Abstract
Vancomycin-resistant Enterococcus faecium (VREfm) is a leading cause of healthcare-associated infection. Reservoirs of VREfm are largely assumed to be nosocomial although there is a paucity of data on alternative sources. Here, we describe an integrated epidemiological and genomic analysis of E. faecium associated with bloodstream infection and isolated from wastewater. Treated and untreated wastewater from 20 municipal treatment plants in the East of England, United Kingdom was obtained and cultured to isolate E. faecium, ampicillin-resistant E. faecium (AREfm), and VREfm. VREfm was isolated from all 20 treatment plants and was released into the environment by 17/20 plants, the exceptions using terminal ultraviolet light disinfection. Median log10 counts of AREfm and VREfm in untreated wastewater from 10 plants in direct receipt of hospital sewage were significantly higher than 10 plants that were not. We sequenced and compared the genomes of 423 isolates from wastewater with 187 isolates associated with bloodstream infection at five hospitals in the East of England. Among 481 E. faecium isolates belonging to the hospital-adapted clade, we observed genetic intermixing between wastewater and bloodstream infection, with highly related isolates shared between a major teaching hospital in the East of England and 9/20 plants. We detected 28 antibiotic resistance genes in the hospital-adapted clade, of which 23 were represented in bloodstream, hospital sewage, and municipal wastewater isolates. We conclude that our findings are consistent with widespread distribution of hospital-adapted VREfm beyond acute healthcare settings with extensive release of VREfm into the environment in the East of England.
Vancomycin-resistant Enterococcus faecium (VREfm) is a leading cause of healthcare-associated infection and particularly affects critically ill and immunocompromised patients (Sievert et al. 2013). This problem has been driven by the emergence and global dissemination of successful lineages belonging to the hospital-adapted E. faecium clade designated as A1 (previously known as clonal complex 17 [CC17] by multilocus sequencing typing [MLST]) (Willems et al. 2005; Arias and Murray 2013; Lebreton et al. 2013; Guzman Prieto et al. 2016). Clade A1 isolates are characterized by chromosomally mediated ampicillin and fluoroquinolone resistance, high genomic plasticity, and the accumulation of horizontally acquired genes encoding virulence factors and antibiotic resistance (Willems et al. 2001; Leavis et al. 2004, 2006; Hegstad et al. 2010; Palmer et al. 2010). This includes resistance to vancomycin, the drug of choice for ampicillin-resistant E. faecium (AREfm) infection (Howden et al. 2013; Raven et al. 2017). In contrast to clade A1, an animal-associated clade A2 and a distantly related human commensal (or community-associated) clade B are rarely associated with invasive infection or vancomycin resistance (Lebreton et al. 2013; Raven et al. 2016).
Carriage of nosocomial pathogens precedes infection, and effective strategies to prevent infection are built on an understanding of how and from where people acquire their infecting organism. Genomic studies of nosocomial E. faecium infection have confirmed that transmission of E. faecium clade A1 is common (Howden et al. 2013; Pinholt et al. 2017; Raven et al. 2017; van Hal et al. 2017). A key unknown, however, is whether the development of carrier status is solely attributable to acquisition in hospitals or whether external reservoirs in the wider community and the environment could contribute to the background carriage rate. There is also a lack of information regarding rates of VREfm carriage in community populations, including those receiving healthcare and otherwise healthy populations. Municipal wastewater represents a surrogate reservoir containing pooled bacteria from human populations, the geographical spacing of which means that these receive waste from a differing case-mix, including plants that directly receive hospital waste and those that do not, in addition to being located in urban and rural settings (Cai et al. 2014).
Here, we describe a study that aimed to use wastewater to generate indirect evidence for the extent to which healthcare-associated E. faecium is disseminated in the community. This took an integrated approach that combined microbiological, epidemiological, and bacterial whole-genome sequence data and compared the genetic relatedness and presence of antibiotic resistance and putative virulence genes in E. faecium from sewage and patients with bloodstream infection in the same geographic region.
Results
Isolation of E. faecium from wastewater
A cross-sectional survey was conducted between June 2014 and January 2015 to isolate and quantify E. faecium in untreated and treated wastewater obtained from 20 municipal treatment plants in the East of England, half of which were not in direct receipt of hospital waste (Fig. 1A). Treated samples were obtained at the point of release into surface waters. Ampicillin-resistant E. faecium (a surrogate marker for hospital-adapted lineages) and VREfm were isolated from untreated wastewater at all 20 plants. AREfm and VREfm were isolated from 18 and 17 treated samples, respectively, the negative samples being from plants using terminal ultraviolet light decontamination.
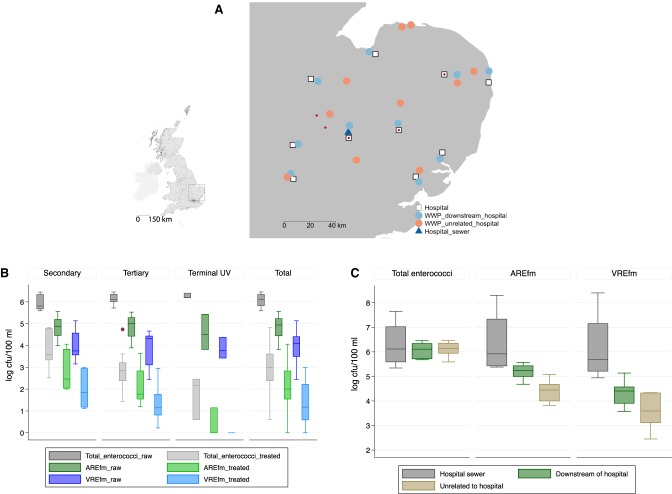
Figure 1.
