Abstract
Drosophila NonA and its mammalian ortholog NONO are members of the Drosophila behavior and human splicing (DBHS) family. NONO also has a strong circadian connection: it associates with the circadian repressor protein PERIOD (PER) and contributes to circadian timekeeping. Here we investigate NonA, which is required proper levels of evening locomotor activity as well a normal free running period in Drosophila. NonA is associated with the positive transcription factor CLOCK/CYCLE (CLK/CYC), interacts directly with complexin (cpx) pre-mRNA and upregulates gene expression including the gene cpx. Downregulation of cpx expression in circadian neurons phenocopies NonA downregulation, whereas cpx overexpression rescues the nonA RNAi phenotypes, indicating that cpx is an important NonA target gene. As the cpx protein contributes to proper neurotransmitter and neuropeptide release in response to calcium, these results and others indicate that this control is important for the normal circadian regulation of locomotor activity.
INTRODUCTION
Circadian rhythms, an evolutionary adaptation of most animals to the rotation of the earth, is dictated by a conserved transcription/translation feedback loop. In Drosophila melanogaster, the basic Helix-Leucine-Helix (bHLH) heterodimeric transcription factors CLK and CYC directly activate hundreds of genes during the daytime, including period (per) and timeless (tim) (Crane and Young, 2014). Their gene products, PER and TIM respectively, are transcriptional repressors that accumulate during the nighttime and undergo orchestrated phosphorylation mediated by several kinases. Accumulation of phosphorylated PER and TIM, as well as their translocation into the nucleus, gradually reduces the binding of CLK/CYC to the promoters of their target genes, thereby repressing their transcriptional activity (Crane and Young, 2014; Hardin, 2011). Light exposure at the end of the night and parallel mechanisms in constant darkness trigger TIM and PER degradation, which releases the CLK/CYC complex for the next round of transcription.
A similar circadian repression mechanism is employed by mammals. The transcription factor CLK/BMAL1 is orthologous to CLK/CYC and activates the transcription of several direct target genes including period (per) and cryptochrome (cry) (Takahashi, 2017). They encode the transcription repressors PER and CRY, which recruit corepressors to the chromatin and negatively regulate CLK/BMAL1 activity during a 24-hour cycle (Kume et al., 1999). One circadian corepressor is the SIN3A/HDAC1 complex, which contributes to CLK/BMAL1 repression by deacetylation of lysine 9 of histone3 (H3K9) and lysine 5 of histone 4 (H4K5) residues (Duong et al., 2011). Furthermore, recruitment of SIN3A/HDAC1 requires the polypyrimidine tract–binding protein–associated splicing factor (PSF), which also belongs to the DBHS family (Duong et al., 2011).
NONO is another member of this family and is found along with PSF in a large complex associated with mammalian PERIOD proteins (Brown et al., 2005; Duong et al., 2011). NONO like PFS binds directly to the period1 promoter as well as to many other promoters, recruits SIN3A/HDAC1 and represses transcription (Amelio et al., 2007; Duong et al., 2011; Kowalska et al., 2013; Park et al., 2013). However, there is also evidence that NONO plays a positive role in transcriptional activation, by synergizing with other co-activators and nascent RNA. For example, NONO knockdown leads to reduced levels of direct CLK/BMAL1 target gene expression (Brown et al., 2005), and NONO interacts with photoreceptor transcription factors to activate rhodopsin expression (Yadav et al., 2014). More recently, NONO was shown to regulate the abundance of synaptic RNAs in mouse hippocampus (Mircsof et al., 2015). This mechanistic uncertainty notwithstanding, the association of NONO with PER appears relevant to circadian rhythms as NONO downregulation results in markedly attenuated rhythmicity and altered periods of circadian gene reporters (Brown et al., 2005; Kowalska et al., 2013). Moreover, mutation of NonA, the Drosophila ortholog of NONO, results in attenuated tim mRNA expression and behavioral arrhythmicity, suggesting a role for NonA in Drosophila circadian rhythms that parallels the role of NONO in mammalian rhythms (Brown et al., 2005).
NonA must function within the ~150 central circadian neurons that govern circadian locomotor activity rhythms of Drosophila. These neurons all express high levels of core clock proteins, which undergo synchronized circadian oscillations. Despite this common feature, the central circadian neurons are anatomically and functionally distinct (Helfrich-Forster et al., 2007). For example, the ventral lateral neurons (LNvs) are known as the morning (M) cells because they govern the morning peak of locomotor activity (Grima et al., 2004; Stoleru et al., 2004). These M cells are also the only central circadian neurons expressing the pigment-dispersing factor (PDF) and are important for maintaining circadian period in constant darkness as well as synchronizing and communicating information from the M cells to other circadian neurons (Collins et al., 2014; Liang et al., 2016, 2017; Renn et al., 1999b; Stoleru et al., 2004; Stoleru et al., 2005). In contrast to the M cells, the evening cells (E cells), which consist of the 6 LNds and the PDF-negative 5th s-LNv, dictate circadian locomotor activity in the evening and in constant light (Picot et al., 2007; Stoleru et al., 2004). More recently, we discovered these E cells as a major source of rhythmicity and locomotor activity (Guo et al., 2014). However, the molecular mechanisms that underlie how the E cells control circadian evening activity are still unclear.
It has been known that the core central clock sits upstream of at least some circadian-relevant features of neuropeptide function. For example, pdf mRNA and peptide are undetectable in adult small LNvs neurons (s-LNvs) in clk and cyc mutants. Furthermore, PDF staining intensity undergoes circadian cycling in the dorsal termini of the s-LNvs, which is abolished in per01 and tim01 loss of function mutants (Renn et al., 1999b). Similarly, circadian neuron expression and possibly release of the PDF and ion transport peptide (ITP) are under circadian control (Gunawardhana and Hardin, 2017; Hermann-Luibl et al., 2014). The word “possibly” indicates that cycling staining intensity in neuron terminals is usually interpreted to indicate circadian control of secretion and/or vesicle fusion (Fernandez et al., 2008; Gorostiza et al., 2014). Yet there is little direct information that addresses neuropeptide release from circadian neurons.
In Drosophila as in other eukaryotes, the protein Complexin (CPX) positively stimulates Ca2+ dependent evoked release of neurotransmitter and also functions as a clamp to inhibit spontaneous release in the absence of Ca2+ (Buhl et al., 2013). It probably functions similarly to regulate neuropeptide release from dense core vesicles (E Levitan, personal communication.) In this study, we show that cpx pre-mRNA is a target of NonA. NonA also physically interacts with CLK during active transcription, suggesting that NonA physically bridges the circadian transcription machinery to nascent RNAs including cpx. Downregulation of NonA in circadian neurons inhibits circadian evening activity anticipation and modestly lengthen the free-run period, and a very similar phenotype results from downregulation of cpx mRNA; either overexpression of CPX or PDF knockdown rescues this evening activity defect. Interestingly, NonA and CPX knockdowns inhibit circadian neuronal activity, whereas knockdown of PDF alone does the opposite, namely, it stimulates neuronal activity. The results taken together suggest that CLK helps brings NonA to cpx pre-mRNA and positively regulates cpx gene expression. Proper CPX levels are necessary for proper PDF release and for generating wild-type levels of locomotor activity and a normal free-running period.
RESULTS
CLK Physically Interacts with NonA
To identify CLK-associated proteins, we generated a 3XFLAG-CLK-14.8-HBH transgenic fly strain containing a full-length genomic sequence with the CLK protein sequence (Menet et al., 2010) flanked by an N-terminal 3XFLAG and a C-terminal biotin recognition sequence (Fig. 1A). Extracts were made from this strain with entrained flies collected at different times of day. Because the circadian regulation of clock gene transcription is linked to the circadian regulation of CLK/CYC chromatin association, we used a nuclear extract protocol at times of active CLK gene transcription, ZT10-ZT14-ZT18, but a whole cell extract protocol at ZT02 when the complex is predominantly free from chromatin (Menet et al., 2010; Yu et al., 2006). The CLK complex at the above four time-points was purified with a simple two-step affinity purification protocol as previously described (Luo et al., 2012), i.e., anti-FLAG immunoprecipitation and elution with 300 ng/μl 3XFLAG peptide, which was immediately subjected to a streptavidin pull-down. Bead-bound proteins were extensively washed, subjected to on-gel tryptic digestion and detected by mass spectrometry.
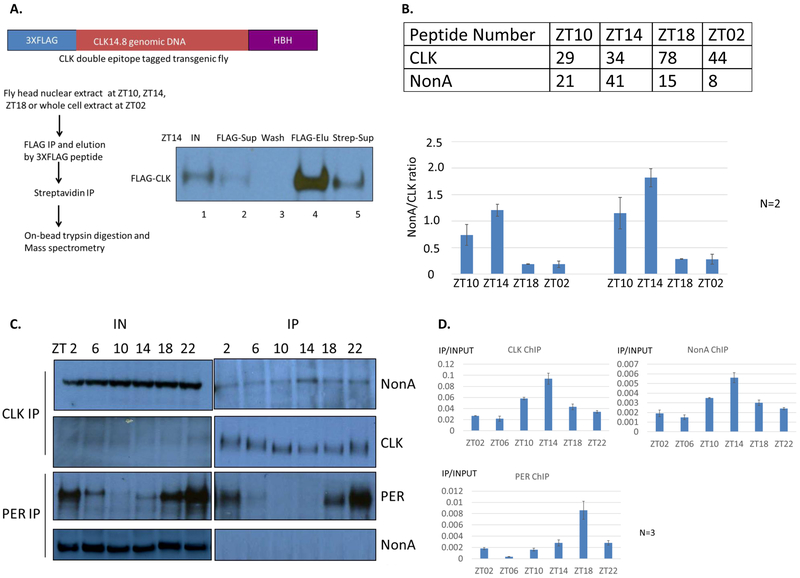
Figure 1. NonA Physically Interacts with CLK in Fly Head Extracts.
