Abstract
Some pathogens and pests deliver small RNAs (sRNAs) into host cells to suppress host immunity. Conversely, hosts also transfer sRNAs into pathogens and pests to inhibit their virulence. Although sRNA trafficking has been observed in a wide variety of interactions, how sRNAs are transferred, especially from hosts to pathogens and pests, is still unknown. Here, we show that host Arabidopsis cells secrete exosome-like extracellular vesicles to deliver sRNAs into fungal pathogen Botrytis cinerea. These sRNA-containing vesicles accumulate at the infection sites and are taken up by the fungal cells. Transferred host sRNAs induce silencing of fungal genes critical for pathogenicity. Thus, Arabidopsis has adapted exosome-mediated cross-kingdom RNA interference as part of its immune responses during the evolutionary arms race with the pathogen.
Small RNAs (sRNAs) are short regulatory RNAs that silence genes with complementary sequences (1). Within an animal, sRNAs can be transported by extracellular vesicles, specific transmembrane proteins, high-density lipoprotein complexes, or gap junctions (2). Within a plant, sRNAs move from cell to cell, presumably through plasmodesmata, and travel systemically through vasculature (3). Remarkably, sRNAs also move between hosts and interacting organisms and induce gene silencing, a phenomenon called cross-kingdom/organism RNA interference (RNAi) (4–10). But how do mobile sRNAs travel across the boundaries between organisms of different taxonomic kingdoms? The gastrointestinal parasite Heligmosomoides polygyrus secretes exosomes, a class of extracellular vesicles, to transport microRNAs (miRNAs) into mammalian cells to suppress host immunity (7). By contrast, the mechanism by which sRNAs are transported from hosts to interacting pathogens and pests is totally unknown.
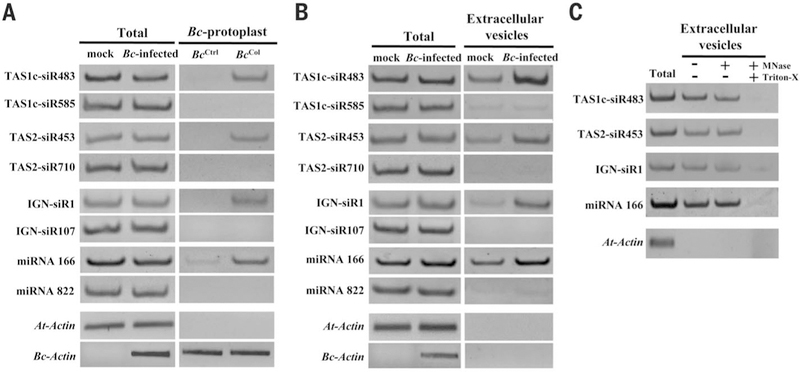
To investigate how host sRNAs move into interacting fungal cells and identify plant endogenous transferred sRNAs, we used the Arabidopsis–Botrytis cinerea pathosystem that displays bidirectional sRNA trafficking and cross-kingdom RNAi (6, 8, 11, 12). Because the cell wall compositions of plants and fungi are different, we developed a sequential protoplast purification method to isolate pure fungal cells from infected tissues (fig. S1). sRNA profiling of purified B. cinerea protoplasts identified 42 Arabidopsis sRNAs in all three biological replicates, using 40 normalized reads per million total reads as a cutoff (table S1). We performed sRNA profiling on total RNAs of Arabidopsis leaves as a control. Although the more abundant sRNAs were more likely to be transported (table S2), there is a clear selection in transferred sRNAs. Among the 42 transferred Arabidopsis sRNAs, 25 were lowly abundant and not among the top 100 sRNAs in the total sRNA libraries (tables S2 and S3). Despite being generated from the same TAS1c mRNA precursor and belong to the top 20 most abundant sRNAs from the total sRNA libraries, trans-acting small interfering RNA locus 1C-derived siRNA483 (TAS1c-siR483) but not TAS1c-siR585 was enriched in the fungal protoplasts (tables S2 and S3). Similarly, although TAS2-siR710 had 30 times higher reads in the total sRNA libraries than TAS2-siR453 that derived from the same TAS2 precursor, only TAS2-siR453 was present in the fungal protoplasts. Furthermore, Arabidopsis heterochromatic sRNAs that derived from intergenic regions, such as IGN-siR1 but not IGN-siR107, were enriched in B. cinerea cells, although IGN-siR107 accumulated to a higher level in the total sRNA libraries (tables S2 and S3). These results were validated by means of sRNA reverse transcription polymerase chain reaction (RT-PCR) analysis (Fig. 1A). Thus, host cells transferring endogenous sRNAs into fungal cells are not simply through concentration-dependent diffusion but possibly through a more selective process.
