Abstract
Background
Gut symbiotic microbiota plays a critical role in nutrient supply, digestion, and absorption. The bamboo snout beetle, Cyrtotrachelus buqueti, a common pest of several bamboo species, exhibits high lignocellulolytic enzyme activity and contains various CAZyme genes. However, to date, no studies have evaluated the role of gut symbiotic microbiota of the snout beetle on bamboo lignocellulose degradation. Therefore, the present study investigated the role of gut symbiotic microbiota of C. buqueti on bamboo lignocellulose degradation.
Results
Gut symbiotic microbiota of female (CCJ), male (XCJ), and larvae (YCJ) beetles was used to treat bamboo shoot particles (BSPs) in vitro for 6 days. Scanning electron microscopy (SEM) revealed significant destruction of the lignocellulose structure after treatment, which was consistent with the degradation efficiencies of CCJ, XCJ, and YCJ for cellulose (21.11%, 17.58% and 18.74%, respectively); hemicellulose (22.22%, 27.18% and 34.20%, respectively); and lignin (19.83%, 24.30% and 32.97%, respectively). Gut symbiotic microbiota of adult and larvae beetles was then identified using 16sRNA sequencing, which revealed that four microbes: Lactococcus, Serratia, Dysgonomonas and Enterococcus, comprise approximately 84% to 94% of the microbiota. Moreover, the genomes of 45 Lactococcus, 72 Serratia, 86 Enterococcus and 4 Dysgonomonas microbes were used to analyse resident CAZyme genes. These results indicated that gut symbiotic microbiota of adult and larvae C. buqueti is involved in the lignocellulose degradation traits shown by the host.
Conclusions
This study shows that the gut symbiotic microbiota of C. buqueti participates in bamboo lignocellulose degradation, providing innovative findings for bamboo lignocellulose bioconversion. Furthermore, the results of this study will allow us to further isolate lignocellulose-degrading microbiota for use in bamboo lignocellulose bioconversion.
Electronic supplementary material
The online version of this article (10.1186/s13068-019-1411-1) contains supplementary material, which is available to authorized users.
Keywords: Lignocellulose degradation, Bamboo, Gut symbiotic microbiota, Cyrtotrachelus buqueti, 16sRNA-sequencing
Background
Insects are the largest group of organisms on earth, and most insect species feed on plants. Some insects, such as termites and beetles, consume plant cell walls and are involved in the degradation of lignocellulose and other types of biomass; thereby contributing to lignocellulose bioconversion and energy utilisation [1, 2]. To date, many insects, including termites, wood-feeding roaches, beetles, wood wasps, leaf-shredding aquatic insects, silverfish and leaf-cutting ants, have been shown to exhibit lignocellulose degradation ability [1].
In phytophagous insects, such as termites, beetles and wood bees, lignocellulose digestion requires cooperation between insects and symbiotic microorganisms in the insect gut, especially bacteria, protozoa, fungi, and yeasts [3]. In Costelytra zealandica (New Zealand grass grub), various hindgut bacteria participate in lignocellulose degradation [4] and some lignocellulose-degrading bacteria have been isolated from the larvae of the scarab beetle Pachnoda marginata [5]. Moreover, bacteria exhibiting cellulose hydrolysis activity have been isolated from other insects, such as Ips pini (North American pine engraver), Dendroctonus frontalis (Southern pine beetle), Saperda vestita (Linden borer) [6] and Tipula abdominalis (giant crane fly) [7]. Symbiotic gut microbes of the fungus-cultivating termites Macrotermes annandalei, Odontotermes yunnanensis and Macrotermes barneyi possess various lignocellulolytic enzyme genes [8]. A recent study identified 111 glycoside hydrolase (GH) family genes among the symbiotic microbes of Macrotermes natalensis [9]. Taken together, these studies show that there is significant involvement of insect symbiotic microbes in lignocellulose degradation.
16S rRNA sequencing has been widely applied to the detection and identification of gut microorganisms in insects. Gut bacteria in many phytophagous insects, including termites [10, 11], Rhynchophorus ferrugineus (palm weevil) [12] and Dendroctonus armandi (Chinese white pine beetle) [13] have been identified by 16S rRNA sequencing and show that the symbiotic bacteria primarily belong to Escherichia sp., Serratia sp., Pantoea sp., Acinetobacter sp., Salmonella sp., Pseudomonas sp., Shigella sp., Staphylococcus sp., Klebsiella sp., Enterobacter sp., Erwinia sp., Vibrio sp., Proteus sp. and Bacillus sp. Of these identified bacteria, Serratia sp. [14], Pseudomonas sp. [15] and Bacillus sp. [16] exhibited lignocellulose degradation activity.
The bamboo snout beetle, Cyrtotrachelus buqueti, is a serious pest of bamboo species, which include Phyllostachys pubescens, Neosinocalamus affinis, Bambusa textilis and Dendrocalamus farinosus [17]. Cyrtotrachelus buqueti also exhibits high lignocellulolytic enzyme activity [18]. However, to date, no studies have investigated the role of snout beetle gut microbiota in lignocellulose degradation, as has been done for termites [8] and other species of beetles [4–7]. The present study investigates the role of adult and larval C. buqueti gut bacteria using 16sRNA sequencing. Adult and larval C. buqueti were shown to carry similar loadings of lignocellulose-degrading bacteria. Furthermore, the degradation of bamboo shoots (Bambusa emeiensis) by snout beetle gut microbiota was investigated in vitro, and the results revealed that the microbiota of both adult and larval C. buqueti has lignocellulose-degrading ability.
Results
Gut structure in different age groups and microbial colonisation of the gut paunch
In insects, the gut is divided into foregut, midgut, and hindgut [19]. Although the foregut is mostly involved in mechanical treatment [20], delignification occurs in the midgut [21] and symbiotic microorganisms exist mainly in the hindgut, where the biomass is degraded [19].
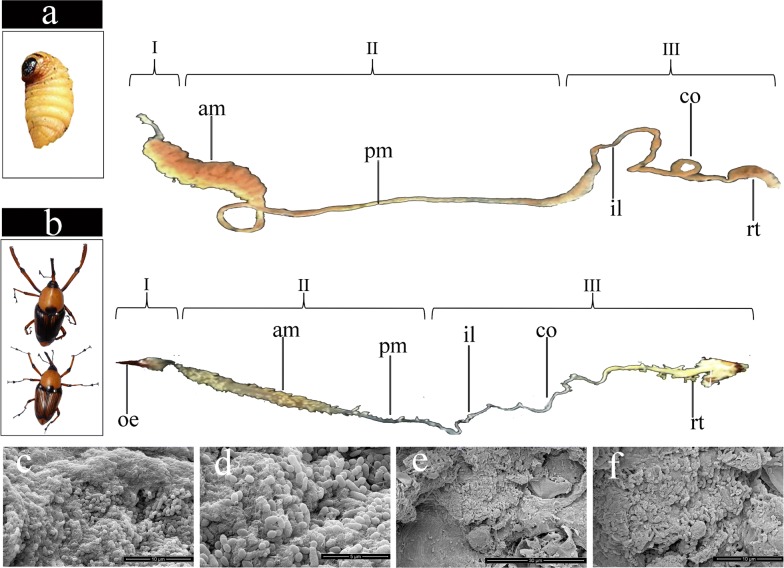
In the present study, the intestinal tract of the C. buqueti was analysed. Results showed that it comprises a foregut, a large midgut comprising an anterior and a posterior midgut, and a hindgut comprising a paunch, an ileum and a colon (Fig. 1a, b). The total length of the larval intestinal tract was 21.25 ± 0.55 cm, which corresponds to 3.8 times its body length. The foregut represented 3.33–3.7% of the total length, whereas the midgut represented 52–58%, and the hindgut represented 38–44%. On the other hand, the total length of the adult intestinal tract was 9.83 ± 0.84 cm, which corresponds to 2.4 times of its body length. The foregut represented 13–20% of the total length, the midgut represented 28–32%, and the hindgut represented 51–55%.
Fig. 1.
Intestinal tract structure in adult and larvae of C. buqueti. Morphological characteristics of larvae (a) and adults (b). I: foregut; II: midgut; III: hindgut; oe: oesophagus; am: anterior midgut; pm: posterior midgut; il: ileum; Co: colon; rt: rectum; Scanning electron micrographs global (c) and detailed (d) views of the adult intestinal tract. Scanning electron micrographs global (e) and detailed (f) views of the larvae intestinal tract
The distribution of microbial cells colonising the gut paunch was analysed by SEM (Fig. 1c–f). In adults and larvae, the gut paunch was mostly inhabited by rods and cocci, indicating the existence of symbiotic microbiota in the gut of adult and larval C. buqueti.
SEM, BSPs components and culture reducing sugar content after treatment
In insects that digest lignocellulose, gut symbiotic microbes are ubiquitous and involved in lignocellulose degradation in various degrees [4]. To investigate the degradation efficiency of the gut symbiotic microbes in C. buqueti, the gut symbiotic microbiota of female (CCJ) and male (XCJ) adults and larvae (YCJ) beetles was extracted and used for in vitro degradation of bamboo shoot particles (BSPs) for 6 days.
