Significance
Growing evidence suggests that the new family of human ATPases, MORCs, is fundamentally important. Clinical observations directly link MORCs to a number of immunological disorders, Down syndrome, and cancer; however, little is known about the mechanism of action of MORCs at the molecular level. Here, we used a set of complementary approaches to pinpoint the mechanism underlying the catalytic function of MORC3, one of the members of this family. We show that an epigenetic reader, the CW domain of MORC3, prevents binding of the adjacent catalytic domain to DNA in the autoinhibited, off state. MORC3 autoinhibition is released when the CW domain binds to histone H3 tail.
Keywords: MORC3, ATPase, CW, histone, chromatin
Abstract
Microrchidia 3 (MORC3) is a human protein linked to autoimmune disorders, Down syndrome, and cancer. It is a member of a newly identified family of human ATPases with an uncharacterized mechanism of action. Here, we elucidate the molecular basis for inhibition and activation of MORC3. The crystal structure of the MORC3 region encompassing the ATPase and CW domains in complex with a nonhydrolyzable ATP analog demonstrates that the two domains are directly coupled. The extensive ATPase:CW interface stabilizes the protein fold but inhibits the catalytic activity of MORC3. Enzymatic, NMR, mutational, and biochemical analyses show that in the autoinhibited, off state, the CW domain sterically impedes binding of the ATPase domain to DNA, which in turn is required for the catalytic activity. MORC3 autoinhibition is released by disrupting the intramolecular ATPase:CW coupling through the competitive interaction of CW with histone H3 tail or by mutating the interfacial residues. Binding of CW to H3 leads to a marked rearrangement in the ATPase–CW cassette, which frees the DNA-binding site in active MORC3 (on state). We show that ATP-induced dimerization of the ATPase domain is strictly required for the catalytic activity and that the dimeric form of ATPase–CW might cooperatively bind to dsDNA. Together, our findings uncovered a mechanism underlying the fine-tuned regulation of the catalytic domain of MORC3 by the epigenetic reader, CW.
Microrchidia 3 (MORC3) is a member of a new family of human ATPases. Originally identified as an autoantigen in inflammatory myopathies (1–3), it has since been linked to other autoimmune disorders, Down syndrome, and cancer (4–7). Studies using mouse models show that knockout of Morc3 is perinatally lethal, and partial loss of Morc3 results in changes in bone calcium homeostasis and up-regulation of inflammatory pathways (8). In cells, MORC3 has been found to localize to promyelocytic leukemia nuclear bodies (PML-NBs) and is implicated in the recruitment of p53 to PML-NBs to induce cellular senescence (9, 10). It has also been reported that MORC3 associates with gene promoters enriched in the histone mark H3K4me3 throughout the genome (11) and mediates gene silencing when bound to SUMO-2 (12). Growing evidence suggests a role of MORC3 in essential nuclear processes, yet our understanding of physiological and pathological activities of this protein remains very limited.
The MORC family consists of four human proteins, and all MORCs, including MORC3, contain a gyrase, Hsp90, histidine kinase, and MutL (GHKL)-type ATPase domain, CW-type zinc finger, and at least one coiled-coil region (13, 14) (Fig. 1A). The CW domain of MORC3 functions as an epigenetic reader that binds to histone H3 tail and shows preference for methylated lysine 4 of H3 (H3K4me) (4, 11, 15, 16). This interaction is important for MORC3 subcellular localization, because mutations in the CW domain abrogate MORC3 recruitment to chromatin and disrupt its accumulation in NBs (4, 10).
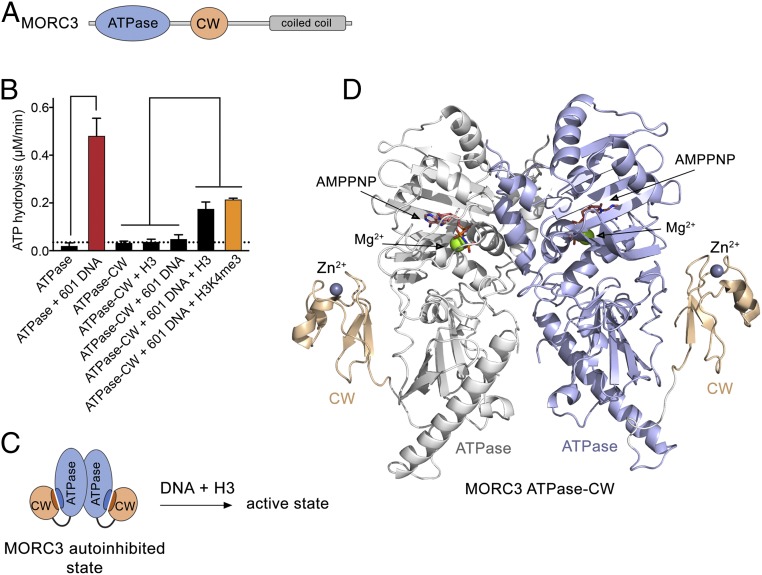
Fig. 1.
Structural basis for MORC3 autoinhibition. (A) MORC3 architecture. (B) Rates of ATP hydrolysis by the indicated domains of MORC3. Error represents SD of at least three separate experiments. To eliminate binding of the ATPase domain to bacterial DNA during sample preparation, the purified proteins were subjected to DNase treatment followed by extensive washes with high-salt buffer. We note that without the additional treatment with DNase and high salts, the ATPase domain shows a high background-level activity due to stimulation by bound bacterial DNA (4). (C) A schematic showing that DNA and H3 are required to activate the MORC3 ATPase–CW cassette. (D) The crystal structure of the dimeric MORC3 ATPase–CW/AMPPNP complex. The ATPase and CW domains are shown as a ribbon with the ATPase domain colored blue in monomer A and white in monomer B, and the CW domain colored wheat. The magnesium (green) and zinc (gray) atoms are shown as spheres. The AMPPNP molecules are in stick representation and colored dark salmon.
A second function of the MORC3 CW domain has been identified more recently. In addition to acting as a histone reader through recognizing the H3 tail, CW associates with the adjacent ATPase domain and modulates its DNA-dependent catalytic activity (4). The crystal structure of the mouse Morc3 ATPase–CW construct bound to a nonhydrolyzable ATP analog and H3K4me3 peptide has demonstrated that the ATPase domain forms a dimer, and mass spectrometry analysis revealed that this dimerization is ATP-dependent (11). Intriguingly, formation of PML-independent nuclear domains of MORC3 was found to rely on its ATPase activity (10). Collectively, data accumulated to date indicate that MORC3 is a chromatin-binding modulator whose biological role and mechanism of action remain unclear.
Here, we present the structural and molecular basis for switching the catalytic function of MORC3 off and on. We show that in the autoinhibited state, the CW domain of MORC3 couples to the ATPase domain and blocks its binding to DNA, which is required for the catalytic activity of MORC3. A competitive binding of CW to histone H3 tail alleviates the autoinhibition, allowing the association with DNA and stimulating the catalytic activity. We demonstrate that ATP-induced dimerization of the ATPase domain is necessary for ATP hydrolysis and that the dimeric form of ATPase–CW might cooperatively bind to dsDNA.