Fig. 1. Plant endogenous sRNAs are transferred into fungal cells via extracellular vesicles.
(A) TAS1c-siR483, TAS2-siR453, IGN-siR1, and miRNA166 were detected by means of sRNA semiquantitative RT-PCR in B. cinerea protoplasts purified from B. cinerea–infected Col-0 Arabidopsis (BcCol). As a negative control, cultured B. cinerea mixed with uninfected leaves was subjected to the same procedure (BcCtrl). (B) These host sRNAs were also in Arabidopsis extracellular vesicles. (C) Arabidopsis sRNAs were detected in micrococcal nuclease–treated extracellular vesicles. In (A) to (C), TAS1c-siR585, TAS2-siR710, IGN-siR107, miRNA822, and Actin genes were used as controls. The “total” lane indicates total RNAs from leaves. Similar results were obtained from two biological replicates.
Extracellular vesicles are implicated in systemic sRNA transport in animals (13). To test whether plants secrete vesicles to transfer sRNAs into fungal cells, we profiled vesicle sRNAs isolated from the extracellular apoplastic fluids of infected leaves. In all three biological replicates, TAS1c-siR483, TAS2-siR453, and IGN-siR1 accumulated to higher levels than TAS1c-siR585, TAS2-siR710, and IGN-siR107, respectively (Fig. 1B and tables S2 and S4). This was similar to fungal protoplast results. Of the 42 transferred host sRNAs, 31 (73.8%) were present in vesicle libraries (table S4). These results support a positive correlation between the sRNA profiles from extracellular vesicles and the fungal cells. Nuclease protection assay showed that transferred host sRNAs were protected from nuclease digestion unless vesicles were ruptured with Triton X-100 (Sigma, St. Louis, Missouri) (Fig. 1C) (7, 14), confirming that transferred sRNAs are indeed inside the vesicles instead of bound to the surface. These findings indicate that plant cells secrete extracellular vesicles to transfer sRNAs into fungal cells.
Animal extracellular vesicles are classified into exosomes, shedding microvesicles and apoptotic bodies by their specific protein markers and origins (13), whereas plant extracellular vesicles remain undefined. Because exosomes are involved in transferring miRNAs between animal cells (14, 15) and from the parasite H. polygyrus to its mammalian host (7), we sought to determine whether plants also use exosome-like vesicles to transfer sRNAs. Mammalian tetraspanins, such as CD63, are specific exosome markers (16). Arabidopsis has 17 TETRASPANIN (TET)–like genes (17), but only closely related TET8 and TET9 are induced through B. cinerea infection (fig. S2A) (18). TET8 and TET9 are structurally similar to mammalian CD63 (fig. S2B) (17).