Geographic origin of E. faecium isolates and wastewater counts. (A) Map of 20 wastewater treatment plants in the East of England tested for E. faecium, 10 of which were directly downstream from acute hospitals belonging to the Cambridge University Hospitals NHS Foundation Trust (CUH) referral network. (WWP) Wastewater plant. Red dots indicate the source of bloodstream isolates. (Inset) Map of the United Kingdom, with square denoting the East of England. (B) Log10 counts of E. faecium recovered from 100 mL of wastewater under increasing antibiotic selection for untreated and treated wastewater according to water treatment type: secondary (n = 7), tertiary (n = 10), UV light treatment (n = 3), and for all plants (n = 20). All comparisons between treated and untreated wastewater counts were statistically significant (P < 0.001), as were comparisons of count reductions according to treatment type for each of total enterococci, AREfm, and VREfm (P = 0.02, P < 0.001, and P = 0.001, respectively). (C) Median log10 counts of E. faecium recovered from 100 mL of wastewater under increasing antibiotic selection from untreated wastewater from the main CUH sewer on four separate occasions, from 10 wastewater plants located downstream from acute hospitals, and from 10 wastewater plants unrelated to acute hospital waste. Boxes represent interquartile ranges, whiskers 1.5 times the interquartile range, and dots outside values (B,C).
Wastewater treatment was associated with an average reduction in count of 3.0, 2.7, and 2.5 log10 cfu/100 mL for all enterococci, AREfm, and VREfm, respectively (comparison of untreated versus treated wastewater, P < 0.001 in all cases) (Fig. 1B). The median count of VREfm in untreated and treated wastewater was 12,463 and 14 cfu/100 mL, respectively (P < 0.001). The log10 reduction was not dependent on E. faecium counts in untreated wastewater but varied by treatment process. The highest level of treatment processes used at the plants was secondary (n = 7), tertiary (n = 10), or terminal ultraviolet light decontamination (n = 3) (see Supplemental Methods and Supplemental Table S1 for further details). Compared with secondary treatment, tertiary treatment reduced AREfm and VREfm by a further 0.8 and 0.7 log10 cfu/100 mL on average, respectively. UV light reduced the concentration of AREfm and VREfm by a further 2.4 and 2.0 log10 cfu/100 mL, respectively (P < 0.001 and 0.002, respectively).
Four sewage samples were also obtained for culture from the main hospital sewer at the Cambridge University Hospitals NHS Foundation Trust (CUH, a major teaching hospital in the East of England) between September 2014 and December 2015. AREfm and VREfm were isolated from all four samples. Bacterial counts on selective agar for AREfm and VREfm were roughly equivalent to the total enterococcal count on nonselective agar (median log10 cfu/100 mL 5.9, 5.7, and 6.1, respectively) (Fig. 1C), indicating that drug-resistant E. faecium predominated. This contrasted with untreated municipal wastewater, where the counts of AREfm and VREfm were 5.7% (IQR 2.9%–14.5%) and 1.1% (IQR 0.4%–2.6%) of the counts of total enterococci, respectively (Fig. 1B). Median log10 counts of AREfm and VREfm in 100 mL of untreated wastewater from plants that were in direct receipt of hospital sewage were significantly higher than plants that were not (5.2 vs. 4.4, P < 0.001, and 4.3 vs. 3.5, P = 0.008, respectively), although total enterococcal counts were not significantly different (6.0 vs. 6.1, P = 0.60) (Fig. 1C).
Comparing the genomes of E. faecium from wastewater and invasive disease
We selected 423 E. faecium isolates for sequencing (383 from treatment plants and 40 from the CUH sewer) (see Supplemental Methods for further details of isolation and selection). In silico MLST of the 383 isolates from treatment plants resolved 93 sequence types (STs), including 28 novel STs (including two previously nontypeable isolates due to the absence of pstS assigned to ST1478) (Carter et al. 2016; Raven et al. 2016), indicating a high genetic diversity (STs are listed in Supplemental Tables S2, S3). E. faecium from nonselective agar was the most diverse (60 STs, 92 isolates, 0.65 ST/isolate), compared with 29 STs for E. faecium selected for ampicillin resistance (112 isolates, 0.26 ST/isolate) and 17 STs for E. faecium selected for vancomycin resistance (179 isolates, 0.09 ST/isolate) (Fig. 2A; Supplemental Table S3). In silico MLST of the 40 isolates from the hospital sewer resolved 15 STs, the majority of which were either ST18 (n = 10) or ST80 (n = 10) and included four novel STs (Supplemental Table S2). These findings are consistent with specific subsets of STs being associated with drug resistance.
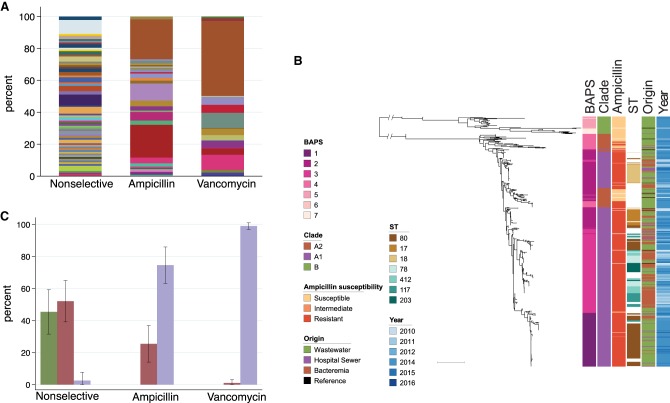
Figure 2.