Results and Discussion
CW-Dependent Autoinhibition of MORC3.
The CW domain of MORC3 has been shown to associate with the ATPase domain and negatively regulate its DNA-dependent catalytic activity (4). To explore the idea of MORC3 autoinhibition, here we measured the rates of ATP hydrolysis of the ATPase domain, either isolated or linked to the CW domain (ATPase–CW), using a phosphate release-based enzymatic assay (Fig. 1B and SI Appendix, Fig. S1). We found that the ATPase activity of the isolated ATPase domain is hardly detectable; however, addition of 147-bp 601 Widom DNA at a 1:1 molar ratio resulted in a substantial ∼24-fold increase in the rate of ATP hydrolysis, confirming that the catalytic activity of the ATPase domain is DNA-dependent. In contrast, the linked ATPase–CW cassette was catalytically inactive either in the presence or absence of 601 DNA, supporting the idea of autoinhibition. Histone H3 tail peptide (residues 1 to 12 of H3), a ligand of the CW reader, was incapable of alleviating the autoinhibition, but a combination of 601 DNA and H3 peptide induced a robust ATPase activity. The catalytic activity of ATPase–CW was further increased when trimethylated H3K4me3 peptide was tested in combination with 601 DNA. This result points to an essential role of histone recognition by CW in relieving the inhibition, because CW exhibits higher selectivity to H3K4me3 than to unmodified H3 (4, 11, 15, 16). Collectively, these data suggest that synergistic interactions of the ATPase domain with DNA and of the CW domain with H3 tail are necessary to release autoinhibition and activate the catalytic function of MORC3 (Fig. 1C).
Structural Basis for MORC3 Autoinhibition.
To elucidate the molecular mechanism of MORC3 autoinhibition, we determined the crystal structure of ATPase–CW (amino acids 1 to 455 of human MORC3) in complex with the nonhydrolyzable ATP analog adenylyl-imidodiphosphate (AMPPNP) to a 2.4-Å resolution (Fig. 1D and Table 1). The structure revealed that ATPase–CW forms a symmetric dimer with the large ∼2 × 103 Å2 surface being buried between two monomers. The ATPase domain adopts a GHKL fold, consisting of 10 α-helices and 12 β-strands, and superimposes with the ATPase domain in the H3K4me3-bound state of ATPase–CW [Protein Data Bank (PDB) ID code 5ix1] (11) with an rmsd of 0.6 Å, indicating that the presence of H3K4me3 induces only small changes in the ATPase domain. Strikingly, the CW domain in the autoinhibited state of MORC3 is packed against the ATPase domain and cannot be aligned with the CW domain in the H3K4me3-bound state of ATPase–CW (see below). The entire linker connecting the ATPase and CW domains showed a well-defined electron density and therefore was unambiguously defined (Fig. 1D).
Table 1.
Data collection and refinement statistics for the MORC3 ATPase–CW cassette
| MORC3 ATPase–CW (PDB ID code 6O1E) | |
| Data collection | |
| Space group | P3121 |
| Cell dimensions | |
| a, b, c, Å | 119.51, 119.51, 76.88 |
| α, β, γ, ° | 90, 90, 120 |
| Wavelength | 1.0 |
| Resolution,* Å | 42.93–2.41 (2.50–2.41)* |
| Rpim | 0.011 (0.383) |
| I/σ, I | 50.4 (1.8) |
| CC1/2 | 1.000 (0.827) |
| Completeness, % | 99.9 (100) |
| Redundancy | 43.9 (41.9) |
| Refinement | |
| Resolution, Å | 42.93–2.41 |
| No. reflections | 24,733 |
| Rwork/Rfree | 0.1786/0.2318 |
| No. atoms | 3,613 |
| Protein | 3,522 |
| AMPPNP | 31 |
| Mg | 1 |
| Zn | 1 |
| Water | 58 |
| B factors | 76.59 |
| Protein | 76.84 |
| AMPPNP | 62.08 |
| Mg | 50.33 |
| Zn | 66.84 |
| Water | 70.25 |
| Rms deviations | |
| Bond lengths, Å | 0.012 |
| Bond angles, ° | 1.37 |
Values in parentheses are for the highest-resolution shell. Datasets were collected from a single crystal.
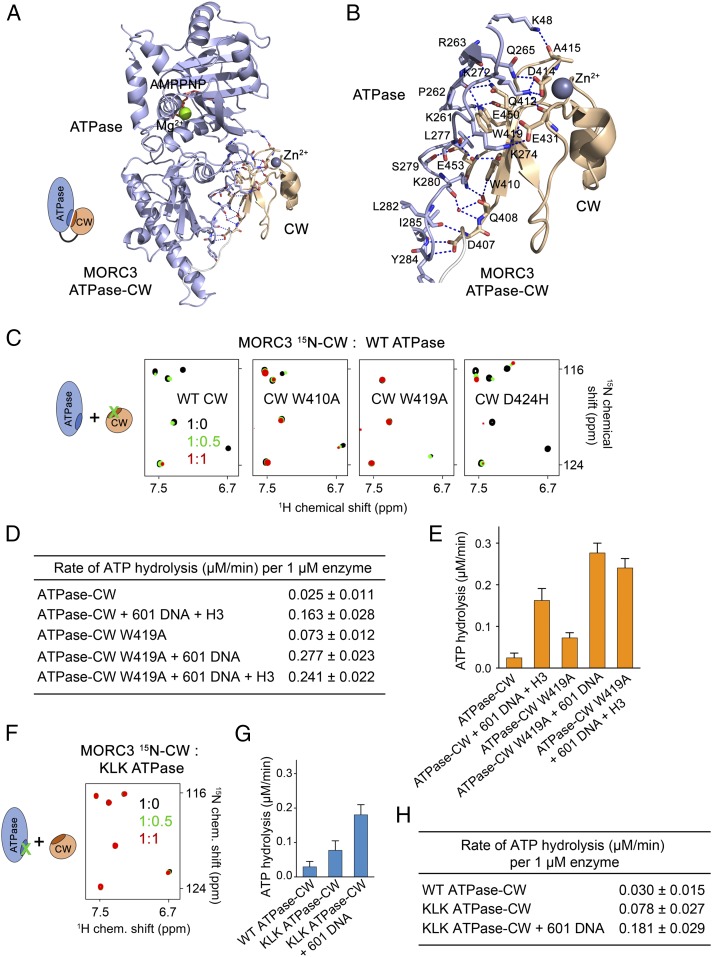
A substantial interface between the ATPase and CW domains in the autoinhibited state buries ∼860 Å2 of the surface area (Fig. 2 A and B). The CW domain is in direct contact with primarily two regions of the ATPase domain. One region contains a loop linking α8 and β8, and another region is a loop between β8 and β9. The residues K261, P262, R263, and Q265 of the α8–β8 loop and K272, K274, K280, Y284, and I285 of the β8–β9 loop in the ATPase domain form an extensive network of hydrogen bonds and salt bridges with the residues D407, Q408, Q412, D414, E431, E450, and E453 of the CW domain. In addition, an N-terminal residue of the ATPase domain, K48, donates a hydrogen bond to the backbone carbonyl group of A415, and the hydrophobic side chain of L277 is caged between the aromatic rings of W410 and W419 of CW.