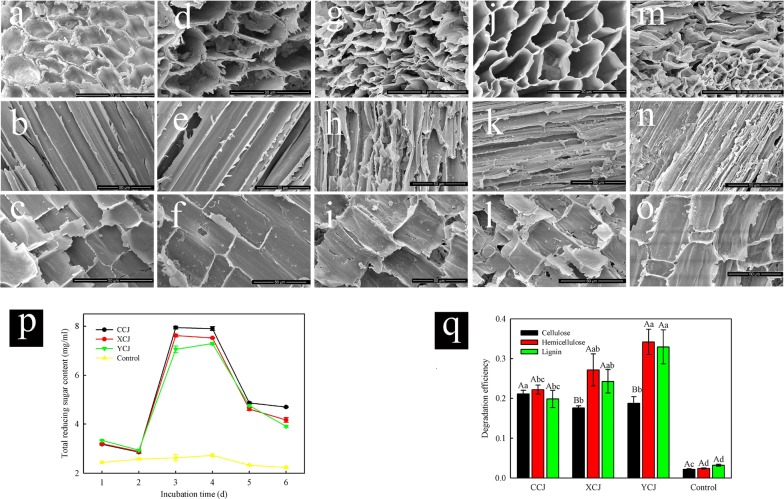
Changes in the fibre microstructure of plant raw materials can be accurately observed using SEM. Following in vitro exposure to the intestinal tract microbiota of female, male, and larval beetles, the BSPs were observed using SEM. The results showed that the surface of lignocellulose from untreated, control BSPs was rough, showing gully-like surface features and a dense structure (Fig. 2a–f). However, no significant differences were observed between raw (Fig. 2a–c) and control (Fig. 2d–f) samples. On the other hand, after 72 h of treatment, the cell walls of BSPs became thinner with an enlarged cellulosic cavity (Fig. 2g, j, m) and many cracks appeared on the surface (Fig. 2h, k, n). Moreover, compared to what is shown in Fig. 2c (Fig. 2i, l, o) new dimples or holes were found on the surface of BSPs. These results are consistent with previous reports [22, 23]. A comparison between the pre- and post-treatment microstructure of BSPs revealed that its structure was significantly destroyed by treatment.
Fig. 2.
Scanning electron microscopy (SEM), bamboo shoot particles (BSPs) components and reducing sugar content in cultures after in vitro treatment. a–c SEM for raw BSPs; d–f SEM for BSPs in the control group; g–i SEM for BSPs after 72 h of treatment in CCJ; j–l SEM for BSPs after 72 h of treatment in XCJ; m–o SEM for BSPs after 72 h of treatment in YCJ; p amount of reducing sugar in the culture after 6 days of treatment; q cellulose, hemicellulose and lignin degradation efficiency of BSPs after 6 days of treatment. CCJ: gut symbiotic microbiota of female; XCJ: gut symbiotic microbiota of male; YCJ: gut symbiotic microbiota of larvae. Descriptive data were expressed as mean ± Standard Error of Mean; the different normal letters indicated significant difference at different developmental stage at 0.05 level (n = 3); the different capital letters indicated significant difference in different components at 0.05 level (n = 3)
Since the reducing sugars in the culture medium was mainly derived from the hydrolysis of cellulose and hemicellulose in BSPs, the reducing sugar content was determined to reflect the degree of conversion of lignocellulose. The results showed small content changes during the first day of digestion, which increased henceforth. The maximum value was reached on day three in adults and on day 4 in larvae, subsequently decreasing until day 6 (Fig. 2p). The increase of reducing sugars indicated the degradation of cellulose and hemicellulose [24].
We then determined the degradation efficiencies of lignocellulose from BSPs after 6 days of treatment. As shown in Fig. 2q, it was found that cellulose, lignin, and hemicellulose were partially removed by the gut symbiotic microbiota. The degradation efficiencies of CCJ, XCJ, and YCJ digestions were: 21.11%, 17.58% and 18.74%, respectively, for cellulose; 22.22%, 27.18% and 34.20%, respectively, for hemicellulose and 19.83%, 24.30% and 32.97%, respectively, for lignin (Fig. 2q). These results showed that gut symbiotic microbiota degrading cellulose, lignin, and hemicellulose in BSPs could be cultured in vitro and then applied to the pretreatment and hydrolysis of bamboo lignocellulose.
Determination of lignocellulolytic enzyme activity
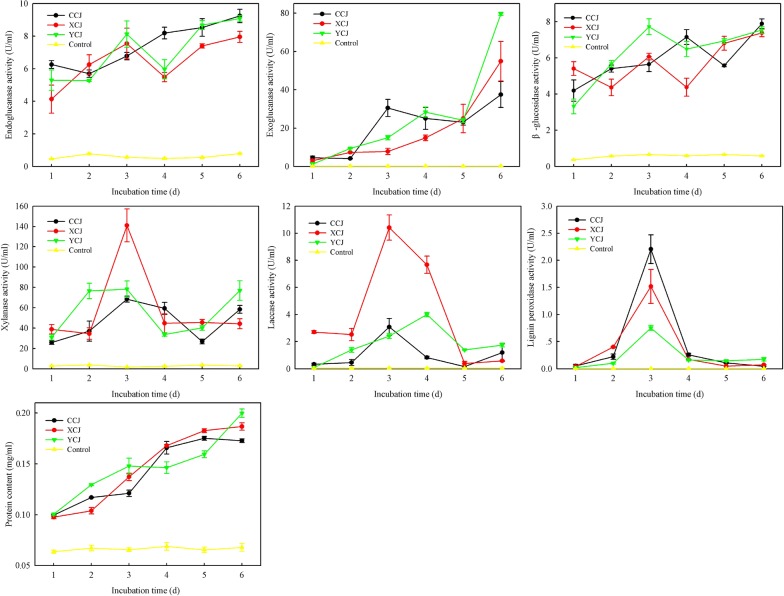
To investigate the mechanism of the degradation of BSPs lignocellulose in vitro, the activities of the lignocellulolytic enzymes present in the gut symbiotic microbiota of adults and larvae, such as endoglucanase, β-glucosidase, xylanase, exoglucanase, laccase and lignin peroxidase, were determined. As presented in Fig. 3, the activity of cellulase, including endoglucanase, β-glucosidase and exoglucanase increased with treatment, whereas that of xylanase, laccase and lignin peroxidase first increased and subsequently decreased. To investigate whether the higher enzyme activity was caused by an increase of protein secretion, we determined the secreted protein content during the course of the treatment. The amount of secreted proteins was found to increase continuously (Fig. 3), indicating that the higher enzyme activity was partially caused by increased protein secretion.
Fig. 3.
The lignocellulolytic enzyme activities of C. buqueti displayed by gut symbiotic microbiota 1, 2, 3, 4, 5 and 6 days after being co-cultured with BSP in vitro
Gas chromatography–mass spectrometry (GC–MS) of lignin degradation products of BSP in vitro
GC–MS has been shown to effectively detect lignin degradation products [24]. We used GC–MS to identify the type of low-molecular-weight products obtained from the cultures after six days of treatment. The total ion chromatograms (TIC) of compounds extracted from control and treated samples revealed that several small molecular aromatic metabolites occurred only in treated samples. Such was the case of phenol (RT 7.22), phenylethyl alcohol (RT 10.71), 2-methoxyphenol (RT 9.85), 4-ethylphenol (RT 12.36) and p-cresol (RT 9.97), which are considered as the basic units of the lignin polymer (Table 1; Additional file 1: Figure S1). This result indicates that lignin in BSPs was degraded to produce phenolic compounds, with the guaiacyl structure unit as the main body. Furthermore, bond breakage between the methoxyl groups linked to the benzene ring of guaiacyl produces one-substituted aromatics, such as phenol and dimethyl phenol.
Table 1.
Identification of metabolites as trimethylchlorosilane (TMS) derivatives from BSP samples
| Retention time | Compound | Controla | Treatedb | Treatedc | Treatedd |
|---|---|---|---|---|---|
| 4.33 | 4-Benzyloxy-3-methoxy-2-nitro-benzaldehyde | − | − | − | + |
| 4.90 | Phenyl-butanedioic acid | − | − | − | + |
| 7.22 | Phenol | − | + | + | + |
| 7.49 | Succinic acid | − | − | + | − |
| 9.85 | 2-Methoxy-phenol | − | + | + | + |
| 9.97 | p-Cresol | − | − | + | − |
| 10.71 | Phenylethyl alcohol | − | − | + | − |
| 12.36 | 4-Ethyl-phenol | − | + | − | − |
| 15.08 | 4-Ethyl-2-methoxy-phenol | − | − | + | + |
aNon-inoculated (control) BSP samples
bCCJ degraded BSP samples
cXCJ degraded BSP samples
dYCJ degraded BSP samples
Bamboo lignin contains guaiacyl (G), syringyl (S) and p-hydroxyphenyl (H) units, belonging to the G–S–H type [25]. It was previously reported that most bamboo cell walls are mainly composed of guaiacyl units at the early stage of lignification [26]. In the present study, BSPs were degraded to produce phenolic compounds; with guaiacyl structure unit as the main body, indicating that the G unit of BSPs was degraded by the beetles’ gut symbiotic microbiota.
