Significance
It is proposed that chromatin modifiers can regulate transcription through different mechanisms sometimes referred as “canonical” (toward chromatin) and “noncanonical” (toward other proteins). However, their relative contribution to the overall function of a given chromatin modifier is often enigmatic. We focused on the Polycomb complex PRC2 to investigate this question. Our results indicate that the canonical activity of PRC2 is largely predominant, if not exclusive, and that the particular pattern of PRC2 mutations in cancer is due to proliferation-dependent redundancy between the two enzymatic subunits of the complex.
Keywords: chromatin, cancer, Polycomb, EZH2
Abstract
Genetic mutations affecting chromatin modifiers are widespread in cancers. In malignant peripheral nerve sheath tumors (MPNSTs), Polycomb repressive complex 2 (PRC2), which plays a crucial role in gene silencing, is inactivated through recurrent mutations in core subunits embryonic ectoderm development (EED) and suppressor of zeste 12 homolog (SUZ12), but mutations in PRC2’s main catalytic subunit enhancer of zeste homolog 2 (EZH2) have never been found. This is in contrast to myeloid and lymphoid malignancies, which harbor frequent loss-of-function mutations in EZH2. Here, we investigated whether the absence of EZH2 mutations in MPNST is due to a PRC2-independent (i.e., noncanonical) function of the enzyme or to redundancy with EZH1. We show that, in the absence of SUZ12, EZH2 remains bound to EED but loses its interaction with all other core and accessory PRC2 subunits. Through genetic and pharmacological analyses, we unambiguously establish that EZH2 is functionally inert in this context, thereby excluding a PRC2-independent function. Instead, we show that EZH1 and EZH2 are functionally redundant in the slowly proliferating MPNST precursors. We provide evidence that the compensatory function of EZH1 is alleviated upon higher proliferation. This work reveals how context-dependent redundancies can shape tumor-type specific mutation patterns in chromatin regulators.
It is estimated that over 25% of the most frequently mutated genes in cancers encode chromatin regulators (1, 2). While some of these mutations are recurrent in a wide range of cancers, many others are found in specific tumor types, suggesting a context-dependent function. This is well illustrated in the case of Polycomb repressive complex 2 (PRC2), a chromatin-modifying complex involved in maintaining transcriptional repression. PRC2 contains several essential subunits: embryonic ectoderm development (EED), suppressor of zeste 12 homolog (SUZ12), retinoblastoma-binding protein 4/7 (RBBP4/7), and two paralogous enzymatic subunits enhancer of zeste homolog 1 and 2 (EZH1 and EZH2). EZH1 and EZH2 assemble into alternative PRC2 complexes with similar composition that both catalyze methylation of lysine 27 on histone 3 (H3K27) (3, 4). In addition, several accessory subunits associate with PRC2, assist in its recruitment, and/or modulate its enzymatic activity (5). Methylation of H3K27 is essential for Polycomb-mediated silencing (6). Alterations of the PRC2 complex have been reported across different malignancies, and each alteration displays striking tumor type specificity (reviewed in ref. 7). Gain-of-function mutations in EZH2 have been reported in follicular lymphoma, diffuse large B cell lymphoma, and a small subset of melanoma, while functionally similar mutations affecting EZH1 have been found in autonomous thyroid adenomas. In contrast, recurrent loss-of-function mutations in PRC2 genes occur in myeloid malignancies, T cell acute lymphoblastic leukemia (T-ALL), and malignant peripheral nerve sheath tumors (MPNSTs) (7).
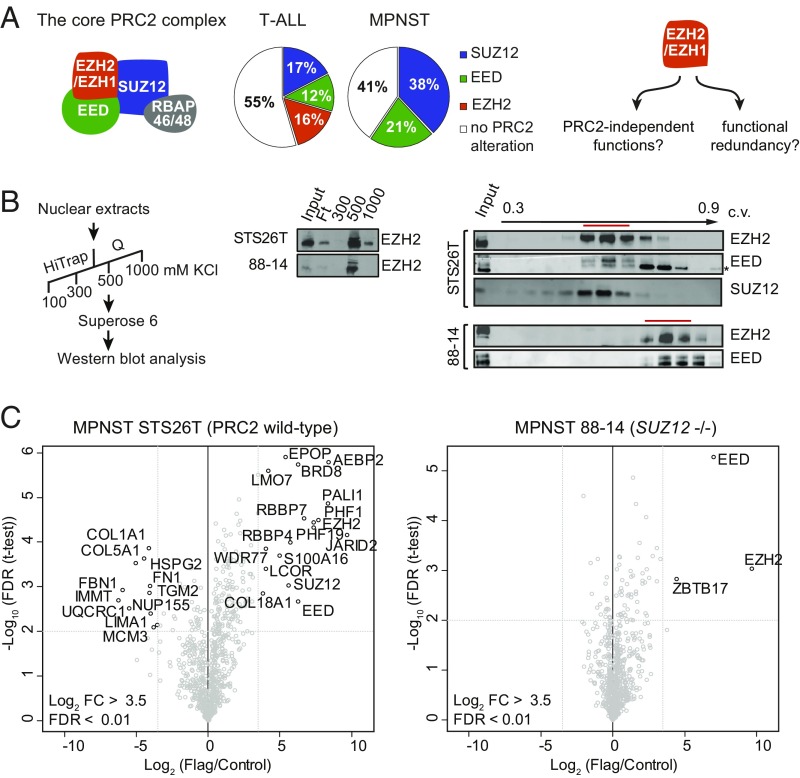
MPNSTs are aggressive soft-tissue sarcomas that develop either sporadically or from preexisting benign tumors called plexiform neurofibromas in patients with neurofibromatosis type 1 (NF1) (Online Mendelian Inheritance in Man 162200). Previous studies have identified recurrent biallelic mutations in EED and SUZ12 that result in a complete loss of H3K27me3 in over 50% of MPNSTs (8–10). Loss of PRC2 function as well as co-occurring inactivation of NF1 and CDKN2A tumor suppressor genes are considered to be the most significant diagnostic markers of MPNST in the revised World Health Organization 2016 classification of tumors of the central nervous system (11). Surprisingly, among 121 MPNST samples analyzed across five studies (8–10, 12, 13), no mutations were detected in EZH1 or EZH2 despite the high prevalence of lesions in EED and SUZ12. This mutation signature is in sharp contrast to the spectrum of PRC2 mutations found in myeloid malignancies and T-ALL, where mutations in EZH2 occur at high frequency (Fig. 1A). The absence of mutations in EZH1 and EZH2 in MPNST raises the possibility that the enzymatic subunits might have PRC2-independent functions that are required for MPNST development. Several studies have indeed suggested that EZH1 and/or EZH2 can function independently of their enzymatic activity within PRC2 (14–16). An alternative reason for the absence of EZH2 and EZH1 mutations in MPNSTs might come from their potential functional redundancy. The two enzymes have indeed been shown to be partially redundant in several cell types (3, 17, 18).
