Significance
We have completed a screen of ∼106 small molecules to identify compounds that induce cell death in multipotent glioblastoma multiforme (GBM) cancer stem cells (CSCs). This resulted in the identification of a hit class (RIPGBM) that was found to induce apoptosis in GBM CSCs in a cell type-selective manner. Metabolite profiling experiments led to the identification of a proapoptotic derivative of RIPGBM (cRIPGBM), which was found to be selectively formed in GBM CSCs. Mechanistic studies revealed that cRIPGBM induces apoptosis by binding to receptor-interacting protein kinase 2 (RIPK2) in a mode that results in the formation of a proapoptotic RIPK2/caspase 1 complex. In a physiologically relevant orthotopic intracranial GBM CSC tumor xenograft mouse model, RIPGBM was found to significantly inhibit in vivo tumor formation.
Keywords: glioblastoma, phenotypic drug screening, chemical genetics, target identification, receptor-interacting protein kinase 2
Abstract
Glioblastoma multiforme (GBM; grade IV astrocytoma) is the most prevalent and aggressive form of primary brain cancer. A subpopulation of multipotent cells termed GBM cancer stem cells (CSCs) play a critical role in tumor initiation, tumor maintenance, metastasis, drug resistance, and recurrence following surgery. Here we report the identification of a small molecule, termed RIPGBM, from a cell-based chemical screen that selectively induces apoptosis in multiple primary patient-derived GBM CSC cultures. The cell type-dependent selectivity of this compound appears to arise at least in part from redox-dependent formation of a proapoptotic derivative, termed cRIPGBM, in GBM CSCs. cRIPGBM induces caspase 1-dependent apoptosis by binding to receptor-interacting protein kinase 2 (RIPK2) and acting as a molecular switch, which reduces the formation of a prosurvival RIPK2/TAK1 complex and increases the formation of a proapoptotic RIPK2/caspase 1 complex. In an orthotopic intracranial GBM CSC tumor xenograft mouse model, RIPGBM was found to significantly suppress tumor formation in vivo. Our chemical genetics-based approach has identified a drug candidate and a potential drug target that provide an approach to the development of treatments for this devastating disease.
Stem cell mechanisms have been established to play critical roles in the development, progression, and recurrence of multiple cancer types (1–5). One such case is the infiltrative brain cancer glioblastoma multiforme (GBM). For recurrent GBM, even with aggressive treatment, the median survival rate is presently 12–15 mo (6, 7). GBM cancer stem cells (CSCs) were among the first cancer stem cell populations to be isolated and characterized from solid tumors (8–11). These cells share features in common with neural stem cells (NPCs), namely the expression of NSC markers (e.g., Nestin and SOX2), the capacity for self-renewal, and the ability to differentiate and give rise to cell types of glial and neuronal lineages in response to inductive cues (8, 9, 11, 12). The highly infiltrative nature of GBM tumors is attributed to the ability of GBM CSCs to migrate within the brain, a feature also shared with NSCs (13). Further, GBM CSCs are thought to contribute to drug resistance (14, 15) and have even been demonstrated to have the potential to give rise to endothelial cells that enable tumor vascularization (16). Thus, efforts to develop new therapeutic strategies for the treatment of GBM have recently focused on targeting this stem cell population (17).
Here we have used an unbiased large-scale screening approach to identify drug-like small molecules that induce apoptosis in GBM CSCs in a cell type-selective manner. The use of expanded populations of proliferative nonstem/multipotent GBM cells for such screens is of limited utility, as such cell lines fail to recapitulate the in vitro and in vivo properties, including drug sensitivity, of the original tumor (9, 18–22). In contrast, in vitro and in vivo preclinical models using cultured human tumor-derived GBM CSCs more accurately recapitulate the biology of the disease (9, 15, 23–26). Under defined serum-free adherent culture conditions, these patient-derived GBM CSCs can be expanded as stable cell lines that retain their in vitro differentiation potential, as well as their in vivo engraftment, tumor formation, and migration potential (17). In the present study, we have used patient-derived GBM CSC cultures to identify a potential drug candidate for the treatment of this devastating disease.
Results
RIPGBM Is a Selective Inducer of Apoptosis in GBM CSCs.