Fig. 2.
Autoinhibition is released through disrupting the ATPase:CW interface. (A) A ribbon diagram of the monomer A of ATPase–CW as in Fig. 1F. The ATPase:CW interface residues are shown in stick representation. Dashed lines indicate hydrogen bonds and salt bridges. (B) A zoomed-in view of the ATPase:CW interface shown in A. (C) Superimposed 1H,15N TROSY-HSQC spectra of the 15N-labeled WT and mutated MORC3 CW domain collected upon titration with the unlabeled ATPase domain. Spectra are color-coded according to the protein-to-protein molar ratio. A schematic (Left) specifies that the interfacial residues in CW were mutated. D424H mutant, control. (D and E) Rates of ATP hydrolysis by the MORC3 ATPase–CW cassette, WT and W419A mutant. Error represents SD of at least three separate experiments. (F) Superimposed 1H,15N TROSY-HSQC spectra of 15N-labeled MORC3 CW collected upon titration with the unlabeled K274E/L277E/K280E mutant of His-ATPase (residues 1 to 392 of MORC3). Spectra are color-coded according to the protein-to-protein molar ratio. A schematic (Left) specifies that the interfacial residues in the ATPase domain were mutated. (G and H) Rates of ATP hydrolysis by the MORC3 ATPase–CW cassette, WT and KLK mutant. Error represents SD of three separate experiments.
Disrupting the ATPase:CW Interface Releases Autoinhibition.
To gain insight into the inhibitory mechanism, we generated a set of CW and ATPase mutants and tested their reciprocal interactions in 1H,15N transverse relaxation optimized spectroscopy (TROSY) experiments. Titration of the unlabeled wild-type (WT) ATPase domain into uniformly 15N-labeled WT CW caused chemical shift perturbations (CSPs) and substantial broadening and disappearance of CW signals, corroborating the direct and robust association of the two domains that results in formation of a large, >40-kDa complex (Fig. 2). However, when WT ATPase domain was titrated into the W410A or W419A mutant of CW, only minor changes in NMR spectra of CW were observed (Fig. 2C). These results demonstrated that in contrast to the WT CW domain, the mutants, particularly W419A, do not bind well to the ATPase domain and that both tryptophan residues are necessary for the interaction. Likewise, the K274E/L277E/K280E mutant of the ATPase domain (referred to as KLK) did not induce CSPs in WT CW, indicating that its ability to associate with CW is lost and that overall the β8–β9 loop of the ATPase domain is required for the association (Fig. 2F).
To determine how the ATPase:CW interface residues affect the enzymatic activity of MORC3, we incorporated mutations that abrogate interaction of CW with the ATPase domain, namely W419A and KLK, in the linked ATPase–CW cassette and measured rates of ATP hydrolysis of the mutant proteins. The ATPase–CW W419A mutant showed an approximately threefold increase in its intrinsic catalytic activity compared with the activity of WT ATPase–CW (0.073 µM⋅min−1 vs. 0.025 µM⋅min−1) (Fig. 2 D and E). Addition of 601 DNA to the ATPase–CW W419A reaction mixture stimulated the enzymatic activity to 0.277 µM⋅min−1. We note that this activity is higher than the activity of WT ATPase–CW in the presence of both H3 peptide and DNA (0.163 µM⋅min−1), which indicates that without autoinhibition, the mutated ATPase–CW cassette becomes essentially constitutively active. Since the W419A mutant is impaired in histone H3 binding, adding H3 peptide to the reaction mixture had little effect on the rate of catalysis (Fig. 2 D and E). Similarly, the intrinsic catalytic activity of the KLK ATPase–CW mutant was increased ∼2.5-fold compared with the activity of WT ATPase–CW (0.078 µM⋅min−1 vs. 0.030 µM⋅min−1) and was further stimulated by 601 DNA (Fig. 2 G and H). Altogether, these results demonstrate that MORC3 autoinhibition is released by disrupting the intramolecular binding of CW to the ATPase domain.
CW Stabilizes the MORC3 ATPase Domain.
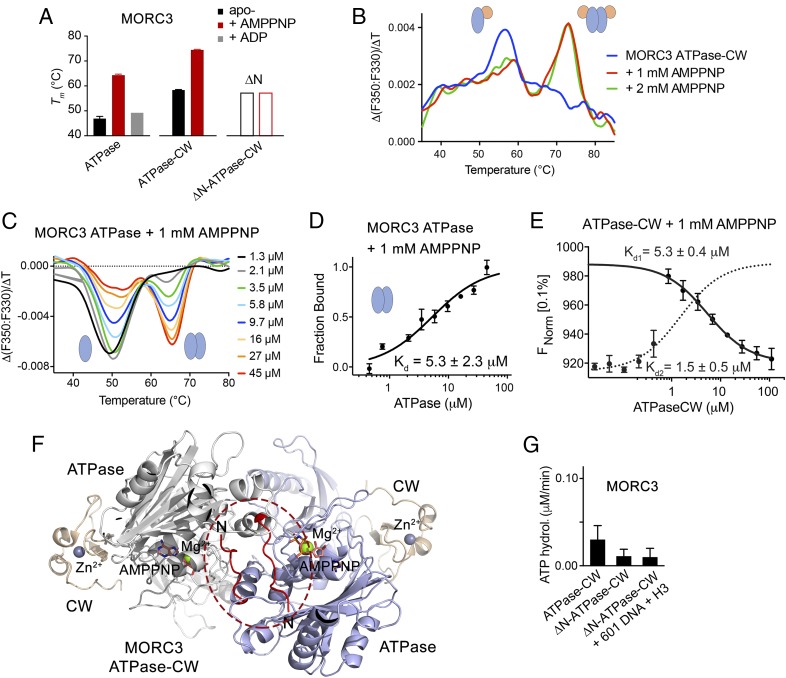
To characterize the autoinhibition state in detail, we monitored the thermal stability of the MORC3 ATPase domain and the ATPase–CW cassette in nano differential intrinsic tryptophan scanning fluorimetry (DSF) assays (Fig. 3 A–C and SI Appendix, Fig. S2). The melting temperature of the isolated ATPase domain (47 °C) was increased by 11 °C in the ATPase–CW cassette (58 °C), pointing to a substantial stabilizing effect of the CW domain (Fig. 3A, compare black bars). Further stabilization of the ATPase–CW fold was observed upon addition of 1 mM AMPPNP (Fig. 3A, red bars and Fig. 3B). The melting temperature in this case was raised by another 16 °C to 74 °C that in agreement with findings on stimulation of MORC3 dimerization by ATP (10, 11) suggested formation of the ATPase–CW dimer. In support, protein cross-linking experiments showed formation of the ATPase–CW dimer in the presence of AMPPNP but not ADP, a product of ATP hydrolysis (SI Appendix, Fig. S3).
