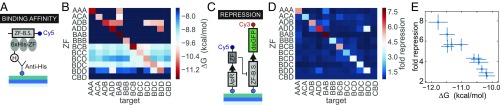
Significance
Understanding basic mechanisms and engineering new systems in biology are hampered by the challenge of quantifying and manipulating numerous molecular components and their interactions. In this work, we present an approach to tackle these difficulties, by combining a cell-free system with a microfluidic platform for high-throughput measurements. We apply the system to comprehensively characterize a library of synthetic zinc-finger transcription factors, which are common building blocks of transcriptional regulatory networks. We subsequently use the knowledge gained to engineer highly specific, tunable, and strong cooperative repressors, which can be applied to carry out logical computation on promoters.
Keywords: cell-free synthetic biology, biophysics, transcriptional regulation, gene regulatory network, synthetic transcription factors
Abstract
Gene-regulatory networks are ubiquitous in nature and critical for bottom-up engineering of synthetic networks. Transcriptional repression is a fundamental function that can be tuned at the level of DNA, protein, and cooperative protein–protein interactions, necessitating high-throughput experimental approaches for in-depth characterization. Here, we used a cell-free system in combination with a high-throughput microfluidic device to comprehensively study the different tuning mechanisms of a synthetic zinc-finger repressor library, whose affinity and cooperativity can be rationally engineered. The device is integrated into a comprehensive workflow that includes determination of transcription-factor binding-energy landscapes and mechanistic modeling, enabling us to generate a library of well-characterized synthetic transcription factors and corresponding promoters, which we then used to build gene-regulatory networks de novo. The well-characterized synthetic parts and insights gained should be useful for rationally engineering gene-regulatory networks and for studying the biophysics of transcriptional regulation.
Cell-free systems have emerged as versatile and efficient platforms for rapid engineering, characterization, and implementation of genetic networks. It has been demonstrated that linear genetic cascades (1), logic gates (2), and oscillators (3–5) could be implemented and characterized in cell-free systems and that networks engineered in cell-free systems function in cells with remarkably similar characteristics, indicating that cell-free systems accurately emulate the cellular environment (4, 6). Besides these examples in molecular-systems engineering and characterization of complex biological systems, cell-free systems provide a viable starting point for the bottom-up synthesis of artificial cells (7, 8). Work is progressing in establishing critical cellular subsystems, including DNA replication (9), metabolism (10), ribosome synthesis (11), membrane synthesis (12), and protein structures (13). Gene-regulatory networks (GRNs) are one such critical subsystem, and here we demonstrate de novo bottom-up engineering and comprehensive characterization of synthetic GRNs in a cell-free system.
GRNs execute the genome and thus play a central role across all domains of life. Due to their importance and ubiquity, GRNs have been intensely studied, and considerable progress is being made in deciphering components, topologies, and general mechanisms of GRNs, although a complete mechanistic understanding is still lacking. Because GRNs perform many sophisticated cellular tasks, synthetic biologists use GRNs to engineer new systems (14) such as logic gates (15), toggle switches (16), band-pass filters (17), and oscillators (18). Nonetheless, past and current efforts in engineering GRNs have shown that rational design is not yet possible and that engineering GRNs still heavily relies on trial-and-error and high-throughput screening approaches (15). The inability to rationally design GRNs is in part due to the aforementioned lack of complete mechanistic understanding and because basic GRN components such as transcriptional regulators and promoters are often neither fully characterized nor standardized. A corollary of the lack of an in-depth mechanistic understanding of these systems is that individual components are not yet readily composable. Nature provides a plethora of potential transcriptional regulators, but the number that have been tested and characterized remains rather limited. Most engineered GRNs make use of naturally occurring transcription factors, making it difficult to robustly engineer GRNs with such a nonstandard set of proteins (19). A library of well-characterized, synthetic transcription factors could alleviate many of these problems by providing a set of standardized transcription factors that are based on the same basic structural framework and whose function can be extended by generating fusion proteins in a plug-and-play format.
Native GRNs use a wide range of transcription factors that can be categorized into several structural families. The family with the largest number of members is the zinc-finger (ZF) family, followed by homeodomain, basic helix–loop–helix, and basic leucine-zipper (LZ) families (20). ZFs are of interest in biology, as they represent the largest class of transcriptional regulators and are involved in diverse biological functions. ZFs are also appealing for bottom-up engineering, as they consist of well-defined subunits that, in combination, determine DNA sequence specificity (21, 22). Many resources are therefore available that provide sequence-specificity information for a large number of native (23) and engineered (24) ZF transcription factors. An additional advantage is that ZFs are small [264 bp, 10.6 kDa (Zif268)] compared with other engineerable transcriptional regulators, such as TALE [e.g., 1,161–2,397 bp, 39.9–82.6 kDa, DNA binding domain only (25)] or dCas9 (4,107 bp, 158.3 kDa), so that the coding sequence for ZFs is easily obtainable and modifiable. Due to their small size and simple structure, ZFs can be readily expressed both in vivo and in vitro. Synthetic ZFs have already been successfully used as activators in Saccharomyces cerevisiae (26) and human cells (27). Here, we engineer and explore the use of synthetic ZF transcriptional regulators as ideal building blocks for bottom-up design and implementation of cell-free GRNs.
In this paper, we took advantage of an existing synthetic ZF library (28) to generate a well-characterized resource of transcriptional repressors and corresponding synthetic promoters that can be used for bottom-up design, implementation, and characterization of GRNs in cell-free systems. While the mechanism of action of the simplest prokaryotic repression is competitive inhibition (29), it has long been appreciated that both cis modifications to the promoter, such as operator position (30), and basal promoter strength (31), as well as trans modifications to the transcription factor itself strongly affect repression (32, 33). These interdependencies result in a large experimental space with many degrees of freedom. To tackle this complexity, we developed a microfluidics-based method capable of performing 768 cell-free transcription–translation (TX-TL) reactions on a single device. The ability to rapidly generate ZF repressor and promoter variants by using fast-PCR assembly and the use of our high-throughput microfluidic device allowed us to perform a comprehensive characterization of repressors and promoters. We investigated the effects of binding-site position, binding-site affinity, binding-site combinations, and cooperative interactions between the repressors on transcriptional repression performance. We generated quantitative position weight matrices (PWMs) for four ZF repressors with MITOMI (34), which allowed us to rationally tune binding-site affinity and promoter output. Finally, we used the parts library and insights acquired in this study to engineer logic gates, showing that de novo synthetic GRNs can be rationally engineered by using a bottom-up approach. The transcription factor/promoter parts library, data, and methods described here provide a resource that should facilitate efforts to build synthetic GRNs, serve as a viable approach for building GRNs for use in artificial cells, and establish an experimental platform for studying the biophysics of transcriptional regulation.
Results
Design and Characterization of a Microfluidic Device for High-Throughput Cell-Free Experiments.
The design space of even a single TF–promoter pair is large, encompassing different binding-site affinities, binding-site positions, binding-site sequences, and binding-site combinations. This complexity necessitates high-throughput methods capable of the functional characterization of hundreds to thousands of engineered variants. Current approaches in cell-free synthetic biology primarily rely on standard microtiter plates, which require a minimal reaction volume of 5–10 L. Such relatively large volumes quickly become cost-limiting in terms of how much cell-free reaction solution and DNA are required to perform the assays. Researchers recently made use of an acoustic liquid-handling robot that reduced reaction volumes to 2 L in a 384-well plate format (35). Here, we repurposed the MITOMI platform, a microfluidic device originally developed for high-throughput molecular-interaction analysis (34, 36), and applied it to the high-throughput characterization of cell-free genetic networks. The repurposed device performs 768 cell-free reactions and reduces volumes by 4 orders of magnitude to 690 pL per reaction.
