serine proteases. Rattlesnakes possess multiple members of each of these gene families, and the steps taken above appeared to underestimate the total number of copies in the C. viridis genome. Therefore, for each of these families, we performed an empirical annotation using the Fgenesh++ (Solovyev et al. 2006) protein similarity search.
To detect potential tandem duplication events in venom gene families, we used LASTZ (Harris 2007) to align the genomic regions containing PLA2, SVMP, and SVSP genes to themselves. We used program defaults, with the exception of the “hspthresh” command, which we set to 8000. This was done to only return very high similarity matches between compared sequences. We then performed Bayesian phylogenetic analyses to further evaluate evidence of tandem duplication and monophyly among members of the PLA2, SVMP, and SVSP venom gene families. We generated protein alignments of venom genes with their most similar homologs, which we identified using tBLASTx searches between venom genes and our whole gene set using MUSCLE (Edgar 2004) with default parameters, with minor manual edits to the alignment to remove any poorly aligned regions, and analyzed protein alignments using BEAST2 (Bouckaert et al. 2014).
Gene expression of annotated genes was compared between the venom gland and multiple body tissues. To test for significant expression differences between venom gland and body tissues, we performed pairwise comparisons between combined venom gland (i.e., 1-d venom gland, 3-d venom gland, and unextracted venom gland) and body (all other tissues, except for accessory venom gland) tissue sets using an exact test of the binomial distribution estimated in edgeR, integrating tagwise dispersion (Robinson and Oshlack 2010). Genes with differential expression at an FDR value ≤0.05 were considered significant.
To identify candidate transcription factors regulating venom gene expression, we searched the genome annotation for all genes included on the UniProt (http://www.uniprot.org) reviewed human transcription factor database. Twelve candidate transcription factors included in this list were found to be significantly up-regulated in the venom gland (Supplemental Tables S12, S13). Because four transcription factors of the NFI family each showed evidence of venom gland-specificity, we tested that their binding motifs are also upstream of venom genes by quantifying the number of predicted NFI binding sites using predictive searches analogous to those used for ESR1 (detailed above) in the 1-kb upstream region of each venom gene. We also searched for proximity of GRHL1 binding sites to venom gene regions, as well as all nonvenom genes. Here, we did not confine our search only to promoter regions. To test for enrichment of NFI binding sites in the upstream regions of venom genes, we divided the number of predicted binding sites upstream of venom genes by the total length of upstream regions and compared this value to the analogous proportion for upstream regions of all nonvenom genes using a Fisher's exact test (Supplemental Table S13). We performed a similar analysis for GRHL1 at each interval size, again comparing the density of predicted GRHL1 binding sites within intervals of venom genes to nonvenom genes (Supplemental Table S13).
Venom gene contact domains were identified using contact frequency heat maps from venom gland Hi-C, and CTCF binding sites were again predicted using the PoSSuM Search approach detailed above, using the conserved vertebrate CTCF position weight matrix. Because each PSSM has a different probability distribution based on the relative frequencies of observed binding and the length of the element, we precalculated the complete probability distribution for each PSSM using PoSSuMdist. We then used the resulting distribution in conjunction with relative base frequencies for the genome calculated using PoSSuMfreqs to identify putative binding sites exceeding a significance threshold. This threshold necessarily varied for different PSSMs but was never higher than P < 1 × 10−5.
Data access
The genome assembly has been deposited at DDBJ/ENA/Genbank (https://www.ncbi.nlm.nih.gov/nuccore/PDHV00000000) under accession number PDHV02000000. The Chicago and Hi-C data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA413201. This database also contains the previously published genome assembly (CroVir2.0) published in Pasquesi et al. (2018). The genome resequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA476794. The RNA-seq data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJNA477004.
Supplementary Material
Acknowledgments
Support for this work was provided by National Science Foundation (NSF) grant DEB-1655571 to T.A.C. and S.P.M., NSF grant IOS-655735 to T.A.C., a Research Dissemination and Faculty Development grant from the University of Northern Colorado to S.P.M., and NSF DDIG grants to D.R.S. and T.A.C. (DEB-1501886) and to D.C.C. and T.A.C. (DEB-1501747).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.240952.118.
