Abstract
The role of chromosome rearrangements in driving evolution has been a long-standing question of evolutionary biology. Here we focused on ruminants as a model to assess how rearrangements may have contributed to the evolution of gene regulation. Using reconstructed ancestral karyotypes of Cetartiodactyls, Ruminants, Pecorans, and Bovids, we traced patterns of gross chromosome changes. We found that the lineage leading to the ruminant ancestor after the split from other cetartiodactyls was characterized by mostly intrachromosomal changes, whereas the lineage leading to the pecoran ancestor (including all livestock ruminants) included multiple interchromosomal changes. We observed that the liver cell putative enhancers in the ruminant evolutionary breakpoint regions are highly enriched for DNA sequences under selective constraint acting on lineage-specific transposable elements (TEs) and a set of 25 specific transcription factor (TF) binding motifs associated with recently active TEs. Coupled with gene expression data, we found that genes near ruminant breakpoint regions exhibit more divergent expression profiles among species, particularly in cattle, which is consistent with the phylogenetic origin of these breakpoint regions. This divergence was significantly greater in genes with enhancers that contain at least one of the 25 specific TF binding motifs and located near bovidae-to-cattle lineage breakpoint regions. Taken together, by combining ancestral karyotype reconstructions with analysis of cis regulatory element and gene expression evolution, our work demonstrated that lineage-specific regulatory elements colocalized with gross chromosome rearrangements may have provided valuable functional modifications that helped to shape ruminant evolution.
The extent that chromosome rearrangements provide a substrate for natural selection during animal evolution and adaptation is still an outstanding question in evolutionary biology (White 1968). Whole-genome comparisons among mammals and birds point to regions in genomes where the order of orthologous sequences can be maintained for tens of millions of years of evolution (homologous synteny blocks [HSBs]), often demarcated by evolutionary breakpoint regions (EBRs) where the order of orthologous sequences differs among species. Several studies have shown that HSBs are enriched for evolutionary conserved sequences and genes related to basic organismal development (Larkin et al. 2009; Farré et al. 2016), whereas EBRs are clustered in regions with a high number of repetitive elements and segmental duplications (Murphy et al. 2005; Bailey and Eichler 2006; Ma et al. 2006; Kehrer-Sawatzki and Cooper 2007; Larkin et al. 2009; Farré et al. 2011) and genes related to lineage-specific biology (Elsik et al. 2009; Groenen et al. 2012; Ullastres et al. 2014; Farré et al. 2016). The exact reason behind the genomic content differences between HSBs and EBRs is unknown, as well as the potential functional (nonmechanistic) role of chromosome rearrangements in genome evolution.
It is well established that chromosome rearrangements can have both direct and indirect effects on genomes at the molecular level. Indirect effects include the suppression of recombination within the rearranged region (Navarro and Barton 2003; Joron et al. 2011; Farré et al. 2013) due to incomplete pairing during meiosis, which could lead to the accumulation of genetic incompatibilities and, in some cases, speciation (Brown and O'Neill 2010). EBRs can disrupt coding sequences or alter gene expression of adjacent genes by separating them from their regulatory elements, bringing new regulatory sequences, moving the genes to different regulatory domains, or reconfiguring chromatin interactions by rearranging topologically associating domains (TADs) (Cande et al. 2009; Puig et al. 2015; Krefting et al. 2018; Lazar et al. 2018). Although the relationship between chromosome rearrangements and changes in gene expression has been demonstrated in some species (Marquès-Bonet et al. 2004; Giannuzzi et al. 2014; Fuller et al. 2016), these studies analyzed the genomic and epigenomic features in a reference genome framework, and assumed that they might be representative of ancestral states, without attempting any explicit ancestral reconstruction or inferring the origin of the genomic or epigenomic features. Herein we reconstructed the ancestral chromosome organization for several mammalian ancestral clades to investigate the functional role of EBRs in the context of changes that occurred for 60 million years (My) in the lineage leading to cattle.
We used ruminants as a model clade for this study because they show a high diversity of karyotypes varying from only three pairs of chromosomes in the Indian muntjac (Wurster and Benirschke 1970) to 35 pairs in gray brocket (Frohlich et al. 2017). Ruminants demonstrate highly diverged phenotypes, ranging from adaptations facilitating survival from extreme drought and heat (gemsbok) to high altitude (Tibetan antelope). Moreover, this clade comprises around 150 species, including most of the economically important livestock species (cattle, sheep, goat, buffalo, and yak) and some of the most iconic wildlife (giraffe). Ruminants are Cetartiodactyls, which include other even-toed ungulates (pigs, camels, and hippopotamuses) and cetaceans (whales and dolphins). The evolutionary success of ruminants is in part due to a very specialized digestive tract, characterized by a four-chambered stomach, making them capable of feeding on relatively low nutritional vegetation. Ruminant species have been widely studied to characterize their carbohydrate and lipid metabolism, which is different than in other mammals (for review, see Nafikov and Beitz 2007). Within ruminants, two infraorders are recognized (Fig. 1): tragulids and pecorans. Chevrotains, the only extant representatives of tragulids, display a less developed stomach (Langer 2001); whereas pecorans represented by the other five ruminant taxonomic families are considered higher ruminants (Decker et al. 2009). Ruminant chromosomes have been previously studied at low resolution using traditional cytogenetics and genetic maps (Slate et al. 2002; Murphy et al. 2005; Kulemzina et al. 2009, 2011), as well as sequenced genomes (Elsik et al. 2009). These studies showed that ruminant karyotypes are populated with multiple inter- and intrachromosomal rearrangements enriched for lineage-specific transposable elements (TEs), in contrast to other mammalian groups, such as primates, that had a higher percentage of intrachromosomal rearrangements in their recent evolutionary history (Kim et al. 2017).
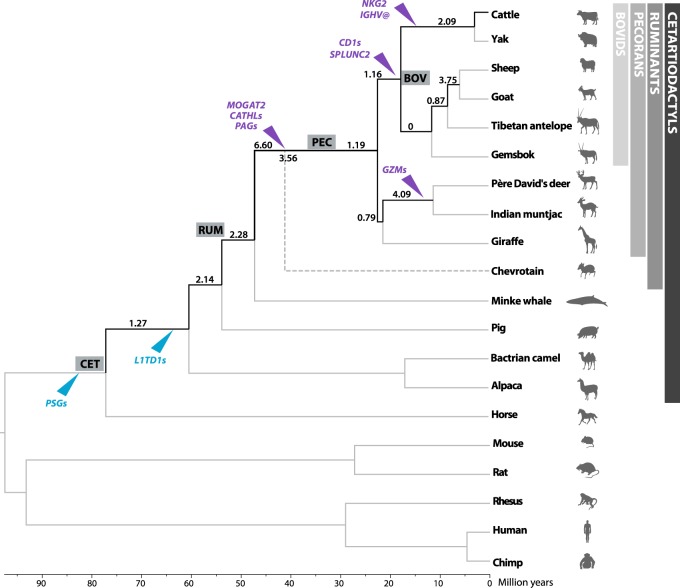
Figure 1.
Phylogenetic tree of the species and reconstructed ancestors. Numbers on branches from the cetartiodactyl ancestor (CET) to the cattle lineage are the evolutionary breakpoint rates (the number of evolutionary breakpoints per 1 My), and labels in italics are significantly different from the mean rearrangement rate across all branches. The dotted line leading to chevrotain represents the split of Pecora from the other ruminants, whereas the number crossing the line is the combined ruminant/pecoran evolutionary breakpoint rate. Arrowheads indicate gene family expansions (purple) and contractions (blue) in each branch. Details of the new genomes used can be found in Supplemental Table S1. Additional details of the reconstructed phylogenetic trees, rearrangements rates, and gene family expansions and contractions are shown in Supplemental Figure S1 and Supplemental Tables S5 and S6. (CET) Cetartiodactyl ancestral node; (RUM) ruminant ancestral node; (PEC) pecoran ancestral node; (BOV) bovid ancestral node.
Until recently, ancestral chromosome reconstruction lacked sufficient resolution and coverage to analyze individual EBRs or to study the evolution of chromosomes. However, with the development of the DESCHRAMBLER algorithm, these problems have been tackled (Kim et al. 2017). In this study, combining in silico (DESCHRAMBLER) and fluorescence in situ hybridization (FISH) techniques, we described four ancestral karyotypes in the lineage leading to cattle: Cetartiodactyl, Ruminant, Pecoran, and Bovidae. The aim of this study was to utilize these reconstructed ancestral chromosome structures to assess the functional contribution of chromosomal rearrangements to the evolution of Ruminants. We achieved this by integrating the results from ancestral reconstruction with data from selective sequence constraint (conserved noncoding elements [CNEs]) in Cetartiodactyls, functional constraint (putative enhancers), and gene expression among species with established ancestral and derived states of local genome structures.
Results
Patterns of gross chromosome evolution: from the cetartiodactyl ancestor to cattle
Using 19 mammalian genomes, we reconstructed the most likely karyotype structures of four ancestors leading to cattle: Cetartiodactyl, Ruminant, Pecoran, and Bovidae (Figs. 1, 2) and then determined the chromosome rearrangements along each lineage. We analyzed the relationships among the boundaries of EBRs or HSBs that are present in multiple species (msHSBs) with genetic, epigenetic, and transcriptomic data.
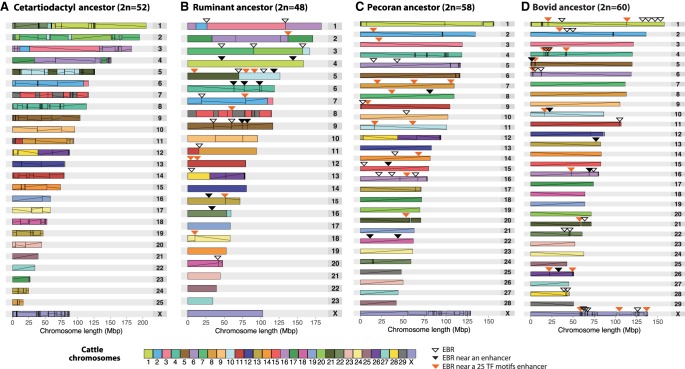
Figure 2.
