Significance
We demonstrate that multivariate patterns of activity in the human hippocampus during the recognition and cued mental replay of long-term sequence memories contain temporal structure information in the order of seconds. By using an experimental paradigm that required participants to remember the durations of empty intervals between visually presented scene images, our study provides evidence that the human hippocampus can represent elapsed time within a sequence of events in conjunction with other forms of information, such as event content. Our findings complement rodent studies that have shown that hippocampal neurons fire at specific times during the empty delay between two events and suggest a common hippocampal neural mechanism for representing temporal information in the service of episodic memory.
Keywords: hippocampus, CA1, episodic memory, time, functional magnetic resonance imaging
Abstract
There has been much interest in how the hippocampus codes time in support of episodic memory. Notably, while rodent hippocampal neurons, including populations in subfield CA1, have been shown to represent the passage of time in the order of seconds between events, there is limited support for a similar mechanism in humans. Specifically, there is no clear evidence that human hippocampal activity during long-term memory processing is sensitive to temporal duration information that spans seconds. To address this gap, we asked participants to first learn short event sequences that varied in image content and interval durations. During fMRI, participants then completed a recognition memory task, as well as a recall phase in which they were required to mentally replay each sequence in as much detail as possible. We found that individual sequences could be classified using activity patterns in the anterior hippocampus during recognition memory. Critically, successful classification was dependent on the conjunction of event content and temporal structure information (with unsuccessful classification of image content or interval duration alone), and further analyses suggested that the most informative voxels resided in the anterior CA1. Additionally, a classifier trained on anterior CA1 recognition data could successfully identify individual sequences from the mental replay data, suggesting that similar activity patterns supported participants’ recognition and recall memory. Our findings complement recent rodent hippocampal research, and provide evidence that long-term sequence memory representations in the human hippocampus can reflect duration information in the order of seconds.
Space and time are significant dimensions of our episodic memories (1). However, while much is known about the neural substrates that contribute to spatial cognition and memory (2–4), relatively little is known about how the brain, in particular the medial temporal lobe (MTL), processes temporal information in the service of episodic memory. The discovery of rodent hippocampal time cells (5–7), which fire at specific moments during the empty delay between two events, suggests a potential hippocampal mechanism for representing the temporal structure of memories (8). Crucially, however, it is unclear whether a similar hippocampal mechanism supports human memory.
Because time cells in the hippocampus (HPC) of the rodent signal the passage of time in the order of seconds, one would expect that a similar neural mechanism in humans would lead to the human HPC representing temporal duration information in the order of seconds in the context of episodic memory. To our knowledge, however, there is no existing evidence for this. No work has examined human HPC involvement in memory for temporal durations in the order of seconds within the context of long-term memory. While recent human investigations have focused on HPC contributions to the representation of temporal order, context, and distance (9–12), there remains little insight into the hypothesized involvement of the human HPC in memory for temporal durations that spans seconds. Furthermore, studies that have investigated duration memory using short-term working memory tasks (13–16) have often observed HPC involvement in relatively long (i.e., greater than ∼90 s) but not shorter durations, which runs counter to the proposed characteristics of time cells and the fact that studies (5–7) have observed moment-to-moment time cell firing during delay periods that are less than 20 s in duration. Notably, because the HPC is suggested to be critical for sequence processing and the temporal binding of temporally discrete events (17–21), the involvement of this structure in representing temporal durations may be contingent on such information being embedded within a set of contiguous sequence events. Suggestive of this, we recently demonstrated using functional magnetic resonance imaging (fMRI) that human HPC activity is sensitive to changes in the durations of short intervals within event sequences (22, 23). Importantly, however, because these findings were in the context of a trial-unique match–mismatch working memory paradigm, their relevance to long-term memory is unknown and it remains to be seen whether human HPC activity contains an abstract code for duration information contained within sequences.
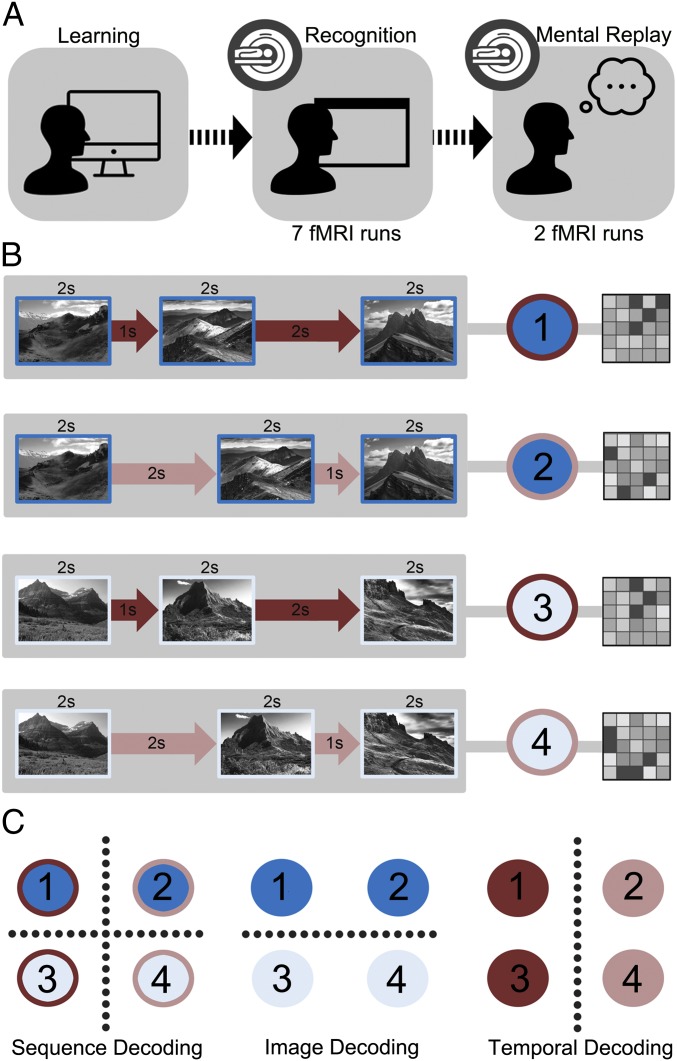
To bridge this gap between the rodent and human literature, and determine whether the human HPC represents duration information in support of long-term memory, the present study employed a sequence-based memory paradigm with high-resolution fMRI (Fig. 1A). Participants first learned four distinct event sequences, each 9 s in length. Image identity (two series of three scene images) and temporal duration (two series of intervals separating each image) were manipulated orthogonally in a 2 × 2 factorial design (Fig. 1B). Thus, each individual sequence could only be remembered by combining information about the images and the temporal structure. During fMRI scanning, participants were then administered a recognition test for these sequences and were also cued to mentally replay each sequence. (Our use of the term “mental replay” is intended to capture the requirement of participants to play out each sequence in their mind in as much pictorial and temporal detail as possible during the recall task, and is distinct from the term “hippocampal replay,” which is often used to refer to the sequential reactivation of HPC place cells associated with recent spatial experience, during sleep or wakefulness when the animal is stationary.) We explored whether the manipulation of image identity and temporal duration resulted in differential multivoxel patterns in the HPC. Specifically, we predicted that if sequence representations in the HPC contained information about temporal durations in the order of seconds, then a classifier trained on data from this region would be successful in decoding individual sequences (Fig. 1C).
Fig. 1.
