Abstract
Despite advances in understanding the molecular pathogenesis of acute myeloid leukaemia (AML), overall survival rates remain low. The ability to predict treatment response based on individual cancer genomics using computational modeling will aid in the development of novel therapeutics and personalize care. Here, we used a combination of genomics, computational biology modeling (CBM), ex vivo chemosensitivity assay, and clinical data from 100 randomly selected patients in the Beat AML project to characterize AML sensitivity to a bromodomain (BRD) and extra-terminal (BET) inhibitor. Computational biology modeling was used to generate patient-specific protein network maps of activated and inactivated protein pathways translated from each genomic profile. Digital drug simulations of a BET inhibitor (JQ1) were conducted by quantitatively measuring drug effect using a composite AML disease inhibition score. 93% of predicted disease inhibition scores matched the associated ex vivo IC50 value. Sensitivity and specificity of CBM predictions were 97.67%, and 64.29%, respectively. Genomic predictors of response were identified. Patient samples harbouring chromosomal aberrations del(7q) or −7, +8, or del(5q) and somatic mutations causing ERK pathway dysregulation, responded to JQ1 in both in silico and ex vivo assays. This study shows how a combination of genomics, computational modeling and chemosensitivity testing can identify network signatures associating with treatment response and can inform priority populations for future clinical trials of BET inhibitors.
Keywords: BET inhibitor, JQ1, Computational modeling, AML, Drug response, Genetics
1. Introduction
Acute myeloid leukaemia (AML) is a complex heterogeneous disease characterized by uncontrolled proliferation of immature myeloid blasts and bone marrow failure [1]. Standard treatment for AML consists of cytarabine-based chemotherapy, with hematopoietic stem cell transplant as the only potentially curative option. The 5-year overall survival (OS) rate for patients younger than the age of 60 is approximately 40%, while 5-year OS for patients older than 60 is less than 20% [2,3]. Unfortunately, a majority of patients relapse, further complicating clinical care.
Studies suggest that AML may arise due to a series of genetic alterations that accumulate with age [2]. With the advent of next-generation sequencing technologies, the complex amalgam of cytogenetic, genetic, and epigenetic alterations present at diagnosis and/or relapse of AML are now better defined. Although chromosomal abnormalities occur in more than 50% of adult AML patients, most recurrent alterations are rare and found in less than 10% of patients [1]. Commonly mutated genes include FLT3, NPM1, DNMT3A, IDH1, RUNX1, and cKIT, among others [1,4,5]. Certain cytogenetic abnormalities are used for prognostic and diagnostic purposes. For example, t(8;21)(q22;q22), t (15;17)(q22;q12) and inv(16)(p13.1;q22) alterations are favorable risks associated with better patient outcomes. Other more common alterations such as deletion or monosomy of chromosome 5 or 7 are poor risk factors associated with resistance to therapy and worse overall survival. Deletion of chromosome 5 occurs in approximately 10–20% of de novo AML cases [5]. A recent study suggests that loss of chromosome 5 may be an early event that leads to additional genetic alterations, including amplification of chromosome 8 [4]. Such heterogeneity complicates the prognosis and treatment of AML for these patients.
Epigenetic alterations are also considered key players in the progression of AML. The process by which leukaemia stem cells aberrantly self-renew and propagate the disease has been linked to changes in regulatory chromatin modifications [6]. Novel therapies that target these epigenetic modifiers such as demethylating agents (decitabine, azacitidine) and histone deacetylase inhibitors (panobinostat) have shown some promise in leukemia, other hematological malignancies, as well as solid tumors [7,8]. A new class of epigenetic therapy include the BET inhibitors (iBETs). The BET protein family consists of 3 ubiquitously expressed proteins, BRD2, BRD3, BRD4, and the testis-specific protein BRDT. As chromatin scaffolds, they recruit elements of the positive transcriptional elongation factor b (P-TEFb) complexes to RNA polymerase II (RNA Pol II) to initiate transcriptional elongation. In AML and other hematological cancers, these BET proteins have been found to preserve aberrant chromatin states, thereby increasing transcription of known oncogenes including c-MYC [9].
Using an RNAi screen, Zuber et al identified BRD4 as a chromatin modifier critical for tumor growth in an AML mouse model. The study demonstrated that suppression of BRD4 using shRNAs, or the small-molecule inhibitor JQ1, led to robust anti-leukemic effects in vitro and in vivo [10]. Since the discovery of JQ1 as the first BET inhibitor with both differentiation and specific anti-proliferative effects on human squamous carcinoma, new derivatives and inhibitors have been generated [11]. Since then, several BET inhibitors have shown promise in AML and other hematological malignancies both in vitro and in early phase clinical trials (Table 1) [12–17].
Table 1.
Summary of BET inhibitors in clinical trials for hematological malignancies.
| Summary of BET inhibitors in Clinical Trials | |||
|---|---|---|---|
| BET Inhibitor | Indication | Clinical Status | Reference |
| CPI-0610 | Leukemia; Lymphoma; Multiple myeloma; Myelodysplastic syndromes; Myeloproliferative disorders | Phase I | NCT02158858 [11] |
| OTX 015 | Acute Myeloid Leukemia; Diffuse Large B-cell Lymphoma; Acute Lymphoblastic Leukemia; Multiple Myeloma | Phase I | NCT01713582; NCT02698189 [13] |
| RO 6870810; TEN 010 | AML; myelodysplastic syndrome | Phase I | NCT02308761 |
| INCB-054329 | Advanced malignancies | Phase I/II | NCT02431260 |
| ABBV-075 | Advanced malignancies | Phase I | NCT02391480 |
| FT 1101 | Hematological malignancies | Phase I | NCT0254387 [15] |
| GSK525762; I-BET-762 | Relapsed, refractory hematological malignancies, solid tumors | Phase I/II | NCT01943851 [12] |
| PLX-51107 | AML, myelodysplasia, solid tumors | Phase I | NCT02683395 [16] |
The mechanisms that mediate sensitivity to the iBETs remain broad. In addition to downregulation of c-MYC, BET inhibitors have been shown to affect additional transcriptional regulators including FOSL1 and E2F target genes [18]. Wild-type NPM1 has been shown to inhibit BRD4 activity. However, approximately 35% of AML patients harbor a mutation in NPM1c, which leads to the release of BRD4 and upregulation of the core transcriptional program which facilitates leukemia development. Treatment of NPM1c AML cells with a BET inhibitor can restore BRD4 inhibition, reducing BRD4 recruitment to chromatin and downregulating expression of critical oncogenes such as c-MYC [19]. Mutations in FLT3 are also common in AML, yet treatment with FLT3 tyrosine kinase inhibitors (TKI) is often associated with resistance. However, combining the BET inhibitor JQ1 with a FLT3 TKI, ponatinib, was highly synergistic and enhanced cell death in AML cell line models as well as human CD34 + AML blast progenitor cells [20]. These studies highlight the pleiotropic effects of BET inhibitors and their potential benefit to treat the heterogeneous nature of AML.