Fig. 1.
Loss of SUZ12 leads to a dramatic reduction of EZH2 interactome. (A, Left) Schematic representation of the core PRC2 complex formed around EZH1 or EZH2 enzymatic subunits. (A, Middle) Pie charts displaying the relative proportions of PRC2 alterations among early T cell precursor acute lymphoblastic leukemia (T-ALL) (as reported in ref. 35) and 121 MPNST samples analyzed across five studies for which whole-genome sequencing or targeted sequencing of EED, SUZ12, EZH1, and EZH2 is available. (A, Right) Alternative hypotheses explaining the absence of EZH1/2 mutations in MPNST. (B, Left) SUZ12-mutated 88-14 and SUZ12-WT STS26T nuclear extracts were purified by successive steps summarized in the scheme. (B, Middle) Western blot analysis of EZH2 distribution in the different fractions obtained by anion exchange chromatography performed on 88-14 and STS26T nuclear extracts. (B, Right) EZH2 and EED Western blot analysis of the fractions eluted from gel filtration chromatography performed on the STS26T and 88-14 500 mM fractions. Western blot analysis of SUZ12 was additionally performed on STS26T extracts. A red line highlights the peak of elution, and the asterisk indicates an unspecific cross-reactivity of EED antibody. c.v., column volume; Ft, flow-through. (C) Mass spectrometry analysis of EZH2 interactome in STS26T (Left) and 88-14 (Right) stably expressing a Flag-tagged version of EZH2. Volcano plots represent mass spectrometry analysis of Flag-EZH2 cells compared with control cells.
In principle, redundancy between EZH1 and EZH2 or a PRC2-independent function for these proteins could both explain why they are not found mutated in MPNST (Fig. 1A). Through biochemical and genetic approaches, we unambiguously demonstrate that EZH1/2 do not regulate transcription independently of PRC2. Furthermore, we find that EZH1 and EZH2 display a remarkable degree of redundancy in the cells from which MPNST develop, providing an explanation for why alterations of the cognate genes are not selected for during MPNST development. We further provide evidence that the rate of cell proliferation is a major modulator of EZH2/EZH1 ratio and consequently the ability of EZH1 to compensate for loss of EZH2. These results suggest that context-dependent redundancies within chromatin-modifying complexes can shape mutational signatures in cancer.
Results
In the Absence of SUZ12, EZH2 Forms a Residual Complex with EED.
To evaluate putative functions of EZH2 independent of an intact PRC2 core complex, we first analyzed the biochemical properties of the enzyme in the context of loss of SUZ12. For this purpose, we compared a SUZ12-mutated MPNST cell line (88-14) and a PRC2 wild-type cell line (STS26T). EZH2 protein accumulation appears much lower in the absence of SUZ12 (Fig. 1B, top Western blot; compare signal in the input fraction), consistent with previous reports showing that PRC2 constituents stabilize each other (5). To investigate the functionality of the residual EZH2 protein, we subjected nuclear extracts from 88-14 and STS26T to anion exchange chromatography followed by size exclusion chromatography (Fig. 1B). During anion-exchange chromatography, EZH2 elutes at 500 mM salt regardless of SUZ12 presence (Fig. 1B, Top). Following size exclusion chromatography, EZH2 and EED coeluted in both cases; however, the elution pattern is shifted toward a smaller molecular weight in the absence of SUZ12 (Fig. 1B, Bottom). These results suggest that, upon loss of SUZ12, EZH2 remains part of a smaller complex containing EED.
To determine whether EZH2 alone, or the residual complex with EED, could form new interactions in the absence of SUZ12, we characterized the interactome of EZH2 by mass spectrometry in 88-14 and STS26T cells. We overexpressed a Flag-tagged version of EZH2 in both cell lines and performed anti-Flag immunoprecipitation followed by quantitative proteomics (PXD012547, ref. 19). For the analysis of mass spectrometry data, we chose a low enrichment cutoff to avoid missing weak interactors. As expected, in PRC2 wild-type cells, EZH2 pulled down the core PRC2 components (EED, SUZ12, RBBP4, and RBBP7) along with its well-established cofactors (JARID2, AEBP2, PHF1/19, PALI1, and EPOP) (5, 20) (Fig. 1C, Left). In contrast, in SUZ12-mutant cells, EZH2 remained bound only to EED and lost its interaction with all accessory PRC2 subunits (Fig. 1C, Right). ZBTB17A, a potential EZH2 interactor that appeared near the enrichment cutoff, was not validated by coimmunoprecipitation experiments (SI Appendix, Fig. S1A). To ascertain that the collapse of EZH2 interactome in 88-14 cells is due to the absence of SUZ12, we reexpressed SUZ12 in 88-14 cells. As shown in SI Appendix, Fig. S1B, the binding of the PRC2 cofactor AEBP2 to EZH2 was restored in the presence of SUZ12. In addition, expression of other key PRC2 cofactors was equivalent in 88-14 and STS26T cells (SI Appendix, Fig. S1C), indicating that these two cell lines have a similar environment for the regulation of PRC2. The massive loss of interaction between EZH2 and PRC2 cofactors in SUZ12-mutant cells is in agreement with recent in vitro evidence suggesting that SUZ12 acts as a platform for the recruitment of these subunits (21, 22).