(A) Structure of experimental paradigm. (B) Participants were required to learn, recognize and mentally replay (i.e., recall) four distinct sequences within a 2 × 2 factorial design (two sets of three images and two sets of interval durations were used). For fMRI analyses, each entire sequence (presented during the recognition task or mentally replayed) was modeled as a single event to create a single t-statistic map. (C) Classification analyses explored four-way sequence, two-way image, and two-way temporal decoding of recognition fMRI data using a leave-one-run-out cross-validation method. Four-way sequence decoding was conducted on the replay data using a cross-classification approach (i.e., train on recognition data, test on replay data).
Results
During fMRI scanning, participants were presented with the sequences that they had learned across two prescanning sessions (∼24 h and immediately before scanning), and were required to identify each one with a button press. Participants exhibited good recognition memory: the mean accuracies across participants for the four sequences were 0.88 (SD = 0.11), 0.86 (SD = 0.10), 0.81 (SD = 0.19), and 0.85 (SD = 0.10), respectively, and performance did not differ significantly across sequences [F(3, 48) = 1.27, P = 0.29]. To ensure that participants made their recognition memory judgments on the entirety of each sequence, we also included challenging catch trials (11.11% of trials) in which the two intervals within the presented sequence were identical. Participants were required to refrain from responding on the catch trials, and were able to do so on the majority of these trials (mean accuracy = 0.71, SD = 0.20).
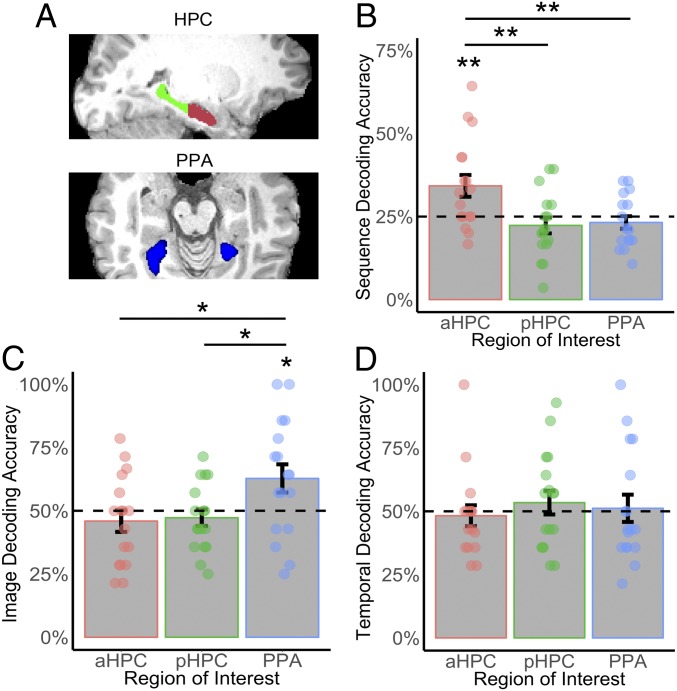
To examine the representations underlying successful sequence recognition, we analyzed multivoxel patterns of activity associated with correct trials. An average voxel-wise response map based on t-statistics was generated for each sequence type (i.e., all images and durations within a 9-s sequence) and acquisition run, incorporating correctly recognized trials only (mean 17.64 of 112 trials excluded per participant due to an incorrect or missing response) (Methods). Given our a priori hypothesis, we focused our analyses on the HPC, using separate anterior (aHPC) and posterior (pHPC) regions-of-interest (ROI) to take into consideration the functional and anatomical distinctions along the longitudinal axis of this structure (4, 24, 25) (Fig. 2A). The parahippocampal place area (PPA) was also chosen as an ROI, given our use of scene images and the involvement of this cortical region in processing scene information (26, 27).
Fig. 2.
A priori ROI results for classification analyses on recognition task data (train and test on recognition data using leave-one-run-out cross-validation). (A) The aHPC (red), pHPC (green), and PPA (blue) were used as ROIs. (B) Sequence decoding accuracy. (C) Image decoding accuracy. (D) Temporal decoding accuracy. Dashed lines indicate chance performance. Error bars depict SE, **P < 0.01, *P < 0.05.
First, we investigated whether multivoxel patterns in our selected ROIs reflected sequence-specific neural activity (i.e., combined event and temporal duration information). To this end, we used a four-way classifier to determine whether it was possible to decode individual sequences (“sequence decoding”). An ANOVA of classification performance across participants and ROIs revealed a significant effect of region [F(2, 45) = 6.73, P = 0.0033], driven by above-chance classification in the aHPC [t(16) = 2.82, P = 0.0020, one-tailed], but not the pHPC [t(16) = −1.10, P = 0.85, one-tailed] or the PPA [t(15) = −0.72, P = 0.75, one-tailed] (Fig. 2B). Sequence decoding accuracy was also significantly greater in the aHPC compared with the pHPC [t(16) = 3.13, P = 0.0070, two-tailed] and PPA [t(15) = 2.63, P = 0.013, two-tailed], highlighting sequence-specific neural activity in the aHPC but not the other ROIs.
Next, we examined whether successful decoding could be achieved based on scene (“image decoding”) or temporal duration (“temporal decoding”) information only (i.e., two-way decoding). An ANOVA of mean image decoding accuracy revealed a significant effect of region [F(2, 45) = 4.13, P = 0.036], with classification being significantly above chance in the PPA [t(15) = 2.08, P = 0.027, one-tailed], but not in the aHPC [t(16) = −0.94, P = 0.816, one-tailed] or pHPC [t(16) = −0.83, P = 0.79, one-tailed]. Two-way image decoding accuracy in the PPA was also significantly higher compared with aHPC [t(15) = 2.14, P = 0.037, two-tailed] and pHPC [t(15) = 2.21, P = 0.027, two-tailed] (Fig. 2C). In contrast, an ANOVA of mean temporal decoding accuracy revealed that there was no significant effect of region [F(2, 45) = 0.34, P = 0.75], with classification performance not being reliable for any of the ROIs (all ts ≤ 0.73, Ps ≥ 0.22, one-tailed) (Fig. 2D). Thus, activity in the PPA only captured image information, with none of the ROIs signaling temporal information alone. Crucially, these two-way classification findings highlight that successful decoding in the aHPC was dependent on the conjunction of image content and temporal structure information.
While we took steps to equate our images on low-level properties and demonstrated that the two differing stimulus onsets did not trigger differing hemodynamic responses (Methods), we also sought to discount the possibility that classification performance could arise due to differences in stimuli or presentation timing that were inherent to our experimental manipulations. To this end, we examined the possibility that our four sequences may differ in their overall univariate response level. This analysis did not reveal a difference in overall activity, making it unlikely that our classification performance reflected differential univariate activity associated with each sequence.
In the light of significant four-way sequence decoding in the HPC, we employed a searchlight analysis to identify the most informative voxels within this structure. A three-voxel radius searchlight was applied in conjunction with a four-way classifier to decode individual sequences across a bilateral MTL ROI, which also allowed us to examine the potential involvement of other mnemonic structures within this region. Converging with our ROI-based classification findings, this revealed a single significant cluster in the right aHPC, in the region of the CA1 subfield [P < 0.05 familywise error-corrected; 26 voxels; peak voxel P = 0.019, x = 29, y = −11, z = −24] (Fig. 3A). Notably, exploring our searchlight findings further using a P ≤ 0.001 uncorrected threshold revealed an additional cluster in the right lateral entorhinal cortex (39 voxels; peak voxel P = 0.001 uncorrected, x = 21, y = −1, z = −38) (Fig. 3B). No other clusters were observed in the MTL.
Fig. 3.
