Significance
Heart failure is a major cause of mortality in humans. In a heart failure model induced by pressure overloading, we previously reported that the RNA binding protein RBFox2 was greatly diminished, leading to impaired excitation–contraction (EC) coupling. We have now pursued the mechanism for RBFox2 ablation-induced EC-coupling defects, revealing that RBFox2 depletion and resultant cardiac stress induce a set of microRNAs, including miR-34a, which directly targets the critical EC coupler Jph2 to further stress the heart. Significantly, systematic administration of a miR-34a antagomir is able to attenuate RBFox2 ablation-induced heart failure. These findings suggest a critical role of the RBFox2-miR-34a-Jph2 axis in causing progressive failure, which may be therapeutically intervened with a microRNA antagomir.
Keywords: RBFox2, miR-34a, Jph2, EC coupling, heart failure
Abstract
Heart performance relies on highly coordinated excitation–contraction (EC) coupling, and defects in this critical process may be exacerbated by additional genetic defects and/or environmental insults to cause eventual heart failure. Here we report a regulatory pathway consisting of the RNA binding protein RBFox2, a stress-induced microRNA miR-34a, and the essential EC coupler JPH2. In this pathway, initial cardiac defects diminish RBFox2 expression, which induces transcriptional repression of miR-34a, and elevated miR-34a targets Jph2 to impair EC coupling, which further manifests heart dysfunction, leading to progressive heart failure. The key contribution of miR-34a to this process is further established by administrating its mimic, which is sufficient to induce cardiac defects, and by using its antagomir to alleviate RBFox2 depletion-induced heart dysfunction. These findings elucidate a potential feed-forward mechanism to account for a critical transition to cardiac decompensation and suggest a potential therapeutic avenue against heart failure.
Heart failure is a leading cause of human mortality worldwide and has been a focus of intense biomedical research. It is well known that both physiological and pathological factors, such as aging, high blood pressure, coronary heart diseases, diabetes, and obesity contribute to heart failure (1, 2). Although a variety of transcription factors and signaling molecules have been implicated in this disease process (3), accumulating evidence suggests the involvement of a large number of miRNAs in both heart aging and failure (4). A few such miRNAs have been characterized in details with respect to their targets in the heart (5). Of particular interest is miR-34a, which is induced in aged hearts of both human and mice as well as in hearts stressed by pressure overloading and myocardial infarction (6–8). To study this important and highly disease-relevant miRNA, multiple targets have been identified, including the phosphatase 1 nuclear targeting subunit (PNUTS) involved in the regulation of apoptosis, DNA damage, telomere shortening (7), BCL-2, and CYCLIN D1 critical for triggering apoptosis and cell cycle arrest (9). However, it has remained unclear whether miR-34a directly regulates any cardiac-specific gene to compromise heart performance during the onset and/or progression of heart failure.
Recently, dysregulated RNA binding proteins (RBPs) have emerged as important drivers to heart failure. We have a long-standing interest in the RBFox family of proteins in gene regulation, which have been extensively studied as splicing regulators (10, 11). Our recent research on the two members (RBFox1 and RBFox2) of this important RBP family in the heart not only demonstrated their functional requirement for heart development, but also elucidated their protective roles against heart failure (12, 13). Interestingly, we found in the pressure-overload mouse heart that RBFox1 (12) was down-regulated at the mRNA level whereas RBFox2 (13) was diminished at the protein level. As expected from their roles in splicing regulation, we detected prevalent changes in alternative splicing of cardiac genes that are linked to heart failure phenotypes in both RBFox1 and RBFox2 knockout (KO) mouse models.
During detailed characterization of conditional RBFox2 KO mice, we noted that genetic ablation of this critical RBP in the heart caused severe defects in excitation–contraction (EC) coupling, which is critical for transmitting electronic signals to the contraction apparatus in cardiac muscle (14). However, it has remained unclear whether the observed EC-coupling defects resulted from some critical mRNA isoform switches or from other unknown dysfunctions induced in the RBFox2-deficient heart. It is of particular interest to note from our more recent study revealing a direct and repressive role of RBFox2 in transcription, which caused transcriptional derepression of a large number of miRNAs, including miR-34a, in the RBFox2-deficient heart (14). These findings suggest a potential regulatory paradigm where RBFox2 may function in conjunction with altered miRNA expression to regulate critical cardiac-specific genes.
In the current investigation, we specifically focused on elucidating the functions of RBFox2 and induced miR-34a in EC coupling. This led to the discovery that induced miR-34a specifically targets Jph2, which encodes an EC coupler that links T-tubules with the sarcoplasmic reticulum (SR) to coordinate the electronic signal on the membrane with the contractile apparatus within the cardiomyocyte (15, 16). As JPH2 down-regulation is a signature event induced in response to pressure overloading (16), our findings suggest a potential feed-forward loop that may directly contribute to the transition from compensation to decompensation characterized by myocardial insufficiency (3). Importantly, breaking this regulatory axis with a miR-34a antagomir was found to significantly alleviate the disease phenotype in the RBFox2-deficient heart, suggesting a potential therapeutic strategy against the progression of heart failure.
Results
RBFox2 Ablation-Induced Heart Failure and the Structural Basis for the Phenotype.
