Significance
Extracellular vesicles (EVs) are cell-derived nanosized particles medicating cell−cell communication and transferring biology information molecules like nucleic acids to regulate human health and diseases. Conventional methods for EV surface marker profiling can’t tell the differences in the quantity and phenotypes of EV secretion between cells. To address this need, we developed a microchip platform for profiling an array of surface markers on EVs from large numbers of single cells, enabling more-comprehensive monitoring of cellular communications. Single-cell EV secretion assay led to previously unobserved cell heterogeneity underlying EV secretion, which might open up avenues for studying cell communication and cell microenvironment in both basic and clinical research.
Keywords: single-cell analysis, extracellular vesicle, cellular heterogeneity, antibody barcodes
Abstract
Extracellular vesicles (EVs) are important intercellular mediators regulating health and diseases. Conventional methods for EV surface marker profiling, which was based on population measurements, masked the cell-to-cell heterogeneity in the quantity and phenotypes of EV secretion. Herein, by using spatially patterned antibody barcodes, we realized multiplexed profiling of single-cell EV secretion from more than 1,000 single cells simultaneously. Applying this platform to profile human oral squamous cell carcinoma (OSCC) cell lines led to a deep understanding of previously undifferentiated single-cell heterogeneity underlying EV secretion. Notably, we observed that the decrement of certain EV phenotypes (e.g., CD63+EV) was associated with the invasive feature of both OSCC cell lines and primary OSCC cells. We also realized multiplexed detection of EV secretion and cytokines secretion simultaneously from the same single cells to investigate the multidimensional spectrum of cellular communications, from which we resolved tiered functional subgroups with distinct secretion profiles by visualized clustering and principal component analysis. In particular, we found that different cell subgroups dominated EV secretion and cytokine secretion. The technology introduced here enables a comprehensive evaluation of EV secretion heterogeneity at single-cell level, which may become an indispensable tool to complement current single-cell analysis and EV research.
Extracellular vesicles (EVs), including exosome, microvesicle, etc., are critical components in the cell microenvironment, regulating intercellular communications and transferring biology information molecules like cytosolic proteins, lipids, and nucleic acids (1–6). Due to their relatively stable duration in the circulation system, they have shown great potential to be used as noninvasive diagnostic markers for disease progression (7–9) or treatment (2, 3). Thus, detection and stratification of EVs, based on their sizes (10), morphologies (11), molecular compositions (12, 13), etc., is crucial to increase our understanding of EVs and may bring applications in biomedicine. Among different molecular components involved in EV functionalities, proteomic surface markers provide direct targets for intercellular communication mediated by EVs (14, 15). A variety of methods have been reported for profiling protein markers on EVs’ surface, such as ELISA (16), Western blotting (15), flow cytometry (10), imaging (17), etc., from the population of EVs (15) down to single-vesicle level (10, 17). However, these measures are still at population cell level, which averaged EV secretion from different cellular sources and obscured cell-to-cell heterogeneity in quantity/phenotypes of EV secretion and their related functions (18–22). Nanowell-based (23, 24) and tetraspanin-based pH-sensitive optical reporters (25) for single-cell EV secretion assay have been developed to address the need, but with limited proteomic parameters (≤2) for EVs from every single cell, which is not sufficient to dissect EV secretion heterogeneity comprehensively. A technology that can profile an array of surface markers on EVs from large numbers of single cells is still lacking and will help to address a host of important biological questions ranging from intertumor and intratumor diversity to the cell−cell communication network, and will be of great value to clinical applications like personalized diagnostics and medicine.
Herein, we demonstrate a microchip platform for multiplexed profiling of single-cell EV secretion to address the critical need for technologies to dissect the communication spectrum of tumor cells mediated by EVs. The multiplexed profiling was realized with antibody barcodes, which is a reliable, reproducible technology previously adopted for blood testing (26), single-cell proteomic analysis (27–31), and immunotherapy monitoring (32–34). We applied the platform to profile human oral squamous cell carcinoma (OSCC) cell lines, and patient samples, which revealed previously unobserved secretion heterogeneity and identified that the decrement of certain EV phenotypes (e.g., CD63+EV) were associated with the invasive potential of both OSCC cell lines and primary OSCC cells. Besides, we also realized the simultaneous profiling of EV secretion and cytokine secretion from the same single cells for a deep understanding of cellular organizations and uncovering the correlation between different types of intercellular communication mediators.
Results
Platform for Multiplexed Profiling of Single-Cell EV Secretion.