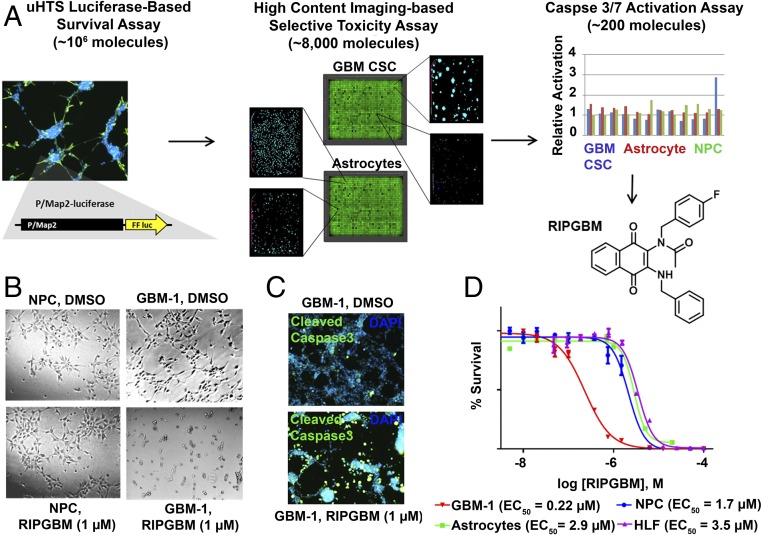
We have previously described an adapted system for the adherent in vitro expansion of patient-derived GBM CSCs that was successfully used to perform a kinomewide lentiviral RNAi screen in 384-well assay format (24). Importantly, these primary cell lines retain stem cell-like characteristics and differentiation properties, as well as the ability to engraft and form tumors that recapitulate the highly heterogeneous and infiltrative characteristics of high-grade gliomas in relevant rodent disease models (24). Here, we used one of these GBM CSC lines, termed GBM-1, to establish a robust 1,536-well format luciferase-based survival assay and completed a large-scale screen of ∼106 drug-like small molecules (1 μM) with the goal of identifying novel compounds that are selectively toxic to chemoresistant GBM CSCs (Fig. 1A).
Fig. 1.
A cell-based phenotypic screening approach identifies the small molecule RIPGBM, which induces apoptosis in GBM CSCs in a cell type-selective manner. (A) Schematic representation of the screening approach used to identify molecules that selectively induce apoptosis in GBM CSCs and structure of RIPGBM. (B) Bright-field image of GBM CSCs (GBM-1) or nondiseased human NPCs treated with RIPGBM (1 μM) for 96 h. (C) Immunofluorescent analysis of caspase 3 cleavage in GBM CSCs (GBM-1) following treatment with RIPGBM (1 μM) for 24 h. (D) Cell survival curves for GBM CSCs (GBM-1), human NPCs, primary human astrocyte cells, and primary HLFs treated with RIPGBM for 48 h. Values shown are mean ± SD.
Confirmed primary hits (∼8,000, plate-based robust Z-score ≤−3) were evaluated by using a laser scanning cytometer-based cell death imaging assay (Acumen eX3; TTP Labtech; Fig. 1A). Selective cytotoxicity was determined by evaluating the primary hits at two concentrations (5 μM and 1 μM), using a panel of two patient-derived GBM CSC lines (GBM-1 and GBM-5, ref. 24) and 3 nondiseased cell types [primary human astrocytes, WA09 human ES cell-derived NPCs, and primary human lung fibroblasts (HLFs)]. Compounds found to kill GBM CSC lines with greater than fivefold toxicity index compared with control cell types were further characterized. A caspase 3/7 activation assay (Caspase-Glo 3/7; Promega) was used to evaluate the mechanism of induced cell death. The most potent and selective compound identified from these assays, termed RIPGBM (Fig. 1A), was found to selectively induce apoptosis in GBM CSC cell lines with an observed EC50 of ≤500 nM and a selectivity index of at least fivefold compared with control cell types (Fig. 1 B–D and SI Appendix, Table S1). For comparison, the observed EC50 for the standard-of-care drug temozolomide (TMZ) used to treat GBM is ≥20 μM for the same GBM CSC lines (SI Appendix, Table S1). Moreover, human ES-derived NSCs are at least twofold more sensitive to TMZ than GBM CSCs (SI Appendix, Fig. S1). The latter observation is in agreement with published data showing that GBM CSCs are resistant to chemotherapeutic agents (27, 28). We confirmed the ability of RIPGBM to induce apoptosis in GBM CSCs by immunofluorescent analysis using a cleaved caspase 3-recognizing antibody (Fig. 1C).
RIPGBM Is Converted to an Apoptosis-Inducing Derivative Selectively in GBM CSCs.