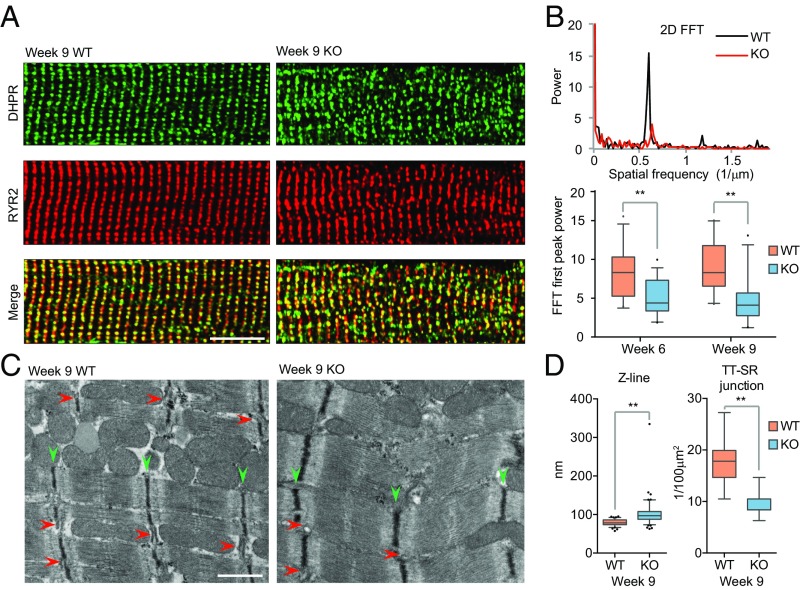
We recently documented that RBFox2 ablation in developing embryos by crossing floxed RBFox2 mice with a Mlc2v-Cre transgenic mouse induced severe EC-coupling defects detected by high-resolution confocal microcopy as part of the heart failure phenotype (13). To further understand the mechanism underlying such EC-coupling defects, we examined the structural integrity of the T-tubule, a key subcellular structure for EC coupling. By using a fluorescent dye that specifically illuminates the cell membrane, we observed a dramatic disarray of T-tubules with disrupted colocalization between the voltage-dependent long-lasting- (L)-type calcium channel [dihydropyridine receptor (DHPR)], which is responsible for triggering calcium transient from the SR via calcium-induced calcium release (17), and the ryanodine receptor RYR2, a critical calcium release channel on the SR to induce contraction (18) (Fig. 1A). FFT on spatially converted T-tubule images showed that the power of the first peak, an indicator of T-tubule ordering (19), was markedly reduced in the RBFox2−/− heart compared with those from the WT littermate controls (Fig. 1B). By transmission electron microscopy (TEM), we found that RBFox2−/− cardiomyocytes showed irregular sarcomeres with broadened Z bands (green arrowheads in Fig. 1C) and reduced junctions between TT-SR where calcium instructs EC coupling (red arrowheads in Fig. 1C) as, respectively, quantified (Fig. 1D). These data established the structural basis for the observed EC-coupling defects in the RBFox2−/− heart.
Fig. 1.
T-tubules and SR (TT-SR) disorganization in RBFox2 KO mice. (A) Double staining for DHPR (green) and RYR2 (red) on isolated wild-type (WT) and RBFox2−/− (KO) cardiomyocytes from 9 wk-old mice. (Scale bar, 10 μM.) (B) Power spectrum (Top) retrieved from two-dimensional fast Fourier transformation (FFT) of images in A. The first peak at the spatial frequency of ∼0.6 μm−1 corresponds to the ∼2 μm interval of the T-tubule in WT cardiomyocytes. The second and third harmonic peaks observable in WT were absent in KO mice. Statistics of the FFT first peak power (Bottom) at two different postnatal ages; n = 27 for WT at week 6; n = 30 for KO at week 6; n = 21 for WT at week 9; n = 22 for KO at week 9, **P < 0.01. (C) Disrupted ultrastructure of the contractile apparatus in WT and RBFox2−/− cardiomyocytes detected by TEM. Green arrowheads, Z line; red arrowheads, TT-SR junctions. (Scale bar, 200 nM.) (D) Statistics of the Z-line width (Left) and the number of TT-SR junctions (Right) were quantified from randomly selected TEM images; n = 81 from 3 WT hearts, n = 109 from 3 KO hearts, **P < 0.01.
The Heart Failure Phenotype Linked to Down-Regulation of the EC-Coupler Jph2.
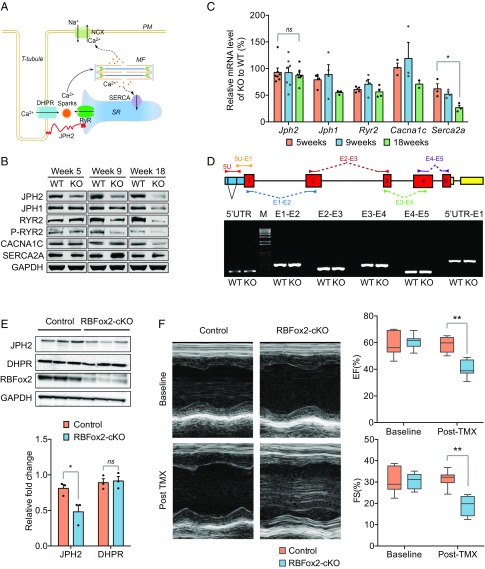
To investigate the underlying cause(s) at the molecular level, we focused on a set of key components involved in calcium handling in primary cardiomyocytes (Fig. 2A), including CACNA1C, a major subunit of the voltage-dependent L-type calcium channel DHPR in T-tubules (20); RYR2, a critical calcium channel on the SR for releasing calcium to the cytoplasm to trigger contraction (21); JPH2, a main linker between T-tubules and the SR critical for maintaining nanometer scale interactions between DHPR and RYR2 (22); and SERCA2A, a calcium pump on the SR for calcium recycling back to the SR during relaxation (23). Interestingly, we noted that the JPH2 protein level was first reduced at week 5 before the appearance of any detectable gross pathology (13), whereas all other key players we examined were altered only subsequently (Fig. 2B). In contrast, the mRNA levels of these genes quantified by RT-qPCR were relatively consistent with only Serca2a showing a significant decline after 18 wk, indicative of an indirect effect (Fig. 2C). These observations suggest a post-transcriptional mechanism for the reduction of JPH2 in the heart. Because heart-specific knockdown of Jph2 by shRNA caused a nearly identical phenotype (24, 25), these results suggest that induced Jph2 down-regulation is a causal event for the EC-coupling defect observed in the RBFox2−/− heart.
Fig. 2.
Contraction defects linked to JPH2 down-regulation in both global and conditional RBFox2 KO mice. (A) Diagram of a T-tubule, highlighting JPH2 in connecting TT-SR. NCX, a Na+/Ca2+ exchange channel on the plasma membrane (PM); MF, myofilaments; SERCA, a Ca2+ pump on the surface of SR; RyR2, Ryanodine receptor 2 for Ca2+ release from the SR; DHPR, a voltage-dependent L-type calcium channel at the T-tubule. (B) Western blotting analysis of key proteins involved in EC coupling at three different postnatal ages and comparison between WT and global RBFox2 KO cardiomyocytes. (C) Statistics of RT-qPCR analysis of gene expression of the genes described in B; for Jph2 mRNA, n = 7 at each time point, ns, not significant; for Serca2a mRNA, n = 3 at each time point, *P < 0.05. (D) Semiquantitative RT-PCR analysis of Jph2 splicing in WT and global RBFox2 KO cardiomyocytes from 9 wk-old mice. The gene structure and the locations of primer pairs are indicated on top of the agarose gel. 5U, 5′UTR; M, 1 kb DNA ladder marker; E1, exon 1; E2, exon 2; E3, exon 3; E4, exon 4; E5, exon 5. (E) Western blotting analysis of JPH2 expression in control and tamoxifen (TMX)-induced RBFox2 KO hearts (Top). GAPDH served as loading control. Relative fold changes in JPH2 protein levels (Bottom) were normalized to GAPDH and quantified; n = 3 hearts for each group, *P < 0.05, ns, not significant. (F) Left ventricle M-mode echocardiodiagram imaging (Left) and quantification of EF (Right Top) and FS (Right Bottom) at the baseline or 2 wk after TMX induction; n = 11 for each group, **P < 0.01.