A. Schematic flow chart showing purification of the CLK protein complex from fly head extracts. CLK protein is pull down efficiently by two-step IP by western blotting. Only ZT14 time point was shown here. Lane 1: input. Lane 2: supernatant after 1st FLAG IP. Lane 3: bead wash. Lane 4: protein elution from FLAG beads. Lane 5: supernatant after 2nd streptavidin IP. B. More NonA peptides were identified in the CLK complex at ZT10 and ZT14. CLK and NonA peptide number detected by mass spectrometry (average of two experiments) (Top). The ratio of NonA to CLK peptide numbers (left bottom). The ratio of NonA to CLK peptide numbers normalized to their molecular weights (right bottom). C. Co-IP of NonA with CLK by western blotting. Note that the maximal interaction of NonA and CLK at ZT14. The immunoprecipitates were assayed by anti-FLAG (CLK) and anti-PER, anti-NonA Western blotting. D. Anti-CLK, anti-NonA, anti-PER ChIP assay at the tim E-box. Note that CLK and NonA have the maximal binding on tim E-box at ZT14. Y-axis represents the IP signals relative to the input signals assayed by Q-PCR at the tim E-box. Averages of three experiments are shown.
CLK was efficiently captured by both anti-FLAG and Streptavidin IPs at all time points (data not shown); a representative western blot experiment from a ZT14 extract is shown (Fig. 1A; compare lane 2 to lane 1, lane 5 to lane 4). Other expected proteins were also identified by mass spectrometry. For example and consistent with previous studies (Menet et al., 2010), the CLK partner CYC was identified at all 4 time points, substantial PER and TIM were identified at ZT18, and PER but not TIM is still associated with CLK at ZT02 (data not shown). Importantly, we found substantial NonA peptides in the CLK complex at all 4-time points (Fig. 1B top). By this peptide number criterion, NonA was the top CLK-associated RNA binding protein.
To generate a very approximate ratio of NonA to CLK molecules in the complex and how that ratio changes with time, we divided the NonA peptide number by the CLK peptide number and then normalized for their molecular weights. This should help accommodate the somewhat variable CLK peptide numbers at the different time points as well as the substantial difference in molecular weight between CLK and NonA. The estimated ratio of NonA to CLK molecules is large, between 0.5 and 2.0, at ZT10 and ZT14. This is when active circadian transcription is at or near maximal levels. There is much less-CLK associated NonA at ZT18 and ZT02, when there is considerably less circadian transcription (Fig. 1B bottom).
To validate the interaction of CLK and NonA, we assayed NonA by a simple one step coimmunoprecipitation with anti-FLAG antibody. Consistent with the mass spectrometry results (Fig. 1B), NonA is maximally recovered at ZT14 (Fig. 1C by inspection; also by calculation, data not shown). Importantly, there is no detectable NonA recovered with an anti-PER antibody at any time point (Fig. 1C), unlike what one might have anticipated from the mammalian literature (see Introduction and Discussion).
Consistent with an optimal CLK association during periods of maximal circadian transcription, NonA and CLK exhibit similar binding patterns to the tim E-box by chromatin immunoprecipitation; both show peak binding at ZT14, whereas PER binding appears to parallel transcriptional repression and is delayed as previously shown in mammals and flies (Koike et al., 2012; Menet et al., 2010) (Fig. 1D). The results taken together suggest that NonA promotes rather than inhibits circadian gene expression.
NonA is Important for Period Maintenance and Evening Locomotor Activity
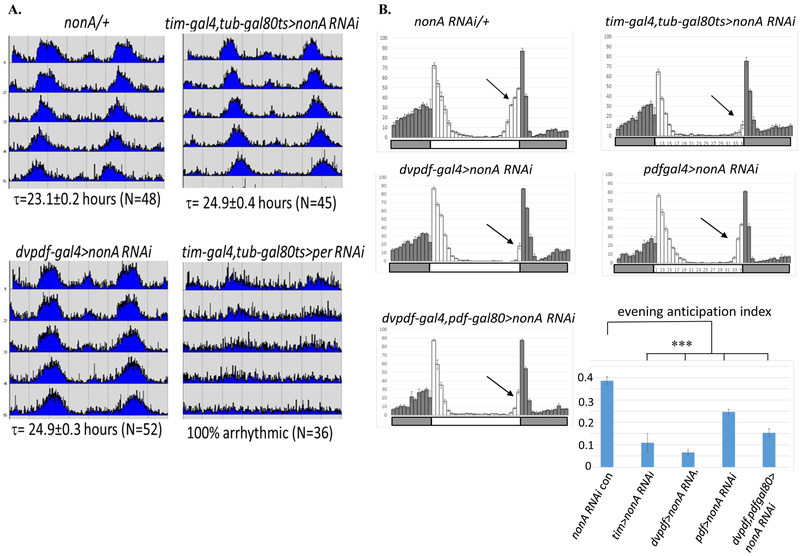
NonA is important for fly courtship behavior and locomotor activity rhythms, but the homozygous NonA loss of function mutant causes lethality and infertility (Brown et al., 2005; Rendahl et al., 1996; Rendahl and Hall, 1996). To investigate in more detail the role of NonA in circadian rhythms, we took advantage of the GAL4-UAS system and several NonA RNAi constructs to selectively knock down the expression of NonA within circadian neurons. We screened several nonA RNAi lines from VDRC and Bloomington fly stocks and found that combining single nonA RNAi constructs with the tim-gal4 driver resulted in modest circadian phenotypes (data not shown). Our screen strategy incorporated the tubulin-gal80ts system to bypass developmental effects (McGuire et al., 2003). To enhance efficacy, we recombined the two RNAi nonA constructs (V26442, V33901) (Dietzl et al., 2007) with the strongest circadian phenotypes and expressed them with different circadian GAL4 drivers at 29°C.
Conditional knockdown of NonA in all adult circadian neurons with the tim-gal4 driver lengthened circadian period by 1.8hrs. NonA knockdown with the narrower dvpdf-gal4, expressed in both morning and evening circadian neurons (Guo et al., 2014), had a comparable effect (Fig. 2A and Table 1). Surprisingly, we observed a similar long period when NonA is knocked down only in evening neurons (dvpdf-gal4,pdf-gal80), whereas the effect is more modest with the pdf-gal4 driver (Table 1). Although NonA is essential for cell survival, all nonA RNAi flies tested here exhibit a high rhythmicity percentage and index (Table 1). In addition, NonA downregulation with the dvpdf-gal4 driver had no discernable effect on the presence or normal morphology of M- and E-cells (Suppl. Fig.1). As a control, we knocked down per expression with a per RNAi construct and the tim-gal4 driver; this resulted in arrhythmic behavior as expected for a strong per loss of function phenotype (Fig. 2A).
Figure 2. Downregulation of NonA Lengthens Circadian Free-run Period and Inhibits Evening Activity Anticipation.
A. nonA RNAi knockdown by tim-gal4, tubgal80ts or dvpdf-gal4 resulted in modest long period. Flies were entrained for 3–4 d in 12-hrs-light-12-hrs-dark (LD) cycles and released into constant darkness (DD) for at least 5 days at 29°C. τ: free-run period. Average activity actograms under DD1-5 were shown for nonA RNAi/+, tim-gal4, tubgal80ts>NonA RNAi, dvpdf-gal4>NonA RNAi flies (Levine et al 2012). tim-gal4, tubgal80ts>per RNAi as a control for the driver. Note: Knockdown of NonA by tim-gal4 causes lethality of flies. B. Evening activity anticipation is inhibited in the nonA RNAi mutants. Flies were entrained as A. Average activity plots at 4 LD cycles were shown. Evening locomotor activity (anticipation) was dramatically inhibited by the tim-gal4, tub-gal80ts and dvpdf-gal4 drivers (black arrows). The inhibition in the pdf-gal4> nonA RNAi stain is modest probably due to its weaker driver. Evening anticipation index is calculated as the percentage of the total activity 3 hours before light-off relative to total evening activity. Statistical analysis was done by 2-tailed, unpaired Student's t tests. Compared to the control, the evening anticipation indexes in all RNAi strains tested are significantly reduced (***p<0.001). LD cycles light-on (white bar), light-off (gray bar).
Table 1. Summary of Free-run Periods and Evening Activity Inhibition of nonA RNAi Knockdown by Different Circadian Drivers.
