Significance
Taking advantage of two markers critically associated with ATP metabolism, CD39 ectonucleotidase and the adenosine deaminase binding factor CD26, we found that natural regulatory T cells (nTregs) are heterogenous, composed of five major structurally and genetically distinct cell subsets, each representing a stage of nTreg maturation. Three major outcomes are foreseeable from these studies: (i) immunologically, microenvironmental factors dictate nTreg developmental evolution and govern their distinct functional capacity; (ii) clinically, the CD39/CD26 profile is a useful blood biomarker, as illustrated for dermatomyositis, rheumatoid arthritis, and acute myelogenous leukemia; and (iii) therapeutically, FOXP3 Tregs transdetermined from TH0 cells are an optimal source of T cells for nTreg-based adoptive immunotherapy because they do not produce IL-17.
Keywords: nTregs, FOXP3 regulatory transcript, CD39 regulatory receptor, adenosine deaminase-binding CD26, microenvironmental cytokines
Abstract
Natural regulatory T cells (nTregs) ensure the control of self-tolerance and are currently used in clinical trials to alleviate autoimmune diseases and graft-versus-host disease after hematopoietic stem cell transfer. Based on CD39/CD26 markers, blood nTreg analysis revealed the presence of five different cell subsets, each representing a distinct stage of maturation. Ex vivo added microenvironmental factors, including IL-2, TGFβ, and PGE2, direct the conversion from naive precursor to immature memory and finally from immature to mature memory cells, the latest being a no-return stage. Phenotypic and genetic characteristics of the subsets illustrate the structural parental maturation between subsets, which further correlates with the expression of regulatory factors. Regarding nTreg functional plasticity, both maturation stage and microenvironmental cytokines condition nTreg activities, which include blockade of autoreactive immune cells by cell–cell contact, Th17 and IL-10 Tr1-like activities, or activation of TCR-stimulating dendritic cell tolerization. Importantly, blood nTreg CD39/CD26 profile remained constant over a 2-y period in healthy persons but varied from person to person. Preliminary data on patients with autoimmune diseases or acute myelogenous leukemia illustrate the potential use of the nTreg CD39/CD26 profile as a blood biomarker to monitor chronic inflammatory diseases. Finally, we confirmed that naive conventional CD4 T cells, TCR-stimulated under a tolerogenic conditioned medium, could be ex vivo reprogrammed to FOXP3 lineage Tregs, and further found that these cells were exclusively committed to suppressive function under all microenvironmental contexts.
Immunosuppressive T cells contribute to multiple functions, such as tolerance to self-antigens (Ags) and to foreign Ags generated during pregnancy and food digestion (1). These vital suppressive activities are carried out by different CD4 and CD8 regulatory T cell types (2–5) through an apparent redundancy of regulatory mechanisms (6). Dysfunction of regulatory T cells (Tregs) leads to severe chronic pathologies, including autoimmune diseases (2), viral infections (7), cancers (8), allergies (9), and graft-versus-host disease (GVHD) (10).
Among suppressive T cell populations, three critical conventional regulatory T cell types have been structurally and functionally well identified: type 1 T regulatory (Tr1) cells, suppressive Ag-specific HLA-E–restricted CD8 T cells, and CD4 natural Tregs (nTregs). Tr1 cells are characterized by their release of IL-10 (IL-10 Tr1) to control overinflammation/danger during adaptive immune reactions (11). Given that these cells originate from different activated T cell types, including Th1, Th2, Th17, Treg, and CD8 T cells, IL-10 Tr1 cells should be considered not as a specific T cell lineage but rather as activated T cells skewing their functional differentiation under overinflammation/danger to a reprogrammed suppressive Tr1 function, thereby preventing immunopathogenesis (12). The suppressive Ag-specific HLA-E–restricted CD8+ T cells, which lyse by cell contact activated CD4+ T cell targets expressing the stimulating peptide Ag–HLA-E complex (13), express CD44, CD122, and killer cell immunoglobulin-like receptors.