A searchlight analysis for four-way sequence decoding (train and test on recognition data using leave-one-run-out cross-validation) was conducted across a bilateral MTL mask, revealing: (A) a significant cluster of voxels in the right aHPC in the region of CA1 at P < 0.05 small volume corrected; and (B) an additional cluster of voxels in the right lateral entorhinal cortex at P ≤ 0.001 uncorrected (thresholded at P < 0.005 uncorrected for display purposes). Clusters are rendered on a MNI-152 template (R, right hemisphere), and participant group probabilistic map of hippocampal subfields (thresholded at 50%) is shown in A, Insets.
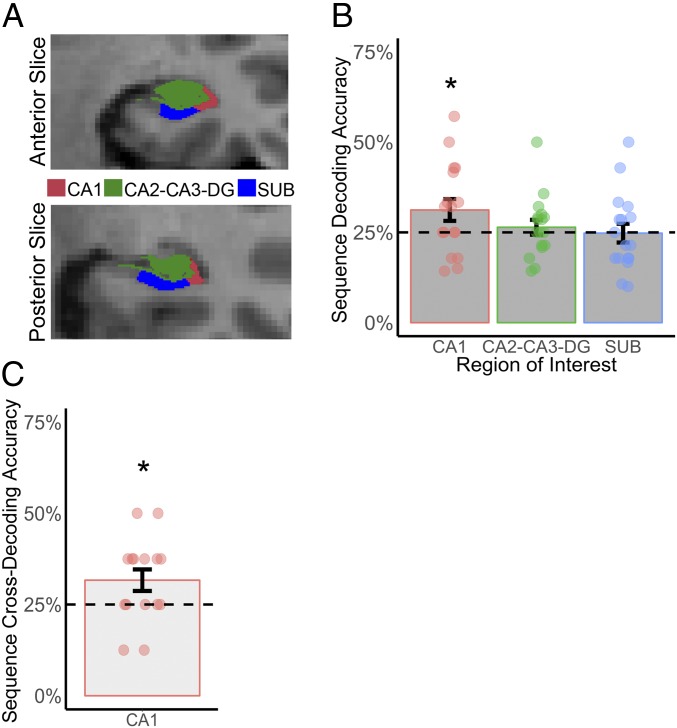
Our searchlight findings highlighted the possibility of wider differences across aHPC subfields in the representation of long-term sequence information. To investigate this, we conducted a post hoc analysis to examine four-way sequence decoding accuracy across participant-specific subfield ROIs, including the anterior CA1, CA2-CA3-dentate gyrus (CA2-CA3-DG), and subiculum (Fig. 4A). While there was no overall effect of subfield in sequence decoding accuracy [F(2, 46) = 1.77, P = 0.23], classification accuracy was significantly above chance in the anterior CA1 [t(16) = 2.06, P = 0.021, one-tailed], but not the anterior CA2-CA3-DG [t(16) = 0.69, P = 0.24, one-tailed] or anterior subiculum [t(16) = −0.083, P = 0.531, one-tailed] (Fig. 4B).
Fig. 4.
(A) Anterior CA1, CA2-CA3-DG, and subiculum ROIs in a representative participant. (B) Four-way sequence decoding accuracy of recognition task data (train and test on recognition data using leave-one-run-out cross-validation). (C) Four-way cross-decoding of replay data in anterior CA1 (train on recognition data, test on replay data). Dashed line indicates chance performance. Error bars depict SE, *P < 0.05.
Finally, we examined the multivoxel patterns of activity during cued mental replay of individual sequences. On each trial, participants were asked to mentally replay a target sequence in response to a visual cue, in as much detail and temporal accuracy as possible. Once complete, participants indicated the vividness of their mental replay on a 1–4 scale (4 = high vividness; 1 = low vividness) via a button press (mean = 2.81, SD = 0.70). Participants’ mean response times for sequences 1–4 were 10.45 s (SD = 2.85), 10.50 s (SD = 3.09), 11.06 s (SD = 2.77), and 10.14 s (SD = 2.29), respectively. Similar to the recognition task data, a single t-statistic map was created for each trial, with trials without a response excluded (mean 2.27 of 32 trials excluded per subject). These t-statistic maps were then averaged for each sequence type within each data-acquisition run. Because the same neural representations should be reinstated during recognition and recall memory (28, 29), we hypothesized that a four-way classifier trained on the recognition data should be able to successfully decode individual sequences using data from the recall task. This is indeed what was found. In view of our searchlight findings we focused on anterior CA1 activity and found that cross-decoding accuracy was significantly above chance [t(14) = 2.26, P = 0.022, one-tailed] (Fig. 4C), suggesting that distinct sequence representations within the anterior CA1 underpinned both recognition and recall memory. There was no significant relationship between participants’ average vividness ratings and cross-decoding accuracy (r = −0.091, P = 0.746).
Discussion
Using a sequence memory paradigm together with high-resolution fMRI, we have provided compelling evidence that human HPC long-term memory sequence representations are sensitive to temporal duration information in the order of seconds. Distinct individual sequences consisting of varying image and temporal duration information could be decoded successfully from multivoxel patterns of activity in the aHPC during recognition memory as well as cued mental replay. Critically, successful decoding cannot be explained by differences in low-level stimulus properties, varying hemodynamic responses elicited by differing stimulus onsets, or overall univariate activity, strongly indicating that the successful classification reflects the distinct images and temporal structure of each sequence. These findings complement electrophysiological studies that have demonstrated that rodent HPC time cells represent the temporal structure of events in the order of seconds (5–7), and point toward a similar neural mechanism for representing temporal information in the service of episodic memory in the human HPC.
Our finding that the most informative voxels underlying the four-way decoding of individual sequences reside in CA1 aligns with earlier research that first identified time cells in the same subfield in rodent HPC (6, 7). In this work, different ensembles of CA1 neurons were observed to fire at specific time points during a short interval between two events, effectively bridging the gap between these events. It is plausible, therefore, that the distinct multivoxel patterns of activity associated with each individual sequence in the present study reflect, in part, activity from different neuronal populations signaling the temporal structure within each sequence. Notably, we did not observe successful decoding of temporal structure alone (i.e., two-way classification), suggesting that the human HPC does not code for temporal information in isolation and extends previous rodent work by demonstrating the importance of the HPC in representing conjunctions of information, including temporal durations, in the service of memory (see below for further discussion). Moreover, while rodent studies have examined the characteristics of time cells in the dorsal HPC, our significant four-way sequence classification findings were specific to aHPC, which is suggested to be the human homolog of rodent ventral HPC (24). Although no rodent work has, to our knowledge, identified time cell-like neurons within the rodent ventral HPC, it is interesting to note that a number of human fMRI studies have implicated the human aHPC in processing other aspects of temporal structure in the context of other types of mnemonic tasks. For example, aHPC activity has been associated with processing temporal regularities in statistical learning (i.e., two objects occurring regularly in succession) (30, 31) as well as memory for the temporal distance of autobiographical memories over a period of a month (11). Finally, while our findings point toward a role for CA1 in the context of our experimental paradigm, we cannot discount the contribution of other regions in the representation of duration information in episodic memory. Indeed, recent work has found evidence for time cells beyond CA1, including CA3 (32), as well as the representation of longer time intervals in the order of hours in CA2 (33), suggesting that the representation of temporal information in long-term memory may not be limited to one subfield.