We previously characterized a large splicing program regulated by RBFox2 in the cardiac muscle and linked the altered splicing program to postnatal heart remodeling (13). Here we asked whether Jph2 down-regulation in the RBFox2−/− heart might result from a defect in Jph2 splicing by semiquantitative RT-PCR using a set of primers to scan all exon–exon junctions in Jph2 pre-mRNA. Unexpectedly, we found no difference in either expression or alternative splicing between WT and RBFox2−/− cardiomyocytes (Fig. 2D). This observation suggests a new mechanism by which the JPH2 protein might be down-regulated at a post-translational level in the RBFox2−/− heart.
Repaid JPH2 Down-Regulation in Adult Mice in Response to Transient RBFox2 Ablation.
Because RBFox2 was ablated in an early embryonic stage (E8.5) in Mlc2v-Cre mice, it remained unclear whether compromised JPH2 levels in the adult heart might result from the immediate consequence of RBFox2 ablation or from a cascade of events manifested due to the absence of RBFox2 in early embryos. We thus examined the specific effect of RBFox2 KO in the mature heart by using TMX to induce RBFox2 ablation in the adult mouse heart (e.g., ∼2 mo after birth). By crossing the floxed RBFox2 mice with an inducible cardiac-specific Myh6-MerCreMer mouse (26, 27), we obtained homozygous RBFox2-cKO mice (SI Appendix, Fig. S1A), and upon TMX administration for a week, we confirmed marked RBFox2 reduction at both mRNA (SI Appendix, Fig. S1B) and protein (Fig. 2E) levels. Noninvasive echocardiography recorded significant decreases in both ejection fraction (EF) and fractional shortening (FS), indicative of greatly compromised left ventricle contractility (Fig. 2F). In addition, we detected mRNA induction of two key heart failure biomarkers, the atrial natriuretic factor (Anf) and the brain natriuretic protein (Bnp) (SI Appendix, Fig. S1C).
Consistent with our early findings (13), the TMX-induced RBFox2-cKO heart showed marked chamber dilation but without obvious cardiac hypertrophy (SI Appendix, Fig. S1D), which was also evidenced by a slightly decreased, rather than increased, heart weight/body weight ratio compared with the control heart (SI Appendix, Fig. S1E). These data suggest that compromised heart function resulted directly from RBFox2 deficiency rather than indirectly from aggregated developmental defects. We further validated key induced splicing events on Pdlim5, Mef2a, and Ldb3 genes in this induced RBFox2 ablation model as we observed earlier in RBFox2-Mlc2v-Cre mice (SI Appendix, Fig. S1F). Importantly, we also detected JPH2 down-regulation in the TMX-treated RBFox2-cKO heart (Fig. 2E), which correlated with the contraction defect detected by echocardiography and significantly reduced EF and FS values deduced from such echocardiographic measurements (Fig. 2F). Together, these data strongly suggest RBFox2 deficiency caused the reduction of JPH2, which contributed to the contraction defect in the adult heart. The RBFox2-cKO mice also afforded us a second genetic model to study RBFox2-regulated gene expression.
Induction of miR-34a in RBFox2−/− Cardiomyocytes.
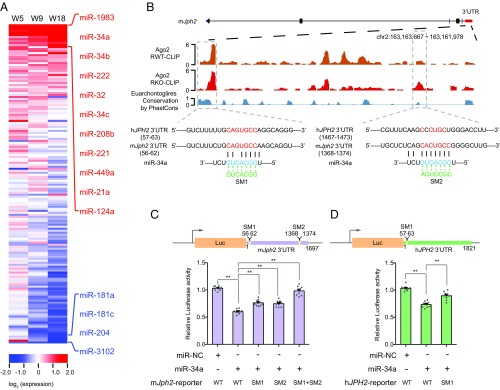
To investigate how RBFox2 might regulate gene expression beyond splicing control, we had an important clue from our previous study showing transcriptional derepression of a large number of miRNAs in RBFox2−/− cardiomyocytes (14), suggesting that an induced miRNA(s) might account for unaltered Jph2 mRNA but reduced JPH2 protein we now uncovered. To systematically investigate changes in miRNA expression in cardiomyocytes, we used RT-qPCR to profile 160 miRNAs previously detected in the mouse heart (28, 29) (SI Appendix, Table S1). We detected the induction of a large subset of miRNAs in RBFox2−/− cardiomyocytes at week 5, and observed persistent elevated expression of these miRNAs in later time points (Fig. 3A). Importantly, this set of highly induced miRNAs includes miR-34a, which has been previously linked to both age-linked and pressure–overloading-induced heart failure (6, 7), indicating a potential combinatory mechanism (i.e., transcription depression without RBFox2 and stress signaling) responsible for elevating this stress-induced miRNA. We also confirmed the marked induction of miR-34a in RBFox2-cKO mice (see Fig. 5A below). In comparison, another set of miRNAs initially showed little difference at week 5 but became progressively repressed at later stages, indicative of potential secondary effects.
Fig. 3.
