Significance
Colorectal cancer (CRC) is a heterogeneous disease, with significant variation in genotype and phenotype within each individual tumor. This intratumor heterogeneity emerges during tumor development due to clonal evolution and in part can explain therapy resistance in CRC. However, a detailed understanding of the spatiotemporal development of tumors underlying cancer evolution and intratumor heterogeneity remains absent. Here, we use lineage-tracing experiments of human CRC cells transplanted into immunocompromised mice, in combination with computational modeling, to study the growth mode of CRC. We found that the clonal position is crucial for clonal outgrowth. This demonstrates that, in addition to the genetic composition, the environment and the geometry of tumor growth play a significant role in shaping tumor evolution.
Keywords: colorectal cancer, tumor growth, intratumor heterogeneity, cancer evolution, cancer stem cells
Abstract
Cancer evolution is predominantly studied by focusing on differences in the genetic characteristics of malignant cells within tumors. However, the spatiotemporal dynamics of clonal outgrowth that underlie evolutionary trajectories remain largely unresolved. Here, we sought to unravel the clonal dynamics of colorectal cancer (CRC) expansion in space and time by using a color-based clonal tracing method. This method involves lentiviral red-green-blue (RGB) marking of cell populations, which enabled us to track individual cells and their clonal outgrowth during tumor initiation and growth in a xenograft model. We found that clonal expansion largely depends on the location of a clone, as small clones reside in the center and large clones mostly drive tumor growth at the border. These dynamics are recapitulated in a computational model, which confirms that the clone position within a tumor rather than cell-intrinsic features, is crucial for clonal outgrowth. We also found that no significant clonal loss occurs during tumor growth and clonal dispersal is limited in most models. Our results imply that, in addition to molecular features of clones such as (epi-)genetic differences between cells, clone location and the geometry of tumor growth are crucial for clonal expansion. Our findings suggest that either microenvironmental signals on the tumor border or differences in physical properties within the tumor, are major contributors to explain heterogeneous clonal expansion. Thus, this study provides further insights into the dynamics of solid tumor growth and progression, as well as the origins of tumor cell heterogeneity in a relevant model system.
Solid malignancies result from the accumulation of genetic aberrations that provide cells with a clonogenic advantage over their environment; for example, by promoting proliferation or reducing cell death (1–3). However, our incomplete knowledge of the quantitative effects of these oncogenic events and the fundamental dynamics of tumor expansion have so far precluded a thorough understanding of the dynamics of tumor evolution. For example, it remains unresolved what the effective population size is that drives long-term tumor expansion and progression (4, 5). Do rare cancer stem cells exist, or are all cells capable of driving tumor growth? In addition, the impact of the geometry of tumor expansion on clonogenic outgrowth is a topic of great relevance (6). In contrast to hematological malignancies, cells in solid cancers directly compete for space and nutrients. Furthermore, the dynamics of tissue turnover and the geometry of competing clones are predicted to directly impact on evolutionary trajectories (7, 8). Intratumor heterogeneity, which contributes to resistance to therapies and poor outcome, is a direct consequence of the concepts introduced above and a better understanding of these is essential to improve patient outcomes (9, 10). Recently, it was suggested in the big-bang model of colorectal cancer (CRC) evolution that spatial separation of competing clones results in a largely neutral competition, and that the variation in clone sizes within cancers reflects the age of the clone rather than the relative clonogenic advantage of the unique molecular properties of that lineage (11). However, this model did not consider the possible heterogeneity in clone sizes that could result from a heterogeneous clonogenicity instilled by the specific geometry of the tumor tissue and its microenvironment.
Here we set out to investigate the impact of the environment on clone size variation in primary xenograft models of human CRC. We employed the lentiviral gene ontology (LeGO) method to spatially trace clone lineages within tumors by their unique red-green-blue (RGB) color-coding (12). This improves on previous barcoding studies from which spatial information is absent (13, 14). We found that injection of homogenous populations of cancer cells results in extensive heterogeneity in clonogenic outgrowth with large clones located close to the tumor surface. Our results are in line with two recent studies that suggested that clonal outgrowth predominantly occurs at the tumor-leading edge and that cell external rather than intrinsic properties determine the clonogenic potential (7, 8). We expand on our previous work that utilized short-term lineage tracing only, to study and explain the complete growth dynamics of established tumors (8). Importantly, using computational simulations in conjunction with detailed clone size measurements, we conclude that the full clone size heterogeneity is defined by cell-extrinsic features, and thus no evidence of an intrinsic hierarchy was found in established CRC tissue. Additionally, we found that clonal dispersal is limited and that the number of clones remains constant during tumor growth. Taken together, these findings provide critical insights to the commonly employed s.c. xenograft assay and indicate that spatial location and time of emergence of a clone is an important but until now under-recognized force in colon cancer evolution and heterogeneity.
Results
Multicolor Clonal Tracing in Colorectal Cancer.