Experiments were performed and analyzed as in Fig. 2. Evening anticipation index, periods with error bar (SEM) and numbers of flies, rhythmicity percentage and rhythmicity index were shown. The lowest evening anticipation index indicates the strongest inhibition of evening anticipation.
| Genotype | Evening anticipation index |
Period±SEM | Rhythmicity percentage |
Rhythmicity index |
|---|---|---|---|---|
| nonA RNAi/+ | 0.386±0.017 | 23.1±0.2(N=48) | 87.5% | 0.42±0.11 |
| tim-gal4, tub-gal80>nonA RNAi | 0.110±0.040 | 24.9± 0.4(N=45) | 93.3% | 0.37±0.09 |
| dvpdf-gal4>nonA RNAi | 0.066±0.011 | 24.9± 0.3(N=52) | 86.5% | 0.41±0.1 |
| pdf-gal4>nonA RNAi | 0.247±0.011 | 24.5±0.2(N=56) | 91.1% | 0.45±0.12 |
| dvpdf-gal4;pdf-gal80>nonA RNAi | 0.567±0.025 | 24.9± 0.2(N=65) | 92.3% | 0.39±0.12 |
| tim-gal4, tub-gal80ts/+ | 0.304± 0.010 | 24.1± 0.2(N=42) | 90.5% | 0.45±0.13 |
| dvpdf-gal4/+ | 0.376±0.013 | 23.9± 0.2(N=30) | 86.7% | 0.40±0.16 |
| pdf-gal4/+ | 0.356± 0.017 | 23.8±0.3 (N=24) | 95.8% | 0.39±0.1 |
| dvpdf-gal4,pdf-gal80/+ | 0.327± 0.018 | 23.7±0.2 (N=29) | 100% | 0.43±0.09 |
| dvpdf-gal4, tubgal80ts | 0.328±0.023 | 23.8±0.2(N=36) | 88.9% | 0.36±0.05 |
| dvpdf-gal4, tubgal80ts >nonA RNAi | 0.102±0.016 | 24.6±0.1(N=52) | 86.5% | 0.40±0.12 |
| tim-gal4, tub-gal80ts>per RNAi | 0.257± 0.013 | Arrhythmic(N=36) | 0% | NA |
We noticed that down-regulation of NonA with most of these circadian drivers (especially tim-gal4, dvpdf-gal4, and dvpdf-gal4,pdf-gal80) dramatically reduced circadian evening anticipation, the major locomotor activity event under LD conditions. Normally, evening activity gradually increases before lights off at ZT12 and is followed by the startle event that occurs at lights off (Fig. 2B). To rule out potential developmental effect of NonA knockdown, we performed a similar experiment with the dvpdf-gal4, tub-gal80ts driver. Adult-specific downregulation of NonA also significantly inhibits evening anticipation and slightly lengthens the free-run period (Suppl. Fig. 2 and Table 1). There is a more modest effect on the evening anticipation of NonA knockdown in PDF neurons (Fig. 2B). This is similar to the weaker period effect of PDF neuron knockdown (Table 1); this may be due to the weaker pdf driver relative to the dvpdf-gal4 driver (Fig. 2A). These effects on circadian period as well as on evening locomotor activity presumably reflect the contribution of NonA to circadian neuron gene expression.
RNA Targets Identified by nonA RNAi Knockdown and the TRIBE Method
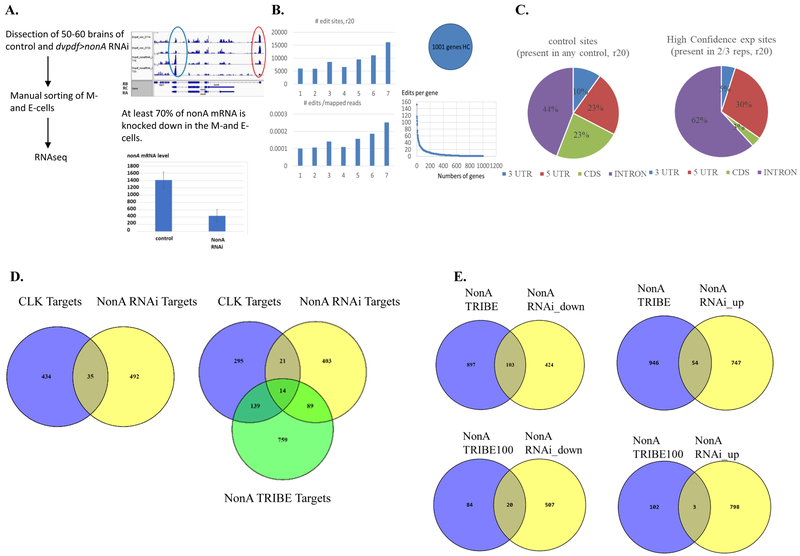
To find potential RNA targets of NonA in circadian neurons, we GFP-labeled morning and evening circadian neurons with the dvpdf-gal4 driver and manually sorted these cells at two different time points (ZT14 and ZT23), from the control and the nonA RNAi knockdown strains. Cell sorting, mRNA isolation and library preparation for RNA-seq were as described (Abruzzi et al., 2017). Successful knockdown is evident by inspection (Fig. 3A left) and was approximately 70% effective by calculation (Fig. 3A right).
Figure 3. Cpx pre-mRNA is a Bona Fide NonA Target.
A. Flow chart for manually sorting GFP-labeled dvpdf cells from fly brains. Flies were entrained for 4 LD cycles at 29°C and collected at ZT14 and ZT23. Brains were dissociated and GFP-labeled dvpdf (M- and E-) cells were collected according to Nagoshi et al 2012. RNAseq libraries were prepared according to Abruzzi et al 2017. RNAseq showed that about 70% nonA mRNA is reduced in the M- and E-cells of dvpdf>nonA RNAi line (red oval and the right graph). RNA libraries were 3’ end biased. The blue oval validates the over-expression of nonA dsRNAi replicon in the neurons. B. NonA TRIBE analysis was done from fly brains collected at ZT14. An inducible pan-neuronal driver was used to drive the expression of UAS-nonA-ADAR due to the toxicity of the fusion protein. 4 controls and 3 experiments were done as follows: 1 and 2: elev-gs with RU486; 3, 4: UAS-nonA-ADAR with RU486; 5,6,7: elev-gs>UAS-nonA-ADAR with RU486. Nascent RNAs were extracted from 30 brains and libraries were prepared and analyzed according to McMahon et al 2016. Total edit sites and edits per million mapped reads were show the left. The high confidence (HC) edit was defined as those with at least 20 reads and 10% editing in 2/3 of three experiment group (5,6,7). Negative control edit sites are those present in any of the 4 control groups with at least 20 reads and 10% editing. 1001 genes were identified as HC NonA TRIBE targets within which 639 genes have at least two editing sites. C. The quantification of the location of NonA TRIBE edits according to (McMahon et al., 2016) in nascent RNAs show that NonA preferably binds introns. There is a modest increase of editing percentage (18%) when NonA-ADAR is expressed in the neurons. D. The overlap of CLK direct targets from fly heads and transcripts down-regulated by nonA RNAi in M- and E-cells and NonA TRIBE targets as in Fig. 3C. We intersected transcripts downregulated in the NonA RNAi mutant and CLK direct targets (Abruzzi et al 2011) and found there is only 32 genes shared by these two groups (top graph). There are 14 genes overlapping within CLK targets, NonA RNAi targets and NonA TRIBE targets (bottom graph). E. Intersection of NonA TRIBE targets and RNAi targets suggests that NonA preferably activates gene expression in circadian neurons. There is about 20% percent of NonA TRIBE targets whose transcripts are also downregulated in the nonA RNAi mutant. Only 3% of TRIBE top100 targets whose transcripts are upregulated by the RNAi.
Assuming a positive role of NonA in transcription, we focused on transcripts at least two-fold reduced by NonA knockdown, at both time points and in both replicas. GO ontology analysis indicates that these 527 genes are enriched in synapse assembly, signal transduction and transmembrane transport functions (Table 2). We also found 801 genes whose gene expression is upregulated in the nonA mutant, but there is no significant pathway enrichment. Please see Supplemental excel Table 1 for the full lists of genes.
Table 2. About 527 Genes are Down-regulated in the nonA RNAi Mutant by 2 Folds or More.
Gene expression of M- and E cells in the dvpdf>nonA RNAi mutant were analyzed by cufflinks and its FPKM normalized to the expression levels in the control. DAVID GO ontology analysis shows genes are enriched in the pathways of synapse assembly, signaling transduction, receptor and transporter activity. Also see the supplemental excel table 1 for full list of genes affected by nonA RNAi.
| GO Term | p-value | Gene lists |
|---|---|---|
| synapse assembly | 4.88E-05 | nej, comm, Sema-1a, cg18405, Pdk1, mtg, jeb, pros, Gad1, AGO1, cpx, dlg1, Gs2 |
| Signal transduction | 2.29E-04 | Mam, CG3894, rdgB, Gp150, cg5820, pan, Hs2st, Dh31, rdgA, trc, Galpha49B, Pkc53E, W, Dh44, N, Arr2, CG42541, PGRP, Pvf3, Hs6st, CG6954, Pdk1, lin, Ast, Btk29A, nej, Gyc32E, Tab2, NorpA, ogre, Src64B, Pde9, Ilk, shakB, Eip78C, Arr1, mthl2, tkv, Npc2a, Rab9, Smox, fz2, ltd, ush, aop, H, l(2), tid, gt, ex, Ggamma1, Mpk2, CG8641, cngl, hpo, Plc21C, R, Sema-1a, cg18405, CalpA, Gyc76C, Ggamma30A, 18w, Dgk, Nplp2 |
| cell surface receptor signaling pathway | 9.49E-04 | nej, mam, rdgB, Tab2, norpA, Gp150, Ilk, pan, Dh31, mthl2, tkv, rdgA, Npc2a, Smox, fz2, ush, Galpha49B, H, l(2)tid, Dh44, gt, N, ex, Ggamma1, sty, PGRP, Hs6st, Pvf3, Sema-1a, CalpA, Pdk1, lin, Ast, Ggamma30A, Dgk, Nplp2 |
| sodium ion transmembrane transporter activity | 6.98E-04 | CG1732, Ndae1, CG7708, CG8791, NaCP60E, Picot, CG32669, nrv2, Nckx30C, Eaat1 |
To further narrow down these gene lists and to identify NonA direct targets (RNAs bound by NonA), we took advantage of our recently developed TRIBE approach (McMahon et al., 2016). TRIBE combines the catalytic domain of RNA editing enzyme ADAR and an RNA binding protein (RBP) of interest. The fusion protein is expressed in specific cells or tissues, and additional RNA editing events identify strong candidate RBP targets. We therefore expressed NonA-ADAR in fly brains with an inducible pan-neuronal driver (elev-gs). Given previous results with NonA in tissue culture cells (McMahon et al., 2016), we assayed nascent RNA from brains of both parent control and nonA-ADAR overexpression strains. We used brains because there is no straightforward way to assay nascent RNA from small numbers of discrete neurons.