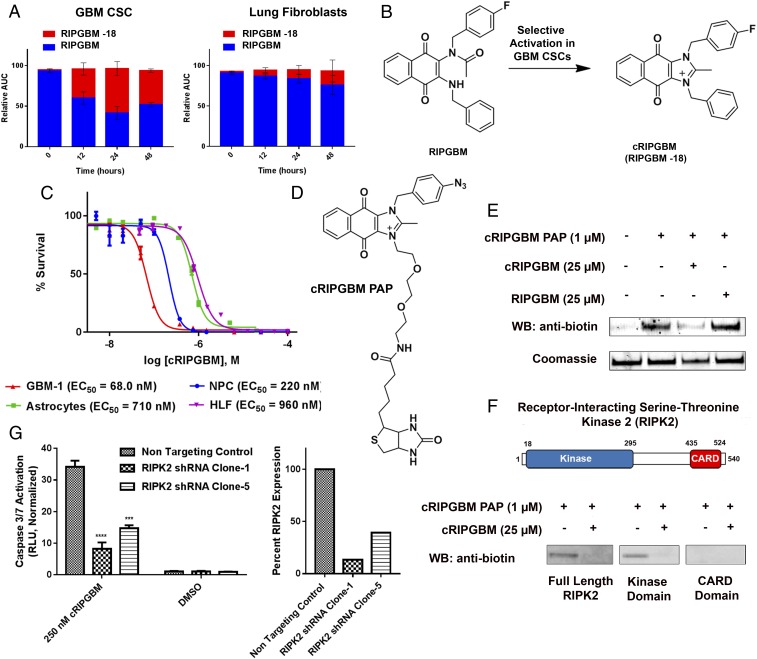
Quinone-containing drugs represent a large and diverse class of antitumor agents approved for clinical use. For many of these drugs, cell type-selective reduction to reactive hydroquinone species has been shown to play a key role in their antitumor activity (29). As the naphthoquinone core of RIPGBM is found in various substrates for quinone oxidoreductase enzymes (e.g., NQO1), which generate hydroquinone or semihydroquinone species and are frequently up-regulated in various cancer cell types (30), we hypothesized that either of these events could lead to the selective formation of a proapoptotic species in GBM CSCs. We used an MS-based metabolite identification approach to evaluate whether RIPGBM undergoes selective conversion to a +1 (i.e., semihydroquinone) or +2 (i.e., hydroquinone) species in GBM CSCs. Cell pellets and growth media of GBM CSC and control cell types were extracted following drug treatment and subjected to quantitative high-resolution Orbitrap LC-MS analysis (Thermo Fisher Scientific). The formation of +1 or +2 species was not observed from samples derived from either cell type. However, RIPGBM was found to undergo significant conversion to a −18 species, potentially corresponding to a dehydration event, in GBM CSCs (∼50% in 24 h), and this conversion was found to occur selectively in diseased cells (Fig. 2A). An accurate mass measurement of 411.1506 m/z (SI Appendix, Fig. S2) and MS2 fragmentation data (SI Appendix, Fig. S3) were consistent with the cyclized imidazolium species, termed cRIPGBM, shown in Fig. 2B. In parallel experiments, it was found that, whereas RIPGBM is relatively stable in culture media (t1/2 > 4 d), addition of the reducing agent NaBH4 results in near-instantaneous formation of the −18 cRIPGBM species (SI Appendix, Fig. S4). Presumably, reduction of the quinone moiety makes the benzylic amine more nucleophilic, resulting in its addition to the acetamido group and subsequent loss of water. The putative cyclic derivative was synthesized and evaluated against a panel of GBM CSCs and nondisease cell lines. The cyclized species cRIPGBM was found to induce apoptosis in GBM CSCs with enhanced potency compared with the parent compound following 48 h of drug treatment (e.g., EC50 = 68 nM vs. 220 nM in GBM-1), but with reduced selectivity compared with control cells (Fig. 2C and SI Appendix, Table S1). As such, the mechanism by which RIPGBM selectively induces cell death in GBM CSCs likely involves a redox-dependent prodrug-like process involving cell type-dependent formation of a proapoptotic species.
Fig. 2.
A metabolite of RIPGBM induces apoptosis in GBM CSCs by interacting with RIPK2. (A) Orbitrap MS-based metabolite identification studies in GBM-1 (GBM CSC) or primary HLF cells incubated with RIPGBM (1 μM) for 0, 12, 24, or 48 h. (B) Structure of the cyclized RIPGBM metabolite cRIPGBM generated in GBM CSCs. (C) Cell survival curves for GBM CSCs (GBM-1), human NPCs, primary human astrocyte cells, and HLFs treated with cRIPGBM for 48 h. (D) Structure of PAP reagent cRIPGBM-PAP. (E) In vitro binding of cRIPGBM-PAP to recombinant human full-length RIPK2 protein in the presence or absence of competition using underivatized cRIPGBM or RIPGBM. (F) Domain structure of RIPK2 and in vitro binding of cRIPGBM-PAP to recombinant full-length, truncated kinase domain, or truncated CARD domain human RIPK2 protein. (G) cRIPGBM-induced apoptosis in GBM-1 GBM CSCs following shRNA-mediated RIPK2 gene knockdown. Values shown are mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001).
cRIPGBM Targets RIPK2 in GBM CSCs.
Structure–activity relationship studies revealed sites off the RIPGBM core tolerant of modification. This information was used to design and synthesize photoactivatable affinity probe (PAP) reagents. MS-based proteomic target identification studies involving incubation and photo-cross-linking with live GBM CSCs using cRIPGBM-PAP (Fig. 2D), which retained activity (EC50) within fivefold of that of the parent molecule, resulted in the identification of receptor-interacting serine-threonine kinase 2 (RIPK2) as a candidate protein target for cRIPGBM (SI Appendix, Fig. S5). Based on its known role in regulating apoptosis and the degree of identified peptide coverage observed for this potential biomolecular target (SI Appendix, Fig. S5B), RIPK2 was explored in detail to examine its role in the mechanism of cRIPGBM-induced apoptosis.