Ideograms of the reconstructed ancestors relative to cattle chromosomes: (A) cetartiodactyl, (B) ruminant, (C) pecoran, (D) bovid. The vertical lines inside each chromosome reconstruction demarcate individual HSBs, whereas the diagonal lines indicate their orientation compared to the cattle genome. Arrowheads indicate evolutionary breakpoint regions (EBRs) associated with ruminant or cetartiodactyl enhancers (black), including the 25 TF motif enhancers (orange) and those not associated to any enhancers (white). Comparison of EBR positions with positions of enhancers is described in the “Functional constraint of enhancers” section.
Ancestral karyotype reconstructions
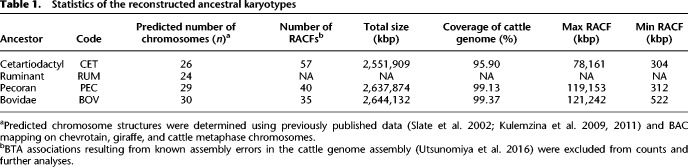
For the cetartiodactyl ancestor, 57 reconstructed ancestral chromosome fragments (RACFs) were produced using DESCHRAMBLER, spanning 95.9% of the cattle genome sequence. This compares with the 40 and 35 RACFs reconstructed for the pecoran and bovidae ancestors, respectively, each covering >99% of the cattle genome sequence (Table 1).
Table 1.
Statistics of the reconstructed ancestral karyotypes
We compared the pecoran and cetartiodactyl RACFs with the previously published pecoran (Slate et al. 2002), ruminant (Kulemzina et al. 2011), and cetartiodactyl (Kulemzina et al. 2009) karyotypes built from genetic or cytogenetic maps. The cetartiodactyl RACFs contained all previously proposed syntenic associations of cattle chromosomes (Kulemzina et al. 2009). Two additional detected associations (BTA4/22, BTA22/27) are now known assembly artifacts in the cattle genome (Utsunomiya et al. 2016) and were discarded from the reconstruction. We merged the RACFs using data from Kulemzina et al. (2009) as a guide, resulting in 26 reconstructed cetartiodactyl ancestral chromosomes (Table 1; Supplemental Table S3).
Twenty-five of 40 pecoran RACFs exactly matched the pecoran ancestral chromosomes proposed by Slate et al. (2002). The remaining RACFs represented fragments of published pecoran ancestral chromosomes and were merged to produce a final set of 29 reconstructed chromosomes (Table 1).
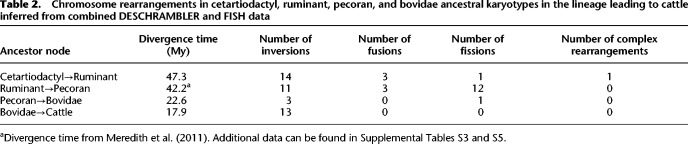
To determine the putative organization of the ancestral ruminant karyotype, we compared the reconstructed pecoran and cetartiodactyl chromosomes with syntenic inferences derived from FISH of 160 cattle bacterial artificial chromosome (BAC) clones onto the metaphase chromosomes of chevrotain (Java chevrotain, Tragulus javanicus) and two other ruminant species: giraffe (Giraffa camelopardalis) and cattle (Bos taurus) (Supplemental Fig. S2; Supplemental Table S4; Supplemental Methods). Four interchromosomal rearrangements differentiate the ancestral ruminant and cetartiodactyl karyotypes, whereas 15 additional interchromosomal rearrangements characterize the pecoran ancestor, in agreement with previous publications (Kulemzina et al. 2011). For the first time, intrachromosomal rearrangements were also identified, with 14 inversions assigned to the ruminant ancestor and an additional 11 inversions classified as pecoran-specific (Fig. 2; Table 2; Supplemental Fig. S2).
Table 2.
Chromosome rearrangements in cetartiodactyl, ruminant, pecoran, and bovidae ancestral karyotypes in the lineage leading to cattle inferred from combined DESCHRAMBLER and FISH data
Of 35 RACFs reconstructed for the bovidae ancestor, 27 matched previously published bovidae ancestral chromosomes (Slate et al. 2002; Balmus et al. 2007), whereas the remaining eight were fragments of three ancestral chromosomes that we subsequently merged to produce a final set of 30 bovidae ancestral chromosomes (Table 1; Supplemental Table S3).
Evolutionary breakpoint regions and multispecies homologous synteny blocks
We identified and classified genomic intervals that flank EBRs as well as mammalian msHSBs found in the cattle genome. Pairwise HSBs were defined at 300-kbp resolution for rearrangement detection following our previous work on mammals (Supplemental Table S2; Kim et al. 2017), and msHSBs were defined as overlapping intervals of several species HSBs. From the 2.1 Gbp of the cattle sequence assigned to chromosomes, 1.6 Gbp (76.28%) were found in mammalian msHSBs (Supplemental Table S2). The EBRs were classified using our previously published method (Farré et al. 2016) utilizing a phylogenetic tree that was constructed on the basis of genomic data from this study (Fig. 1; Supplemental Fig. S1). A total of 1699 EBRs were assigned to all phylogenetic nodes, of which 78 and 33 were putative ruminant lineage- and cattle-specific EBRs, respectively (Supplemental Table S5). Only 162 EBRs that occurred from the split of cetartiodactyls to cattle were included in further analyses. The average length of these EBRs was 15.3 kbp, spanning a total of 2.7 Mbp (0.15%) of the cattle genome. Using cattle BACs placed on the cattle, giraffe, and chevrotain chromosomes, we further classified 58/78 putative ruminant EBRs into 33 ruminant- and 25 pecoran-specific EBRs (Supplemental Table S5).
Chromosome rearrangements in the lineage leading to cattle
Only two chromosomes from the reconstructed cetartiodactyl ancestor were found intact in the cattle genome (BTA25 and BTA27), and nine ancestral chromosomes underwent intrachromosomal rearrangements only (Fig. 2). Inversions were the predominant type of chromosome rearrangements detected in the lineage leading from the cetartiodactyl ancestor to the ruminant ancestor, with only four interchromosomal rearrangements detected. In contrast, interchromosomal rearrangements (12 fissions and three fusions) have shaped the pecoran ancestral karyotype, from the ancestor of all ruminants to the karyotype of all pecoran ruminants (Table 2).
After summing inter- and intrachromosomal EBRs, we estimated the rearrangement rate for each ancestral node as the number of EBRs per million years and found a higher rate of rearrangements in the lineage leading from the cetartiodactyl to the ruminant ancestor around 47 Mya (6.60 EBRs/My) (Fig. 1), compared to a slower rate in the ruminant lineage after the split of tragulids (chevrotains), 25 Mya (1.19 EBRs/My) (Fig. 1; Supplemental Table S5). The pattern that emerges from our results is of two different rates and a shift in the type of chromosome rearrangement during cetartiodactyl/ruminant genome evolution: (1) a faster rate in the branch leading to the ruminant ancestor, characterized by multiple intrachromosomal rearrangements, and (2) a slower rate in the branch leading to the pecoran ancestor after the split of tragulids, with an increase in interchromosomal changes.
Selective sequence and functional constraint in cetartiodactyl and ruminant genomes
To identify links between clade-specific chromosome rearrangements and gene birth/deaths in the lineage leading to cattle, we first identified and then compared positions and frequency of these events with the positions of evolutionary stable (msHSBs) and dynamic (EBRs) genome intervals. We found no significant association of gene expansions or contractions with EBRs in the lineage leading to cattle (Supplemental Table S6) when we used cattle gene annotations as a reference.
Selective sequence constraint
We identified ∼1.59 million conserved elements (CEs) of ≥50 bp (Methods) covering 11.3% of the cattle genome sequence using a multiple alignment of nine cetartiodactyl species. About 46.5% were found in coding regions of cattle genes, whereas the remaining 53.5% were intronic or intergenic, representing conserved noncoding elements (CNEs). To trace the evolution of CNEs in the lineage leading to cattle and their distribution compared to EBRs and msHSBs, we separated CNEs present only in ruminant genomes from those that were cetartiodactyl- or mammal-specific. The ruminant-specific CNEs covered 13.18 Mbp (0.57%) of the cattle genome, whereas the cetartiodactyl and mammalian CNEs covered 74.32 Mbp (2.79%) and 54.09 Mbp (2.34%), respectively (Supplemental Table S7).
Previous studies have shown that some CNEs originated from retrotransposons or other TEs, which have been exapted and since come under selective constraint (Lindblad-Toh et al. 2011). We found that mammalian CNEs were enriched in ancestral TEs (including Eulor, MERs, and UCONs), whereas ruminant-specific CNEs were enriched in ruminant-specific TEs (LTR31B_BT, SINE2-1_BT, and L1-2_BT, with enrichment of 3.94-, 1.6-, and 1.2-fold, respectively) (FDR < 0.05) (Supplemental Table S9).