This strategy identified a large number of NonA TRIBE targets (about 1001 genes, Fig. 3B, also see Supplemental Excel table 2), of which 639 have at least two editing events. Despite the high levels of endogenous editing in brain RNA, we observed a slight increase in total editing events in the experimental group (Fig. 3B left), and they are mostly in introns (Fig. 3C) as expected (McMahon et al., 2016). Because NonA interacts with CLK, we compared these genes with the CLK direct target from fly heads (Abruzzi et al., 2011) and NonA RNAi downregulation targets in M- and E-cells. We only found 14 NonA TRIBE targets which are also CLK direct targets and NonA RNAi downregulation targets (Fig. 3D). We suspect that the small overlap may reflect at least in part the different tissues used in the assays, i.e., fly heads vs neurons. Interestingly, we also discovered there are more NonA TRIBE targets whose gene expression is downregulated than upregulated by NonA RNAi knockdown in E-and M-cells (Fig. 3E). This is particularly evident in examining the top 100 NonA TRIBE target genes (Fig. 3E bottom), which have a strikingly higher overlap with downregulated than upregulated genes.
cpx pre-mRNA is a NonA Direct Target in Circadian Neurons and CPX is Required for Evening Locomotor Activity.
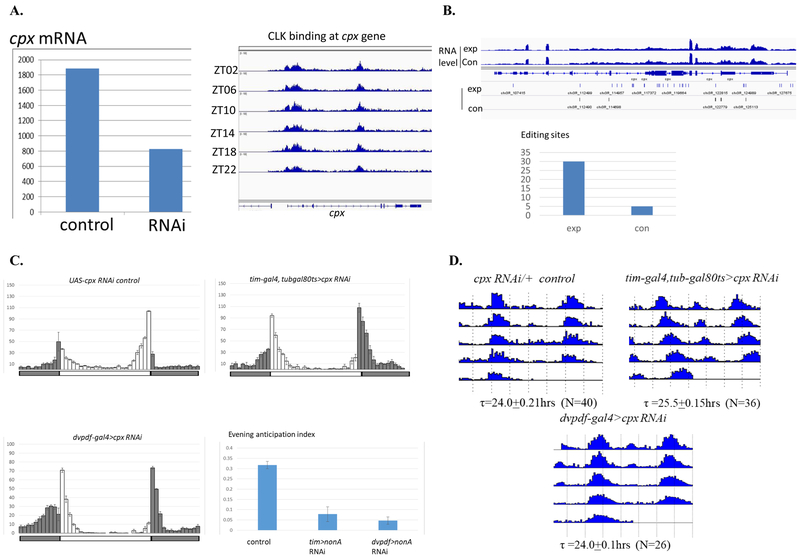
We focused our subsequent efforts on the cpx gene, because more than half of cpx mRNA was reduced by NonA knockdown in M- and E-cells, and CLK binds to the cpx gene locus (Fig. 4A, left and right respectively). Indeed, cpx is a strong TRIBE target: it has 30 editing sites located in intron and exon regions, a 6-fold increase over the number of cpx editing sites present in the control RNA; this comparison suggest that NonA strongly interacts with cpx pre-mRNA (Fig. 4B).
Figure 4. CPX is a Strong NonA TRIBE Target and Required for Evening Activity Anticipation.
A. RNA-seq analysis shows cpx mRNA is reduced by about 2 folds in the M- and E-cells of dvpdf>nonA RNAi line, Y axis: the FPKM reads from one reprehensive replica in the driver control and RNAi samples (left). Anti-CLK ChIP-seq suggests that CLK binds at the cpx gene locus probably with the aid of NonA and cpx RNA (right). B. CPX is a NonA TRIBE target. Top graph shows comparable cpx mRNA expression levels in the control and nonA-ADAR strains. Middle graph shows there is a dramatic increase in edit sites at the cpx locus in the nonA-ADAR strain. Each solid bar indicates an editing event. Bottom graph showed the total editing sites on the cpx transcripts as in B. C: Downregulation of CPX by the tim-gal4, tub-gal80ts or dvpdf-gal4 drivers inhibits evening activity anticipation. Experiments and behavior analysis were performed as in Figure 2. D. Down-regulation of CPX by the tim-gal4, tub-gal80ts driver lengthens the free-run period whereas CPX knockdown in the dvpdf cells has no period effect.
Strikingly, adult-specific downregulation of CPX in circadian neurons with tim-gal4 dramatically inhibits evening activity and results in a modestly long circadian period, i.e., a phenocopy of the nonA RNAi phenotypes (Fig. 4C and 4D, respectively). Interestingly, downregulation of cpx mRNA with dvpdf-gal4 or an adults-specific driver dvpdf-gal4,tubgal80ts also inhibits evening anticipation (Fig. 4C, Suppl. Fig. 3). Although the period is barely changed by the CPX knockdown with dvpdf-Gal4 (Fig. 4D), the data taken together indicate that CPX as well as NonA promotes circadian evening activity under normal LD conditions.
How does CPX work to influence circadian evening activity? CPX binds to the SNARE complex and stimulates Ca2+-evoked neurotransmitter release following an action potential, and CPX also inhibit spontaneous synaptic vesicle fusion in the absence of Ca2+ (Buhl et al., 2013). We therefore reasoned that NonA and CPX might stimulate evening activity by influencing neuropeptide or neurotransmitter release in circadian neurons. Because it was recently shown that the circadian neuropeptide PDF could inhibit Ca2+ levels in evening cells and delay evening locomotor activity (Liang et al., 2016, 2017), we directly addressed this possibility.
PDF signaling is required for NonA/CPX-mediated evening activity anticipation.
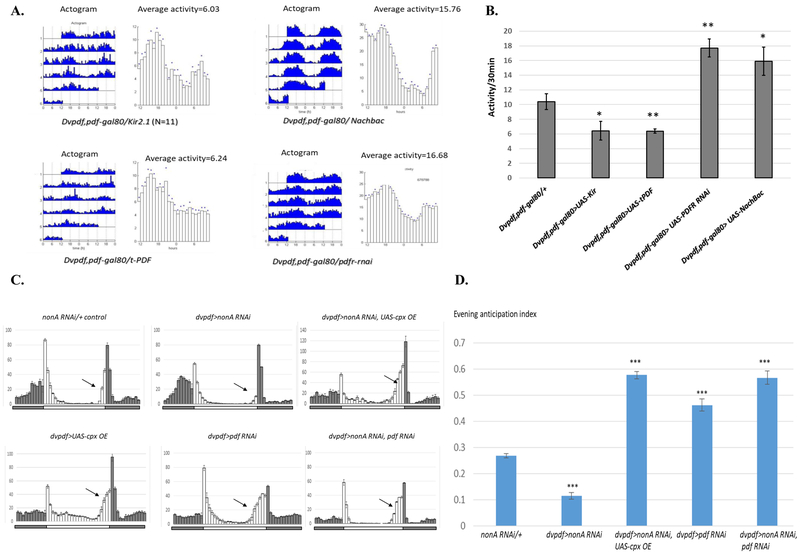
We first assayed the response of E cell-derived locomotor activity to manipulations of PDF. Consistent with an inhibition of evening activity by PDF, expression of t-PDF (tethered PDF; (Choi et al., 2012) in evening cells has a similar effect on the overall locomotor activity pattern as expression of the inhibitory potassium channel Kir2.1. In contrast, down-regulation of the PDF receptor in evening cells stimulated overall locomotor activity, comparable to the positive effect of the sodium channel Nachbac (Fig. 5A and 5B).
Figure 5. PDF Signaling is Required for the NonA/CPX-Mediated Evening Locomotor Activity.
A. PDF signaling inhibits overall activity under constant darkness. Overexpression of t-PDF (inhibitory) or knockdown of PDF receptor(excitatory) showed opposite effects on overall locomotor activity. B Quantification of average mean activities as in Fig. 5A. C. Either CPX over-expression or PDF knockdown rescues the evening activity anticipation defect in the nonA RNAi mutant. Experiments were performed as in Fig. 2. Average activity actograms under 4 LD cycles were shown. Arrow shows the evening locomotor anticipation. D. Quantification of Fig. 5B. Note that that cpx overexpression or pdf RNAi knockdowns significantly increases evening anticipation index and therefore rescues the evening defect of the nonA RNAi mutant. Compared to the control, the evening anticipation index in all RNAi strains tested is significantly reduced (*** p<0.001).