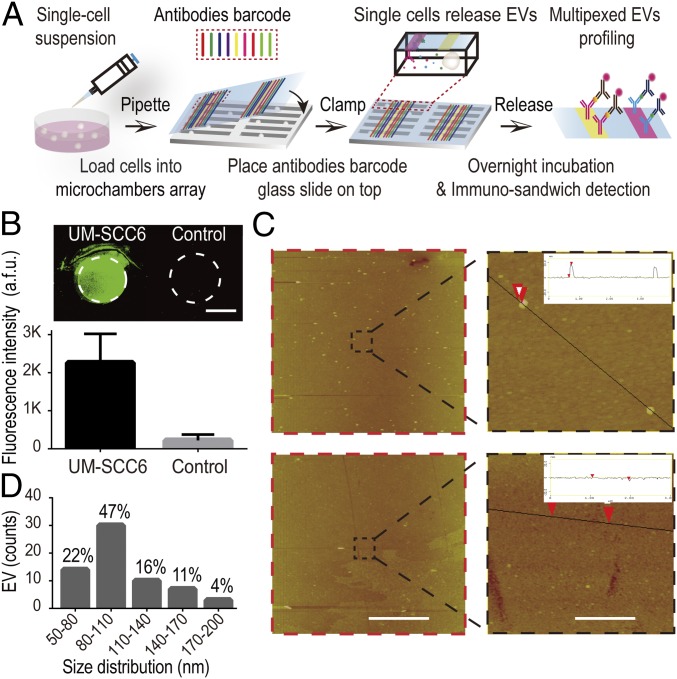
The platform to realize multiplexed single-cell EV secretion detection (Fig. 1A) is modified from previously reported devices (31), and combines two functional components: a high-density microchamber array and a spatially resolved antibody barcode glass slide. The high-density microchamber array (SI Appendix, Fig. S1A) accommodates 6,343 identical units for isolating and concentrating EVs secreted from exactly a single cell (dimension of each microchamber: width 40 μm, length 1,440 μm, depth 30 μm). The volume of each microchamber is around 1.7 nL, which corresponds to 5 × 105 cells per mL cell density, comparable to the cell density typically used in bulk experiments. Due to the drastic decrease in liquid volume, the concentration of detection targets will be concentrated as much as 103 to 105 times compared with population measurements (1.7 nL vs. 10 μL to 200 μL), which ensures high-sensitivity detection. We designed and fabricated the accompanying microchip with highly parallel microchannel array to pattern spatially resolved antibody barcodes onto a poly-L-lysine glass slide, which can accommodate up to nine different antibodies (with each antibody stripe 40 μm in width) for multiplexed profiling (SI Appendix, Fig. S1B; the consumption of each antibody for patterning is only 3 μL of volume at 250 μg/mL). The antibody patterning can be finished within 4 h with excellent uniformity (fluorescein isothiocyanate-labeled BSA coating: coefficient of variance of <5% in 2 cm × 5.5 cm area; SI Appendix, Fig. S2).
Fig. 1.
Platform for multiplexed profiling of single-cell EV secretion. (A) Schematic illustration of the workflow for multiplexed profiling of single-cell EV secretion. Images of two functional components are shown in SI Appendix, Fig. S1. (B) EV detection results on anti-human CD81 antibody-coated spot with UM-SCC6 cells conditioned medium and control sample: blank cell culture medium supplemented with 10% ultracentrifuged FBS. (Scale bar: 1 mm.) (C) AFM characterization of fluorescence detection regions shown in B. [Scale bars: 10 μm (Left) and 1 μm (Right).] Insets show the cross-section view of the captured vesicles and the control. (D) The size distribution of EVs captured on the anti-CD81 antibody-coated surface.
We verified antibody-based EV capture/detection principle at bulk level (SI Appendix, Fig. S3). Antibodies targeting human CD81 and CD63 for EVs were used to form a detectable immuno-sandwich, in which both proteins are tetraspanins highly expressed in EVs for reliable EV marker proteins (14, 15). Here we used two different antibodies to recognize two antigens on each vesicle to form sandwich assembly: One antibody is for EV capture (e.g., CD9, CD81, EpCAM), and the other one is biotinylated anti-CD63 antibody for fluorescence detection. This double-positive detection strategy based on recognition of different epitopes can eliminate the crosstalk from soluble molecules to ensure the detection specificity and has been widely used in EV research (16, 35). We obtained positive fluorescence signals with conditioned medium from human oral squamous carcinoma cells (UM-SCC6) (Fig. 1B). Atomic force microscope (AFM) characterization confirmed the fluorescent signals were from EVs (Fig. 1C). The diameter of the captured particles ranged from 50 nm to 200 nm, suggesting EVs captured covered both exosomes (size: 50 nm to 150 nm) and microvesicles (size: 100 nm to 1,000 nm) (Fig. 1D). Consistent with fluorescence results, we didn’t capture any particles from exosome-depleted cell culture medium sample. Similar results were obtained using a scanning electron microscope to characterize the EVs captured on the CD63 antibody-coated spots, which further validated the antibody-based capture strategy for EV detection (SI Appendix, Fig. S4). We also demonstrated that multiple EVs could be profiled on micrometer-sized antibody stripes (SI Appendix, Fig. S5), suggesting the feasibility to use antibody barcodes for multiplexed EV detection.
Multiplexed Single-Cell Profiling Reveals Complex Heterogeneity Underlying EV Secretion.
We then used the platform to profile the EV secretion with human OSCC (SCC25) to assess its single-cell detection sensitivity (36). Forty thousand cells (200 μL at 2 × 105 cells per mL density) was pipetted directly onto the hydrophilic microchamber array (oxygen plasma-treated). When enclosed by putting an antibody barcode glass slide on the top, more than 1,000 single cells were captured (1386 ± 276, n = 12, accounting for 20.5% of total microchambers; SI Appendix, Fig. S6), ensuring high-throughput analysis and statistical significance. The proteomic parameters for EV surface marker profiling used in this study include CD63, CD9, CD81, EpCAM, and HSP70. With the combination of surface markers used here, the EVs captured from the same single cells can be further categorized into five subgroups: CD63+EV, CD9+CD63+EV, CD81+CD63+EV, EpCAM+CD63+EV, and HSP70+CD63+EV. Before single-cell experiments, we conducted validation experiments to confirm that the cell secretion profile is not altered by the polydimethylsiloxane (PDMS) surface compared with secretion from cells cultured in a conventional tissue culture plate (SI Appendix, Figs. S7 and S8). Besides, the single cells trapped in microchambers were still viable after being incubated for 18 h (SI Appendix, Fig. S9) before conducting sandwich detection procedures to transform captured EVs into detectable fluorescence signals. A representative detection result from SCC25 cells was shown in Fig. 2A, from which we observed fluorescent positive square spots intersecting CD63/CD81/CD9 antibody barcodes with a signal-to-noise ratio (SNR) of ≥3, demonstrating EVs with different surface proteins from the same single cells were reliably detected.