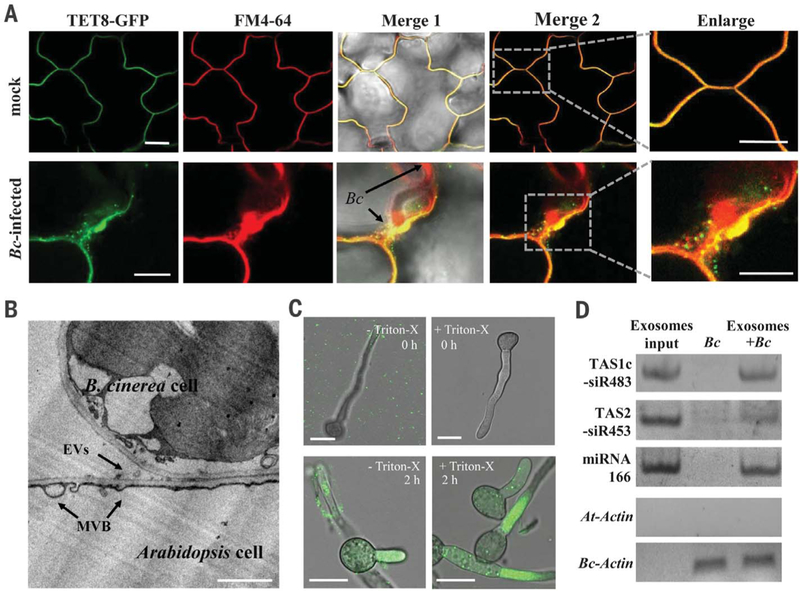
In transgenic plants expressing TET8-GFP under its native promoter, TET8 accumulated at the fungal infection sites (Fig. 2A and fig. S3). Mammalian exosomes are derived from multivesicular bodies (MVBs) (13, 19). We found that Arabidopsis MVB marker Rab5-like guanosine triphosphatase (GTPase) ARA6 (20) was enriched at infection sites (fig. S4) and partially colocalized with TET8 (fig. S5), suggesting that TET8-associated vesicles are likely derived from MVBs. By using transmission electron microscopy, we observed MVBs fusing with plasma membrane to release the extracellular vesicles at the infection sites (Fig. 2B and fig. S6A). Furthermore, TET8-associated vesicles are secreted because numerous green fluorescent protein (GFP)–labeled apoplastic vesicles were observed from TET8-GFP plants but none from ARA6-GFP plants (fig. S6B). TET8, but not ARA6, was detected in extracellular vesicles (fig. S6C) because ARA6 localizes to MVB outer membranes that fused to and remained on plasma membranes. Thus, TET8-labeled extracellular vesicles can be considered plant exosomes.
Fig. 2. Tetraspanin-associated exosomes are responsible for transferring plant sRNAs to fungal cells.
(A) B. cinerea induces accumulation of TET8-GFP–labeled vesicles at the sites of infection. Short staining of FM4–64 shows extracellular membrane structures and plasma membranes. (B) Transmission electron microscopic image of Arabidopsis extracellular vesicles (EVs) near the B. cinerea infection sites. Scale bars, 1 mm. (C) TET8-GFP–labeled exosomes were taken up by B. cinerea within 2 hours of co-incubation. Scale bars, (A) and (C), 10 μm. (D) Plant sRNAs were detected in B. cinerea after uptake of the exosomes.
To test whether plant exosomes can be taken up by fungal cells, we incubated B. cinerea cells with isolated TET8-GFP–labeled exosomes in vitro. TET8-GFP was detected in fungal cells within 2 hours and remained in the fungal cells after rupturing exosomes with Triton X-100 (Fig. 2C), indicating that fungal cells can efficiently take up plant exosomes. Furthermore, the exosome-carried sRNAs were detected in the fungal cells (Fig. 2D). These results support that plants secrete exosomes to transfer sRNAs into fungal cells.
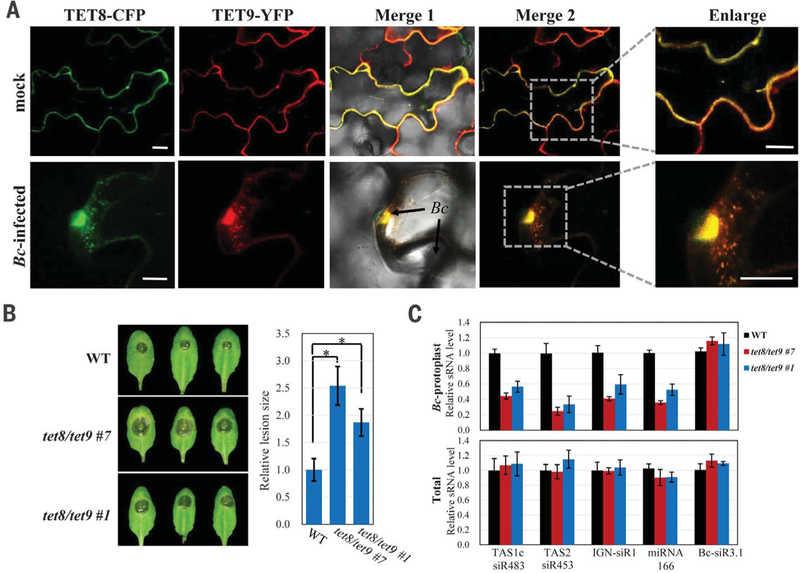
Animal tetraspanin proteins often function together and form specific membrane microdomains (21). We found that TET8-CFP and TET9-YFP were colocalized to the infection sites (Fig. 3A and fig. S7, A and B). TET8-TET9 association was confirmed with reciprocal coimmunoprecipitation when transiently coexpressed in Nicotiana benthamiana (fig. S7C). To explore the physiological role of TET8 and TET9, we examined their loss-of-function mutants. Both the tet8 transferred DNA knockout mutant (17) and two independent tet9 CRISPR frame-shift lines that we generated displayed weak but consistent enhanced susceptibility to B. cinerea as compared with the wild type and the TET8 complementary lines (fig. S8). We generated the tet8tet9 double mutants by knocking down TET9 expression with artificial miRNA in the tet8 background (fig. S9). Double mutants showed pronounced enhanced susceptibility (Fig. 3B), suggesting that TET8 and TET9 have partial functional redundancy. Furthermore, transferred host sRNA levels in the fungal cells isolated from the tet8tet9 mutants were reduced (Fig. 3C), supporting that TET8-and TET9-associated exosomes contribute to plant immunity against fungal infection by transferring host sRNAs into fungal cells.