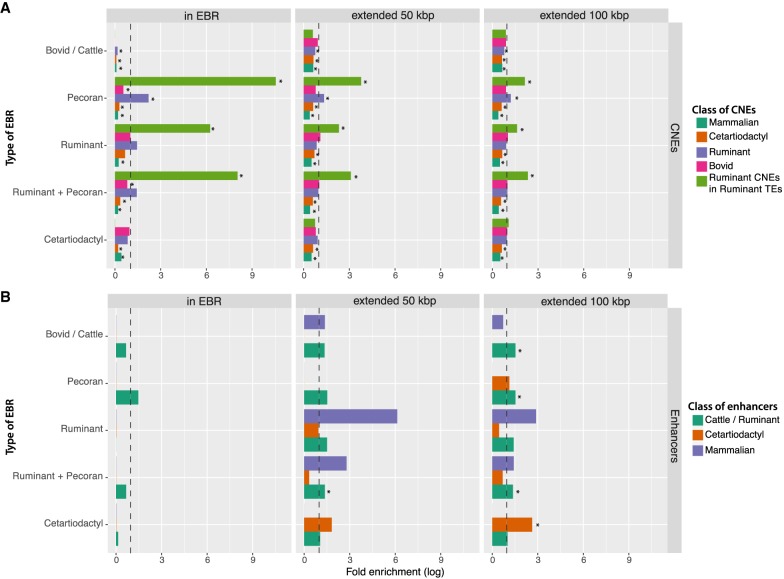
Consistent with the previous findings in mammals and birds (Larkin et al. 2009; Damas et al. 2017), CNEs were significantly depleted in EBRs (Fig. 3A; Supplemental Fig. S3) and enriched in mammalian msHSBs (Supplemental Table S11). However, when we focused on ruminant-specific CNEs with sequences that overlap with ruminant-specific TEs (Supplemental Fig. S3), these CNEs were highly enriched in ruminant-specific EBRs (8.1-fold enrichment, FDR = 0.0001) (Fig. 3A), whereas overall, ruminant-specific TEs had only 1.7-fold enrichment in ruminant-specific EBRs (FDR = 0.0001). As expected, both enrichments decreased with increasing distance from the EBR boundaries, suggesting that they reflect genetic events occurring at ruminant-specific chromosome rearrangement boundaries (Fig. 3A; Supplemental Fig. S3).
Figure 3.
Association of different types of EBRs with conserved noncoding elements (CNEs) and functional enhancers. (A) Fold enrichment of the CNEs inside EBRs and within 50 and 100 kbp of the different types of EBRs. (B) Fold enrichment of the functional enhancers. Asterisks mark statistically significant enrichments (FDR < 0.05). Dotted lines demarcate a fold enrichment of one. Additional data can be found in Supplemental Figure S3.
Functional constraint of enhancers
Using published ChIP-seq data for two histone modifications (H3K4me3 and H3K27ac) in the liver of 20 mammals (Villar et al. 2015), we defined putative liver enhancers in six species (human, mouse, dog, cattle, pig, and beaked whale) as genomic regions with peaks of H3K27ac only. To investigate possible links between the distribution and evolution of putative enhancers and structural chromosome evolution in the lineage leading to cattle, we translated all the putative liver enhancer coordinates to the cattle genome coordinates and defined three sets of enhancers: (1) enhancers conserved and active in all mammals, (2) enhancers conserved and active only in cetartiodactyl genomes (pig, beaked whale, and cattle), and (3) enhancers active only in cattle. From all the 31,372 enhancers found in the cattle genome, 15,387 were unique to cattle (group 3); 481 and 232 were conserved in cetartiodactyl and all mammalian genomes, respectively (Supplemental Table S10). The remaining 15,272 enhancers found in the cattle genome could not be assigned confidently to any group using stringent criteria (Methods). Enhancers were not found enriched inside cetartiodactyl lineage- and clade-specific EBRs when compared to the rest of the genome. However, when EBRs were extended from ±50 kbp to 1 Mbp (Supplemental Fig. S3), cattle-specific enhancers were enriched in surrounding areas with a peak at ±50 kbp of ruminant and pecoran EBRs (1.46× fold enrichment, FDR = 0.03) (Fig. 3B), whereas cetartiodactyl enhancers were enriched in neighboring areas with a peak at ±100 kbp of cetartiodactyl-specific EBRs (2.63× fold enrichment, FDR = 0.04) (Fig. 3B; Supplemental Fig. S3).
To investigate the relationship between gene regulatory changes in the lineage leading to cattle and changes in the landscape of enhancers found in or ±50 kbp from boundaries of chromosomal rearrangements, we scanned all enhancer regions for occurrences of potential transcription factor binding sites (TFBSs). We used a computational model developed earlier (Yokoyama et al. 2014) to assign a “branch of origin” to the 3,832,385 TF motifs identified in mammalian, cetartiodactyl, or cattle enhancers. This way we identified clusters of TFBSs within classified enhancers, where transcription factors (TFs) cooperatively bind to the enhancers (for review, see Long et al. 2016).
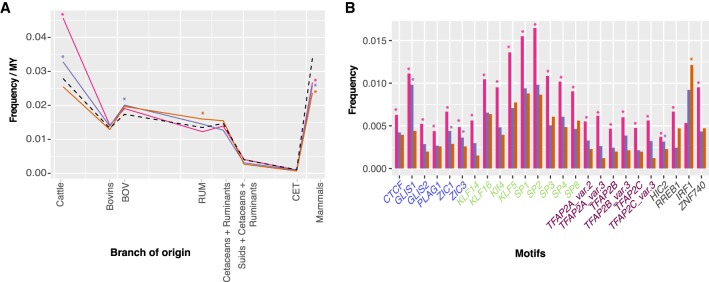
Overall, mammalian enhancers from our classified set were enriched for ancestral TFBSs (30.1% of all TFBSs were classified as mammalian, goodness-of-fit test, Bonferroni-corrected P-value <0.01), whereas enhancers only present in cattle were enriched in TFBSs that were assigned to branches after the split of ruminants and whales from pigs (Supplemental Table S12). Moreover, the enhancers found near EBRs had clear signatures of corresponding evolutionary events: Enhancers close to bovid-cattle lineage EBRs contained more TFBSs that originated after the split of bovids from cervids (42.76% of all TFBSs, P-value <0.01), whereas enhancers near ruminant-specific EBRs, found only in the ancestor of all ruminants, contained more TFBSs that formed after the split of ruminants from cetaceans (75.03%, P-value <0.01) (Fig. 4A; Supplemental Table S14). We further identified 25 TFs that had significantly more TFBSs present in enhancers near ruminant-lineage EBRs (i.e., all EBRs that appeared after the split of ruminants from cetaceans and present in the lineage leading to cattle) than expected from uniform distribution (goodness-of-fit test, Bonferroni-corrected P-value <0.05), including most of the members of three TF families: AP-2s, Three-zinc finger Krüppel-related factors, and More than 3 adjacent zinc finger factors (Fig. 4B; Supplemental Table S13).
Figure 4.
Association of transcription factor binding sites (TFBSs) with the different types of EBRs and their branches of origin. (A) Frequency of motifs in enhancers near each type of EBR according to their branch of origin. The frequency has been normalized by branch length of each classification. The dotted line corresponds to the total frequency of each branch of origin. (B) TFs with a different frequency of motifs in each lineage-specific EBR type. It shows the frequency of each motif in enhancers found in or ±50 kbp of EBRs. The TF motifs are colored according to their TF family: Blue TFs are part of the More than 3 adjacent zinc fingers; green TFs belong to the Three-zinc finger Krüppel-related factors; mauve TFs are in the AP-2 family; whereas gray are part of other TF families. The pink, purple, and orange lines in both A and B correspond to bovid-to-cattle lineage, ruminant- and cetartiodactyl-specific EBRs, respectively. Color-coded asterisks, according to the type of EBR, show significantly different frequencies (goodness-of-fit P-value <0.05). Additional data can be found in Supplemental Tables S13 and S14.
Lineage-specific EBRs were previously found to be enriched for TEs that are active in the same lineage (Supplemental Table S8; Larkin et al. 2009; Farré et al. 2016). Focusing on the 25 TFBS motifs enriched in the ruminant-lineage EBRs, using a permutation test we found that they were preferentially located inside ruminant-specific TEs (such as SINE2-2_BT, BOV-A2, and L1_Art, with 1.62×, 1.51×, and 1.28× fold enrichment, respectively) (Supplemental Table S15) compared to enhancers found distant from ruminant-lineage EBRs (FDR < 0.05), suggesting that the insertion of these TEs might have influenced the distribution of TFBSs in enhancers near ruminant-lineage EBRs similar to their contribution to formation of novel CNEs.
Evolution of gene expression in EBRs and in the rest of the genome
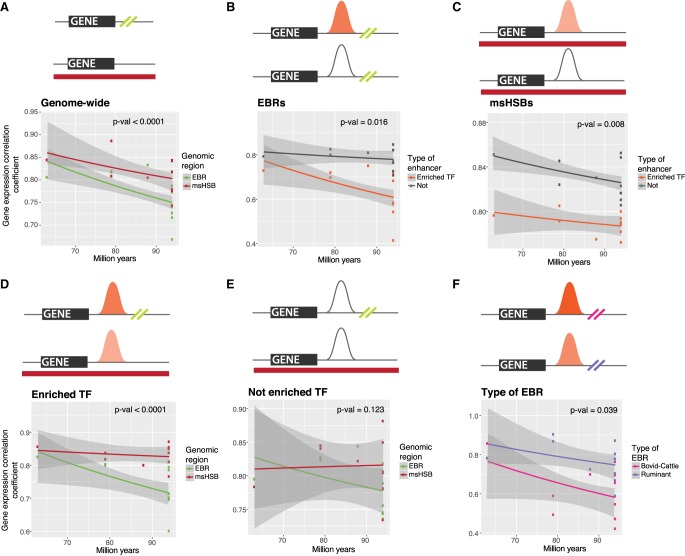
To further investigate if evolutionary structural rearrangements in ruminant chromosomes are associated with differences in expression levels of orthologous genes between species, we compared expression divergence of one-to-one orthologs for five species (Berthelot et al. 2018). These species were selected from the 20 species with available liver RNA-seq data to ensure the highest number of genes with no missing expression data and representing ruminant cetartiodactyls (cattle), nonruminant cetartiodactyls (pig), and noncetartiodactyl (human, mouse, and cat) lineages. We investigated whether genes found in or within 50 kbp of ruminant-lineage EBRs (including ruminant-, pecoran-, or bovidae-to-cattle lineage EBRs) have a lower evolutionary similarity of expression between species, as measured by expression correlation, compared to the genes found in mammalian msHSBs (Methods) for all the five species. Of 11,327 genes with liver expression data available for all five species, 112 genes were within or ±50 kbp of ruminant-lineage EBRs (Methods), whereas 1948 were found in mammalian msHSBs. After matching each gene near an EBR to a gene in an msHSB, we found that genes in/near the ruminant-lineage EBRs exhibited significantly lower cross-species expression correlation than genes in msHSBs, indicating that the genes in/near EBRs have a more diverged gene expression in liver between species than genes in msHSBs (100 iterations, Wilcoxon signed-rank test P-value < 0.0001) (Fig. 5A). This difference was due to a lower correlation of expressions observed in the pairwise comparisons of species involving cattle (Wilcoxon signed-rank test P-value < 0.0001) than in the comparisons not involving cattle. To rule out the possible random effect of a small gene set (112 genes in/near EBRs), we randomly selected and compared expression divergence between two sets of 100 genes with similar average levels of expression in msHSBs, repeated the process 2000 times, and found no significant differences in the expression divergence for any pairwise comparisons of different species (P-value = 0.37). Similarly, two subsets of 100 genes were selected from non-EBR regions and the same comparison was performed. After 2000 iterations no significant differences in the expression divergence were found (P-value = 0.27).