In addition to unsuccessful temporal structure decoding, our classifier was also not able to decode image information alone on the basis of activity within the HPC. This contrasts with the PPA, where significant two-way image decoding was observed, in line with a role for this region in scene processing (26, 27). The lack of successful two-way decoding in the HPC highlights that it is the combination of image and temporal information that is critical for HPC involvement and is consistent with a role for the HPC in associative memory (34, 35) and the representation of conjunctive information (36, 37). Related to this, while human studies have typically failed to observe a role for the HPC in the judgment of single durations in the order of seconds (13–16) (see also ref. 38 for comparable findings in rodent behavioral pharmacological work), recent fMRI work has suggested that short-duration information must be embedded within a sequence to elicit the involvement of this structure (22, 23), in keeping with the notion that the HPC plays an important role in sequence memory and the binding of discontiguous events (17–21). For example, we recently used fMRI to scan participants while they made match–mismatch judgments of short sequences of scene images presented before (study phase) and after (test phase) a jittered 3.5-s delay (23). In support of the idea that the hippocampus represents temporal duration information within sequences, there was a significant change in study-test HPC pattern similarity when the temporal durations within a sequence were altered. The present study provides a significant advance beyond this work by demonstrating the involvement of the HPC in representing duration information in conjunction with other types of information (such as image content in this study and temporal order) in the context of long-term sequence memory. Our ability to decode individual sequences provides evidence that the HPC incorporates temporal duration information into sequence representations, making it unlikely that classification performance is driven merely by domain-general mnemonic processing. Specifically, successful performance on the current paradigm likely necessitates HPC-dependent pattern completion (39–41), in which conjunctive sequence information is retrieved in response to previously learned information (i.e., the presentation of a sequence during the recognition task or a presented cue during the mental replay task). This mechanism alone, however, is unlikely to account for a 9-s unfolding of a sequence-specific pattern of activity in the HPC. Instead, our finding of significant four-way sequence classification likely reflects the detection of distinct conjunctive information associated with each sequence.
A key strength of the present study is that we examined HPC patterns of activity associated with the recognition and cued mental replay of sequences. Our finding of successful cross-classification between these two forms of memory retrieval reinforces our interpretation that the observed anterior CA1 activity during successful sequence recognition reflects sequence-specific representations and that these same representations contributed to cued mental replay. Of note, because we manipulated the temporal structure of the sequence memoranda by adjusting the duration of the empty intervals within each sequence, participants were required to encode and retrieve the amount of time between successive image presentations. This design resembles, to a certain extent, the examination of HPC time cell activity during the empty interval between events (6, 42, 43) and differs from recent fMRI paradigms in which the passage of time is intertwined with other factors such as the number of events that have transpired between two time points (12) or the context (e.g., same or different) in which two events have occurred (10). Thus, our study provides evidence that human HPC activity can represent elapsed time in conjunction with other information within a sequence of events in the context of long-term memory and supports the idea of temporal representation in the HPC, in which both the order and durations of events are encoded (44).
Interestingly, our searchlight analysis also revealed a cluster of voxels in lateral entorhinal cortex in association with four-way sequence decoding. This cluster did not survive our a priori-corrected statistical threshold (P < 0.05 small volume correction for bilateral MTL; maximum P = 0.001 uncorrected) but is in line with recent work demonstrating the representation of temporal information in this region in rodents and humans (45–47). Although our present paradigm is unable to highlight the distinct contributions of the lateral entorhinal cortex compared with the HPC, one possibility is that distinct HPC sequence representations reflecting specific events and temporal durations may be supported by input from entorhinal cortex. Indeed, it has been suggested that temporal representations in CA1 may be driven by connections with the entorhinal cortex (48) and recent theoretical frameworks posit that direct (monosynaptic) and indirect (through CA3 and DG via the trisynaptic pathway) entorhinal–CA1 connections are important in establishing temporal sequence representations through experience (49).
A potential limitation of our experimental paradigm is that the manner of sequence learning differs from how everyday episodic memories are typically encoded. Specifically, participants were asked to pay careful attention to the temporal structure of four sequences across many repetitions, which is in contrast to everyday episodic memories that arise from a one-time experience and for which temporal information is processed implicitly. While it is important to stress that our paradigm does have the advantage of assessing memory retrieval without requiring participants to make explicit judgments about temporal duration (i.e., our use of mental replay), it remains to be seen whether our findings can be generalized to more naturalistic memory paradigms in which temporal information is learned incidentally after minimal exposure.
To summarize, we show that multivoxel patterns of HPC activity are sensitive to the temporal structure of a series of successive events that unfold over seconds. Furthermore, this information is stable over multiple stimulus exposures, and can even be decoded from activity associated with active mental replay. These findings highlight the importance of human aHPC, and in particular CA1, in the representation of temporal durations in the order of seconds in long-term sequence memory.
Methods
Participants.
Eighteen neurologically healthy individuals participated in the study (eight female, mean age = 27.00, SD = 5.87), each with normal or corrected-to-normal vision and no history of neurological illness. All subjects gave written informed consent before participation. One subject’s data were subsequently removed due to poor behavioral performance during scanning (multiple runs with 0 trials correct), leaving a final sample of 17 subjects (eight female, mean age = 27.18, SD = 6.00). Because one of our experimental conditions required the mental replay of learned sequences, we assessed participants’ ability to visualize a mental image by administering the Vividness of Visual Imagery Questionnaire (50), a 16-item self-report questionnaire that instructs participants to visually image specific details of specific visual scenes and to record the vividness of the imagery on a five-point scale (1 = “perfectly clear and as vivid as normal vision” to 5 = “no image at all, you only know you are thinking of an object”). No participant received a score greater than 3.75 and the group mean was 2.43 (SD = 0.69). This work received approval from the University of Toronto (#27455) and York University (#2016–291) Ethics Boards.
Experimental Paradigm.
The experimental paradigm consisted of three phases: prescan learning, a scanned recognition task, and finally, scanned active mental replay. Stimuli in the learning and recognition phases were identical and consisted of six grayscale scene images (1,000 × 750 pixels) depicting mountain landscapes presented in the center of a black screen. To ensure that classification of fMRI data was not driven by low-level image properties, all images were normalized using the SHINE toolbox (51). Specifically, luminance histograms were equated using a method that maximized the image quality by optimizing structural similarity to the original image (52). No scene stimuli were presented during the active mental replay condition. Instead, participants were cued to recall these images from memory (see Scanned Mental Replay, below). All experimental tasks were programmed in E-Prime (v2.2; Psychology Software Tools). Learning took place outside of the scanner, with stimuli presented on a 15-inch laptop computer (1,600 × 900-pixel resolution). During scanning, stimuli were projected (1,920 × 1,080-pixel resolution) on a screen at the head of the MRI bore and viewed via a mirror attached to the head coil.
Prescan learning.
Participants took part in one learning session ∼24 h before scanning and another session immediately before entering the scanner. The goal was for participants to learn four different event sequences. Each sequence was 9,000 ms in duration and composed of three scene images separated by two intervals of distinct length. A 2 × 2 factorial design was implemented in which there were two unique sets of three scene images (presented in a fixed order, with each scene shown for 2,000 ms), and two orders of interval duration (1,000 ms followed by 2,000 ms, or vice versa) (Fig. 1A). Our motivation for using the minimum number of sequences necessary for a 2 × 2 factorial design (i.e., 4) was to maximize behavioral performance as well as the amount of fMRI data collected per sequence within a single scanning session. All scene stimuli were counterbalanced across participants.
At the start of the first learning session, participants were administered a viewing task in which they were exposed to a sequence on each trial and asked to pay attention to the content of the images and the durations of the intervals. On each trial, a number (1–4) was first displayed denoting the identity of the sequence to be shown (1,000 ms), and then the sequence was presented. Each sequence was shown six times in total, in a pseudorandomized order, with an intertrial interval (ITI) of 1,000 ms. After this, participants were given an active training task that was designed to encourage memorization of each of the event sequences. On each trial, a sequence was presented, followed by the word “Event?” for 2,000 ms, during which participants indicated which sequence they had just seen using one of four preassigned keys with their right hand. The correct sequence number was then shown for 1,000 ms regardless of the participant’s button press and the next trial started after a 1,000-ms ITI. Importantly, catch trials were also included to make sure that participants were paying attention to both intervals in each sequence. On these trials, the intervals were of the same duration (i.e., both 1,000 or 2,000 ms) and participants were instructed to withhold from responding. There was a blank feedback screen for these trials. Trials were presented in blocks of 10, consisting of 2 trials for each sequence and 2 catch trials pseudorandomly ordered. The task ran until participants got all 10 trials within a block correct.