The process involves the synthesis of DNA parts, followed by microarraying and incorporation into microfluidic unit cells, where they serve as templates in cell-free TX-TL reactions (Fig. 1A). To expedite the synthesis of large libraries of DNA parts, we used an assembly PCR strategy to generate linear DNA templates with different promoter regions upstream of an eGFP-Del6-229 (deGFP) gene. A microarray robot was used to spot the linear templates onto an epoxy-coated glass slide, on top of which the polydimethylsiloxane (PDMS) device was aligned. Immobilizing DNA within each reaction chamber first required surface patterning in the assay section of each unit cell, resulting in a circular area of neutravidin, to which biotinlyated DNA can bind. Once DNA was surface-immobilized, cell-free extract was flowed into the device, and the unit cells were isolated from one another while the TX-TL reactions occurred. A detailed schematic of the experimental procedure is shown in SI Appendix, Fig. S1A.
Fig. 1.
High-throughput microfluidic cell-free reactions. (A) A schematic overview of the experimental design, including synthesis of DNA templates, DNA spotting, chip alignment, surface functionalization followed by DNA immobilization, and on-chip cell-free TX-TL reactions. (B) Fluorescence images of Cy3-, -, and deGFP expressed for a range of dsDNA:ssDNA ratios. (C) Quantification of surface-immobilized DNA () as a function of free DNA in solution (). (D) deGFP expression at the final time point as a function of concentration. All values represent means SD (). (E) deGFP expression measured over time in all unit cells. (F) Schematic of a promoter library design and on-chip experimental throughput, followed by the deGFP expression for all single base mutations from position −47 to −7 of the promoter. RFU, relative fluorescence units.
Controlling the precise amount of DNA in each unit cell is important for quantitative experiments. By simply varying the concentration of spotted biotinylated DNA templates, we were unable to precisely control DNA concentration on-chip. We thus developed an approach based on spotting a mixture of single-stranded biotinylated DNA oligos (ssDNA) and double-stranded DNA templates (dsDNA). The amount of DNA immobilized on the surface reached saturation at a concentration of 100 nM spotted DNA (SI Appendix, Fig. S2). We therefore held the total concentration of spotted DNA above this saturation point. Changing the ratio of dsDNA:ssDNA gave rise to a linear correlation between the concentration of dsDNA free in solution () and dsDNA bound to the surface () and was insensitive to the total amount of DNA deposited during spotting (Fig. 1 B and C). This approach allowed us to immobilize DNA over a wide concentration range, which gave rise to corresponding levels of expressed deGFP (Fig. 1 D and E). The results obtained with the high-throughput microfluidic device are reproducible with a global normalized root-mean-square deviation of 14%, not only when a single dsDNA template is used, but also for more complex experiments requiring multiple templates in each unit cell (SI Appendix, Fig. S3). Furthermore, a subset of on-chip measurements was carried out in standard microwell plate reactions, showing good correlation (SI Appendix, Fig. S4).
To demonstrate the high-throughput capabilities of our microfluidic chip, we created and characterized a library based on the Escherichia coli promoter. We synthesized 124 promoter variants that covered all possible single-base mutations within the −47 to −7 region of the promoter (Fig. 1F). Cell-free reactions for each promoter were run in six replicates on a single chip and yielded deGFP expression profiles revealing the impact of each mutation on protein expression (Fig. 1F and SI Appendix, Fig. S3A). As expected, mutations within the −10 and −35 boxes affected deGFP expression most strongly, and the results are comparable to previous results obtained by an in vivo analysis of the lac promoter (37).
Protein synthesis eventually stopped in cell-free batch reactions, as seen in the saturation dynamics in time-course measurements (Fig. 1E); this is fundamentally different from cellular steady-state protein levels which result from balancing production with degradation and dilution rates. In this work, we report end-point batch reaction values and derived quantities such as fold repression. It is thus important that the end-point values correspond to protein production rates. While the relationship between the initial rate of deGFP production and its final saturated level may be complex, we observed a linear relationship between the two quantities under our experimental conditions (SI Appendix, Fig. S5). This is an important validation of our use of end-point protein levels and linearly derived quantities such as fold repression as proxies for synthesis rates and their ratios.
ZF Repressor and Promoter Library Design.
Using the characterization of the promoter as a starting point, we applied our chip to the in-depth characterization of synthetic ZFs for use as transcriptional repressors. We adopted a ZF design based on Zif268, a three-finger Cys2–His2 protein. A large ZF repressor library can be generated by combinatorially shuffling a small number of individual ZF domains (Fig. 2A). We used ZF proteins drawn from a 64-member library that we previously synthesized and characterized (28) (SI Appendix, Fig. S6).
Fig. 2.
ZF repressor and promoter design. (A) Our repressor design is based on the Zif268 protein, whose three ZFs (F1–F3) each recognizes a nucleotide triplet. We created a combinatorial library of repressors by shuffling individual ZFs, starting from four initial ZF proteins (here labeled with the codes AAA–DDD). (B) We designed a library of repressible promoters based on the promoter. To test the effectiveness of repression, we designed promoters containing single and dual sites with variable spacing, as well as engineering direct cooperativity between ZF proteins, which can be mediated by PDZ–ligand or LZ interactions.
The affinity of a ZF repressor to DNA can be improved by increasing the number of finger domains (38–41). The same effect can also be achieved by engineering dimerizing ZFs that bind cooperatively. An early example used structure-based design to engineer a two-finger ZF which dimerized via a LZ motif to form a four-finger complex (42, 43). Three-finger ZFs have also been dimerized by using PDZ domains (26). Cooperative interactions are of interest because they potentially increase the nonlinearity of regulation, as well as decrease nonspecific binding compared with extended arrays of ZFs. To study cooperative interactions, we built several different ZFs fused to either PDZ or LZ domains (Fig. 2B).
In parallel, we designed corresponding repressible promoter libraries. As we used an E. coli cell-free system (44), we based our promoter designs on the strong promoter in combination with transcription and translation elements optimized for E. coli cell-free expression (45). Previous work has shown that the most effective position for transcriptional repression is the space between the −35 and −10 boxes (30); we thus generated a library with consensus ZF binding sites (ZFBSs) inserted into this location. Additionally, we built promoters with a second ZFBS upstream of the −35 box, allowing us to study the effect of multiple noncooperative and cooperative ZFBSs (Fig. 2B). The promoters drive expression of a deGFP reporter, a GFP protein previously optimized for cell-free translation (46). All constructs were built and tested by using linear DNA templates generated by PCR in concordance with recommended guidelines for cell-free expression (45).
Repression with Single and Multiple Binding Sites.
We performed an in-depth characterization of 11 synthetic ZFs by assessing their repressive capacity in cell-free reactions and by measuring their respective dissociation constants () with MITOMI. We used MITOMI to measure the values for each ZF against all possible target promoters. By localizing presynthesized his-tagged ZFs to the surface of each unit cell, we were able to measure the binding of DNA sequences spanning the promoter region, including the ZF binding site (Fig. 3A and SI Appendix, Fig. S1B). We obtained standard Gibbs free energies, , for each ZF–target promoter complex (Fig. 3B). A range of binding strengths was observed for the respective consensus ZF binding sequences, as well as low-affinity off-target binding. The CBD ZF was included as a negative control, as it does not bind to its own predicted binding site or any of the other targets.