In in vitro binding assays involving purified recombinant proteins, cRIPGBM- PAP was observed to interact with full-length RIPK2 protein in a concentration-dependent manner at concentrations of 100 nM and greater (apparent Kd of ∼2.3 µM; SI Appendix, Fig. S5 C and D). A 25-fold molar excess of cRIPGBM was found to inhibit this interaction, demonstrating the specificity of this interaction (Fig. 2E). Consistent with playing a relevant role in the induction of apoptosis, RIPK2 consists of an N-terminal autophosphorylation kinase domain and a C-terminal caspase 1 recruitment CARD domain separated by a domain of unknown function (Fig. 2F) (31). In vitro binding experiments suggest that cRIPGBM interacts with the kinase domain of this protein (Fig. 2F). However, the compound was not found to inhibit kinase activity at concentrations lower than 10 µM in an RIPK2 enzymatic assay (SI Appendix, Fig. S6). To further validate RIPK2 as the molecular target of cRIPGBM in GBM CSCs, we measured compound-induced cytotoxicity and caspase activation in cells in which RIPK2 levels were reduced by using shRNA. Consistent with the mechanistic relevance of this target, as demonstrated by the observed reduction of cRIPGBM-induced caspase 3/7 activation in GBM CSCs (Fig. 2G), RIPK2 suppression resulted in a significant decrease in drug sensitivity. The acyclic parent compound RIPGBM was not found to interact with RIPK2 (Fig. 2E). These results suggest that it is the cyclized derivative cRIPGBM that is responsible for RIPK2-dependent induction of apoptosis.
cRIPGBM Induces Caspase 1-Mediated Apoptosis.
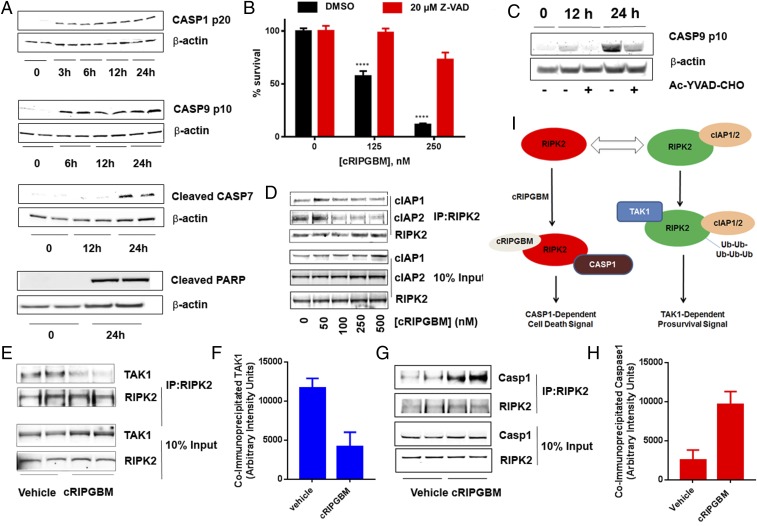
To elucidate the downstream mechanism of action by which cRIPGBM induces apoptosis, we assessed caspase activation in GBM CSCs. Compound treatment (250 nM) resulted in a time-dependent activation of caspase 1, caspase 9, and caspase 7, as well as PARP cleavage (Fig. 3A), providing further evidence that cRIPGBM induces cell death via an apoptotic mechanism. Correspondingly, the pan-caspase inhibitor Z-VAD-FMK significantly reduced compound-mediated GBM CSC death (Fig. 3B). PARP cleavage occurred in a time-dependent manner concomitant with the onset of DNA fragmentation as determined by using a TUNEL assay (SI Appendix, Fig. S7). In addition, we established that caspase 1 activation is upstream of caspase 9, caspase 7, and PARP cleavage by observing that pretreatment with the caspase 1 inhibitor Ac-YVAD-CHO effectively blocks caspase 9 cleavage in cRIPGBM-treated GBM CSCs (Fig. 3C).