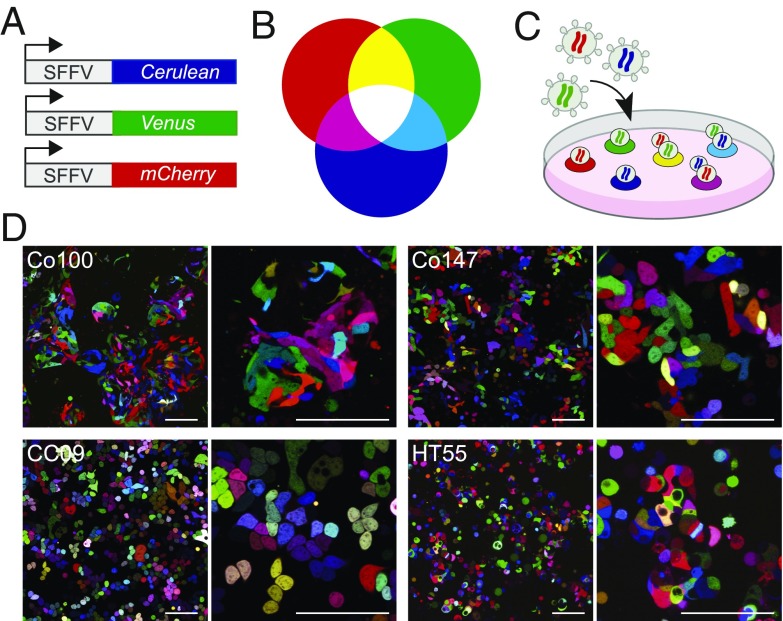
To study the clonal dynamics that drive expansion of CRC tissue in situ, we transduced a series of three primary colon cancer cultures (Co100, Co147, and CC09) and one serum-cultured cell line (HT55) with the LeGO vector set (Fig. 1 A–C). Following titration of the virus, we obtained RGB (red, green, blue) marked cultures in which cells were labeled with a wide range of unique colors that allows for clonal tracking (Fig. 1D). Importantly, by monitoring a series (n = 10) of single cell clones by fluorescence-activated cell sorting (FACS) in time, we confirmed the stability of the expression of the LeGO vectors and resulting color, as well as the overall neutrality of the integration events (SI Appendix, Fig. S1). Next, we injected the cultures using different injection volumes of Matrigel s.c. in immunocompromised (nude) mice. We found that the injection volume had an important impact on the resulting clone configuration. Larger injection volumes (100 µL) resulted in diffuse clonal expansion within the Matrigel plug, and clones simply expanded until they made contact rather than being in direct competition early after injection (SI Appendix, Fig. S2A). Therefore, to resemble clonal growth dynamics of established colon cancer in our xenograft model, we selected the smallest injection volume (50 µL) for the follow-up experiments.
Fig. 1.
Clonal tracing by employing the LeGO vector set. (A) Schematic overview of the LeGO system, which includes three vectors containing a constitutively active promotor. Each vector encodes for a different fluorescent protein, from top to bottom: Cerulean (blue), Venus (green), and mCherry (red). (B) Theoretical model of the LeGO system whereby mixing of the three basic colors red, blue, and green leads to the generation of the whole spectrum of rainbow colors. (C) Transduction of cells with the LeGO system facilitates clonal tracking by marking each cell with a different color. Two main factors underlie the different color expressions per cell; vector copy number and stochastic expression depending on the vector insertion site in the genome. (D) Representative images of LeGO-transduced colon cancer cell cultures with three different vectors. (Top) Co100 (Left) and Co147 (Right). (Bottom) CC09 (Left) and HT55 (Right). (Scale bars, 100 µM.)
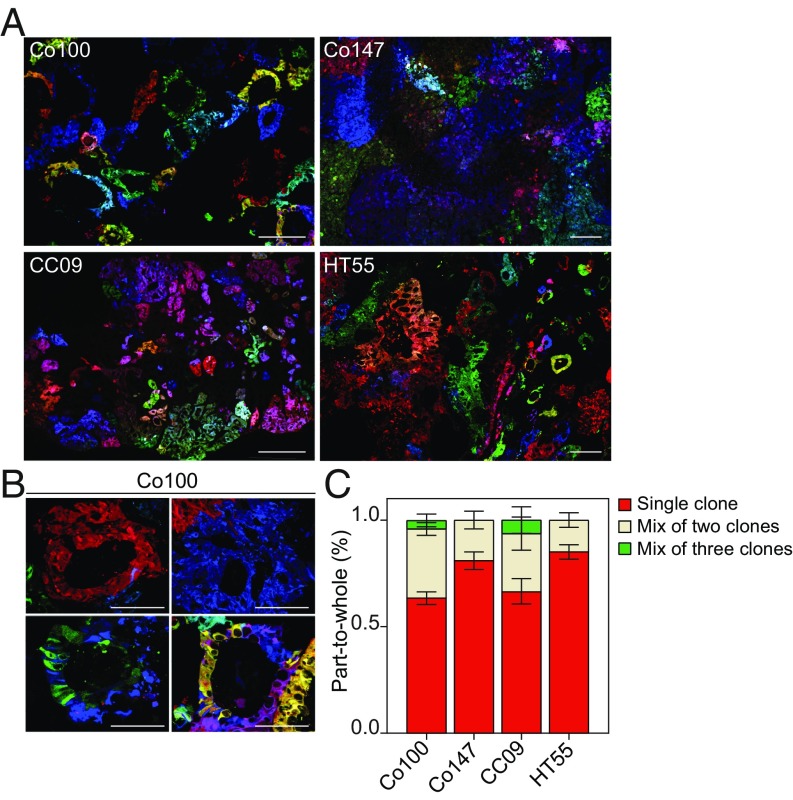
Analysis of small xenografts of ∼300 mm3 showed a clear demarcation of individual clones in all evaluated models (Fig. 2A). We define a clone as a region of identical color representing the offspring of an individual injected cell. Although multiple clones can represent with similar colors, we estimate that we can visually separate ∼96 hues, and combining the RGB marking with spatial information allows for robust identification of clones originating from the moment of injection. The various tumor models presented distinct morphologies. Whereas Co100 and HT55 xenografts showed a well-differentiated morphology with evident glandular structures separated by murine stroma, Co147 and CC09 instead were moderate/poorly differentiated, presenting with large tissue regions without glandular differentiation (Fig. 2A). In all models, clonal dispersal was limited, and only rarely were regions with a mixture of multiple clones in 2D sections detected (Fig. 2 B and C). While the fraction of mixed clones is probably higher if all three dimensions are considered, it indicates that competition between clones in CRC is mostly the result of parallel expansion at distinct rates rather than the result of direct competition within glandular structures, something that remained elusive before. Also, the limited spread of clonally related cells throughout larger cancers revealed that the motility of cancer cells within xenografts is rather limited. These findings are in line with the results of multiregion sequencing analysis that indicate that private mutations are often detected in separate tumor regions and show that the LeGO xenografts are appropriate model systems to study colon cancer growth and progression (11, 15, 16).