Figure 5.
Gene expression correlation comparisons of genes in EBRs and msHSBs. (A) Pairwise correlation coefficients plotted against evolutionary distance for pairs of species with genes ±50 kbp of EBRs (olive green) and genes in msHSBs with the same distribution of mean expression levels across species (red), showing that genes in/near EBRs have more evolutionarily diverged expression patterns than genes in msHSBs. (B) Correlation coefficients of genes near EBRs with 25 TF motif enhancers (orange) compared to genes near EBRs without 25 TF motif enhancers (gray), suggesting that the 25 TF motif enhancers might contribute to the differences. (C) Correlation coefficients of genes in msHSBs with 25 TF motif enhancers (orange) compared to genes in msHSBs without 25 TF motif enhancers (gray). Genes near EBRs with 25 TF motif enhancers (D) or without 25 TF motif enhancers (E) in their regulatory regions were compared to matching genes in msHSBs, showing that the 25 TF motif enhancers in EBRs have a stronger effect on gene expression than the same type of enhancers in msHSBs. This effect was not observed for other types of enhancers. (F) Comparison between ruminant- (purple) and bovid-to-cattle lineage (pink) EBRs for expression of genes with 25 TF motif enhancers, suggesting that a higher number of motifs for the 25 TFs correlated with a more diverged gene expression. Lines correspond to linear regression trends with 95% confidence intervals in gray shading. P-values were obtained using Wilcoxon rank-sum test. Shading of the 25 TF motif enhancers represents the mean number of TF motifs in enhancers in each genomic region, ranging from a mean of 14.25 (pale orange) to 24.91 (dark orange) motifs in 25 TF motif enhancers. Additional data can be found in Supplemental Figure S6.
To investigate whether the differences in expression of genes in/near EBRs might be related to changes in cis regulatory regions, we focused on genes putatively regulated by liver enhancers enriched for one or more of the 25 TF motifs overrepresented in EBRs (Fig. 4B). We first checked if these 25 TFs were expressed in cattle liver and found that at least 21 of them were either highly expressed (in the 75% quantile of expression) or expressed, representing 84% of the 25 TFs; and only 68.6% of the annotated cattle genes were found expressed in cattle liver. One of the 25 TFs was absent from the gene annotation used by Berthelot et al. (2018), and only three were not found expressed (ZIC1, ZIC3, and SP8). Genome-wide, a total of 3990 genes contained enhancers with one or more of the 25 TFs in their regulatory domains, whereas 7337 genes did not have these types of enhancers. We found that genes regulated by these types of enhancer(s) had significantly lower cross-species correlation of expression than genes without these enhancers (Wilcoxon signed-rank test P-value = 0.0009). Our results imply that the TFBS landscape of enhancers, their position relative to EBRs, or both factors could contribute to different levels of expression of orthologous genes between species (at least in liver). The 970 genes in msHSBs that had enhancers enriched for one or more of the 25 TF motifs indeed demonstrated significantly lower correlation of cross-species expression for the pairwise comparisons involving cattle when compared to the 978 genes in msHSBs without these types of enhancers (Fig. 5C). The same pattern was observed for the 58 genes with enhancers enriched for one or more of the 25 TF motifs found near the ruminant-lineage EBRs when compared to the remaining genes near EBRs with other types of enhancers (Wilcoxon signed-rank test P-value = 0.008) (Fig. 5B). As expected, the enhancers enriched for one or more of the 25 TFs in/near ruminant-lineage EBRs had a higher fraction of the 25 TF motifs than the same type of enhancers in msHSBs (mean of 24.14 and 14.26 TFBSs in enhancers in/near EBRs and msHSBs, respectively; P-value <0.0001) (Supplemental Fig. S6). Genes near the most recently appearing EBRs (bovid- and cattle-specific), that on average contained the highest number of enhancers enriched in the 25 TF motifs (mean of 28.98 TFBSs for bovid enhancers P-value < 0.0001) (Supplemental Fig. S7), demonstrated a lower correlation of cross-species expression (due to the cattle data, the only species in the analysis with rearranged chromosome structures in bovid- or cattle-specific EBRs) than genes near this type of enhancers in the more ancient ruminant EBRs (P-value = 0.001) and in msHSBs (P-value = 0.03, Fig. 5D–F). In addition, a higher fraction of genes near all ruminant EBRs contained enriched enhancers in their cis regulatory domains if compared to msHSBs (31% and 21%, respectively, χ2 test = 23.0) (P-value < 0.0001). This suggests that the 25 TF motif enhancers could account for the differences in gene expression between cattle and other species, and the proximity of these enhancers to EBRs might strengthen the effect and expand it to a larger number of genes due to a higher gene density in EBRs compared to the rest of the genome.
Gene pathways associated with gross chromosome rearrangements
Finally, to identify the gene pathways associated with chromosome rearrangements in the evolution of ruminants and cetartiodactyls, we analyzed which Gene Ontology (GO) terms were enriched in msHSBs and in EBRs (Methods).
Mammalian msHSBs were enriched in genes related to developmental process, biological adhesion, and meiosis I (including SPO11, RAD51, and ATM genes), among other GO terms (Supplemental Fig. S4) consistent with our previous findings (Larkin et al. 2009). On the other hand, when we investigated GO enrichment of genes in or surrounding lineage-specific EBRs (±50 kbp consistent with our enhancer analysis), we found that genes linked to inflammatory response (including SAA1, SAA3, and SAA4) and MHC class II protein complex were enriched in cetartiodactyl EBRs (FDR < 0.01) (Supplemental Fig. S5). Genes with prostaglandin receptor activity (such as PTGER2 and PTGDR) and serine-type endopeptidase activity (including granzyme B, GZMB) were enriched in ruminant-specific EBRs; whereas pecoran-specific EBRs contained genes involved in the protein-lipid complex (CLU and PCYOX1, FDR < 0.01) (Supplemental Fig. S5).
Focusing on the 58 genes in/near EBRs with enhancers enriched in one or more of the 25 TF motifs in their regulatory domains, six are involved in metabolic process, with three genes related to lipid metabolism (STARD4 related to cholesterol binding, ACOX3 involved in fatty acid metabolism, and NSFL1C in lipid binding); two genes are linked to glutamate (DGLUCY) and glucose (GHRL) metabolism. Four genes are connected to inflammatory response (JAM3, IRAK2, PTGRD, and ELF3), whereas one is involved in erythrocyte maturation (EPB42), one in hematopoiesis (MKNK2), and one in coagulation (SERPINA5).
Discussion
Using a combination of computational and cytogenetic techniques, we reconstructed the chromosomal structure of four cetartiodactyl ancestors in the lineage leading to cattle. We then utilized the reconstructed karyotype structures to trace chromosome rearrangements and their relationship to variations in genomic-feature landscapes. Using liver as a representative tissue, we provided novel lines of evidence supporting the hypothesis that differences in gene expression among cetartiodactyl and other mammalian species might be related to cis regulatory landscape modifications particularly for the genes found near recent evolutionary breakpoint regions.
A combination of genomic, computational, and cytogenetic approaches allowed us for the first time to reconstruct detailed chromosome structures of the Cetartiodactyl, Ruminant, Pecoran, and Bovidae ancestors, which were highly consistent with reconstructions based on FISH comparisons (Slate et al. 2002; Kulemzina et al. 2009, 2011). Our study also included intrachromosomal rearrangements absent from earlier cytogenetic reconstructions and linked reconstructed structures to cattle genome sequence, thus allowing for functional analyses. Consistent with the findings in other mammalian clades (Ferguson-Smith and Trifonov 2007), we observed a shift from a high rate of chromosome changes characterized by mostly intrachromosomal modifications in the lineage leading to the ruminant ancestor to a slower rate characterized by mostly interchromosomal modifications in the pecoran ancestor. As the rearrangement rates were calculated using the branch length of phylogenetic trees, they might change depending on node age estimation; however, the shift from intra- to interchromosomal rearrangements is independent from differences in phylogenetic node dating. Among other reasons, this shift could be related to a huge expansion of BovB transposable elements in the lineage leading to the ruminant ancestor (Adelson et al. 2009; Gallus et al. 2015) after the split from other cetartiodactyls about 45 Mya. Expansions of BovB elements would provide extra opportunities for nonallelic recombination in the ruminant ancestor germ cells. This is supported by our finding of enrichment for ruminant-specific EBRs with BovB transposable elements, whereas pecoran-specific EBRs were not enriched for BovB repeats, suggesting that BovB elements could be modified or methylated (Carbone et al. 2009) in the pecoran lineage and therefore not used as templates for aberrant intrachromosomal rearrangements.