CD4 nTregs, first identified by Sakaguchi et al. in 1995 (2), were initially characterized by their thymic developmental origin, their CD25 high phenotype, and, after TCR stimulation, their cell-cell contact-mediated suppressive activity toward activated immune cells, including autoreactive ones, thus implicating them in the control of self-tolerance. Since then, numerous reports have extended our basic knowledge of these cells, showing that human nTregs are characterized by the high mean fluorescence intensity (MFI) expression of the master FOXP3 transcript (14), which also may be expressed by activated CD4+ (15) and CD8+ (16) T cells. Further work has also shown that nTregs produce not only anti-inflammatory IL-10 (11), but also its proinflammatory IL-17 counterpart (17, 18). Remarkably, these FOXP3 lineage Tregs can be induced in culture by appropriate stimulation of naive CD45RA+CD4+ T cells (TH0) in the presence of IL-2 and TGFβ (iTregs) (19, 20). Most importantly, the regulatory function of these cells in the control of self-tolerance has prompted experiments and clinical trials based on the transfer of nTreg populations to treat GVHD or autoimmune pathologies (21).
The results concerning the adoptive transfer of crude nTreg preparations, although encouraging, remain not fully satisfactory, and treatment failures have been attributed to inflammatory complications triggered by the capacity of nTregs to behave as proinflammatory Th17 cells (17, 18). The need to significantly improve nTreg preparations for a more effective therapeutic benefit was further confirmed by two sets of experimental data pointing to a functional heterogeneity of the FOXP3 Treg populations currently administered in adoptive immune therapy. On one hand, in a study on nTregs carried out in healthy and lupus-affected individuals (12), we reported that the suppressive activities of human blood nTregs, as tested by the current suppressive assay (22) (ie, following stimulation by polyclonal anti-CD3 Abs), varied according to the stimulatory environmental conditions. When the assay was performed in a culture medium corresponding to an in vivo quiescent stromal tissue (steady state), nTreg cells inhibited activation and proliferation of their cocultured autologous cell targets by a cell-cell contact-mediated mechanism, while in culture conditions mimicking the in vivo inflammation associated with adaptive reactions to pathogenic antigens, this cell–cell contact-mediated suppressive effect was inhibited, but nTregs unexpectedly released IL-10 (12). Furthermore, in an overinflammatory conditioned medium comprising inflammatory proteins, including C3b and C5b complement proteins, these cells acted as IL-10 Tr1-like cells, turning off proliferation of activated T cells (12). On the other hand, we considered the critical impact of adenosine (ADO) on nTreg immunosuppressive mechanisms (23, 24). Given that ectonucleotidase CD39 and adenosine deaminase (ADA) are two enzymes involved in the metabolism of extracellular ATP, the major source of cell energy that further generates ADO, we analyzed the expression of CD39 and CD26 on nTregs, with CD26 serving as a surrogate marker of ADA (25). We found that only a percentage of nTregs in human peripheral blood mononuclear cells (PBMCs) express either or both of these markers.
These data prompted the present research, carried out on human blood FOXP3 Tregs ex vivo at a resting stage and following polyclonal or antigen-specific stimulation occurring under the various microenvironmental conditions prevailing in vivo. The microenvironmental context of TCR stimulation includes (i) quiescent context (steady state), mimicked ex vivo by a weak CD3 TCR stimulation; (ii) acute inflammation ex vivo, promoted by strong CD3 and CD28 stimulation in the presence of IL-2 (26); (iii) proinflammation triggered by an additional stimulation with IL-1β, IL-6, and IL-23 cytokines (27); (iv) overinflammation/danger occurring ex vivo in presence of additional inflammatory complement protein (C3b and C5b) signals to the CD46 receptor (28); and (v) a tolerogenic context triggered in vitro by additional TGFβ, PGE2, and rapamycin (rapa) (19).
The findings that we report here show that, based on CD39/CD26 expression, human blood nTregs are divided into distinct subtypes, each representing a phase of nTreg life cycle, and that microenvironmental context governs the maturation of these cells. This nTreg CD39/CD26 profile is stable over time for an individual but variable interindividually, making it as a valuable biomarker to consider in chronic inflammatory diseases. We also highlight the relationship linking nTregs and IL-10 Tr1 CD4+ T cells. In contrast to nTregs, ex vivo Tregs expressing FOXP3, reprogrammed (transdetermined) from naive conventional CD4 T (TH0) cells (iTregs) (29), are seen to exclusively exert a regulatory activity independent of the microenvironmental context.