Fig. 3.
cRIPGBM activates caspase 1-mediated apoptotic signaling in GBM CSCs by modulating the interaction of RIPK2 with TAK1 and caspase 1. (A) Time-dependent cRIPGBM (250 nM)-induced cleavage of caspase 1, caspase 9, caspase 7, and poly(ADP-ribose) polymerase (PARP) in GBM-1 GBM CSCs. (B) cRIPGBM-induced cell death in GBM-1 GBM CSCs following treatment with the pan-caspase inhibitor Z-VAD (20 μM). (C) cRIPGBM-induced (250 nM) caspase 9 cleavage in GBM-1 GBM CSCs following treatment with the caspase 1-selective inhibitor Ac-YVAD-CHO (20 μM). (D) Coimmunoprecipitation of cIAP1 and cIAP2 with anti-RIPK2 antibody in GBM-1 GBM CSCs following treatment with cRIPGBM. (E) Coimmunoprecipitation of TAK1 with anti-RIPK2 antibody in GBM-1 GBM CSCs following treatment with cRIPGBM (250 nM). (F) Quantification of TAK1 levels coimmunoprecipitated by using an anti-RIPK2 antibody in GBM-1 GBM CSCs following treatment with cRIPGBM (250 nM). (G) Coimmunoprecipitation of caspase 1 with anti-RIPK2 antibody in GBM-1 GBM CSCs following treatment with cRIPGBM (250 nM). (H) Quantification of caspase 1 levels coimmunoprecipitated by using an anti-RIPK2 antibody in GBM-1 GBM CSCs following treatment with cRIPGBM (250 nM). (I) Schematic representation of the proposed mechanism of action for cRIPGBM-induced apoptosis in GBMB CSCs. Values shown are mean ± SD (*P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001).
Ubiquitination is a known key modification that regulates the ability of RIPK2 to act as a prosurvival or proapoptotic molecule (32, 33). Specifically, K63-ubiquitinated RIPK functions as a scaffold for the assembly of protein complexes that activate prosurvival signaling pathways (34–36). The E3 ubiquitin ligases cIAP1 and cIAP2 have been previously shown to interact with and promote RIPK2 ubiquitination in various cell lines (33, 37, 38). Coimmunoprecipitation assays were used to determine if cRIPGBM treatment alters its interaction with cIAP1 and/or cIAP2 in GBM CSCs. Compound treatment reduced RIPK2 binding to cIAP1 (Fig. 3D) and significantly reduced binding to cIAP2 in a dose-dependent manner (Fig. 3D), which suggests that cIAP1 and cIAP2 are endogenous regulators of RIPK2 ubiquitination in GBM CSCs.
cRIPGBM Acts as a Molecular Switch That Modulates RIPK2 Binding Partners.
On the basis of these observations, as well as previously established mechanisms that have been established for RIPK2 (34, 35), we hypothesized that cRIPGBM promotes cell death by modulating RIPK2 ubiquitination status, which could impact its interactions with a prosurvival molecule to favor its interaction with a proapoptotic adaptor protein. To test this notion, we determined if compound treatment alters RIPK2 binding partners by using coimmunoprecipitation experiments. Previous studies have established that K63-ubiquitinated RIPK2 associates with the prosurvival TAK1 complex (34, 35). Consistently, TAK1 was found to coimmunoprecipitate with RIPK2 under basal conditions in GBM CSCs (Fig. 3E). In cRIPGBM-treated cells, the interaction between RIPK2 and TAK1 was significantly decreased (Fig. 3 E and F). Conversely, because cRIPGBM treatment induces caspase 1-dependent cell death, we determined whether drug treatment correlated with an increased association between RIPK2 and caspase 1. Indeed, treatment with cRIPGBM for 6 h was found to result in a significant enhancement of RIPK2–caspase 1 interaction (Fig. 3 G and H). Taken together, these data suggest that cRIPGBM induces apoptosis in GBM CSCs by a mechanism that involves its interaction with RIPK2 in a mode that results in decreased association with TAK1 and increased association with and activation of caspase 1, which can lead to downstream activation of a caspase 1-mediated apoptotic signaling cascade (Fig. 3I). Additional future work will be required to determine a detailed understanding of how cRIPGBM/RIPK2 interaction impacts RIPK2 function in normal and disease cell types.
RIPGBM Inhibits Tumor Growth in a Patient-Derived GBM CSC Intracranial Xenograft Model.
The observed lack of brain exposure for cRIPGBM following oral or i.p. dosing precluded characterization of its in vivo activity using a physiologically relevant intracranial tumor model. However, encouragingly, the parent acyclic cell type-selective prodrug form of the molecule (RIPGBM), which would likely be the preferred drug candidate based on its selectivity profile, was found to have reasonable brain exposure properties following oral administration (brain Cmax = 540 nM; brain t1/2 = 1.5 h; 20 mg/kg orally; SI Appendix, Table S2). Further, in a 1-wk mouse toxicity study, oral delivery of RIPGBM was well tolerated with no overt toxicity observed at doses as high as 100 mg/kg twice per day based on body weight loss (SI Appendix, Fig. S8) and terminal hematology (SI Appendix, Tables S3 and S4), blood chemistry (SI Appendix, Table S5), and anatomical findings.