Due to the diverse mechanism behind leukemogenesis as well as the pleiotropic mechanisms mediating sensitivity to iBETS, not all cell lines and patients respond in the same manner or achieve the same depth of response. Therefore, success of these iBETs lies, in part, on the ability to identify patients likely to respond to targeted therapies before initiating therapy. Predictive simulation is an emerging technology in the era of personalized medicine. By performing next-generation sequencing and subsequently translating the genomic aberrations into patient-specific network maps of activated and inactivated protein pathways, a patient-specific cancer avatar can be created. After performing digital drug simulation on these avatars, sensitivity to specific therapies can be calculated in silico. Such an approach was shown to predict drug sensitivity in eight models of glioblastoma patient-derived tumor cells exposed to ten therapeutics with 75% accuracy [21]. Another retrospective study on three cohorts of myelodysplastic syndrome (MDS) patients accurately predicted drug responses to standard of care therapies (azacitidine, decitabine, and lenalidomide) with ≥80% accuracy [22]. One of the major advantages to using cancer avatars is the ability to assess tumor cell sensitivity to FDA-approved and investigational therapies by modeling their unique mutational profiles, broadening the therapeutic options for refractory patients and avoiding potentially ineffective regimens. This method also allows clinicians, researchers, and pharmaceutical companies to simulate digital clinical trials to gain perspective on molecular criteria for identifying sensitive and resistant profiles.
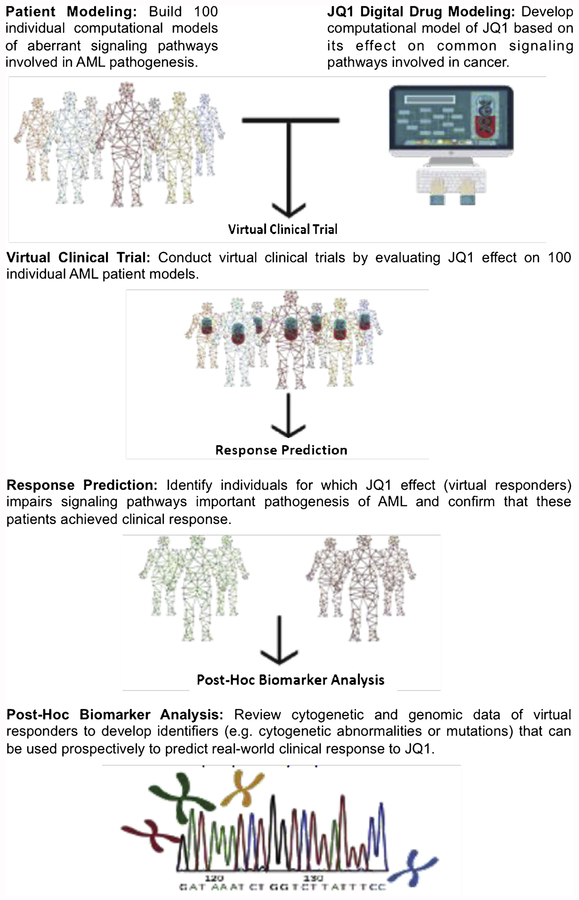
This study has created simulated computational models of AML patient genomics to create a digital clinical trial to evaluate the cytotoxic effect of JQ1 on primary AML samples. Patients included in this virtual trial participated in the BEAT AML study spearheaded by our collaborators at Oregon Health Sciences University. Patient information including cytogenetics and whole-exome sequencing data were entered into a computational biology modeling (CBM) software system to generate patient-specific network maps, or cancer avatars. Using these patient-specific cancer avatars, we simulated in silico drug sensitivity assays to JQ1 to predict which patient samples will be sensitive to the drug. We compared the CBM drug sensitivity predictions with ex vivo drug sensitivity data of the primary AML cells treated with JQ1. Additionally, we correlated genomic abnormalities identified in this patient cohort to in situ and ex vivo JQ1 sensitivity to discover bio-markers and molecular aberrations that may be used prospectively to predict clinical response to JQ1 (Fig. 1).
Fig. 1.
Schema for the Retrospective Virtual Clinical Trial.
Schematic illustrates the study design and methods. 100 randomly selected patients form the BEAT AML project were modelled using CBM on which efficacy of the JQ1 digital drug model was evaluated. Predicted responses to JQ1 were compared with ex vivo chemosensitivity assay to determine prediction correlation and accuracy. Post-hoc biomarker analysis was done to determine genomic predictors of JQ1 response.
2. Materials and methods
2.1. Patient samples
All patients gave consent to participate in the Beat AML cores study (NCT01728402), which had the approval and guidance of the Institutional Review Board at Oregon Health & Science University (OHSU), University of Utah, University of Texas Medical Center (UT Southwestern), Stanford University, University of Miami (UM), University of Colorado (UC), University of Florida (UF), National Institutes of Health (NIH), Fox Chase Cancer Center and University of Kansas (KUMC). Samples were sent to the coordinating center (OHSU; IRB#9570) where they were coded and processed. Access to patient data and all analyses for this study were approved by the UF IRB (#201601364). Patients provided bone marrow and/or peripheral blood, along with a skin punch biopsy as germline control.
Mononuclear cells were isolated by Ficoll-gradient centrifugation from freshly obtained bone marrow aspirates or peripheral blood draws and plated into assays within 24 h. All samples were analysed for clinical characteristics and drug sensitivity [47].
2.2. Whole exome, custom capture validation sequencing
Whole exome sequencing was performed on bone marrow and/or peripheral blood, along with a skin punch biopsy using Illumina Nextera Rapid Capture Exome capture probes. Custom capture validation probes were assembled by Roche Sequencing Solutions. Sequencing was performed on an Illumina HiSeq 2500.