Results
CD39 CD26 Markers Help Delineate Structural Phenotypic and Genetic Heterogeneity of nTregs in Human Blood.
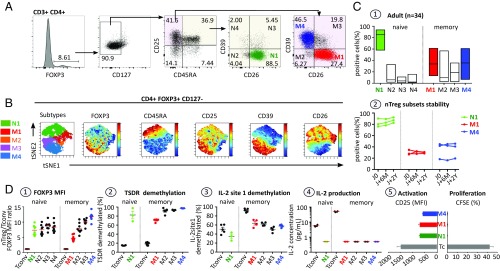
At a resting stage, nTregs developed in the thymus are currently characterized by their high expression levels of CD25, low expression levels of CD127, and expression of the master transcript FOXP3 (14) and demethylated TSDR (30) and are not able to synthesize IL-2, rendering them functionally anergic (12, 31). In the present study, nTregs were identified in healthy human blood using intracellular FOXP3. The cells were then analyzed for the expression of CD127, CD25, and CD45RA in combination with the functional markers CD26 and CD39 (Fig. 1A). On viSNE analysis, CD4+FOXP3+CD127−/low nTregs exhibited various expression levels of CD45RA, CD25, CD39, and CD26 markers, supporting nTreg heterogeneity (Fig. 1B). Furthermore, based on CD39 and CD26 expression, the FOXP3+CD127−/lowCD25+ nTreg population comprises five major subsets in healthy adult PBMCs: naive CD45RA+CD26+CD39− (N1) and memory RA-CD26+CD39− (M1), RA-CD26−CD39− (M2), RACD26+CD39+ (M3), and RA-CD26−CD39+ (M4) (Fig. 1 B and C1). All five subsets exhibited high FOXP3 expression (Fig. 1 D1), associated with a very high demethylation level of TSDR (Fig. 1 D2), and, in contrast to memory conventional T cells (Tconvs), contained a relatively low demethylation level of CpG site 1 in the IL-2 promoter, essential for inducing IL-2 production on TCR activation (Fig. 1 D3) (26). Moreover, nTreg subset cells were functionally in an anergic state, given that after CD3 stimulation, they were unable to synthesize IL-2 (Fig. 1 D4), lost their CD25 activating marker, and did not proliferate (Fig. 1 D5). These characteristics include these subsets in the nTreg lineage definition. Importantly, whereas nTreg subset distribution is stable in each adult individual for a >2-y period (Fig. 1 C2), it varies interindividually (Fig. 1 C1). It also appears that nTreg distribution is not sex-dependent (SI Appendix, Fig. S1, 1) but does vary slightly with age from the newborn (SI Appendix, Fig. S1, 2) to the elderly (SI Appendix, Fig. S1, 3). Newborn blood was enriched in nTregs with a naive phenotype (75.31%), a frequency decreasing to 32.04% in adults and 18.77% in the elderly (SI Appendix, Fig. S1, 4). Conversely, the frequency of memory nTregs increased steadily with age, from 24.69% to 67.96–81.23% (SI Appendix, Fig. S1, 5).
Fig. 1.
FOXP3 nTreg heterogeneity in healthy human PBMCs. (A) Flow cytometry gating strategy for identifying the major nTreg subsets in human PBMCs based on FOXP3, CD127, CD25, CD45RA, CD26, and CD39 markers. (B) After initial gating on the CD3+CD4+FOXP3+CD127− nTreg population, the gated cells were clustered using viSNE (Cytobank). Cells are color-coded according to the expression level of FOXP3, CD45RA, CD25, CD39, and CD26 markers. (C1) Boxplots illustrating the distribution of the four nTreg subsets based on the expression of CD39 and CD26 in naive and memory nTreg compartments. (C2) Longitudinal analysis of nTreg subset frequencies in three individuals for a >2-y period. (D) Phenotypic, epigenetic, and physiological characteristics of FACS-sorted nTreg subsets. (D1) Summary plot of the MFI ratio of FOXP3 expression on Treg subsets to Tconvs. (D2 and D3) Scatterplot indicating FOXP3-TSDR (D2) and IL-2 CpG site 1 demethylation status (D3) of the five major FACS-sorted nTreg subsets (N1, M1–M4) and the two conventional T cells (naive and memory) as assessed by bisulfite pyrosequencing. Carboxyfluorescein succinimidyl ester (CFSE)-labeled nTreg subsets (N1, M1, and M4) and Tconvs (4 × 104 per well) were stimulated with a low dose of plate-bound anti-CD3 mAb (pbαCD3; 0.5 µg/mL) in the presence of irradiated feeder. (D4) IL-2 concentration in culture supernatant from 40 h-stimulated Tconv cells and nTreg subsets as measured by ELISA. (D5) T cell activation status and T cell proliferation were evaluated by the MFI of CD25 and the CFSE dilution assay, respectively. Data are expressed as mean ± SEM.