Structure of bacterial communities
To investigate the bacterial diversity in the larval and adult C. buqueti guts, a bacterial 16S rRNA gene clone library was constructed by PCR. The whole DNA was extracted from the whole intestinal tracts of the adults and larvae. The larval and adult guts were labelled as ‘YWG’ and ‘AWG’, respectively. A total of 1,957,125 raw reads from 15 YWGs and 15 AWGs were characterised by Illumina high-throughput sequencing. After quality trimming, 1,889,645 high-quality clean tags were obtained and binned into 31,196 operational taxonomic units (OTUs) (Table 2).
Table 2.
Estimated richness and diversity of bacterial communities in the gut of adult and larval Cyrtotrachelus buqueti obtained from pyrosequencing analysis
| Sample | Number of reads | Number of OTUs | Species richness | Goods coverage | Community diversity | PD whole tree | ||
|---|---|---|---|---|---|---|---|---|
| ACE | Chao1 | Shannon | Simpson | |||||
| AWG1 | 31,150 | 182 | 278.8226 | 289.2500 | 0.9977 | 0.6883 | 2.4760 | 9.3095 |
| AWG2 | 56,943 | 859 | 1066.7443 | 1049.8792 | 0.9948 | 0.8003 | 3.7530 | 22.9572 |
| AWG3 | 60,119 | 923 | 1233.6556 | 1244.2368 | 0.9935 | 0.7983 | 3.6131 | 22.7947 |
| AWG4 | 52,573 | 1101 | 1330.6848 | 1321.6359 | 0.9932 | 0.8031 | 3.8502 | 27.3003 |
| AWG5 | 65,985 | 974 | 1282.3685 | 1264.5150 | 0.9941 | 0.6436 | 3.1327 | 24.3539 |
| AWG6 | 37,952 | 841 | 1203.8728 | 1144.7186 | 0.9896 | 0.6455 | 3.3916 | 21.7401 |
| AWG7 | 50,506 | 1059 | 1384.7723 | 1364.1053 | 0.9919 | 0.7939 | 4.0728 | 26.7679 |
| AWG8 | 62,913 | 987 | 1200.7457 | 1172.2312 | 0.9945 | 0.4227 | 2.5382 | 26.0306 |
| AWG9 | 62,582 | 733 | 1080.6907 | 1077.8824 | 0.9945 | 0.8028 | 3.4679 | 20.8508 |
| AWG10 | 43,560 | 876 | 1302.5937 | 1272.2903 | 0.9906 | 0.7623 | 3.6795 | 24.8618 |
| AWG11 | 60,729 | 1097 | 1453.0054 | 1438.9048 | 0.9927 | 0.6030 | 3.5085 | 33.2195 |
| AWG12 | 48,588 | 910 | 1312.9882 | 1270.3208 | 0.9909 | 0.7812 | 3.9470 | 24.6801 |
| AWG13 | 57,739 | 930 | 1240.5902 | 1321.8898 | 0.9927 | 0.7341 | 3.6324 | 25.3036 |
| AWG14 | 66,286 | 853 | 1326.6953 | 1390.7027 | 0.9937 | 0.7559 | 3.2992 | 26.4275 |
| AWG15 | 30,357 | 470 | 625.4256 | 602.0000 | 0.9935 | 0.7590 | 3.6334 | 14.5617 |
| Mean of YWG | 52,532 | 853 | 1154.9104 | 1148.3042 | 0.9932 | 0.7196 | 3.4664 | 23.4106 |
| YWG1 | 89,632 | 1290 | 1529.4723 | 1562.8947 | 0.9957 | 0.8009 | 4.2393 | 29.2471 |
| YWG2 | 73,877 | 1234 | 1510.1208 | 1567.0368 | 0.9948 | 0.7527 | 3.6564 | 27.8022 |
| YWG3 | 85,232 | 1328 | 1561.7405 | 1618.0491 | 0.9958 | 0.7966 | 4.1156 | 30.2476 |
| YWG4 | 64,352 | 999 | 1194.9094 | 1244.0476 | 0.9957 | 0.6055 | 2.9833 | 22.9895 |
| YWG5 | 75,328 | 1079 | 1407.4523 | 1419.0066 | 0.9953 | 0.4898 | 3.0009 | 26.8487 |
| YWG6 | 80,143 | 989 | 1275.2326 | 1275.1728 | 0.9954 | 0.5682 | 3.0797 | 25.9651 |
| YWG7 | 106,571 | 1410 | 1630.6420 | 1665.7977 | 0.9966 | 0.7790 | 4.0954 | 30.8279 |
| YWG8 | 91,679 | 1518 | 1955.5009 | 1987.6239 | 0.9941 | 0.6591 | 3.7524 | 34.5241 |
| YWG9 | 95,196 | 1416 | 1691.5796 | 1732.2813 | 0.9952 | 0.8102 | 4.3674 | 31.9566 |
| YWG10 | 71,581 | 1405 | 1871.9408 | 1866.4194 | 0.9926 | 0.8188 | 4.6616 | 30.3750 |
| YWG11 | 65,622 | 1129 | 1637.4775 | 1608.9306 | 0.9930 | 0.6933 | 3.9235 | 27.3060 |
| YWG12 | 66,211 | 1097 | 1457.2757 | 1471.6908 | 0.9940 | 0.7594 | 3.9203 | 25.9297 |
| YWG13 | 59,952 | 1312 | 1730.7135 | 1699.8066 | 0.9917 | 0.8682 | 5.0228 | 32.0469 |
| YWG14 | 61,949 | 987 | 1218.5840 | 1319.5909 | 0.9944 | 0.7178 | 3.9469 | 23.7491 |
| YWG15 | 81,818 | 1208 | 1508.3660 | 1559.4024 | 0.9944 | 0.8290 | 4.5729 | 27.4453 |
| Mean of YWG | 77,942 | 1226 | 1545.4005 | 1573.1834 | 0.9946 | 0.7299 | 3.95589 | 28.4841 |
| P value | 9.2251E−06 | 4.1182E−05 | 4.4533E−04 | 1.4405E−04 | 3.2404E−02 | 7.9292E−01 | 1.8278E−02 | 4.8929E−03 |
OTU: operational taxonomic units; PD: phylogenetic diversity
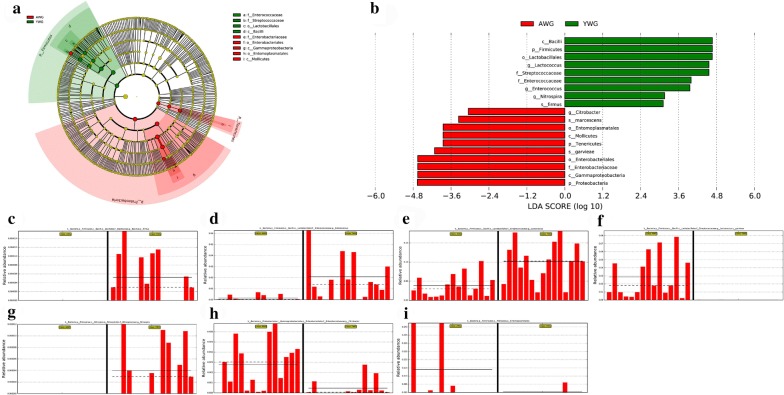
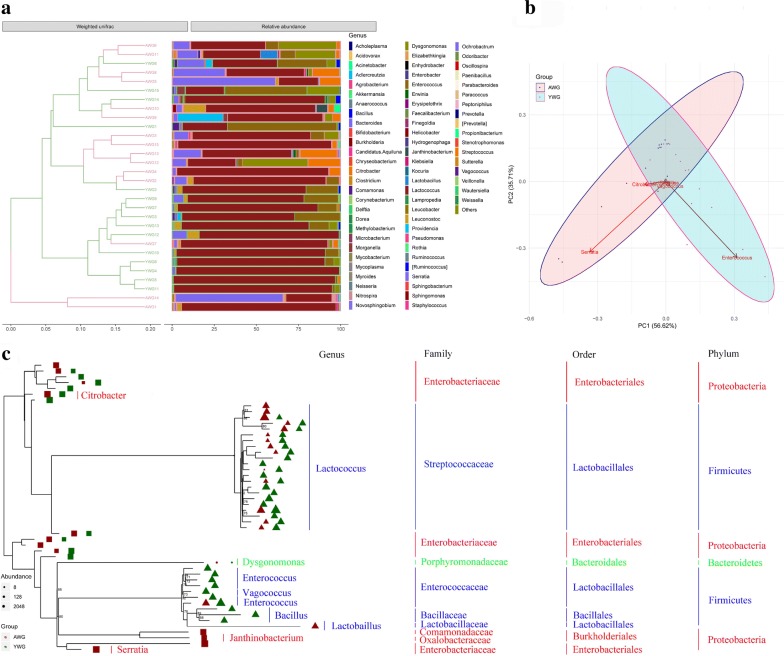
At the phylum level, a total of 14 prokaryotic phyla were identified, among which five were common to both groups (Fig. 4a, b). In both groups, Proteobacteria (38.6–73.0%) and Firmicutes (19.2–59.8%) were the most abundant microbial communities. The two most increased phyla in the AWG group were Proteobacteria and Tenericutes; which, compared with the YWG group, had a relative abundance increase of 89% and 3095%, respectively. On the contrary, two phyla, Firmicutes and Fibrobacteres, decreased by 63% and 68%, respectively (Additional file 2: Table S1). At the genus level, besides unclassified OTUs, 74 genera were detected, among which 29 were common to both groups (Fig. 4c, d). The abundance of all genera in each group is shown in Additional file 3: Table S2. Lactococcus (59.1–72.3%) was consistently abundant in both groups. Thirty-four genera were detected only in the AWG group, whereas 11 were detected only in the YWG group. Non-metric multi-dimensional scaling (NMDS) plots and similarity analysis (ANOSIM) (p < 0.05) (Additional file 4: Figure S2) revealed a divergence of the community structure in the AWG and YWG groups.