Fig. 2.
Multiplexed profiling of SCC25 single-cell EV secretion revealed previously unobserved secretion heterogeneity. (A) Representative images showing the raw data of multiplexed single-cell EV profiling, including fluorescence detection images (partial and enlarged), and corresponding cells in microchambers (red circled are single cells). (B) Scatter plots showing five-plexed EV secretion profiling from SCC25 single cells (n = 1,264 and 1,417 for zero cells and single cells, respectively). The fluorescence intensity and secretion frequency were normalized with the average plus 2 SD of the zero-cell data. **P < 0.005; ***P < 0.0005; ****P < 0.00005; ns, not significant (P > 0.05). (C) Histograms showing secretion intensity distribution of different EVs (CD63+EV, CD81+CD63+EV, and CD9+CD63+EV). (D) Polyfunctionality analysis of SCC25 single cells (fraction of cells to secrete multiple EVs simultaneously). (E) Scatter plots showing the change of CD9+CD63+EV secretion frequency with the increased number of cells per microchamber. (F) Heatmap showing EV−EV correlation in single cells.
Single-cell data were normalized based on mean fluorescence intensity plus 2 times the SD of all zero-cell microchambers as thresholds to define positive secretion events (31). SCC25 single-cell EV secretion results were presented as scatter plots in Fig. 2B, which provides direct insights to understand EV secretion heterogeneity: (i) Not all cells can secrete EVs; for example, only around 6.2% cells secreted CD81+CD63+EV. (ii) Intensity distribution within these EV secreting cells revealed that a very small number of cells could secrete ∼10 times more than average secretion, indicating the presence of outliers or “super EV secretors” within cell population (Fig. 2C). (iii) Cells secreted EVs with preference within different surface markers; for example, around 23.5% of SCC25 cells secreted CD9+CD63+EV, while we could barely see HSP70+CD63+EV secretion at single-cell level. (iv) A small fraction of SCC25 cells could secrete multiple cytokines or EVs simultaneously (Fig. 2D); for example, only ∼2.7% of single cells can secrete EVs with more than three different combinations of surface markers at the same time, further confirming the presence of super EV secretors within the cell population. Collectively, these observations present the complex heterogeneity underlying EV secretion, which is difficult to profile with population measurements. Interestingly, we found the percentage of cells with positive EV secretion would increase with more cells in each microchamber (Fig. 2E), suggesting EV secretion is also mediated with paracrine signaling, which is in agreement with other reports (37). We also saw that these EV phenotypes were weakly correlated via linear regression analysis of the correlation coefficient between EVs (Fig. 2F). It is noted that we observed similar secretion results when single cells were cultured in microchips for 6 and 12 h, indicating the intrinsic nature of cell heterogeneity in EV secretion (SI Appendix, Fig. S10). Moreover, with the increased incubation time, secretion frequencies of some EVs gradually increased (e.g., CD9+CD63+EV), and a larger portion of cells started to secrete more than one type of EV simultaneously.
Decreased Single-Cell EV Expression in Invasive Tumor Cells.
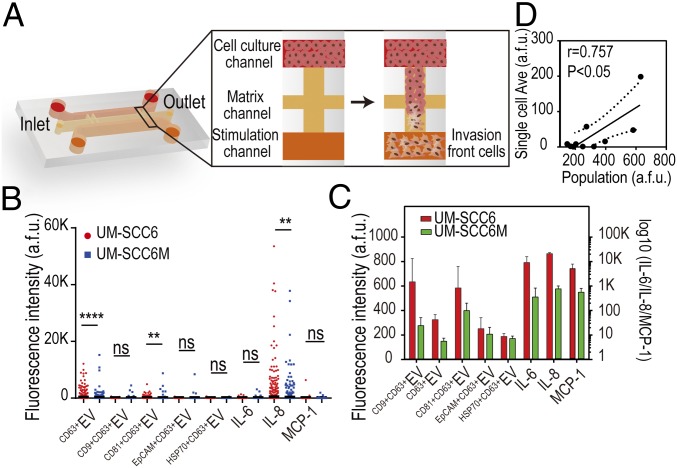
We then applied the platform to profile EVs derived from tumor cells with different migratory properties at single-cell level to uncover the correlation between EV secretion and a cell’s invasive behavior. A subgroup of UM-SCC6 cells with high invasion behavior in Matrigel matrix (named UM-SCC6M) was obtained by three rounds of isolation of invasion front of UM-SCC6 cells in an H-shaped microfluidic chip (Fig. 3A). Detailed isolation procedures have been reported previously (38). To dissect the multidimensional spectrum of intercellular communications, here we profiled five-plexed EV secretion (CD63, CD9, CD81, EpCAM, and HSP70) along with three-plexed proteins secretion (IL-6, IL-8, and MCP-1) simultaneously from each single cell to provide a direct correlation between different types of intercellular messengers (SI Appendix, Tables S1 and S2). Titration tests with recombinant proteins, EV standard, and antibody crosstalk tests were completed to validate technical validity (SI Appendix, Figs. S11–S14). Interestingly, we found that UM-SCC6M cells, which are active in the invasion, were less active in secretion for both EVs and proteins, compared with UM-SCC6 cells (Fig. 3B). Specifically, 12.3% of UM-SCC6 single cells secreted CD63+EV, while 4.3% of UM-SCC6M single cells secreted CD63+EV; 10.9% of UM-SCC6 single cells secreted CD9+CD63+EV vs. 0.9% for UM-SCC6M single cells; 10.6% of UM-SCC6 single cells were positive in IL-8 secretion, while only 5.7% of UM-SCC6M single cells secreted IL-8. Previous studies have demonstrated that tetraspanins CD63 and CD9 are metastasis suppressors, highly expressed in the early stages of different cancers [e.g., melanoma (39, 40) and carcinoma (41, 42)] and decreased in advanced stages. Our results showed the CD63 and CD9 expression is also decreased on tumor cell-derived EV surfaces when cells are in the invasive state, which has never been observed previously at single-cell level. We also saw the similar trend in cell population assay (Fig. 3C) and a reasonable degree of correlation between single-cell results and cell population measurements (Fig. 3D; Pearson r = 0.76, P < 0.05), despite significant differences in assay conditions between them.