Fig. 2.
Clonal tracing in colorectal xenografts by LeGO. (A) Tumors derived from injected LeGO-transduced colon cancer cultures are shown. (Top) Co100 (Left) and Co147 (Right). (Bottom) CC09 (Left) and HT55 (Right). (Scale bar, 200 µM.) (B) Representative images of different clones in a LeGO-transduced Co100 tumor containing one or multiple clones intermingled. The Top shows monoclonal clones and the Bottom shows, respectively, from left to right, a mixture of two and three clones. (Scale bars, 200 µM.) (C) Bar graph depicting the percentage of clones that is monoclonal or a mixture of two or three clones within a tumor derived from the indicated cell lines as observed in 2D sections. Error bars represent SEM, a minimum of n = 5 tumors were analyzed per line.
Effective Population Size of Colorectal Cancers.
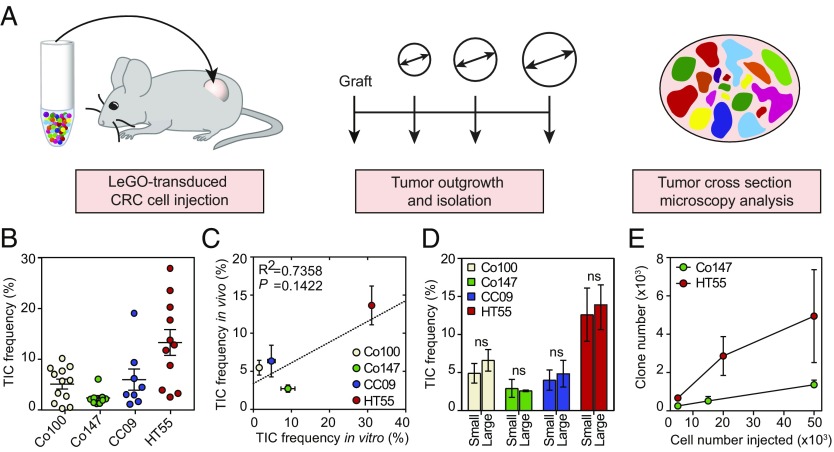
Previous work has revealed that not all CRC cells have an equal ability to initiate tumor growth in immunocompromised mice. It has been established that cells that express markers of immature cell types such as AC133, or that present with high Wnt pathway activity, have a superior capacity to induce colon cancer xenografts (17–19). These studies have mostly been performed using limiting dilution assays, in which a decreasing number of cells is injected, which then allows for the calculation of the proportion of cells within a population capable of initiating xenograft growth. However, it remains unclear how the reduced cell numbers impact on the ability to initiate tumors; for example, due to the lack of para- and juxtacrine signaling input. Using our model system, we now have the ability to directly determine the proportion of injected cells that contributes to tumor initiation using an equal number of injected cells (Fig. 3A). By high-resolution analysis of xenograft tissue, and quantification of the number of clones, we could estimate the number of injected cells that actively grew out. We found that in vivo dispersal of cells is limited (Fig. 2), and this allowed us to identify each clone that resulted from the expansion of an injected cell as a connected region of cells with the same color. We found that the percentage of injected cells contributing to tumor initiation ranged between ∼2–20%. The highest proportion of tumor-initiating cells (TICs) was detected in the serum cultured cell line (HT55), and the primary cell cultures displayed more limited clonogenic potential during the initiation phase (Fig. 3B). Additionally, comparison of the TIC frequency in the LeGO model versus the in vitro limiting dilution assay revealed only a weak correlation between both methods for determining the clonogenicity of tumor cells (Fig. 3C). We found that the limiting dilution assay could both under- and overestimate the clonogenic cell frequency. This indicates that in some models, injection of a larger cell number suppresses outgrowth of cells, as for example in Co147, while in another model the coinjected cancer cells promote outgrowth of cells (e.g., Co100). Hence, we suggest that the interpretation of data derived from an in vitro assay about the clonogenic capacity of tumor cells should be done with caution. Importantly, the estimated proportion of cells initiating tumor growth was independent of the tumor volume analyzed (Fig. 3D) and actual number of cells injected (Fig. 3E), indicating that clones that contribute to tumor initiation permanently reside in the tumor tissue, and are not lost due to competition for example, making this assay robust to analyze different time points or tumor volumes.