Of importance, the FOXP3 approach enabled us to identify, in conventional CD4+ T cells, three other FOXP3 lineage subsets present at low frequency in healthy PBMCs (SI Appendix, Table S1). The first of these is a subset bearing the CD8 marker (0.12% of CD4+ T cells), and the second is a subset that exhibits an abnormally low CD25 expression level (0.5% of CD4+ T cells). Both of these nTreg subsets exhibit otherwise similar nTreg phenotypic and genetic characteristics. The third subset, corresponding to 1.2% of the CD4+ T cells, is characterized by expression of CD127 marker and differs genetically from nTregs by low demethylated levels of TSDR associated with a lower level of FOXP3 expression than seen in nTregs. This was anticipated, given that FOXP3 is a regulatory transcript known to be expressed by activated memory Tconvs following their TCR stimulation (15).
In summary, based on CD39/CD26 markers, the human blood nTreg population can be subdivided into five major subsets in which expression of FOXP3 is a necessary but not sufficient characteristic to define nTregs. Moreover, FOXP3 regulatory transcripts also may be expressed by memory CD4+CD127+ Tconvs, although at lower levels than by nTregs, in healthy human blood.
Each nTreg Subset Corresponds to a Structurally Well-Defined Stage of Maturation.
The microenvironmental context of TCR stimulation governs nTreg subset parental maturation.
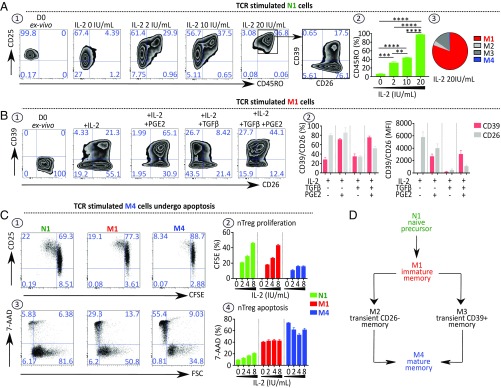
nTreg subsets from PBMCs were sorted and cultured separately in different conditioned media. As shown in Fig. 2 A1, naive N1 cells, cultured in the presence of different IL-2 doses, express the CD25 marker, lose their anergic state, and convert to memory cells (Fig. 2 A2) exhibiting a CD26+CD39− M1 profile (Fig. 2 A3). When TCR-stimulated and cultured with IL-2, memory M1 cells convert into M4 cells in the presence of TGFβ plus PGE2. PGE2 enhances the CD39 marker (25–67%), while TGFβ favors the loss of cell surface CD26 marker (72–30%) (Fig. 2 B1 and B2). Fig. 2C shows that, following TCR stimulation in the presence of IL-2, M4 cells, being at an advanced stage of differentiation, proliferate less (Fig. 2 C1 and C2) and are more susceptible to apoptosis (Fig. 2 C3 and C4) compared with N1 and M1 cells after a 4 d-culture.
Fig. 2.