Genetic characterization of E. faecium isolates from wastewater and bloodstream infections. (A) MLST sequence types of 383 E. faecium isolates recovered from wastewater according to antibiotic selection during isolation. Nonselective: no selection for antibiotic resistance; ampicillin: ampicillin-resistance selection; vancomycin: vancomycin-resistance selection. (B) Midpoint rooted maximum-likelihood tree based on SNPs in 1336 core genes of 620 E. faecium (383 wastewater, 40 hospital sewage, 187 from bloodstream, and 10 reference isolates) labeled by hierarchical Bayesian cluster (1–7), clade (B, A2, and A1), ampicillin-susceptibility, commonest STs, origin, and year of isolation. Scale bar, ∼10,000 SNPs. (C) Proportion of each E. faecium clade recovered from wastewater according to antibiotic selection during isolation. A single isolate of clade A2 recovered on the vancomycin selective plate was vancomycin-susceptible. Error bars, standard deviation.
We then compared the genomes of the 423 wastewater E. faecium isolates with 187 E. faecium isolates associated with bloodstream infection in patients in the East of England between 2010 and 2016. Phylogenetic reconstruction using SNPs in the core genome of these 610 isolates together with 10 nonstudy genomes from open access databases demonstrated that the population divided into two major lineages, which were consistent with clade B (43 isolates), and clade A (577 isolates) (Fig. 2B). Hierarchical Bayesian clustering resolved seven BAPS (Bayesian Analysis of Population Structure) groups, of which three corresponded to clade B and four to clade A, the latter including BAPS-2 corresponding to the ST17/18 lineage, BAPS-3 corresponding to ST78, and BAPS-1 predominantly corresponding to ST80 (Fig. 2B).
BAPS-4 had several distinctive features, including genomes that resided on long branches (indicating distantly related isolates) and ampicillin-susceptible isolates (consistent with clade A2). Furthermore, this BAPS group was paraphyletic (located in two different positions in the tree). The distinction between A1 and A2 was resolved by constructing a phylogeny based on core genome mapping to the Aus0004 strain (Lam et al. 2013) after excluding the most basal A2 isolates (designated A2.1) (Supplemental Fig. S1). These findings suggested that clade A2 was heterogeneous and contained a lineage (designated here as A2.2) that resided between A2.1 and A1, including AREfm isolates belonging to STs associated previously with carriage by domestic dogs (e.g., ST19, ST192, ST266, ST401) (de Regt et al. 2012), although distinct from other ST192 isolates that clearly resided in clade A1. These findings are indicative of a highly dynamic genome with recombination between lineages that have been considered to have different associations (i.e., hospital- versus animal-associated). Trees of clades A2 and B are shown in Supplemental Figure S2.
Having identified clades A1, A2, and B, we plotted each clade against the three different culture media used for bacterial isolation (Fig. 2C). This confirmed that ampicillin-susceptible E. faecium resided in clades B and A2, AREfm resided in clades A2 and A1, and VREfm was exclusive to clade A1. We observed that clade A2 isolates constituted approximately half of E. faecium in untreated sewage. None of the plants knowingly received farm effluent, suggesting that these “animal-associated” lineages may be commonly carried by people in the community. Hospital-adapted lineages of clade A1 were present in treatment plants in both urban and rural settings and were not restricted to wastewater plants downstream from hospitals.
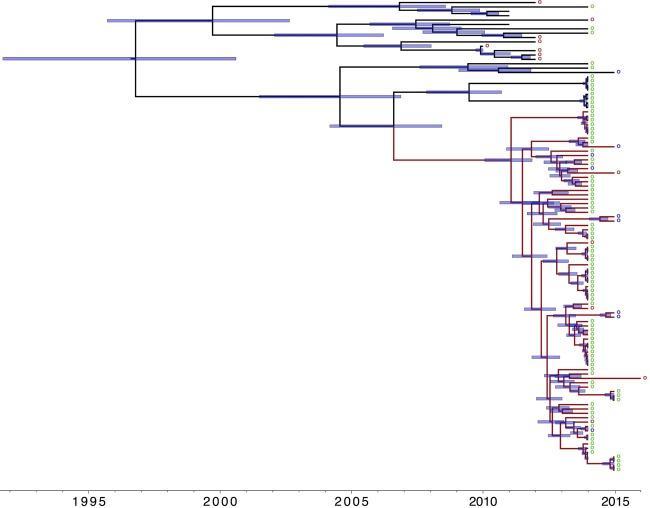
We then focused our analysis on the 481 isolates in clade A1 to determine the comparative phylogeny and gene content between isolates from wastewater and patients with bloodstream infection. A maximum-likelihood phylogeny following removal of recombination and mobile elements showed a diverse population interspersed with clonal expansions (Fig. 3A). Numerous clonal expansions were noted, with closely related isolates shared between two or more locations, including wastewater treatment plants (located downstream and unrelated to hospitals), hospital sewage, and/or invasive isolates. The largest cluster (termed C1) contained 72 isolates (15% of the total), including 64 from 15/20 wastewater treatment plants, four from CUH sewage, and four bloodstream isolates from CUH in 2014–15. The majority of isolates (68/72) in this cluster belonged to ST80. The substitution rate of the branch leading to this large cluster (calculated based on isolates shown in red in Fig. 3A) was 9.21 SNP/genome/yr (3.23 × 10−6 SNPs/site/yr), estimating the most recent common ancestor of C1 and related isolates at ∼9.4 yr (95% highest posterior density [HPD], 7.6–11.8 yr), with 83/91 (91%) having diverged 4.9 yr ago (95% HPD, 4.1–5.9 yr) (Fig. 4). This suggests their recent emergence and dissemination in our geographic region, including isolates that caused invasive disease.