Fig. 3.
Dimerization of MORC3 is required for ATPase activity. (A) Tms derived from DSF for the ATPase domain, ATPase–CW cassette, and N-terminally truncated ATPase–CW of MORC3 in the absence (black) or presence (red) of 1 mM AMPPNP or ADP (gray). Error bars represent an SEM based on two separate experiments. (B and C) AMPPNP-induced MORC3 ATPase–CW (B) or ATPase (C) dimerization monitored by DSF. (D) MST binding curve used to determine Kd for the MORC3 ATPase dimerization. Error represents SD between three separate experiments; Kd values ± SD are shown. (E) MST binding curves used to determine Kds for the MORC3 ATPase–CW dimerization (solid line) and ATPase–CW/H3 interaction (dashed line). Error represents SD between three separate experiments; Kd values ± SD are shown. (F) Ribbon diagram of the ATPase–CW dimer as in Fig. 1E with the N terminus (residues 9 to 16) colored red and encircled by a red oval. (G) Rates of ATP hydrolysis by the MORC3 ATPase–CW cassette, WT and ∆N mutant. Error represents SD of at least three separate experiments.
The ATPase domain alone was also able to dimerize, which was efficiently promoted by AMPPNP (Fig. 3 C and D and SI Appendix, Fig. S4A). The dissociation constant (Kd) measured by microscale thermophoresis (MST) for dimerization of the ATPase domain in the presence of AMPPNP was found to be 5.3 ± 2.3 μM, and a similar value, 5.3 ± 0.4 μM, was obtained for dimerization of ATPase–CW (Fig. 3 D and E). Of note, titration of ATPase–CW into fluorescein-labeled histone H3 peptide in the presence of AMPPNP yielded a biphasic MST signal (Fig. 3E). Fitting the high-affinity phase (dotted line) yielded a Kd of ∼1.5 μM, whereas fitting the low-affinity phase (solid line) yielded a Kd of ∼5.3 μM. While the former value agrees with the affinity of CW for H3 peptide (SI Appendix, Fig. S4B), the latter value is consistent with the ATPase domain dimerization. Overall, these data suggest that the interaction of CW with H3 and the ATPase domain dimerization are largely independent processes.
ATPase Dimerization Is Required for ATP Hydrolysis.
The dimeric structure of the MORC3 ATPase–CW cassette shows that the N-terminal part of each monomer is anchored in a cleft of another monomer (Fig. 3F, in red and encircled), in a manner similar to that observed in the structure of ATPase–CW in complex with H3K4me3 (11). To define the impact of dimerization on the catalytic activity of MORC3, we generated a mutant lacking 16 N-terminal residues (ATPase–CWΔN) and tested it in DSF and cross-linking assays. The ATPase–CWΔN mutant was neither stabilized nor cross-linked upon addition of AMPPNP, implying that this mutant lost its dimerization function (Fig. 3A, open bars and SI Appendix, Figs. S2E and S3). Importantly, ATPase–CWΔN showed no ATPase activity at all, even in the presence of both DNA and H3 peptide, thus indicating that dimerization of the ATPase domain is strictly required for ATP hydrolysis (Fig. 3G).
Dimeric ATPase Binds to Double-Stranded DNA.
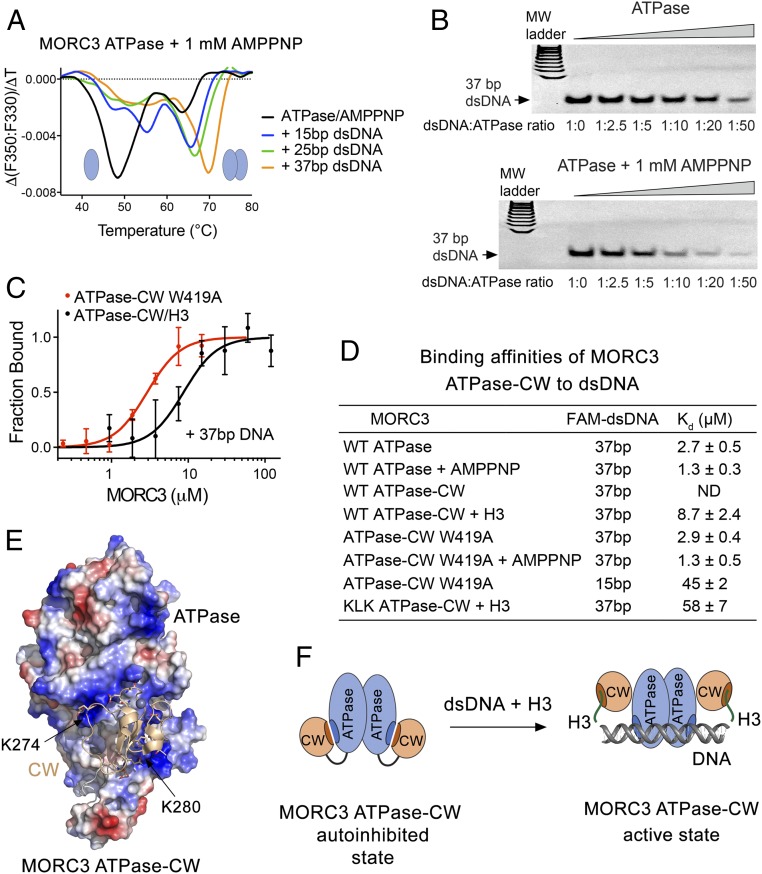
Because both DNA binding and dimerization are necessary for the catalytic activity of MORC3, we explored the relationship between the two functions. DSF experiments showed that dsDNA substantially shifts monomer–dimer equilibrium for the ATPase domain toward the dimeric state and in addition stabilizes the protein (Fig. 4A). The ATPase domain was predominantly in a monomeric state in buffer supplemented with 1 mM AMPPNP (Fig. 4A, black line). Adding a 15-bp dsDNA fragment, however, greatly increased the dimer fraction, and both monomeric and dimeric states of ATPase were stabilized (Fig. 4A, blue line). The thermal stability particularly of the monomer was increased by ∼8 °C. Addition of a longer, 25-bp dsDNA fragment transformed nearly all of the monomeric ATPase domain to the dimeric form and stabilized the dimer by ∼3 °C (Fig. 4A, green line). However, 37-bp dsDNA produced the most notable effect, not only further shifting equilibrium toward the dimer formation but also considerably, by ∼6 °C, stabilizing it (Fig. 4A, orange line and SI Appendix, Fig. S2D). Together, these findings suggest that two ATPase domains in the dimer might cooperatively bind to the 37-bp dsDNA, whereas 25- and 15-bp dsDNA could be too short to simultaneously accommodate both ATPase domains. In agreement, in the absence of AMPPNP and therefore efficient dimerization, 37-bp dsDNA showed a stabilization effect that was similar to the stabilization effect caused by 15-bp dsDNA (Fig. 4A and SI Appendix, Fig. S2C). In further support, the AMPPNP-induced dimer of the ATPase domain bound to 37-bp dsDNA tighter than the ATPase monomer in EMSA assays (Fig. 4B).