Fig. 4.
Bacterial operational taxonomic units (OTUs) composition in adult and larvae beetles (AWG and YWG). a Venn diagram showing the coincidence of phyla between the groups. b Phylum-level comparison of bacterial OTUs between the groups. c Venn diagram showing the overlap of genera between groups. d Genus-level comparison of bacterial OTUs between the groups
Diversity and richness of microbial communities
The YWG had the highest species richness (Chao 1, ACE and OTUs) (Table 2). A significant difference was observed between the groups regarding the diversity of microbial communities as determined by the Shannon index (p < 0.05), whereas the Simpson index revealed no significant difference (Table 2; Additional file 5: Figure S3). Differentially represented OTUs were analysed via linear discriminant analysis (LDA) effect size (LEfSe), a statistical measure used in metagenomic biomarker discovery (Fig. 5a, b). Genera such as Lactococcus, Enterococcus and Nitrospira and only one species—the Bacillus firmus—increased in YWG, whereas one genus and two species increased in AWG (Fig. 5c–i).
Fig. 5.
LEfSe analysis of the two groups. a Cladogram of the two groups. b LDA score histogram. The relative abundance of c Bacillus firmus, d Enterococcus, e Lactococcus, f Lactococcus garvieae, g Nitrospira, h Citrobacter and i Entomoplasmatales. Solid and dashed lines indicate mean and median, respectively. The x-axis in c–i represented the samples: AWG1-15 and YWG1-15
UniFrac analysis was performed to compare the degree of phylogenetic overlap in the microbial communities of AWG and YWG (Fig. 6a). Moreover, a total of six OTUs with maximum abundance, including Lactococcus, Enterococcus, Bacillus, Citrobacter, Vagococcus and Serratia, were used for principal component analysis and clearly separated into two groups. Specifically, the difference of PC1 was caused mostly by Enterococcus and Serratia, which were positively correlated. Additionally, the Enterococcus was positively correlated with the other OTUs except Citrobacter, whereas the Serratia was positively correlated with all OTUs (Fig. 6b). Maximum likelihood (ML) analysis of the 50 detectable OTUs showed that the significantly increased OTUs in the YWG group belonged to the families: Enterobacteriaceae, Streptococcaceae, Enterococcaceae, and Bacillaceae subdivision 4 (Fig. 6c).
Fig. 6.
a Weighted UPGMA tree. b Beta diversity of the samples. c Maximum likelihood tree of 50 detectable OTUs (the relative abundance > 1% in the given sample). The complete 16S rRNA gene sequences of the corresponding species in the RDP database were used to construct the tree. Red indicates the OTUs in the AWG group, and green indicates the OTUs in the YWG group. Only the OTUs with significant differences (p < 0.05) in relative abundance are shown behind the branches. The size of the symbol indicates the relative abundance of OTUs
Functional analysis of microbial communities
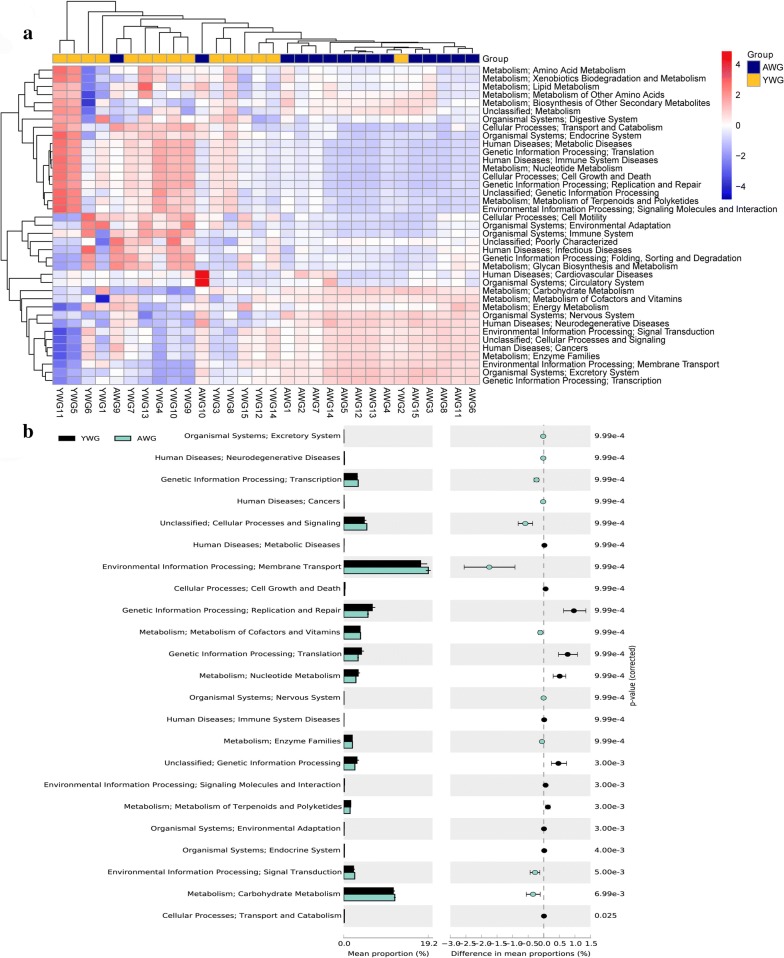
In this study, PICRUSt was used to predict the microbial community metagenome [27]. At the KEGG L2 level, in AWG, OTUs were mainly enriched in metabolic pathways, such as amino acid metabolism, lipid metabolism, xenobiotics biodegradation and metabolism and human diseases. In YWG, the OTUs were enriched in energy metabolism, environment adaption, nucleotide metabolism and molecular signalling and interaction (Fig. 7a). The functions of each bacterial community in AWG and YWG were significantly different for all pathways (Fig. 7b).
Fig. 7.
KEGG pathways enriched in adults and larvae of insects (AWG and YWG). a Cluster of pathways in AWG and YWG. b The relative pathways abundance was compared between AWG and YWG
Moreover, PICRUSts predicted an enrichment of the relative abundance of genes relevant to lignocellulose degradation, genes encoding the carbohydrate-active enzymes (CAZymes), including glycoside hydrolases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), and auxiliary activities (AAs) (Additional file 6: Figure S4). Among the CAZyme family genes included on the prediction, many were closely related to lignocellulose degradation.
Composition and lignocellulose degradation potential of the core degradative microbiota
16sRNA-sequencing showed that Lactococcus, Serratia, Dysgonomonas, and Enterococcus represented about 84–94% of the microbiota, comprising the core digestive microbiota of AWG and YWG (Additional file 7: Figure S5). To further explore the potential role of Lactococcus, Serratia, Dysgonomonas, and Enterococcus in lignocellulose degradation, we compared their CAZymes content with that of Cytophaga hutchinsonii ATCC 33406, a model bacterium for lignocellulose bioconversion [28]. This comparison was based on the public CAZy databases (Table 3; Additional file 8: Table S3). Since no Dysgonomonas genome was available in the CAZy database, we manually annotated the CAZyme-coding sequences in the genome of the genus available to date in the NCBI Genome, i.e., that of Dysgonomonas mossii DSM 22836, Dysgonomonas capnocytophagoides DSM 22835, Dysgonomonas macrotermitis and Dysgonomonas gadei ATCC BAA-286. Compared with the genome of C. hutchinsonii ATCC 33406, the genomes of Dysgonomonas contained more GHs and CEs (Table 3; Additional file 8: Table S3). To evaluate the ability of Lactococcus to degrade lignocellulose in the intestine, we examined the CAZyme content of the 45 Lactococcus genomes currently available in the CAZy database (Additional file 8: Table S3). The 45 genomes present lower numbers of CAZyme than the C. hutchinsonii genome (Table 3; Additional file 8: Table S3). However, the examined Lactococcus genomes were mostly enriched in GH1 and GH13, which putatively exhibit (among others) β-glucosidase and β-galactosidase activity [29] and α-amylase [30], respectively (Additional file 8: Table S3). For Serratia, 72 genomes were examined, which showed a higher capacity to degrade carbohydrates, including lignocellulose, than C. hutchinsonii, due to a higher number of CAZyme families and genes in the genome (Table 3; Additional file 8: Table S3). Moreover, the GHs of Serratia were enriched in GH1, GH2, GH3, GH4, GH13, GH18 and GH23, and more AAs were performed. Eighty-six Enterococcus genomes were compared with the C. hutchinsonii genome, which exhibited more families and number, especially in GHs (Table 3; Additional file 8: Table S3). The examined Enterococcus genomes were mostly enriched in GH1, GH13 and GH73, which putatively exhibit (among others) β-glucosidase and β-galactosidase activity [22], α-amylase [23] and lysozyme [31], respectively (Additional file 8: Table S3).