Fig. 3.
Single-cell EV secretion in invasive tumor cells. (A) Illustration of the isolation of invasion front of UM-SCC6 cells in a microchip. (B) Comparative analysis of secretion frequencies between UM-SCC6 (n = 1,263) and UM-SCC6M (n = 1,512) single cells. **P < 0.005; ****P < 0.00005; ns, not significant (P > 0.05). (C) Eight-plexed secretion profiles from cell populations. (D) Correlation of EV secretion levels between single-cell averages and cell population measurements (the dashed line shows the 95% confidence interval).
Multiplexed Profiling of Single-Cell Secretion of OSCC Patient Samples.
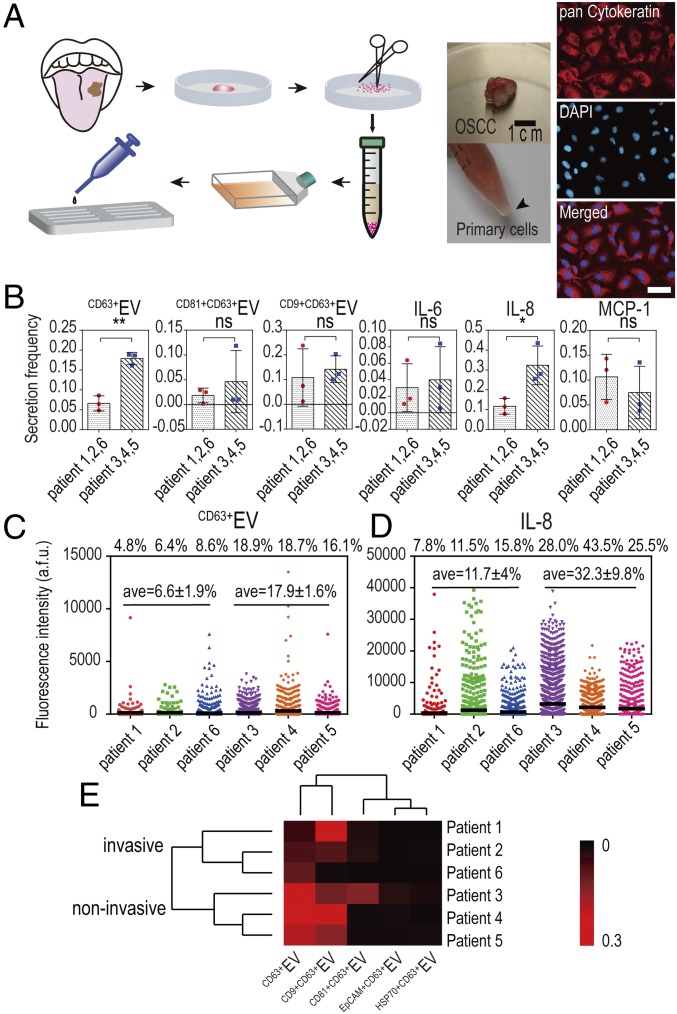
To further demonstrate potential applications of a single-cell EV analysis platform to clinical samples, we profiled six primary ex vivo tissues from OSCC patients to discern metastatic tumor-derived EVs associated with EV secretion (SI Appendix, Fig. S15 and Table S3). The fresh OSCC tumor tissues from surgery were disassociated, purified into primary tumor cell suspensions, and verified with epithelial malignancy marker pan Cytokeratin immunostaining (43) (Fig. 4A). Based on clinical records (SI Appendix, Table S3), patients 1, 2, and 6 were diagnosed as metastatic, while patients 3 to 5 were nonmetastatic. We applied the single-cell analysis workflow to primary tumor cells and generated excellent EV secretion signals and similar background comparable to the results from cell lines. Each primary tumor cell sample from a patient exhibited similar secretion signatures as OSCC cell lines; for example, they were relatively strong in CD9+CD63+EV and IL-8 secretions (Fig. 4B), while attenuated in EpCAM+CD63+EV and HSP70+CD63+EV secretions. Notably, we found averaged CD63+EV secretion in metastatic patients 1, 2, and 6 (average = 6.6 ± 1.9%) were significantly decreased, compared with nonmetastatic patients 3 to 5 (average = 17.9 ± 1.6%) (P = 0.0013; Fig. 4C), which confirmed our observation from the UM-SCC6 cell line that CD63+EV secretion decreased in advanced stage of OSCC. Besides, we also found that tumor cells from invasive OSCC patients showed decreased IL-8 secretion (P = 0.0275; Fig. 4D), while CD9+CD63+EV didn’t show significant differences between invasive and noninvasive patients, suggesting the heterogeneity between cell lines and primary cells. Unsupervised hierarchical clustering of the single-cell secretion profiles successfully resolved the differential EV secretion pattern between invasive patients and noninvasive patients (Fig. 4E), demonstrating the potential value of multiplexed single-cell EV profiling for cancer metastasis diagnostics.
Fig. 4.