References
- Alföldi J, Di Palma F, Grabherr M, Williams C, Kong L, Mauceli E, Russell P, Lowe CB, Glor RE, Jaffe JD, et al. 2011. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477: 587–591. 10.1038/nature10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backström N, Forstmeier W, Schielzeth H, Mellenius H, Nam K, Bolund E, Webster MT, Öst T, Schneider M, Kempenaers B. 2010. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res 20: 485–495. 10.1101/gr.101410.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker RJ, Bull JJ, Mengden GA. 1972. Karyotypic studies of thirty-eight species of North American snakes. Copeia 257 10.2307/1442486 [DOI] [Google Scholar]
- Beckstette M, Homann R, Giegerich R, Kurtz S. 2006. Fast index based algorithms and software for matching position specific scoring matrices. BMC Bioinformatics 7: 389 10.1186/1471-2105-7-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Skaletsky H, Cho TJ, Brown L, Locke D, Chen N, Galkina S, Pyntikova T, Koutseva N, Graves T. 2017. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat Genet 49: 387–394. 10.1038/ng.3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10: e1003537 10.1371/journal.pcbi.1003537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. 2015. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet 48: 427–437. 10.1038/ng.3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, et al. 2013. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience 2: 10 10.1186/2047-217X-2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado AS, Yandell M. 2008. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res 18: 188–196. 10.1101/gr.6743907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Harrison RA, Wüster W, Wagstaff SC. 2009. Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC Genomics 10: 564 10.1186/1471-2164-10-564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casewell NR, Huttley GA, Wuster W. 2012. Dynamic evolution of venom proteins in squamate reptiles. Nat Commun 3: 1066 10.1038/ncomms2065 [DOI] [PubMed] [Google Scholar]
- Castoe TA, de Koning APJ, Hall KT, Card DC, Schield DR, Fujita MK, Ruggiero RP, Degner JF, Daza JM, Gu WJ, et al. 2013. The Burmese python genome reveals the molecular basis for extreme adaptation in snakes. Proc Natl Acad Sci 110: 20645–20650. 10.1073/pnas.1314475110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn MJ, Tickle C. 1999. Developmental basis of limblessness and axial patterning in snakes. Nature 399: 474–479. 10.1038/20944 [DOI] [PubMed] [Google Scholar]
- Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC, Sun Z, Huang SC, Sanborn AL, Machol I, Shamim M. 2016. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci 113: E4504–E4512. 10.1073/pnas.1609643113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B. 2016. Chromatin domains: the unit of chromosome organization. Mol Cell 62: 668–680. 10.1016/j.molcel.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. 2009. Biased gene conversion and the evolution of mammalian genomic landscapes. Ann Rev Genomics Hum Genet 10: 285–311. 10.1146/annurev-genom-082908-150001 [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fane M, Harris L, Smith AG, Piper M. 2017. Nuclear factor one transcription factors as epigenetic regulators in cancer. Int J Cancer 140: 2634–2641. 10.1002/ijc.30603 [DOI] [PubMed] [Google Scholar]
- Fujita MK, Edwards SV, Ponting CP. 2011. The Anolis lizard genome: an amniote genome without isochores. Genome Biol Evol 3: 974–984. 10.1093/gbe/evr072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble T, Castoe TA, Nielsen SV, Banks JL, Card DC, Schield DR, Schuett GW, Booth W. 2017. The discovery of XY sex chromosomes in a Boa and Python. Curr Biol 27: 2148–2153.e4. 10.1016/j.cub.2017.06.010 [DOI] [PubMed] [Google Scholar]
- Geffeney S, Brodie ED, Ruben PC. 2002. Mechanisms of adaptation in a predator-prey arms race: TTX-resistant sodium channels. Science 297: 1336–1339. 10.1126/science.1074310 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. 2011. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat Biotechnol 29: 644 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat Rev Genet 17: 33–46. 10.1038/nrg.2015.2 [DOI] [PubMed] [Google Scholar]