Table 3.
Total number of putative CAZy genes, including glycoside hydrolase (GH), carbohydrate esterase (CE), auxiliary activity (AA) and polysaccharide lyase (PL) genes in selected genomes
| GHa | CEa | AAa | PLa | |
|---|---|---|---|---|
| 45 Lactococcus genomes | 8–23b 9–51c |
0–3 0–6 |
0–1 0–2 |
0–1 0–2 |
| 72 Serratia genomes | 2–27 3–90 |
0–5 0–6 |
0–2 0–4 |
0–6 0–12 |
| 86 Enterococcus genomes | 15–34 28–121 |
1–6 1–11 |
0–1 0–8 |
0–3 0–4 |
| 4 Dysgonomonas genomes | 43–55 131–223 |
7–8 23–28 |
0–0 0–0 |
0–3 0–6 |
| C. hutchinsonii ATCC 33406 | 19 50 |
7 14 |
0 0 |
3 3 |
This table also contains total number of CAZy genes number in selected genomes for the four genera of the core digestive microbiota of the insect and of Cytophaga hutchinsonii ATCC 33406, a model bacterium for cellulose degradation [21]. For more details on the CAZyme content, see Additional file 7: Table S2
C. hutchinsonii Cytophaga hutchinsonii. Data retrieved from the CAZy databases (http://www.cazy.org)
aCAZyme class
bTotal number of CAZyme families in the given class
cTotal number of CAZyme genes in the given class
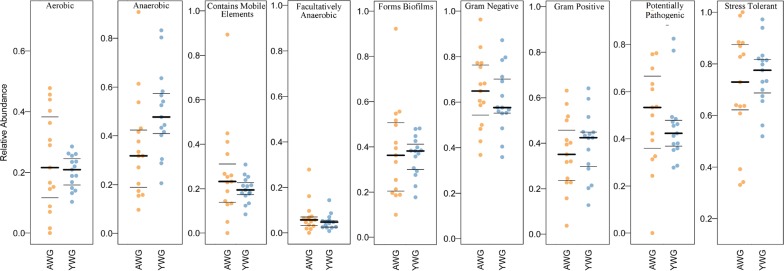
Microbiota phenotype prediction using BugBase
BugBase (https://bugbase.cs.umn.edu/) is an algorithm that predicts the organism-level coverage of functional pathways; as well as biologically interpretable phenotypes, such as oxygen tolerance, Gram staining, and pathogenic potential within complex microbiota using either whole-genome shotgun or marker gene sequencing data [32]. However, no significant differences were observed in the relative abundance of all classifications (Fig. 8; Additional file 9: Table S4), indicating similarities in the bacterial phenotype of AWG and YWG. To date, few reports have focused on degradation by microbes, such as Coriolus versicolor [33] and Galactomyces sp. CCZU11-1 [34]. These predicted results will, therefore, expand the current knowledge on gut symbiotic microbiota of the beetle and support the culture of lignocelluloses-degrading bacteria in vitro.
Fig. 8.
BugBase analyses, based on the NGS dataset. The outcome is grouped according to the modules AWG and YWG (X-axis). The relative abundance is given on the Y-axis. ‘Mobile elements’ refers to bacteria, most probably carrying mobile elements
Discussion
The symbiotic microbiota of insects includes bacteria, protozoa, fungus and archaea, among which bacteria are the most representative. Symbiotic bacteria exist in Coleoptera, Blattaria, Isoptera, Diptera, Hemiptera, Mallophaga, and Anoplura [35] and the symbiotic bacteria of insects primarily belong to the phyla: Proteobacteria, Bacteroidetes, Firmicutes, Actinomycetes, Spirochetes, and Verrucomicrobia [36].
Gut symbiotic microbes play an important role in nutrient supply, digestion and absorption. Bashir et al. [37] isolated 42 cellulose-degrading bacteria from the gut of termites, pill bugs, and stem borers. Dantur et al. [38] isolated 118 cellulose-degrading bacteria from the larval intestine of Diatraea saccharalis, and Manfredi et al. [39] isolated 233 cellulose-degrading bacteria from Spodoptera frugiperda and Diatraea saccharalis, among which Bacillus and Paenibacillus were the most prevalent. Peterson et al. [40] used antibiotics to clarify the role of symbiotic bacteria on cellulose degradation by Reticulitermes flavipes. Furthermore, Shotorkhoft et al. [41] isolated three bacteria exhibiting ligninase activity from Microcerotermes diversus and discussed their ability to degrade wheat straw.
In the present study, we used 16sRNA sequencing to identify gut symbiotic bacteria in C. buqueti. Results showed that Lactococcus, Serratia, Dysgonomonas, and Enterococcus represented approximately 84–94% of the microbiota. Robert et al. [42] found that the most common cellulolytic strains isolated corresponded to the Ruminococcus and Enterococcus species found in the human colon. Shil et al. [43] showed that Enterococcus species participates in lignocellulose degradation by insects. In the present study, BSPs were treated with gut symbiotic microbiota, which revealed that the degradation efficiency of CCJ, XCJ and YCJ was 21.11%, 17.58% and 18.74%, respectively, for cellulose; 22.22%, 27.18% and 34.20%, respectively, for hemicellulose; and 19.83%, 24.30% and 32.97%, respectively, for lignin, indicating that symbiotic s degrade the cellulose, hemicellulose, and lignin of BSPs. Moreover, gut symbiotic microbiota has the potential to be cultured in vitro and at large scale; and then applied to the pretreatment and hydrolysis of bamboo lignocellulose. BugBase (https://bugbase.cs.umn.edu/) was used to predict microbiota phenotypes, which allowed the expansion of current knowledge regarding gut symbiotic microbiota of C. buqueti, with the potential to support in vitro cultures of lignocellulose-degrading bacteria.
Conclusions
The present study investigated the bamboo lignocellulose-degrading ability of gut symbiotic microbiota of C. buqueti. The in vitro hydrolysis assay revealed bamboo lignocellulose-degrading efficiencies in CCJ, XCJ, and YCJ of 21.11%, 17.58% and 18.74% for cellulose; 22.22%, 27.18% and 34.20% for hemicellulose, and 19.83%, 24.30% and 32.97% for lignin. Results from the 16sRNA-sequence identified four microbes, namely Lactococcus, Serratia, Dysgonomonas and Enterococcus representing approximately 84–94% of the microbiota in this system. These microorganisms exhibited abundant CAZyme genes and lignocellulose-degrading ability. Finally, predicted results of BugBase support the in vitro culturing of lignocellulose-degrading bacteria of the gut, such as C. buqueti. This study has enriched our knowledge of bamboo lignocellulose-degrading microbiota, which can be applied to bamboo lignocellulose bioconversion.
Methods
Collection of insect samples
Adult and larval C. buqueti were collected in August 2018 in the Muchuan City, Sichuan Province, China (N103°98′, E28°96′). All adults were collected at the 3rd day after emergence [44]. Intestines were extracted from 15 individual adults (males and females) and 15 individual larval beetles, and then immediately stored in liquid nitrogen to ensure gut microbiome stabilisation until subjection to DNA extraction and 16sRNA-sequencing.
In vitro assay of bamboo shoot particles (BSPs) degradation by gut symbiotic microbiota
In the present study, BSPs at the early stage of lignification were used. BSPs were prepared by drying to a constant weight at 65 °C, crushing to particles using a pulverising machine, and straining through a 40-mesh sieve. Gut symbiotic microbiota of adult and larval beetles was prepared by grounding and mixing, respectively. The mixed suspension was then cultured in liquid medium 1 (pH 7.2) composed of 0.04 g yeast extract, 0.1 g malt extract, 2 g CaCO3 and 10 g BSPs for 15 d. Two millilitres of the culture was added into a second liquid medium (pH 7.2) composed of 0.5 g yeast extract, 0.5 g malt extract, 0.5 g tryptone, 0.5 g NaCl, 0.2 g KH2PO4, 0.13 g MgSO4·7H2O and 0.5 g CaCl2. The assay was then performed following the steps listed in Table 4.
Table 4.