Fig. 3.
Clone numbers are stable during tumor growth. (A) Schematic model of the experimental setup for clonal outgrowth quantifications per LeGO-transduced colon cancer cell line. A fixed cell number was injected s.c. for each cell line and tumors were isolated at different tumor volumes. (B) Graph showing the TIC frequency of Co100-, Co147-, CC09-, and HT55-derived tumors. Each dot represents one tumor (a minimum of n = 8 per cell line), error bars represent SEM. (C) Graph showing the correlation between the average TIC frequency in an in vitro limiting dilution assay versus in vivo LeGO experiments for the indicated cell lines. Pearson correlation. Error bars represent SEM. (D) Bar graph showing the TIC frequency of Co100-, Co147-, CC09-, and HT55-derived tumors for two subgroups; tumors indicated as small having an average volume of 170 mm3 and tumors indicated as large having an average volume of 525 mm3. Error bars represent SEM, ns, not significant, Student’s t test. (E) Graph showing the number of clones as determined by image analysis for Co147 and HT55 xenografts (minimum of n = 3 tumors per injected cell number for each cell line) that were derived from injections with different cell numbers, error bar represents SEM.
Growth Dynamics of Colorectal Cancer Tissue.
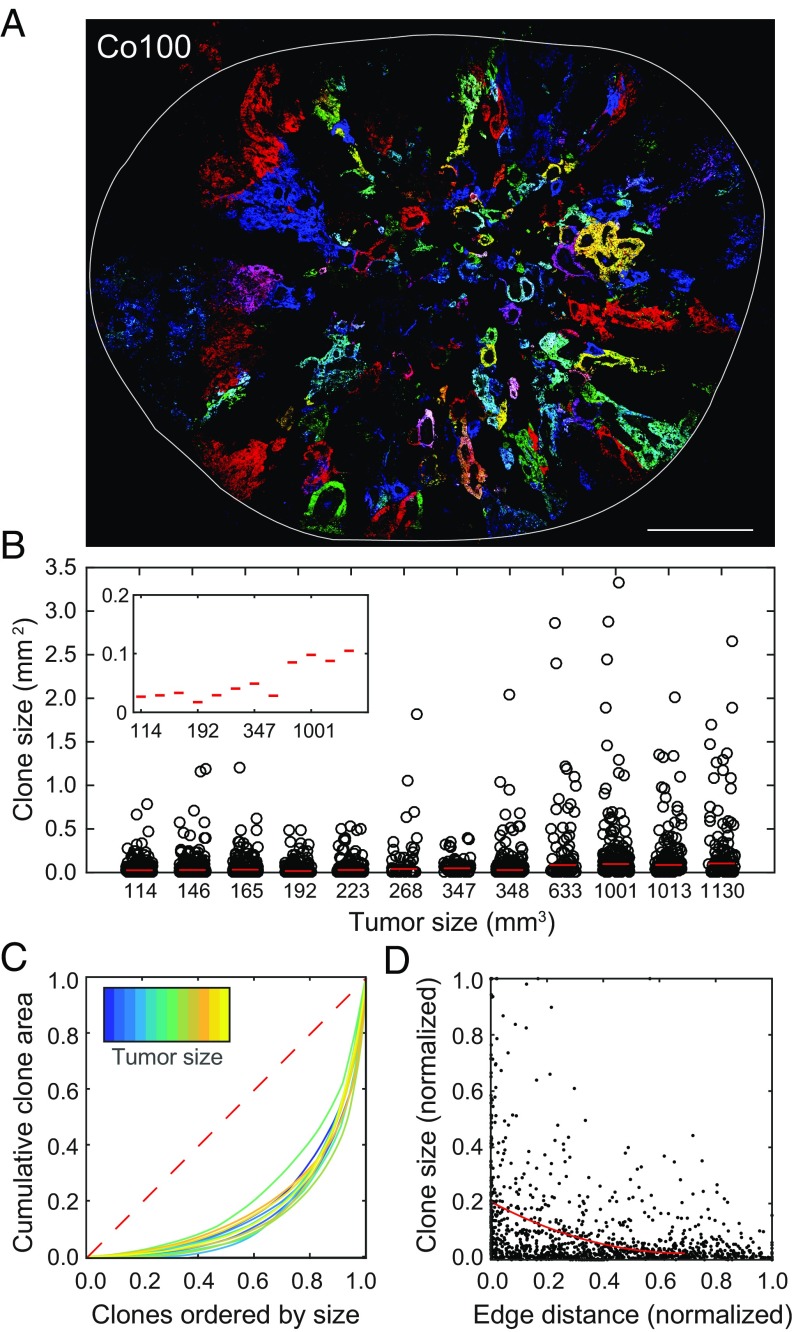
To elucidate the underlying dynamics of colon cancer tissue expansion, we mixed LeGO cultures with nontransduced cultures. This had the benefit that LeGO clones were better separated and allowed us to use a semiautomated image analysis pipeline to quantify the clone sizes within the whole xenograft tissues (Fig. 4A and Material and Methods). Analysis of hundreds of clones within tumors of different sizes revealed that on average the median clone size increased as expected in an expanding tissue (Fig. 4B). More interestingly, we detected that the heterogeneity in clone sizes was very large, and many clones remained small and did not seem to significantly contribute to tumor expansion (Fig. 4B and SI Appendix, Fig. S3). When plotting the relation between the proportion of clones that contribute to which fraction of tumor volume, we indeed detected that a small number of clones is responsible for the majority of the tumor growth (Fig. 4C). Furthermore, in larger tumors, the trend toward a relatively small number of clones driving tumor expansion is increased (Fig. 4C). It has been observed previously, by using genetic clonal tracing strategies in solid tumors, that not all cells contribute equally to cancer growth. In those studies, this heterogeneity was attributed to the intrinsic differences in clonogenic potential of cells, resulting from different cell states, i.e., stem cells vs. differentiated cells (13, 14). We now have the ability to evaluate this by studying the configuration of clones within the tissue. As is immediately apparent from the images from whole LeGO xenograft sections, there is a clear relationship between the position of the clone and its size in all cancer models studied (Fig. 4 A and D and SI Appendix, Fig. S2 A–D). Larger clones are predominantly located at the xenograft edges, implying that competition for an optimal location instead of the intrinsic properties of clones defines which clones drive expansion in this model. This implies that before clones get into direct competition, i.e., before an established tumor has formed from the injected cells, the heterogeneity in clone sizes is much smaller. Indeed, in very small tumors, where clones are not yet in contact, all clones appear to expand equally (SI Appendix, Fig. S2A). These results are in line with two recent studies that used short-term lineage-tracing strategies to confirm that clonal proliferation is most abundant at the leading edge of cancers (7, 8). Additionally, we have generated pancreatic ductal adenocarcinoma (PDAC) xenografts from LeGO-transduced cultures, and confirmed that PDACs show very similar growth dynamics as the CRC xenografts (SI Appendix, Fig. S4). To test if the leading edge might differ intrinsically from the center, we first confirmed that our cell lines contain genetic variations on the copy number level as described recently