Despite EBRs being associated with segmental duplications in previous studies in primates (Kehrer-Sawatzki and Cooper 2007; Larkin et al. 2009), we did not observe an association between EBRs detected in our ancestral genomes and expansions of gene families in the corresponding ruminant lineages (Fig. 1), with only 15 of 144 gene family expansions being within or close to ruminant-, bovid-, and cattle-specific EBRs. This might imply that most EBRs in the cetartiodactyl lineage colocalize with duplications of nongenic sequences, or alternatively, that it is the consequence of the high resolution that EBRs were defined in our study. Nevertheless, one of the gene family expansions colocalizing with ruminant EBRs is the pregnancy-associated glycoprotein family (PAGs), a ruminant-specific gene family related to the function of ruminant placenta. We cannot, however, completely rule out the misidentification of some gene families or incorrect node assignment of the gene expansions, because seven of the genomes that we used were assembled to scaffold-level and their gene annotations were modeled in silico and not supported by RNA-seq experimental validation. Therefore, further studies including experimentally validated gene annotations will be required to completely account for the possible association of gene family expansions and EBRs in the cetartiodactyl lineage. Overall, and although we used high-quality assemblies, our approach to detect ancestral chromosome structures and rearrangements is resistant to structural misassemblies found in individual genomes because detection of ancestral EBRs and msHSBs is based on more than one genome, minimizing the impact of individual assembly structural errors (e.g., those fixed in the newer versions of the human, mouse, and cattle genomes) proving robustness of our results.
Several studies on insect and bird genomes have shown that chromosome rearrangements can modify the regulatory landscape by moving regulatory elements to new locations and creating new regulatory sequences (Cande et al. 2009; Puig et al. 2015; Farré et al. 2016; Damas et al. 2017). We observed enrichment of ruminant-specific conserved noncoding elements (CNEs) originating from TEs active in the ruminant lineage (8.1× fold compared to the rest of the cattle genome) near ruminant-specific EBRs, suggesting that EBRs may contribute to changes in regulation of the nearby genes during evolution by providing high-density of TE sequences as material for new CNEs, including regulatory elements. These results are in agreement with a recent report of primate-specific CNEs associated to primate-specific TEs (Trizzino et al. 2017), suggesting that this could be a general pattern of gene regulation change in mammalian evolution. Further support for the hypothesis that EBRs might contribute to changes in gene regulation comes from the observed enrichment of active lineage-specific enhancers in close proximity to the EBRs formed in the same lineage.
Transcription factors often work in clusters to bind TFBSs within enhancers to regulate target genes (Bradley et al. 2010; Long et al. 2016). Also, changes in types of TFBSs within enhancers during the course of evolution may lead to changes in the corresponding enhancers’ regulatory activity and specificity (Long et al. 2016). Consistent with EBRs being hotspots of gene regulatory changes in cetartiodactyls, we observed that putative liver enhancers near ruminant EBRs were highly enriched for a set of 25 TF binding motifs, possibly strengthening EBR-associated enhancers relative to enhancers containing the same TFBSs in the rest of the genome. Most of these TFs belong to three TF families: Activating enhancer-binding protein 2 family (AP-2s, including six of the nine members), Three-zinc finger Krüppel-related factors (KLFs, with nine of the 16 members), and More than 3 adjacent zinc finger factors (with six of the 31 members). Members of the KLF family are involved in adipogenesis in liver (KLF4 and KLF14) (for review, see Swamynathan 2010), a process that is different in ruminants compared to other mammals (Nafikov and Beitz 2007; Laliotis et al. 2010). It has been proposed that a high proportion of lineage-specific regulatory sequences are derived from TEs because they are a source of suboptimal TFBSs, which could be turned into additional/new TFBS to develop new or change older enhancers (Chuong et al. 2016; Sundaram and Wang 2018). Our data suggest that this could be achieved relatively easily for enhancers found in EBR areas because these intervals are enriched in lineage-specific TEs. In support of this theory, the 25 TF motifs were strongly associated with ruminant-specific TEs. Therefore, our data point to ruminant-specific TEs being used in evolution as possible regulatory elements in two ways: They may be co-opted as ruminant-specific CNEs near lineage-specific EBRs, and/or they can provide new TFBSs in enhancers with more such TFBSs located near EBRs.
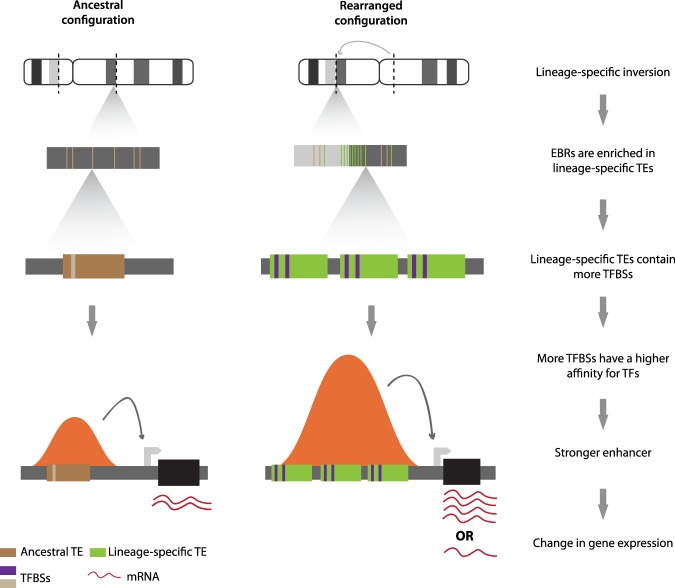
The correlation of gene expression levels in liver for genes found near ruminant lineage (ruminant-specific and bovid-cattle) EBRs and mammalian msHSBs indicates that these genes have significantly different expression profiles, driven largely by divergent cattle expression profiles. However, when we analyzed genes without any of the 25 TF motifs in their regulatory regions, this difference was not observed, suggesting that enhancers enriched for the 25 TF motifs might be the major factor causing differences in the expression profiles in liver cells between species, rather than other factors. This was further supported by significant differences in the correlations of expression observed for genes near ruminant lineage EBRs that contained 25 TF motif enhancers when compared with those genes near the EBRs that did not have such enhancers in their regulatory domains. On the other hand, genes regulated by 25 TF motif enhancers found in mammalian msHSBs also had significantly different expression profiles when compared to other genes in msHSBs without such enhancers nearby. This suggests that the 25 TF motif enhancers may affect gene expression regardless of their location near EBRs. However, comparison of gene expression correlations among genes found near 25 TF motif enhancers in ruminant-lineage EBRs and mammalian msHSBs demonstrated that those genes near EBRs had a significantly lower expression correlation (i.e., more divergence) than the genes found in msHSBs, suggesting that 25 TF motif enhancers near EBRs had a stronger influence on changes in gene expression in liver cells. Finally, 25 TF motif enhancers found near more recent bovidae EBRs had a significantly stronger influence on gene expression correlation than older 25 TF motif enhancers found near ruminant ancestral EBRs, containing significantly fewer TFBSs for the 25 TFs. This implies that an introduction of novel TFBSs in evolution might affect gene expression genome-wide, but genes in/near lineage-specific EBRs will be more affected by this process than genes in msHSBs. Therefore, a more dynamic regulatory turnover in/near EBRs might be associated with stronger changes in expression for nearby genes that, in turn, may serve as a substrate for shaping lineage-specific phenotypes in evolution (Fig. 6).
Figure 6.
A model for the evolution of chromosome rearrangements with gene expression divergence by means of lineage-specific transposable elements (TEs). Chromosome rearrangement boundaries (EBRs) are enriched for lineage-specific TEs. These TEs harbor a higher number of TFBSs than ancestral TEs; therefore, they have a higher affinity for TFs and a stronger influence in gene expression and regulation than those found elsewhere in the genome. This leads to a higher differential expression for orthologous genes between species with and without the gross genomic rearrangement. Brown and green boxes represent ancestral or lineage-specific TEs, respectively. Purple bars represent TFBSs, and black boxes represent genes. Orange bell-shaped curves represent peaks of H3K27ac as functional enhancers, with the height of the bell proportional to the strength of the enhancer.
Overall, our data point to an effect of lineage-specific TEs in changing gene expression and regulation in cetartiodactyl genomes, with lineage-specific EBRs being the genomic regions where this effect is most profound. The insertion of these TEs might promote chromosome rearrangements by means of nonallelic homologous recombination as found in primates (Bailey and Eichler 2006) or colocalize with EBRs due to both events happening in regions of active, open chromatin (Berthelot et al. 2015; Farré et al. 2015). Regardless of the exact mechanism, EBRs are hotspots of lineage-specific changes in gene expression, which could then be used by natural selection to develop new phenotypes (Fig. 6). Indeed, we found multiple key genes related to ruminant biology near ruminant-specific and more recent EBRs, as well as ruminant-specific gene families (PAGs) expanded in these areas. Our study answers important questions about the evolution of chromosome structures, gene expression, and phenotypes. However, studies on other clades, involving chromosome-level assemblies and expression data from multiple tissues, are required to prove that our findings are indeed a general pattern of mammalian evolution.
Methods
Genome data
The genome assemblies of 19 mammalian species were used in this study. Human (Homo sapiens, hg19), chimp (Pan troglodytes, panTro4), Rhesus macaque (Macaca mulatta, rheMac3), mouse (Mus musculus, mm9), rat (Rattus norvegicus, rno4), dog (Canis familiaris, canFam3), horse (Equus caballus, equCab2), pig (Sus scrofa, susScr3), Minke whale (Balaenoptera acuturostatta, balAcu1), Père David's deer (Elaphurus davidianus, Milu1.0), sheep (Ovis aries, oviAr3), and cattle (Bos taurus, bosTau6) were downloaded from the UCSC Genome Browser. Bactrian camel (Camelus bactrianus, Ca_bactrianus_MBC_1.0), Tibetan antelope (Pantholops hodgsonssi, PHO1.0), goat (Capra hircus, CHIR_1.0), and yak (Bos grunniens, BosGru_v2.0) assemblies were downloaded from NCBI. Alpaca genome assembly was provided by NHGRI sequencing performed at Washington University (Vicugna pacos, GCA_000164845.3, vicPac2). We included the newly sequenced genomes of gemsbok (Oryx gazella) (Farré et al. 2019), giraffe (Giraffa camelopardalis), and Indian muntjac (Muntiacus muntjak) (Supplemental Table S1). Although newer assembly versions are available for outgroup human and mouse genomes, using a total of 19 genome assemblies in our work minimized the impact of possible individual misassemblies in these genomes on our reconstructions. Structures of ancestral ruminant and pecoran chromosomes were also independently supported by FISH verification of individual breakpoint regions and chromosome structures (see below).