Fig. 4.
DNA-binding property of MORC3 ATPase–CW. (A) MORC3 ATPase binding to the indicated double-stranded DNA fragments in the presence of AMPPNP monitored by DSF. (B) EMSA with 37-bp dsDNA in the presence of increasing amounts of ATPase with or without AMPPNP. (C) Binding curves used to determine binding affinities of the MORC3 ATPase–CW W419A mutant for DNA by MST. Error represents SD between three separate experiments. (D) Binding affinities of the MORC3 ATPase or ATPase–CW cassette, WT and mutants, to the indicated dsDNAs; Kd values ± SD are shown. ND, not detected. (E) Electrostatic surface potential of the ATPase domain (in the ATPase–CW cassette) is colored blue and red for positive and negative charges, respectively. The CW domain is shown as a ribbon with selected negatively charged D and E residues depicted in stick form. (F) A model of MORC3 activation through the bivalent synergistic interactions with H3 and DNA.
The DNA-Binding Site of ATPase Is Blocked by CW During Autoinhibition.
To explore the interaction with DNA, we determined binding affinities of the MORC3 ATPase–CW cassette to various-length dsDNA by MST (Fig. 4 C and D). MST measurements showed no detectable interaction between ATPase–CW and 37-bp dsDNA, supporting the idea that CW inhibits the DNA-binding activity of the ATPase domain. However, addition of H3 peptide induced a robust interaction of ATPase–CW with 37-bp dsDNA (Kd 8.7 μM), demonstrating that inhibition is released through a competitive binding of CW to the H3 peptide. The ATPase–CW W419A mutant, in which the interaction between ATPase and CW and therefore autoinhibition is compromised, bound to 37-bp dsDNA tighter (Kd 2.9 μM), and this binding was further augmented in the presence of AMPPNP that stimulates dimerization (Kd 1.3 μM) (Fig. 4 C and D). Notably, these interactions were comparable in strength to the respective interactions of the ATPase domain alone, further supporting the idea that CW blocks the DNA-binding site of ATPase (Fig. 4D). Shortening the dsDNA length to 15 bp reduced the binding affinity of ATPase–CW W419A, indicating that 15-bp DNA may not be long enough for cooperative binding of the dimeric cassette.
The electrostatic surface potential of the ATPase domain reveals large positively charged patches that could interact with negatively charged DNA (Fig. 4E). Indeed, DNA binding of the ATPase–CW K274E/L277E/K280E mutant in the presence of H3 peptide was reduced approximately sixfold compared with the binding of WT ATPase–CW in the presence of H3 peptide (Fig. 4D; Kd 58 and 8.7 μM, respectively). The mutated residues, K274 and K280, are located in two positively charged patches that are blocked by CW in the autoinhibited state. We concluded that the K274-to-K280 region of the ATPase domain is involved in the interactions with both CW and DNA. Furthermore, our data suggest a direct correlation between the DNA-binding and catalytic activities of the ATPase–CW cassette. In the presence of 601 DNA, the rate of ATP hydrolysis of the KLK ATPase–CW mutant with diminished DNA-binding activity was ∼1/3 lower than that of the fully active in the DNA binding W419A mutant (Fig. 2 D and H). Altogether, these results reinforce the idea of MORC3 autoinhibition and suggest that CW abolishes the DNA-stimulated catalytic activity through physically blocking the DNA-binding site of the ATPase domain (Fig. 4F).
Mechanism of MORC3 Activation.
Structural overlay of the ATPase–CW cassette in the autoinhibited state and bound to H3K4me3 peptide (11) shows that while the ATPase domain superimposes well, the CW domain is rotated by almost 180° over its axis and is displaced by over 30 Å (Fig. 5A). In the autoinhibited state, CW (wheat color in Fig. 5A) is tightly packed against the ATPase domain with the histone-binding site facing the ATPase domain. In contrast, in the active state, the histone-binding site of CW (magenta color in Fig. 5A) is flipped away from the ATPase domain. This conformation of CW allows the bound peptide to be fully solvent-exposed and the opposite side of CW to loosely contact the ATPase domain (11). We found that the histone-binding site of CW is occupied by the α8–β8 loop and β8–β9 loop residues of the ATPase domain in the autoinhibited state (Fig. 2B). Strikingly, while there is no apparent similarity between amino acid sequences of H3K4me3 and the α8–β8/β8–β9 loops of the ATPase domain, a number of electrostatic and hydrophobic contacts involving side chains are conserved in the two complexes (Fig. 5B). Intriguingly, a leucine residue of ATPase (L277) occupies the aromatic cage formed by W410 and W419, which normally accommodates trimethylated lysine (K4me3) of the histone peptide. Furthermore, mutation of W410 or W419 disrupts binding of CW to either H3K4me3 or the ATPase domain, suggesting that much like caging of trimethylated lysine through cation–π and hydrophobic interactions, caging of the leucine hydrophobic side chain between the two aromatic moieties is critical.
Fig. 5.