Fig. 3. TET8 and TET9 coordinately regulate sRNA secretion and host immunity.
(A) TET8-CFP and TET9-YFP colocalized in vesicles that accumulated at the site of fungal infection. Scale bars, 10 μm. (B) Enhanced susceptibility to B. cinerea was observed in two independent tet8tet9 double mutant lines. Relative lesion sizes were measured at 2 days after infection. Error bars indicate the SD of more than 10 leaves. The asterisks indicate significant difference [analysis of variance (ANOVA) Dunnett’s multiple comparisons, P < 0.01]. (C)Accumulation of host transferred sRNAs was decreased in the purified fungal protoplasts isolated from the infected tet8tet9 mutants as compared with that from the wild type. In fungal protoplasts and total RNA samples, Bc-Actin and At-Actin were used as internal controls, respectively. The B. cinerea sRNA Bc-siR3.1 serves as a control.
To determine whether transferred sRNAs affect fungal virulence, we infected the Arabidopsis triple mutant dcl2/3/4 that impairs the biogenesis of trans-acting siRNAs (tasiRNAs) and heterochromatic siRNAs and the rdr6 mutant that is compromised in tasiRNA formation (22). Both dcl2/3/4 and rdr6–15 mutants were more susceptible to B. cinerea as compared with wild type (Fig. 4, A and B), suggesting that transferred sRNAs inhibit B. cinerea infection. We hypothesize that transferred sRNAs suppress fungal pathogenicity by targeting fungal virulence genes. Of the transferred Arabidopsis sRNAs, 21 have predicted B. cinerea target genes (table S5), with a bias toward vesicle-trafficking pathways (7 out of 32 target genes) (fig. S10). We performed functional analysis on TAS1c-siR483 and TAS2-siR453, which showed selective accumulation in the fungal cells (Fig. 1). TAS1c-siR483 target two B. cinerea genes BC1G_10728 and BC1G_10508, and TAS2-siR453 target BC1G_08464, all of which are involved in vesicle-trafficking pathways. BC1G_10728 encodes vacuolar protein sorting 51 (Bc-Vps51) (23), and its homologous gene plays a key role in Candida albicans virulence (24). BC1G_10508 encodes the large subunit of the dynactin complex (Bc-DCTN1). DCTN binds to kinesin II and dynein and coordinates vesicle trafficking (25). BC1G_08464 encodes a suppressor of actin (SAC1)–like phosphoinositide phosphatase that regulates secretory membrane trafficking (26). As expected, these predicted target genes were down-regulated after infection (fig. S11A), but not in B. cinerea collected from the infected dcl2/3/4 and rdr6 mutants (Fig. 4C). The co-expression assay of host sRNAs and fungal targets in N. benthamiana confirmed that silencing of fungal genes was indeed triggered by host sRNAs, and silencing was abolished when sRNA target sites were mutated (fig. S11B). Fungal Argonaute proteins have ribonuclease activities that cleaves sRNA targets (10, 27). The cleavage products of fungal targets guided by host sRNAs were detected in fungal cells isolated from the infected tissue (fig. S12), indicating that plant sRNAs silence fungal target genes through mRNA cleavage.