2.3. Ex vivo functional screen
Small-molecule inhibitors, purchased from LC Laboratories (Woburn, MA, USA) and Selleck Chemicals (Houston, TX, USA), were reconstituted in DMSO and stored at −80 °C. Inhibitors were distributed into 384-well plates prepared with a single agent/well in a 7-point concentration series ranging from 10 μM to 0.0137 μM for each drug. The final concentration of DMSO was ≤0.1% in all wells; plates were stored at −20 °C and thawed immediately prior to use. Primary mononuclear cells were plated across inhibitor panels within 24 h of collection. Cells were seeded into 384-well assay plates at 10,000 cells/well in RPMI-1640 media supplemented with fetal bovine serum (10%), L-glutamine, penicillin-streptomycin and β-mercaptoethanol (10−4 M). After three days of culture at 37 °C in 5% CO2, MTS reagent (CellTiter96 AQueous One; Promega Madison, WI, USA) was added, optical density was measured at 490 nm [47].
2.4. AML computation biology model (CBM)
The computational biology model (CBM) used in this study is an extensively validated, comprehensive network of signalling, metabolic, epigenetic and transcriptional regulatory pathways underlying cancer physiology [21–25]. The network is created through a rigorous work-flow of manually curating and aggregating published experimental data and representing the functionality of the genes, proteins and interactions mathematically, using ordinary differential equations [21]. The CBM coverage includes pathway networks underlying many cancer phenotypes including growth factor, cytokine and chemokine signaling pathways, transcriptional, post-transcriptional, translational and post-translational regulation, epigenetic regulation, cell cycle machinery, oxidative and ER stress, protein homeostasis including proteasomal machinery and autophagy, DNA repair pathways, apoptotic cascade and TP53 signaling, metabolic pathways, angiogenic and immune-suppressive pathways, among others. The CBM includes about 112 central pathways, over 75,000 reactions, and 3300 cancer specific-genes including comprehensive coverage of the kinome, transcriptome, proteome and metabolome. This extensively integrated network that makes up the CBM can be used to predict a patient’s response to a single drug or a combination of drugs. Both prospective and retrospective validations have been shown in studies of glioblastoma multiform (GB), multiple myeloma (MM), myeloproliferative neoplasms (MPN) and MDS [22–26].
2.5. Creation of AML profiles
Bone marrow samples and clinical data from 100 AML patients from the BEAT AML project (Leukemia and Lymphoma Society) were obtained and underwent conventional cytogenetic, whole exome sequencing, and an ex-vivo drug sensitivity assay. Genomic aberrations were interpreted for phenotypic implications (i.e., gain of function (GOF) versus loss of function (LOF)). The cytogenetic segments related to deletions, gains, translocations, or derivatives, were interpreted as amplifications (AMP) and deletions (DEL) of the genes residing in those segments. The genes found on the loci of these affected regions of the chromosome are extracted from the human reference genome at ENSEMBL. The complete list of genes is matched with the CBM to identify those that are to be represented in the model. All genes that have coverage in the model (Suppl. table 5) are included in the input file that is used to create the patient cancer avatar. Genes reported to have a gain in copy number due to chromosomal amplifications are interpreted as being over-expressed at the gene expression level, while those genes in the deleted segments are considered a loss of copy number and are interpreted as having a knock-down in the model.
For mutation signatures, the gene variants with known functional impact and therapeutic implication are searched in the public domain and are recorded in a mutation library. Mutational signatures are processed through our internal variant calling workflow that utilizes DbNFSP, a database that uses multiple prediction algorithms including SIFT, FATHMM, Mutation Assessor, LRT, Mutation Taster, PROVEAN, MetaSVM, and others, to determine if the gene mutation will have a functional impact on the protein, which will be classified as either deleterious or non-deleterious based on a concordance of more than 5 algorithms [27–31]. A deleterious mutation in an oncogene is assumed to be a GOF mutation at the protein activity level, or a LOF if present in a tumor suppressor gene. Frameshift and missense mutations are assumed to cause a loss of protein function except in those cases where there is experimental evidence that the mutation causes a gain of function.
Finally, this input file is overlaid on the control model (non-tumorigenic baseline) by indicating the gene mutations, amplifications, deletions and translocations, and the profile is simulated as per the rules outlined above to create a dynamic disease state. Protein network maps are created for each patient profile based on their input data and disease specific biomarkers that are unique to each profile.
2.6. Digital drug model
JQ1 was used as a representative BET inhibitor. A digital drug model of JQ1 was created for CBM by programming its mechanism of action inhibiting the iBET target isoforms BRD4, BRD2 and BRD3 in the network, and the resultant effects on specific pathways and biomarkers determined from published literature [11,32,33].
2.7. Simulation and analysis
Using the digital drug model of JQ1, virtual applications of JQ1 were applied to each patient’s disease via computer simulation in a dose respondent fashion. The efficacy of JQ1 for each patient was measured as a function of disease inhibition score (DIS) – the degree to which crucial cancer signalling pathways and phenotypes were repressed. DIS is a composite of the percentage impact on proliferation and viability index with the drug in reference to the untreated disease network. The proliferation index is an average function of the active CDK-cyclin complexes that define cell cycle checkpoints and is determined by calculating contributions in the biomarkers CDK4-CCND1, CDK2-CCNE, CDK2-CCNA and CDK1-CCNB1. A viability index based on survival and apoptosis markers is also generated for each patient. The biomarkers constituting the survival index include AKT1, BCL2, MCL1, BIRC5, BIRC2 and XIAP, while the apoptosis index comprises the pro-apoptotic markers of caspases, Puma and cleaved PARP. Viability of a cell is calculated as a ratio of survival index/apoptosis index, and the weightage of each biomarker is adjusted to achieve a maximum correlation with experimental trends of the endpoint from peer-reviewed studies.
2.8. Statistics
Raw absorbance values were adjusted to a reference blank value, and then used to determine cell viability (normalized to untreated control wells). The concentration of inhibitor required to inhibit cell growth by 50% (IC50) was calculated using a non-linear regression analysis.
Computational JQ1 drug sensitivity predictions were compared against JQ1 IC50 values obtained from experimental ex vivo testing. For ex vivo results, an IC50 < 2.70μM was considered sensitive and an IC50 > 2.70μM was considered resistant to JQ1 therapy. This threshold was determined based on the Cmax concentration derived from pharmacokinetic studies of JQ1.11 For CBM results, a DIS ≥ 30% was classified as sensitive (responder), and a DIS < 30% was classified as resistant (non-responder). Post-hoc analysis of virtual responders and non-responders were performed to determine all unique cytogenetic and genomic identifiers between the two groups. Correlation between actual response and predicted response was assessed by using a 2 × 2 contingency table to calculate PPV, NPV, sensitivity and specificity, and significance was calculated using a Chi-square test.