Fig. 3.
ZF binding affinities, functional repression, and orthogonality. (A) Schematic depicting the MITOMI assay used to determine TF–DNA binding affinities. (B) Affinity orthogonality matrix of values for all ZFs vs. all possible DNA targets. (C) Schematic depicting the linear templates used to test functional repression in on-chip cell-free TX-TL reactions. (D) Fold repression orthogonality matrix for all ZFs vs. all possible targets. (E) Fold repression values vs. measured for all ZF–promoter consensus pairs. The fold-repression data were collected from a single chip, and all values represent means SD (). The error bars shown for the values represent the 95% confidence interval for the fit to a single binding-site model.
To test whether the relative binding strength of each ZF related to functional gene repression, we implemented cell-free TX-TL reactions screening the same matrix of ZFs vs. promoters. Each microfluidic unit cell contained a linear template encoding the ZF to be tested and a second linear template encoding deGFP downstream of a promoter with a single ZF binding site (Fig. 3C). Binding of the expressed ZF to the target promoter would lead to down-regulation of deGFP expression. A common measure of repression performance is fold repression, or the ratio of unrepressed to repressed expression levels. Unrepressed measurements were obtained by coexpressing the target promoter template with the nonbinding template to control for loading effects (47). Despite some off-target binding observed by MITOMI, functional repression of all ZF–target pairs was almost perfectly orthogonal (Fig. 3D), with one exception: the repression of promoter by . However the general trend of weak off-target affinities translated to no or minimal off-target repression, resulting in functional repression only for cognate pairs. Furthermore, on-target fold repression directly correlated with the measured MITOMI affinity values (Fig. 3E). Using two high-throughput microfluidic techniques, we were able to characterize the binding affinity, repressive strength, and orthogonality of synthetic transcription factor–promoter pairs.
Promoters with a single ZF binding site achieved low to medium fold repression levels in the range of 1.5–7 (Fig. 4A). We tested whether placing an additional binding site upstream of the −35 box could further improve fold repression levels. While fold repression is a convenient measure used to describe the functionality of a given repressor–promoter pair, for applying these repressors in genetic networks, it is important to also consider basal promoter strength (unrepressed state) and leak (repressed state). These quantities are also shown in Fig. 4, where we observed that variation in binding-site sequence led to variations in basal promoter strength; this variation increased upon inclusion of the second binding site upstream of the −35 box. At the same time, the average leak from the repressed state decreased for the dual site library, resulting in higher fold-repression values. Overall, fold repression improved for almost all two-binding-site promoters, with the best promoters achieving a fold repression level of 7–10 (Fig. 4B). These results showed that good repression levels can be achieved by synthetic ZF repressors with either single or double binding-site promoters in a cell-free system.
Fig. 4.
Effect of binding-site number and position on repression. (A and B) Shown are endpoint unrepressed and repressed levels for the single-binding-site library (A) and the dual-site library (B). Within each library, the data are rank-ordered by fold repression values. Data were collected from two separate chips, and all values represent means SD (). (C) A single BCB site was shifted up to 10 bp upstream and 7 bp downstream of the −35 box; position values are given by the number of nucleotides separating the 9-bp site from the −35 box. (C, Lower) The binding affinities of the ZF to its target site remain approximately constant, irrespective of target site position. Data from C, Upper were measured from a single chip, and all values represent means SD (). The error bars shown for the values represent the 95% confidence interval of the fit to a single binding-site model. RFU, relative fluorescence units.
Next, we characterized the effect of binding-site position on repression strength. We generated a library of promoters containing a single ZF binding site that was placed in various positions relative to the −35 box. The best fold repression was achieved by positioning binding sites directly proximal to the −35 box, in the range of −2 to +4 bps relative to the start and end of the −35 box, respectively. We also observed that repression was sensitive to single base-pair shifts in position. For instance, the site at the +5 position was effectively nonfunctional compared with repressing neighboring sites at +4 and +6, and the site at the −5 position exhibited significantly stronger repression than its neighbors at −4 and −6. Based on the crystal-structure alignment of ZF and RNA polymerase (RNAP) bound to DNA containing the binding site at position +5, we note that it is possible for both proteins to bind simultaneously with minimal steric interference. To ascertain that the observed repression strengths were not due to changes in binding-site affinity of the ZF, as each binding site is located in a different sequence context, we measured the binding affinity of the ZF repressor to each promoter using MITOMI. The results showed only minor differences in affinity across all promoters, suggesting that the ZF repressor bound to these promoters with equal strength. Promoter repression thus appears to be primarily a function of the ability of the ZF to sterically hinder and compete with RNAP. These data are consistent with an occlusion mechanism whereby RNAP binding is competitively inhibited by ZF binding (29), and the effectiveness of the competition is dependent on the relative positions of ZF and RNAP on the promoter.
Engineering Cooperativity.
We showed that incorporating a second binding site can result in improved fold repression. However, engineering certain types of genetic circuits often requires an additional increase in the nonlinear response, as well as a decrease in the leak for a given promoter–TF pair. Nonlinearity can be increased by introducing cooperativity via protein–protein interactions. We implemented two different protein-interaction domains previously demonstrated to successfully dimerize ZFs.
PDZ domains enable natural protein–protein interactions by binding specific C-terminal peptide sequences with micromolar affinity (26). We took advantage of this interaction to engineer cooperativity by linking to a mammalian -syntrophin PDZ domain and to its corresponding cognate C-terminal peptide ligand (VKESLV). Furthermore, we linked with a noncognate ligand (VKEAAA) to use as a noncooperative control.
The second type of interaction we explored was dimerization by linking and to GCN4 LZ domains. The GCN4 LZ has been used in a structure-based design to enable homodimerization of two-finger ZFs (42), and we thus also tested this existing structure. In both cases, a mutated LZ was used as a negative control.
Preliminary studies on a plate reader demonstrated that ZFs containing interaction domains exhibited significantly increased fold repression and decreased leak (Fig. 5 A and B). Whereas two noncooperative repressors gave a maximum fold repression of 6, this value was increased to 30 for PDZ and 16 for LZ-mediated cooperativity. Concurrently, leak values decreased fourfold from ∼4,000 to 1,000 relative fluorescence units. One critical parameter affecting PDZ cooperativity was the choice of linker, with an optimized glycine–serine linker vastly outperforming a rigid proline linker. The two-finger LZ transcriptional repressor also performed very well, achieving a fold repression ratio of 28.
Fig. 5.
Engineering cooperativity. (A and B) Comparison of unrepressed and repressed (Repr.) levels (A), as well as fold repression (B), for different cooperative (coop.) ZF designs. Three-finger ZFs dimerized by using either PDZ–ligand or GCN4 LZ domains are shown. ZFs were linked to interaction domains by using proline (–PDZ*) or optimized glycine–serine linkers (–PDZ, –LZ). Additionally, two-finger ZFs were dimerized by using LZs (AA-LZ). Data in A and B were taken from plate-reader experiments; all values represent means SD (). (C–E) The dose–response is shown for individual ZF repressors as well as noncooperative (−) and cooperative (+) pairs of ZFs, for the three-finger PDZ and two- and three-finger LZ designs. The maximum a posteriori values as well as 2-SD boundaries of the model predictions are represented as solid lines and shaded regions, respectively. Data in C–E were measured on a single chip; all values represent means SD (). (F–H) Shifting the binding site upstream of the −35 box resulted in periodic modulation of the fold repression for the cooperative designs (F), as well as in the ratio between the cooperative and noncooperative fold repressions (G), likely due to periodic changes in relative ZF positioning (H). All data were collected from a single chip, and all values represent means SD (). RFU, relative fluorescence units.