Microenvironmental context of TCR stimulation governs nTreg subset parental maturation. (A) N1 cells convert into M1 cells after ex vivo stimulation. (A1) Representative dot plots showing expression of CD25, CD45RO, CD26, and CD39 by N1 cells stimulated for 4 d, as described in Fig. 1 D5, with increasing doses of IL-2. (A2) Histograms indicating the percentage of CD45RO expressed by stimulated N1 cells (n = 3). (A3) Pie chart indicating the frequency of each memory nTreg subset in the 4-d culture of N1 cells stimulated with 20 IU/mL IL-2 (n = 4). (B) M1 cells convert into M4 cells ex vivo when stimulated as above in the presence of IL-2, TGFβ, and PGE2. (B1) Representative dot plots showing CD26 and CD39 expression by M1 cells stimulated in the presence of IL-2 with or without PGE2 (1 μM) and with or without TGFβ (5 ng/mL). (B2) Histograms indicating the percentage of stimulated M1 cells expressing CD26 and CD39 and their MFI (n = 3). (C) M4 cells represent a no-return differentiation stage. CFSE-labeled nTreg subsets were stimulated as indicated above. (C1) Representative dot plots depicting CD25 expression and CFSE dilution of 4-d cultured nTreg subsets. (C2) Histograms indicating the percentage of proliferating cells in stimulated cell cultures (n = 4). (C3 and C4) Representative dot plots (C3) and histograms (C4) showing the percentage of 7-AAD+ stimulated nTreg subsets (n = 4). (D) Diagram of the parental maturation process of the nTreg population. Data are expressed as mean ± SEM. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Fig. 2D briefly schematizes the parental maturation process of the nTreg population in healthy individuals. Naive precursor (N1) subset cells progress through immature memory (M1) and then to mature memory (M4) via either transient CD26− (M2) or CD39+ (M3) subsets.
The maturation of nTreg subsets is correlated with expression of regulatory markers.
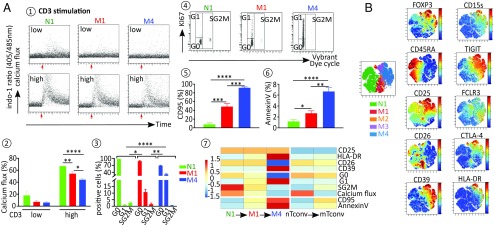
To explore the parental maturation link between the three N1, M1, and M4 nTregs, patterns of calcium responses, cell cycle status activation, and maturation markers were investigated in each subset and compared with expression of regulatory markers. Calcium influx analysis showed that intracellular calcium response to low or high anti-CD3 stimulation is greater in N1 precursors than in M1 and M4 subsets (Fig. 3 A1 and A2). Cell cycle profile analysis by KI-67/Vybrant staining also showed that the N1 subset contains a higher percentage of cells in G0 and S/G2/M phases compared with M1 and M4 subsets. In contrast, the percentage of cells in G1 phase gradually increases from the N1 subset to the M4 subset (Fig. 3 A3 and A4). The expression analysis of nTreg maturation markers by flow cytometry showed that CD25 and HLA-DR (Fig. 3B) are more highly expressed in the M4 subset than in the M1 and N1 subsets. Moreover, in contrast to N1 precursors, memory nTreg subsets exhibit high levels of annexin V, CD95, characterizing a senescent stage mature M4 subset expressing relatively greater amounts of these markers than immature M1 cells (Fig. 3 A5 and A6). The heat map Fig. 3 A7 shows the variation in these nTreg maturation markers. Furthermore, FOXP3 regulatory transcript, TSDR demethylation levels, and regulatory markers, particularly CD15S, TIGIT, CTLA-4, and GARP, are more intensely marked in mature memory M4 subsets (Fig. 3B and SI Appendix, Fig. S2). Variations in these regulatory markers are illustrated in a heat map in SI Appendix, Fig. S2.
Fig. 3.
Expressions of regulatory markers are correlated with nTreg cell cycle evolution. (A) Flow cytometry analysis of biological characteristics of nTreg subsets. Calcium mobilization induced in nTreg subsets after TCR stimulation with either low (0.5 µg/mL) or high (5 µg/mL) dose of anti-CD3 Ab was measured as the ratio of indo violet to indo blue. (A1 and A2) Representative dot plot (A1) and mean ± SEM (A2) of the percentage of responding cells in each nTreg subset. (A3 and A4) nTreg subset cell cycle status (G0/G1/SG2M) as assessed by the costaining of KI-67 and DNA. Mean ± SEM (A3) and representative dot plots (A4). (A5 and A6) Expression of CD95 (A5) and Annexin V (A6) in each nTreg subset expressed as mean % ± SEM. (A7) Heatmap representation of the foregoing data. The columns represent the cell subsets N1, M1, M4, nTconv, and mTconv. The color of each row represents the fold change of the expression of the marker compared with the mean expression level; the degree of change is shown in the scale. (B) After initial gating on the CD3+CD4+FOXP3+CD127− nTreg population, the gated cells were clustered using viSNE. Cells are color-coded according to the expression levels of FOXP3, CD45RA, CD25, CD39, CD26, CD15s, TIGIT, FCLR3, CTLA4, and HLA-DR markers. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Collectively, these data suggest that nTreg maturation from precursor naive N1 to immature memory M1 and then to mature memory M4 subsets is associated with increased expression of specific regulatory markers.