If CPX is downstream of (epistatic to) NonA, overexpression of CPX might rescue some of the NonA knockdown phenotypes. Indeed, CPX expression with the dvpdf driver rescued the evening activity defect of the nonA RNAi mutant strain (Fig. 5C and 5D). Notably, expression of CPX with this driver dramatically increased evening activity, (Fig. 5C and 5D) (see in discussion). These results together suggest that cpx pre-mRNA is indeed an important downstream target of NonA in circadian neurons.
Because PDF is reported to inhibit Ca2+ levels in E-cells and evening locomotor activity (Liang et al., 2016, 2017), we next asked whether PDF activity influences the NonA knockdown phenotypes. Interestingly, knockdown of PDF in addition to NonA with the dvpdf driver results in a complete rescue of the NonA knockdown evening anticipation phenotype; otherwise put, pdf RNAi rescues the evening activity defect of nonA RNAi (Fig.5C and 5C). A similar rescue phenotype was observed with a pdf and cpx double RNAi knockdown (Suppl. Fig. 3) Consistent with expectation from the classical pdf mutant phenotype (Renn et al., 1999b), the PDF knockdown or CPX over-expression causes an advance in the evening activity peak (Fig. 5C and Suppl. Fig. 5). We also noticed that NonA downregulation modestly enhances morning anticipation whereas either PDF downregulation or CPX overexpression almost eliminates morning anticipation, which will be discussed below (Fig.5C and Suppl. Fig. 4). The results also indicate that NonA and PDF have opposite effects on evening anticipation and suggest that the NonA knockdown phenotype requires PDF for its reduced evening activity phenotype.
Distinct Effects on Circadian Neuronal Activity by NonA, CPX and PDF.
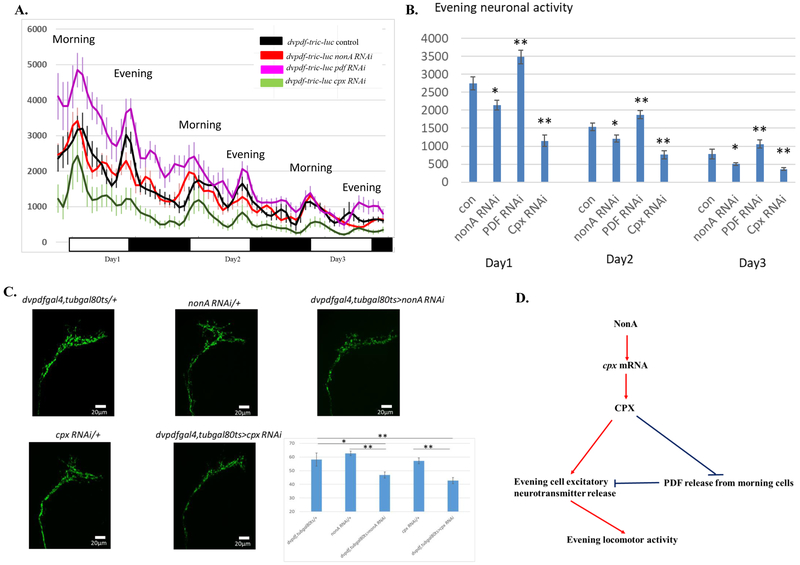
Downstream locomotor activity is controlled by circadian neuron neuronal activity (Cavey et al., 2016). We reasoned therefore that the inhibition of evening anticipation by NonA knockdown may arise from reduced E cell neuronal activity. To address directly the effects of NonA, CPX and PDF on neuronal activity, we took advantage of an in vivo calcium reporter (Tric-LUC) system, which utilizes a Calmodulin (CAM)-CAM-binding peptide interaction to rapidly activate calcium-dependent luciferase transcription (Gao et al., 2015; Guo et al., 2017). The LUC reporter activity expressed with the dvpdf-gal4 driver is therefore a rough surrogate of morning and evening cell neuronal activity.
Consistent with previous observations (Guo et al., 2017), the two most prominent luciferase peaks coincide well with the morning and evening locomotor activity peaks in the control (Fig. 6A black). Although there is no significant difference in morning LUC activity between the NonA RNAi knockdown and control strains, the evening LUC activity is significantly reduced by NonA knockdown (Fig.6A and 6B, Suppl. Fig. 6). Interestingly, downregulation of CPX dramatically inhibits morning as well as evening LUC activity (Fig. 6A, green), whereas PDF knockdown modestly increases overall neuronal activity (Fig.6A, magenta). The data therefore indicate that NonA and CPX both positively regulate E cell neuronal activity, CPX may have additional roles in regulating basal calcium activity whereas PDF inhibits both E- and M-neuronal activity. Although the regulation of PDF release by NonA/CPX may account for much of the neuronal activity and evening anticipation phenotypes, it is likely that more complex regulatory interactions account for the molecular and behavioral consequences of these NonA and CPX manipulations within circadian neurons (see Discussion). Indeed, adult-specific downregulation of NonA or CPX with the dvpdf-gal4, tub-gal80ts driver reduces PDF signal in the M-cell dorsal projections. This is consistent with the enhanced PDF release suggested by the behavioral phenotypes (Fig. 6C).
Figure 6. Circadian Neuronal Activity in the nonA RNAi, cpx RNAi and pdf RNAi Mutants Assayed by a Tric-LUC Reporter.
A. Evening neuronal activity is reduced in the nonA or cpx RNAi mutants whereas PDF downregulation increases evening neuronal activity. Flies were entrained under 3-4 LD cycles at 29°C and luciferase activity was recorded in a topcounter monitor. Data was analyzed by Matlab software as described in the method. Y-axis: LUC activity. X-axis: days under LD. B. Quantification of evening and morning peak neuronal activity from the control and RNAi mutants. 3 LD cycles of peak activity were averaged and were plot for the control and RNAi strains Note that nonA RNAi knockdown significantly inhibited evening neuronal activity compared to the driver control. The cpx RNAi mutant exhibited reduced morning and evening neuronal activity relative to the driver control whereas PDF knockdown increases the activity (*P<0.05, **P<0.01). C. PDF signals at the PDF cells dorsal projections are reduced in the nonA and cpx RNAi driven by the dvpdf-gal4, tubgal80ts driver. The RNAi and control lines were entrained for 4 LD cycles at 29°C and PDF immunostaining were performed according to Menegazzi et al 2013. scale bar: 20 μm. Statistical analysis was done by one-way Anova with post-hoc Turkey HSD. * p<0.05, ** p<0.01. D. Model of how NonA and CPX contribute to circadian evening locomotor activity. NonA and CPX may coordinate the PDF-dependent and PDF-independent mechanisms to regulate circadian evening activity.
DISCUSSION
We address in this study the circadian function of NonA, a Drosophila DBHS family member involved in various aspects of transcription, metabolism and especially RNA processing. Downregulation of NonA in all adult circadian neurons gives rise to very specific circadian phenotypes, namely, the modest longer free-running period in constant darkness and the almost complete absence of evening anticipation. Quite similar phenotypes resulted from NonA knockdown with more targeted circadian drivers, namely, dvpdf-gal4 and dvpdf-gal4;pdf-gal80 (Fig. 2). These phenotypes more convincingly reflect an impact on core timekeeping than those of flies harboring a NonA P-element mutation. They were arrhythmic in constant darkness and hyperactive in a light-dark cycle (Brown et al., 2005). We suspect that these less specific phenotypes were from developmental effects of the P-element mutant and from ubiquitous expression of NonA throughout the fly brain, i.e., from outside the adult CLK network.
How does NonA function to help maintain proper circadian timing and activity? The mammalian homolog NONO is associated with PER, but we could not detect a comparable NonA-PER interaction from fly head extracts. NonA is rather associated with the positive transcription factor CLK (Fig. 1B). Consistent with this finding are the similar temporal patterns of CLK and NonA binding to the tim promoter region by ChIP; maximal binding of both is at ZT14. This is about the peak time of tim and per transcription and four hours prior to the peak time of maximal PER binding to the same promoter region. All these data suggest a positive role of NonA in transcription (Fig. 1D). Interestingly, characterization of NONO knockdown cell lines also suggested a positive contribution of NONO to mammalian circadian transcription despite the association of NONO with the repressor protein PER (Brown et al., 2005).
Like many proteins functioning in transcription and RNA processing, the binding of NonA to the tim promoter region by ChIP may be via a direct DNA interaction and/or indirectly via another DNA-binding protein like CLK. Because NonA does not have a DNA binding region like its mammalian ortholog NONO, but still has two RRMs (RNA recognition motifs) (Peng et al., 2002), we propose that NonA associates initially with DNA binding proteins and is then transferred to nascent RNAs, to promote downstream RNA processing events.
NonA-TRIBE further supports direct binding of NonA to RNA with about 1000 additional transcripts edited in fly brains. Consistent with previous NonA-TRIBE results from tissue culture cells (McMahon et al., 2016), most NonA binding sites are located within nascent RNA introns. Consistent with the mass spectrometry results (Fig. 1B), these TRIBE data suggest that NonA functions on chromatin and also identify a large set of putative NonA direct target genes within neurons.