Altogether, these analyses show that loss of SUZ12 destabilizes the PRC2 interactome, leaving EZH2 and EED as the only members of a residual PRC2 complex.
EZH1/2 Do Not Regulate Transcription Independently of PRC2.
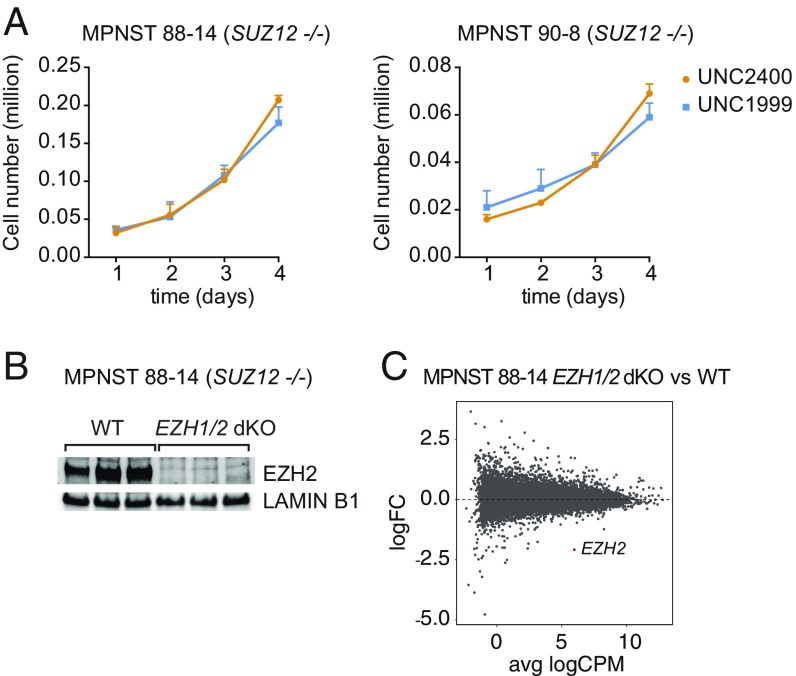
Several studies have suggested that EZH2 might function independently of canonical PRC2 activity (14–16). SUZ12-mutant MPNST cell lines represent an ideal system to rigorously investigate such noncanonical functions because the canonical PRC2 function is inactivated. Since EZH1, the paralogous enzyme of EZH2, can in some instances compensate for loss of EZH2, we assessed the functionality of both enzymes together. We analyzed the impact of inhibiting EZH1/2 enzymatic activity on cell proliferation, using the small-molecule inhibitor UNC1999 or its inactive analog UNC2400 (23). As shown in Fig. 2A, treatment of two different SUZ12-null MPNST cell lines did not impact cell proliferation. However, this observation does not exclude a role for the residual PRC2 complex in controlling gene expression and/or functions of EZH2 that do not rely on its catalytic activity.
Fig. 2.
EZH2 does not retain a specific function in the absence of SUZ12 in MPNST cells. (A) Proliferation assays in two SUZ12-null MPNST cell lines in presence of UNC1999 dual EZH1/2 inhibitor or of UNC2400, its control inactive compound (mean ± SD; n = 3). (B) Western blot analysis EZH2 protein expression in three independent wild-type and EZH1/2 dKO 88-14 clones. (C) Scatterplot showing log2fold-change (logFC) expression between wild-type and EZH1/2 dKO 88-14 cells versus average log2 counts per million (logCPM). EZH2, the only differentially expressed transcript, is highlighted in red.
To address this question, we genetically inactivated EZH1 and EZH2 in the SUZ12-null 88-14 cell line. Loss of EZH2 in three independent clones was confirmed by Western blot (Fig. 2B). We introduced frameshift-inducing mutations in the EZH1 gene to exclude a potential compensation upon loss of EZH2 (SI Appendix, Fig. S2A). These mutations do not affect proliferation as shown by the lack of consequences of restoring EZH2 expression in 88-14 EZH1/2 double knockout (dKO) (SI Appendix, Fig. S2B). We performed RNA sequencing on EZH1/2 wild-type and EZH1/2 dKO clones. Strikingly, with the exception of EZH2, no genes were found significantly differentially expressed [false discovery rate (FDR) < 0.05] between control and EZH1/2 dKO cells (GSE118186, ref. 24) (Fig. 2C).
These results strongly argue against a PRC2-independent function for EZH2 in the context of MPNST.
EZH2 Functions as Part of a Canonical PRC2 Complex in Androgen-Independent LNCaP-abl Cells.
Our findings in the context of MPNST contrast with those reported in androgen-independent prostate cancer (AIPC), where biochemical and functional evidence points to a PRC2-independent role for EZH2, mediating gene activation as part of a distinct complex comprising the androgen receptor (16).