RNA Sequencing Analysis Confirmed both nTreg Subsets Heterogeneity and Parental Maturation.
Heterogeneity of nTreg populations.
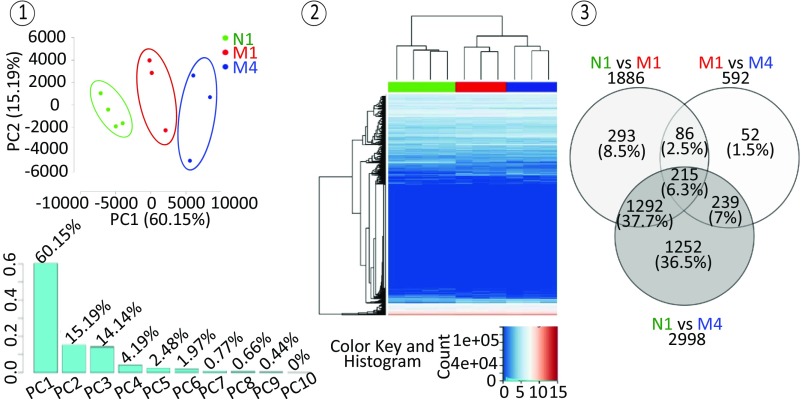
To characterize the N1, M1, and M4 nTreg populations at a transcriptomic level, RNA sequencing experiments were performed on 10 nTreg total RNA samples (four N1, three M1 and three M4), which generated RNA expression data of 25,313 genes in transcripts per kilobase million (TPM). Principal component analysis performed on these data revealed a first component explaining 60.15% of the total variance of the transcriptome among the samples, which is sufficient to separate them into their three respective groups of N1, M1, and M4 (Fig. 4, 1). These results were confirmed by unsupervised hierarchical clustering analysis of RNA sequencing data, which showed the clustering of the samples in three distinct groups corresponding to N1, M1, and M4, with M1 and M4 samples showing transcriptomic profiles closer to each other than to those of N1 samples (Fig. 4, 2). Further differential expression analysis among the three groups revealed 1,886, 2,998, and 592 differentially expressed genes with a greater than twofold change (Benjamini–Hochberg-adjusted P < 0.05) between N1 and M1, between N1 and M4, and between M1 and M4, respectively, including 215 differentially expressed genes between the three groups (Fig. 4, 3). Thus, the RNA sequencing results demonstrated the transcriptomic heterogeneity of nTregs.
Fig. 4.
RNA sequencing analysis confirmed both nTreg subset heterogeneity and parental maturation. N1, M1, and M4 nTreg populations show distinct transcriptomic profiles. (1) Principal component analysis performed on whole transcriptome data of 10 nTreg samples obtained by RNA sequencing experiments including 25,313 genes in TPM. (2) A 2D heatmap representation of unsupervised hierarchical clustering of nTreg whole transcriptome data using log2-transformed TPM data. N1, M1, and M4 samples are labeled in green, red, and blue, respectively. (3) Venn diagram of differentially expressed genes with a greater than twofold change (Benjamini–Hochberg-adjusted P < 0.05) among N1, M1, and M4 nTreg populations.
Parental maturation of nTreg subsets.
The RNA sequencing supervised analysis confirms that each nTreg subset tested represents a maturation stage in nTreg life, even though in resting nTreg cells, expression levels of mRNA and corresponding protein are not systematically parallel (32). The analysis focused on the mRNA expression of markers each characterizing a different phase of a T cell life. As shown in SI Appendix, Fig. S3, N1 to M1 to M4 maturation is reflected in mRNA expression levels of markers corresponding to cell activation, proliferation, functional regulatory differentiation, and senescence (SI Appendix, Table S2). Interestingly, in this table of 40 relevant markers, KI-67 was associated with cell cycle phases G1, S, G2, and M but was blocked in phase G0 (33) and was included in the activation phase but not in the IL-2–dependent proliferation phase, given nTregs’ intrinsic inability to produce IL-2 required for proliferation. Owing to the low number of tested individuals (three per subset), a trend, but not a significant difference, in the mRNA expression level of markers was most often observed. However, individuals of each subset most often exhibited the same mRNA maturation profile, although each at a variable magnitude.
nTreg Subset Physiology Is Critically Conditioned by the Microenvironmental Context of Their Stimulation.