3. Results
3.1. Correlation summary of predictive vs. ex-vivo outcomes
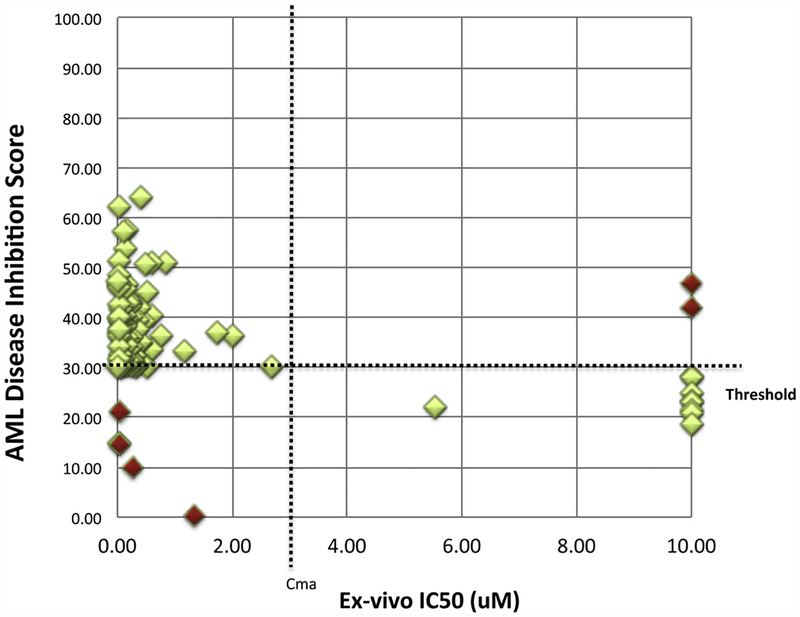
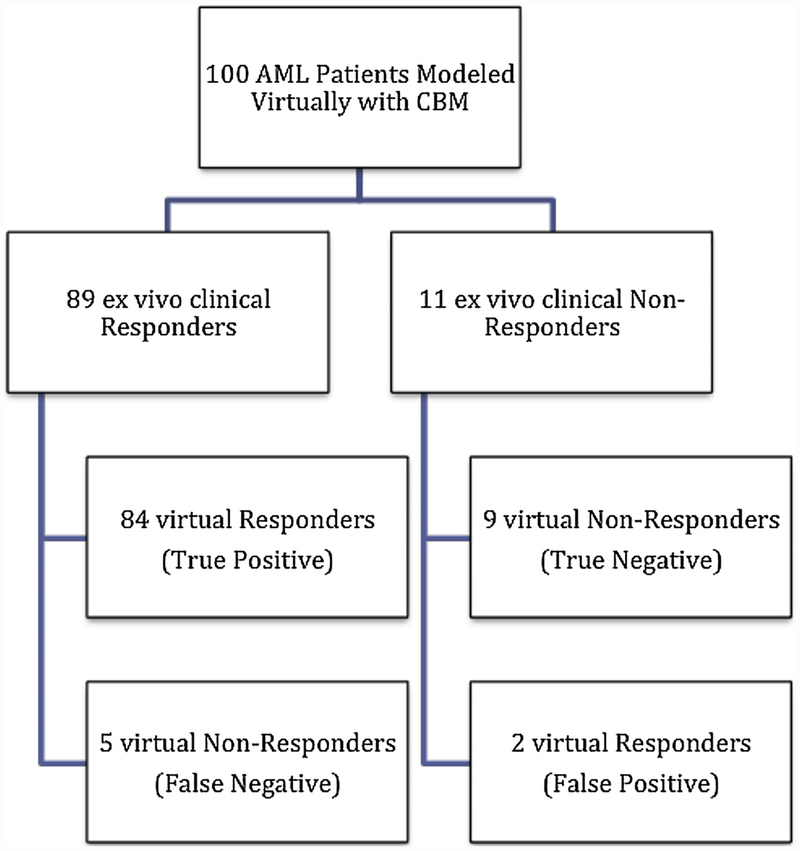
As part of the BEAT AML study, ex vivo drug sensitivity assays were performed on 100 patient’s sample from the BEAT AML study. Patient characteristics details are included in supplementary Table 1. CBM derived AML DIS for each patient were compared to experimentally determined ex vivo sensitivity towards JQ1 (Fig. 2, Suppl. Table 2 Table 2). For ex vivo results, an IC50 less than 2.70 μM for JQ1 was considered sensitive and an IC50 greater than 2.70 μM was considered resistant to JQ1 therapy as indicated by the vertical Cmax dotted line on the plot. For predictive CBM results, patients with a DIS ≥ 30% were classified as sensitive, whereas patients with a DIS < 30% were classified as resistant, indicated by the horizontal % threshold dotted line on the plot. A schema of patient-response is depicted in Fig. 3. Experimental sensitivity to JQ1 closely matched with virtual predictions using the CBM model. Of the 100 patients modeled from the BEAT AML project, 89 patients’ ex vivo results were considered sensitive to JQ1 and 11 were considered resistant. CBM correctly predicted 84 of 89 sensitive profiles to JQ1, and 5 of these incorrect predictions were false negatives. CBM accurately predicted 9 out of the 11 patient samples that showed resistance to JQ1, with the two mismatched predictions being false positive. As shown in Table 2, the CBM predictions of JQ1 sensitivity are highly accurate with positive predictive value (PPV) of 97.67%, negative predictive value (NPV) of 64.29%, and an overall accuracy of 93%. As calculated, specificity and sensitivity of our CBM model is 81.82% and 94.38%, respectively.
Fig. 2.
Illustration of AML disease inhibition score associations with ex vivo JQ1 IC50 values. A scatterplot representing ex vivo determined IC50 (X-axis) and virtually simulated disease inhibition scores of JQ1 (Y-axis). Cut-offs to determine sensitivity and resistance were determined empirically. For CBM response predictions (Y-axis), 30% disease inhibition was chosen as the threshold for response (horizontal threshold dotted line). A disease inhibition score > 30% is classified as a responder, or sensitive to JQ1, while a disease inhibition score < 30% is classified as a non-responder, or resistant to JQ1. For ex vivo IC50 values (X-axis), the Cmax of JQ1 was calculated from a previous study and used as the threshold for response. [11] (Vertical Cmax dotted line) IC50 < 2.7μM was considered sensitive, and IC50 > 2.7μM was considered resistant. The green diamonds indicate the responder and non-responder predictions that matched with the ex vivo response, while the red diamonds indicate false negatives and false positive predictions. Green diamonds (light) vs. red diamonds (Dark) (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Fig. 3.