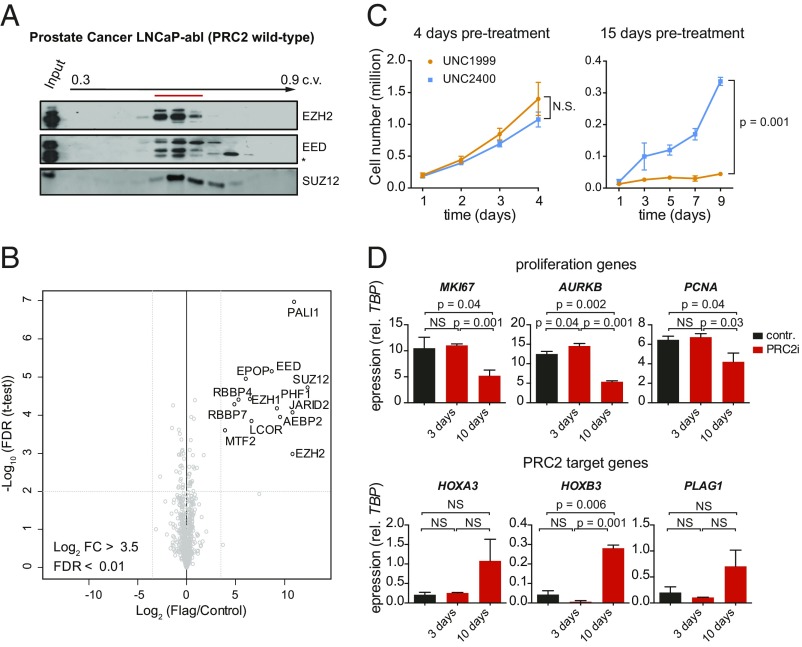
To understand the basis of this discrepancy, we repeated the biochemical characterization of PRC2 in the LNCaP-abl cell line, an androgen-independent derivative of the LNCaP prostate cancer cell line used as model of AIPC (16, 25). In contrast to Xu et al., we observed coelution of EZH2 with EED following size exclusion chromatography, a pattern identical to that observed in STS26T cells (Fig. 3A and SI Appendix, Fig. S3A; compare with Fig. 1B). Analysis of EZH2 migration on a native gel further confirmed that it is found in a single high–molecular-weight complex slightly smaller than the recombinant complex where all subunits are tagged (SI Appendix, Fig. S3B). Moreover, mass spectrometry analysis of the EZH2 interactome in LNCaP-abl cells recovered all known core and accessory PRC2 subunits but did not reveal additional partners such as the androgen receptor despite the low stringency used in the analysis (PXD012547, ref. 19) (Fig. 3B). These biochemical analyses therefore do not support the existence of measurable noncanonical composition of PRC2 in this AIPC cell line. Of note, we obtained similar findings for the OVCAR8 cell line (SI Appendix, Fig. S3C), in which EZH2 has also been reported to have an unusual elution pattern on a size exclusion column (26).
Fig. 3.
EZH2 acts as part of a canonical PRC2 complex in LNCaP-abl cells. (A) Western blot analysis of EZH2 and EED in the fractions eluted from gel filtration chromatography performed on the 500 mM fraction. A red line highlights the peak of elution, and the asterisk indicates an unspecific cross-reactivity of EED antibody. c.v., column volume. (B) Mass spectrometry analysis of EZH2 interactome in LNCaP-abl cells stably expressing Flag-tagged version of EZH2. (C) Proliferation assay in LNCaP-abl treated with 1 μM UNC1999 or UNC2400 from the indicated times after the beginning of drug treatment. Mean ± SD; n = 2; P values from two-tailed t tests on the final time points are shown. (D) RT-qPCR analysis of proliferation markers (Top) and classical PRC2 target genes (Bottom) in control-treated (UNC2400) cells or at different times after PRC2 inhibitor treatment. Mean ± SD; n = 2; P values from one-tailed t tests are shown for differences that are statistically significant. NS, not significant.
These results prompted us to further investigate EZH2 contribution to LNCaP-abl growth. We measured cell proliferation upon EZH2 enzymatic inhibition since the PRC2-independent function of EZH2 has been shown to require an intact catalytic domain (16). We pretreated cells with UNC1999 or UNC2400 and performed cell growth assays beginning after either 4 or 15 d of continued treatment. Efficient inhibition of EZH1/2 catalytic activity was verified by Western blot for the trimethylated form of H3K27 (H3K27me3; SI Appendix, Fig. S3D). In contrast to the dramatic effects observed with siRNA against EZH2 on cell growth as early as 48 h after transfection, pharmacological inhibition of PRC2 only impaired LNCaP-abl growth after the first week of treatment (Fig. 3C). This delay has been reported in other models (23, 27, 28) and attributed to the stability of H3K27me3 and the time required to achieve full dilution of the histone mark through cell divisions.
Importantly, the transcriptomic data generated following siEZH2 treatment (16) revealed a prominent cell-proliferation signature among down-regulated genes (i.e., genes that are proposed to be regulated by EZH2 in a noncanonical fashion; SI Appendix, Fig. S3E). Indeed, following PRC2 inhibition, expression of cell proliferation markers is diminished but only at 10 d posttreatment (Fig. 3D, Top). This is the same kinetics as the one observed for the derepression of classical PRC2 target genes (Fig. 3D, Bottom) raising the possibility that down-regulation of cell proliferation genes is an indirect consequence of the more global transcriptional alterations resulting from EZH2 inhibition.
Thus, together with our biochemical analyses, the kinetics of cell growth suppression and of gene expression changes upon PRC2 inhibition suggests a predominantly canonical function for EZH2 in LNCaP-abl cells.