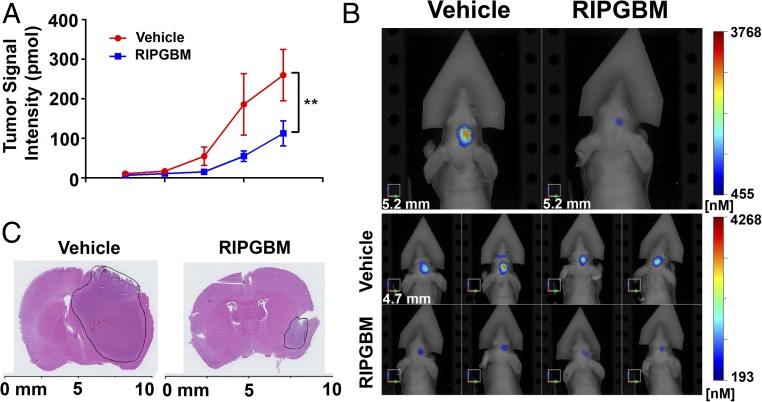
Given the selectivity and pharmacologic properties of RIPGBM, we sought to assess its efficacy in vivo by using a clinically relevant, patient-derived GBM intracranial xenograft model. In contrast to non-CSC glioma cell lines, which fail to form tumors that recapitulate the heterogeneous highly infiltrative nature of the parent tumor (9, 21), intracranial xenotransplantation of CSCs derived from GBM tumors results in the formation of tumors that recapitulate the invasive nature of the human disease (8, 9, 24). A highly aggressive GBM patient-derived line (GBM-39), which robustly recapitulates the heterogeneous and highly infiltrative nature of GBM tumors in vivo (39, 40), was engineered to express the IR fluorescent protein 720 (IRFP720) to allow noninvasive and quantitative assessment of orthotopic tumor growth by fluorescence molecular tomography (FMT) imaging. Oral administration of RIPGBM (50 mg/kg orally twice daily) to mice bearing GBM39 IRFP720 intracranial xenografts resulted in a significant inhibition of tumor growth, as monitored by FMT imaging (Fig. 4 A and B), which was grossly associated with decreased tumor size, as assessed by H&E staining (Fig. 4C). Taken together, these results demonstrate that oral delivery of a well-tolerated dose of RIPGBM results in substantial anti-GBM activity in vivo. Thus, with improvements in potency and pharmacokinetic properties, this compound series has the potential to provide a new therapeutic approach for patients with GBM.
Fig. 4.
RIPGBM reduces GBM tumor growth in an orthotopic intracranial xenograft model. (A) GBM39 patient-derived neurosphere cells engineered to stably express the IR protein 720 (IRFP 720) were orthotopically injected into 5-wk-old nu/nu mice. Mice were treated with vehicle or RIPGBM 50 mg/kg orally twice daily (n = 8 for vehicle and n = 6 for RIPGBM). (B) Representative FMT images of mice at week 5. (C) Representative H&E stains of mouse brains from mice at week 5. Values shown are mean ± SD (*P < 0.05 and **P < 0.01).
Discussion
We have identified a small molecule, RIPGBM, that selectively induces apoptosis in GBM CSCs in vitro and significantly decreases tumor size in vivo in a physiologically relevant, patient-derived intracranial xenograft mouse model. The cell-type selectivity of this prodrug molecule appears to be derived, at least in part, from selective redox-dependent bioactivation in GBM CSCs, which leads to the formation of a proapoptotic molecule termed cRIPGBM, as well as sensitivity to RIPK2-induced apoptosis. Additional target identification studies have yet to reveal a potential activating enzyme for the RIPGBM prodrug molecule. As such, it remains unclear if cyclization is dependent on a cell type-specific enzymatic conversion or altered cellular redox potential. Indeed, antioxidant response pathways (e.g., NRF2-dependent pathways) are frequently found to be induced in diverse cancer cell types as a result of oxidative stress and mutations within the tumor microenvironment (41). Pharmacological data suggest that the mechanism of action of cRIPGBM-induced apoptosis involves its direct interaction with RIPK2. This results in decreased association with TAK1 and increased association with caspase 1, leading to downstream activation of a caspase 1-mediated apoptotic signaling cascade. Interestingly, RIPK2-dependent caspase 1-induced apoptosis has previously been demonstrated to play an essential role in hypoxia and ischemia-induced neuronal cell death (42), which is consistent with the ability of RIPK2 to act as a key molecular switch that can control prosurvival vs. proapoptotic signaling pathways in neural cell types including GBM CSCs. Given the high rate of GBM tumor relapse following surgery, which results from the therapeutic resistance of GBM CSCs, the observed sensitivity of GBM CSCs to RIPK2-induced apoptosis and the ability to control this molecular switch with an identified small molecule has significant implications for the development of new therapies for GBM. In theory, such a molecule could not only decrease the rate of tumor regrowth, but also spare nontarget cells, including normal neural cell populations, thus lowering the side effects observed with standard-of-care treatments for GBM.
Methods
Cell Culture.