JPH2 down-regulation by induced miR-34a. (A) Heat map of the relative expression of 160 miRNAs determined by RT-qPCR in global RBFox2 KO cardiomyocytes relative to WT at week 5, 9, and 18. The data were sorted based on the mean value of miRNA levels at three different ages with those that showed >1.75-fold changes as highlighted on the right (red for increased expression; blue for decreased expression). (B) Ago2 binding events in the 3′UTR of mouse Jph2. The predicted miR-34a target regions are highlighted in the dashed boxes. The Bottom shows the predicted base pairing in the seed region between miR-34a and its target site and the mutations introduced in the seed regions. The first predicted miR-34a targeting site near the 5′ end is conversed between mouse and human JPH2 genes, but the second near the 3′ end is unique to mouse Jph2. (C and D) The luciferase assays of the reporters containing the full-length mouse Jph2 3′UTR (C) or human JPH2 3′UTR (D) in response to miR-34a overexpression with or without mutations in the miR-34a seed target regions; n = 9 for each group, *P < 0.05, **P < 0.01.
We next determined potential miRNA targets by performing duplicated Ago2 CLIP-seq to compare the miRNA targeting landscape in response to RBFox2 ablation in the mouse heart (13) (SI Appendix, Fig. S2A). The resulting Ago2 CLIP-seq data were globally concordant even when comparing the Ago2 CLIP-seq data from WT and RBFox2-ablated hearts (SI Appendix, Fig. S2B). Focusing on Jph2 in the mouse, we noted two strong Ago2 peaks in the 3′UTR of Jph2, which showed a degree of detectable induction upon RBFox2 ablation, and inspection of the underlying sequences suggests that both might be directly targeted by miR-34a based on potential seed base-pairing interactions (Fig. 3B). We further noted that the sequence underlying the first Ago2 peak was conserved between mice and humans, whereas the second Ago2 peak appeared unique to the mouse gene. These observations raised the possibility that induced miR-34a might directly target Jph2 to cause its down-regulation in RBFox2−/− cardiomyocytes.
miR-34a Targets the 3′UTR of Both Mouse and Human JPH2 Genes.
To validate the direct action of miR-34a on Jph2, we first cloned the full-length mouse Jph2 3′UTR into a luciferase reporter, and to test the functionality of the predicted miR-34a target sites, we also introduced point mutations in the seed region of both potential target sites, either individually or in combination (Fig. 3B, Bottom). Upon transfecting these reporters in HeLa S3 cells with or without a miR-34a mimic, we found that transfected miR-34a indeed repressed the WT reporter (Fig. 3C). Mutations in either seed reduced the effect, and the combined mutations largely abolished the effect (Fig. 3C). These data supported the regulation of mouse Jph2 by miR-34a and confirmed the required base-pairing interactions in both seed regions.
Given significant sequence divergence in the 3′UTRs of Jph2 between mice and humans (SI Appendix, Fig. S3A), we extended this analysis to human JPH2, focusing on the relatively conserved target beneath the first Ago2 peak. We similarly cloned the full-length human JPH2 3′UTR into the luciferase reporter and tested both WT and corresponding mutations in response to the transfected miR-34a mimic in HeLa S3 cells. We found that the relatively conserved sequence underlying the first Ago2 peak was indeed responsive to the transfected miR-34a mimic; however, mutations in the seed base-pairing reduced but not fully obligated the responsiveness to miR-34a (Fig. 3D), implying that human JPH2 may carry another target sequence(s) for miR-34a. We also cloned the core miR-34a target sequences from both mouse and human JPH2 genes and showed that both were sufficient to mediate the response to transfected miR-34a (SI Appendix, Fig. S3 B and C). Together, these data suggest that both mouse and human JPH2 genes are subjected to regulation by miR-34a.
Direct Contribution of miR-34a to JPH2 Down-Regulation in the Heart.
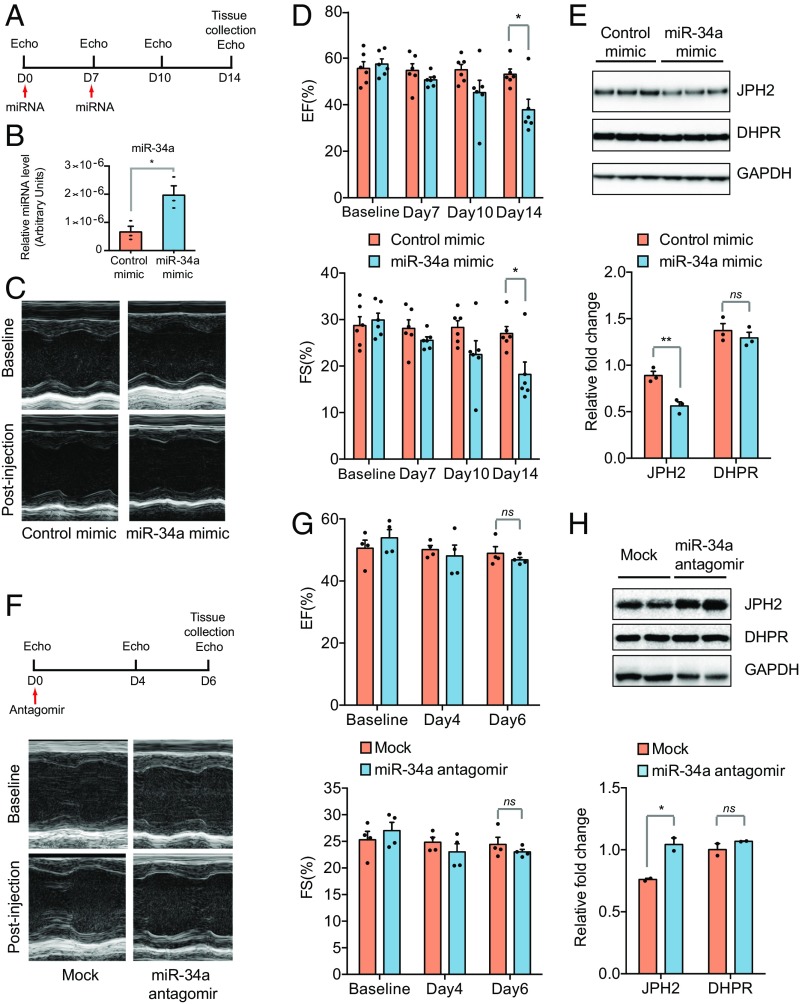
The data presented above revealed a critical RBFox2-miR-34a-Jph2 pathway for heart function. To substantiate a key contribution of this circuit to heart failure, we asked whether increased miR-34a alone would be sufficient to imitate the heart failure phenotype observed in the RBFox2−/− heart. We i.v. injected control and miR-34a mimic into WT mice at day 0 and again at day 7 and performed echocardiography analysis at day 0, 7, 10, and 14 postinjection (Fig. 4A). We confirmed an increased miR-34a level relative to mock injection in the heart even 14 d after injection (Fig. 4B), and echocardiography revealed a clear decrease in cardiac contractility (Fig. 4C) as indicated by the quantified reduction of EF and FS along the time (Fig. 4D). Notably, the miR-34a mimic alone was insufficient to invoke alternative splicing of Mef2a or Ldb3 as observed in the RBFox2−/− heart (SI Appendix, Fig. S4A); however, the injected miR-34a mimic clearly caused a reduction of JPH2 to a measurable degree as indicated by the quantified Western blotting data (Fig. 4E). Conversely, we injected a specific miR-34a antagomir. Unlike the miR-34a mimic, the miR-34a antagomir did not cause a detectable cardiac phenotype in the WT heart (Fig. 4 F and G), and consistent with its expected effect, we detected an increase in JPH2 expression (Fig. 4H). As a control, we examined DHPR, another important T-tubule component, which showed little changes at the protein level in mice treated with either miR-34a mimics or antagomir. These data demonstrated that endogenous miR-34a is titrating the level of JPH2 expression in the heart, and augmentation of miR-34a alone is sufficient to impair heart function.