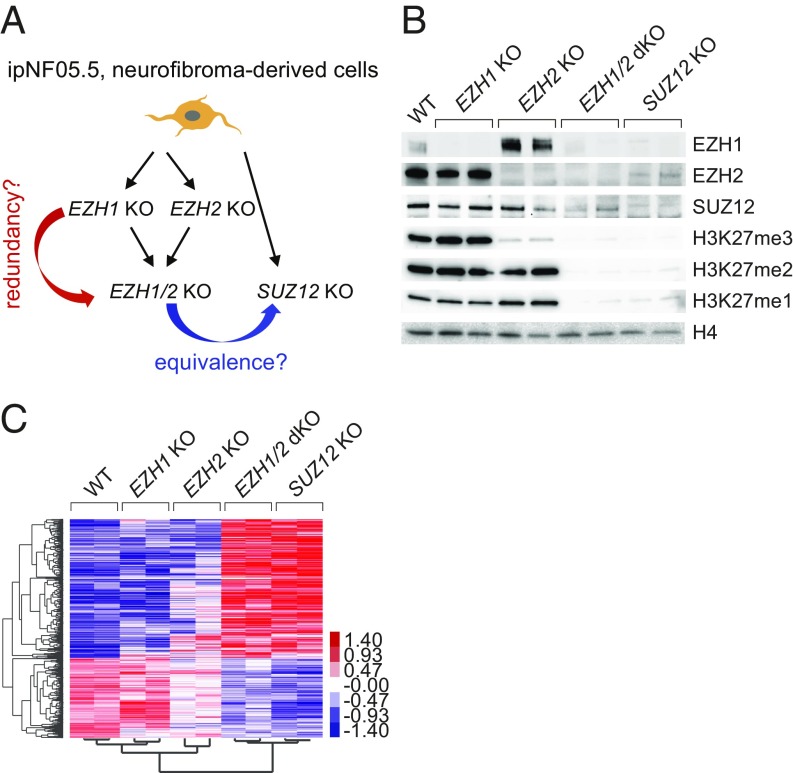
Pronounced Redundancy Between EZH1 and EZH2 in Neurofibroma Cells.
Considering the lack of evidence for PRC2-independent role of EZH2 that would explain why it is not found to be mutated in MPNST, we investigated the alternative hypothesis that the absence of mutations affecting EZH2 might reflect compensation by EZH1. We inactivated EZH1 and EZH2 separately or in combination in the ipNF05.5 cell line derived from a plexiform neurofibroma (29), corresponding to the tumor type from which MPNSTs arise. The resulting mutant cells were compared with SUZ12-mutant cells derived from the same cell line (GSE118186, ref. 24) (Fig. 4A). Loss of SUZ12 in ipNF05.5 cells led to transcriptional up-regulation of 813 genes (SI Appendix, Fig. S4A; FDR < 0.05). As expected, a majority of these genes are marked by H3K27me3 in wild-type ipNF05.5 cells (SI Appendix, Fig. S4B). Gene ontology analysis indicated that up-regulated genes were significantly enriched for developmental regulators (SI Appendix, Fig. S4C), as has been found in PRC2-mutant MPNST samples (9).
Fig. 4.
Acute redundancy between EZH1 and EZH2 in a model of plexiform neurofibroma. (A) Schematic of the different isogenic KO cell lines established from the ipNF05.5 plexiform neurofibroma cell line and of the experimental rationale. (B) Western blot showing EZH1, EZH2, SUZ12, and the various degrees of H3K27 methylation in wild-type ipNF05.5 and the different mutant clones indicated on Top. Each KO condition is represented by two independent clones. Histone H4 is used as a loading control. (C) Heatmap of centered, log-transformed values (counts per million) of all differentially expressed genes, each KO condition versus WT. Genes and samples were clustered using Spearman rank correlation.
We then analyzed the impact of individual or combined loss of EZH1 and EZH2 on the methylation of H3K27 (Fig. 4B; also see SI Appendix, Fig. S4D for quantification of Western blot signals). While loss of EZH1 did not impact global levels of H3K27me1, me2, or me3, loss of EZH2 led to a marked reduction of H3K27me3 with only modest effects on H3K27me1 and H3K27me2. Combined loss of both enzymes or loss of SUZ12 leads to a comparable acute loss of all three methylation levels. We next assessed transcriptional changes by RNA-seq (Fig. 4C and SI Appendix, Fig. S4 E and F). Strikingly, deletions of either EZH1 or EZH2 caused only subtle changes in gene expression, with no differentially expressed genes in EZH1-mutant cells and only 11 in EZH2-mutant cells. In contrast, combined loss of EZH1 and EZH2 led to up-regulation of 629 genes, indicative of a high degree of redundancy between the two enzymes. Consistently, inhibition of PRC2 activity in EZH2 KO ipNF05.05 using the general PRC2 inhibitor A-395 (30) led to a robust de-repression of PRC2 target genes (SI Appendix, Fig. S4H). These data support the hypothesis that the absence of EZH2 (or EZH1) mutations in MPNST is a consequence of the high redundancy between the two enzymes in neurofibroma cells.
Remarkably, transcriptional changes in EZH1/EZH2 dKO and SUZ12 KO were highly correlated (GSE118186, ref. 24) (Fig. 4C and SI Appendix, Fig. S4I). Direct comparison of the two mutant conditions did not uncover any significantly differentially expressed genes (SI Appendix, Fig. S4J), demonstrating that combined loss of EZH1 and EZH2 is equivalent to loss of SUZ12. Together with our observations in the MPNST and AIPC models, these results in neurofibroma cells argue against a PRC2-independent function for EZH1/2.
Redundancy Between EZH1 and EZH2 Is Modulated by Cell Proliferation Rate.
Previous studies have found that functional compensation between EZH1 and EZH2 is context dependent, depending on tissue type and developmental stages (17, 31). Furthermore, as mentioned above, mutations in EZH2 are selected for in T-ALL and in myeloid malignancies, suggesting that EZH1 cannot fully compensate for loss of EZH2 in these cell types. The circumstances under which EZH1 and EZH2 compensate for each other remain unclear. We and others have previously shown that EZH2 expression is driven by cell proliferation (3, 18, 32), a process that ensures H3K27me3 homeostasis (18). As shown in SI Appendix, Fig. S5A, analysis of publicly available RNA-seq data from The Cancer Genome Atlas database indicates that the positive correlation between EZH2 transcript levels and cell proliferation (assessed by MKI67 proliferation marker) is a general property that extends across various cancer types. In contrast, EZH1 levels show no positive correlation to cell proliferation, suggesting that the EZH2/EZH1 ratio and hence redundancy between the two enzymes is mainly controlled by cell proliferation rate.