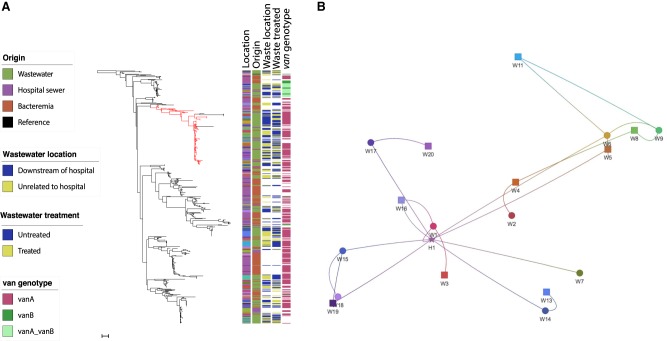
Figure 3.
Distribution of clade A1 E. faecium and clonal expansion of dominant clusters. (A) Maximum-likelihood tree based on SNPs in the core genes of 481 clade A1 E. faecium isolates colored by geographical location in relation to receipt of hospital sewage, origin, wastewater plant location in relation to hospitals, treated or untreated wastewater, and presence of van genes. Branch leading to and including the dominant cluster is colored in red. Scale bar, ∼55 SNPs. (B) Phylogenetic network analysis showing relatedness of E. faecium based on place of origin displayed as shapes corresponding to geographical coordinates. Circles: wastewater treatment plants located downstream from hospitals; squares: wastewater treatment plants unrelated to hospital waste; star: Cambridge University Hospitals (bloodstream isolates). The edges (lines) link isolates that were very closely related (within 5 SNPs in the core genome).
Figure 4.
Timeline of emergence of dominant E. faecium cluster. Maximum clade credibility tree of branch leading to and including the dominant cluster from BEAST analysis (n = 108 isolates). Branch in red includes all isolates of cluster (and neighboring isolates). Blue bars indicate 95% highest probability density (HPD) values. (Right) Circles denote origin of isolates. Green: wastewater; purple: CUH sewage; red: bloodstream infection. Time bar (yr) shown at the bottom.
The majority of wastewater treatment plants each contained a genetically diverse E. faecium clade A1 population based on core genome pairwise SNP distance (Supplemental Fig. S3a). This diversity was comparable to the diversity reported for isolates associated with bloodstream infection from across the United Kingdom (Raven et al. 2016) and was not affected by the treatment process (median SNP distance 251 vs. 242 in raw and treated wastewater, respectively, P = 0.54) (Supplemental Fig. S3b). In contrast, the minimum SNP distance between untreated and treated wastewater samples was zero for 14 of 18 evaluable locations (range 0–10), suggesting persistence of some strains at each plant following treatment (Supplemental Fig. S3c). Closely related isolates were also detected between different wastewater treatment plants (Supplemental Fig. S3d).
Comparison of relatedness between E. faecium from wastewater and bloodstream infection showed that the minimum SNP distance between any bloodstream to wastewater isolate did not differ significantly for plants located downstream from hospitals versus plants unrelated to hospitals (median 35 vs. 46, P = 0.09) (Supplemental Fig. S3e). Network analysis using a strict 5-SNP cut-off (consistent with less than 1 yr of evolution based on this study and previous estimates [Raven et al. 2017]) revealed geographical clustering of wastewater treatment plants. Bloodstream isolates from CUH formed a direct network with 9/20 plants (three unrelated to hospital waste), indicating recent dissemination of invasive and wastewater E. faecium across the region (Fig. 3B).
Genes encoding antibiotic resistance and virulence
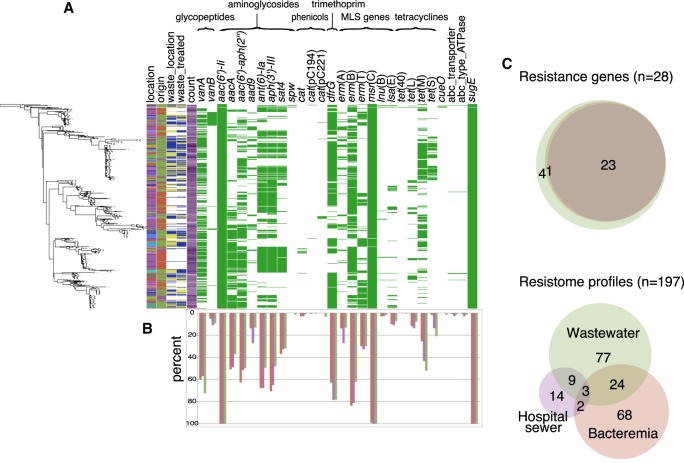
Having established close genetic relatedness between isolates from different reservoirs and locations based on the core genome, we analyzed the pangenome of clade A1 isolates for the presence of antibiotic, metal, and biocide resistance genes (Fig. 5A,B). Of 28 different resistance genes detected, 23 (82%) were represented in bloodstream, hospital sewage, and municipal wastewater isolates. Municipal wastewater isolates harbored the greatest diversity, including spw, cat(pC221), tet(40), and cueO that confer spectinomycin, chloramphenicol, tetracycline, and copper resistance, respectively, which were exclusive to wastewater isolates (Fig. 5C, top). There were 197 different antibiotic resistance gene combinations, with only three profiles shared between all three reservoirs and greater diversity detected in wastewater (Fig. 5C, bottom). The median number of genes detected was 9 (interquartile range [IQR] 8–10), 9 (IQR 9–11), and 8 (IQR 7–11) for bloodstream, hospital sewage, and municipal wastewater isolates, respectively. An analysis of putative virulence genes led us to detect 32 genes that were present in wastewater, hospital sewage, and bloodstream isolates (Supplemental Fig. S4). No gene was exclusively present in wastewater or bloodstream isolates based on a pangenome analysis (Supplemental Fig. S5).