To investigate cooperativity in more detail, we measured dose–response curves by titrating repressor DNA concentration. To keep a fixed load on the TX-TL machinery, the total ZF DNA concentration was kept constant by adding DNA coding for a nonbinding ZF control (). Fig. 5C shows dose–response curves of and separately, together with those for the cooperative pair, + , and the noncooperative pair, + . An increase in the steepness of the dose–response curve was observed as we proceeded from a single ZF to two noncooperatively interacting ZFs, and finally to two cooperatively interacting ZFs. Similar results were obtained for the LZ designs (Fig. 5 D and E). The effect of cooperativity can be quantified by determining the sensitivity (SI Appendix, Fig. S7), which measures the steepness of the dose–response curve (48), as well as the effective Hill coefficient, which is obtained by fitting phenomenological Hill functions (SI Appendix, Fig. S8). The results of this analysis are shown in SI Appendix, Table S1. We observe that cooperativity increased sensitivity by nearly with respect to the noncooperative repression, as well as slightly increasing the Hill coefficient.
We sought to understand this behavior quantitatively by developing a thermodynamic model that relates protein expression to the equilibrium occupancy of the promoter by RNAP (49). We extended the standard competitive model of repression to include a term for the interaction between repressor and RNAP, which is quantified by an effective interaction energy. As this energy tends to large positive values, DNA binding by either RNAP or the repressor is exclusive, and the model tends toward that of competitive inhibition. As the energy approaches zero, both RNAP and DNA can bind simultaneously, resulting in leaky expression at full repressor occupancy. This extension to the model was motivated by our results that a ZF with a fixed binding affinity represses with varying efficiency, depending on the position of the binding site; the changing RNAP–ZF interaction energy therefore provides a simple description of this effect. We fit the model to the dose–response curves using Markov chain Monte Carlo (MCMC) sampling (SI Appendix, Fig. S9), allowing us to consistently extract the posterior probability distributions of all parameters, which consist of fixed effective dissociation constants of each individual ZF, as well as the effective energies describing ZF–RNAP and ZF–ZF interactions. The fits are shown in Fig. 5 C–E as solid lines and shading, which represent the mean and 2-SD boundaries for model predictions, respectively. The values of all fitted parameters are given in SI Appendix, Table S2, and a full description of the model is given in Materials and Methods. We found physically sensible values for all our parameters; in particular, the cooperative interaction energies for PDZ–ligand ( kcal/mol) and LZ ( kcal/mol) were consistent with literature values for similar domains [ to −10 kcal/mol (50, 51)].
Since the location of the ZF binding site, and hence the relative positioning of ZF and RNAP, is an important determinant of repression efficiency, it is likely that the relative positioning of the and binding sites would also determine their ability to interact and subsequently alter their repressive strength. Keeping the binding-site position fixed, we shifted the binding site further and further upstream. If the two ZFs are positioned on the promoter such that the cooperative PDZ–ligand interaction is unfavorable, we would expect fold repression to be similar to that of the noncooperative ZFs. In other words, the ratio between the cooperative and the noncooperative fold repression, a quantity we call the cooperativity ratio, should go to unity when the PDZ–ligand interaction cannot occur.
We observed an effect due to this variation of spacing between the two binding sites (Fig. 5F), and this behavior corresponded to the relative orientation of the PDZ–ligand domains. As the binding site was shifted, rotated around the DNA, modulating its alignment with . The cooperativity ratio fell to 1 when the interaction was unfavorably aligned, but increased again as the domains began to realign (Fig. 5G). The cartoon in Fig. 5H shows the predicted orientations of the two ZFs as the left-hand site is shifted. The ability of the ZFs to interact over distances of a few tens of base pairs is likely due to extension of the long, flexible glycine–serine linker used to join the and the PDZ domain. It is unlikely that DNA bending plays a significant role at these distances, due to dsDNA’s much longer persistence length of 150 bp.
We incorporated into our model a phenomenological exponential decay of interaction energies with distance, both between the two ZFs as well as between the ZF and the RNAP. Additionally, the ZF–ZF interaction energy was modulated by a periodic function at the frequency of the DNA helical pitch (10.5 bp per turn). Using previously inferred parameters for energies and values from the dose–response measurements, we performed a fit to determine the decay constant and phase shift; the results are shown as solid lines and shading in Fig. 5 F and G and in SI Appendix, Table S2. Fitting a model with an explicit position dependence for the binding sites illustrates the importance of site positioning for functional repression. More generally, while simplistic, our model fits demonstrate that it is possible to understand cell-free gene expression in terms of thermodynamic occupancy.
Affinity Tuning.
To test whether fold repression levels could be precisely and predictively tuned, we investigated the effect of varying binding-site affinity. To rationally tune binding-site affinity, we first generated quantitative PWMs for three ZFs: , , and , covering the 9-bp core sequence plus three flanking bases on either side (Fig. 6A and SI Appendix, Fig. S10 A and B). The sequence logo determined for is in concordance with the consensus sequence determined by bacterial one-hybrid and in vitro SELEX assays (52, 53). Based on our PWMs, we designed a library of promoters that included a single binding site at a fixed position between the −35 and −10 boxes, with single or double mutations within or outside the core binding sequence. As binding-site affinity decreased, we observed corresponding decreases in fold repression for all ZFs tested (Fig. 6B and SI Appendix, Fig. S10C). By converting our macroscopically measured values into microscopic interaction energies , we found that the fold-repression data could be described by the same thermodynamic model presented in the previous section.
Fig. 6.
Tuning repression by changing binding-site affinity. (A) Schematic of the MITOMI assay used for measuring the binding affinity of a ZF to a given DNA target. (A, Left) The sequence logo and PWM for , where the core sequence is designated by positions 4–12. (B) The relationship between fold repression and values for . The fold repression data were collected from three separate chips, and all values represent means SD (). (C) Schematic of the MITOMI surface used for measuring the binding affinity of the homodimer to a given DNA target. (C, Left) The sequence logo and PWM for , where the core sequence is designated by positions 3–12. (D) The relationship between fold repression and values for both the cooperative (coop.) and noncooperative (non-coop.) variants of . The fold repression data were collected from a single chip, and all values represent means SD (). (E) Fold repression vs. values for the – heterodimer pair. In E, Left, the values refer to the arising from the specific change made to the ADD binding site, whereas in E, Right, the values are associated with the BCB binding site. The fold repression data were collected from a single chip, and all values represent means SD (). In all cases, the error bars shown for the values represent the 95% confidence interval for the fit to a single binding-site model; the solid lines are maximum a posteriori values from thermodynamic model fits; and the shaded region represents a 2-SD boundary.
Mutating either a single base outside the core site, or one core position of low information content (high entropy), enabled fine tuning of fold repression, whereas a single mutation in the core site of high information content strongly decreased fold repression. Two core mutations decreased fold repression to baseline levels. Fold repression was therefore precisely tunable over the entire dynamic range by modulating binding-site affinity, and the affinity changes required to achieve tuning were relatively small. Affinity changes of to 1 kcal/mol were sufficient to cover the entire dynamic range for each ZF repressor tested. The results are in line with previous findings that promoter tuning in S. cerevisiae can be accomplished by relatively subtle affinity changes in a single binding site created by mutations in flanking or single-core site mutations of high entropy (54). They also correspond to recent results obtained in E. coli (55).