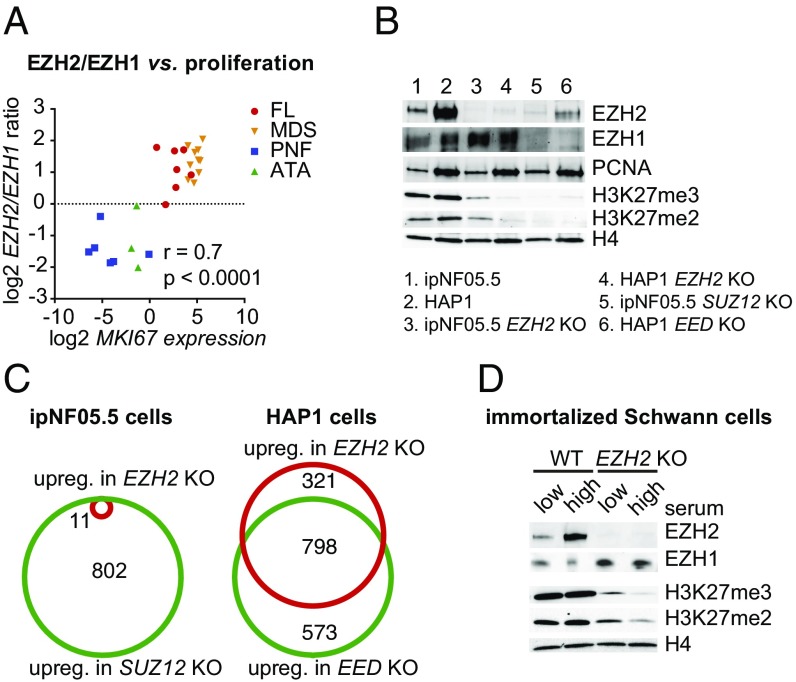
To directly evaluate the link between EZH2/EZH1 ratio and cell proliferation in tumor types subject to mutation affecting PRC2 genes, we analyzed tumor samples of autonomous thyroid adenoma (ATA) and plexiform neurofibroma (PNF), two tumor types in which EZH2 is never found mutated, as well as myelodysplastic syndrome (MDS) and follicular lymphoma (FL), in which EZH2 mutations occur. Using digital droplet PCR, we quantitatively measured EZH1, EZH2, and MKI67 transcript abundance. Fig. 5A shows the strong correlation between EZH2/EZH1 ratio and MKI67 expression (Spearman r = 0.7, P < 0.0001). Samples are distributed in two separate clusters, ATAs and PNFs forming a cluster characterized by low MKI67 expression and low EZH2/EZH1 ratio, while MDSs and FLs samples form a cluster characterized by high MKI67 and high EZH2/EZH1 ratio. Thus, these observations reveal a striking association between proliferation status, EZH2/EZH1 ratio, and the occurrence of EZH2 mutations, the latter being found only in highly proliferative tumors.
Fig. 5.
Redundancy between EZH1 and EZH2 is dependent on the proliferation rate. (A) Scatterplot of EZH2/EZH1 transcript ratio versus MKI67 transcript abundance as determined by digital droplet PCR in the tumor samples indicated on the Right. (B) Western blot showing the comparative abundance of EZH1, EZH2, the PCNA proliferation marker, and H3K27me2 and H3K27me3 in ipNF05.5 versus HAP1 cells in the indicated wild-type or KO conditions. H4 is used as a loading control. (C) Venn diagrams showing the overlap between genes up-regulated upon complete PRC2 inactivation (SUZ12 or EED KO) and upon EZH2 KO in ipNF05.5 and the highly proliferative HAP1 cells. (D) Western blot showing EZH1, EZH2, H3K27me2, and H3K27me3 levels in wild-type or EZH2 KO immortalized Schwann cells grown in low proliferation medium (low serum) or high proliferation medium (high serum).
We next sought to directly assess whether redundancy between EZH1 and EZH2 is indeed alleviated upon higher proliferation. To this aim, we compared the consequence of deleting EZH2 in ipNF05.5 cells and HAP1 cells, which have a much higher proliferation rate (12-h doubling time compared with 30 h for ipNF05.5 cells). The level of EZH1 is similar between the two cell lines, but EZH2 is much more abundant in HAP1 cells than in ipNF05.5 cells and is paralleled by a high level of the PCNA proliferation marker (Fig. 5B). Interestingly, substantial levels of H3K27me2 and me3 remain in EZH2 KO ipNF05.5 cells, while loss of EZH2 in HAP1 cells results in acute loss of H3K27me2 and me3, similar to that obtained upon deletion of EED (Fig. 5B; also see SI Appendix, Fig. S5B for quantification of Western blot signals). Accordingly, loss of EZH2 in HAP1 cells led to transcriptional derepression of a number of genes, similar to KO of the core PRC2 component EED, but has moderate transcriptional consequences in ipNF05.5 cells (Fig. 5C). The differential effect of deleting EZH2 in ipNF05.5 and HAP1 cells is further illustrated at genes that are regulated by PRC2 in both cell lines (SI Appendix, Fig. S5C).
To further ascertain that the association between cell proliferation and EZH2/EZH1 redundancy is not a bias resulting from analyzing different cell types, we investigated the impact of direct manipulation of cell proliferation rate in the context of a defined cell type. We performed a KO of EZH2 in an immortalized Schwann cell line (29) and grew wild-type and EZH2 KO cells in either low-serum (slow-proliferation) or high-serum (high-proliferation) medium. As expected, upon high proliferation, EZH2 levels strongly increased while EZH1 expression remain constant (Fig. 5D). Interestingly, in the absence of EZH2, higher proliferation led to a decrease of H3K27me2/me3 levels (Fig. 5D; also see SI Appendix, Fig. S5D for quantification of Western blot signals). This experiment thus demonstrates that EZH1’s ability to compensate for loss of EZH2 is inversely proportional to the rate of cell proliferation.