To identify a subset of these NonA direct targets that might function within circadian neurons and contribute to the knockdown phenotypes, we also overlapped these data with the large number of circadian neuron transcripts with levels strongly affected by NonA RNAi knockdown (Fig. 3). The TRIBE data intersect much more successfully with genes that decrease in their mRNA levels. About 20% of the top 100 TRIBE targets are downregulated in the nonA RNAi knockdown, but less than 3% of these top 100 TRIBE targets are upregulated (Fig. 3E). This suggests that the direct effects of NonA on transcription are mostly positive and not surprisingly that there a large number of NonA TRIBE targets for which NonA may participate in their RNA splicing or export rather than transcription.
Because of the association of NonA with CLK (Fig. 1), we examined the overlap between a previous CLK direct target data set (Abruzzi et al 2011), NonA RNAi targets and NonA TRIBE targets. The gene cpx meets all of these criteria, and downregulation of CPX in circadian neurons largely phenocopies the nonA RNAi results, namely, a longer circadian period and dramatically reduced evening anticipation (Fig. 4). Remarkably, CPX overexpression alone has increased evening anticipation (Fig.5) and its addition to nonA RNAi within circadian neurons rescues the evening anticipation and period defect of the nonA RNAi (Fig. 4). These genetic data indicate that cpx is a major circadian NonA direct target gene.
CPX plays a positive role in calcium-mediated release of neurotransmitter-containing vesicles as well as a negative role in inhibiting spontaneous vesicle release (Buhl et al., 2013; Jorquera et al., 2012). Because a similar evening anticipation phenotype occurs when vesicle release is blocked in circadian neurons by expression of the tetanus toxin light chain (UAS-TNT) (Guo et al., 2014), NonA RNAi knockdown may decrease evening anticipation by similarly inhibiting neurotransmitter release from E cells. Which neurotransmitter’s release from E-cells could be affected by NonA knockdown? Previous data showed that E cells secrete acetylcholine (Johard et al., 2009), choline acetyltransferase (ChAT) mRNA is enriched in E-cells, and specific knockdown of ChAT in E-cells enhances total sleep and inhibits evening locomotor activity (Abruzzi et al., 2017); notably, acetylcholine is a major excitatory neurotransmitter in the fly brain. Because cpx RNAi phenocopied the inhibition of evening anticipation by nonA RNAi and because CPX overexpression rescued the nonA RNAi mutant effect, we suggest that downregulation of NonA inhibits evening activity in part via its downregulation of CPX and subsequent inhibition of calcium-mediated release of E cell acetylcholine or of other E cell excitatory neurotransmitters (Fig. 6D model). We note that NonA may also affect the acetylcholine pathway independent of CPX-mediated neurotransmitter vesicle release. For example, nicotinic acetylcholine receptor (nAchR) RNA is ranked as the No. 2 NonA TRIBE target, and downregulation of NonA dramatically reduces nAChR mRNA levels in E- and M-cells (data not shown).
In contrast to the similar evening activity anticipation effects of the nonA and cpx mutants, their effects on circadian period are more variable, due perhaps to multiple and different targets in E and M cells. For example, NonA knockdown in E cells lengthens free-run period whereas the knockdown in M (PDF) cells has little effect (Table 1). This suggests that under normal conditions E cell-NonA and CPX contribute to period determination, consistent with recent studies that circadian period is determined by multiple independent oscillators in M cells and E cells via PDF signaling (Guo et al., 2014; Yao and Shafer, 2014).
What function does PDF play in the evening phenotype of the NonA/CPX RNAi knockdowns? Our data indicate that continuous activation of PDF signaling in constant darkness inhibits overall activity (Fig. 5A). This is consistent with recent observations of Taghert and colleagues: they showed that PDF application decreases the Ca2+ level of both E-cells and M-cells, suggesting that PDF inhibits E and M cell neuronal activity (Liang et al., 2017). Consistent with this observation, downregulation of PDF rescues the inhibition of evening anticipation of the nonA or cpx RNAi mutant. These results suggest that NonA and CPX also regulate PDF release. Indeed, their adult-specific knockdown gave rise to less PDF signal at the M-cell dorsal projections, which may reflect more PDF release. As CPX inhibits spontaneous vesicle release, we suggest that this inhibition may also apply to dense core vesicles and PDF release, i.e., NonA/CPX knockdown enhances PDF spontaneous release. This will lead to an inhibition of evening anticipation activity, which is also consistent with the phase advance of evening activity in pdf01 mutant flies (Renn et al., 1999a).
To address more directly neuronal activity, we assayed M and E cell TRIC-LUC activity in wake-behaving flies in nonA, cpx or pdf RNAi genotypes. Luciferase activity is calcium-dependent and should therefore be a proxy for relative neuronal activity (Gao et al., 2015; Guo et al., 2017). Both NonA and CPX knockdowns reduced evening neuronal activity, consistent with the hypothesis that NonA and CPX both function to increase E cell neuronal activity and therefore normal evening anticipation. The CPX effect is more dramatic, consistent with the idea that CPX has a more direct effect on neuronal activity and that it may regulate the release of multiple neurotransmitters and neuropeptides. pdf RNAi in contrast increased evening neuronal activity. The proposed inhibition of PDF release from M cells by NonA/CPX endogenous function would then have a positive effect on evening anticipation and inhibit morning anticipation, consistent with the observations (Fig. 2B, Fig. 4C, Fig. 5C and Fig. 6C). However, the mechanism by which PDF inhibits E cell activity is uncertain. PDF may act indirectly, for example by stimulating the release of inhibitory neurotransmitters such as glycine (Frenkel et al., 2017), or the E cell PDF receptor may be coupled to inhibitory signaling such as Gi. Consistent with this possibility, it is known that not all PDR receptor coupling is to Gs (Agrawal et al., 2013).
A similar logic applies to the effects of pdf and cpx RNAi on morning neuronal activity, which are even stronger than their effects on evening activity. Auto-stimulation of M cell PDFR by PDF may explain the stronger effect on morning activity (Fig 6D. the model). The observation that manipulation of NonA, CPX and PDF have different effect on neuronal activity suggests that neuronal activity and its behavioral consequences are balanced between M cells and E cells (Fig. 6D). In any case, more work is required to validate this model and especially to substantiate the roles of spontaneous as well as evoked neuropeptide release in circadian locomotor activity rhythms.
START Methods
Fly Stocks
All gal4, gal80 and gal80ts drivers used in this study have been described previously: timgal4 (Kaneko et al., 2000), timgal4,tubgal80ts (Luo et al., 2012), pdfgal4 (Stoleru et al., 2004), dvpdfgal4 (Bahn et al., 2009), dvpdfgal4,pdfgal80 (Guo et al., 2014). 3XFLAG-CLK14.8-HBH transgenic fly was generated by injecting yw embryos with pCasPer4.0 3XFLAG-CLK14.8-HBH plasmid. The pCasPer4.0 3XFLAG-CLK14.8-HBH plasmid was constructed previously (Kadener et al., 2008) with the following modification: the sequence encoding 3XFLAG peptides was inserted in-frame before the ATG codon of CLK 14.8 kb fragment and the sequence for HBH tag (Tagwerker et al., 2006) was inserted before the stop codon. The UAS-NonA-ADAR transgenic fly was generated according to McMahan et al 2016: The nonA coding sequence fused to the sequence encoding for ADAR catalytic domain was cloned into a modified pJFRC7-20x UAS construct (Addgene #26220) and injected by BestGene. per RNAi 31286 from Bloomington stocks. nonA RNAi strains: V26441, V26442, V33901, V100723 from VDRC stocks, 52933, 56944, 61279 from Bloomington stocks. cpx RNAi V21477 from VDRC stocks. UAS-cpx 39473 from Bloomington stocks. dvpdf-gal4>UAS-Tric-LUC (Guo et al., 2017).
Fly Entrainment and Locomotor Activity and Statistical Analysis.
Flies were raised on standard cornmeal/agar medium supplemented with yeast. Locomotor activity of young male flies (aged 3–5 days) was entrained and monitored for at least 4 days with in LD conditions at 29°, followed by at least 5 days in DD using Trikinetics Drosophila activity monitors (Waltham MA). Behavior analyses were performed by MATLAB 2014 with a signal processing toolbox(Levine et al., 2002). 4 LDs average group activity was also generated and analyzed with MATLAB as in Guo et al (Guo et al., 2014). The evening anticipation index is calculated as the sum activity for the last 3 hours before light-off divided by the total evening activity excluded the startle effects. The average of group activity and evening anticipation index for 4 LD cycles were shown. All statistical analysis was conducted using Microsoft Excel software. Normally distributed data were analyzed with 2-tailed, unpaired Student's t tests. Differences between groups were considered significant if the probability of error was less than 0.05 (p<0.05).