Single-cell secretion analysis of primary tumor cells from OSCC patients. (A) Overview of disassociating patient surgery specimens into primary tumor cells suspension (verified with pan Cytokeratin immunostaining) and it’s follow-on procedures to apply primary cells to the microchamber array for single-cell analysis. (Scale bar: 50 μm.) (B) Comparative analysis of individual secretions among invasive and noninvasive patients. **P < 0.005; ns, not significant (P > 0.05). (C and D) Scatter plots showing the distribution of individual secretions (CD63+EV and IL-8) in six patients (n = 974, 1,351, 1,801, 914, 842, and 1,285, respectively, for patients 1 through 6). (E) Clustering of six patient samples based on secretion frequencies of all EV parameters.
Single-Cell Secretion Functional Phenotyping.
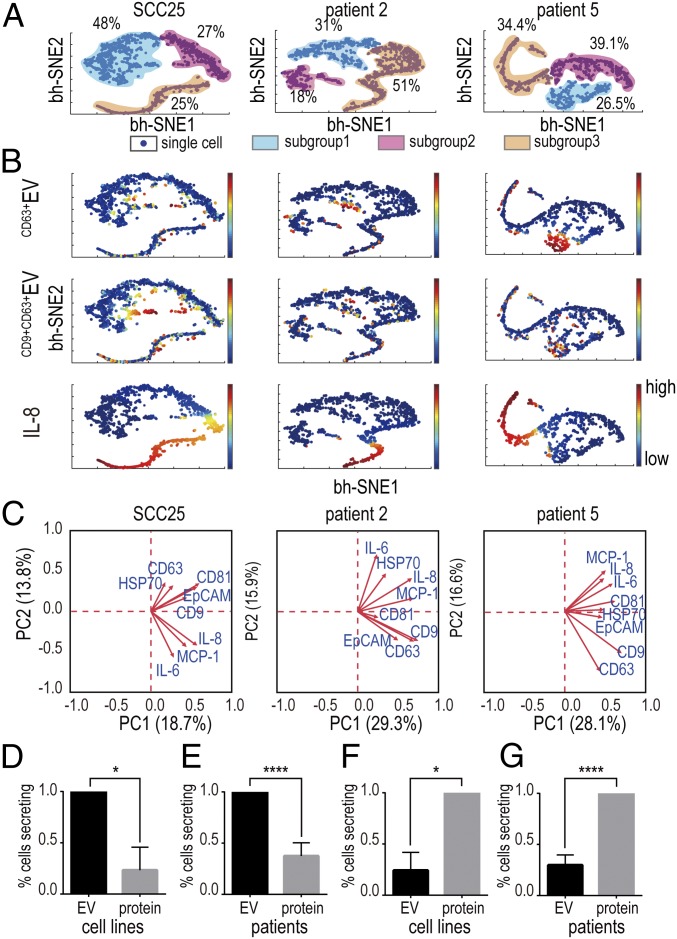
We then mapped all of the single-cell data from OSCC cells using t-Distributed Stochastic Neighbor Embedding (t-SNE)–based visualization (viSNE) (44) to reveal their functional organizations (Fig. 5A). We saw that these cells generally gave rise to structured clusters: group 1 is mainly distinguished with EV secretion, like CD9+CD63+EV and CD63+EV; group 3 dominated proteins secretion, mostly for IL-8; and group 2 accommodates both EVs secretion and proteins secretion, but with much-attenuated frequency. Interestingly, similar functional organizations were revealed in both OSCC cell lines and patient samples (Fig. 5A and SI Appendix, Figs. S16–S23), demonstrating that the functional architecture of population cells is relatively stable across both cell lines and primary cells (28). Visualizing the distribution of each EV or protein in viSNE maps (Fig. 5B and SI Appendix, Figs. S16–S23), we further observed that the cells that were positive for protein secretion were less likely to secrete EVs simultaneously, suggesting the EV and protein secretions were generally dominated by different cell subsets within the population. We also used principal component analysis (PCA), another commonly used technique to reduce the dimensionality of the data by capturing maximal covariation, to analyze the same single cells. Despite the differences between linear (PCA) and nonlinear (viSNE) dimensionality reduction algorithms (44), we found that both OSCC cell lines and primary cells were also separated into tiered clusters in PCA plots contributed by different cytokines and EVs (Fig. 5C and SI Appendix, Fig. S24). For example, SCC25 cells were separated to be a cytokine-secreting cluster and two EV-secreting clusters. Cytokine-secreting cells were dictated by IL-6, IL-8, and MCP-1, while two EV-secreting clusters were contributed by CD63/HSP70-secreting cells and CD81/CD9/EpCAM-secreting cells differently. Patient 2 cells were clustered into subgroups dictated by IL-6/HSP70, IL-8/MCP-1, and CD63/CD9/CD81/EpCAM, respectively, in which the second subgroup dominated cytokine secretion and the third subgroup contributed the most EV secretion. Collectively, both t-SNE−based visualized clustering and PCA demonstrated the existence of tiered structures in OSCC cells and differential secretion profiles among OSCC cell subpopulations.
Fig. 5.
Multidimensional single-cell secretion analysis delineated OSCC cellular functional organization. (A) Visualized clustering analysis with viSNE revealed functional subgroups in OSCC cell line and patient samples. (B) Distribution of individual secretors (CD63+EV, CD9+CD63+EV, and IL-8) in single-cell−derived viSNE plots. (C) PCA analysis of single cells confirms the OSCC cells were resolved into structured clusters contributed differently by cytokines or EVs. (d and E) Probability of EV secreting cells secreting proteins simultaneously: (D) cell lines, n = 3; (E) patients, n = 6. (F and G) Probability of proteins secreting cells to secrete EVs simultaneously: (F) cell lines, n = 3; (G) patients, n = 6. *P < 0.05 and ****P < 0.00005 by paired t test.