Establishing the reconstructed ancestral chromosome fragments (RACFs) and ancestral karyotypes
We used the cattle genome as a reference to reconstruct the ancestral RACFs. First, we aligned 19 mammalian genomes to the cattle genome using LastZ (https://github.com/lastz/lastz), and transformed them into chains and nets using the UCSC Kent Utilities (Kent et al. 2002). From the 19 pairwise alignments, only extant genomes assembled to chromosomes or with high scaffold N50 (>3 Mbp) were included in the reconstructions to maximize the reconstructed karyotypes’ coverage and to minimize their fragmentation, representing 12 genomes. Syntenic fragments (SFs) of at least 300 kbp in length were used as input for the RACF reconstructions and the phylogenetic tree defined in this study. By using our previously published ancestral karyotype reconstruction algorithm, DESCHRAMBLER (Kim et al. 2017), we defined the bovidae, the pecoran, and the cetartiodactyl RACFs. The RACFs were then merged to reconstruct ancestral chromosomes using our FISH data and previously published data as a framework (Slate et al. 2002; Kulemzina et al. 2009, 2011). Orientation of the RACFs in the ancestral chromosomes was established by comparing it to extant species and outgroups. The ruminant ancestral karyotype was inferred using the pecoran and the cetartiodactyl ancestral chromosomes combined with data on FISH on chevrotain, giraffe, and cattle metaphase chromosomes. We selected BACs flanking the structural differences between pecoran and cetartiodactyl ancestral chromosomes as detected from our reconstructions (Supplemental Fig. S2). When chevrotain only maintained the same chromosome configuration as the cetartiodactyl ancestor, and giraffe and cattle showed a different hybridization pattern, the chromosome configuration was considered pecoran-specific (Supplemental Fig. S2). Instead, when all three species showed the same hybridization pattern, but different from the reconstructed cetartiodactyl ancestor, the configuration was considered ruminant-specific.
Evolutionary breakpoint region and multispecies homologous synteny block detection
Alignments of nine ruminants and 11 outgroup genomes were performed against cattle genome using SatsumaSynteny, part of the Satsuma package (Grabherr et al. 2010). Syntenic fragments were defined using three sets of parameters to detect genome rearrangements that are ≥500 kbp, ≥300 kbp, and ≥100 kbp in the cattle genome with SyntenyTracker (Donthu et al. 2009). To detect and classify the EBRs, we used the EBR classification algorithm with our phylogenetic tree and a reuse threshold of 20 (Farré et al. 2016). After the EBRs were classified, EBRs were sorted by the confidence score provided by our algorithm, and those with the lowest 5% scores were removed from further analysis. Only EBRs assigned to ancestral nodes or to the cattle genome were included in further analyses.
Mammalian msHSBs were defined as the regions of reference chromosomes that had no EBRs or uncertain (unclassified) breakpoint regions detected in any of the species. Rates of chromosome rearrangement (EBRs/My) were calculated using the number of EBRs detected for each phylogenetic branch divided by the estimated length of each branch (in My) of the tree. To compare the rearrangement rates, we calculated the t-test statistics for a given branch as the difference between the rate in this branch and the mean rate across all the tree and normalized for the standard error. P-values were corrected for false discovery rate (FDR) using the p.adjust function from the R package (R Core Team 2018).
Cell culture and chromosome preparation
Metaphase chromosomes of Java mouse deer or chevrotain (Tragulus javanicus) and giraffe were obtained from cultured fibroblast cell lines provided by Prof. Ferguson-Smith. Briefly, cells were incubated at 37°C and 5% CO2 in Alpha MEM (Gibco), supplemented with 15% Fetal Bovine Serum (Gibco), 5% AmnioMAX-II (Gibco), and antibiotics (ampicillin 100 μg/mL, penicillin 100 μg/mL, amphotericin B 2.5 μg/mL). Metaphases were obtained by adding colcemid (0.02 mg/mL) and EtBr (1.5 mg/mL) to actively dividing culture. Hypotonic treatment was performed with KCl (3 mM) and NaCit (0.7 mM) for 20 min at 37°C and followed by fixation with 3:1 methanol—glacial acetic acid fixative. Metaphase chromosome preparations were made from fixed cultures, as described previously (Yang et al. 2000). G-banding on metaphase chromosomes for FISH was performed using standard procedure (Seabright 1971).
Selection and preparation of BAC clones for FISH
Cattle BAC clones from the CHORI-240 library were used. At least two BAC clones were selected for each EBR detected in the ruminant ancestor. BAC DNA was isolated using the Plasmid DNA Isolation Kit (Biosilica) and amplified with the GenomePlex Whole Genome Amplification (WGA) Kit (Sigma-Aldrich). Labeling of BAC DNA was performed using the GenomePlex WGA Reamplification Kit (Sigma-Aldrich) by incorporating biotin-16-dUTP (Roche) or digoxigenin-dUTP (Roche). Two-color FISH experiments on G-banded metaphase chromosomes were performed as described by Yang and Graphodatsky (2017). Digoxigenin-labeled probes were detected using Antidigoxigenin-Cy3 (Jackson ImmunoResearch), whereas biotin-labeled probes were identified with avidin-FITC (Vector Laboratories) and anti-avidinFITC (Vector Laboratories, catalog number BA-0300). Images were captured and processed using Videotest 2.0 Image Analysis System and a Baumer Optronics CCD Camera mounted on an Olympus BX53 microscope (Olympus). Cattle BACs were first validated on cattle metaphase spreads and then hybridized onto chevrotain and giraffe chromosomes to determine if an EBR was formed in ruminant or pecoran lineages (Supplemental Fig. S2).
Conserved noncoding element detection
The same pairwise alignments constructed for the detection of RACFs were transformed into multiple alignment format (MAF) files using UCSC Kent utilities (Kent et al. 2002). Then, we used the MULTIZ package to create five multiple alignments (Blanchette et al. 2004): (1) including only ruminant genomes; (2) including ruminants and whale; (3) including ruminants, whale, and pig; (4) including all cetartiodactyls; and (5) including all mammalian species analyzed. For each multiple alignment, we defined the conserved elements (CEs) using phastCons (Siepel et al. 2005). We estimated a neutral model for nonconserved sites with phyloFit (Siepel et al. 2005) and set the parameters as --target-coverage 0.3 --expected-length 20 --rho=0.3 after three runs of phastCons for the ruminant multiple alignment. The same parameters were used for the rest of multiple alignments.
Once the CEs were defined for each multiple alignment, using BEDTools (Quinlan et al. 2009), we removed those elements shorter than 50 bp to minimize the probability that a genomic sequence is not under purifying selection (Lindblad-Toh et al. 2011) and excluded those elements overlapping cattle coding regions (RefSeq and Ensembl gene predictions) and ESTs to finally obtain the conserved noncoding elements (CNEs). Then, to find the ruminant-specific CNEs, we removed from the ruminant multiple alignment the CNEs overlapping other CNE sets.
Functional conservation of putative enhancers in liver
Using previously published ChIP-seq data on two histone modifications in liver of 20 mammalian species (Villar et al. 2015), we selected the species whose genome was assembled at chromosome level (cattle, pig, dog, human, and rat), and the cetacean with the highest N50 (beaked whale, Mesoplodon bidens). First, we defined the enhancer peaks as the regions of the genome containing H3K27ac marks but not H3K4me3 marks. Then, using reciprocal liftOver with a minimum match of 0.5 (Kent et al. 2002), we translated their coordinates to the cattle genome, and defined three sets of functionally conserved enhancers: (1) mammalian, as enhancers peaks found in orthologous regions in all the species included in the analysis; (2) cetartiodactyl, present in only pig, whale, and cattle; and (3) cattle lineage, as those only present in cattle.
For each type of enhancer, we used a custom Python script (Supplemental Code) to scan the cattle sequence for TFBS motifs known to be functional in mammals (Mathelier et al. 2014) with a P-value cutoff of 0.0001 calculated by TFM-Pvalue (Touzet and Varré 2007). We then scanned TFBS motifs within ±100 bp orthologous sequence centered on cattle TFBS motifs using the same P-value cutoff and our multiple sequence alignment, and assigned a “branch of origin” to each of the TFBS in cattle using a birth and death model (Yokoyama et al. 2014). A multinomial goodness-of-fit test was used to determine if the frequency of a given TFBS motif deviated from the population of all enhancer types, and a post-hoc analysis using a binomial test was implemented to establish which enhancer type was statistically different from the rest. The same approach was used to establish which motifs were associated to enhancers closer to EBRs.
Association of genomic and epigenomic features with EBRs and msHSBs
Using the Genomic Association Test (GAT) (Heger et al. 2013), we computed the significance of overlap between several genomic and epigenomic features with EBRs. We used GAT to estimate the significance based on 10,000 simulations of the regions in all cattle chromosomes and an FDR ≤ 0.05. For each set of EBRs (cetartiodactyl, ruminant, pecoran, and bovidae), we calculated the association of all the features inside the EBRs and extending the EBRs for 50 kbp, 100 kbp, 200 kbp, and 1 Mbp. The cutoff distance for further analyses was set empirically to ±50 kbp (for more information, see Supplemental Fig. S3). The same approach was used to determine the association of TEs with enhancers, CNEs, and EBRs.