Schema representing JQ1 responders and non-responders. Delineation of virtual and ex vivo profiles sensitive and resistant to JQ1.
Table 2.
Statistical Summary of AML Patients’ Predicted Response to BET Inhibitor JQ1.
| Sensitivity | Specificity | PPV | NPV | Accuracy | |
|---|---|---|---|---|---|
| BET Inhibitor (JQ1) | 96.67% (95% CI: 92.2999.33) |
64.29% (95% CI: 42.3681.51) |
94.38% (95% CI: 87.3798.15) |
81.82% (95% CI: 48.2297.72) |
93.00% |
3.2. Patient-specific CBM analysis for responders and non-responders to JQ1
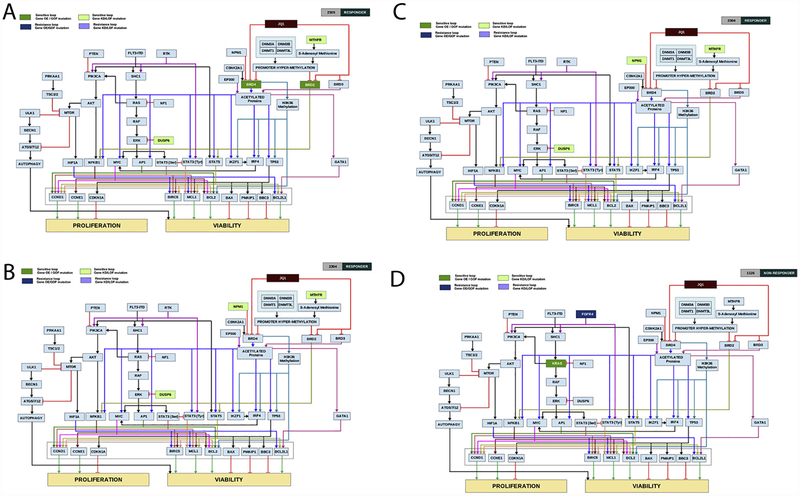
Representative computer-simulated patient network maps are shown in Fig. 4A–D. Profile 2305 was virtually simulated with increasing concentrations of JQ1 in silico. CBM predicted a 50% decrease in the profile’s AML disease inhibition, suggesting sensitivity to JQ1. This profile had an ex vivo IC50 of 0.04 μM. (Suppl Table 2) The mechanism of drug sensitivity was determined to be due to a gain of function of BRD2 and BRD4, key targets of JQ1 [9]. This patient profile also possessed loss of Dual specificity phosphatase 6 (DUSP6) activity, which promotes ERK activation, ultimately leading to an increase in its transcriptional target, c-MYC, another key mediator of JQ1 drug impact [34]. Additionally, a loss of function in the enzyme methylene tetrahydrofolate reductase (MTHFR) was observed in this patient profile. MTHFR has been shown to affect heterochromatin maintenance and can lead to the hypermethylation of specific genes [35,36]. In a similar manner, abrogation of MTHFR may impact the expression of BRD2 and BRD4, further sensitizing this patient to the iBET JQ1 (Fig. 4A).
Fig. 4.
A-D: Computer-simulated patient-specific network maps and digital response to JQ1. Patient specific network maps were generated using CBM modeling. Patient 2305(A) and 2304 (B) are representative examples of profiles predicted to be sensitive to JQ1 as measured by the in-silico AML disease inhibition score. Patient 4006 (C) and 1126 (D) are representative examples of profiles predicted to be resistant to JQ1 as measured by the in-silico AML disease inhibition score. Boxes highlighted in light green represent gene mutations leading to protein loss of function or knock-down contributing to drug sensitivity. Boxes highlighted in darker green represent gene mutations leading to protein gain of function or over-expression contributing to drug sensitivity. Boxes highlighted in purple represent gene mutations leading to loss of function or knock-down of proteins contributing to drug resistance, and boxes highlighted in dark blue represent gene mutations leading to protein gain of function or over-expression contributing to drug resistance. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
Patient ID 2304 had a different set of aberrations that supported the response of this profile to JQ1 (Fig. 4B). This patient had a LOF aberration in nucleophosmin (NPM1), a gene commonly mutated in one third of adult AML patients. Loss of NPM1 has been shown to activate a BRD4-dependent core transcriptional program in AML [19]. This patient also had a LOF of DUSP6 and MTHFR genes that would further enhance iBET sensitivity. Profile 2304 had an ex vivo IC50 of 0.16 μM and a DIS of 57.54. (Suppl. Table 2)
Conversely, the patient profile 4006 is classified as resistant to JQ1 in silico as the AML disease inhibition was only reduced by 25% and this profile had an ex vivo IC50 of 10 μM. (Suppl. Table 2) CBM identified resistance due to a LOF of EP300. EP300 functions as a histone acetylase that impacts gene transcription by recruiting bromodomain proteins, including BRD4 [37,38]. Loss of EP300 function also results in loss of BRD4, therefore making this profile less sensitive to JQ1. Additionally, amplification of AMPK (PRKAA1) was observed in this patient profile and may indirectly affect sensitivity to JQ1 (Fig. 4C). A recent study found that AMPK induces a pro-survival autophagic response after treatment with JQ1, and inhibiting AMPK could increase JQ1-mediated apoptosis [39].
Patient ID 1126 is also predicted to be a non-responder by CBM at DIS of 20.61 and ex vivo IC50 of 10 μM. Lack of response in this profile can be explained due to presence of a GOF mutation in Fibroblast growth factor receptor 4 (FGFR4) gene that can activate a parallel resistance pathway. (Fig. 4D) FGFR4 aberrations have been shown to cause resistance to chemotherapy in other cancers including colorectal cancer and could mediate this resistance through activating parallel RTK pathway loops thus overcoming BET inhibition [40,41].
3.3. Identification of common chromosomal aberrations with patient-specific response to JQ1
As part of the BEAT AML study, conventional cytogenetic analysis was provided for each patient sample (Suppl. table 1 & 3). Of the 100 patients included in the present study, 18 patients presented one or more of the common cytogenetic abnormalities including monosomy 7, 7q-del, trisomy 8, 5q-del, 19p-amp, or 22q-del. As shown in Table 3, associations of some of these chromosomal aberrations with response to JQ1 were analysed. Patient samples harboring chromosomal aberrations del(7q) or monosomy 7 (n = 4), trisomy 8 (n = 10) or del(5q) (n = 7) responded to JQ1 in both in silico and ex vivo sensitivity assays. Post-hoc analysis shows that the patients with trisomy 7, 7q-del, trisomy 8, or 5q-del would have a higher likelihood of sensitivity to JQ1 (p = 0.041) or other BET inhibitors. However, a single patient sample with del(22q) was not sensitive to JQ1.