Figure 5.
Distribution of antimicrobial resistance genes in hospital-adapted E. faecium from bloodstream infections, hospital, and municipal sewage. (A) Presence of antibiotic, metal, and biocide resistance genes grouped by antibiotic family in clade A1 E. faecium isolates shown against the core SNP maximum likelihood tree (left-hand side). Colors in left-hand columns correspond to those in Figure 3. Antimicrobial resistance gene columns: green, present; white, absent. (B) Frequency of detection of each gene in wastewater, hospital sewage, and bloodstream isolates (green: wastewater; purple: hospital sewer; red: bloodstream). (C) Venn diagrams showing the degree of overlap between individual and combined resistance gene profiles in the three reservoirs. (Upper panel) Innermost circle: bloodstream; middle circle: hospital sewer; outer circle: wastewater.
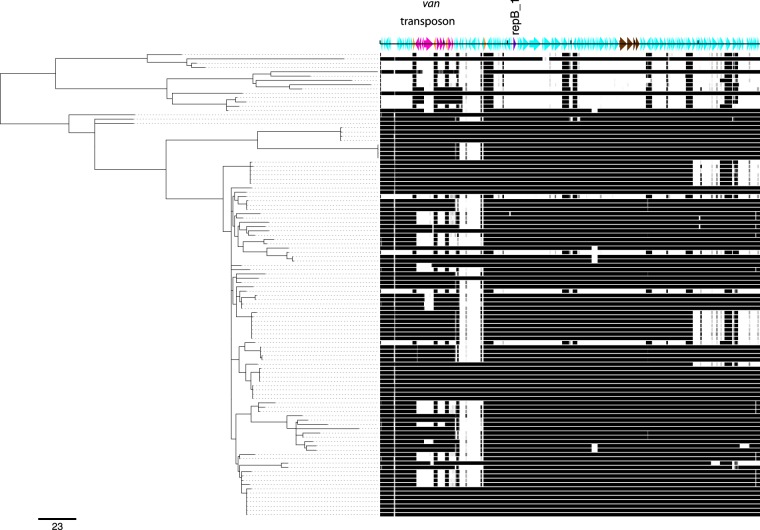
We investigated the genetic context of the vanA transposon for 108 isolates belonging to the lineage leading to the dominant cluster C1 investigated above, of which 80 contained the vanA gene (see Methods). Ninety-four isolates had reads mapping at >80% (65 at >90%) to a completely sequenced plasmid from a dominant clinical cluster of bloodstream isolates circulating in CUH since 2007 (Raven et al. 2017) that contained a novel rep gene and two genes encoding putative cell-surface proteins with Cna protein B-type domains but no additional resistance genes. Absence of vanA genes was associated either with loss of this plasmid or with loss of the vanA transposon and an approximately 9.5-kb downstream region flanked by IS1216E (Fig. 6).
Figure 6.
Reads mapped against vanA reference plasmid EC503 showing common plasmid backbone present in bloodstream and wastewater isolates. Left-hand side: maximum-likelihood tree of branch leading to dominant clade (n = 108). Scale bar, 23 SNPs. The density of the horizontal bars corresponds to the read coverage, with black indicating presence and white absence. (Top) Turquoise arrows, coding sequences: pink, vanA transposon genes; orange, IS1216E; purple, novel repB_1 gene; brown, genes encoding for novel putative cell-surface proteins (Cna protein B-type domains) and associated sortases. Markings shown at 20,000 bp. Vancomycin susceptibility was associated with lack of plasmid or lack of vanA transposon (with or without downstream 3′ region). Full sequence and annotation files of plasmid EC503 are available in Supplemental File S2.
Discussion
This systematic genomic survey of E. faecium in wastewater indicates that hospital-adapted lineages of VREfm are widespread in wastewater across the East of England, adding to previous studies restricted to wastewater collected downstream from hospitals (Novais et al. 2005; Caplin et al. 2008; Leclercq et al. 2013; Varela et al. 2013). Wastewater treatment did not prevent downstream environmental contamination with these lineages, with the majority of plants releasing drug-resistant E. faecium into the environment. Stronger controls are required to prevent these organisms being freely released into the water system. Ultraviolet light was used at three study plants and was effective in decontaminating wastewater prior to release. This represents a ready solution that would reduce environmental contamination with VREfm and drug-resistant bacteria more generally.
The use of culture media containing incremental antibiotic selective pressure allowed us to sample the diversity of E. faecium and quantify and obtain sufficient numbers of resistant lineages that were present in wastewater as a minority of the overall population. The observation that AREfm and VREfm counts were higher in untreated wastewater from plants in direct receipt of hospital sewage versus those that were not is predictable and consistent with the finding that the relative proportion of VREfm in wastewater from the CUH hospital main sewer was high. However, the detection of VREfm at all treatment plants is an important finding and consistent with widespread dissemination of drug-resistant lineages in the community. Potential reservoirs include people with past or on-going healthcare contact, including hospital patients and those in long-term care facilities (Brodrick et al. 2016). Given the scale of VREfm detection, including from plants in rural areas, it is possible that VREfm carriage extends to the wider healthy population. This could include healthcare workers, family contacts of people receiving healthcare, and people with no direct or indirect healthcare-associated contact. Potential sources in the wider community include the environment and the food chain. There is a lack of data on rates of VREfm carriage by healthy populations, with recent data from our group suggesting that VREfm in the food chain differs genetically from human and wastewater VREfm (Gouliouris et al. 2018). Further systematic studies are needed to identify community carriage.