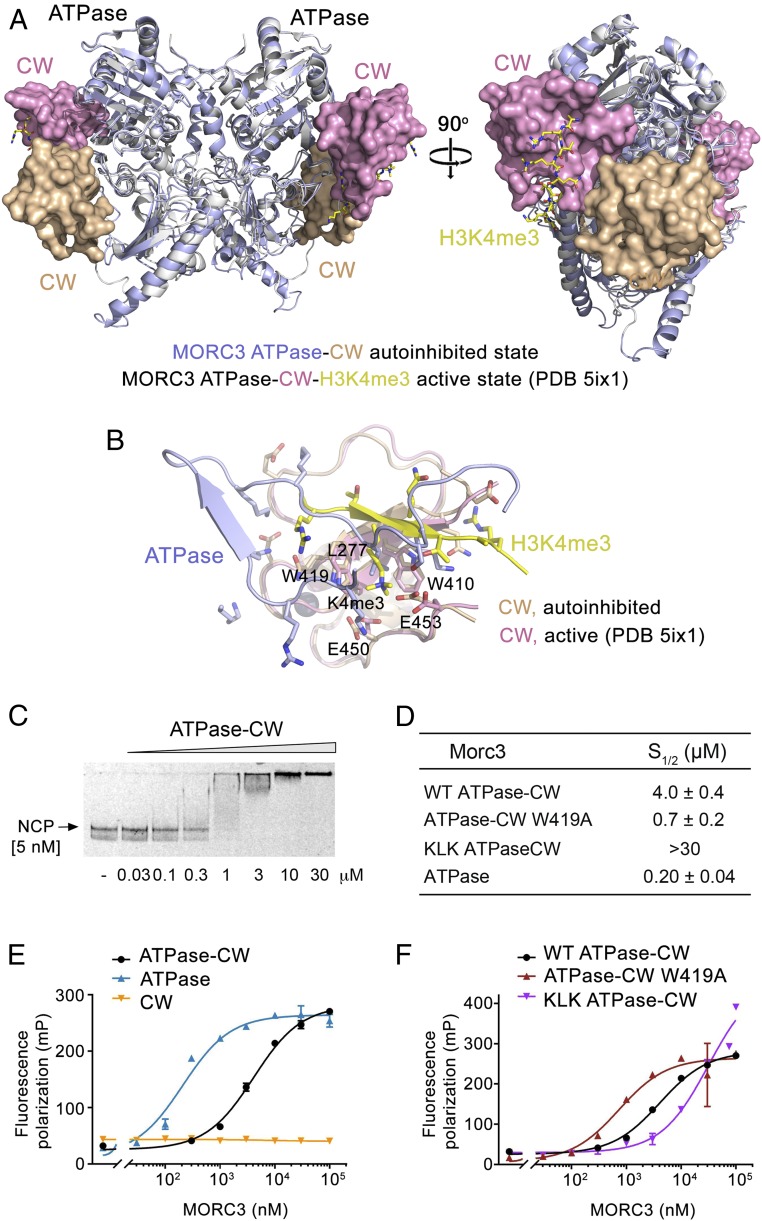
MORC3 ATPase–CW binds to the nucleosome. (A) Structural overlay of the autoinhibited (this study; light blue and wheat) and active (PDB ID code 5ix1; white and magenta) (11) states of the MORC3 ATPase–CW cassette. The ATPase and CW domains are shown in ribbon and surface representations, respectively. H3K4me3 peptide in the active state of MORC3 is shown in stick form (yellow). (B) Structural overlay of the CW domain in the autoinhibited state (wheat) bound to the α8–β8 and β8–β9 loops of the ATPase domain (light blue) with the CW domain in the active state (PDB ID code 5ix1; magenta) bound to the H3K4me3 peptide (yellow). (C) EMSA with NCPs in the presence of increasing amounts of ATPase–CW. (D–F) Binding affinities (D) and binding curves (E and F) for the interactions of the indicated MORC3 constructs with NCP as measured by fluorescence polarization. Error bars represent an SD based on three separate experiments.
We next assessed the mechanism for MORC3 activation using nucleosome core particles (NCPs) that contain both ligands for the ATPase–CW cassette: histone H3 and DNA. Although ATPase–CW does not interact with 601 DNA (4), it binds well to the intact nucleosome reconstituted with 601 DNA in EMSA experiments (Fig. 5C). Fluorescence anisotropy measurements yielded a binding affinity of 4 μM for ATPase–CW (Fig. 5 D and E). Binding of the isolated ATPase domain to the same nucleosome was 20-fold tighter (Kd 200 nM), indicating that the interaction of CW with unmodified H3 tail is not robust enough to fully eliminate autoinhibition. We predict that engagement of ATPase–CW with the H3K4me3-containing NCP would release the autoinhibition to a greater extent.
In further support, binding affinity of the ATPase–CW W419A mutant to the nucleosome, measured by fluorescence anisotropy, was found to be 730 nM (Fig. 5 D and F). As the ATPase domain in this mutant is more available for binding to DNA, it associated with the nucleosome ∼5.5-fold tighter compared with WT ATPase–CW but ∼3.5-fold weaker compared with the isolated ATPase domain. The latter indicates that the disruption of the ATPase:CW interface via substituting W419 with alanine is incomplete. On the contrary to the W419A mutation that increases DNA binding of the ATPase–CW W419A cassette, the KLK mutation that reduces DNA binding also reduced the NCP-binding ability of the KLK ATPase–CW cassette in fluorescence anisotropy experiments (Fig. 5 D and F).
Concluding Remarks.
In this study, we report the mechanism by which the catalytic activity of MORC3 is switched on and off. The direct intramolecular coupling of CW to the ATPase domain of MORC3 blocks binding of the ATPase domain to DNA, resulting in an autoinhibited, catalytically inactive state of MORC3. The synergistic interactions of CW with histone H3 tail and of the ATPase domain with DNA, required to alleviate autoinhibition and stimulate the catalytic function of MORC3, indicate that (i) the ATPase-binding interface and the H3-binding site of CW overlap, and (ii) the CW-binding interface and the DNA-binding site of the ATPase domain also overlap at least in part. Our findings illuminate the intricate molecular mechanism that implicates the epigenetic reader, the CW domain, in the direct regulation of the adjacent catalytic domain.
Is the autoinhibition mechanism conserved among other members of the MORC family? Given the high sequence and domain architecture similarity between MORC3 and MORC4, it is likely that the ATPase activity of MORC4 is autoregulated by its histone-recognizing CW domain. However, architecture of two other members of this family, MORC1 and MORC2, differ from that of MORC3/4. Recent biochemical and structural studies have shown that MORC2 has a distinctly different mechanism for targeting of chromatin (17, 18). MORC2 contains a CW domain that does not bind to histone tails and a coiled-coil (CC) insertion between the ATPase domain and the CW domain that is involved in binding to DNA (16, 18). In contrast to the mechanism by which MORC3 binds to chromatin and is activated, it has been demonstrated that following DNA damage, MORC2 is phosphorylated on Ser739, and this phosphorylation is required for the association of MORC2 with chromatin and its ATPase activity (17).
Structural comparison of the ATPase–CW cassette of MORC3 and the ATPase–CC–CW cassette of MORC2 (18) corroborates different mechanisms of action. Although the CW domains of both proteins are packed against the ATPase domains in a similar way, many ATPase:CW interfacial residues are not conserved and form distinct patterns of hydrogen bonding, electrostatic, and hydrophobic contacts (Fig. 2 A and B and SI Appendix, Fig. S5). While both DNA binding and dimerization are required for the ATPase activity of MORC3, mutation of the MORC2 residues that disrupt the dimerization interface results in an increased rate of ATP hydrolysis (18). Furthermore, MORC3 and MORC2 have different DNA-binding sites. The ATPase surface that engages CW in MORC3 is also involved in binding to DNA, whereas the distal end of the CC is responsible in part for DNA binding in MORC2 (18). Consequently, the CW domain does not inhibit the DNA-binding activity of MORC2, whereas the CW domain of MORC3 does. The suppression of the DNA-binding ability, which is crucial for the catalytic function of MORC3, suggests a highly controlled autoregulatory mechanism that allows MORC3 to be activated at certain chromatin regions, such as H3K4me3-enriched loci.
What is precisely the ATPase hydrolysis by MORC3 required for in the cell? MORC3 and homologous plant and animal MORC proteins have been implicated in gene silencing (11), and it will be essential in future studies to elucidate the role of autoinhibition in this function. Another important question is whether MORCs target specific DNA sequences or regulate particular genes. Understanding these physiological functions of MORCs will have a profound impact on our understanding of the pathological implications associated with this family of human ATPases.