Table 3.
Correlation of cytogenetic aberrations with patient-specific response to JQ1.
| BET Inhibitor (JQ1) - Patients with Chromosome aberrations | ||||||||
|---|---|---|---|---|---|---|---|---|
| Patient ID | Drugs Tested | Clinical Response | Simulation Response | Correlation Status | 7q-del / Monosomy 7 | Trisomy 8 | 5q-del | 19p-amp 22q-del |
| 1953 | JQ1 | Responder | Responder | MATCH | Y | Y | Y | |
| 2236 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2500 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2612 | JQ1 | Responder | Responder | MATCH | Y | |||
| 4006 | JQ1 | Non-Responder | Non-Responder | MATCH | Y | Y | Y | |
| 2633 | JQ1 | Responder | Responder | MATCH | Y | Y | ||
| 2079 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2443 | JQ1 | Responder | Responder | MATCH | Y | |||
| 1924 | JQ1 | Responder | Responder | MATCH | Y | |||
| 1727 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2704 | JQ1 | Responder | Responder | MATCH | Y | |||
| 4324 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2690 | JQ1 | Responder | Responder | MATCH | Y | |||
| 4043 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2452 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2315 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2426 | JQ1 | Responder | Responder | MATCH | Y | |||
| 2333 | JQ1 | Responder | Responder | MATCH | Y | |||
3.4. Identification of common somatic aberrations with patient-specific response to JQ1
The insights gained from CBM predictive JQ1 response analysis in patient disease networks are supported by the frequency of occurrence of somatic mutations in the drug sensitizing pathways. Mutations in genes that are linked to ERK pathway dysregulation in the disease network including KRAS (n = 7), NRAS (n = 23), DUSP6 (n = 9) and NF1 (n = 6), were present predominantly in the responder profiles. FLT3 (n = 30) and NPM1 (n = 9) mutations were also highly frequent in the responder profiles in this cohort (Suppl. table 4).
4. Discussion
There is a significant need to develop novel treatment strategies for AML and to personalize patient care. One approach uses computational modeling to predict treatment response based on patient-specific tumor genomics. The present study leveraged the use of CBM technology to predict response to the iBET JQ1 in AML patients, and validated the predictions by measuring JQ1-induced cytotoxicity on primary patient samples in an ex vivo drug sensitivity assay. Additionally, the in silico CBM technology provided novel insight into the mechanisms that mediate sensitivity and resistance and aids in the identification of potential biomarkers that predict response to iBET.
By integrating cytogenetic, genomic, and transcriptomic data from 100 patients participating in the BEAT AML study, we predicted these patients’ response to the iBET JQ1 using CBM technology. Our data shows high accuracy between digitally derived DIS and experimentally determined ex vivo IC50 values for JQ1, with a reported accuracy of 93% (Table 2, Suppl. Table 2). Additionally, the transparency of the CBM network provides insight into the molecular mechanisms mediating a response to JQ1. For example, profile 2305, which was predicted to be a responder (Fig. 4A), showed enhanced activity of key targets of JQ1 and pro-tumorigenic proteins including BRD2 and BRD4 and loss of function of proteins expected to play an anti-tumorigenic function such as DUSP6 and MTHFR [34,36] In another responder profile 2304, the NPM1 mutation along with MTHFR loss enhanced BRD4 activity and sensitized the profile to BET inhibition (Fig. 4B). An example of a non-responsive patient, profile 4006 (Fig. 4C) suggests that loss of the his-tone acetylase EP300 may be responsible for lack of sensitivity to JQ1 due to its role in regulating BRD4 [37]. FGFR4 gain in another profile 1126 could be the reason for non-response to JQ1 due to activation of a parallel resistance pathway [40,41]. Due to the complexity of interactions between signalling pathways, additional validation of the roles these proteins play in mediating iBET sensitivity are necessary. However, these proteins, and others, that are highlighted by the CBM technology may help identify biomarkers that mediate response to iBETs including JQ1.
The results of this study also suggest that trisomy 8 is one of the cytogenetic aberrations associated with a positive response to JQ1 (Table 3). The gene coding for the oncogenic transcription factor, MYC, is located on chromosome 8. An increase in MYC expression has been associated with increased cancer cell proliferation and survival due to its pleiotropic effects on cell signalling pathways. For example, MYC can activate IRF4, MCL1, and Cyclin D1 expression, regulating cell death and cell cycle pathways [42,43]. Importantly, MYC was described as an important target of bromodomain proteins, including BRD4, and inhibition of bromodomain proteins by JQ1 leads to a significant decrease in MYC expression [32]. The ability of iBETs to modulate MYC expression has implications across a multitude of cancer types. MYC is also induced in profiles with dysregulated ERK [44,45] linked to mutations in ERK regulators KRAS, NRAS, DUSP6 and NF1 genes in JQ1 responder profiles as evident from the gene matrix (Suppl. table 4).
Other chromosomal abnormalities associated with a response to JQ1 include del-5q and monosomy 7. Genes present within these chromosomal regions may also mediate sensitivity to JQ1 such as NPM1 (chromosome 5) and EZH2 and KMT2C (chromosome 7), as they have been implicated in the regulation of bromodomain proteins. For example, loss of NPM1 activity increased BRD4 activity [19]. Additionally, a recent study found that sensitivity to iBETs can be enhanced by loss of EZH2 function [46].
Thus, through this CBM analysis of AML patients’ genetics and predicting JQ1 sensitivity using an AML- specific in silico approach, we have identified genomic aberrations with molecular mechanisms of sensitivity towards iBETs. The ability of the CBM to predict multiple genetic factors contributing to drug response is unique and nicely complements ex vivo and clinical studies for identifying genetic signatures for drug responses. The predictions however are based on interpretation and translation of genomic inputs for creating and understanding the patient disease characteristics. The false predictions could be due to incorrect interpretations of unknown deleterious variants or incomplete genomic data.