Fig. 4.
Contraction defects in miR-34a mimic-transduced WT mice. (A) Scheme for miR-34a mimic administration and echocardiographic analysis at different day points. (B) Quantitative analysis of miR-34 expression in control and miR-34 mimic-transduced mice; n = 3 for each group, *P < 0.05. (C and D) Representative M-mode echocardiograph (C) and quantification (D) of EF (Top) and FS (Bottom) at the baseline, day 7, 10, and 14 after miR-34a mimic administration; n = 6 in each group, *P < 0.05. (E) Western blotting analysis of JPH2 (Top) in the control and miR-34a mimic administrated WT hearts. Relative fold changes in JPH2 protein levels (Bottom) were normalized to loading control GAPDH and quantified; n = 3 hearts, **P < 0.01, ns, not significant. (F and G) Representative M-mode echocardiograph (F) and quantification (G) of EF (Top) and FS (Bottom) at the baseline, day 4 and 6 after the control or miR-34a antagomir administration in WT mice; n = 4 in each group, ns, not significant. (H) Western blotting analysis of JPH2 expression in the heart (Top) in the control and miR-34a antagomir administrated WT mice. Shown is one of two independent experiments. Relative fold changes in JPH2 protein levels (Top) were normalized to loading control GAPDH and quantified (Bottom); n = 4 for each sample, *P < 0.01, ns, not significant.
Functional Rescue of RBFox2 Ablation-Induced Heart Defects by miR-34a Antagomir.
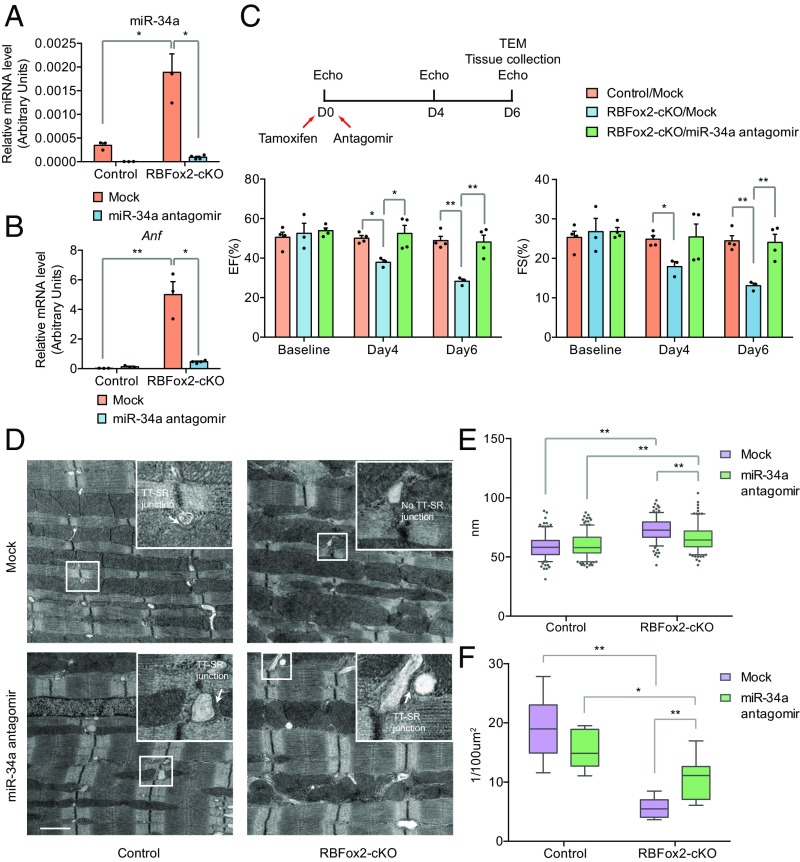
To further demonstrate that the newly elucidated RBFox2-miR-34a-Jph2 pathway critically contributed to the overall cardiac defect in the RBFox2−/− heart, we next performed a functional rescue experiment by breaking the pathway with the miR-34a antagomir, taking advantage of the rapidly induced cardiac phenotype in RBFox2-cKO mice where multiple miRNAs, including miR-34a, were induced as in the RBFox2−/− heart (Fig. 5A and SI Appendix, Fig. S4B). We first confirmed that the administrated antagomir was able to reduce the miR-34a levels in both WT and RBFox2-cKO hearts at day 6 after antagomir injection (Fig. 5A). As expected, the miR-34a antagomir suppressed the induction of the heart failure-associated biomarker Anf (Fig. 5B) and improved cardiac performance in the RBFox2-cKO heart as indicated by the rescued EF and FS levels measured by echocardiography (Fig. 5C). Moreover, TEM detected improved sarcomere organization at day 6 of TMX treatment (Fig. 5D). Quantitative analysis of the TEM images showed that, although blocking basal miR-34a had no effect on sarcomere organization in the WT heart, the miR-34a antagomir significantly suppressed increased the Z line width in the RBFox2-cKO myocardium (Fig. 5E). More importantly, the miR-34a antagomir was effective in restoring the number of TT-SR junctions in the RBFox2-cKO heart (Fig. 5F). Collectively, these data demonstrated that the induced miR-34a expression critically contributed to the heart failure phenotype in the RBFox2-ablated heart and blockage of this miRNA was insufficient to rescue the phenotype.