Whole-genome sequence-based comparison of E. faecium from wastewater and patients with significant infection is essential for the accurate determination of relatedness between bacteria from disease and nondisease reservoirs. The core genome phylogenies generated here demonstrated that these were genetically intermixed and detected numerous highly related clusters that contained isolates from patients and wastewater. Wastewater isolates from nine treatment plants (including those that did not receive hospital sewage) were closely related to E. faecium associated with bloodstream infection at CUH. Furthermore, isolates from patients and nonhuman sources shared the majority of antibiotic resistance and virulence genes investigated. These observations build on our culture-based findings and provide strong evidence for the widespread dissemination of highly related healthcare-associated drug-resistant E. faecium lineages.
In conclusion, our findings highlight the challenges of controlling healthcare-associated dissemination of VREfm. The extent to which isolates from humans and wastewater were related indicates that wastewater could be used for the surveillance of circulating VREfm lineages. The risk posed to human health from extensive release of VREfm into the environment is uncertain but could be controlled by improving wastewater decontamination both at the hospital and municipal waste level. Both the risk of release and the benefit derived from preventing this require further study.
Methods
Wastewater sampling and microbiology
We performed a cross-sectional survey of 20 municipal wastewater treatment plants in the East of England between June 2014 and January 2015. Ten plants directly received sewage from acute NHS Hospital Trusts (median distance between plant and respective hospital: 5.3 km [range 3.3–9.6 km] downstream), and 10 plants did not directly receive hospital sewage. Treatment plant characteristics are shown in Supplemental Table S1 and described in Supplemental Methods. In brief, all plants processed wastewater using primary treatment (sedimentation of large particles in primary settlement tanks) and secondary treatment, which consisted of activated sludge and/or biological trickling filter beds. Ten plants utilized an additional tertiary step, such as lagooning (reed beds) or sand filtration. Wastewater effluent from three plants with discharge into sensitive coastal waters (shellfish production areas) was disinfected by a terminal ultraviolet light step. Untreated and treated wastewater were obtained from each plant. Grab samples of 0.5 L were collected at each sampling point and mixed into 1 L sterile bottles containing 18 mg sodium thiosulphate (Sigma-Aldrich). Additional sampling was performed at the hospital sewer (main septic tank) of CUH, when a single 1 L wastewater sample was obtained at four spaced time points between September 2014 and December 2015.
Bacterial quantification in wastewater was achieved by the membrane filtration method adapted from previously described protocols (Figueras et al. 2000) using the EZ-Stream vacuum pump and EZ-Fit Manifold system (Merck Millipore). Membranes were placed onto Slanetz-Bartley (SB) agar (Oxoid); ampicillin-selective medium BBL Enterococcosel agar (BD) supplemented with 30 mg/L ampicillin (Sigma-Aldrich); and vancomycin-selective medium Brilliance VRE chromogenic agar (Oxoid). Bacteria were identified to the species level using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS; Biotyper version 3.1, Bruker Daltonics). Antimicrobial susceptibility testing was determined using the P607 card on the VITEK2 instrument (bioMérieux) (Supplemental Fig. S6). More details of sample processing are provided in Supplemental Methods.
Bacteria for genome sequencing
Up to 20 colonies (isolates) were selected from primary cultures from each plant for genome sequencing (equal numbers from untreated and treated wastewater). Final selection was based on diversity of antibiotic resistance patterns (100 of the 110 possible patterns were included), which we used as a rough surrogate for genetic diversity. A further 10 isolates were selected from each CUH sewer sample using the same criteria. In all, 428 wastewater isolates were sequenced (388 from treatment plants and 40 from CUH sewers), of which five were subsequently excluded based on sequence quality metrics. Supplemental Table S2 provides the details of origin and isolation year for each isolate. The clinical isolate collection used here contained 187 E. faecium associated with bloodstream infection in 187 patients, as follows: 140 patients at CUH between 2010–2012, 24 patients at four further hospitals in the East of England between 2010–2012, and 23 consecutive cases of E. faecium bacteremia in hematology patients at CUH between May 2014 and April 2016. Isolates from 2010–2012 had already been sequenced, and data were downloaded from the Wellcome Sanger Institute. The remainder were sequenced as part of this project. DNA extraction and library preparation were performed as previously described (Raven et al. 2017). DNA libraries were sequenced using the Illumina HiSeq platform (Illumina Inc.). Further details can be found in Supplemental Methods.
Phylogenetic analyses
A phylogenetic analysis was undertaken combining 620 genomes (423 wastewater, 187 clinical, and 10 nonstudy publicly available complete E. faecium genomes (see Supplemental Table S2 for accession numbers). Genomes were aligned to define a core and accessory genome using Roary (Page et al. 2015). The resulting alignment of 100,136 core single nucleotide polymorphisms (SNPs) was used to create a phylogeny with RAxML (Stamatakis 2014) and to produce a tree-independent hierarchical Bayesian clustering with hierBAPS (Cheng et al. 2013). Further details are provided in the Supplemental Methods. Multilocus sequence typing was determined from genome assemblies using MLST Check (Page et al. 2016) with novel alleles and STs deposited in the pubMLST website (https://pubmlst.org/efaecium/). Absence of the pstS gene in two isolates was confirmed by SRST2 (Inouye et al. 2014).