On completion of the active training task, participants practiced a simulated run of the temporally jittered fMRI recognition task, to ensure they understood the fMRI task. On each trial, a white fixation cross first appeared on screen for 2,000 ms. This cross then turned red for 1,000 ms to signal the upcoming presentation of a sequence, which followed immediately. A white fixation cross was then presented for 3,000 ms, and this subsequently turned red for 1,500 ms. During this red fixation cross, participants were asked to indicate which sequence they had just seen. No feedback was provided and a jittered ITI of 4,700 ms (SD = 1,800 ms) then ensued. There were 18 trials in total (4 trials for each of the 4 sequences, plus 2 catch trials) and each sequence was equally likely to follow every other sequence.
For the second learning session that was conducted just before scanning, participants were required to repeat all tasks from the first day. This included the initial sequence-exposure task, the active training task, and finally a practice run of the scanned recognition task.
It is important to highlight that before the start of the first learning session, all participants were explicitly instructed not to use any verbal strategies during the course of the experiment, including labeling the scenes or durations, or counting time. A debriefing session following the final fMRI scan indicated that all subjects bar one adhered to these instructions. This sole participant’s data were, in any case, not included in the statistical analyses due to poor behavioral performance.
Scanned recognition.
Participants completed seven fMRI runs of the recognition task (Fig. 1B) with run order counterbalanced across participants. As in the prescan practice task, there were 18 trials in each run (4 trials for each of the 4 sequences, plus 2 catch trials) and each sequence was equally likely to follow every other sequence. Responses were made using a four-button MR-safe response box placed in the right hand.
Scanned mental replay.
After the sequence-recognition task, participants underwent two additional fMRI runs during which they were required to recall and mentally replay the four learned sequences with as much image detail and as temporally accurate as possible. Each trial started with a 1,000-ms red fixation cross followed by a 1000-ms cue (nos. 1–4) informing the participant which of the four sequences they were required to recall. Participants were then given a 20-s window to mentally replay the target sequence while keeping their eyes open. To mark the completion of their mental replay, participants made a button press that also indicated the vividness of their mental replay on a 1–4 scale, with vividness referring to both the fidelity of the images and the temporal information recalled (4 = high vividness; 1 = low vividness). Each fMRI run consisted of 16 trials in total (4 trials for each of the 4 sequences presented in a pseudorandom order, with each cue equally likely to follow every other cue). Data from 15 of the 17 participants were included in the statistical analysis of the mental replay data: one participant misunderstood task instructions and did not provide any vividness button presses, and mental replay data collection was incomplete for another participant who had to exit the scanner early.
Scanning Procedure.
All imaging data were acquired at the MRI Facility of York University (Keele campus, Toronto, ON, Canada) using a 3T Siemens Tim Trio MRI scanner and a 32-channel head coil. Eleven sets of functional data series (nine experimental, two localizer) were collected from each participant using a T2*-weighted echo-planar imagining sequence with 25 oblique slices acquired in an interleaved order [slice thickness = 1.75 mm, interslice distance = 0 mm, voxel size = 1.5 × 1.5 × 1.75 mm, TR = 2000 ms, TE = 34 ms, matrix size = 128 × 128, field-of-view (FOV) = 192 mm, FA = 78°]. This high-resolution slice acquisition plan yielded a partial volume for each participant, which was angled parallel to the long axis of the hippocampus and captured the temporal and occipital lobes. Each experimental run for the sequence recognition task lasted for 432 s, during which 216 volumes were acquired. Each experimental run for the sequence replay task lasted 360 s and consisted of 180 volumes. Finally, each functional localizer run lasted for 408 s, yielding 204 volumes. The first four scans of each run were discarded to take into consideration the time for the MR signal to reach equilibrium. In addition, each participant received a high-resolution T1-weighted MPRAGE scan (slices =192; voxel size= 1 mm3, TR = 2,300 ms; TE = 2.62 ms; FA = 9°; matrix size = 256 × 256), which was used for registration purposes and HPC delineation (including anterior, posterior, and subfield demarcation).
Neuroimaging Analyses.
Data preprocessing.
All neuroimaging data were preprocessed using FEAT (FMRI Expert Analysis Tool) v6.00 and additional tools from FSL (FMRIB software library; https://fsl.fmrib.ox.ac.uk/fsl/fslwiki) (53). Preprocessing steps included motion correction using MCFLIRT (54), brain extraction using BET (55), grand mean scaling, and high-pass temporal filtering (cut-off frequency of 100 s). No spatial smoothing was applied to the experimental data for multivariate pattern analyses. For univariate analysis of the experimental data, a 3-mm full-width half-maximum (FWHM) Gaussian smoothing kernel was applied. Functional localizer data were smoothed with a 6-mm FWHM Gaussian kernel. Each participant’s functional data were coregistered to their respective high-resolution 3D anatomical scan using a linear transformation as implemented by FLIRT (54) as well as to the Montreal Neurological Institute (MNI) 152 template using FNIRT. Each registration step was visually inspected for accuracy.
Multivariate pattern analyses.
Each event sequence was modeled as a single 9,000-ms event in our general linear model (GLM) and thus, a single-parameter estimate was derived for each sequence. Because we aimed to examine the multivariate patterns of activity associated with each sequence, we wanted to ensure before data collection that any differences in blood-oxygen level-dependent activity would not be driven simply by differences in the hemodynamic response elicited by the shifting onset of the second image in each sequence, a consequence of our interval manipulation. To this end, we simulated the hemodynamic response associated with the two sets of interval durations and found that they were highly correlated (P < 0.0001 correlation), such that any difference between the conditions would be dwarfed by typical noise levels in fMRI data. This precludes the possibility that hemodynamic response sensitivity to differences in event time course are sufficient for classification.
All multivariate analyses were run in native space. Per trial parameter estimates were generated using an iterative least-squares single GLM approach (56) in FEAT. For each trial, a GLM was implemented that included a regressor for that trial as well as another regressor for all other trials. For the sequence-recognition task, the entire sequence presentation (i.e., 9,000 ms) was modeled as a single trial. For the replay task, trial duration was defined as the amount of time that had elapsed from the end of the sequence cue to the participant’s vividness-rating button press, which indicated the end of their mental replay. Each predictor and its temporal derivative were convolved with a double-gamma hemodynamic response function and FILM prewhitening was applied, which included temporal autocorrelation correction. Motion parameters estimated by MCFLIRT (six in total) were also entered as nuisance covariates. Per trial parameter estimate maps were then converted to t-statistic maps to suppress the contribution of noisy voxels that can have potentially high β-estimates (57). This resulted in 112 t-statistic maps for each subject for the sequence recognition task (28 maps per condition, excluding catch trials). For sequence recall, there were 32 t-statistic maps for each subject (8 maps per condition, excluding catch trials). All maps were then spatially realigned to the first map from the recognition-task data to ensure that head movement across runs was minimized (because multivariate analyses involved cross-validation across runs). Per trial t-statistic maps associated with incorrect trials were then removed (i.e., an inaccurate response provided during the recognition task or a failure to provide a vividness button press during the replay task) and the remaining maps were averaged across trials for each condition and within each run to create patterns with improved signal-to-noise ratio. Due to insufficient numbers, incorrect trials were not analyzed further [i.e., a number of participants made very few errors (≤5) on the recognition task or possessed no trials at all on the recall task with a missing button press, and incorrect trials were often distributed unequally across the four different sequences].