The microenvironmental conditions dictate the nTreg subset functional differentiation as assessed by cytokine production.
We explored the functional capacity of each nTreg subset by measuring their ability to produce the proinflammatory and anti-inflammatory cytokines IL-17 and IL-10 at both the mRNA and protein levels. At a resting stage, after stimulation with PMA/ionomycin, mature memory M4 cells produce very limited amounts of IL-17 and IL-10, while both naive precursor N1 and immature memory M1 cells have the capacity to produce the two cytokines (SI Appendix, Fig. S4A). Of interest, N1 and M1 cells express increased levels of IL-17 (transcript and protein) compared with nTconvs and mTconvs. Moreover, while the production of IL-10 mRNA is similar in all subset cells, M1 cells display increased IL-10 protein levels.
In addition, IL-17 and IL-10 production were also measured after in vitro activation both intracellularly by flow cytometry (SI Appendix, Fig. S4B) and in culture supernatants by ELISA (SI Appendix, Fig. S4C). Under steady state, mimicked in culture by a TCR stimulation (anti-CD3 mAb) in the presence of minimal amounts of IL-2, M1 subset cells expressed and released minimal amounts of IL-10. In this setting, neither N1 nor M4 subset cells produced cytokines. Under acute inflammation, mimicked in vitro by anti-CD3/CD28 stimulation in the presence of IL-2, immature memory M1 subset cells produced the two tested cytokines. Of note, in proinflammatory conditioned medium, mimicked in vitro by the additional presence of IL-1β and IL-6, only the M1 subset expressed and secreted IL-17 at levels comparable to mTconvs. Under overinflammatory/danger conditions, mimicked in vitro by adding a stimulation of CD46 (the complement C3b/C4b natural receptor) (14, 28), the M1 subset displayed and released marked amounts of IL-10 and also, but at a lower level, IL-17. In summary, ex vivo cytokine production by nTregs is governed by their microenvironmental medium and to a lesser degree by their maturational stage. Among nTregs, only the M1 immature subset secretes notable amounts of cytokines, comparable to those secreted by memory CD4+ T helper cells, with IL-17 highly produced under proinflammation conditions and IL-10 highly produced in an overinflammation/danger context.
The microenvironment of TCR stimulation conditions nTreg regulatory activity.
Ex vivo polyclonal TCR stimulation.