Measures of gene expression divergence analysis
The one-to-one gene orthologous expression level data for five species, normalized between species using the median of ratios to the geometric means, were obtained from a previous publication (Berthelot et al. 2018; Supplemental Fig. S8). Gene expression divergence was measured as Spearman's correlation coefficients of orthologous gene expression between pairs of species. Genes were then labeled as in/near EBRs (if they were within an EBR or at ±50 kbp) or in msHSBs, as well as with/without 25 TF motif enhancers. The relative divergence of two gene subsets was compared using the correlations within each subset across all pairs of species or pairs including only ruminant species using Wilcoxon paired rank-sum test in R. Because comparing the evolutionary stability of expression for subsets of genes originating from tissue samples of different origins (species) could be affected by overall gene expression levels in individual samples (Berthelot et al. 2018), for each gene near a ruminant-lineage EBR, we identified a gene in a mammalian msHSB with the most similar average expression level estimated from all five species and then performed pairwise comparison of correlation of expression for such gene pairs for 20 possible combinations of different species (Pereira et al. 2009; Berthelot et al. 2018). To control these confounding effects, we matched genes one-to-one to control genes with similar expression using the MatchIt library with a caliper option of 0.1 and the “nearest” method (Ho et al. 2007). For comparisons in which the number of genes in one set was 10 times lower than in the other set, we matched genes one-to-one using 100 permutations. The correlation coefficients across all pairs of species of each subset of genes were compared using Wilcoxon rank-sum test for paired data.
Gene Ontology enrichment analysis
The basic Gene Ontology file (go-basic.obo) was downloaded from The Gene Ontology Consortium (Ashburner et al. 2000; The Gene Ontology Consortium 2017), and the Gene Ontology annotations with Ensembl IDs using QuickGO from EMBL-EBI on February 2016. We used an hypergeometric test to analyze the GO enrichment of several traits, implemented in the Perl module GO::TermFinder (Boyle et al. 2004). The GO enrichments were visualized using R.
For the GO enrichment analysis of gene family expansions in ruminants, we created a background list of all known protein coding genes in cattle from Ensembl BioMart. For GO enrichment analysis of gene family contractions, a background list of protein coding genes in human was used. For the Gene Ontology enrichment in msHSBs and EBRs, sequence coordinates of all protein coding genes in cattle genome were obtained. We assigned genes from the background list to EBRs and msHSBs based on overlaps of gene coordinates in cattle chromosomes following the procedures described previously (Larkin et al. 2009). For the identification of functional categories of genes overrepresented in msHSBs, we considered msHSBs ≥ 1.5 Mbp in the cattle genome to avoid genes that could be located in proximity to EBRs, as done previously (Larkin et al. 2009). To evaluate GO enrichment in and near EBRs, we considered genes that were located within or ±50 kbp from EBR boundaries.
Next, for the enrichment analysis of CNEs and enhancers, we applied the proximal distance rule implemented in GREAT (McLean et al. 2010), stating that “gene regulatory domains extend two directions from the proximal promoter of the nearest gene (−5 kbp/+1 kbp from the transcription starting site), but no more than 1Mbp.” Using all the protein coding genes in cattle, we defined the GREAT domains and created a background list containing the domains with at least one CNE. A false discovery rate (FDR) of 0.05 was used as a significance threshold in all the preceding analyses.
Data access
Multispecies genome alignments, conserved noncoding elements (CNEs), evolutionary breakpoint regions (EBRs), and classified enhancers can be accessed in our public UCSC track hub (http://sftp.rvc.ac.uk/rvcpaper/ruminantsHUB/hub.txt) (Raney et al. 2014). Ancestral karyotype reconstructions and homologous synteny blocks (HSBs) can be visualized in Evolution Highway (http:// eh-demo.ncsa.uiuc.edu/ruminants). The script to scan for TFBS motifs can be found as Supplemental Code and also at https:// github.com/zocean/Marta_TFBS_tracing.
Supplementary Material
Acknowledgments
We thank Jon Bakh for early access to the Minke whale genome and Professor Malcolm Ferguson-Smith for the chevrotain cell line. This work was funded by grants BB/P020062/1 and BB/J010170/1 from the Biotechnology and Biological Sciences Research Council (D.M.L.), Russian Foundation for Basic Research (RFBS) grants 17-00-00147 (D.M.L.) and 17-00-00146 (A.S.G.) as part of 17-00-00148 (K), Russian Science Foundation (RSF) grant 16-14-10009 (A.S.G.), National Institutes of Health grant R01HG007352 (J.M.), and the Ministry of Science, ICT, and Future Planning of Korea grant 2014M3C9A3063544 (J.K.). S.J.O'B. was supported, in part, by the Russian Science Foundation grant (project No. 17-14-01138). This manuscript was prepared while W.E.J. held a National Research Council Research Associateship Award at the Walter Reed Army Institute of Research. The published material reflects the views of the authors and should not be construed to represent those of the Department of the Army or the Department of Defense.
Author contributions: M.F. and D.M.L. designed and performed the analyses and prepared the first draft of the paper. J.K. performed ancestral karyotype reconstructions; A.A.P., A.I.K., and A.S.G. performed and analyzed FISH data; Y.Z. and J.M. performed the analysis of TFBS age in enhancers; J.L.J. and A.V.K. obtained and cultivated cattle BACs; and P.L.P., W.E.J., W.C.W., and S.J.O. provided alpaca genome. Q.L., Y.Z., and Y.X. performed sequencing, genome assembly, and annotation of the three new ruminant genomes. G.Z. supervised the sequencing and assembly. O.A.R. handled DNA sample preparation. M.F., J.K., J.M., H.A.L., and D.M.L. interpreted the results and wrote the final version of the paper.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239863.118.
References
- Adelson DL, Raison JM, Edgar RC. 2009. Characterization and distribution of retrotransposons and simple sequence repeats in the bovine genome. Proc Natl Acad Sci 106: 12855–12860. 10.1073/pnas.0901282106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Eichler EE. 2006. Primate segmental duplications: crucibles of evolution, diversity and disease. Nat Rev Genet 7: 552–564. 10.1038/nrg1895 [DOI] [PubMed] [Google Scholar]
- Balmus G, Trifonov VA, Biltueva LS, O'Brien PC, Alkalaeva ES, Fu B, Skidmore JA, Allen T, Graphodatsky AS, Yang F, et al. 2007. Cross-species chromosome painting among camel, cattle, pig and human: further insights into the putative Cetartiodactyla ancestral karyotype. Chromosome Res 15: 499–515. 10.1007/s10577-007-1154-x [DOI] [PubMed] [Google Scholar]
- Berthelot C, Muffato M, Abecassis J, Roest Crollius H. 2015. The 3D organization of chromatin explains evolutionary fragile genomic regions. Cell Rep 10: 1913–1924. 10.1016/j.celrep.2015.02.046 [DOI] [PubMed] [Google Scholar]
- Berthelot C, Villar D, Horvath JE, Odom DT, Flicek P. 2018. Complexity and conservation of regulatory landscapes underlie evolutionary resilience of mammalian gene expression. Nat Ecol Evol 2: 152–163. 10.1038/s41559-017-0377-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF, Roskin KM, Baertsch R, Rosenbloom K, Clawson H, Green ED, et al. 2004. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14: 708–715. 10.1101/gr.1933104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder—open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20: 3710–3715. 10.1093/bioinformatics/bth456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC, Tonkin LA, Biggin MD, Eisen MB. 2010. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol 8: e1000343 10.1371/journal.pbio.1000343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JD, O'Neill RJ. 2010. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet 11: 291–316. 10.1146/annurev-genom-082509-141554 [DOI] [PubMed] [Google Scholar]
- Cande JD, Chopra VS, Levine M. 2009. Evolving enhancer-promoter interactions within the tinman complex of the flour beetle, Tribolium castaneum. Development 136: 3153–3160. 10.1242/dev.038034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone L, Harris RA, Vessere GM, Mootnick AR, Humphray S, Rogers J, Kim SK, Wall JD, Martin D, Jurka J, et al. 2009. Evolutionary breakpoints in the gibbon suggest association between cytosine methylation and karyotype evolution. PLoS Genet 5: e1000538 10.1371/journal.pgen.1000538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351: 1083–1087. 10.1126/science.aad5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, O'Connor R, Farré M, Lenis VPE, Martell HJ, Mandawala A, Fowler K, Joseph S, Swain MT, Griffin DK, et al. 2017. Upgrading short-read animal genome assemblies to chromosome level using comparative genomics and a universal probe set. Genome Res 27: 875–884. 10.1101/gr.213660.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker JE, Pires JC, Conant GC, McKay SD, Heaton MP, Chen K, Cooper A, Vilkki J, Seabury CM, Caetano AR, et al. 2009. Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci 106: 18644–18649. 10.1073/pnas.0904691106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donthu R, Lewin HA, Larkin DM. 2009. SyntenyTracker: a tool for defining homologous synteny blocks using radiation hybrid maps and whole-genome sequence. BMC Res Notes 2: 148 10.1186/1756-0500-2-148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, Elnitski L, Guigo R, et al. 2009. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science 324: 522–528. 10.1126/science.1169588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Bosch M, López-Giráldez F, Ponsà M, Ruiz-Herrera A. 2011. Assessing the role of tandem repeats in shaping the genomic architecture of great apes. PLoS One 6: e27239 10.1371/journal.pone.0027239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Micheletti D, Ruiz-Herrera A. 2013. Recombination rates and genomic shuffling in human and chimpanzee—a new twist in the chromosomal speciation theory. Mol Biol Evol 30: 853–864. 10.1093/molbev/mss272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Robinson TJ, Ruiz-Herrera A. 2015. An Integrative Breakage Model of genome architecture, reshuffling and evolution: the Integrative Breakage Model of genome evolution, a novel multidisciplinary hypothesis for the study of genome plasticity. BioEssays 37: 479–488. 10.1002/bies.201400174 [DOI] [PubMed] [Google Scholar]