5. Conclusions
In this study, we ran a virtual clinical trial to evaluate the efficacy of the BET inhibitor JQ1 in patients with AML. Our study successfully validated the accuracy of our CBM technology using ex vivo drug sensitivity data. Additionally, by integrating known cytogenetic and genomic alterations specific for each patient, this CBM technology can elucidate novel biomarkers that may predict patient sensitivity to JQ1. Virtual clinical trials on larger datasets will help further elucidate the inclusion and exclusion criteria for response to iBETs and other such first-in-class therapeutics used in specific disease settings.
Supplementary Material
Acknowledgements
Supported in part by The Leukemia & Lymphoma Society Beat AML Program and the NIH (1U01CA217862–01; 1U54CA224019–01; 3P30CA069533–18S5) and Knight Cancer Institute. This work was also partially supported by the Harry T. Mangurian Foundation (F022243), the Gateway for Cancer Research Foundation (G-16–700), and the Gatorade Foundation administered by the UF Department of Medicine. CRC received a Scholar in Clinical Research award from the Leukemia & Lymphoma Society (0725–14).
Disclosures
Research support for J.W.T. is received from Aptose, Array, AstraZeneca, Constellation, Genentech, Gilead, Incyte, Janssen, Seattle Genetics, Syros, Takeda; J.W.T. is a co-founder of Leap Oncology. TA, SV, RV, SG, AVL, VPS, AKA, AT, are employees of Cellworks.
Abbreviations:
- AML
Acute myeloid leukemia
- BRD
bromodomain
- BET
bromodomain extra-terminal
- iBET
BET inhibitor
- CBM
computational biology modelling
- OS
overall survival
- TKI
tyrosine kinase inhibitor
- DIS
disease inhibition score
Footnotes
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.leukres.2018.11.010.
References
- [1].Khwaja A, Bjorkholm M, Gale RE, Levine RL, Jordan CT, Ehninger G, et al. , Acute myeloid leukaemia, Nat. Rev. Dis. Primers 10 (March 2) (2016) 16010. [DOI] [PubMed] [Google Scholar]
- [2].Saultz JN, Garzon R, Acute myeloid leukemia: a concise review, J. Clin. Med 5 (March (3)) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Foran JM, Do cytogenetics affect the post-remission strategy for older patients with AML in CR1? Best Pract. Res. Clin. Haematol 30 (December (4)) (2017) 306–311. [DOI] [PubMed] [Google Scholar]
- [4].Qian Z, Joslin JM, Tennant TR, Reshmi SC, Young DJ, Stoddart A, et al. , Cytogenetic and genetic pathways in therapy-related acute myeloid leukemia, Chem. Biol. Interact 184 (March (1–2)) (2010) 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ganguly BB, Banerjee D, Agarwal MB, Impact of chromosome alterations, genetic mutations and clonal hematopoiesis of indeterminate potential (CHIP) on the classification and risk stratification of MDS, Blood Cells Mol. Dis 69 (March) (2018) 90–100. [DOI] [PubMed] [Google Scholar]
- [6].Dick JE, Stem cell concepts renew cancer research, Blood 112 (December (13)) (2008) 4793–4807. [DOI] [PubMed] [Google Scholar]
- [7].Dombret H, Seymour JF, Butrym A, Wierzbowska A, Selleslag D, Jang JH,et al. , International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with > 30% blasts, Blood 126 (July (3)) (2015) 291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Quintas-Cardama A, Santos FP, Garcia-Manero G, Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia, Leukemia 25 (February (2)) (2011) 226–235. [DOI] [PubMed] [Google Scholar]
- [9].Braun T, Gardin C, Investigational BET bromodomain protein inhibitors in early stage clinical trials for acute myelogenous leukemia (AML), Expert Opin. Investig. Drugs 26 (July (7)) (2017) 803–811. [DOI] [PubMed] [Google Scholar]
- [10].Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. , RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia, Nature 478 (August (7370)) (2011) 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. , Selective inhibition of BET bromodomains, Nature 468 (December (7327)) (2010) 1067–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Siu KT, Ramachandran J, Yee AJ, Eda H, Santo L, Panaroni C, et al. , Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma, Leukemia 31 (August (8)) (2017) 1760–1769. [DOI] [PubMed] [Google Scholar]
- [13].Chaidos A, Caputo V, Gouvedenou K, Liu B, Marigo I, Chaudhry MS, et al. , Potent antimyeloma activity of the novel bromodomain inhibitors I-BET151 and IBET762, Blood 123 (January (5)) (2014) 697–705. [DOI] [PubMed] [Google Scholar]
- [14].Odore E, Lokiec F, Cvitkovic E, Bekradda M, Herait P, Bourdel F, et al. , Phase I population pharmacokinetic assessment of the oral bromodomain inhibitor OTX015 in patients with haematologic malignancies, Clin. Pharmacokinet 55 (March (3)) (2016) 397–405. [DOI] [PubMed] [Google Scholar]
- [15].Bui MH, Lin X, Albert DH, Li L, Lam LT, Faivre EJ, et al. , Preclinical characterization of BET family bromodomain inhibitor ABBV-075 suggests combination therapeutic strategies, Cancer Res 77 (June (11)) (2017) 2976–2989. [DOI] [PubMed] [Google Scholar]
- [16].Millan DSAMM, Barr KJ, Cardillo D, Collis A, Dinsmore CJ, Escobedo JA, Foley KP, Herbertz T, Hubbs S, Kauffman GS, Kayser-Bricker KJ,Kershaw MT, et al. , FT-1101: a structurally distinct Pan-BET bromodomain inhibitor with activity in preclinical models of hematologic malignancies, Blood (2015) 126. [Google Scholar]
- [17].Grieselhuber NRMS, Orwick S, Harrington BK, Goettl VM, Walker AR,Bhatnagar B, Mims AS, Klisovic RB, Vasu S, Blum W, Blachly JS, Lucas DM, et al. , The novel BET inhibitor PLX51107 has in vitro and in vivo activity against acute myeloid leukemia, Blood (2016) 128. [Google Scholar]
- [18].Jung M, Gelato KA, Fernandez-Montalvan A, Siegel S, Haendler B, Targeting BET bromodomains for cancer treatment, Epigenomics 7 (3) (2015) 487–501. [DOI] [PubMed] [Google Scholar]
- [19].Dawson MA, Gudgin EJ, Horton SJ, Giotopoulos G, Meduri E, Robson S, et al. , Recurrent mutations, including NPM1c, activate a BRD4-dependent core transcriptional program in acute myeloid leukemia, Leukemia 28 (2) (2014) 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, et al. , BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD, Mol. Cancer Ther 13 (October (10)) (2014) 2315–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pingle SC, Sultana Z, Pastorino S, Jiang P, Mukthavaram R, Chao Y, et al. , In silico modeling predicts drug sensitivity of patient-derived cancer cells, J. Transl. Med 21 (May 12) (2014) 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Drusbosky L, Medina C, Martuscello R, Hawkins KE, Chang M, Lamba JK, et al. , Computational drug treatment simulations on projections of dysregulated protein networks derived from the myelodysplastic mutanome match clinical response in patients, Leuk. Res 52 (January) (2017) 1–7. [DOI] [PubMed] [Google Scholar]