Fig. 4. Transferred host sRNAs silence fungal virulence genes and suppress fungal pathogenicity.
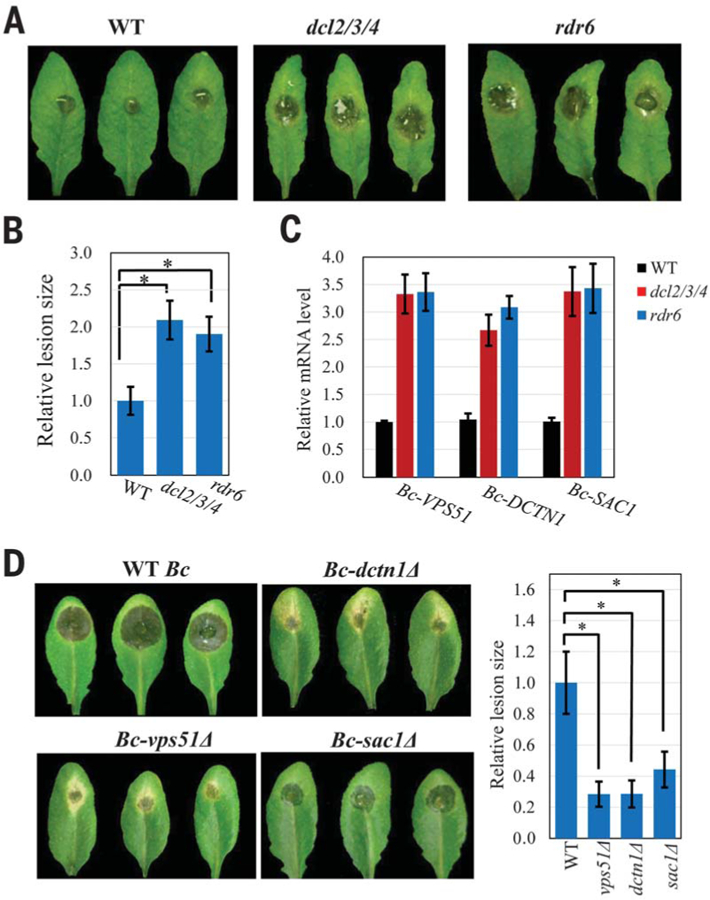
(A and B) compared with the wild type. Relative lesion sizes were measured at 2 days after infection. (C) Fungal target genes of transferred sRNAs were derepressed in B. cinerea collected from the dcl2/3/4 and rdr6 mutants. (D) Mutant B. cinerea strains with deletions in the targets of transferred host sRNAs displayed reduced virulence. Relative lesion sizes were measured at 3 days after infection. In (B) and (D), error bars indicate the SD over 10 leaves. The asterisk indicates significant difference (ANOVA Dunnett’s multiple comparisons; P < 0.01).
We further assessed whether these vesicle-trafficking target genes are important for fungal virulence by generating vps51∆, dctn1∆, and sac1∆ deletion mutant strains (fig. S13A). All mutant strains showed reduced virulence on Arabidopsis (Fig. 4D). The sac1∆ strain showed no reduction of growth on media (fig. S13B), suggesting that SAC1 has a direct role in fungal virulence and does not act by disrupting fungal growth. Thus, vesicle-trafficking pathways are important for fungal virulence. To further confirm that transferred host sRNAs promote host immunity, we generated transgenic Arabidopsis lines that either overexpress TAS1c-siR483 or TAS2-siR453 or knockdown both TAS1c-siR483 and TAS2-siR453 (figs. S14A and S15A). The over-expression lines of TAS1c-siR483 and TAS2-siR453, but not other unrelated sRNA, displayed reduced susceptibility to B. cinerea (fig. S14, B to D), and B. cinerea from infected tissue showed reduced target gene expression (fig. S14E). Consistently, the short tandem target mimic knockdown lines displayed increased susceptibility to B. cinerea (fig. S15B), and target gene expression was elevated (fig. S15C). These findings further support that transferred host sRNAs contribute to host immunity by silencing fungal genes.