- Farré M, Narayan J, Slavov GT, Damas J, Auvil L, Li C, Jarvis ED, Burt DW, Griffin DK, Larkin DM. 2016. Novel insights into chromosome evolution in birds, archosaurs, and reptiles. Genome Biol Evol 8: 2442–2451. 10.1093/gbe/evw166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farré M, Li Q, Zhou Y, Damas J, Chemnick LG, Kim J, Ryder OA, Ma J, Zhang G, Larkin DM, et al. 2019. A near-chromosome-scale genome assembly of the gemsbok (Oryx gazella): an iconic antelope of the Kalahari desert. Gigascience 8: 1–8. 10.1093/gigascience/giy162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson-Smith MA, Trifonov V. 2007. Mammalian karyotype evolution. Nat Rev Genet 8: 950–962. 10.1038/nrg2199 [DOI] [PubMed] [Google Scholar]
- Frohlich J, Kubickova S, Musilova P, Cernohorska H, Muskova H, Vodicka R, Rubes J. 2017. Karyotype relationships among selected deer species and cattle revealed by bovine FISH probes. PLoS One 12: e0187559 10.1371/journal.pone.0187559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller ZL, Haynes GD, Richards S, Schaeffer SW. 2016. Genomics of natural populations: how differentially expressed genes shape the evolution of chromosomal inversions in Drosophila pseudoobscura. Genetics 204: 287–301. 10.1534/genetics.116.191429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallus S, Kumar V, Bertelsen MF, Janke A, Nilsson MA. 2015. A genome survey sequencing of the Java mouse deer (Tragulus javanicus) adds new aspects to the evolution of lineage specific retrotransposons in Ruminantia (Cetartiodactyla). Gene 571: 271–278. 10.1016/j.gene.2015.06.064 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2017. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res 45: D331–D338. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannuzzi G, Migliavacca E, Reymond A. 2014. Novel H3K4me3 marks are enriched at human- and chimpanzee-specific cytogenetic structures. Genome Res 24: 1455–1468. 10.1101/gr.167742.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Russell P, Meyer M, Mauceli E, Alfoldi J, Di Palma F, Lindblad-Toh K. 2010. Genome-wide synteny through highly sensitive sequence alignment: Satsuma. Bioinformatics 26: 1145–1151. 10.1093/bioinformatics/btq102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groenen MA, Archibald AL, Uenishi H, Tuggle CK, Takeuchi Y, Rothschild MF, Rogel-Gaillard C, Park C, Milan D, Megens HJ, et al. 2012. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 491: 393–398. 10.1038/nature11622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger A, Webber C, Goodson M, Ponting CP, Lunter G. 2013. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics 29: 2046–2048. 10.1093/bioinformatics/btt343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho D, Imai K, Stuart E. 2007. Matching as nonparametric preprocessing for reducing model dependence in parametric causal inference. Polit Anal 15: 199–236. 10.1093/pan/mpl013 [DOI] [Google Scholar]
- Joron M, Frezal L, Jones RT, Chamberlain NL, Lee SF, Haag CR, Whibley A, Becuwe M, Baxter SW, Ferguson L, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477: 203–206. 10.1038/nature10341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Cooper DN. 2007. Structural divergence between the human and chimpanzee genomes. Human Genet 120: 759–778. 10.1007/s00439-006-0270-6 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The Human Genome Browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Farré M, Auvil L, Capitanu B, Larkin DM, Ma J, Lewin HA. 2017. Reconstruction and evolutionary history of eutherian chromosomes. Proc Natl Acad Sci 114: E5379–E5388. 10.1073/pnas.1702012114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krefting J, Andrade-Navarro MA, Ibn-Salem J. 2018. Evolutionary stability of topologically associating domains is associated with conserved gene regulation. BMC Biol 16: 87 10.1186/s12915-018-0556-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulemzina AI, Trifonov VA, Perelman PL, Rubtsova NV, Volobuev V, Ferguson-Smith MA, Stanyon R, Yang F, Graphodatsky AS. 2009. Cross-species chromosome painting in Cetartiodactyla: reconstructing the karyotype evolution in key phylogenetic lineages. Chromosome Res 17: 419–436. 10.1007/s10577-009-9032-3 [DOI] [PubMed] [Google Scholar]
- Kulemzina AI, Yang F, Trifonov VA, Ryder OA, Ferguson-Smith MA, Graphodatsky AS. 2011. Chromosome painting in Tragulidae facilitates the reconstruction of Ruminantia ancestral karyotype. Chromosome Res 19: 531–539. 10.1007/s10577-011-9201-z [DOI] [PubMed] [Google Scholar]
- Laliotis GP, Bizelis I, Rogdakis E. 2010. Comparative approach of the de novo fatty acid synthesis (lipogenesis) between ruminant and non ruminant mammalian species: from biochemical level to the main regulatory lipogenic genes. Curr Genomics 11: 168–183. 10.2174/138920210791110960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer P. 2001. Evidence from the digestive tract on phylogenetic relationships in ungulates and whales. J Zool Syst Evol Res 39: 77–90. 10.1046/j.1439-0469.2001.00158.x [DOI] [Google Scholar]
- Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA. 2009. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res 19: 770–777. 10.1101/gr.086546.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar NH, Nevonen KA, O'Connell B, McCann C, O'Neill RJ, Green RE, Meyer TJ, Okhovat M, Carbone L. 2018. Epigenetic maintenance of topological domains in the highly rearranged gibbon genome. Genome Res 28: 983–997. 10.1101/gr.233874.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482. 10.1038/nature10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Prescott SL, Wysocka J. 2016. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167: 1170–1187. 10.1016/j.cell.2016.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Zhang L, Suh BB, Raney BJ, Burhans RC, Kent WJ, Blanchette M, Haussler D, Miller W. 2006. Reconstructing contiguous regions of an ancestral genome. Genome Res 16: 1557–1565. 10.1101/gr.5383506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquès-Bonet T, Cáceres M, Bertranpetit J, Preuss TM, Thomas JW, Navarro A. 2004. Chromosomal rearrangements and the genomic distribution of gene-expression divergence in humans and chimpanzees. Trends Genet 20: 524–529. 10.1016/j.tig.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Mathelier A, Zhao X, Zhang AW, Parcy F, Worsley-Hunt R, Arenillas DJ, Buchman S, Chen CY, Chou A, Ienasescu H, et al. 2014. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res 42: D142–D147. 10.1093/nar/gkt997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28: 495–501. 10.1038/nbt.1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith RW, Janečka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334: 521–524. 10.1126/science.1211028 [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, et al. 2005. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309: 613–617. 10.1126/science.1111387 [DOI] [PubMed] [Google Scholar]
- Nafikov RA, Beitz DC. 2007. Carbohydrate and lipid metabolism in farm animals. J Nutr 137: 702–705. 10.1093/jn/137.3.702 [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH. 2003. Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science 300: 321–324. 10.1126/science.1080600 [DOI] [PubMed] [Google Scholar]
- Pereira V, Waxman D, Eyre-Walker A. 2009. A problem with the correlation coefficient as a measure of gene expression divergence. Genetics 183: 1597–1600. 10.1534/genetics.109.110247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig M, Casillas S, Villatoro S, Caceres M. 2015. Human inversions and their functional consequences. Brief Funct Genomics 14: 369–379. 10.1093/bfgp/elv020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan R, Graf M, Mason I, Lumsden A, Kiecker C. 2009. Complex and dynamic patterns of Wnt pathway gene expression in the developing chick forebrain. Neural Dev 4: 35 10.1186/1749-8104-4-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna: https://www.R-project.org/. [Google Scholar]
- Raney BJ, Dreszer TR, Barber GP, Clawson H, Fujita PA, Wang T, Nguyen N, Paten B, Zweig AS, Karolchik D, et al. 2014. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics 30: 1003–1005. 10.1093/bioinformatics/btt637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabright M. 1971. A rapid banding technique for human chromosomes. Lancet 2: 971–972. 10.1016/S0140-6736(71)90287-X [DOI] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J, Van Stijn TC, Anderson RM, McEwan KM, Maqbool NJ, Mathias HC, Bixley MJ, Stevens DR, Molenaar AJ, Beever JE, et al. 2002. A deer (subfamily Cervinae) genetic linkage map and the evolution of ruminant genomes. Genetics 160: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Wang T. 2018. Transposable element mediated innovation in gene regulatory landscapes of cells: re-visiting the “gene-battery” model. Bioessays 40: 1700155 10.1002/bies.201700155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamynathan SK. 2010. Krüppel-like factors: three fingers in control. Hum Genomics 4: 263–270. 10.1186/1479-7364-4-4-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touzet H, Varré JS. 2007. Efficient and accurate P-value computation for position weight matrices. Algorithms Mol Biol 2: 15 10.1186/1748-7188-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res 27: 1623–1633. 10.1101/gr.218149.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullastres A, Farré M, Capilla L, Ruiz-Herrera A. 2014. Unraveling the effect of genomic structural changes in the rhesus macaque—implications for the adaptive role of inversions. BMC Genomics 15: 530 10.1186/1471-2164-15-530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya AT, Santos DJ, Boison SA, Utsunomiya YT, Milanesi M, Bickhart DM, Ajmone-Marsan P, Solkner J, Garcia JF, da Fonseca R, et al. 2016. Revealing misassembled segments in the bovine reference genome by high resolution linkage disequilibrium scan. BMC Genomics 17: 705 10.1186/s12864-016-3049-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. 10.1016/j.cell.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ. 1968. Models of speciation. New concepts suggest that the classical sympatric and allopatric models are not the only alternatives. Science 159: 1065–1070. 10.1126/science.159.3819.1065 [DOI] [PubMed] [Google Scholar]
- Wurster DH, Benirschke K. 1970. Indian Momtjac, Muntiacus muntiak: a deer with a low diploid chromosome number. Science 168: 1364–1366. 10.1126/science.168.3937.1364 [DOI] [PubMed] [Google Scholar]
- Yang F, Graphodatsky AS. 2017. Animal probes and ZOO-FISH In Fluorescence in situ hybridization (FISH) (ed. Liehr T), pp. 323–346. Springer, Berlin. [Google Scholar]
- Yang F, Graphodatsky AS, O'Brien PC, Colabella A, Solanky N, Squire M, Sargan DR, Ferguson-Smith MA. 2000. Reciprocal chromosome painting illuminates the history of genome evolution of the domestic cat, dog and human. Chromosome Res 8: 393–404. 10.1023/A:1009210803123 [DOI] [PubMed] [Google Scholar]
- Yokoyama KD, Zhang Y, Ma J. 2014. Tracing the evolution of lineage-specific transcription factor binding sites in a birth-death framework. PLoS Comput Biol 10: e1003771 10.1371/journal.pcbi.1003771 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.