Given that a single ZF binding site could be mutated to yield varying levels of repression, we investigated whether the same tuning could be applied to cooperative s. We measured the binding affinity of the homodimer vs. a library of DNA targets that consisted of all single-point mutations for the 10-bp core binding sequence plus two flanking bases on either side. The resulting sequence logo and PWM reveal the symmetric binding profile of the homodimer (Fig. 6C). Mutating a single binding site within the −35 and −10 boxes led to a change in repression levels that reflected the measured s for both the cooperative and noncooperative variants (Fig. 6D). As the two 6-bp binding sequences overlap, mutating a single base within the core site leads to a finer tuning of fold repression in comparison with the three-finger ZFs. Furthermore, we extended binding-site tuning to the – heterodimer pair, taking advantage of the PWMs generated for and . Implementing a subset of mutations to each ZF binding site yielded a range of fold repression values not only for the single ZF but also for the cooperative and noncooperative ZF pairs (Fig. 6E). As the affinity of one ZF is reduced, we see that the fold repression observed for the cooperative and noncooperative cases tends to the fold repression measured for the second ZF, whose binding site remains constant.
Logic-Gate Construction.
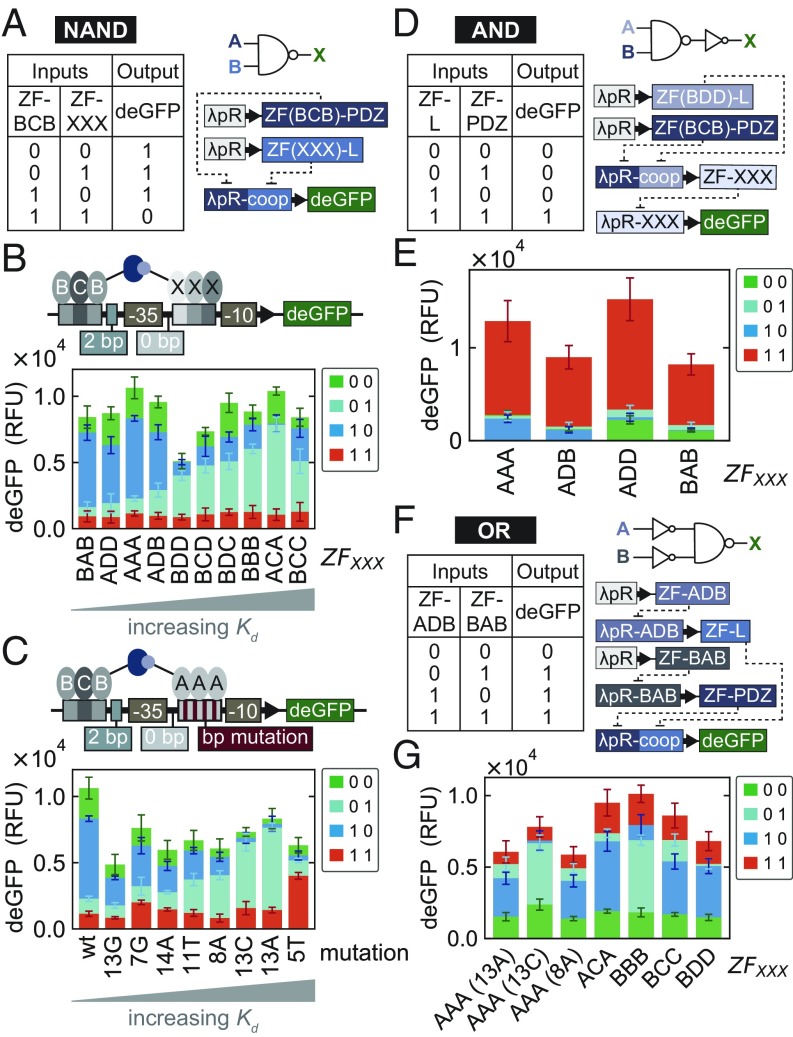
Having established a well-characterized resource of transcriptional repressors and promoters, we applied them to designing logic gates. By combining two cooperative ZF repressors on a single promoter, we were able to create NAND gates, which are of particular interest, as they are functionally complete. An effective NAND gate should have low output only when both inputs are present (Fig. 7A). We therefore placed the binding site for a strongly binding ZF () 2 bp upstream of the −35 box and second binding site for different ZFs between the −35 and −10 boxes. cannot strongly repress by itself at the −2 position, and the second ZF should also not strongly repress on its own. Only when both ZFs are bound to the promoter should they strongly repress, which can be achieved by including a cooperative interaction between the two ZFs. Using this general design, we tested NAND gates for in combination with the remaining ZFs (Fig. 7B). As expected, NAND gate performance improved as the affinity of the decreased. For instance, the combination of and gave rise to a functional NAND gate, whereas a combination with did not, due to the high affinity of , which led to functional repression even when only was present.
Fig. 7.
Logic gates. (A) Truth table, logic-gate symbol, and biological network design for constructing a NAND logic gate. (B) NAND gate design combining with all other values. (B, Lower) The output for each NAND gate tested. (C) The same NAND gate design as in A except that only and are used as inputs and the binding-site affinity is rationally adjusted to yield a functional NAND gate. The deGFP outputs for all NAND gates were measured from a single chip, where all values presented correspond to the mean SD (). (D) Truth table, logic gate symbols, and biological network design for constructing an AND logic gate. (E) The output for each AND gate tested. All output values were measured from a single chip and represent the mean SD (). (F) Truth table, logic gate symbols, and biological network design for constructing an OR logic gate. (G) The output for each OR gate tested. When is used as part of the NAND gate, the mutation of the binding site is indicated in parentheses. All output values were measured from a single chip and represent the mean SD (). RFU, relative fluorescence units.
Since we showed that binding affinity could be precisely tuned (Fig. 6), we tested whether we could improve our nonfunctional NAND gates. Based on the PWM measured for , we mutated the binding-site sequence in the NAND gate promoter and showed that we could achieve tuning in this context as well (SI Appendix, Fig. S10D). We then investigated the effect of tuning the binding site for all possible input combinations and showed that the NAND gate improved as we weakened binding affinity (Fig. 7C). Mutations +1C and +1A gave rise to functional NAND gates. Decreasing the binding-site affinity increased the output when only was present; however, when the mutation resulted in a of greater than kcal/mol (), the cooperative binding output also suffered. Our synthetic ZF repressors can thus be used to build functional NAND gates, which can additionally be rationally optimized and precisely tuned by modifying binding-site affinities.
As a final example, we generated compound logic gates by combining NAND and NOT logic gates as linear cascades to create AND and OR gates. We created an AND gate by appending a NOT gate to the output of a NAND gate (Fig. 7D). Specifically, we combined the – NAND gate with four different ZFs. Each AND gate was tested and yielded the expected outputs (Fig. 7E). We then generated OR logic gates by prepending two NOT gates in front of different NAND gates to invert the inputs (Fig. 7F). We used and as the two NOT gate inverters and a set of NAND gates, all of which gave rise to functional OR gates (Fig. 7G).