- Gronostajski RM. 2000. Roles of the NFI/CTF gene family in transcription and development. Gene 249: 31–45. 10.1016/S0378-1119(00)00140-2 [DOI] [PubMed] [Google Scholar]
- Harris RS. 2007. “Improved pairwise alignment of genomic DNA.” PhD thesis, The Pennsylvania State University, State College, PA. [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MAM, Delany ME. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432: 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Ikeda N, Chijiwa T, Matsubara K, Oda-Ueda N, Hattori S, Matsuda Y, Ohno M. 2010. Unique structural characteristics and evolution of a cluster of venom phospholipase A2 isozyme genes of Protobothrops flavoviridis snake. Gene 461: 15–25. 10.1016/j.gene.2010.04.001 [DOI] [PubMed] [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schütz F, Daish T, Grützner F, Kaessmann H. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol 10: e1001328 10.1371/journal.pbio.1001328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchove CM, Luna MSA, Zablith MB, Lazari MFM, Smaili SS, Yamanouye N. 2008. α1-adrenoceptors trigger the snake venom production cycle in secretory cells by activating phosphatidylinositol 4,5-bisphosphate hydrolysis and ERK signaling pathway. Comp Biochem Physiol A Mol Integr Physiol 150: 431–437. 10.1016/j.cbpa.2008.04.607 [DOI] [PubMed] [Google Scholar]
- Kim M, McGinnis W. 2011. Phosphorylation of Grainy head by ERK is essential for wound-dependent regeneration but not for development of an epidermal barrier. Proc Natl Acad Sci 108: 650–655. 10.1073/pnas.1016386108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackessy SP. 2008. Venom composition in rattlesnakes: trends and biological significance. In The biology of rattlesnakes (ed. Hayes WK, et al. ), pp. 495–510. Loma Linda University Press, Loma Linda, CA. [Google Scholar]
- Mackessy SP. 2010. The field of reptile toxinology. In Handbook of venoms and toxins of reptiles (ed. Mackessy SP), pp. 3–23. CRC Press, New York. [Google Scholar]
- Marin R, Cortez D, Lamanna F, Pradeepa MM, Leushkin E, Julien P, Liechti A, Halbert J, Brüning T, Mössinger K. 2017. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res 27: 1974–1987. 10.1101/gr.223727.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara K, Tarui H, Toriba M, Yamada K, Nishida-Umehara C, Agata K, Matsuda Y. 2006. Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci 103: 18190–18195. 10.1073/pnas.0605274103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor RE, Romanov MN, Kiazim LG, Barrett PM, Farré M, Damas J, Furguson-Smith M, Valenzuela N, Larkin DM, Griffin DK. 2018. Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat Commun 9: 1883 10.1038/s41467-018-04267-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmo E. 2005. Rate of chromosome changes and speciation in reptiles. Genetica 125: 185–203. 10.1007/s10709-005-8008-2 [DOI] [PubMed] [Google Scholar]
- Organ CL, Godínez Moreno R, Edwards SV. 2008. Three tiers of genome evolution in reptiles. Integr Comp Biol 48: 494–504. 10.1093/icb/icn046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquesi GI, Adams RH, Card DC, Schield DR, Corbin AB, Perry BW, Reyes-Velasco J, Ruggiero RP, Vandewege MW, Shortt JA, et al. 2018. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat Commun 9: 2774 10.1038/s41467-018-05279-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry BW, Card DC, McGlothlin JW, Pasquesi GI, Hales NR, Corbin AB, Adams RH, Schield DR, Fujita MK, Demuth JP, et al. 2018. Molecular adaptations for sensing and securing prey, and insight into amniote genome diversity, revealed by the garter snake genome. Genome Bio Evol 10: 2110–2129. 10.1093/gbe/evy157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. 2016. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res 26: 342–350. 10.1101/gr.193474.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez F, Bhardwaj V, Arrigoni L, Lam KC, Grüning BA, Villaveces J, Habermann B, Akhtar A, Manke T. 2018. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 9: 189 10.1038/s41467-017-02525-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice ES, Kohno S, John JS, Pham S, Howard J, Lareau LF, O'Connell BL, Hickey G, Armstrong J, Deran A, et al. 2017. Improved genome assembly of American alligator genome reveals conserved architecture of estrogen signaling. Genome Res 27: 686–696. 10.1101/gr.213595.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 11: R25 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta DR, Lemmon AR, Margres MJ, Aronow K. 2012. The venom-gland transcriptome of the Eastern diamondback rattlesnake (Crotalus adamanteus). BMC Genomics 13: 312 10.1186/1471-2164-13-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saviola AJ, Pla D, Sanz L, Castoe TA, Calvete JJ, Mackessy SP. 2015. Comparative venomics of the Prairie Rattlesnake (Crotalus viridis viridis) from Colorado: identification of a novel pattern of ontogenetic changes in venom composition and assessment of the immunoreactivity of the commercial antivenom CroFab®. J Proteomics 121: 28–43. 10.1016/j.jprot.2015.03.015 [DOI] [PubMed] [Google Scholar]
- Secor S, Diamond J. 1998. A vertebrate model of extreme physiological regulation. Nature 395: 659–662. 10.1038/27131 [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smeds L, Kawakami T, Burri R, Bolivar P, Husby A, Qvarnström A, Uebbing S, Ellegren H. 2014. Genomic identification and characterization of the pseudoautosomal region in highly differentiated avian sex chromosomes. Nat Commun 5: 5448 10.1038/ncomms6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AF, Hubley R. 2015. RepeatModeler Open-1.0. 2008–2015. http://www.repeatmasker.org.
- Smit AFA, Hubley R, Green P. 2015. RepeatMasker Open-4.0. 2013–2015. Institute for Systems Biology. http://www.repeatmasker.org.
- Solovyev V, Kosarev P, Seledsov I, Vorobyev D. 2006. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol 7: S10 10.1186/gb-2006-7-s1-s10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikulnath K, Nishida C, Matsubara K, Uno Y, Thongpan A, Suputtitada S, Apisitwanich S, Matsuda Y. 2009. Karyotypic evolution in squamate reptiles: Comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res 17: 975–986. 10.1007/s10577-009-9101-7 [DOI] [PubMed] [Google Scholar]
- Stanke M, Morgenstern B. 2005. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33: W465–W467. 10.1093/nar/gki458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting SB, Caddy J, Hislop N, Wilanowski T, Auden A, Zhao L-l, Ellis S, Kaur P, Uchida Y, Holleran WM. 2005. A homolog of Drosophila grainy head is essential for epidermal integrity in mice. Science 308: 411–413. 10.1126/science.1107511 [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol 11: e1001643 10.1371/journal.pbio.1001643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonk FJ, Casewell NR, Henkel CV, Heimberg AM, Jansen HJ, McCleary RJR, Kerkkamp HME, Vos RA, Guerreiro I, Calvete JJ, et al. 2013. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc Natl Acad Sci 110: 20651–20656. 10.1073/pnas.1314702110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss SR, Kump DK, Putta S, Pauly N, Reynolds A, Henry R, Basa S, Walker JA, Smith JJ. 2011. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res 21: 1306–1312. 10.1101/gr.116491.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, Arnold AP, Hillier LW, Künstner A, Searle S, White S, Vilella AJ, Fairley S, et al. 2010. The genome of a songbird. Nature 464: 757–762. 10.1038/nature08819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CC, Boussau B, Romiguier J, Jarvis ED, Ellegren H. 2014. Evidence for GC-biased gene conversion as a driver of between-lineage differences in avian base composition. Genome Biol 15: 549 10.1186/s13059-014-0549-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Wang ZJ, Li QY, Lian JM, Zhou Y, Lu BZ, Jin LJ, Qiu PX, Zhang P, Zhu WB, et al. 2016. Evolutionary trajectories of snake genes and genomes revealed by comparative analyses of five-pacer viper. Nat Commun 7: 13107 10.1038/ncomms13107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wiens J. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol Phylogenet Evol 94: 537–547. 10.1016/j.ympev.2015.10.009 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.