A priori ROI classification.
Separate HPC ROIs were created for each participant using FSL’s FIRST automatic segmentation tool (58). The resultant ROIs were visually inspected and edited by hand if necessary to ensure that the HPC was correctly identified, according to published criteria (59). Given the proposed anatomical and functional differences along the longitudinal axis of the HPC (4, 24, 25), these bilateral subject-specific HPC ROIs were then further segmented into anterior (mean = 1082.00 voxels, SD = 140.54) and posterior (mean = 909.82 voxels, SD = 171.09) segments manually, using the uncal apex as an anatomical landmark.
The PPA area was defined for each subject using two independent functional localizer scans in which subjects passively viewed photos of faces, objects, scenes, and scrambled versions of those same stimuli. Thirty-two gray-scale stimuli were presented within each block, each for 400 ms (interstimulus interval 50 ms). Each run included four blocks of faces, four blocks of objects, and four blocks of scenes, all separated by blocks of scrambled images of each category. A blank screen (1,000 ms) was also presented between each block. In subsequent analyses, for each subject a predictor was convolved with a double-gamma model for each stimulus category (scenes, objects, faces) and scrambled versions of each (i.e., six explanatory variables in total). Parameter estimates were created for each regressor as well as for the standard linear contrast of scenes > (faces + objects). The resulting parameter estimate images for each participant were combined in a fixed-effects analysis and thresholded at P < 0.001 uncorrected to identify a contiguous cluster of voxels in the parahippocampal gyrus (60) (mean = 1015.18 voxels, SD = 357.21). One subject did not have a sufficient number of active voxels in the PPA (even at a liberal threshold). Therefore, this subject was not included for all statistical analyses that involved the PPA as an ROI. All ROIs were binarized and coregistered to each subject’s functional data before multivariate classification analyses.
Multivariate pattern analyses were used to assess whether distributed patterns of activity during sequence recognition in any of the ROIs described above could reliably distinguish between sequences based on: (i) image and temporal information (sequence decoding: four unique sequences defined by event and temporal structure); (ii) image information only (image decoding: the two sets of scene images); or (iii) temporal information only (temporal decoding: the two sets of intervals separating the scene images). A linear discriminant analysis (LDA) classifier was used in conjunction with a leave-one-run-out cross-validation method as implemented in CoSMo MVPA (61). For cross-classification analyses, the LDA classifier was trained on seven runs of recognition data and tested on two runs of mental replay data using an anterior CA1 ROI (see HPC Subfield Analysis, below). For each ROI, classification accuracies for each participant were entered into a one-sample t test (one-tailed) against chance. Chance was defined as one/(number of classes), which equated to 50% for the two-way classification analyses (event and temporal structure decoding) and 25% for the four-way classification (individual sequence decoding). Classification accuracies were also compared across regions by using an ANOVA and follow-up pairwise comparisons (see Statistical Tests, below) (requests for data and/or analysis code should be addressed to the corresponding author).
Searchlight analysis.
We used a spherical multivariate searchlight approach (62) to localize the most informative voxels involved in the classification of individual sequences in the HPC. To allow us to explore whether other informative voxels were present in MTL structures beyond the HPC, we applied this analysis to an MTL ROI. This ROI was created using the Oxford–Harvard cortical and subcortical atlases encompassing the HPC, anterior parahippocampal gyrus, and posterior parahippocampal gyrus bilaterally, and thresholded at 50%. This ROI was subsequently realigned to each participant’s native functional space (mean = 2862.24 voxels, SD = 217.82). A three-voxel radius sphere was then centered on every voxel within this ROI and for each sphere, an LDA classifier was applied to obtain a decoding accuracy that was assigned to the center voxel. This procedure resulted in decoding accuracy maps that characterized representations of sequence information across the MTL and each participant’s accuracy map was then normalized to MNI space (1-mm template). Group-level significance testing was conducted using permutation tests as implemented by the “Randomise” function in FSL (63) and threshold-free cluster enhancement (TFCE) using default values for all options other than number of permutations (n = 10,000) (64). A voxel-wise significance threshold of P < 0.05 FWE (small volume corrected) was used.
HPC subfield analysis.
HPC subfields—including CA1, CA2, CA3, DG, and subiculum—were extracted for each subject using FreeSurfer 6.0.0 (https://surfer.nmr.mgh.harvard.edu/) (65) and the HPC subfield segmentation atlas (66) built on ultrahigh resolution (∼0.1 mm isotropic) ex vivo MRI data. This produces subregion volume estimates that closely match volumes derived from histological investigations (66) and has reliable test–retest segmentations (67). The resultant ROIs were visually inspected to ensure that subregions were correctly identified, according to published criteria (68). Consistent with previous studies (30, 69), CA2, CA3, and DG were combined into a single ROI because these subfields could not be adequately distinguished at our functional resolution (1.5 × 1.5 × 1.75 mm). Classification analyses for recognition were then conducted on patterns of activity within CA1 (mean = 336.82 voxels, SD = 30.59), CA2-CA3-DG (mean = 272.29 voxels, SD = 38.87), and the subiculum (91.17 voxels, SD = 16.73) in aHPC, with cross-classification on active mental replay data focused on the anterior CA1 given the searchlight and recognition task subfield classification findings.
Statistical tests.
Because not all classifier accuracies for decoding analyses met parametric assumptions (i.e., data significantly deviated from a normal distribution), statistical tests involved nonparametric procedures. For one-sample t tests, we used a bootstrapping procedure to determine statistical significance (70). For each test, the original classification accuracies across the 17 participants (15 for mental replay analysis) were tested against chance to obtain a parametric t value. Seventeen datasets (15 for mental replay analysis) were then randomly sampled with replacement 10,000 times and an absolute t value was calculated for each sample. A bootstrapped P value was then obtained by calculating the proportion of resampled absolute t values that was higher than the original t value (i.e., two-tailed test). For repeated-measures ANOVAs, statistical significance was determined using randomization procedures. For the ANOVAs this involved computing the parametric F values, permuting the data within each participant, and recomputing the F values on the permuted data. This procedure was then repeated 10,000 times to yield a permuted (null) distribution of F values. We then assessed each of the original F values relative to the permuted distribution to compute the probability of obtaining an F value that is equal to or larger than the original F value. For pairwise t tests, differences between conditions were first calculated for each participant and a group mean was calculated. This procedure was then repeated 10,000 times with random sign-flips on the data to yield a null distribution of absolute differences. The original mean difference was then compared with this null distribution to estimate the likelihood of obtaining a difference equal to or greater than this value (i.e., two-tailed test). All statistics were run using custom scripts in R v3.3.3 (https://www.R-project.org/).
Univariate analysis.
We ran additional analyses to rule out any overall univariate differences in activity between sequences. Each run of the preprocessed, smoothed data from the scanned recognition task for each participant was submitted to a GLM, with the different sequences specified as predictors and convolved with a double-gamma model of the hemodynamic response function. Parameter estimates were generated for contrasts between sequences, which included an effect of event [e.g., (sequence 1 + sequence 2) vs. (sequence 3 + sequence 4)] and temporal structure [e.g., (sequence 1 + sequence 3) vs. (sequence 2 + sequence 4)], as well as an interaction effect [e.g., (sequence 1 − sequence 2) vs. (sequence 3 − sequence 4)]. The parameter estimates for each run for each participant were then combined in a fixed-effects analysis and the resulting statistical images were subsequently combined in a higher-level group analysis. Significance was assessed using TFCE and Randomise (default settings with 10,000 permutations), and a relatively liberal threshold of P < 0.1 (small volume corrected within bilateral HPC or PPA) was applied to explore potential differences in univariate activity across the different sequences.