Discussion
GRNs are of central importance in both native and engineered systems. They integrate, compute, and transduce input signals, leading to specific changes in gene expression. Many components contribute to the function of GRNs, and transcription factors and their interaction with promoters are core players. Due to the complexity of even a single transcription factor–promoter interaction, it has proven difficult to quantitatively study these systems in vitro or in vivo. Although the development of new technologies is steadily enabling progress in this area, our understanding of GRNs remains limited, as exemplified by our inability to predict in vivo gene-expression levels in essentially any organism and the difficulty associated with de novo engineering of GRNs. Although methods exist for high-throughput in vitro characterization of transcription-factor-binding specificities (34, 56–58), and medium- to high-throughput approaches are used to understand gene regulation in vivo (33, 54, 55, 59), both approaches have limitations. Both an advantage and disadvantage of in vitro methods is that they generally include only the smallest number of components necessary, i.e., a transcription factor, dsDNA target, and a defined buffer solution. In vivo methods are, on the other hand, convoluted by cellular complexity. Furthermore, generating and analyzing defined libraries in vivo remains labor-intensive and difficult. Here, we explored the use of a cell-free TX-TL system to build and characterize GRNs in an environment that bridges the gap between in vitro and in vivo methods. This cell-free approach also has the advantage of allowing complex assays to be performed in high-throughput, in a well-controlled and accessible environment. As a consequence, the ability to study functional transcriptional regulation in an in vitro system has allowed us to delve into much greater depth than comparable in vivo methods have been able to achieve (54, 60, 61).
We chose to build GRNs from the bottom up using ZF transcription factors for several reasons. First, in regard to GRN engineering, researchers have long been hampered by the relatively small number and poor characterization of available transcriptional regulators. Khalil et al. (26) have engineered ZF regulators, showing that they are viable tunable transcriptional regulators in vivo. We built on this concept, generating additional ZF regulators and interaction domains. More importantly, we quantified the binding-energy landscapes of several synthetic ZF regulators and were able to show that repression can be precisely tuned with small changes in affinity. These small changes were achieved by mutating the flanking bases lying outside of the consensus core sequence or by mutating one consensus core base of low information content. Hitherto, only coarse tuning has been accomplished through varying the number of consensus-sequence binding sites, leading to rather large differences in output (26, 27). The ability to predictively and precisely tune expression levels as demonstrated here is important in engineered GRNs, where individual nodes of the network need to be matched in expression levels. For example, we show here that the ability to precisely adjust individual binding-site affinities is crucially important for optimizing logic gate function.
With the advent of TALEs and dCas9, ZFs might be considered outdated technology, but there are a number of reasons why ZF TFs remain an appealing tool for GRN engineering. ZFs have several advantages, such as small size, relatively easy gene synthesis, and good expressability. The biggest advantage of dCas9 and TALEs is their programmability, allowing them to be precisely targeted to any DNA sequence. Conversely, for ZFs, it remains relatively difficult to rationally design a particular binding-site preference. For genome editing and in vivo targeting approaches, in which the target sequence is defined and immutable, programmability is crucial. In the context of bottom-up GRN design, this ability becomes less important as target sequences can be easily adjusted to a particular TF specificity. We argue that it is actually more important to be in possession of a well-characterized TF binding-energy landscape that can be obtained for ZF TFs using current methods (28).
A second argument in support of using ZF transcription factors over TALEs and dCas9 is the simple but important fact that ZFs are native transcriptional regulators and the most abundant class of transcriptional regulators in vivo. Cas9, to the best of our knowledge, has not been shown to be involved in gene regulation in native systems, while TALEs are injected into plant host cells to modulate gene expression by pathogenic bacteria (62). If cell-free approaches are to be used to understand the function of native systems, it is important to build GRNs with native transcription factors. For example, the protein–DNA interaction kinetics are very different in that dCas9 (63) and TALE (64) tend to have very slow DNA dissociation rates, while native transcriptional regulators have fast dissociation rates (65), which may make engineering dynamic GRNs using TALEs and dCas9 difficult.
To improve fold repression and to add more control over the system, we engineered cooperative binding into our ZF TFs by including PDZ or LZ protein–protein interaction domains. These interactions improved repression from 10-fold to up to 30-fold and were functional for both two- and three-finger ZFs. We showed that the relative placement of binding sites for two cooperative TFs is a major determinant of interaction capacity and, consequently, repression strength. Repression was achieved when the TFs were located on the same face of the DNA, and repression strength followed the helical twist of DNA. Cooperative interactions consequently allowed us to engineer functionally complete NAND gates. In all cases, we were able to explain our data with thermodynamic models. Combining these models with binding-energy landscapes thus provides a viable and useful approach to rationally engineer GRNs.
One outstanding problem encountered during this study is the issue of composability. Although transcription-factor binding sites were only introduced in regions outside the −10 and −35 boxes of the original promoter, many of the synthetic promoters had considerably different baseline (nonrepressed) expression levels. In the future, it will clearly be important to better understand and predict basal promoter strength from the underlying sequence, which would lead to models that allow introduction of transcription-factor binding sites without affecting basal promoter output. Here, we have seen that basal promoter strength itself can be finely tuned over a relatively large range of expression levels (Fig. 1). It should therefore be possible to adjust promoter strength as desired: We demonstrate a basic example of this idea by tuning the basal expression level of a repressible promoter (SI Appendix, Fig. S11). Ultimately, understanding the outcome of multiple base changes in close context with each other remains a complex issue. Evaluating a greater number of sequences and systematically addressing all factors affecting transcription efficiency, similar to the approach taken by Cambray et al. (66) toward translation, could lead to an improved understanding of promoter-sequence design principles.
To characterize and measure our synthetic ZF transcription factors and promoters in detail, we repurposed a high-throughput microfluidic device that allowed us to measure 768 cell-free reactions in parallel. Eliminating cloning and transformation steps by relying on PCR-based assembly strategies allowed us to measure a large number of defined transcription-factor and promoter variants. Over 13,000 on-chip cell-free TX-TL reactions were performed, encompassing replicates for 2,000 unique reactions. We furthermore took >8,000 MITOMI measurements to provide binding-energy landscapes for four synthetic ZF transcription factors. Together, these technologies allowed us to establish a quantitative and in-depth dataset and insights into transcriptional regulation that should be of general interest. The approach taken here nonetheless does not per se require these state-of-the-art technologies and is easily transferable to standard laboratory equipment. Cell-free lysate can now be easily and cheaply generated, yielding sufficient material so that medium-scale screens in 384-well plates are feasible (44). Commercial liquid-handling equipment can also be used to scale up throughput. Binding-energy landscapes can be generated by many approaches, including PBMs (56), MITOMI (67), SELEX-seq (57), and HiP-FA (58). While our binding-energy landscapes are based on direct affinity measurements, it may be sufficient to use PWMs from indirect measurements as found in other high-throughput techniques.
Rapid progress is being made in the development and application of cell-free synthetic biology. Cell-free systems are being used to tackle fundamental problems in molecular engineering and are being applied to molecular diagnostics (68), therapeutics (69), and synthesis (70) and are even being used for educational purposes (71). Cell-free systems are an appealing alternative to cellular systems, as they eliminate many of the complexities associated with working with cells. Cell-free systems are also a rapid prototyping platform for engineering molecular systems destined to be applied in cellular hosts (4). As engineered systems become more complex, it will become increasingly important that a large number of standardized characterized components become available. It will be equally important to develop a comprehensive mechanistic understanding of these components and systems to allow parts to be standardized and rationally assembled without requiring extensive trial-and-error cycles or large screens, which may not be feasible for large systems. As work progresses on cellular subsystems such as gene regulation, DNA replication, ribosome biogenesis, metabolic networks, and membrane and protein superstructures, it will be intriguing to contemplate whether it may be possible to integrate these individual systems to create a synthetic cell or cell-like mimic. Work in this area will not only provide tools and methods aiding the engineering of synthetic systems, but is likely to provide insights into the function of native systems as well. Before being used as tools for protein synthesis and synthetic biology, cell-free systems have already had a rich history in deciphering fundamental aspects of biochemistry, including DNA replication (72) and the genetic code (73). It is likely that they will continue to provide fundamental insights into complex systems such as transcriptional regulation.