Methods
Protein Expression and Purification.
The human MORC3 CW domain (amino acids 401 to 455) was cloned into a pGEX 6p-1 vector. MORC3 ATPase (amino acids 1 to 392) and ATPase–CW (amino acids 1 to 455) constructs were cloned into a pET28a vector. Proteins were expressed in Rosetta2 (DE3) pLysS or BL21 (DE3) RIL in LB or minimal media supplemented with 15NH4Cl. Protein expression was induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside for 16 h at 16 °C. The ATPase and ATPase–CW constructs were purified on Ni-NTA agarose beads (QIAGEN) in 20 mM Tris⋅HCl (pH 7.5), 300 mM NaCl, 5 mM MgCl2, and 2 mM DTT. GST–CW proteins were purified on glutathione Sepharose 4B beads (GE Healthcare) in 20 mM Tris⋅HCl (pH 7.0) buffer supplemented with 100 mM NaCl and 2 mM DTT. The GST tag was cleaved overnight at 4 °C with PreScission protease. Unlabeled proteins were further purified by size-exclusion chromatography and concentrated in Millipore concentrators. All mutants were generated by site-directed mutagenesis using the Stratagene QuikChange mutagenesis protocol, grown, and purified as wild-type proteins.
ATPase Activity Assays.
The ATPase assays were performed using the EnzChek Phosphate Assay Kit (Molecular Probes). The reactions were carried out on 1.0 µM MORC3 His-ATPase or His-ATPase–CW, in the presence and absence of 1 µM 601 DNA or 50 µM unmodified H3K4me0 or H3K4me3 peptide (1 to 12) in a buffer containing 50 mM Tris⋅HCl (pH 7.5), 50 mM NaCl, 1 mM MgCl2, 0.1 mM sodium azide, 200 µM MESG, and 1 U of PNP. The reaction was started by adding 2 mM ATP to the mixture at room temperature, and the release of inorganic phosphate was monitored by measuring the absorbance at 360 nm on a NanoDrop 2000c spectrophotometer (Thermo Scientific). In the presence of inorganic phosphate, produced by the hydrolysis of ATP to ADP, MESG is enzymatically converted to ribose 1-phosphate and MESG by PNP, resulting in a shift in the wavelength absorbance from 330 nm for MESG to 360 nm for the product. The rate of ATP self-hydrolysis was measured in samples prepared in parallel replacing the MORC3 protein with the buffer and then subtracted from each set of measurements. Error was calculated as the SD of at least three separate experiments.
NMR Experiments.
NMR experiments were carried out at 298 K on a Varian INOVA 900-MHz spectrometer. NMR samples contained 0.1 mM uniformly 15N-labeled WT or mutated CW in 10 mM Hepes (pH 7.2) buffer supplemented with 100 mM NaCl, 1 mM TCEP, and 8% D2O. Binding was characterized by monitoring CSPs in the proteins induced by WT or mutated ATPase domain.
X-Ray Crystallography.
The His tag in His-ATPase–CW (1 to 455) was cleaved overnight at 4 °C with TEV protease. Cleaved protein was further purified by size-exclusion chromatography, concentrated to 9 mg/mL, and incubated with 2 mM AMPPNP on ice for 2 h in a buffer containing 10 mM Tris⋅HCl (pH 7.2), 150 mM NaCl, 2 mM MgCl2, and 2 mM DTT. Good-quality diffracting crystals were obtained at room temperature by the hanging-drop vapor-diffusion method in 0.1 M Hepes (pH 7.5), 10% (wt/vol) PEG8000, and 8% (vol/vol) ethylene glycol. Data collection was performed at the ALS 4.2.2 beamline. HKL3000 was used for indexing, scaling, and data reduction. Solution was found by the single-wavelength anomalous dispersion method with Zn anomalous signal. Manual model building was performed using Coot (19), and the structure was refined using Phoenix Refine (20). The final structure was verified by MolProbity (21). The X-ray diffraction and structure refinement statistics are summarized in Table 1.
Nucleosome Preparation.
Human H2A, H2B, H3.2, and H4 histone proteins were expressed in Escherichia coli BL21 DE3 pLysS cells, separated from inclusion bodies, and purified using size-exclusion and ion-exchange chromatography, as described previously (22). Histones were then mixed together in 7 M guanidine HCl, 20 mM Tris⋅HCl (pH 7.5), and 10 mM DTT in appropriate molar ratios and refolded into octamers by slow dialysis into 2 M NaCl, 20 mM Tris⋅HCl (pH 7.5), 1 mM EDTA (pH 8.0), and 2 mM β-mercaptoethanol. The octamer was purified from tetramers and dimers by size-exclusion chromatography. The octamer was then mixed with a 75% excess of 147-bp 601 Widom nucleosome-positioning sequence DNA fluorescein-labeled on the 5′ end in 2 M NaCl, 5 mM Tris (pH 8.0), and 0.5 mM EDTA (pH 8.0), and NCPs were reconstituted by slow desalting dialysis into 5 mM Tris (pH 8.0) and 0.5 mM EDTA (pH 8.0). Finally, the NCPs were separated from free DNA via sucrose gradient purification.
Fluorescence Polarization.
Fluorescence polarization measurements were carried out by mixing increasing amounts of MORC3, WT and mutants, with 5 nM NCPs in 75 mM NaCl, 15 mM Tris⋅HCl (pH 7.5), 0.00625% Tween 20, and 5 mM DTT. The samples were loaded into a Corning round-bottom polystyrene plate and allowed to incubate at 4 °C for 20 min. The polarization measurements were acquired with a Tecan Infinite M1000 Pro plate reader by exciting at 470 nm and measuring polarized emission at 519 nm with 5-nm excitation and emission bandwidths. The fluorescence polarization was calculated from the emission polarized parallel and perpendicular to the polarized excitation light as described previously (23, 24). The data were then fit to a noncooperative binding isotherm to determine the dissociation constants.
Differential Scanning Fluorimetry.
Samples of 10 µL MORC3 constructs at 5 µM or the indicated concentrations in the presence or absence of 1 mM AMPPNP or ADP were prepared in a buffer containing 10 mM Tris⋅HCl (pH 7.2), 150 mM NaCl, 2 mM MgCl2, and 0.5 mM TCEP, and then loaded into glass capillaries (NanoTemper) by capillary action. Intrinsic protein fluorescence at 330 and 350 nm was monitored between 35 and 90 °C in the Tycho instrument (NanoTemper), and the Tm values were calculated using the accompanying software by taking the turning point of the first derivative of the F350:F330 ratio as a function of temperature.
Fluorescent Microscale Thermophoresis Binding Assay.