Western blot, Immunoprecipitation and Mass Spectrometry
Transgenic 3XFLAG-CLK14.8-HBH flies were entrained at 3-4 LD cycles at 25°C and collected on dry ice at different time points. Fly head extracts were prepared by homogenization in lysis buffer (20 mM Tris-HCl at pH 7.5, 10% glycerol, 1% NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM DTT) supplemented by protease inhibitor cocktail and phosphatase inhibitor cocktail. Extracts were subjected to mild sonication with a bioruptor 300 (Diagenode). Clear and denatured lysates were resolved by NuPAGE Novex 3%−8% Tris-Acetate gel (Invitrogen). Protein transfer was performed by using the iBlot dry blotting system (Invitrogen). Protein bands were visualized by an ECL reagent kit according to the manufacturer's manual. For CLK immunoprecipitation, 25 μL of M2 anti-FLAG beads (Sigma) were incubated with extracts for 2 h at 4°C. For PER immunoprecipitation, 1μl of PER antibody were incubated with extracts overnight at 4°C followed by adding 30 μl of Dyna Protein G beads (Invitrogen) for two additional hours. Proteins were eluted by 1× SDS loading buffer for 5 min at 95°C. To Purify CLK protein complex, crude nuclei extracts using10 ml of fly heads at ZT10, ZT14 and ZT18 or whole cells extracts at ZT02 were prepared as previously (Luo et al., 2012) and subject to mild sonication (8× 30 sec on, 30 sec off) at 4°C using bioruptor 300 (Diagenode). Clear extracts were incubated with 0.5 ml M2 anti-FLAG agarose beads (Sigma) for 2 hours at 4°C. The M2 beads were extensively washed and the CLK protein complex were eluted by 1ml lysis buffer supplemented with 300 μg/ml 3xFLAG peptides. The elutes were immediately subject to 2nd step IP by incubating with 100 μl streptavidin high-performance agarose beads (GE healthcare) for 2 hours at 4°C. The streptavidin beads were extensively washed with lysis buffer and then 1xPBS buffer and subject to on-beads tryptic digestion. Protein peptides were identified by the Center for Mass Spectrometry (CMS) facility at Boston College and Harvard Taplin Biological Mass Spectrometry Facility. Antibodies used for western blotting were as follows: anti-M2 FLAG (Sigma f1804), anti-NonA (Reim et al., 1999), anti-PER (Dembinska et al., 1997).
Chromatin Immunoprecitation (ChIP) and ChIP-Sequencing
Crude nuclei were prepared from 1ml fly heads of 3XFLAG-CLK14.8-HBH flies (Luo et al., 2012) with slight modifications: Fly heads were quickly ground into powder on dry ice and homogenized in 5 vol of homogenization buffer (10 mM Hepes-KOH at pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 0.8 M sucrose, 0.5 mM EDTA, 1 mM DTT, 1× protease inhibitor, 1× phosphatase inhibitor cocktail) at 4°C. Homogenates were loaded on equal volumes of sucrose cushion buffer (with 1.0 M sucrose and 10% glycerol in the homogenization buffer) and centrifuged in a HB-6 rotor (Sorvall) at 11,000 rpm for 10 min at 4°C. Pelleted nuclei were cross-linked in 1% formaldehyde 1xPBS buffer for 15 min at room temperature and quenched with 0.125M glycine. Crosslinked nuclei were extensively washed with cold 1xPBS and sonicated in 0.5 mL of lysis buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 10% glycerol, 1% SDS) for 20 cycles with the high-power setting at 30 sec on, 30 sec off. Sonicated chromatin was diluted at least 10 folds with IP buffer (20 mM Tris-HCl at pH 7.5, 150 mM NaCl, 10% glycerol, 1% NP40, protease inhibitor tablet). Anti-FLAG, anti-PER and anti-NonA ChIPs and Q-PCR were performed as previously described (Menet et al., 2010; Yu et al., 2006). For ChIP-seq, 30 μl of DNA from each IP or 50ng of Input DNA were used to generate ChIP-seq libraries according to illumine ChIP-seq protocol. After adaptor ligation, DNA samples were separated by 2% agarose TAE gel and gel slices corresponding to 250-450 bps were recovered and purified using Qiagen gel purification kit. ChIP-seq reads were mapped to the Drosophila genome (dm3) using bowtie and visualized by IGV genome browser (Menet et al., 2014).
RNA-seq of NonA RNAi and NonA TRIBE
GFP labeled M- and E-neurons (dvpdf-gal4>UAS-GFP, nonA RNAi and dvpdf-gal>UAS-GFP) were manually sorted by dissociating fly brains with papain and followed by pipette trituration. RNA-seq libraries from M- and E-neurons were generated as described in (Abruzzi et al., 2015). For NonA TRIBE, fly brains were dissected from young flies (elev-gs-gal4>UAS-nonA-ADAR). Nascent RNA were prepared according to previous study (Khodor et al., 2011). Analysis of RNA editing event was performed as previously (McMahon et al., 2016; Rodriguez et al., 2012).
Fly Brain Immunocytochemistry
Whole flies after 4 LD entrainment were fixed in 1XPBS with 4% paraformaldehyde supplemented with 0.5% Triton X-100 for 2 hours 45 mins at room temperature. Flies were washed with 1XPBS with 0.5% Triton X-100 and dissected in 1XPBS. Fly brains were blocked in 10% goat serum and incubated with primary antibodies for two nights. Mouse anti-PDF (DSHB, 1:1000) and chicken anti-GFP antibody (Abcam Cat # ab13970, 1:1000) were used. After washing with PBS-Triton X 100 4, the brains were incubated with mouse or chicken Alexa Fluor 488 secondary antibodies (1:500, Invitrogen Cat#: A-11001) for 3 hours. Brains were imaged at χ 20 on a Leica SP5 confocal microscope. Images are maximum projections of Z sections. Qualification of the PDF cell dorsal projections was analyzed with Photoshop CC. Statistical analysis was done by one-way Anova with post-hoc Turkey HSD.
In Vivo Tric-Luciferase Assays
Young flies from dvpdf-gal4>Tric-LUC, dvpdf-gal4>Tric-LUC, nonA RNAi, dvpdf-gal4>Tric-LUC, cpx RNAi, dvpdf-gal4>Tric-LUC, pdf RNAi were loaded onto white 96-well Microfluor 2 plates (Thermo Fisher Scientific) containing 5% sucrose and 2% agar food and 20 mM D-luciferin potassium salt (Gold Biotechnology). Luciferase activity from live flies were recorded in a TopCount NXT luminescence counter (PerkinElmer). Luminescence counts were collected for 3 days in LD cycles at 27.5 °C and analyzed as previously (Guo et al., 2017).
Supplementary Material
Supplemental excel table 1. Gene lists affected by nonA RNAi knockdown in M and E cells. Related to Figure 3.
Supplemental excel table 2. High confidence NonA TRIBE targets. Related to Figure 3.
Luo et al demonstrated that NonA is required for normal circadian locomotor activities in Drosophila. NonA is associated with transcription factor CLK, interacts with cpx pre-mRNA and upregulates its gene expression. NonA and CPX coordinate to regulate neuropeptide release and neuronal activity in circadian neurons.
Drosohila NonA interacts with CLOCK.
NonA is required for normal circadian locomotor activities.
cpx RNA is a NonA target.
NonA and CPX regulate circadian neuropeptides/neurotransmitters release.
Acknowledgements:
We thank Steve Brown, Sebastian Kadener, Orie Shafer, Fernanda Ceriani, Troy Littleton, E Levitan and Meghana Holla for communications or comments on the project as well as current Rosbash lab members for comments and discussion. The work was supported by the Howard Hughes Medical Institute and by a NIH EUREKA grant (DA037721) to M.R, and NIH grant 1R01GM117004 and 1R01GM118431-01A1 to E. W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
The Authors declare that there are no conflicts of interests.