The hospital-adapted lineage (A1) was defined by phylogenetic methods (see Supplemental Methods for details) and a phylogenetic tree of 481 genomes created using RAxML based on mapping to the Aus0004 genome (ENA accession number CP003351). Pairwise SNP distances were calculated from the core genome using a custom script available as Supplemental Code, and at https://github.com/simonrharris/pairwise_difference_count. Timeline reconstruction of the largest cluster was performed using BEAST v1.8.2 (Drummond et al. 2012), as described in the Supplemental Methods.
Analysis of antimicrobial resistance, virulence genes, and plasmids
Acquired genes encoding antimicrobial resistance were detected using a manually curated and updated version of the ResFinder database (compiled in 2012) (Zankari et al. 2012; additional genes are listed in Supplemental Table S4). A database of putative virulence genes was created based on E. faecium genes from the Virulence Factor Database (VFDB) (http://www.mgc.ac.cn/cgi-bin/VFs/genus.cgi?Genus=Enterococcus) (Chen et al. 2016) and supplemented after a literature review (Supplemental Table S5). The raw genomic reads were queried for resistance and virulence genes using Antibiotic Resistance Identification by Assembly (ARIBA) v2.5.0 (https://github.com/sanger-pathogens/ariba) (Hunt et al. 2017) with default length and match thresholds of 95% and 90%, respectively. Further processing of the output data is described in the Supplemental Methods. Plasmid analysis was performed using plasmidSPAdes (Antipov et al. 2016) and BLASTN, with details provided in the Supplemental Methods.
Statistical analyses were performed using STATA v13.1 (StataCorp LLC), as described in Supplemental Methods.
Ethical approval
The study protocol was approved by the CUH Research and Development Department (ref: A093285) and the National Research Ethics Service East of England Ethics Committee (ref: 12/EE/0439 and 14/EE/1123).
Data access
Sequence data from this study have been submitted to the European Nucleotide Archive (ENA; www.ebi.ac.uk/ena) under the accession numbers listed in Supplemental Table S2. The MATLAB script described in Supplemental Methods is available in Supplemental File S1.
Competing interest statement
J.P. and S.J.P. are paid consultants to Next Gen Diagnostics. S.J.P. is a consultant to Specific.
Supplementary Material
Acknowledgments
We thank the staff at the wastewater treatment plants and at Cambridge University Hospitals Estates Department for assistance in sample collection. We also thank the library construction, sequencing, and core informatics teams at the Wellcome Trust Sanger Institute for assistance with Illumina sequencing. This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health or Wellcome Trust. This work was supported by a Wellcome Trust Research Training Fellowship (to T.G. [103387/Z/13/Z]), a Wellcome Trust Sir Henry Postdoctoral Fellowship (to C.L. [110243/Z/15/Z] and F.C. [201344/Z/16/Z]), and an ERC grant (742158) (to J.C.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.232629.117.
Freely available online through the Genome Research Open Access option.
References
- Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32: 3380–3387. 10.1093/bioinformatics/btw493 [DOI] [PubMed] [Google Scholar]
- Arias CA, Murray BE. 2013. The rise of the Enterococcus: beyond vancomycin resistance. Nat Rev Microbiol 10: 266–278. 10.1038/nrmicro2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodrick HJ, Raven KE, Harrison EM, Blane B, Reuter S, Török ME, Parkhill J, Peacock SJ. 2016. Whole-genome sequencing reveals transmission of vancomycin-resistant Enterococcus faecium in a healthcare network. Genome Med 8: 4 10.1186/s13073-015-0259-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Ju F, Zhang T. 2014. Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl Microbiol Biotechnol 98: 3317–3326. 10.1007/s00253-013-5402-z [DOI] [PubMed] [Google Scholar]
- Caplin JL, Hanlon GW, Taylor HD. 2008. Presence of vancomycin and ampicillin-resistant Enterococcus faecium of epidemic clonal complex-17 in wastewaters from the south coast of England. Environ Microbiol 10: 885–892. 10.1111/j.1462-2920.2007.01507.x [DOI] [PubMed] [Google Scholar]
- Carter GP, Buultjens AH, Ballard SA, Baines SL, Tomita T, Strachan J, Johnson PDR, Ferguson JK, Seemann T, Stinear TP, et al. 2016. Emergence of endemic MLST non-typeable vancomycin-resistant Enterococcus faecium. J Antimicrob Chemother 71: 3367–3371. 10.1093/jac/dkw314 [DOI] [PubMed] [Google Scholar]
- Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44: D694–D697. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30: 1224–1228. 10.1093/molbev/mst028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Regt MJA, van Schaik W, van Luit-Asbroek M, Dekker HAT, van Duijkeren E, Koning CJM, Bonten MJM, Willems RJL. 2012. Hospital and community ampicillin-resistant Enterococcus faecium are evolutionarily closely linked but have diversified through niche adaptation. PLoS One 7: e30319 10.1371/journal.pone.0030319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29: 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueras MJ, Borrego J, Pike EB, Robertson W, Ashbolt N. 2000. Sanitary inspection and microbiological water quality. In Monitoring bathing waters: a practical guide to the design and implementation of assessments and monitoring programmes (ed. Bartram J, Rees G), Chap. 8. World Health Organization, New York. [Google Scholar]