Altogether, our analyses show that EZH1 cannot compensate for loss of EZH2 under high proliferation, suggesting that proliferation is a major factor underlying the redundancy between EZH1 and EZH2. We propose that the proliferative index is a key constraint underlying the PRC2 mutation pattern that is selected for in the course of tumorigenesis (SI Appendix, Fig. S5E). In tumors characterized by a low proliferation index such as PNFs, mutations in EZH2 will not be selected for because of the redundancy with EZH1. In tumor types characterized by a much higher proliferative index such as myeloid malignancies and T-ALL, EZH2 becomes predominant relative to EZH1, and thus loss of EZH2 can be selected for. Interestingly, this simple model also accounts for gain-of-function mutations that selectively occur on EZH1 in the slow proliferating ATAs or on EZH2 in the more proliferative FL and diffuse large B cell lymphoma types of lymphomas.
Discussion
The PRC2 complex is diverted from its normal function in cancer through defined tumor-type specific mutations. These alterations have been suggested to entail canonical as well as noncanonical functions of EZH2, the main PRC2 enzymatic subunit. However, we currently lack a precise understanding of how mutations found in cancer affect each of these activities, thus limiting our ability to develop rational therapeutic approaches. In this study, we investigated the relative contribution of canonical versus noncanonical activities of EZH2 in the regulation of gene expression. MPNST cells represent an ideal system to study such a noncanonical activity since the canonical PRC2 function is absent. In such a context, we provide compelling biochemical and genetic evidence that EZH2 does not regulate transcription. PRC2-independent functions could still be at play in certain cell types; however, our biochemical analysis of EZH2 and the kinetics of cell growth upon EZH2 inhibition in the LNCaP-abl model cell line are more consistent with a canonical function of the enzyme.
We uncover a pronounced functional redundancy between EZH1 and EZH2 in MPNST precursor cells. Combined loss of both enzymes leads to gene expression changes that are indistinguishable from those induced by loss of SUZ12. These findings strongly suggest that the signature of PRC2 mutations in MPNST, that is, the absence of mutations in EZH2/EZH1, stems from the high redundancy between the two enzymes. In this context, the probability of inactivating PRC2 through biallelic mutation of EZH1 and EZH2 (four alleles) would be far smaller than mutation of SUZ12 or EED (two alleles only at most; one allele of SUZ12 often being inactivated simultaneously with the driving mutation in NF1 by a large deletion encompassing both genes).
Our analyses also show that the rate of cell proliferation, which positively controls the expression of EZH2 but not EZH1, is a key parameter modulating the redundancy between the two enzymes, explaining why mutations in EZH2 are favored in high proliferating tumor types. Nonetheless, other independent cues are also likely to participate in modulating the balance between EZH2 and EZH1 as recently shown for EZH1 expression during male germ cell development (31). As in the case of PRC2, mutations in genes encoding subunits of other chromatin-modifying complexes such as SWI/SNF and COMPASS show a certain degree of tumor type specificity (33, 34). It will be interesting to investigate whether similar context-dependent redundancies within these chromatin-modifying complexes underlie such mutation patterns. In conclusion, our study addresses key issues regarding canonical activities of the PRC2 complex and provides insights into the characteristic spectrum of PRC2 mutations found in different cancers.
Materials and Methods
Cell Culture.
SUZ12 mutated 88-14 and SUZ12 wild-type STS26T cell lines were kindly provided by Nancy Ratner, Cincinnati Children’s Hospital Medical Center, Cincinnati. The LNCaP-abl cell line was kindly provided by Zoran Culig, Innsbruck Medical University, Innsbruck, Austria. ipNF05.5 plexiform neurofibroma cell line and immortalized Schwann cell line are described in ref. 29. HAP1 cells were kindly provided by T. Brummelkamp, Oncode Institute, Amsterdam. OVCAR8 cells were kindly provided by Fatima Mechta-Grigoriou, Institut Curie, Paris. Additional details cell culture conditions, transfection, constitutive knockouts, proliferation assays, and downstream biochemical analyses are provided in SI Appendix, Supplementary Methods.
RNA Extraction, RT-qPCR, Digital Droplet PCR, RNA Sequencing, and Data Analysis.
Total RNA was isolated using TRIzol-chloroform extraction and isopropanol precipitation. Additional details on RT-qPCR, digital droplet PCR, RNA sequencing, and data analysis are provided in SI Appendix, Supplementary Methods.
Chromatin Immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed as described previously (3). Additional details on ChIP sequencing and data analysis are provided in SI Appendix, Supplementary Methods.
Patients and Samples.
All patients provided informed consent, and the study was approved by institutional review board and local ethical committees. Project ID CPP17/79, A0296746, and 2015-08-11DC were reviewed by Cochin Hospital institutional review board and CPP Ile-de-France 2 ethics committee APHP, Paris, and project ID BS#2017-311 by the Groupe Thématique de Travail–Hematology Section, Institut Curie, institutional review board and ethics committee of the Hospital Group, Institut Curie, Paris. Additional details on patient samples are provided in SI Appendix, Supplementary Methods.
Data Access.
Next-generation sequencing (NGS) data have been deposited in the GEO database (accession no. GSE118186). Mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD012547.