- [23].Cogle CRTK, Vali S, Kumar A, Singh NK, Tiwari KK, Tyagi A, Abbasi T, Sayeski PP, A novel simulation method for mapping dysregulated pathways and predicting effective therapeutics in the myelodysplastic syndromes, Blood (2014) 124. [Google Scholar]
- [24].Lanzel EA, Paula Gomez Hernandez M, Bates AM, Treinen CN, Starman EE, Fischer CL, et al. , Predicting PD-L1 expression on human cancer cells using next-generation sequencing information in computational simulation models, Cancer Immunol. Immunother 65 (December (12)) (2016) 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Doudican NA, Kumar A, Singh NK, Nair PR, Lala DA, Basu K, et al. , Personalization of cancer treatment using predictive simulation, J. Transl. Med 1 (13) (2015. February) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kobayashi SS, Vali S, Kumar A, Singh N, Abbasi T, Sayeski PP, Identification of myeloproliferative neoplasm drug agents via predictive simulation modeling: assessing responsiveness with micro-environment derived cytokines, Oncotarget 7 (June (24)) (2016) 35989–36001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sim NL, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC, SIFT web server: predicting effects of amino acid substitutions on proteins, Nucleic Acids Res 40 (July (Web Server issue)) (2012) W452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. , A method and server for predicting damaging missense mutations, Nat. Methods 7 (April (4)) (2010) 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Martelotto LG, Ng CK, De Filippo MR, Zhang Y, Piscuoglio S, Lim RS, et al. , Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations, Genome Biol 15 (October (10)) (2014) 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Reva B, Antipin Y, Sander C, Predicting the functional impact of protein mutations: application to cancer genomics, Nucleic Acids Res 39 (Sepember (17)) (2011) e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Choi Y, Sims GE, Murphy S, Miller JR, Chan AP, Predicting the functional effect of amino acid substitutions and indels, PLoS One 7 (10) (2012) e46688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. , BET bromodomain inhibition as a therapeutic strategy to target c-Myc, Cell 146 (September (6)) (2011) 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Philpott M, Yang J, Tumber T, Fedorov O, Uttarkar S, Filippakopoulos P, et al. , Bromodomain-peptide displacement assays for interactome mapping and inhibitor discovery, Mol. Biosyst 7 (October (10)) (2011) 2899–2908. [DOI] [PubMed] [Google Scholar]
- [34].Davis RJ, Transcriptional regulation by MAP kinases, Mol. Reprod. Dev 42 (December (4)) (1995) 459–467. [DOI] [PubMed] [Google Scholar]
- [35].Zhu B, Xiahou Z, Zhao H, Peng B, Zhao H, Xu X, MTHFR promotes heterochromatin maintenance, Biochem. Biophys. Res. Commun 447 (May (4)) (2014) 702–706. [DOI] [PubMed] [Google Scholar]
- [36].Ferlazzo N, Curro M, Zinellu A, Caccamo D, Isola G, Ventura V, et al. , Influence of MTHFR genetic background on p16 and MGMT methylation in oral squamous cell Cancer, Int. J. Mol. Sci 18 (March (4)) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR, BET bromodomain inhibition suppresses the function of hematopoietic transcription factors in acute myeloid leukemia, Mol. Cell 58 (June (6)) (2015) 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang R, You J, Mechanistic analysis of the role of bromodomain-containing protein 4 (BRD4) in BRD4-NUT oncoprotein-induced transcriptional activation, J. Biol. Chem 290 (January (5)) (2015) 2744–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Jang JE, Eom JI, Jeung HK, Cheong JW, Lee JY, Kim JS, et al. , Targeting AMPK-ULK1-mediated autophagy for combating BET inhibitor resistance in acute myeloid leukemia stem cells, Autophagy 13 (April (4)) (2017) 761–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Turkington RC, Longley DB, Allen WL, Stevenson L, McLaughlin K,Dunne PD, Blayney JK, Salto-Tellez M, Van Schaeybroeck S, Johnston P, Fibroblast growth factor receptor 4 (FGFR4): a targetable regulator of drug resistance in colorectal cancer, Cell Death Dis 6 (February 5) (2014) e1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O’Brien S,Gabbasov R, Fink LS, Li Y, Lounsbury N, Abou-Gharbia M, Childers WE, et al. , Resistance to BET Bromodomain inhibitors is mediated by kinome reprogramming in Ovarian Cancer, Cell Rep 16 (August (5)) (2016) 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chen BJ, Wu YL, Tanaka Y, Zhang W, Small molecules targeting c-Myc onco-gene: promising anti-cancer therapeutics, Int. J. Biol. Sci 10 (10) (2014) 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schick M, Habringer S, Nilsson JA, Keller U, Pathogenesis and therapeutic targeting of aberrant MYC expression in haematological cancers, Br. J. Haematol 179 (December (5)) (2017) 724–738. [DOI] [PubMed] [Google Scholar]
- [44].Yasuda T, Sanjo H, Pages G, Kawano Y, Karasuyama H, Pouyssegur J, Ogata M,Kurosaki T, Erk kinases link pre-B cell receptor signaling to transcriptional events required for early B cell expansion, Immunity 28 (April (4)) (2008) 488–490. [DOI] [PubMed] [Google Scholar]
- [45].Tsai WB, Aiba I, Long Y, Lin HK, Feun L, Savaraj N, Kuo MT, Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosucci-nate synthetase, leading to arginine deiminase resistance in melanoma cells, Cancer Res 72 (May (10)) (2012) 2622–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sashida G, Wang C, Tomioka T, Oshima M, Aoyama K, Kanai A, et al. , The loss of Ezh2 drives the pathogenesis of myelofibrosis and sensitizes tumor-initiating cells to bromodomain inhibition, J. Exp. Med 213 (July (8)) (2016) 1459–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tyner JW, et al. , Functional genomic landscape of acute myeloid leukaemia, Nature 17 (October) (2018), 10.1038/s41586-018-0623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.