Deidentified tumor samples classified as GBM were obtained with informed consent from patients undergoing surgery at Stanford Medical Center in accordance with the institutional review boards at Stanford University and The Scripps Research Institute. Specimen-derived cells were cultured at 37 °C and 5% CO2 conditions. GBM CSCs were maintained in Neurobasal medium supplemented with N2, B27, and human basic FGF (20 ng/mL; Life Technologies) and EGF (20 ng/mL; Life Technologies). WA09 human stem cell-derived NPCs (Aruna Biomedical) were cultured following the manufacturer’s instructions. HLFs (IMR-90; American Type Culture Collection CCL-186) were cultured in MEM supplemented with 10% FBS and antibiotic/antimycotic agents. Human astrocytes isolated from the cerebral cortex (no. 1800; ScienCell) were cultured following the manufacturer’s instructions. Further details are provided in SI Appendix, Supplemental Methods.
High-Throughput Screening.
GBM-1 GBM CSCs were plated in complete GBM media as described earlier at a density of 1,000 cells per well in 10 μL and screened (1 μM, 0.1% DMSO) in 1,536-well plates coated with poly-d-lysine (5 μg/mL; Sigma) and laminin (5 μg/mL; Life Technologies). Imaging-based assays were performed by using an Acumen eX3 laser scanning cytometer (TTP Labtech). Hit selection was performed by using plate-based analysis (robust Z-scores <−3 for GBM CSCs and >−2 for control cell types). Further details are provided in SI Appendix, Supplemental Methods.
Metabolite Identification Studies.
GBM CSCs or control cells (200,000 per well) were incubated with RIPGBM (1 µM) for 0–48 h and subjected to LC-MS analysis as described in SI Appendix, Supplemental Methods.
cRIPGBM Protein Target Identification.
GBM-1 GBM CSCs (∼106) were cultured in 100-mm dishes and treated with 100 nM cRIPGBM-PAP for 1 h in the presence or absence of 5 μM cRIPGBM. Bands selectively enriched in samples lacking competition were evaluated by using a previously described method (30). Further details are provided in SI Appendix, Supplemental Methods.
In Vivo Xenograft Animal Model.
Five-week-old female athymic nu/nu mice (Harlan Sprague–Dawley) were intracranially injected with 1 × 105 GBM39 IRFP720 cells in 5 μL of PBS solution as described previously (43). Tumor growth was monitored by using an FMT 2500 Fluorescence Tomography System (PerkinElmer) weekly. Mice were treated with vehicle [10% M-Pyrol, 10% ethanol/Cremophor (1:1, vol/vol), and 80% saline solution] or 50 mg/kg of RIPGBM by oral gavage twice daily, 8 h apart, starting at day 7 postinjection. All procedures were reviewed and approved by the institutional animal use and care committee at the University of California, San Diego. Further details are provided in SI Appendix, Supplemental Methods.
Synthesis.
All chemicals and solvents were obtained from commercial suppliers (Acros and Aldrich) and used without further purification. Unless otherwise indicated, all reactions were run under argon gas. Anhydrous solvents were obtained by passage through an activated alumina column. The 1H and 13C NMR spectra were recorded on a Bruker 400- and 500-MHz spectrometer. Chemical shifts are reported relative to internal CDCl3 (Me4Si, δ 0.0), DMSO-d6 (Me4Si, δ 0.0), and CD3OD (Me4Si, δ 0.0). Further details are provided in SI Appendix, Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Dr. Shoutian Zhu for discussions regarding target identification experiments, Dr. Costas Lyssiotis for general discussions, and the pharmacology group at the California Institute for Biomedical Research for completing pharmacokinetic and mouse toxicity studies. This work was supported by National Cancer Institute/National Institutes of Health Grant R01CA200970 (to L.L.L.). P.S.M. was supported by grants from National Institute for Neurological Diseases and Stroke (NS73831), the Defeat GBM Program of the National Brain Tumor Society, and the Ben and Catherine Ivy Foundation.
Footnotes
Conflict of interest statement: P.S.M. is cofounder of Pretzel Therapeutics, Inc. He has equity and serves as a consultant for the company. P.S.M. also did a one-time consultation for Abide Therapeutics, Inc.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816626116/-/DCSupplemental.