Acknowledgments
We thank all participants for their time and Joy Williams for assisting with data collection at the York MRI Facility. This work was funded by the Natural Sciences and Engineering Research Council of Canada: Graduate Studentship - Doctoral (to S.T.), Postdoctoral Fellowship (to E.B.O.), Summer Research Award (to J.T.), and Discovery Grant (to A.N. and A.C.H.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.R. is a guest editor invited by the Editorial Board.
References
- 1.Tulving E. Episodic and semantic memory. In: Tulving E, Donaldson W, editors. Organization of Memory. Academic; New York: 1972. pp. 381–402. [Google Scholar]
- 2.Moscovitch M, Nadel L, Winocur G, Gilboa A, Rosenbaum RS. The cognitive neuroscience of remote episodic, semantic and spatial memory. Curr Opin Neurobiol. 2006;16:179–190. doi: 10.1016/j.conb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Bird CM, Burgess N. The hippocampus and memory: Insights from spatial processing. Nat Rev Neurosci. 2008;9:182–194. doi: 10.1038/nrn2335. [DOI] [PubMed] [Google Scholar]
- 4.Zeidman P, Maguire EA. Anterior hippocampus: The anatomy of perception, imagination and episodic memory. Nat Rev Neurosci. 2016;17:173–182. doi: 10.1038/nrn.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pastalkova E, Itskov V, Amarasingham A, Buzsáki G. Internally generated cell assembly sequences in the rat hippocampus. Science. 2008;321:1322–1327. doi: 10.1126/science.1159775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MacDonald CJ, Lepage KQ, Eden UT, Eichenbaum H. Hippocampal “time cells” bridge the gap in memory for discontiguous events. Neuron. 2011;71:737–749. doi: 10.1016/j.neuron.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraus BJ, Robinson RJ, 2nd, White JA, Eichenbaum H, Hasselmo ME. Hippocampal “time cells”: Time versus path integration. Neuron. 2013;78:1090–1101. doi: 10.1016/j.neuron.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichenbaum H. Memory on time. Trends Cogn Sci. 2013;17:81–88. doi: 10.1016/j.tics.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsieh L-T, Gruber MJ, Jenkins LJ, Ranganath C. Hippocampal activity patterns carry information about objects in temporal context. Neuron. 2014;81:1165–1178. doi: 10.1016/j.neuron.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ezzyat Y, Davachi L. Similarity breeds proximity: Pattern similarity within and across contexts is related to later mnemonic judgments of temporal proximity. Neuron. 2014;81:1179–1189. doi: 10.1016/j.neuron.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielson DM, Smith TA, Sreekumar V, Dennis S, Sederberg PB. Human hippocampus represents space and time during retrieval of real-world memories. Proc Natl Acad Sci USA. 2015;112:11078–11083. doi: 10.1073/pnas.1507104112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deuker L, Bellmund JL, Navarro Schröder T, Doeller CF. An event map of memory space in the hippocampus. eLife. 2016;5:e16534. doi: 10.7554/eLife.16534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaw C, Aggleton JP. The ability of amnesic subjects to estimate time intervals. Neuropsychologia. 1994;32:857–873. doi: 10.1016/0028-3932(94)90023-x. [DOI] [PubMed] [Google Scholar]
- 14.Noulhiane M, Pouthas V, Hasboun D, Baulac M, Samson S. Role of the medial temporal lobe in time estimation in the range of minutes. Neuroreport. 2007;18:1035–1038. doi: 10.1097/WNR.0b013e3281668be1. [DOI] [PubMed] [Google Scholar]
- 15.Bueti D, Lasaponara S, Cercignani M, Macaluso E. Learning about time: Plastic changes and interindividual brain differences. Neuron. 2012;75:725–737. doi: 10.1016/j.neuron.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 16.Palombo DJ, Keane MM, Verfaellie M. Does the hippocampus keep track of time? Hippocampus. 2016;26:372–379. doi: 10.1002/hipo.22528. [DOI] [PubMed] [Google Scholar]
- 17.Wallenstein GV, Eichenbaum H, Hasselmo ME. The hippocampus as an associator of discontiguous events. Trends Neurosci. 1998;21:317–323. doi: 10.1016/s0166-2236(97)01220-4. [DOI] [PubMed] [Google Scholar]
- 18.Jensen O, Lisman JE. Hippocampal sequence-encoding driven by a cortical multi-item working memory buffer. Trends Neurosci. 2005;28:67–72. doi: 10.1016/j.tins.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Howard MW, Fotedar MS, Datey AV, Hasselmo ME. The temporal context model in spatial navigation and relational learning: Toward a common explanation of medial temporal lobe function across domains. Psychol Rev. 2005;112:75–116. doi: 10.1037/0033-295X.112.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staresina BP, Davachi L. Differential encoding mechanisms for subsequent associative recognition and free recall. J Neurosci. 2006;26:9162–9172. doi: 10.1523/JNEUROSCI.2877-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sellami A, et al. Temporal binding function of dorsal CA1 is critical for declarative memory formation. Proc Natl Acad Sci USA. 2017;114:10262–10267. doi: 10.1073/pnas.1619657114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barnett AJ, O’Neil EB, Watson HC, Lee ACH. The human hippocampus is sensitive to the durations of events and intervals within a sequence. Neuropsychologia. 2014;64:1–12. doi: 10.1016/j.neuropsychologia.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Thavabalasingam S, O’Neil EB, Lee ACH. Multivoxel pattern similarity suggests the integration of temporal duration in hippocampal event sequence representations. NeuroImage. 2018;178:136–146. doi: 10.1016/j.neuroimage.2018.05.036. [DOI] [PubMed] [Google Scholar]
- 24.Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 2014;15:655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- 25.Poppenk J, Evensmoen HR, Moscovitch M, Nadel L. Long-axis specialization of the human hippocampus. Trends Cogn Sci. 2013;17:230–240. doi: 10.1016/j.tics.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Epstein R, Graham KS, Downing PE. Viewpoint-specific scene representations in human parahippocampal cortex. Neuron. 2003;37:865–876. doi: 10.1016/s0896-6273(03)00117-x. [DOI] [PubMed] [Google Scholar]
- 27.Park S, Chun MM. Different roles of the parahippocampal place area (PPA) and retrosplenial cortex (RSC) in panoramic scene perception. NeuroImage. 2009;47:1747–1756. doi: 10.1016/j.neuroimage.2009.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tulving E, Thomson DM. Encoding specificity and retrieval processes in episodic memory. Psychol Rev. 1973;80:352–373. [Google Scholar]
- 29.Norman KA, O’Reilly RC. Modeling hippocampal and neocortical contributions to recognition memory: A complementary-learning-systems approach. Psychol Rev. 2003;110:611–646. doi: 10.1037/0033-295X.110.4.611. [DOI] [PubMed] [Google Scholar]
- 30.Schapiro AC, Kustner LV, Turk-Browne NB. Shaping of object representations in the human medial temporal lobe based on temporal regularities. Curr Biol. 2012;22:1622–1627. doi: 10.1016/j.cub.2012.06.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schapiro AC, Turk-Browne NB, Norman KA, Botvinick MM. Statistical learning of temporal community structure in the hippocampus. Hippocampus. 2016;26:3–8. doi: 10.1002/hipo.22523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salz DM, et al. Time cells in hippocampal area CA3. J Neurosci. 2016;36:7476–7484. doi: 10.1523/JNEUROSCI.0087-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mankin EA, Diehl GW, Sparks FT, Leutgeb S, Leutgeb JK. Hippocampal CA2 activity patterns change over time to a larger extent than between spatial contexts. Neuron. 2015;85:190–201. doi: 10.1016/j.neuron.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichenbaum H, Otto T, Cohen NJ. The hippocampus—What does it do? Behav Neural Biol. 1992;57:2–36. doi: 10.1016/0163-1047(92)90724-i. [DOI] [PubMed] [Google Scholar]