Materials and Methods
Full details are given in SI Appendix. High-throughput cell-free experiments were conducted on a PDMS microfluidic device. DNA templates were assembled by PCR from plasmids and dsDNA gene fragments and spotted onto a glass slide by using a robotic microarrayer. E. coli (BL21 Rosetta) cell-free extract, which contains native enzymes, substrates, and an energy-regeneration mechanism, was used in all experiments. Binding-energy landscapes were obtained by microfluidic MITOMI measurements. Our thermodynamic model was fit to experimental data by using a MCMC method to determine the posterior probability distributions of model parameters. All supporting data and code are available on GitHub (74).
Supplementary Material
Acknowledgments
We thank S. Clamons and M. Yun (Murray laboratory, Caltech) for providing the cell-free TX-TL extract; S. Clamons and R. Murray for helpful discussions; and M. Kabani, E. Pankevich, and S. Bassler for their experimental contributions to this project. This work was supported by Human Frontier Science Program Grant RGP0032/2015; the European Research Council under the European Union’s Horizon 2020 research and innovation program Grant 723106; and the École Polytechnique Fédérale de Lausanne.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.C.J. is a guest editor invited by the Editorial Board.
Data deposition: All supporting data and code are available on GitHub at https://github.com/lbnc-epfl.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816591116/-/DCSupplemental.
References
- 1.Noireaux V, Bar-Ziv R, Libchaber A. Principles of cell-free genetic circuit assembly. Proc Natl Acad Sci USA. 2003;100:12672–12677. doi: 10.1073/pnas.2135496100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin J, Noireaux V. An E. coli cell-free expression toolbox: Application to synthetic gene circuits and artificial cells. ACS Synth Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 3.Niederholtmeyer H, Stepanova V, Maerkl SJ. Implementation of cell-free biological networks at steady state. Proc Natl Acad Sci USA. 2013;110:15985–15990. doi: 10.1073/pnas.1311166110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niederholtmeyer H, et al. Rapid cell-free forward engineering of novel genetic ring oscillators. eLife. 2015;4:e09771. doi: 10.7554/eLife.09771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karzbrun E, Tayar AM, Noireaux V, Bar-Ziv RH. Programmable on-chip DNA compartments as artificial cells. Science. 2014;345:829–832. doi: 10.1126/science.1255550. [DOI] [PubMed] [Google Scholar]
- 6.Chappell J, Jensen K, Freemont PS. Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res. 2013;41:3471–3481. doi: 10.1093/nar/gkt052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwille P, et al. MaxSynBio: Avenues towards creating cells from the bottom up. Angew Chem Int Ed. 2018;57:13382–13392. doi: 10.1002/anie.201802288. [DOI] [PubMed] [Google Scholar]
- 8.Forster AC, Church GM. Towards synthesis of a minimal cell. Mol Syst Biol. 2006;2:1–10. doi: 10.1038/msb4100090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Nies P, et al. Self-replication of DNA by its encoded proteins in liposome-based synthetic cells. Nat Commun. 2018;9:1583. doi: 10.1038/s41467-018-03926-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otrin L, et al. Toward artificial mitochondrion: Mimicking oxidative phosphorylation in polymer and hybrid membranes. Nano Lett. 2017;17:6816–6821. doi: 10.1021/acs.nanolett.7b03093. [DOI] [PubMed] [Google Scholar]
- 11.Jewett MC, Fritz BR, Timmerman LE, Church GM. In vitro integration of ribosomal RNA synthesis, ribosome assembly, and translation. Mol Syst Biol. 2013;9:678. doi: 10.1038/msb.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya A, Brea RJ, Devaraj NK. De novo vesicle formation and growth: An integrative approach to artificial cells. Chem Sci. 2017;8:7912–7922. doi: 10.1039/c7sc02339a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furusato T, et al. De novo synthesis of basal bacterial cell division proteins FtsZ, FtsA, and ZipA inside giant vesicles. ACS Synth Biol. 2018;7:953–961. doi: 10.1021/acssynbio.7b00350. [DOI] [PubMed] [Google Scholar]
- 14.Brophy JAN, Voigt CA. Principles of genetic circuit design. Nat Methods. 2014;11:508–520. doi: 10.1038/nmeth.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nielsen AlecAK, et al. Genetic circuit design automation. Science. 2016;352:aac7341. doi: 10.1126/science.aac7341. [DOI] [PubMed] [Google Scholar]
- 16.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 17.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 18.Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- 19.Stanton BC, et al. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat Chem Biol. 2014;10:99–105. doi: 10.1038/nchembio.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: Function, expression and evolution. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 21.Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 22.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. The New Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najafabadi HS, et al. C2h2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 24.Fu F, Voytas DF. Zinc finger database (ZiFDB) v2.0: A comprehensive database of c2h2 zinc fingers and engineered zinc finger arrays. Nucleic Acids Res. 2013;41:D452–D455. doi: 10.1093/nar/gks1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore R, Chandrahas A, Bleris L. Transcription activator-like effectors: A toolkit for synthetic biology. ACS Synth Biol. 2014;3:708–716. doi: 10.1021/sb400137b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalil AS, et al. A synthetic biology framework for programming eukaryotic transcription functions. Cell. 2012;150:647–658. doi: 10.1016/j.cell.2012.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohmueller JJ, Armel TZ, Silver PA. A tunable zinc finger-based framework for Boolean logic computation in mammalian cells. Nucleic Acids Res. 2012;40:5180–5187. doi: 10.1093/nar/gks142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blackburn MC, Petrova E, Correia BE, Maerkl SJ. Integrating gene synthesis and microfluidic protein analysis for rapid protein engineering. Nucleic Acids Res. 2015;44:e68. doi: 10.1093/nar/gkv1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ptashne M, et al. Autoregulation and function of a repressor in bacteriophage lambda. Science. 1976;194:156–161. doi: 10.1126/science.959843. [DOI] [PubMed] [Google Scholar]
- 30.Cox RS, III, Surette MG, Elowitz MB. Programming gene expression with combinatorial promoters. Mol Syst Biol. 2007;3:1–11. doi: 10.1038/msb4100187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I-I-I-2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lanzer M, Bujard H. Promoters largely determine the efficiency of repressor action. Proc Natl Acad Sci USA. 1988;85:8973–8977. doi: 10.1073/pnas.85.23.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharon E, et al. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol. 2012;30:521–530. doi: 10.1038/nbt.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–237. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 35.Moore SJ, et al. Rapid acquisition and model-based analysis of cell-free transcription-translation reactions from nonmodel bacteria. Proc Natl Acad Sci USA. 2018;115:E4340–E4349. doi: 10.1073/pnas.1715806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Cordero JL, Maerkl SJ. Mechanically induced trapping of molecular interactions and its applications. J Lab Automation. 2016;21:356–367. doi: 10.1177/2211068215578586. [DOI] [PubMed] [Google Scholar]