Design and determination methods of gut symbiotic microbiome degradation of bamboo shoot particles (BSPs) in vitro
| Experimental group | Control group | Incubation time | Temperature | |
|---|---|---|---|---|
| Experimental design | 2 mL culture + 98 mL pH 7.2 liquid medium 2a + 5 g BSPs | 100 mL pH 7.2 liquid medium 2a + 5 g BSPs | 6 days | 37 °C |
| Low-molecular-weight products | Lignocellulose | Surface structure of BSPs | Reducing sugar | |
|---|---|---|---|---|
| Determination index and methods | GC–MS [17] | Van Soest method [38] | SEM | DNS [37] |
aA liquid medium (pH 7.2) composed of 0.5-g yeast extract, 0.5-g malt extract, 0.5-g tryptone, 0.5-g NaCl, 0.2-g KH2PO4, 0.13-g MgSO4·7H2O and 0.5-g CaCl2
As shown in Table 4, the samples were placed into a 250-mL conical flask and incubated under constant-temperature shock at 37 °C and 150 rpm for 6 days. The reaction products were inactivated at 100 °C for 30 min and centrifuged at 13,000 rpm for 10 min, after which the supernatant was collected and dried at 65 °C to a constant weight. The dried deposit was weighed, and the levels of cellulose, hemicellulose, and lignin were determined and used for SEM. The supernatant was used to determine reducing sugar and low-molecular-weight products.
Determination of reducing sugar
Reducing sugar was identified using 3,5-dinitrosalicylic acid [45].
Determination of cellulose, hemicellulose and lignin
The cellulose, hemicellulose, and lignin contents of the dried BSPs were determined using the Van Soest method [46] and the following formulas:
Cellulose, hemicellulose and lignin degradation efficiencies
Scanning electron microscopy (SEM)
BSPs collected 72 h after treatment were analysed using SEM (Hitachi 3400N, Japan), to observe the samples’ surface morphology. Prior to imaging, the samples were sprayed with gold to a thickness of ~ 10 nm using the E-1010 sputtering film coating machine (Japan). The SEM operating current and voltage were 81 mA and 10 kV, respectively.
Determination the lignocellulolytic enzymes activities
From the co-cultures, 2-mL samples were withdrawn at 1, 2, 3, 4, 5 and 6 days for assays determining lignocellulolytic enzyme activity. Endoglucanase (EC 3.2.1.4), exoglucanase (EC 3.2.1.91), β-glucosidase (EC 3.2.1.21), lignin peroxidase (LiP)-like, laccase-like, and xylanase enzyme activities were analysed as previously described by Luo et al. [18]. Briefly, carboxymethyl cellulose (CMC), microcrystalline cellulose (MCC), salicin, veratryl alcohol (VA), 2,2′-azino-bis (ABTS) and xylan were used as substrates to determine endoglucanase, exoglucanase, β-glucosidase, LiP-like, laccase-like and xylanase, respectively. All assays were performed five times.
Gas chromatography–mass spectrometry (GC–MS)
On day 6, control and treatment cultures were collected and centrifuged at 8000 rpm for 20 min to remove biomass. The supernatants were collected and treated using the method described by Raj et al. [47]. Briefly, the supernatants were extracted three times using equal volumes of dichloromethane, collected extracted liquor, dewatered with anhydrous Na2SO4 and filtered and concentrated to ~ 1 mL. Next, 100-µL dioxane and 10-µL pyridine were added to the sample followed by silylation with 50-µL trimethyl silyl (N,O-bis[trimethylsilyl]trifluoroacetamide [BSTFA]/trimethylchlorosilane [TMS] = 99/1 [v/v]). Gas chromatography–mass spectrometry (GC–MS) was analysed according to the procedure described by Chen et al. [48]. The TMS were identified by comparing their mass spectra with the NIST library.
DNA extraction, amplification and sequencing of 16S rRNA encoding gene sequences
DNA from different samples was extracted using the E.Z.N.A. ®Stool DNA Kit (D4015, Omega, Inc., USA) according to the manufacturer’s instructions. PCR amplification was performed in 25 μL of the following reaction mixture: 50-ng DNA, 12.5-μL Phusion Hot start flex 2X Master Mix (NEB, M0536L), 2.5-μL forward primer 338F 5′-ACTCCTACGGGAGGCAGCAG-3′ and 2.5-μL reverse primer 806R 5′-GGACTACHVGGGTWTCTAAT-3′ [49]. The PCR was performed at 98 °C for 30 s, 35 cycles of 98 °C for 10 s, 54 °C for 30 s, 72 °C for 45 s and 72 °C for 10 min. The amplicon pools were prepared for sequencing. The libraries were sequenced on Illumina MiSeq Instrument (Illumina Inc., San Diego, CA, USA) using the 300 bp paired-end protocol.
Sequence analysis
QIIME v1.9.1 [50] was used to further analyse the sequences, using scripts available in the Qiime website (http://qiime.org/). First, potential chimeras were identified in valid sequences using de novo Uchime (usearch v9.0.2132_i86linux32) [51] and removed with Qiime build-in python scripts. The resulting sequences were defined as good sequences. The final good sequences were clustered into OTUs at 97% similarity using the de novo UCLUST algorithm [52]. Taxonomic assignment was then performed using the GreenGenes database (version 13.8) [53], after which an OTU abundance table was constructed using the Qiime python scripts. Multiple sequence alignment was conducted using the PyNAST (v1.2.2) [54] software, and a phylogenetic tree was constructed using FastTree (v2.1.9) [55], to investigate the phylogenetic relationship of different OTUs. To reduce the noise of amplicon datasets and make the data more credible, the OTUs whose abundance was lower than 0.001% of the total were removed, resulting in a modified OTU abundance table [56]. Using the phylogenetic tree and the modified OTU abundance table, alpha diversity (Ace, Chao1, Shannon, Simpson, Observed OTUs, goods_coverage, PD_whole tree) was calculated using the Qiime script, and beta diversity (Bray–Curtis, weighted and unweighted UniFrac) was estimated with the Phyloseq package (v1.20.0) [57]. Both biodiversities were visualised using the R programme (v3.4.1).
Differentially abundant OTUs and microbes
DESeq 2 (v1.16.1) was used to detect differentially abundant OTUs among groups [58]. Padj < 0.01 was considered as statistically significant. DESeq2 tests were performed using the Qiime python script. Differences among microbes regarding abundance of different taxonomic ranks were determined using a Metastats analysis, which is based on a non-parametric t-test [59]. The relative abundance of a microbe in a sample was calculated by determining the read count normalised by the total reads in that sample. Microbes with a relative abundance lower than 1% in all samples were classified into ‘others’. A level of p < 0.05 was considered as statistically significant. Metastats tests were performed using the EDDA R package (v1.10.0) [60].
LEfSe analysis
The LEfSe algorithm was used to identify different abundance biomarkers between the two groups [61]. LEfSe couples robust tests such as Kruskal–Wallis test with quantitative tests such as Wilcoxon rank-sum test. After LDA, the features are ranked by effect size. An effect size threshold above 3 (on a log10 scale) was used for all biomarkers discussed in this study.
Inferred metagenomics and phenotypes
The PICRUSt (v1.1.2) (http://picrust.github.io) was used to predict microbial community metagenome [27]. First, a collection of closed-reference OTUs was obtained from the filtered reads of QIIME v 1.9.1 and by querying the data against the GreenGenes database (http://greengenes.lbl.gov). The OTUs were assigned at 97% identity. PICRUSt was used to predict and derive relative KEGG pathway abundance. Microbial phenotypes were predicted using BugBase (https://bugbase.cs.umn.edu/) [32], a software that relies on the tools PICRUSt, IMG, KEGG and PATRIC.
Statistical analysis
Statistical analyses were performed using SPSS 19.0 (IBM SPSS, Armonk, NY, USA). Descriptive data were expressed as mean ± standard error of mean (SEM). A Student t-test was used to compare the means from two groups. Comparisons of more than two groups were performed using analysis of variance. A level of p < 0.05 indicated a statistically significant difference.
Additional files
Additional file 1: Figure S1. TIC of dichloromethane extract analysed as TMS derivative from control (A) and treatment CCJ (B), XCJ (C) and YCJ (D) in vitro. CCJ: gut symbiotic microbiota of female beetle; XCJ: gut symbiotic microbiota of male beetle; YCJ: gut symbiotic microbiota of beetle larvae.
Additional file 2: Table S1. Microbial composition of the AWG and YWG groups at the phyla level.
Additional file 3: Table S2. Microbial composition of the AWG and YWG groups at the genus level.
Additional file 4: Figure S2. Non-metric multidimensional scaling (NMDS) analysis of the Bray–Curtis similarity coefficients based on the relative abundance of OTUs in the given sample.
Additional file 5: Figure S3. Boxplot analysis comparing the bacterial OTUs between the two groups.
Additional file 6: Figure S4. Relative abundance of PICRUSt-predicted CAZyme genes relevant to lignocellulose degradation. (A) Glycoside hydrolases (GHs). (B) Glycosyl transferases (GTs). (C) Carbohydrate esterases (CEs). (D) Carbohydrate-binding modules (CBMs). (E) Auxiliary activities (AAs).
Additional file 7: Figure S5. Composition of the digestive core microbiota at the genus level.
Additional file 8: Table S3. Number of annotated genes for each GH, CE, PL and AA family in all Lactococcus, Serratia, Enterococcus and Dysgonomonas genomes available at http://www.cazy.org/and in Cytophaga hutchinsonii ATCC 33406. GH, glycoside hydrolase; CE, carbohydrate esterase; AA, auxiliary activity; PL, polysaccharide lyase; CAZyme, carbohydrate-active enzyme.
Additional file 9: Table S4. Relative abundance of the predicted classifications in each sample by BugBase.