- 35.Ranganath C. A unified framework for the functional organization of the medial temporal lobes and the phenomenology of episodic memory. Hippocampus. 2010;20:1263–1290. doi: 10.1002/hipo.20852. [DOI] [PubMed] [Google Scholar]
- 36.Horner AJ, Bisby JA, Bush D, Lin W-J, Burgess N. Evidence for holistic episodic recollection via hippocampal pattern completion. Nat Commun. 2015;6:7462. doi: 10.1038/ncomms8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Backus AR, Bosch SE, Ekman M, Grabovetsky AV, Doeller CF. Mnemonic convergence in the human hippocampus. Nat Commun. 2016;7:11991. doi: 10.1038/ncomms11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacobs NS, Allen TA, Nguyen N, Fortin NJ. Critical role of the hippocampus in memory for elapsed time. J Neurosci. 2013;33:13888–13893. doi: 10.1523/JNEUROSCI.1733-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guzowski JF, Knierim JJ, Moser EI. Ensemble dynamics of hippocampal regions CA3 and CA1. Neuron. 2004;44:581–584. doi: 10.1016/j.neuron.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Leutgeb S, Leutgeb JK. Pattern separation, pattern completion, and new neuronal codes within a continuous CA3 map. Learn Mem. 2007;14:745–757. doi: 10.1101/lm.703907. [DOI] [PubMed] [Google Scholar]
- 41.Hindy NC, Ng FY, Turk-Browne NB. Linking pattern completion in the hippocampus to predictive coding in visual cortex. Nat Neurosci. 2016;19:665–667. doi: 10.1038/nn.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.MacDonald CJ, Carrow S, Place R, Eichenbaum H. Distinct hippocampal time cell sequences represent odor memories in immobilized rats. J Neurosci. 2013;33:14607–14616. doi: 10.1523/JNEUROSCI.1537-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naya Y, Suzuki WA. Integrating what and when across the primate medial temporal lobe. Science. 2011;333:773–776. doi: 10.1126/science.1206773. [DOI] [PubMed] [Google Scholar]
- 44.Howard MW, Eichenbaum H. The hippocampus, time, and memory across scales. J Exp Psychol Gen. 2013;142:1211–1230. doi: 10.1037/a0033621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsao A, et al. Integrating time from experience in the lateral entorhinal cortex. Nature. 2018;561:57–62. doi: 10.1038/s41586-018-0459-6. [DOI] [PubMed] [Google Scholar]
- 46.Montchal ME, Reagh ZM, Yassa MA. Precise temporal memories are supported by the lateral entorhinal cortex in humans. Nat Neurosci. 2019;22:284–288. doi: 10.1038/s41593-018-0303-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bellmund JLS, Deuker L, Doeller CF. 2018. Structuring time in human lateral entorhinal cortex. bioXriv, 10.1101/458133. Preprint, posted November 1, 2018.
- 48.Suh J, Rivest AJ, Nakashiba T, Tominaga T, Tonegawa S. Entorhinal cortex layer III input to the hippocampus is crucial for temporal association memory. Science. 2011;334:1415–1420. doi: 10.1126/science.1210125. [DOI] [PubMed] [Google Scholar]
- 49.Schapiro AC, Turk-Browne NB, Botvinick MM, Norman KA. Complementary learning systems within the hippocampus: A neural network modelling approach to reconciling episodic memory with statistical learning. Philos Trans R Soc B. 2016;372:20160049. doi: 10.1098/rstb.2016.0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marks DF. Visual imagery differences in the recall of pictures. Br J Psychol. 1973;64:17–24. doi: 10.1111/j.2044-8295.1973.tb01322.x. [DOI] [PubMed] [Google Scholar]
- 51.Willenbockel V, et al. Controlling low-level image properties: The SHINE toolbox. Behav Res Methods. 2010;42:671–684. doi: 10.3758/BRM.42.3.671. [DOI] [PubMed] [Google Scholar]
- 52.Wang Z, Bovik AC, Sheikh HR, Simoncelli EP. Image quality assessment: From error visibility to structural similarity. IEEE Trans Image Process. 2004;13:600–612. doi: 10.1109/tip.2003.819861. [DOI] [PubMed] [Google Scholar]
- 53.Smith SM, et al. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 54.Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. NeuroImage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- 55.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mumford JA, Turner BO, Ashby FG, Poldrack RA. Deconvolving BOLD activation in event-related designs for multivoxel pattern classification analyses. NeuroImage. 2012;59:2636–2643. doi: 10.1016/j.neuroimage.2011.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Misaki M, Kim Y, Bandettini PA, Kriegeskorte N. Comparison of multivariate classifiers and response normalizations for pattern-information fMRI. NeuroImage. 2010;53:103–118. doi: 10.1016/j.neuroimage.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watson C, et al. Anatomic basis of amygdaloid and hippocampal volume measurement by magnetic resonance imaging. Neurology. 1992;42:1743–1750. doi: 10.1212/wnl.42.9.1743. [DOI] [PubMed] [Google Scholar]
- 60.Epstein R, Kanwisher N. A cortical representation of the local visual environment. Nature. 1998;392:598–601. doi: 10.1038/33402. [DOI] [PubMed] [Google Scholar]
- 61.Oosterhof NN, Connolly AC, Haxby JV. CoSMoMVPA: Multi-modal multivariate pattern analysis of neuroimaging data in Matlab/GNU octave. Front Neuroinform. 2016;10:27. doi: 10.3389/fninf.2016.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kriegeskorte N, Goebel R, Bandettini P. Information-based functional brain mapping. Proc Natl Acad Sci USA. 2006;103:3863–3868. doi: 10.1073/pnas.0600244103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: A primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith SM, Nichols TE. Threshold-free cluster enhancement: Addressing problems of smoothing, threshold dependence and localisation in cluster inference. NeuroImage. 2009;44:83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 65.Fischl B. FreeSurfer. NeuroImage. 2012;62:774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Iglesias JE, et al. Alzheimer’s Disease Neuroimaging Initiative A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. NeuroImage. 2015;115:117–137. doi: 10.1016/j.neuroimage.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whelan CD, et al. Alzheimer’s Disease Neuroimaging Initiative Heritability and reliability of automatically segmented human hippocampal formation subregions. NeuroImage. 2016;128:125–137. doi: 10.1016/j.neuroimage.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yushkevich PA, et al. Hippocampal Subfields Group (HSG) Quantitative comparison of 21 protocols for labeling hippocampal subfields and parahippocampal subregions in in vivo MRI: Towards a harmonized segmentation protocol. NeuroImage. 2015;111:526–541. doi: 10.1016/j.neuroimage.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stokes J, Kyle C, Ekstrom AD. Complementary roles of human hippocampal subfields in differentiation and integration of spatial context. J Cogn Neurosci. 2015;27:546–559. doi: 10.1162/jocn_a_00736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Efron B, Tibshirani R. Bootstrap methods for standard errors, confidence intervals, and other measures of statistical accuracy. Stat Sci. 1986;1:54–75. [Google Scholar]