for other lines (20, 21). Significant genetic variations were detected between various xenografted tumors as well as the parental line (SI Appendix, Fig. S5A). This shows that genetic diversity is maintained in primary CRC cell cultures employed here, albeit potentially less compared with in situ human cancers. In contrast, we found no significant differences in copy number between the edge and center of the same tumor (SI Appendix, Fig. S5B). Therefore, the spatial difference in growth does not coincide with the observed genetic heterogeneity. Moreover, when cells derived from the center or edge of one tumor are retransplanted, no difference in the growth rate was observed (SI Appendix, Fig. S5C) confirming that larger clones residing at the tumor edge are not endowed with an intrinsic proliferative advantage.
Fig. 4.
Clones located on the tumor edge contribute most to tumor growth. (A) Representative cross-sectional image of xenograft showing a large variety of clone sizes, with larger clones toward the xenograft edge. (Scale bar, 1 mm.) (B) Clone sizes per tumor ordered by tumor size. Individual clones (dots) and median clone size per tumor (line). Inset shows median clone sizes. (C) Cumulative clone size as a fraction of the total cross-sectional area per tumor. The red dashed line displays the theoretical curve for a homogeneous distribution of sizes among clones. (D) Clone size versus distance to edge. Sizes and distances are normalized to the maximum of each xenograft. The red line is a polynomial fit to the clone sizes.
Modeling Colorectal Cancer Growth.
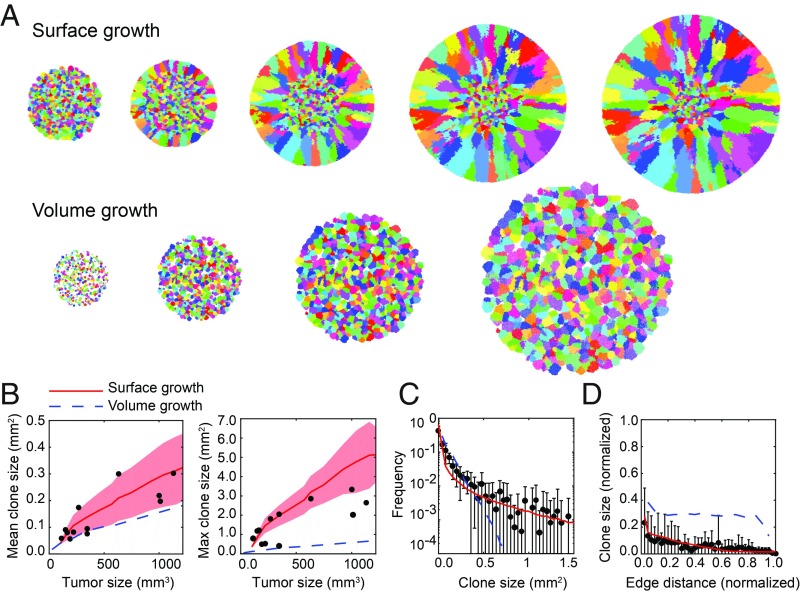
To further support the notion that locations of founding cells rather than cell intrinsic features determine the in vivo clonogenic potential, we developed a cellular automaton model to simulate xenograft expansion with growth either confined to the surface or throughout the whole tumor (Fig. 5A and Movies S1–S3). If neither of the models (volume or surface growth) could explain the data, this would argue in favor of intrinsic differences between cells; for example, as proposed by the cancer stem cell theory. We model tumor growth in 3D as a population of cells that stochastically replicate when they have sufficient free space available for the offspring (SI Appendix, Computer Models), similar to our previous work (6). In the surface growth model, cells replicate only on the surface. In the volume growth model, as the tumor expands, free space is created inside the tumor, which causes it to grow exponentially. The initial tumor conditions are taken to match the xenograft experiments: 10,000 uniquely labeled cells in a volume of 50 µL. Tumor growth is simulated until the maximum size of 1.3 billion cells, which corresponds to a tumor volume of 1.3 cm3, which is well above the maximum tumor volume in our xenograft experiments. For direct comparison with the experimental data, we take virtual 2D sections from the simulated tumors at various positions and quantify clone sizes (SI Appendix, Computer Models). With growth confined to the surface, we found excellent agreement between the simulated clone sizes as a function of the overall tumor volume, and the experimentally observed clone sizes (Fig. 5B). Moreover, the clone size distributions and patterns from the simulated environment-instructed tumor growth were highly similar to the clone patterns observed in the xenografts (Fig. 5C). In contrast, volume growth results in exponentially growing tumors with a different pattern of clonal expansion that does not explain the experimental data (Fig. 5 C and D). The spatiotemporal localization of growth is therefore crucial to explain the data and results in a large heterogeneity of clone sizes even if all clones have an equal growth rate (neutral dynamics). To assess the effect of nonneutrality, we modeled tumor growth with nonuniform growth rates of clones (normal distributed growth rates, mean = 1, SD = 0.05, 0.1, or 0.2). We find that the clone size distribution is very similar to the neutral case (SI Appendix, Fig. S6A). Other biologically realistic alterations of the model (a stem cell hierarchy or extensive cell death during growth) also only induce subtle changes in the growth pattern as revealed by the distribution of clone sizes (SI Appendix, Fig. S6B). Together these results indicate that in situ clonogenicity for clones of equal age is the result of spatial organization of the tissue.