The MST experiments were performed using a Monolith NT.115 instrument (NanoTemper) as described previously (25). All experiments were performed using the purified ATPase domain or ATPase–CW in a buffer containing 10 mM Tris⋅HCl (pH 7.2), 150 mM NaCl, 2 mM MgCl2, and 0.5 mM TCEP. The concentration of fluorophore was 80 and 40 nM for measuring affinities to fluorescein (FAM)-labeled H3-peptide (KE BIOCHEM) and DNA (IDT), respectively. To monitor ATPase dimerization, His-tagged ATPase domain was labeled using a RED-NHS Labeling Kit (NanoTemper), and concentration of the labeled protein was kept at 5 nM. Dissociation constants were determined using a direct binding assay in which increasing amounts of unlabeled binding partner were added stepwise. The measurements were performed at 40 or 80% LED and 40% MST power with 3 s laser on time and 25 s off time. The Kd values were calculated using MO Affinity Analysis software (NanoTemper). To analyze the biphasic process, the curve was fitted using the data points (solid line in Fig. 3E) without using any predetermined parameters, which yielded the Kd1, FNorm(maximum), and FNorm(minimum) (26). The FNorm(maximum) was used then as a fixed parameter while fitting the second curve (dashed line), which yielded the Kd2. All measurements were performed in triplicate.
Cross-Linking Assay.
Proteins (10 μM) were cross-linked in 10 mM Hepes (pH 7.5), 150 mM NaCl, 2 mM MgCl2, 0.5 mM TCEP, and 1 mM AMPPNP or ADP. BS3 cross-linker (Thermo Scientific) was resuspended to 2 mM and used at a concentration of 0.1 mM. Cross-linking was allowed to proceed for 10 min. Cross-linked samples were quenched in 0.1 M Tris (pH 7.5), resolved on SDS/PAGE, and stained with Coomassie blue.
EMSA.
EMSA experiments were performed essentially as described (27). Briefly, increasing amounts of MORC3 His-ATPase domain (2.5 to 50 molar excess) were incubated with 37-bp dsDNA (5 pM) in a DNA-binding buffer containing 20 mM Tris⋅HCl (pH 7.5), 45 mM KCl, 4 mM Mg, 5% glycerol, and 5 mM DTT for 0.5 h on ice. The reaction mixtures were loaded on 5% polyacrylamide gels, and electrophoresis was performed in 0.5 TBE buffer at 120 V for 1 h on ice. Gels were stained with ethidium bromide and visualized with an AlphaImager (Alpha Innotech).
Supplementary Material
Acknowledgments
We thank JaeWoo Ahn for help with experiments. This work was supported by grants from the NIH (GM106416, GM125195, and GM100907 to T.G.K.; GM121966 and GM120582 to M.G.P.) and German Research Foundation DFG CA198/9-1 and 9-2 (to M.C.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code 6O1E).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819524116/-/DCSupplemental.
References
- 1.Gunawardena H, et al. Juvenile Dermatomyositis Research Group, UK and Ireland Autoantibodies to a 140-kd protein in juvenile dermatomyositis are associated with calcinosis. Arthritis Rheum. 2009;60:1807–1814. doi: 10.1002/art.24547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimura Y, et al. The newly identified human nuclear protein NXP-2 possesses three distinct domains, the nuclear matrix-binding, RNA-binding, and coiled-coil domains. J Biol Chem. 2002;277:20611–20617. doi: 10.1074/jbc.M201440200. [DOI] [PubMed] [Google Scholar]
- 3.Ichimura Y, et al. Anti-NXP2 autoantibodies in adult patients with idiopathic inflammatory myopathies: Possible association with malignancy. Ann Rheum Dis. 2012;71:710–713. doi: 10.1136/annrheumdis-2011-200697. [DOI] [PubMed] [Google Scholar]
- 4.Andrews FH, et al. Multivalent chromatin engagement and inter-domain crosstalk regulate MORC3 ATPase. Cell Rep. 2016;16:3195–3207. doi: 10.1016/j.celrep.2016.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling KH, et al. Functional transcriptome analysis of the postnatal brain of the Ts1Cje mouse model for Down syndrome reveals global disruption of interferon-related molecular networks. BMC Genomics. 2014;15:624. doi: 10.1186/1471-2164-15-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Fernández R, Morales M, Avila J, Martín-Vasallo P. Changes in leukocyte gene expression profiles induced by antineoplastic chemotherapy. Oncol Lett. 2012;3:1341–1349. doi: 10.3892/ol.2012.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiorentino DF, et al. Most patients with cancer-associated dermatomyositis have antibodies to nuclear matrix protein NXP-2 or transcription intermediary factor 1γ. Arthritis Rheum. 2013;65:2954–2962. doi: 10.1002/art.38093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jadhav G, Teguh D, Kenny J, Tickner J, Xu J. Morc3 mutant mice exhibit reduced cortical area and thickness, accompanied by altered haematopoietic stem cells niche and bone cell differentiation. Sci Rep. 2016;6:25964. doi: 10.1038/srep25964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, et al. Dynamic regulation of p53 subnuclear localization and senescence by MORC3. Mol Biol Cell. 2007;18:1701–1709. doi: 10.1091/mbc.E06-08-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mimura Y, Takahashi K, Kawata K, Akazawa T, Inoue N. Two-step colocalization of MORC3 with PML nuclear bodies. J Cell Sci. 2010;123:2014–2024. doi: 10.1242/jcs.063586. [DOI] [PubMed] [Google Scholar]
- 11.Li S, et al. Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin. Proc Natl Acad Sci USA. 2016;113:E5108–E5116. doi: 10.1073/pnas.1609709113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosendorff A, et al. NXP-2 association with SUMO-2 depends on lysines required for transcriptional repression. Proc Natl Acad Sci USA. 2006;103:5308–5313. doi: 10.1073/pnas.0601066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li DQ, Nair SS, Kumar R. The MORC family: New epigenetic regulators of transcription and DNA damage response. Epigenetics. 2013;8:685–693. doi: 10.4161/epi.24976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J Am Chem Soc. 2012;134:1982–1985. doi: 10.1021/ja210528v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Family-wide characterization of histone binding abilities of human CW domain-containing proteins. J Biol Chem. 2016;291:9000–9013. doi: 10.1074/jbc.M116.718973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li DQ, et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–1669. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Douse CH, et al. Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms. Nat Commun. 2018;9:651. doi: 10.1038/s41467-018-03045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 23.Gatchalian J, et al. Accessibility of the histone H3 tail in the nucleosome for binding of paired readers. Nat Commun. 2017;8:1489. doi: 10.1038/s41467-017-01598-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tencer AH, et al. Covalent modifications of histone H3K9 promote binding of CHD3. Cell Rep. 2017;21:455–466. doi: 10.1016/j.celrep.2017.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 26.Seidel SA, et al. Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods. 2013;59:301–315. doi: 10.1016/j.ymeth.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein BJ, et al. Bivalent interaction of the PZP domain of BRPF1 with the nucleosome impacts chromatin dynamics and acetylation. Nucleic Acids Res. 2016;44:472–484. doi: 10.1093/nar/gkv1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.