Authors’ contributions
CBL, YQL and YJY designed and performed the experiments; CBL and YJY wrote the manuscript; CBL, YQL, YC, CF, WCL, XMX and HL analysed the data. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank Chengdu Basebiotech Co., Ltd for its assistance in original data processing and related bioinformatics analysis. We also thank other members of the laboratory for suggestions and discussion regarding this work and revision of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The Illumina high-throughput sequencing data are available in the NCBI database Sequence Read Archive (SRA) (BioProject PRJNA503626). The sequencing raw data of the bacteria can be accessed through accession numbers SRR8148938- SRR8148952 and SRR8148220- SRR8148234 in the NCBI database.
Consent for publication
The authors have consented for publication.
Ethical approval and consent to participate
Not applicable.
Funding
This work was supported by Sichuan science and technology programme (2019YFG0139) and the National Natural Science Foundation of China (31470655).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- C. buqueti
Cyrtotrachelus buqueti
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- OTU
operational taxonomic units
- LDA
linear discriminant analysis
- SEM
scanning electron microscopy
- GHs
glycoside hydrolases
- GTs
glycosyltransferases
- CEs
carbohydrate esterases
- CBMs
carbohydrate-binding domains
- PLs
polysaccharide lyases
- AAs
auxiliary activities
- CAZyme
carbohydrate-active enzymes
- NCBI
The National Center for Biotechnology Information
- ABTS
[2,2′-Azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)]
- BSPs
bamboo shoot particles
- CCJ
gut symbiotic microbiota of female
- XCJ
gut symbiotic microbiota of male
- YCJ
gut symbiotic microbiota of larvae
- LEfSe
Linear Discriminant Analysis Effect Size
- TIC
total ion chromatograms
Contributor Information
Chaobing Luo, Email: 13366181512@163.com.
Yuanqiu Li, Email: liyuanqiunord@163.com.
Ying Chen, Email: 946757326@qq.com.
Chun Fu, Email: fuchun421@aliyun.com.
Wencong Long, Email: 236569837@qq.com.
Ximeng Xiao, Email: 807861348@qq.com.
Hong Liao, Email: 113995570@qq.com.
Yaojun Yang, Phone: +86-18608331380, Email: rsyyj@126.com.
References
- 1.Sun JZ, Scharf ME. Exploring and integrating cellulolytic systems of insects to advance biofuel technology. Insect Sci. 2010;17:163–165. doi: 10.1111/j.1744-7917.2010.01348.x. [DOI] [Google Scholar]
- 2.Rizzi A, Crotti E, Borruso L, Jucker C, Lupi D, Colombo M, et al. Characterization of the bacterial community associated with larvae and adults of Anoplophora chinensis collected in Italy by culture and culture-independent methods. Biomed Res Int. 2013;2013:420287. doi: 10.1155/2013/420287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oppert C, Klingeman WE, Willis JD, Oppert B, Jurat-Fuentes JL. Prospecting for cellulolytic activity in insect digestive fluids. Comp Biochem Physiol Part B. 2010;155:145–154. doi: 10.1016/j.cbpb.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 4.Zhang HY, Jackson TA. Autochthonous bacterial flora indicated by PCR-DGGE of 16S rRNA gene fragments from the alimentary tract of Costelytra zealandica (Coleoptera: Scarabaeidae) J Appl Microbiol. 2008;105:1277–1285. doi: 10.1111/j.1365-2672.2008.03867.x. [DOI] [PubMed] [Google Scholar]
- 5.Cazemier AE, Verdoes JC, Reubsaet FAG, Hackstein JH, Van der Drift C, Opden Camp HJM. Promicromonospora pachnodae sp. nov., a member of the (hemi) cellulolytic hindgut flora of larvae of the scarab beetle Pachnoda marginata. Antonie van Leeuwenhoek. 2003;83:135–148. doi: 10.1023/A:1023325817663. [DOI] [PubMed] [Google Scholar]
- 6.Delalibera I, Handelsman JJ, Kenneth FR. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae) Environ Entomol. 2005;34:541–547. doi: 10.1603/0046-225X-34.3.541. [DOI] [Google Scholar]
- 7.Rogers TE, Peterson JB. Analysis of cellulolytic and hemicellulolytic enzyme activity within the Tipula abdominalis (Diptera; Tipulidae) larval gut and characterization of Crocebacterium ilecola gen. nov., sp. nov., isolated from the Tipula abdominalis larval hindut. Insect Sci. 2010;17:291–302. doi: 10.1111/j.1744-7917.2010.01346.x. [DOI] [Google Scholar]
- 8.Ni JF, Tokuda G. Lignocellulose-degrading enzymes from termites and their symbiotic microbiota. Biotechnol Adv. 2013;31:838–850. doi: 10.1016/j.biotechadv.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Poulsen M, Hu HF, Li C, Chen ZS, Xu LH, Otani S, et al. Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc Natl Acad Sci USA. 2014;111:14500–14505. doi: 10.1073/pnas.1319718111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hongoh Y, Deevong P, Hattori S, Inoue T, Noda S, Noparatnaraporn N, et al. Phylogenetic diversity, localization, and cell morphologies of members of the candidate phylum TG3 and a subphylum in the phylum Fibrobacteres, recently discovered bacterial groups dominant in termite guts. Appl Environ Microbiol. 2006;72:6780–6788. doi: 10.1128/AEM.00891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warnecke F, Luginbühl P, Ivanova N, Ghassemian M, Richardson TH, Stege JT, et al. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature. 2007;450:560–565. doi: 10.1038/nature06269. [DOI] [PubMed] [Google Scholar]
- 12.Abrar M, Ya F, Youming H, Zhanghong S. The gut entomotype of red palm weevil Rhynchophorus ferrugineus olivier (Coleoptera: Dryophthoridae) and their effect on host nutrition metabolism. Front Microbiol. 2017;8:2291. doi: 10.3389/fmicb.2017.02291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu X, Wang C, Chen H, Ma J. Differences in the structure of the gut bacteria communities in development stages of the Chinese white pine beetle (Dendroctonus armandi) Int J Mol Sci. 2013;14:21006–21020. doi: 10.3390/ijms141021006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anand AAP, Vennison SJ, Sankar SG, Prabhu DIG, Vasan PT, Raghuraman T, et al. Isolation and characterization of bacteria from the gut of Bombyx mori that degrade cellulose, xylan, pectin and starch and their impact on digestion. J Insect Sci. 2010;10:107. doi: 10.1673/031.010.10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sindhu SS, Dadarwal KR. Chitinolytic and cellulolytic Pseudomonas sp. antagonistic to fungal pathogens enhances nodulation by Mesorhizobium sp. Cicer in chickpea. Microbiol Res. 2001;156:353–358. doi: 10.1078/0944-5013-00120. [DOI] [PubMed] [Google Scholar]
- 16.Trivedi N, Gupta V, Kumar M, Kumari P, Reddy CRK, Jha B. An alkali-halotolerant cellulase from Bacillus flexus isolated from green seaweed Ulva lactuca. Carbohydr Polym. 2011;83:891–897. doi: 10.1016/j.carbpol.2010.08.069. [DOI] [Google Scholar]
- 17.Yang YJ, Wang SF, Gong JW, Liu C, Mu C, Qin H. Relationships among Cyrtotrachelus buqueti larval density and wormhole number and bamboo shoot damage degree. J Appl Ecol. 2009;20:1980. [PubMed] [Google Scholar]
- 18.Luo CB, Li YQ, Liao H, Yang YJ. De novo transcriptome assembly of the bamboo snout beetle Cyrtotrachelus buqueti reveals ability to degrade lignocellulose of bamboo feedstock. Biotechnol Biofuels. 2018;11:292. doi: 10.1186/s13068-018-1291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe H, Tokuda G. Cellulolytic systems in insects. Annu Rev Entomol. 2010;55:609–632. doi: 10.1146/annurev-ento-112408-085319. [DOI] [PubMed] [Google Scholar]
- 20.Bayané A, Guiot SR. Animal digestive strategies versus anaerobic digestion bioprocesses for biogas production from lignocellulosic biomass. Rev Environ Sci Biotechnol. 2011;10:43–62. doi: 10.1007/s11157-010-9209-4. [DOI] [Google Scholar]
- 21.Monlau F, Kaparaju P, Trably E, Steyer JP, Carrere H. Alkaline pretreatment to enhance one-stage CH4 and two-stage H2/CH4 production from sunflower stalks: mass, energy and economical balances. Chem Eng J. 2015;260:377–385. doi: 10.1016/j.cej.2014.08.108. [DOI] [Google Scholar]
- 22.Yang Z, Zhang M, Xin D, Wang J, Zhang J. Evaluation of aqueous ammonia pretreatment for enzymatic hydrolysis of different fractions of bamboo shoot and mature bamboo. Bioresour Technol. 2014;173:198–206. doi: 10.1016/j.biortech.2014.09.109. [DOI] [PubMed] [Google Scholar]
- 23.Li KN, Wang X, Wang JF, Zhang JH. Benefits from additives and xylanase during enzymatic hydrolysis of bamboo shoot and mature bamboo. Bioresour Technol. 2015;192:424–431. doi: 10.1016/j.biortech.2015.05.100. [DOI] [PubMed] [Google Scholar]
- 24.Potumarthi R, Baadhe RR, Nayak P, Jetty A. Simultaneous pretreatment and sacchariffication of rice husk by Phanerochete chrysosporium for improved production of reducing sugars. Bioresour Technol. 2013;128:113–117. doi: 10.1016/j.biortech.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 25.Parameswaran N, Liese W. Ultrastructural aspects of bamboo cells. Cellul Chem Technol. 1980;14:587–609. [Google Scholar]
- 26.Lin J, He X, Hu Y, Kuang T, Ceulemans R. Lignification and lignin heterogeneity for various age classes of bamboo (Phyllostachys pubescens) stems. Physiol Plant. 2010;114:296–302. doi: 10.1034/j.1399-3054.2002.1140216.x. [DOI] [PubMed] [Google Scholar]
- 27.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:1–10. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Li H, Zhou H, Chen G, Liu W. Cellulose and cellodextrin utilization by the cellulolytic bacterium Cytophaga hutchisonii. Bioresour Technol. 2010;101:6432–6437. doi: 10.1016/j.biortech.2010.03.041. [DOI] [PubMed] [Google Scholar]
- 29.McKenna DD, Scully ED, Pauchet Y, Hoover K, Kirsch R, Geib SM, et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016;17:227. doi: 10.1186/s13059-016-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manners DJ. Enzymic synthesis and degradation of starch and glycogen. Adv Carbohydr Chem. 1962;17:371–430. [Google Scholar]
- 31.Blade CCF, Johnson LN, Mair GA, North ACT, Phillips DC, Sarma VR. Crystallographic studies of the activity of hen egg-white lysozyme. Proc R Soc Lond B Biol Sci. 1967;167:378–388. doi: 10.1098/rspb.1967.0035. [DOI] [PubMed] [Google Scholar]
- 32.Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, et al. BugBase predicts organism level microbiota phenotypes. BioRxiv. 2017;1–19.