Fig. 5.
Clonal expansion is highly dependent on clone location. (A) Cross-sections of tumors of increasing size, simulated with stochastic growth confined to the surface (Top) or throughout the entire tumor (volume growth, Bottom). Tumor sizes (in million cells) from left to right for, respectively, the Top and Bottom: 50, 100, 300, 600, 900, and 25, 150, 600, and 1400. (B–D) Comparison of experimental data (black dots) with model predictions of surface (red lines) and volume growth (dashed blue lines). (B) Mean (Left) and maximum (Right) clone size per xenograft. The red shade displays the SD of numerical mean and maximum clone sizes obtained from cross-sections taken at different positions of simulated tumors growing on the surface. (C) Distribution of sectional clone sizes. Error bars represent SD between xenografts. (D) Average clone size versus distance to tumor edge. Clone sizes and distances are normalized to the maximum of each cross-section. Error bars represent SD of the experimental data.
Discussion
Using a color-based clonal tracing method in combination with primary human CRC cultures, we obtained important insights in the dynamics of colon cancer xenograft growth and clonal heterogeneity. Firstly, we revealed that the initiation phase of xenograft growth is dependent on the volume used for injection and the number of cells injected. Controlling these variables is essential for accurate interpretation of TIC assays and clonal competition studies. Secondly, the xenografts from LeGO-transduced primary cultures revealed that clonal dispersal and clonal mixing are limited in colon cancer xenografts. This observation has important implications, as it strengthens the notion that clones in colon cancer tissue expand in parallel when residing in an identical environment. Also, this provides geometrical support to the idea that a large proportion of CRCs display predominantly neutral evolution and that no large selective sweeps follow emergence of novel, more aggressive clones in established cancers.
Previous short-term lineage tracing experiments by us and others indicated that clonal expansion is spatially regulated in established CRC xenograft tissue (7, 8). In these studies, clones expressing one or a few different colors were induced in established tumors. The small number of colors in these experimental systems prevented long-term tracing of individual clones because growth and merging of neighboring clones are hard to distinguish with a limited resolution. In the current study, we have overcome this limitation, which has enabled us to study long-term clonal dynamics in xenografts of established CRCs. The combination of long-term lineage tracing and computational modeling revealed that clone size heterogeneity can be fully explained by externally driven growth at the leading edge of the tumor. In contrast, previous barcoding studies in human xenografts indicated that distinct types of stem-like cells in colon cancer xenografts exhibit distinct repopulating features caused by intrinsic functional differences in the self-renewal and tumor-forming capacity of tumor cells (13, 14). Our data provide a radically different explanation for these observations by showing that these different contributions to tumor expansion following xenograft propagation are spatially orchestrated rather than intrinsically defined. Of note, studying clonal dynamics of early phases of premalignant expansion and conversion toward CRC falls beyond the scope of this study, but in these early stages the clonal dynamics are possibly more defined by genomic differences between clones. In addition, our model system lacks a functional immune system and immune effects are not captured in our study. Notwithstanding, we conclude that within the time frame and spatial scale of our experimental setup, the tumor environment is a dominant factor in shaping CRC growth and progression, as expansion mainly occurs at the tumor edge.