Fig. 5.
Functional rescue of RBFox2 ablation-induced heart failure with miR-34a antagomir. (A and B) RT-qPCR analysis of miR-34a expression (A), and the expression of the heart failure marker Anf (B) in the control and miR-34a antagomir administrated groups in the WT control or RBFox2-cKO hearts; n = 3 for each group except n = 4 for RBFox2-cKO/miR-34a antagomir, *P < 0.05, **P < 0.01. (C) Scheme for miR-34a antagomir administration and echocardiographic analysis (Top) and quantification of EF and FS at the baseline, day 4 and 6 postantagormir administration in control or RBFox2-cKO mice (Bottom); n = 4 for the control/mock; n = 3 for RBFox2-cKO/mock; n = 4 for RBFox2-cKO/miR-34a antagomir, *P < 0.05, **P < 0.01. (D) TEM images of WT or RBFox2-cKO cardiomyocytes treated with the control (Top) or the miR34a antagomir (Bottom). The enlarged white box highlights the TT-SR junction. (Scale bar, 1 μM.) (E and F) Statistics of Z-line width (E) and number of TT-SR junctions (F) were quantified from TEM images captured randomly for each specimen; for the Z line, n = 216 for the control/mock; n = 240 for the control/miR-34a antagomir; n = 181 for the RBFox2-cKO/mock; n = 206 for RBFox2-cKO/miR-34a antagomir; for the TT-SR junction, n = 12 for the control/mock; n = 10 for the control/miR-34a antagomir; n = 8 for the RBFox2-cKO/mock; n = 8 for the RBFox2-cKO/miR-34a antagomir, *P < 0.05, **P < 0.01.
Discussion
RBFox2 Is Directly Relevant to Heart Aging and Disease.
The RBFox family of RNA binding proteins consists of three members, RBFox1 (A2BP1), RBFox2 (RBM9), and RBFox3 (NeuN). Although RBFox3 is exclusively expressed in mature neurons, both RBFox1 and RBFox2 are detectable in multiple tissues, particularly, the brain and heart (30, 31). Relevant to heart development, RBFox1 is little expressed in the embryonic heart, but is dramatically induced in the postnatal heart (32), which is consistent with the demonstrated role of this RBP during heart remodeling (12). In contrast, RBFox2 expression is relatively constant in both embryos and young adults but becomes progressively declined in later developmental stages (32), indicative of its potential contribution to the heart aging process.
Relevant to heart disease, a mutation in RBFox2 has been identified as a risk factor in hypoplastic left heart syndrome, and analysis of the RBFox2 mutation demonstrated its functional impact on a large number of RBFox2-regulated splicing events in the heart, suggesting the causal role of the mutation in cardiomyopathy (33). Study of diabetic cardiomyopathy also reveals a dominant negative isoform of RBFox2, which appears to antagonize the normal function of RBFox2 in regulated splicing (34). These lines of genetic evidence strongly suggest RBFox2 as a disease gene in the heart.
RBFox2 Regulates Gene Expression at Multiple Levels.
Molecular analysis on cell lines and animal models has demonstrated the vital function of RBFox family members as splicing regulators. Interestingly, the functional impact of these RBPs on gene expression has now been expanded beyond their traditional roles in splicing control. For example, RBFox1 has recently been found to enhance mRNA stability in the brain, although the mechanism for such enhancement has remained elusive (35). Moreover, both RBFox2 and RBFox3 have been implicated in binding and modulating pri-miRNA processing (36, 37).
Our recent study also revealed that RBFox2 is broadly involved in transcriptional repression by recruiting Polycomb Complex 2 via nascent RNAs (14). These findings prompted us to investigate disease mechanisms from a broader respective, leading to the discovery that RBFox2 ablation coupled with associated stress induces miR-34a, which, in turn, targets a critical EC-coupler Jph2 to compromise heart performance. The overall cardiac defects likely result from combined effects of RBFox2 ablation at both splicing and miRNA levels.
A Potential Self-Enforcing Mechanism for Heart Failure.
Our current study revealed a protective role of RBFox2 during heart failure. As RBFox2 is a demonstrated sensor to stress (13), reduced RBFox2 in response to the initial insult may induce miR-34a, which targets Jph2, and resulting EC-coupling defects may impose further stress to the heart, thereby further down-regulating RBFox2. Therefore, the elucidated pathway may have the propensity to become manifested, which may be a potential mechanism for progressive down-regulation of RBFox2 during heart aging. This potential feed-forward mechanism may be further enhanced by genetic and environmental factors, leading to heart failure.
Each component in this pathway is likely to have other functions that contribute to the overall heart failure phenotype either associated with aging or induced by other genetic and environmental insults. For example, RBFox2 is known to regulate many genes at both transcriptional and post-transcriptional levels (13, 14, 34). Likewise, increased miR-34a has been shown to target other critical genes, such as PNUTS, to cause myocardial insufficiency (7). Besides miR-34a, another miRNA miR-24 has also been reported to target Jph2 in the heart (38, 39). Therefore, it is likely that the RBFox2-miR-34a-Jph2 pathway may be connected to multiple other branches during the progression of heart failure. Importantly, we have now demonstrated that experimental intervention of this pathway with the miR-34a antagomir is sufficient to alleviate the heart disease phenotype to a significant extent in RBFox2 KO mice, which is in line with the documented benefit of this agent on other heart failure models, together suggesting its clinical potential for treating heart disease.
Materials and Methods
Construction and Analysis of Inducible Cardiac-Specific RBFox2 KO Mice.
The construction of conditional RBFox2 KO mice was previously described (13). Conditional RBFox2-cKO mice were generated by breeding RBFox2 floxed mice with a cardiac-specific inducible model Myh6-MerCreMer mouse. To inactivate RBFox2, TMX was administrated in an animal diet for 1 wk to activate Cre. Mouse pressure-overload surgery was performed as previously described (40) in either C57B/L male animals or RBFox2-cKO mice. For miR-34a mimic and miR-34a antagomir delivery, both miR-34a mimic and miR-34a antagomir were purchased from Thermo Fisher Scientific (#4464088; #4464070). The control animals were injected with corresponding negative controls, also from Thermo Fisher Scientific (#4464078; #4464060). The miR-34a mimic and miR-34a antagomir (4 mg/kg body weight) were delivered via tail vein injection using InvivoFectmine 3.0 (IVF3005; Thermo Fisher Scientific) according to the manufacturer’s protocol. Mice were imaged noninvasively for heart function and morphology by echocardiography at indicated time points. All mice were anesthetized and maintained with 2% isofluorane in 95% oxygen. A Vevo 2100 (FUJIFILM VisualSonics) echocardiography system with a 30 mHz scan head was used to acquire the data. A parasternal long axis and a short axis view were recorded. The short axis view was used to generate M-mode images for analysis of ejection fraction and fraction shortening.