- 33.Zhang X, Xu C, Wang H. Pretreatment of bamboo residues with Coriolus versicolor for enzymatic hydrolysis. J Biosci Bioeng. 2007;104:149–151. doi: 10.1263/jbb.104.149. [DOI] [PubMed] [Google Scholar]
- 34.Dai Y, Zhang HS, Huan B, He Y. Enhancing the enzymatic saccharification of bamboo shoot shell by sequential biological pretreatment with Galactomyces sp. cczu11-1 and deep eutectic solvent extraction. Bioprocess Biosyst Eng. 2017;40:1–10. doi: 10.1007/s00449-016-1669-7. [DOI] [PubMed] [Google Scholar]
- 35.Rosdocimi EM, Mapelli F, Gonella E, Borin S, Crotti E. Microbial ecology-based methods to characterize the bacterial communities of non-model insects. J Microbiol Methods. 2015;119:110–125. doi: 10.1016/j.mimet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Colman DR, Toolson EC, Takacs-Vesbach CD. Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol. 2012;21:5124–5137. doi: 10.1111/j.1365-294X.2012.05752.x. [DOI] [PubMed] [Google Scholar]
- 37.Bashir Z, Kondapalli VK, Adlakha N, Sharma A, Bhatnagar RK, Chandel G, Yazdani SS. Diversity and functional significance of cellulolytic microbes living in termite, pill-bug and stem-borer guts. Sci Rep. 2013;3:2558. doi: 10.1038/srep02558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dantur KI, Enrique R, Welin B, Castagnaro AP. Isolation of cellulolytic bacteria from the intestine of Diatraea saccharalis larvae and evaluation of their capacity to degrade sugarcane biomass. AMB Express. 2015;5:1–11. doi: 10.1186/s13568-015-0101-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manfredi AP, Perotti NI, Martínez MA. Cellulose degrading bacteria isolated from industrial samples and the gut of native insects from Northwest of Argentina. J Basic Microbiol. 2015;55:1384–1393. doi: 10.1002/jobm.201500269. [DOI] [PubMed] [Google Scholar]
- 40.Peterson BF, Stewart HL, Scharf ME. Quantification of symbiotic contributions to lower termite lignocellulose digestion using antimicrobial treatments. Insect Biochem Mol Biol. 2015;59:80–88. doi: 10.1016/j.ibmb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Shotorkhoft A, Mohammadabadi T, Motamedi H, Chaji M, Fazaeli H. Isolation and identification of termite gut symbiotic bacteria with lignocellulose-degrading potential, and their effects on the nutritive value for ruminants of some by-products. Anim Feed Sci Technol. 2016;221:234–242. doi: 10.1016/j.anifeedsci.2016.04.016. [DOI] [Google Scholar]
- 42.Robert C, Bernalier-Donadille A. The cellulolytic microflora of the human colon: evidence of microcrystalline cellulose-degrading bacteria in methane-excreting subjects. FEMS Microbiol Ecol. 2003;46:81–89. doi: 10.1016/S0168-6496(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 43.Shil RK, Mojumder S, Sadida FF, Uddin M, Sikdar D. Isolation and identification of cellulolytic bacteria from the gut of three phytophagus insect species. Braz Arch Biol Technol. 2014;57657:927–932. doi: 10.1590/S1516-8913201402620. [DOI] [Google Scholar]
- 44.Yang H, Su T, Yang W, Yang CP, Lu L, Chen ZM. The developmental transcriptome of the bamboo snout beetle Cyrtotrachelus buqueti and insights into candidate pheromone-binding proteins. PLoS ONE. 2017;12:e0179807. doi: 10.1371/journal.pone.0179807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller GL. Use of dinitrosalicylic acid reagent for the determination of reducing sugar. Anal Chem. 1959;31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 46.Van Soest PJ, Robertson JB, Lewis BA. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. 1991;74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2. [DOI] [PubMed] [Google Scholar]
- 47.Raj A, Reddy MMK, Chandra R. Identification of low molecular weight aromatic compounds by gas chromatography-mass spectrometry (GC–MS) from kraft lignin degradation by three Bacillus sp. Int Biodeterior Biodegrad. 2007;59:292–296. doi: 10.1016/j.ibiod.2006.09.006. [DOI] [Google Scholar]
- 48.Chen YH, Chai LY, Zhu YH, Yang ZH, Zheng Y, Zhang H. Biodegradation of kraft lignin by a bacterial strain Comamonas sp. B-9 isolated from eroded bamboo slips. J Appl Microbiol. 2012;112:900–906. doi: 10.1111/j.1365-2672.2012.05275.x. [DOI] [PubMed] [Google Scholar]
- 49.Ren D, Gong S, Shu J, Zhu J, Rong F, Zhang Z, et al. Mixed Lactobacillus plantarum strains inhibit Staphylococcus aureus induced inflammation and ameliorate intestinal microflora in mice. BioMed Res Int. 2017;7476467. [DOI] [PMC free article] [PubMed]
- 50.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 53.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2009;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Juntao L, Huaien L, Burton CKH, Nagarajan N. EDDA: experimental design in differential abundance analysis. Genome Biol. 2014;15:527. doi: 10.1186/s13059-014-0527-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. TIC of dichloromethane extract analysed as TMS derivative from control (A) and treatment CCJ (B), XCJ (C) and YCJ (D) in vitro. CCJ: gut symbiotic microbiota of female beetle; XCJ: gut symbiotic microbiota of male beetle; YCJ: gut symbiotic microbiota of beetle larvae.
Additional file 2: Table S1. Microbial composition of the AWG and YWG groups at the phyla level.
Additional file 3: Table S2. Microbial composition of the AWG and YWG groups at the genus level.
Additional file 4: Figure S2. Non-metric multidimensional scaling (NMDS) analysis of the Bray–Curtis similarity coefficients based on the relative abundance of OTUs in the given sample.
Additional file 5: Figure S3. Boxplot analysis comparing the bacterial OTUs between the two groups.
Additional file 6: Figure S4. Relative abundance of PICRUSt-predicted CAZyme genes relevant to lignocellulose degradation. (A) Glycoside hydrolases (GHs). (B) Glycosyl transferases (GTs). (C) Carbohydrate esterases (CEs). (D) Carbohydrate-binding modules (CBMs). (E) Auxiliary activities (AAs).
Additional file 7: Figure S5. Composition of the digestive core microbiota at the genus level.
Additional file 8: Table S3. Number of annotated genes for each GH, CE, PL and AA family in all Lactococcus, Serratia, Enterococcus and Dysgonomonas genomes available at http://www.cazy.org/and in Cytophaga hutchinsonii ATCC 33406. GH, glycoside hydrolase; CE, carbohydrate esterase; AA, auxiliary activity; PL, polysaccharide lyase; CAZyme, carbohydrate-active enzyme.
Additional file 9: Table S4. Relative abundance of the predicted classifications in each sample by BugBase.
Data Availability Statement
The Illumina high-throughput sequencing data are available in the NCBI database Sequence Read Archive (SRA) (BioProject PRJNA503626). The sequencing raw data of the bacteria can be accessed through accession numbers SRR8148938- SRR8148952 and SRR8148220- SRR8148234 in the NCBI database.