References
- 1.Pardal R, Molofsky AV, He S, Morrison SJ. Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol. 2005;70:177–185. doi: 10.1101/sqb.2005.70.057. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 3.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 4.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 5.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: Mirage or reality? Nat Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 6.Brennan CW, et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furnari FB, Cloughesy TF, Cavenee WK, Mischel PS. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat Rev Cancer. 2015;15:302–310. doi: 10.1038/nrc3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galli R, et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 9.Lee J, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 12.Nakano I, Kornblum HI. Brain tumor stem cells. Pediatr Res. 2006;59:54R–58R. doi: 10.1203/01.pdr.0000203568.63482.f9. [DOI] [PubMed] [Google Scholar]
- 13.Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 14.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 15.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ricci-Vitiani L, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468:824–828. doi: 10.1038/nature09557. [DOI] [PubMed] [Google Scholar]
- 17.Pollard SM, et al. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 18.Martens T, et al. Inhibition of glioblastoma growth in a highly invasive nude mouse model can be achieved by targeting epidermal growth factor receptor but not vascular endothelial growth factor receptor-2. Clin Cancer Res. 2008;14:5447–5458. doi: 10.1158/1078-0432.CCR-08-0147. [DOI] [PubMed] [Google Scholar]
- 19.Sausville EA, Burger AM. Contributions of human tumor xenografts to anticancer drug development. Cancer Res. 2006;66:3351–3354, discussion 3354. doi: 10.1158/0008-5472.CAN-05-3627. [DOI] [PubMed] [Google Scholar]
- 20.Taillandier L, Antunes L, Angioi-Duprez KS. Models for neuro-oncological preclinical studies: Solid orthotopic and heterotopic grafts of human gliomas into nude mice. J Neurosci Methods. 2003;125:147–157. doi: 10.1016/s0165-0270(03)00043-8. [DOI] [PubMed] [Google Scholar]
- 21.Bonavia R, Inda MM, Cavenee WK, Furnari FB. Heterogeneity maintenance in glioblastoma: A social network. Cancer Res. 2011;71:4055–4060. doi: 10.1158/0008-5472.CAN-11-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Q, et al. A highly invasive human glioblastoma pre-clinical model for testing therapeutics. J Transl Med. 2008;6:77. doi: 10.1186/1479-5876-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wurdak H, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Giannini C, et al. Patient tumor EGFR and PDGFRA gene amplifications retained in an invasive intracranial xenograft model of glioblastoma multiforme. Neuro-oncol. 2005;7:164–176. doi: 10.1215/S1152851704000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cloughesy TF, Cavenee WK, Mischel PS. Glioblastoma: From molecular pathology to targeted treatment. Annu Rev Pathol. 2014;9:1–25. doi: 10.1146/annurev-pathol-011110-130324. [DOI] [PubMed] [Google Scholar]
- 27.Schonberg DL, Lubelski D, Miller TE, Rich JN. Brain tumor stem cells: Molecular characteristics and their impact on therapy. Mol Aspects Med. 2014;39:82–101. doi: 10.1016/j.mam.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bleau AM, et al. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel D, Yan C, Ross D. NAD(P)H:quinone oxidoreductase 1 (NQO1) in the sensitivity and resistance to antitumor quinones. Biochem Pharmacol. 2012;83:1033–1040. doi: 10.1016/j.bcp.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Y, et al. NQO1 overexpression is associated with poor prognosis in squamous cell carcinoma of the uterine cervix. BMC Cancer. 2014;14:414. doi: 10.1186/1471-2407-14-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inohara N, del Peso L, Koseki T, Chen S, Núñez G. RICK, a novel protein kinase containing a caspase recruitment domain, interacts with CLARP and regulates CD95-mediated apoptosis. J Biol Chem. 1998;273:12296–12300. doi: 10.1074/jbc.273.20.12296. [DOI] [PubMed] [Google Scholar]
- 32.Bertrand MJ, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 33.Bertrand MJ, et al. cIAP1/2 are direct E3 ligases conjugating diverse types of ubiquitin chains to receptor interacting proteins kinases 1 to 4 (RIP1-4) PLoS One. 2011;6:e22356. doi: 10.1371/journal.pone.0022356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott DW, et al. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hasegawa M, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vandenabeele P, Bertrand MJM. The role of the IAP E3 ubiquitin ligases in regulating pattern-recognition receptor signalling. Nat Rev Immunol. 2012;12:833–844. doi: 10.1038/nri3325. [DOI] [PubMed] [Google Scholar]
- 37.Bertrand MJ, et al. Cellular inhibitors of apoptosis cIAP1 and cIAP2 are required for innate immunity signaling by the pattern recognition receptors NOD1 and NOD2. Immunity. 2009;30:789–801. doi: 10.1016/j.immuni.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 38.Estornes Y, et al. dsRNA induces apoptosis through an atypical death complex associating TLR3 to caspase-8. Cell Death Differ. 2012;19:1482–1494. doi: 10.1038/cdd.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nathanson DA, et al. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarkaria JN, et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- 41.Sporn MB, Liby KT. NRF2 and cancer: The good, the bad and the importance of context. Nat Rev Cancer. 2012;12:564–571. doi: 10.1038/nrc3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang WH, et al. Fundamental role of the Rip2/caspase-1 pathway in hypoxia and ischemia-induced neuronal cell death. Proc Natl Acad Sci USA. 2003;100:16012–16017. doi: 10.1073/pnas.2534856100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozawa T, James CD. Establishing intracranial brain tumor xenografts with subsequent analysis of tumor growth and response to therapy using bioluminescence imaging. J Vis Exp. 2010:1986. doi: 10.3791/1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.