In this study, we report that plant extracellular vesicles, especially exosomes, play an essential role in cross-kingdom sRNA trafficking between Arabidopsis and the fungal pathogen B. cinerea. Arabidopsis secretes exosomes to deliver host sRNAs into fungal cells to silence virulence-related genes. A parallel is seen in humans: Cross-kingdom trafficking of human miRNAs into the parasite Plasmodium falciparum inhibits its pathogenicity genes, which explains why sickle cell anemia patients, who have elevated transferred miRNAs, are more resistant to malaria (28). It is unclear whether human exosomes are also responsible for sRNA delivery to interacting organisms, and how ubiquitously such sRNA trafficking-mediated defense mechanisms exist in animals and plants. Functional studies of host transferred sRNAs will help identify important virulence genes in interacting pathogens and pests. Furthermore, transgene-derived sRNAs are also delivered into fungal cells by extracellular vesicles (fig. S16) (8). Discovery of exosome function in cross-kingdom RNAi may help develop effective delivery methods of pathogen-targeting artificial RNAs with the goal of controlling plant diseases in various pre- and post-harvesting crops.
Supplementary Material
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/360/6393/1126/suppl/DC1
Materials and Methods
Supplementary Text
References (29–43)
ACKNOWLEDGMENTS
We thank N. Raikhel for the ARA6-GFP seeds and constructive suggestions, L. Boavida for tet8 and TET8-GFP seeds, Z. Ma for the B. cinerea knockout vectors, G. Coaker for the CRISPR-Cas9 vector, and N. Thomas for editing the paper.
Funding: This work was supported by grants from National Institute of Health (R01GM093008), National Science Foundation (IOS1557812), and AES-CE (PPA-7517H) to H.J.
Footnotes
Competing interests: H.J. is the inventor on a patent application (US62/573,546) held/submitted by University of California, Riverside, covering control of fungal pathogens by using RNAi-based approaches.
Data and material availability: All data needed to evaluate the conclusions in the paper are present in the paper or the supplementary materials. The sequencing data sets were deposited in the NCBI database (PRJNA431815).
REFERENCES AND NOTES
- 1.Baulcombe D, Nature 431, 356–363 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Mittelbrunn M, Sánchez-Madrid F, Nat. Rev. Mol. Cell Biol 13, 328–335 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molnar A et al. , Science 328, 872–875 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Weiberg A, Bellinger M, Jin H, Curr. Opin. Biotechnol 32, 207–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang M, Thomas N, Jin H, Curr. Opin. Plant Biol 38, 133–141 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weiberg A et al. , Science 342, 118–123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buck AH et al. , Nat. Commun 5, 5488 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang M et al. , Nat. Plants 2, 16151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahid S et al. , Nature 553, 82–85 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Zhang T et al. , Nat. Plants 2, 16153 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Wang M, Weiberg A, Dellota E Jr., Yamane D, Jin H, RNA Biol 14, 421–428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weiberg A, Jin H, Curr. Opin. Plant Biol 26, 87–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Colombo M, Raposo G, Théry C, Annu. Rev. Cell Dev. Biol 30, 255–289 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Valadi H et al. , Nat. Cell Biol 9, 654–659 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Mittelbrunn M et al. , Nat. Commun 2, 282 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathivanan S, Ji H, Simpson RJ, Proteomics J 73, 1907–1920 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Boavida LC, Qin P, Broz M, Becker JD, McCormick S, Plant Physiol 163, 696–712 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferrari S et al. , Plant Physiol 144, 367–379 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostrowski M et al. , Nat. Cell Biol 12, 19–30, 1–13 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Ebine K et al. , Nat. Cell Biol 13, 853–859 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Levy S, Shoham T, Nat. Rev. Immunol 5, 136–148 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Xie Z, Allen E, Wilken A, Carrington JC, Proc. Natl. Acad. Sci. U.S.A 102, 12984–12989 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bonifacino JS, Hierro A, Trends Cell Biol 21, 159–167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Y, Mittal R, Solis NV, Prasadarao NV, Filler SG, PLOS Pathog 7, e1002305 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steinmetz MO, Akhmanova A, Trends Biochem. Sci 33, 535–545 (2008). [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Bankaitis VA, Prog. Lipid Res 49, 201–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Catalanotto C, Azzalin G, Macino G, Cogoni C, Genes Dev 16, 790–795 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaMonte G et al. , Cell Host Microbe 12, 187–199 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/360/6393/1126/suppl/DC1
Materials and Methods
Supplementary Text
References (29–43)