Several factors can explain the observed growth at the tumor edge. For example, the enrichment for stroma and secretion of stromal factors can drive clonogenic expansion at the leading edge. The increased interstitial pressure within the xenograft centers is also likely to contribute. Importantly, manipulation of these factors could yield new therapeutic avenues to improve treatment; for example, Osteopontin would be an interesting target to further investigate (8). Furthermore, targeting the intercellular machinery associated with clonogenic potential, as a cell state enabled by the environment, is another strategy that could be developed to improve the prognosis of patients with solid cancers.
Materials and Methods
Cell Culture.
Human primary colon cancer cultures were established as described previously (18). Cultures were isolated from patients with colorectal cancer with approval of the medical ethical committee of the Academic Medical Center and University of Palermo. Primary cell lines are cultured in polyHEMA [Poly(2-hydroxyethyl methacrylate; Sigma] coated flasks (Corning) to allow spheroid growth. Advanced DMEM/F12 (Life Technologies) culture medium is used, which is supplemented with N-2 (Life Technologies), l-glutamine, glucose, Hepes, heparin, insulin, epidermal growth factor (EGF), and basic fibroblast growth factor (bFGF) as described previously (18). The primary human PDAC culture 067 was established as described previously (22) and cultured in Iscove’s modified Dulbecco’s medium (Life Technologies) supplemented with 8% FBS and l-glutamine. DLD1 (ATCC) and HT55 (Sanger Institute) cells were cultured in DMEM/F12 (Life Technologies) supplemented with 8% FCS (Life Technologies). Capan-2 (ATCC) was cultured in DMEM (Life Technologies) supplemented with 8% FCS. Cell lines were authenticated by short tandem repeat profiling in combination with mutation analysis and have been regularly tested for mycoplasma infection.
Multicolor Marking.
Cell lines were simultaneously transduced with three different constructs according to a previously published protocol (12). The following lentiviral gene ontology (LeGO) vectors were used: LeGO-C2 (27339), LeGO-V2 (27340), and LeGO-Cer2 (27338) (Addgene). In short, 50,000 single cells were seeded in a 24-well plate in 500 µL culture medium in the presence of 8 μg mL−1 polybrene (Sigma). Lentivirus containing the three vectors was added in a volume that ensured ∼60% transduction rate of each vector. Plates were centrifuged for 1 h at 24 °C and incubated overnight at 37 °C. Transduction rates were analyzed by flow cytometry after 3 d. After transduction, cell lines were passaged in a low dilution and for a maximum of five passages before in vivo use.
Flow Cytometry.
Flow cytometry was performed on a Fluorescence Activated Cell Sorting (FACS) Aria SORP (BD Biosciences) machine with 405-, 488-, and 561-nm lasers. Data were analyzed with the FlowJo (FlowJo LLC) software.
Vector Expression.
Vector integration stability was analyzed by FACS. DLD1 cells were transduced with the LeGO system and then single-cell sorted. Single-cell clone cultures were expanded and passaged twice a week. Upon passaging the expression of Cerulean, Venus and mCherry were analyzed by FACS. Every cell line was analyzed at least twice in a 12-wk follow-up period.
In Vivo Experiments.
The Animal Experimentation Committee at the Academic Medical Center in Amsterdam has approved all in vivo experiments (DEC103181 and DEC102348) and all animal experiments were performed according to the national guidelines. Female nude (Hsd:Athymic Nude-Foxn1nu) mice (6–12 wk old) were obtained from Envigo. Animals were randomly assigned to experimental groups; no blinding was applied for these experiments. Animal exclusion was performed when no tumor growth appeared.
Xenograft Studies.
Xenograft tumors were generated by injecting 10,000 (CC09) or 50,000 (Co100, Co147, and HT55) human colon cancer cells in a mixture of medium and Matrigel (Corning) in a 1:1 ratio with a cell density of around 400–1,000 cell/µL. Cells were injected s.c. into the flanks of nude mice. Tumor growth was measured manually twice a week using a caliper. Mice were killed based on tumor size at various time points to isolate tumors. After isolation tumors were fixed using 4%-paraformaldehyde in PBS solution overnight at 4 °C followed by preservation in a 20% sucrose solution for 12 h at 4 °C. Tumors were split into two equally sized parts and 10 µm-thick frozen tissue sections were collected from the tumor center.
In Vivo Transplantation Assay.
Center and edge (<0.5 mm from tumor border) located cells were isolated from freshly collected xenografts by using razor blades. Immediately after tissue collection, cells were dissociated by using medium containing collagenase (Roche) and hyaluronidase (Sigma) at 37 °C for 1 h. Before injection, cells were filtered using a 70 µM cell strainer, and dead cells were removed by 7-AAD staining (BD Biosciences) by using FACS. For each group, 1,000 cells were injected s.c. into the flanks of nude mice (n = 3), and tumor growth was measured twice a week.
Copy Number Analysis.
DNA was extracted using the NucleoSpin Tissue kit (Bioké) following the manufacturer’s procedure. To extract DNA from the inside and outside of tumors, we first mechanically separated the two regions. Shallow sequencing and data analysis were performed as previously described (23).
Limiting Dilution Assay.
Cells were dissociated and plated in 96-well plates (Corning) using SH800 Cell Sorter (Sony) in a limiting dilution manner at 1, 2, 4, 8, 16, 24, 32, 64, 128, 256 cells per well. Clonal frequency and significance were determined using the Extreme Limiting Dilution Analysis (ELDA) “Limdil” function (24).