To confirm our observation from viSNE and PCA that EV secretor vs. cytokine secretor phenotypes are mutually exclusive, we calculated the conditional probability of EV-positive cells secreting proteins and found EV+ cells were significantly less likely to secrete proteins in both cell lines (SCC25, UM-SCC6, and UM-SCC6M) (P = 0.027 by paired t test) and patients (P < 0.0001) (Fig. 5 D and E). Likewise, protein+ cells were significantly less likely to secrete EVs simultaneously in both cell lines (P = 0.017) and patients (P < 0.0001) (Fig. 5 F and G).
Discussion
Tumor-derived EVs play important roles in tumor metastasis processes (2, 6, 7), which make it vital to obtain more detailed information from these vesicles. However, these EVs were never characterized comprehensively at the single-cell level, due to the lack of available tools. This study introduced an antibody barcode-based platform for high-throughput, multiplexed profiling of single-cell EV secretion. With this platform, we uncovered previously undifferentiated single-cell heterogeneity underlying EV secretion within a phenotypically similar cell population. We then applied the platform to analyze a subgroup of UM-SCC6 cells with high invasion characteristics and observed that some EV expression (e.g., CD63+EV) decreased in metastatic tumor cells. The results were further confirmed with OSCC patient samples. These results demonstrated that our platform could generate critical information to potentially distinguish and quantitate invasive cell states, which can be used to monitor tumor invasiveness and tailor the therapeutic strategy for the individual patient. Also, these EVs that act as intercellular mediators for cell−cell communication in the tumor microenvironment may also be used as therapeutic targets for personalized medicine.
The tumor microenvironment is collectively shaped by complex signaling networks composed of different mediators, including cytokines, EVs, etc. Direct measurement of different mediators from the same single cells was highly desirable to generate information that inspires a deeper understanding of tumor microenvironment and decode the complex signaling network embedded in it. With this platform, we successfully realized multiplexed profiling of two different intercellular communication mediators (five-plexed EVs and three-plexed proteins) simultaneously from the same single cells, which cannot be obtained using other methods. We observed that proteins secretion and EV secretion were dominated by respective cell subgroups within the population, highlighting the unique advantage associated with multidimensional, multiplexed profiling to resolve the correlation between each parameter. This multidimensional analysis strategy may open up avenues for uncovering biology at the single cell level.
Notably, the platform is applicable to different cell types and sources with minimal sample consumption, which makes it especially suitable for rare clinical sample analysis, like circulation tumor cells (45), or fine-needle aspirate (18). The proteomic parameters of EV detection can be further increased if more microchannels are paralleled or spectral encoding is adopted for multicolor detection. When combined with other single-cell analysis technologies or different types of perturbations, the platform could provide more-comprehensive information to map the correlation between different functional mediators in the cellular microenvironment at different biomimetic models (12, 19, 28, 31). We believe this platform holds great potential to become a broadly applicable tool for in-depth EV analysis in both basic and translational research, like tumor biopsies in precision medicine.
Methods
PDMS Microchip Fabrication.
The molds for antibody patterning and single-cell capture were fabricated by photolithography with SU8 3035 (Microchem) and treated with trimethylchlorosilane (Sigma-Aldrich) overnight to facilitate peeling PDMS off the mold. PDMS prepolymer and curing reagent were mixed at 10:1 ratio (RTV 615; Momentive), poured onto the mold, and cured in an oven at 80 °C for 1 h. The PDMS microchip was bonded with a premium-grade microarray glass slide (poly-L-lysine coated; Thermo Fisher) after the inlet and outlet holes were punched out. Then it was baked at 80 °C for an additional 2 h to complete thermal bonding. The PDMS microwell array for single-cell culture was cleaned with ultrasonication in ethanol and blown dry before use.
Flow Patterning Antibody Barcode Glass Slide.
After the PDMS microchips with high-density parallel microchannels were assembled with a poly-L-lysine−coated glass slide, each antibody (SI Appendix, Table S1) was pushed through individual microchannels until completely dry, with 1-psi pressured N2. The antibody barcode glass slide was blocked with 1% BSA (Roche) for 1 h to reduce nonspecific adsorption. Then it was washed with Dulbecco’s phosphate-buffered saline (DPBS), 50/50 DPBS/distilled (DI) water, and DI water sequentially. The antibody slide was spun dry in a slide centrifuge and stored at 4 °C before use.
Cell Culture.
Human oral squamous carcinoma cell line (SCC25) (American Type Culture Collection) was cultured in MEM medium (Gibco; Thermo Fisher Scientific) with 10% FBS (Gibco; Invitrogen), 1% antibiotics (100 U/mL of penicillin G sodium, 100 U/mL of streptomycin) and 1% MEM Non-Essential Amino Acid (Life Technologies). FBS was ultracentrifuged at 100,000 × g at 4 °C for 4 h to deplete exosome in it. The human oral squamous carcinoma cell line (UM-SCC6) (a kind gift from Songling Wang, Capital Medical University) was cultured in DMEM/High Glucose (HyClone) medium with similar conditions as for SCC25. The cells were detached with 0.25% trypsin−0.02% EDTA for 4 min, centrifuged at 300 × g for 5 min, washed, and resuspended in fresh medium before use.
Isolation of Invasion Front Cells from UM-SCC6.