REFERENCES:
- Abruzzi K, Chen X, Nagoshi E, Zadina A, and Rosbash M (2015). RNA-seq Profiling of Small Numbers of Drosophila Neurons. Methods Enzymol 551, 369–386. [DOI] [PubMed] [Google Scholar]
- Abruzzi KC, Rodriguez J, Menet JS, Desrochers J, Zadina A, Luo W, Tkachev S, and Rosbash M (2011). Drosophila CLOCK target gene characterization: implications for circadian tissue-specific gene expression. Genes Dev 25, 2374–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abruzzi KC, Zadina A, Luo W, Wiyanto E, Rahman R, Guo F, Shafer O, and Rosbash M (2017). RNA-seq analysis of Drosophila clock and non-clock neurons reveals neuron-specific cycling and novel candidate neuropeptides. PLoS Genet 13, e1006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal T, Sadaf S, and Hasan G (2013). A genetic RNAi screen for IP(3)/Ca(2)(+) coupled GPCRs in Drosophila identifies the PdfR as a regulator of insect flight. PLoS Genet 9, e1003849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio AL, Miraglia LJ, Conkright JJ, Mercer BA, Batalov S, Cavett V, Orth AP, Busby J, Hogenesch JB, and Conkright MD (2007). A coactivator trap identifies NONO (p54nrb) as a component of the cAMP-signaling pathway. Proceedings of the National Academy of Sciences of the United States of America 104, 20314–20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn JH, Lee G, and Park JH (2009). Comparative analysis of Pdf-mediated circadian behaviors between Drosophila melanogaster and D. virilis. Genetics 181, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Ripperger J, Kadener S, Fleury-Olela F, Vilbois F, Rosbash M, and Schibler U (2005). PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 308, 693–696. [DOI] [PubMed] [Google Scholar]
- Buhl LK, Jorquera RA, Akbergenova Y, Huntwork-Rodriguez S, Volfson D, and Littleton JT (2013). Differential regulation of evoked and spontaneous neurotransmitter release by C-terminal modifications of complexin. Molecular and cellular neurosciences 52, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M, Collins B, Bertet C, and Blau J (2016). Circadian rhythms in neuronal activity propagate through output circuits. Nat Neurosci 19, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi C, Cao G, Tanenhaus AK, McCarthy EV, Jung M, Schleyer W, Shang Y, Rosbash M, Yin JC, and Nitabach MN (2012). Autoreceptor control of peptide/neurotransmitter corelease from PDF neurons determines allocation of circadian activity in drosophila. Cell Rep 2, 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins B, Kaplan HS, Cavey M, Lelito KR, Bahle AH, Zhu Z, Macara AM, Roman G, Shafer OT, and Blau J (2014). Differentially timed extracellular signals synchronize pacemaker neuron clocks. PLoS Biol 12, e1001959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane BR, and Young MW (2014). Interactive features of proteins composing eukaryotic circadian clocks. Annu Rev Biochem 83, 191–219. [DOI] [PubMed] [Google Scholar]
- Dembinska ME, Stanewsky R, Hall JC, and Rosbash M (1997). Circadian cycling of a PERIOD-beta-galactosidase fusion protein in Drosophila: evidence for cyclical degradation. J Biol Rhythms 12, 157–172. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. [DOI] [PubMed] [Google Scholar]
- Duong HA, Robles MS, Knutti D, and Weitz CJ (2011). A molecular mechanism for circadian clock negative feedback. Science 332, 1436–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez MP, Berni J, and Ceriani MF (2008). Circadian remodeling of neuronal circuits involved in rhythmic behavior. PLoS Biol 6, e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel L, Muraro NI, Beltran Gonzalez AN, Marcora MS, Bernabo G, Hermann-Luibl C, Romero JI, Helfrich-Forster C, Castano EM, Marino-Busjle C, et al. (2017). Organization of Circadian Behavior Relies on Glycinergic Transmission. Cell Rep 19, 72–85. [DOI] [PubMed] [Google Scholar]
- Gao XJ, Riabinina O, Li J, Potter CJ, Clandinin TR, and Luo L (2015). A transcriptional reporter of intracellular Ca(2+) in Drosophila. Nat Neurosci 18, 917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza EA, Depetris-Chauvin A, Frenkel L, Pirez N, and Ceriani MF (2014). Circadian pacemaker neurons change synaptic contacts across the day. Curr Biol 24, 2161–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B, Chelot E, Xia R, and Rouyer F (2004). Morning and evening peaks of activity rely on different clock neurons of the Drosophila brain. Nature 431, 869–873. [DOI] [PubMed] [Google Scholar]
- Gunawardhana KL, and Hardin PE (2017). VRILLE Controls PDF Neuropeptide Accumulation and Arborization Rhythms in Small Ventrolateral Neurons to Drive Rhythmic Behavior in Drosophila. Curr Biol 27, 3442–3453 e3444. [DOI] [PubMed] [Google Scholar]
- Guo F, Cerullo I, Chen X, and Rosbash M (2014). PDF neuron firing phase-shifts key circadian activity neurons in Drosophila. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Chen X, and Rosbash M (2017). Temporal calcium profiling of specific circadian neurons in freely moving flies. Proceedings of the National Academy of Sciences of the United States of America 114, E8780–E8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE (2011). Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet 74, 141–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Forster C, Shafer OT, Wulbeck C, Grieshaber E, Rieger D, and Taghert P (2007). Development and morphology of the clock-gene-expressing lateral neurons of Drosophila melanogaster. J Comp Neurol 500, 47–70. [DOI] [PubMed] [Google Scholar]
- Hermann-Luibl C, Yoshii T, Senthilan PR, Dircksen H, and Helfrich-Forster C (2014). The ion transport peptide is a new functional clock neuropeptide in the fruit fly Drosophila melanogaster. J Neurosci 34, 9522–9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johard HA, Yoishii T, Dircksen H, Cusumano P, and Rouyer F (2009). Peptidergic clock neurons in Drosophila: ion transport peptide and short neuropeptide F in subsets of dorsal and ventral lateral neurons. J Comp Neurol 516, 59–73. [DOI] [PubMed] [Google Scholar]
- Jorquera RA, Huntwork-Rodriguez S, Akbergenova Y, Cho RW, and Littleton JT (2012). Complexin controls spontaneous and evoked neurotransmitter release by regulating the timing and properties of synaptotagmin activity. J Neurosci 32, 18234–18245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, and Rosbash M (2008). Circadian transcription contributes to core period determination in Drosophila. PLoS Biol 6, e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Park J, Cheng Y, Hardin P, and Hall J (2000). Disruption of synaptic transmission or clock-gene-product oscillations in circadian pacemaker cells of Drosophila cause abnormal behavioral rhythms. J Neurobiol. 43, 207–233. [DOI] [PubMed] [Google Scholar]
- Khodor YL, Rodriguez J, Abruzzi KC, Tang CH, Marr MT 2nd, and Rosbash M (2011). Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev 25, 2502–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, and Takahashi JS (2012). Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalska E, Ripperger JA, Hoegger DC, Bruegger P, Buch T, Birchler T, Mueller A, Albrecht U, Contaldo C, and Brown SA (2013). NONO couples the circadian clock to the cell cycle. Proceedings of the National Academy of Sciences of the United States of America 110, 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, and Reppert SM (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, and Hall JC (2002). Signal analysis of behavioral and molecular cycles. BMC Neurosci 3, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, and Taghert PH (2016). Synchronous Drosophila circadian pacemakers display nonsynchronous Ca(2)(+) rhythms in vivo. Science 351, 976–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Holy TE, and Taghert PH (2017). A Series of Suppressive Signals within the Drosophila Circadian Neural Circuit Generates Sequential Daily Outputs. Neuron 94, 1173–1189 e1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Li Y, Tang CH, Abruzzi KC, Rodriguez J, Pescatore S, and Rosbash M (2012). CLOCK deubiquitylation by USP8 inhibits CLK/CYC transcription in Drosophila. Genes Dev 26, 2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, and Davis RL (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765–1768. [DOI] [PubMed] [Google Scholar]
- McMahon AC, Rahman R, Jin H, Shen JL, Fieldsend A, Luo W, and Rosbash M (2016). TRIBE: Hijacking an RNA-Editing Enzyme to Identify Cell-Specific Targets of RNA-Binding Proteins. Cell 165, 742–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, and Rosbash M (2010). Dynamic PER repression mechanisms in the Drosophila circadian clock: from on-DNA to off-DNA. Genes Dev 24, 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Pescatore S, and Rosbash M (2014). CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev 28, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mircsof D, Langouet M, Rio M, Moutton S, Siquier-Pernet K, Bole-Feysot C, Cagnard N, Nitschke P, Gaspar L, Znidaric M, et al. (2015). Mutations in NONO lead to syndromic intellectual disability and inhibitory synaptic defects. Nat Neurosci 18, 1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Lee JM, Hwang MY, Son GH, and Geum D (2013). NonO binds to the CpG island of oct4 promoter and functions as a transcriptional activator of oct4 gene expression. Mol Cells 35, 61–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Dye BT, Perez I, Barnard DC, Thompson AB, and Patton JG (2002). PSF and p54nrb bind a conserved stem in U5 snRNA. Rna 8, 1334–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picot M, Cusumano P, Klarsfeld A, Ueda R, and Rouyer F (2007). Light activates output from evening neurons and inhibits output from morning neurons in the Drosophila circadian clock. PLoS Biol 5, e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim I, Mattow J, and Saumweber H (1999). The RRM protein NonA from Drosophila forms a complex with the RRM proteins Hrb87F and S5 and the Zn finger protein PEP on hnRNA. Exp Cell Res 253, 573–586. [DOI] [PubMed] [Google Scholar]
- Rendahl KG, Gaukhshteyn N, Wheeler DA, Fry TA, and Hall JC (1996). Defects in courtship and vision caused by amino acid substitutions in a putative RNA-binding protein encoded by the no-on-transient A (nonA) gene of Drosophila. J Neurosci 16, 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendahl KG, and Hall JC (1996). Temporally manipulated rescue of visual and courtship abnormalities caused by a nonA mutation in Drosophila. J Neurogenet 10, 247–256. [DOI] [PubMed] [Google Scholar]
- Renn SC, Armstrong JD, Yang M, Wang Z, An X, Kaiser K, and Taghert PH (1999a). Genetic analysis of the Drosophila ellipsoid body neuropil: organization and development of the central complex. J. Neurobiol 41, 189–207. [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, and Taghert PH (1999b). A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99, 791–802. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Menet JS, and Rosbash M (2012). Nascent-seq indicates widespread cotranscriptional RNA editing in Drosophila. Molecular cell 47, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Agosto J, and Rosbash M (2004). Coupled oscillators control morning and evening locomotor behaviour of Drosophila. Nature 431, 862–868. [DOI] [PubMed] [Google Scholar]
- Stoleru D, Peng Y, Nawathean P, and Rosbash M (2005). A resetting signal between Drosophila pacemakers synchronizes morning and evening activity. Nature 438, 238–242. [DOI] [PubMed] [Google Scholar]
- Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, and Kaiser P (2006). A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol Cell Proteomics 5, 737–748. [DOI] [PubMed] [Google Scholar]
- Takahashi JS (2017). Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18, 164–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav SP, Hao H, Yang HJ, Kautzmann MA, Brooks M, Nellissery J, Klocke B, Seifert M, and Swaroop A (2014). The transcription-splicing protein NonO/p54nrb and three NonO-interacting proteins bind to distal enhancer region and augment rhodopsin expression. Hum Mol Genet 23, 2132–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, and Shafer OT (2014). The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science 343, 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, and Hardin PE (2006). PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20, 723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental excel table 1. Gene lists affected by nonA RNAi knockdown in M and E cells. Related to Figure 3.
Supplemental excel table 2. High confidence NonA TRIBE targets. Related to Figure 3.