- Gouliouris T, Raven KE, Ludden C, Blane B, Corander J, Horner CS, Hernandez-Garcia J, Wood P, Hadjirin NF, Radakovic M, et al. 2018. Genomic surveillance of Enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. mBio 9: e01780-18 10.1128/mBio.01780-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman Prieto AM, van Schaik W, Rogers MRC, Coque TM, Baquero F, Corander J, Willems RJL. 2016. Global emergence and dissemination of enterococci as nosocomial pathogens: attack of the clones? Front Microbiol 7: 788 10.3389/fmicb.2016.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. 2010. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect 16: 541–554. 10.1111/j.1469-0691.2010.03226.x [DOI] [PubMed] [Google Scholar]
- Howden BP, Holt KE, Lam MMC, Seemann T, Ballard S, Coombs GW, Tong SYC, Grayson ML, Johnson PDR, Stinear TP. 2013. Genomic insights to control the emergence of vancomycin-resistant enterococci. mBio 4: e00412-13 10.1128/mBio.00412-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Mather AE, Sánchez-Busó L, Page AJ, Parkhill J, Keane JA, Harris SR. 2017. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 3: e000131 10.1099/mgen.0.000131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6: 90 10.1186/s13073-014-0090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam MMC, Seemann T, Tobias NJ, Chen H, Haring V, Moore RJ, Ballard S, Grayson LM, Johnson PDR, Howden BP, et al. 2013. Comparative analysis of the complete genome of an epidemic hospital sequence type 203 clone of vancomycin-resistant Enterococcus faecium. BMC Genomics 14: 595 10.1186/1471-2164-14-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis H, Top J, Shankar N, Borgen K, Bonten M, van Embden J, Willems RJL. 2004. A novel putative enterococcal pathogenicity island linked to the esp virulence gene of Enterococcus faecium and associated with epidemicity. J Bacteriol 186: 672–682. 10.1128/JB.186.3.672-682.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavis HL, Willems RJL, Top J, Bonten MJM. 2006. High-level ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J Clin Microbiol 44: 1059–1064. 10.1128/JCM.44.3.1059-1064.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton F, van Schaik W, McGuire AM, Godfrey P, Griggs A, Corander J, Cheng L, Saif S, Young S, Zeng Q, et al. 2013. Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4: e00534-13 10.1128/mBio.00534-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq R, Oberlé K, Galopin S, Cattoir V, Budzinski H, Petit F. 2013. Changes in enterococcal populations and related antibiotic resistance along a medical center-wastewater treatment plant-river continuum. Appl Environ Microbiol 79: 2428–2434. 10.1128/AEM.03586-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novais C, Coque TM, Ferreira H, Sousa C, Peixe L. 2005. Environmental contamination with vancomycin-resistant enterococci from hospital sewage in Portugal. Appl Environ Microbiol 71: 3364–3368. 10.1128/AEM.71.6.3364-3368.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31: 3691–3693. 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Taylor B, Keane JA. 2016. Multilocus sequence typing by blast from de novo assemblies against PubMLST. J Open Source Softw 1: 118 10.21105/joss.00118 [DOI] [Google Scholar]
- Palmer KL, Kos VN, Gilmore MS. 2010. Horizontal gene transfer and the genomics of enterococcal antibiotic resistance. Curr Opin Microbiol 13: 632–639. 10.1016/j.mib.2010.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinholt M, Gumpert H, Bayliss S, Nielsen JB, Vorobieva V, Pedersen M, Feil E, Worning P, Westh H. 2017. Genomic analysis of 495 vancomycin-resistant Enterococcus faecium reveals broad dissemination of a vanA plasmid in more than 19 clones from Copenhagen, Denmark. J Antimicrob Chemother 72: 40–47. 10.1093/jac/dkw360 [DOI] [PubMed] [Google Scholar]
- Raven KE, Reuter S, Reynolds R, Brodrick HJ, Russell JE, Török ME, Parkhill J, Peacock SJ. 2016. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Res 26: 1388–1396. 10.1101/gr.204024.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven KE, Gouliouris T, Brodrick H, Coll F, Brown NM, Reynolds R, Reuter S, Török ME, Parkhill J, Peacock SJ. 2017. Complex routes of nosocomial vancomycin-resistant Enterococcus faecium transmission revealed by genome sequencing. Clin Infect Dis 64: 15–17. 10.1093/cid/ciw872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert DM, Ricks P, Edwards JR, Schneider A, Patel J, Srinivasan A, Kallen A, Limbago B, Fridkin S. 2013. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2009–2010. Infect Control Hosp Epidemiol 34: 1–14. 10.1086/668770 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hal SJ, Espedido BA, Coombs GW, Howden BP, Korman TM, Nimmo GR, Gosbell IB, Jensen SO. 2017. Polyclonal emergence of vanA vancomycin-resistant Enterococcus faecium in Australia. J Antimicrob Chemother 72: 998–1001. 10.1093/jac/dkw539 [DOI] [PubMed] [Google Scholar]
- Varela AR, Ferro G, Vredenburg J, Yanık M, Vieira L, Rizzo L, Lameiras C, Manaia CM. 2013. Vancomycin resistant enterococci: from the hospital effluent to the urban wastewater treatment plant. Sci Total Environ 450–451: 155–161. 10.1016/j.scitotenv.2013.02.015 [DOI] [PubMed] [Google Scholar]
- Willems RJL, Homan W, Top J, van Santen-Verheuvel M, Tribe D, Manzioros X, Gaillard C, Vandenbroucke-Grauls CMJE, Mascini EM, van Kregten E, et al. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357: 853–855. 10.1016/S0140-6736(00)04205-7 [DOI] [PubMed] [Google Scholar]
- Willems RJL, Top J, van Santen M, Robinson DA, Coque TM, Baquero F, Grundmann H, Bonten MJM. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg Infect Dis 11: 821–828. 10.3201/1106.041204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67: 2640–2644. 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.