The matrix channel of the isolation microchip was first loaded with Matrigel (Corning). UM-SCC6 cells were seeded into the cell culture channel in serum-free medium. The cell culture medium containing 20% FBS was introduced into the stimulation channel. Cells that invaded through the matrix channel and migrated into the stimulation channel were termed as the invasion front cells. These invasion front cells were collected by trypsinization and expanded to repeat the steps mentioned above to generate the second round of invasion front cells. The third round of invasion front cells of UM-SCC6 was collected as UM-SCC6M cells.
OSCC Patient Tissue Samples.
Human OSCC patient samples were obtained from the Affiliated Hospital of Dalian Medical University. The collection and use of human samples were approved by the Ethics Committee of Dalian Medical University. Patient primary tissue was first minced with ophthalmic surgical scissors to ∼1 mm3 pieces and then pipetted repeatedly with DPBS containing 2% antibiotics. The tissue was then detached with 0.25% trypsin−0.02% EDTA for 20 min to 40 min at 37 °C, shaking once every 5 min. The tissue was then detached with collagenase I on the shaker until the tissue became flocculent. The tube containing the flocculent tissue was placed in a 37 °C, 5% CO2 incubator for 5 min. The following flocculent precipitate was then spread evenly across the culture dish coated with collagenase I. The culture dish was placed in a 37 °C, 5% CO2 incubator for 1 h, and the culture medium DMEM-HG was added dropwise. The medium was changed periodically until the cells became confluent in the culture dish. Then the cells were detached with 0.25% trypsin−0.02% EDTA and resuspended in fresh medium for the experiment.
Single-Cell EV Secretion Analysis Procedures.
The PDMS microchamber array for single-cell assay was treated with O2 plasma (PDC-32G; Harrick Plasma) for 1 min before single-cell experiment and blocked with cell culture medium (with 10% FBS) to maintain surface hydrophilic, which will facilitate cell loading and minimize nonspecific protein adsorption. Sample cells were prestained with cell viability dye Calcein-AM green at 37 °C for 30 min and resuspended into fresh medium at defined density. The cells were then pipetted onto a microchamber array at 2 × 105 cells per mL cell density, 200 μL per chip. After cells settled down into microchambers within 5 min, an antibody barcode glass slide was imposed onto the top of the microchamber array and clamped together to trap single cells. The microchip trapped with single cells was imaged with a Nikon Eclipse TiE microscope with an automated stage to record the cell number/position information. The clamp was removed after overnight incubation to finish detection procedures. The glass slide was incubated with a mixture of detection antibodies (biotin-IL-8, biotin-IL-6, biotin-MCP-1, biotin-CD63) for 1 h and stained with streptavidin-APC or streptavidin-PE (1:100 dilution; eBioscience) for another 30 min. Then it was washed thoroughly with DPBS, 50/50 DPBS/DI water, and DI water sequentially, and the glass slide was spun dry and scanned with a GenePix 4300A fluorescence scanner (Molecular Devices).
Data Analysis.
The images for single-cell counting (bright field and fluorescence) can be processed in Nikon software (NIS-Elements Ar Microscope Imaging Software) by defining a threshold in combined images to realize automated cell counting. The fluorescence detection image was analyzed with GenePix Pro software (Molecular Devices) by creating and aligning the microchamber array template followed by extraction of mean fluorescence intensity. The cell counts and corresponding fluorescent data would be matched and processed in Excel (Microsoft) and GraphPad Prism. The thresholds to determine positive secretion events were defined as mean +2 × SD of zero-cell data. Heatmaps and unsupervised clustering were generated with software Cluster/Treeview (Eisen Laboratory). The viSNE (Dana Pe’er laboratory) was used to transform complex multiparameter data into 2D categorized maps.
Supplementary Material
Acknowledgments
The project was supported by National Natural Science Foundation of China Grants 21874133 and 21605143, Youth Innovation Promotion Association Chinese Academy of Sciences Grant 2018217, and funds from Dalian Institute of Chemical Physics Grant SZ201601.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814348116/-/DCSupplemental.
References
- 1.Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EL Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 3.van der Pol E, Böing AN, Harrison P, Sturk A, Nieuwland R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev. 2012;64:676–705. doi: 10.1124/pr.112.005983. [DOI] [PubMed] [Google Scholar]
- 4.Février B, Raposo G. Exosomes: Endosomal-derived vesicles shipping extracellular messages. Curr Opin Cell Biol. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 5.György B, et al. Membrane vesicles, current state-of-the-art: Emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo Cicero A, Stahl PD, Raposo G. Extracellular vesicles shuffling intercellular messages: For good or for bad. Curr Opin Cell Biol. 2015;35:69–77. doi: 10.1016/j.ceb.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: Mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shao H, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med. 2012;18:1835–1840. doi: 10.1038/nm.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang KS, et al. Multiparametric plasma EV profiling facilitates diagnosis of pancreatic malignancy. Sci Transl Med. 2017;9:eaal3226. doi: 10.1126/scitranslmed.aal3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tian Y, et al. Protein profiling and sizing of extracellular vesicles from colorectal cancer patients via flow cytometry. ACS Nano. 2018;12:671–680. doi: 10.1021/acsnano.7b07782. [DOI] [PubMed] [Google Scholar]