This study was performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (41). All of the animals were handled according to approved institutional animal care and use committee (IACUC) protocols (S99116 and A3196-01) of the University of California. All surgery was performed under anesthesia, and every effort was made to minimize suffering.
Transmission Electron Microscopy and Imaging of T-Tubules.
Dissected mouse hearts were perfused with 50 mM KCl in Tyrode solution followed by 2% paraformaldehyde and 2% glutaraldehyde. The tissues were processed for FEI Tecnai Spirit G2 BioTWIN transmission electron microscope at the University of California San Diego (UCSD) Transmission Electron Microscopy Core Facility. Quantitative analysis was performed by using the ImageJ software. To image T-tubules, isolated cardiomyocytes were loaded with 10 μM lipophilic membrane marker Di-8-ANEPPS (d-3167; Thermo Fisher Scientific) in Tyrode solution at room temperature for 10 min. Cells were washed twice and imaged under a confocal microscope with a 60× oil objective in the X-Y frame mode. The IDL 5.5 software (Research Systems, Inc.) was used for image processing.
Western Blot and Antibodies.
For Western blotting, isolated cardiomyocytes were lysed to prepare total protein extracts. Total protein samples were resolved in 4–12% NuPAGE Novex Bis-Tris gradient gels (NP0322BOX; Thermo Fisher Scientific) and transferred to a nitrocellulose membrane. After blocking for 1 h with 5% nonfat milk, the membrane was probed with a primary antibody overnight at 4 °C and then with HRP-conjugated anti-mouse or anti-rabbit secondary antibody (7076S; 7074S; Cell Signaling Technology) for 1 h at room temperature. Immunoblot signals were detected by autoradiography after application of the enhanced chemiluminescence substrate. The following primary antibodies were used in the present study: anti-JPH2 (ab79071; Abcam); anti-JPH1 (40–5100; Thermo Fisher Scientific); anti-RYR2 (PA5-38329; Thermo Fisher Scientific); anti-phospho-RYR2 (A0570; Assay Biotech); anti-CACNA1C (ACC-003; Alomone Labs); anti-GAPDH (G9545; Sigma-Aldrich); anti-RBFox2 (ab57154; Abcam); anti-DHPR (ab2864; Abcam); Anti-SERCA2a was from Dr. Ju Chen’s laboratory at the UCSD.
RT-PCR, miRNA Profiling, and Luciferase Assays.
Total RNA from isolated cardiomyocytes was extracted with TRIzol (15596026; Thermo Fisher Scientific) following manufacturer’s instructions, treated with DNase I', and reverse transcribed with SuperScript III (18080051; Thermo Fisher Scientific). Real-time PCR was performed with the miScript SYBR Green PCR Kit (218073; Qiagen) on an Applied Biosystems real-time PCR machine. miRNA-specific primers were designed according to the miRBase database (www.miRbase.org/). The Tm values (melting temperature) of individual primers were adjusted between 50 and 60 °C by adding or deleting several nucleotides from the 5′ end of primers.
Luciferase assay reporters were constructed by cloning PCR-amplified 3′UTRs of mouse and human Jph2 into the Psicheck2 vector between Xho I and Not I sites. The PCR primers used for constructing these luciferase reporters are listed in SI Appendix, Table S2. For transfection, cells were transfected sequentially by using Attractene Transfection Reagent (#301005; Qiagen) with 20 pmol microRNA mimics (Bioneer, AccuTarget miRNA mimic negative control #1, #SMC-2001; AccuTarget Custom Human miRNA mimic, hsa-miR-34a-5p mimic, #SMM-001) and then 5 ng reporters. Luciferase activity was measured 24 h after transfection of plasmids with a dual-luciferase reporter assay kit (E1910; Promega) on a Veritas microplate luminometer (Turner Biosystems).
CLIP-Seq and Data Analysis.
CLIP-seq was performed as previously described (42), and the raw Ago2 CLIP-seq data have been deposited in the public database on the accession number GSE57926. Whole cardiomyocytes from 9-wk-old WT or RBFox2−/− mice were digested with 0.1 U/uL of RQ1 DNase and 2 U/μL of RNaseOUT in lysis buffer (1×PBS buffer with 0.3% SDS, 0.5% deoxycholate sodium and 0.5% Nonidet P-40) for 5 min at 37 °C. Each lysate was processed for CLIP-seq with a mouse monoclonal anti-Ago2 antibody (also called EIF2C2). The sequencing data were processed using tools from several open resources, including the BEDTools (43), the R program (www.r-project.org/), and the bx-python package (bitbucket.org/james_taylor/bx-python/). Tracks of deep sequencing data were generated by using igvtools (https://github.com/bxlab/bx-python). Heat maps were generated by using Java Treeview (jtreeview.sourceforge.net). CLIP-seq tags were mapped to the mouse genome (mm10), and identical reads were removed (M1) or compressed to a maximal of 4 (M1u4). Specific peaks were identified based on the enriched number of reads against randomized background gene by gene as described (11).
Statistical Analyses.
Unless specifically noted, data were collected from technical triplicates of three independent experiments and presented as mean ± SEM. Statistical analyses were performed using unpaired two-tailed Student’s t test or ANOVA when comparing multiple groups. P value < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank members of the X.-D.F., J.C., and Y.W. laboratories for discussion and advice. This work was supported by NIH Grants GM049369, HG004659 (to X.-D.F.), HL122737 (to Y.W.) and National Natural Science Foundation of China Grant 81770227 (to C.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822176116/-/DCSupplemental.