Supplementary Material
Acknowledgments
We thank Pascale Gilardi, Daniel Holoch, and members of the R.M. laboratory for comments on the manuscript. Work in the laboratory of R.M. is supported by the “Association pour la Recherche sur le Cancer” and the Labex “Development, Epigenetics, Epigenetics, and Lifetime Potential.” R.M. and E.P. were supported by Institut Thématique Multi-Organisme Cancer (Grant EpiNF1). The M.V. lab is part of the Oncode Institute, which is partly funded by the Dutch Cancer Society (KWF). A.L. was a recipient of a fellowship from INSERM. High-throughput sequencing was performed by the NGS platform of the Institut Curie, supported by Grants ANR-10-EQPX-03 and ANR10-INBS-09-08 from the Agence Nationale de le Recherche (Investissements d’Avenir) and by the Canceropôle Ile-de-France.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE118186), and in the ProteomeXchange Consortium database via the PRIDE partner repository, https://www.ebi.ac.uk/pride/archive/ (dataset identifier PXD012547).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814634116/-/DCSupplemental.
References
- 1.Shah MA, Denton EL, Arrowsmith CH, Lupien M, Schapira M. A global assessment of cancer genomic alterations in epigenetic mechanisms. Epigenetics Chromatin. 2014;7:29. doi: 10.1186/1756-8935-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Workman P, Al-Lazikani B. Drugging cancer genomes. Nat Rev Drug Discov. 2013;12:889–890. doi: 10.1038/nrd4184. [DOI] [PubMed] [Google Scholar]
- 3.Margueron R, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen X, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holoch D, Margueron R. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem Sci. 2017;42:531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 7.Wassef M, Margueron R. The multiple facets of PRC2 alterations in cancers. J Mol Biol. 2017;429:1978–1993. doi: 10.1016/j.jmb.2016.10.012. [DOI] [PubMed] [Google Scholar]
- 8.De Raedt T, et al. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature. 2014;514:247–251. doi: 10.1038/nature13561. [DOI] [PubMed] [Google Scholar]
- 9.Lee W, et al. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, et al. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahm F, Reuss DE, Giannini C. WHO 2016 classification: Changes and advancements in the diagnosis of miscellaneous primary CNS tumours. Neuropathol Appl Neurobiol. 2018;44:163–171. doi: 10.1111/nan.12397. [DOI] [PubMed] [Google Scholar]
- 12.Brohl AS, Kahen E, Yoder SJ, Teer JK, Reed DR. The genomic landscape of malignant peripheral nerve sheath tumors: Diverse drivers of Ras pathway activation. Sci Rep. 2017;7:14992. doi: 10.1038/s41598-017-15183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sohier P, et al. Confirmation of mutation landscape of NF1-associated malignant peripheral nerve sheath tumors. Genes Chromosomes Cancer. 2017;56:421–426. doi: 10.1002/gcc.22446. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez ME, et al. EZH2 expands breast stem cells through activation of NOTCH1 signaling. Proc Natl Acad Sci USA. 2014;111:3098–3103. doi: 10.1073/pnas.1308953111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee ST, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43:798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Xu K, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ezhkova E, et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wassef M, et al. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes Dev. 2015;29:2547–2562. doi: 10.1101/gad.269522.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jansen PTWC, Margueron R. 2019 Data from “EZH1/2 function mostly within canonical PRC2 and exhibit proliferation-dependent redundancy that shapes mutational signatures in cancer.” PRIDE. Available at www.ebi.ac.uk/pride/archive/projects/PXD012547. Deposited January 29, 2019.
- 20.Conway E, et al. A family of vertebrate-specific Polycombs encoded by the LCOR/LCORL genes balance PRC2 subtype activities. Mol Cell. 2018;70:408–421.e8. doi: 10.1016/j.molcel.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Jiao L, Shubbar M, Yang X, Liu X. Unique structural platforms of Suz12 dictate distinct classes of PRC2 for chromatin binding. Mol Cell. 2018;69:840–852.e5. doi: 10.1016/j.molcel.2018.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasinath V, et al. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science. 2018;359:940–944. doi: 10.1126/science.aar5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Konze KD, et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wassef M, et al. 2019 Data from “EZH1/2 function mostly within canonical PRC2 and exhibit proliferation-dependent redundancy that shapes mutational signatures in cancer.” GEO. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118186. Deposited August 6, 2018.
- 25.Culig Z, et al. Switch from antagonist to agonist of the androgen receptor bicalutamide is associated with prostate tumour progression in a new model system. Br J Cancer. 1999;81:242–251. doi: 10.1038/sj.bjc.6690684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan L, et al. Phosphorylation of EZH2 by AMPK suppresses PRC2 methyltransferase activity and oncogenic function. Mol Cell. 2018;69:279–291.e5. doi: 10.1016/j.molcel.2017.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley WD, et al. EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol. 2014;21:1463–1475. doi: 10.1016/j.chembiol.2014.09.017. [DOI] [PubMed] [Google Scholar]
- 28.Knutson SK, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 29.Li H, Chang L-J, Neubauer DR, Muir DF, Wallace MR. Immortalization of human normal and NF1 neurofibroma Schwann cells. Lab Invest. 2016;96:1105–1115. doi: 10.1038/labinvest.2016.88. [DOI] [PubMed] [Google Scholar]
- 30.He Y, et al. The EED protein-protein interaction inhibitor A-395 inactivates the PRC2 complex. Nat Chem Biol. 2017;13:389–395. doi: 10.1038/nchembio.2306. [DOI] [PubMed] [Google Scholar]
- 31.Mu W, Starmer J, Shibata Y, Yee D, Magnuson T. EZH1 in germ cells safeguards the function of PRC2 during spermatogenesis. Dev Biol. 2017;424:198–207. doi: 10.1016/j.ydbio.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bracken AP, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masliah-Planchon J, Bièche I, Guinebretière J-M, Bourdeaut F, Delattre O. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10:145–171. doi: 10.1146/annurev-pathol-012414-040445. [DOI] [PubMed] [Google Scholar]
- 34.Meeks JJ, Shilatifard A. Multiple roles for the MLL/COMPASS family in the epigenetic regulation of gene expression and in cancer. Annu Rev Cancer Biol. 2017;1:425–446. [Google Scholar]
- 35.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.