- 37.Kinney JB, Murugan A, Callan CG, Jr, Cox EC. Using deep sequencing to characterize the biophysical mechanism of a transcriptional regulatory sequence. Proc Natl Acad Sci USA. 2010;107:9158–9163. doi: 10.1073/pnas.1004290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kamiuchi T, et al. Artificial nine zinc-finger peptide with 30 base pair binding sites. Biochemistry. 1998;37:13827–13834. doi: 10.1021/bi9811112. [DOI] [PubMed] [Google Scholar]
- 39.Kim J-S, Pabo CO. Getting a handhold on DNA: Design of poly-zinc finger proteins with femtomolar dissociation constants. Proc Natl Acad Sci USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore M, Klug A, Choo Y. Improved DNA binding specificity from polyzinc finger peptides by using strings of two-finger units. Proc Natl Acad Sci USA. 2001;98:1437–1441. doi: 10.1073/pnas.98.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pomerantz JL, Wolfe SA, Pabo CO. Structure-based design of a dimeric zinc finger protein. Biochemistry. 1998;37:965–970. doi: 10.1021/bi972464o. [DOI] [PubMed] [Google Scholar]
- 42.Wolfe SA, Ramm EI, Pabo CO. Combining structure-based design with phage display to create new Cys2His2 zinc finger dimers. Structure. 2000;8:739–750. doi: 10.1016/s0969-2126(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 43.Wolfe SA, Grant RA, Pabo CO. Structure of a designed dimeric zinc finger protein bound to DNA. Biochemistry. 2003;42:13401–13409. doi: 10.1021/bi034830b. [DOI] [PubMed] [Google Scholar]
- 44.Sun ZZ, et al. Protocols for implementing an Escherichia coli based TX-TL cell-free expression system for synthetic biology. J Visualized Exp. 2013;79:e50762. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun ZZ, Yeung E, Hayes CA, Noireaux V, Murray RM. Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth Biol. 2014;3:387–397. doi: 10.1021/sb400131a. [DOI] [PubMed] [Google Scholar]
- 46.Shin J, Noireaux V. Efficient cell-free expression with the endogenous E. coli RNA polymerase and sigma factor 70. J Biol Eng. 2010;4:8. doi: 10.1186/1754-1611-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegal-Gaskins D, Tuza ZA, Kim J, Noireaux V, Murray RM. Gene circuit performance characterization and resource usage in a cell-free ‘breadboard’. ACS Synth Biol. 2014;3:416–425. doi: 10.1021/sb400203p. [DOI] [PubMed] [Google Scholar]
- 48.Bintu L, et al. Transcriptional regulation by the numbers: Applications. Curr Opin Genet Development. 2005b;15:125–135. doi: 10.1016/j.gde.2005.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bintu L, et al. Transcriptional regulation by the numbers: Models. Curr Opin Genet Development. 2005;15:116–124. doi: 10.1016/j.gde.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saro D, et al. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry. 2007;46:6340–6352. doi: 10.1021/bi062088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jana NK, Deb S, Bhattacharyya B, Mandal NC, Roy S. A study of energetics of cooperative interaction using a mutant lambda-repressor. Protein Eng. 2000;13:629–633. doi: 10.1093/protein/13.9.629. [DOI] [PubMed] [Google Scholar]
- 52.Meng X, Brodsky MH, Wolfe SA. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wolfe SA, Greisman HA, Ramm EI, Pabo CO. Analysis of zinc fingers optimized via phage display: Evaluating the utility of a recognition code. J Mol Biol. 1999;285:1917–1934. doi: 10.1006/jmbi.1998.2421. [DOI] [PubMed] [Google Scholar]
- 54.Rajkumar AS, Dénervaud N, Maerkl SJ. Mapping the fine structure of a eukaryotic promoter input-output function. Nat Genet. 2013;45:1207–1215. doi: 10.1038/ng.2729. [DOI] [PubMed] [Google Scholar]
- 55.Barnes SL, Belliveau NM, Ireland WT, Kinney JB, Phillips R. Mapping DNA sequence to transcription factor binding energy in vivo. PLoS Comput Biol. 2018;15:e1006226. doi: 10.1371/journal.pcbi.1006226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jung C, et al. True equilibrium measurement of transcription factor-DNA binding affinities using automated polarization microscopy. Nat Commun. 2018;9:1605. doi: 10.1038/s41467-018-03977-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mogno I, Kwasnieski JC, Cohen BA. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res. 2013;23:1908–1916. doi: 10.1101/gr.157891.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amit R, Garcia HG, Phillips R, Fraser SE. Building enhancers from the ground up: A synthetic biology approach. Cell. 2011;146:105–118. doi: 10.1016/j.cell.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Garcia HG, Phillips R. Quantitative dissection of the simple repression input-output function. Proc Natl Acad Sci USA. 2011;108:12173–12178. doi: 10.1073/pnas.1015616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boch Jens, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 63.Boyle EA, et al. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci USA. 2017;114:5461–5466. doi: 10.1073/pnas.1700557114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cuculis L, Abil Z, Zho H, Schroeder CM. TALE proteins search DNA using a rotationally decoupled mechanism. Nat Chem Biol. 2016;12:831–837. doi: 10.1038/nchembio.2152. [DOI] [PubMed] [Google Scholar]
- 65.Geertz M, Shore D, Maerkl SJ. Massively parallel measurements of molecular interaction kinetics on a microfluidic platform. Proc Natl Acad Sci USA. 2012;109:16540–16545. doi: 10.1073/pnas.1206011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cambray G, Guimaraes JC, Arkin AP. Evaluation of 244,000 synthetic sequences reveals design principles to optimize translation in Escherichia coli. Nat Biotechnol. 2018;36:1005–1015. doi: 10.1038/nbt.4238. [DOI] [PubMed] [Google Scholar]
- 67.Maerkl SJ, Quake SR. Experimental determination of the evolvability of a transcription factor. Proc Natl Acad Sci USA. 2009;106:18650–18655. doi: 10.1073/pnas.0907688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keith P, et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 69.Keith P, et al. Portable, on-demand biomolecular manufacturing. Cell. 2016b;167:248–259.e12. doi: 10.1016/j.cell.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 70.Goering AW, et al. In vitro reconstruction of nonribosomal peptide biosynthesis directly from DNA using cell-free protein synthesis. ACS Synth Biol. 2016;6:39–44. doi: 10.1021/acssynbio.6b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stark JC, et al. BioBits bright: A fluorescent synthetic biology education kit. Sci Adv. 2018;4:eaat5107. doi: 10.1126/sciadv.aat5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fuller RS, Kornberg A. Purified dnaA protein in initiation of replication at the Escherichia coli chromosomal origin of replication. Proc Natl Acad Sci USA. 1983;80:5817–5821. doi: 10.1073/pnas.80.19.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nirenberg MW, Heinrich Matthaei J. The dependence of cell-free protein synthesis in E. coli upon naturally occurring or synthetic polyribonucleotides. Proc Natl Acad Sci USA. 1961;47:1588–1602. doi: 10.1073/pnas.47.10.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swank Z, Laohakunakorn N, Maerkl SJ. 2019 doi: 10.1073/pnas.1816591116. Data and code from 2019_Swank_analysis GitHub. Available at https://github.com/lbnc-epfl/2019_Swank_analysis. Deposited December 14, 2018. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.