- 11.Zabeo D, et al. Exosomes purified from a single cell type have diverse morphology. J Extracell Vesicles. 2017;6:1329476. doi: 10.1080/20013078.2017.1329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao H, et al. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917–1950. doi: 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shurtleff MJ, et al. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc Natl Acad Sci USA. 2017;114:E8987–E8995. doi: 10.1073/pnas.1712108114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lötvall J, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the international society for extracellular vesicles. J Extracell Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kowal J, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshioka Y, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun. 2014;5:3591. doi: 10.1038/ncomms4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee K, et al. Multiplexed profiling of single extracellular vesicles. ACS Nano. 2018;12:494–503. doi: 10.1021/acsnano.7b07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullal AV, et al. Cancer cell profiling by barcoding allows multiplexed protein analysis in fine-needle aspirates. Sci Transl Med. 2014;6:219ra219. doi: 10.1126/scitranslmed.3007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heath JR, Ribas A, Mischel PS. Single-cell analysis tools for drug discovery and development. Nat Rev Drug Discov. 2016;15:204–216. doi: 10.1038/nrd.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Bodovitz S. Single cell analysis: The new frontier in ‘omics.’. Trends Biotechnol. 2010;28:281–290. doi: 10.1016/j.tibtech.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colombo M, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013;126:5553–5565. doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 22.Willms E, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep. 2016;6:22519. doi: 10.1038/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu YJ, Cai W, Shih YRV, Lian I, Lo YH. A single-cell assay for time lapse studies of exosome secretion and cell behaviors. Small. 2016;12:3658–3666. doi: 10.1002/smll.201600725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Son KJ, et al. Microfluidic compartments with sensing microbeads for dynamic monitoring of cytokine and exosome release from single cells. Analyst (Lond) 2016;141:679–688. doi: 10.1039/c5an01648g. [DOI] [PubMed] [Google Scholar]
- 25.Verweij FJ, et al. Quantifying exosome secretion from single cells reveals a modulatory role for GPCR signaling. J Cell Biol. 2018;217:1129–1142. doi: 10.1083/jcb.201703206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan R, et al. Integrated barcode chips for rapid, multiplexed analysis of proteins in microliter quantities of blood. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C, et al. A clinical microchip for evaluation of single immune cells reveals high functional heterogeneity in phenotypically similar T cells. Nat Med. 2011;17:738–743. doi: 10.1038/nm.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi Q, et al. Single-cell proteomic chip for profiling intracellular signaling pathways in single tumor cells. Proc Natl Acad Sci USA. 2012;109:419–424. doi: 10.1073/pnas.1110865109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, et al. High-throughput secretomic analysis of single cells to assess functional cellular heterogeneity. Anal Chem. 2013;85:2548–2556. doi: 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue Q, et al. Analysis of single-cell cytokine secretion reveals a role for paracrine signaling in coordinating macrophage responses to TLR4 stimulation. Sci Signal. 2015;8:ra59. doi: 10.1126/scisignal.aaa2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Y, et al. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. Proc Natl Acad Sci USA. 2015;112:E607–E615. doi: 10.1073/pnas.1416756112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue Q, et al. Single-cell multiplexed cytokine profiling of CD19 CAR-T cells reveals a diverse landscape of polyfunctional antigen-specific response. J Immunother Cancer. 2017;5:85. doi: 10.1186/s40425-017-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma C, et al. Multifunctional T-cell analyses to study response and progression in adoptive cell transfer immunotherapy. Cancer Discov. 2013;3:418–429. doi: 10.1158/2159-8290.CD-12-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossi J, et al. Preinfusion polyfunctional anti-CD19 chimeric antigen receptor T cells associate with clinical outcomes in NHL. Blood. 2018;132:804–814. doi: 10.1182/blood-2018-01-828343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang P, He M, Zeng Y. Ultrasensitive microfluidic analysis of circulating exosomes using a nanostructured graphene oxide/polydopamine coating. Lab Chip. 2016;16:3033–3042. doi: 10.1039/c6lc00279j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hasina R, et al. Angiogenic heterogeneity in head and neck squamous cell carcinoma: Biological and therapeutic implications. Lab Invest. 2008;88:342–353. doi: 10.1038/labinvest.2008.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barile L, et al. Extracellular vesicles from human cardiac progenitor cells inhibit cardiomyocyte apoptosis and improve cardiac function after myocardial infarction. Cardiovasc Res. 2014;103:530–541. doi: 10.1093/cvr/cvu167. [DOI] [PubMed] [Google Scholar]
- 38.Li X, et al. Downregulation of miR-218-5p promotes invasion of oral squamous cell carcinoma cells via activation of CD44-ROCK signaling. Biomed Pharmacother. 2018;106:646–654. doi: 10.1016/j.biopha.2018.06.151. [DOI] [PubMed] [Google Scholar]
- 39.Jang HI, Lee H. A decrease in the expression of CD63 tetraspanin protein elevates invasive potential of human melanoma cells. Exp Mol Med. 2003;35:317–323. doi: 10.1038/emm.2003.43. [DOI] [PubMed] [Google Scholar]
- 40.Lupia A, et al. CD63 tetraspanin is a negative driver of epithelial-to-mesenchymal transition in human melanoma cells. J Invest Dermatol. 2014;134:2947–2956. doi: 10.1038/jid.2014.258. [DOI] [PubMed] [Google Scholar]
- 41.Lai X, et al. Decreased expression of CD63 tetraspanin protein predicts elevated malignant potential in human esophageal cancer. Oncol Lett. 2017;13:4245–4251. doi: 10.3892/ol.2017.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huan J, et al. Overexpression of CD9 correlates with tumor stage and lymph node metastasis in esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8:3054–3061. [PMC free article] [PubMed] [Google Scholar]
- 43.Otero M, et al. Human chondrocyte cultures as models of cartilage-specific gene regulation. In: Mitry RR, Hughes RD, editors. Human Cell Culture Protocols. Humana Press; Totowa, NJ: 2012. pp. 301–336. [DOI] [PubMed] [Google Scholar]
- 44.Amir AD, et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol. 2013;31:545–552. doi: 10.1038/nbt.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sinkala E, et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun. 2017;8:14622. doi: 10.1038/ncomms14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.