Imaging.
Frozen tissue sections were imaged by an EVOS FL Cell Imaging System (Thermo Fisher Scientific). Sections were covered with ProLong Gold Antifade Mountant (Thermo Fisher Scientific) to ensure fluorescent signal preservation. Whole tumor sections were scanned for mCherry, Venus, and Cerulean by using the following LED light cubes; TexasRed (excitation 445/45 and emission 510/42 nm), YFP (excitation 500/24 and emission 524/27 nm), and CFP (excitation 585/29 and emission 624/40 nm). For high-resolution imaging, a SP8-X confocal microscope (Leica) with the Leica Application Suite-Advanced Fluorescence software was used.
Image Analysis.
Automated clone size quantification and localization was performed on whole tumor cross-sectional slides imaged by fluorescence microscopy and converted to .tiff file format with a custom written MATLAB program. Boundaries of connected regions with the same color and cross-sections were manually highlighted for accurate tracking of clone position and size. Connected regions with the same color, but separated by >10 cell diameters were considered as separate clones. The number of mixed clones was identified manually.
Spatial Model for Tumor Growth.
We adapted the 3D spatial model we recently introduced for tumor evolution, for direct comparison with the xenograft data (6). In short, in this model tumor cells occupy sites of a regular 3D lattice. To simulate growth, iteratively a random cell which has at least one of the neighboring sites (Von Neumann neighborhood) vacant, replicates to a randomly chosen vacant neighbor site. A detailed description of the computational modeling, a description of the different model versions, and how numerical data are compared with the experimental data can be found in SI Appendix, Computer Models.
Statistical Analysis.
Sample sizes, statistical tests, and definitions of error bars are indicated in the figure legends and calculated using GraphPad Prism 7 or MATLAB. All statistical tests were two-sided. The between-group variances were similar and the data were normally distributed. P values of <0.05 were considered significant.
Data Availability.
Source data for Figs. 4 and 5 and SI Appendix, Fig. S3 is provided in Dataset S1.
Supplementary Material
Acknowledgments
We thank Manouk den Toom for technical assistance. This work is supported by the Amsterdam UMC (Amsterdam), The New York Stem Cell Foundation, Cancer Research UK, and grants from Koningin Wilhelmina Fonds Dutch Cancer Society (Grants UVA2011-4969, UVA2014-7245, and 10529), the Maurits en Anna de Kock Stichting (Grant 2015-2), Worldwide Cancer Research (Grant 14-1164), the Maag Lever Darm Stichting (Grant MLDS-CDG 14-03), the European Research Council (Grant ERC-StG 638193), and ZonMw (Grant Vidi 016.156.308) (to L.V.) and an AG&M PhD talent development grant (to D.M.M.). L.V. is a New York Stem Cell Foundation – Robertson Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813417116/-/DCSupplemental.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Vermeulen L, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 3.Snippert HJ, Schepers AG, van Es JH, Simons BD, Clevers H. Biased competition between Lgr5 intestinal stem cells driven by oncogenic mutation induces clonal expansion. EMBO Rep. 2014;15:62–69. doi: 10.1002/embr.201337799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer. 2014;14:468–480. doi: 10.1038/nrc3744. [DOI] [PubMed] [Google Scholar]
- 5.Clevers H. The cancer stem cell: Premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 6.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamprecht S, et al. Multicolor lineage tracing reveals clonal architecture and dynamics in colon cancer. Nat Commun. 2017;8:1406. doi: 10.1038/s41467-017-00976-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenos KJ, et al. Stem cell functionality is microenvironmentally defined during tumour expansion and therapy response in colon cancer. Nat Cell Biol. 2018;20:1193–1202. doi: 10.1038/s41556-018-0179-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGranahan N, Swanton C. Biological and therapeutic impact of intratumor heterogeneity in cancer evolution. Cancer Cell. 2015;27:15–26. doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber K, Thomaschewski M, Benten D, Fehse B. RGB marking with lentiviral vectors for multicolor clonal cell tracking. Nat Protoc. 2012;7:839–849. doi: 10.1038/nprot.2012.026. [DOI] [PubMed] [Google Scholar]
- 13.Kreso A, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dieter SM, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Kim TM, et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin Cancer Res. 2015;21:4461–4472. doi: 10.1158/1078-0432.CCR-14-2413. [DOI] [PubMed] [Google Scholar]
- 16.Roerink SF, et al. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature. 2018;556:457–462. doi: 10.1038/s41586-018-0024-3. [DOI] [PubMed] [Google Scholar]
- 17.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 18.Todaro M, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell Stem Cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 19.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 20.Ben-David U, et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat Genet. 2017;49:1567–1575. doi: 10.1038/ng.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-David U, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560:325–330. doi: 10.1038/s41586-018-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Damhofer H, et al. Establishment of patient-derived xenograft models and cell lines for malignancies of the upper gastrointestinal tract. J Transl Med. 2015;13:115. doi: 10.1186/s12967-015-0469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheinin I, et al. DNA copy number analysis of fresh and formalin-fixed specimens by shallow whole-genome sequencing with identification and exclusion of problematic regions in the genome assembly. Genome Res. 2014;24:2022–2032. doi: 10.1101/gr.175141.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data for Figs. 4 and 5 and SI Appendix, Fig. S3 is provided in Dataset S1.