References
- 1.Lakatta EG. Age-associated cardiovascular changes in health: Impact on cardiovascular disease in older persons. Heart Fail Rev. 2002;7:29–49. doi: 10.1023/a:1013797722156. [DOI] [PubMed] [Google Scholar]
- 2.Burchfield JS, Xie M, Hill JA. Pathological ventricular remodeling: Mechanisms: Part 1 of 2. Circulation. 2013;128:388–400. doi: 10.1161/CIRCULATIONAHA.113.001878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diwan A, Dorn GW., 2nd Decompensation of cardiac hypertrophy: Cellular mechanisms and novel therapeutic targets. Physiology (Bethesda) 2007;22:56–64. doi: 10.1152/physiol.00033.2006. [DOI] [PubMed] [Google Scholar]
- 4.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lucia C, et al. microRNA in cardiovascular aging and age-related cardiovascular diseases. Front Med (Lausanne) 2017;4:74. doi: 10.3389/fmed.2017.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernardo BC, et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA. 2012;109:17615–17620. doi: 10.1073/pnas.1206432109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boon RA, et al. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 8.Bernardo BC, et al. Silencing of miR-34a attenuates cardiac dysfunction in a setting of moderate, but not severe, hypertrophic cardiomyopathy. PLoS One. 2014;9:e90337. doi: 10.1371/journal.pone.0090337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, et al. MicroRNA-34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res. 2015;117:450–459. doi: 10.1161/CIRCRESAHA.117.305962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao C, et al. RBFox1-mediated RNA splicing regulates cardiac hypertrophy and heart failure. J Clin Invest. 2016;126:195–206. doi: 10.1172/JCI84015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei C, et al. Repression of the central splicing regulator RBFox2 is functionally linked to pressure overload-induced heart failure. Cell Rep. 2015;10:1521–1533. doi: 10.1016/j.celrep.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei C, et al. RBFox2 binds nascent RNA to globally regulate polycomb complex 2 targeting in mammalian genomes. Mol Cell. 2016;62:875–889. doi: 10.1016/j.molcel.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Oort RJ, et al. Disrupted junctional membrane complexes and hyperactive ryanodine receptors after acute junctophilin knockdown in mice. Circulation. 2011;123:979–988. doi: 10.1161/CIRCULATIONAHA.110.006437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beavers DL, Landstrom AP, Chiang DY, Wehrens XH. Emerging roles of junctophilin-2 in the heart and implications for cardiac diseases. Cardiovasc Res. 2014;103:198–205. doi: 10.1093/cvr/cvu151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- 18.Fill M, Copello JA. Ryanodine receptor calcium release channels. Physiol Rev. 2002;82:893–922. doi: 10.1152/physrev.00013.2002. [DOI] [PubMed] [Google Scholar]
- 19.Song LS, et al. Orphaned ryanodine receptors in the failing heart. Proc Natl Acad Sci USA. 2006;103:4305–4310. doi: 10.1073/pnas.0509324103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zühlke RD, Bouron A, Soldatov NM, Reuter H. Ca2+ channel sensitivity towards the blocker isradipine is affected by alternative splicing of the human alpha1C subunit gene. FEBS Lett. 1998;427:220–224. doi: 10.1016/s0014-5793(98)00425-6. [DOI] [PubMed] [Google Scholar]
- 21.Awad SS, Lamb HK, Morgan JM, Dunlop W, Gillespie JI. Differential expression of ryanodine receptor RyR2 mRNA in the non-pregnant and pregnant human myometrium. Biochem J. 1997;322:777–783. doi: 10.1042/bj3220777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: A novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 23.MacLennan DH, Rice WJ, Green NM. The mechanism of Ca2+ transport by sarco(endo)plasmic reticulum Ca2+-ATPases. J Biol Chem. 1997;272:28815–28818. doi: 10.1074/jbc.272.46.28815. [DOI] [PubMed] [Google Scholar]
- 24.Chen B, et al. Critical roles of junctophilin-2 in T-tubule and excitation-contraction coupling maturation during postnatal development. Cardiovasc Res. 2013;100:54–62. doi: 10.1093/cvr/cvt180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds JO, et al. Junctophilin-2 is necessary for T-tubule maturation during mouse heart development. Cardiovasc Res. 2013;100:44–53. doi: 10.1093/cvr/cvt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harmon AW, Nakano A. Nkx2-5 lineage tracing visualizes the distribution of second heart field-derived aortic smooth muscle. Genesis. 2013;51:862–869. doi: 10.1002/dvg.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall ME, Smith G, Hall JE, Stec DE. Systolic dysfunction in cardiac-specific ligand-inducible MerCreMer transgenic mice. Am J Physiol Heart Circ Physiol. 2011;301:H253–H260. doi: 10.1152/ajpheart.00786.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Condorelli G, Latronico MV, Dorn GW., 2nd microRNAs in heart disease: Putative novel therapeutic targets? Eur Heart J. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thum T, Catalucci D, Bauersachs J. MicroRNAs: Novel regulators in cardiac development and disease. Cardiovasc Res. 2008;79:562–570. doi: 10.1093/cvr/cvn137. [DOI] [PubMed] [Google Scholar]
- 30.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–3907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verma SK, et al. Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci Rep. 2016;6:30896. doi: 10.1038/srep30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nutter CA, et al. Dysregulation of RBFOX2 is an early event in cardiac pathogenesis of diabetes. Cell Rep. 2016;15:2200–2213. doi: 10.1016/j.celrep.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rajman M, et al. A microRNA-129-5p/Rbfox crosstalk coordinates homeostatic downscaling of excitatory synapses. EMBO J. 2017;36:1770–1787. doi: 10.15252/embj.201695748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim KK, Yang Y, Zhu J, Adelstein RS, Kawamoto S. Rbfox3 controls the biogenesis of a subset of microRNAs. Nat Struct Mol Biol. 2014;21:901–910. doi: 10.1038/nsmb.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y, et al. Rbfox proteins regulate microRNA biogenesis by sequence-specific binding to their precursors and target downstream Dicer. Nucleic Acids Res. 2016;44:4381–4395. doi: 10.1093/nar/gkw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu M, et al. Mir-24 regulates junctophilin-2 expression in cardiomyocytes. Circ Res. 2012;111:837–841. doi: 10.1161/CIRCRESAHA.112.277418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li RC, et al. In vivo suppression of microRNA-24 prevents the transition toward decompensated hypertrophy in aortic-constricted mice. Circ Res. 2013;112:601–605. doi: 10.1161/CIRCRESAHA.112.300806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, et al. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109:1332–1341. doi: 10.1161/CIRCRESAHA.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 42.Xue Y, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.