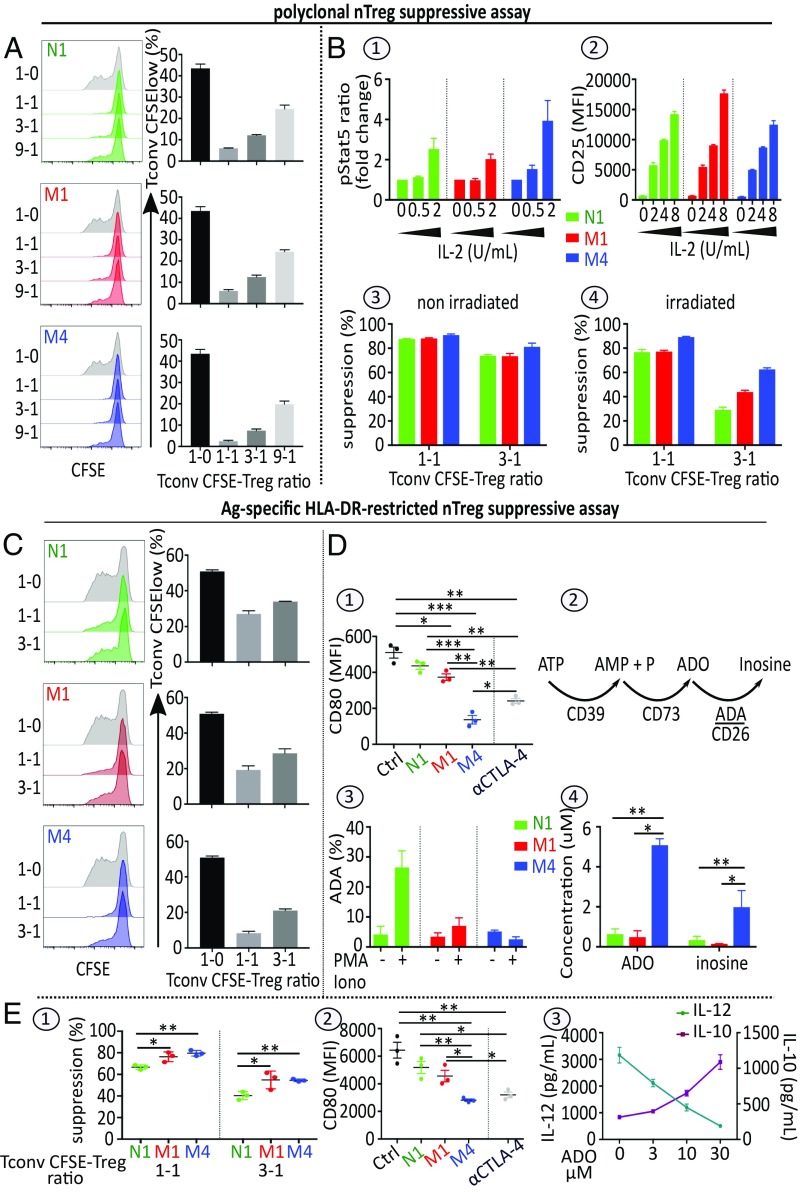
Under steady state, following CD3 stimulation, the three subsets exert their suppressive function by cell-cell contact, as measured by the current nTreg suppressive assay (22). This regulatory activity, which is slightly higher in the M4 subset, is induced chiefly by IL-2 juxtacrine starvation of activated targets (Fig. 5A). On one hand, the M4 subset, exhibiting high level of CD25 (Fig. 1B), efficiently consumes IL-2 supplied by stimulated target cells even at low levels, as assessed by the STAT5 phosphorylation assay (Fig. 5 B1). On the other hand, target cell IL-2 deprivation induced by the N1 and M1 subsets, which display intermediate levels of CD25 at resting stage (Fig. 1B), results from their proliferation in the presence of IL-2. In this context, the N1 and M1 subsets acquire greater CD25 expression (Fig. 5 B2) through positive feedback, thus sequestering more IL-2.
Fig. 5.
Microenvironmental context of TCR stimulation conditions nTreg regulatory activity. (A) nTreg subsets suppressive activity using the standard suppressive assay. 4 × 104 CFSE-labeled Tconvs (TconvCFSE) stimulated as indicated in Fig. 1 D5 were cocultured with nTreg subsets N1, M1, or M4 at different ratios. Proliferation of TconvCFSE was evaluated by the CFSE dilution assay. Representative FACS histograms and mean ± SEM in percentage of TconvCFSE low are shown. (B) Role of IL-2 in nTreg subset suppressive signaling pathways. (B1) pSTAT5 responses in nTreg subsets stimulated with the indicated amount of IL-2 for 15 min. Mean ± SEM of pSTAT5 ratio (MFI at 15 min/MFI at baseline) is shown (n = 3). (B2) CD25 expression in nTreg subsets stimulated as indicated in Fig. 1 D5 in the presence of various amounts of IL-2 for 4 d. Mean ± SEM of MFI values for CD25 are indicated. (B3 and B4) CD3-stimulated nTreg subsets were irradiated. Untreated (B3) and treated (B4) nTregs were then cocultured with Tconv CFSE stimulated as described in Fig. 5A. Mean ± SEM of percentages of suppression are shown (n = 3). (C) nTreg subset suppressive activity following HLA-DR–specific DC stimulation. nTreg N1, M1, or M4 subsets stimulated by iDCs were cocultured with preactivated TconvCFSE at different ratios. The proliferation of TconvCFSE was evaluated by a CFSE dilution assay. Representative FACS histograms and mean ± SEM percentage of Tconv CFSE low are shown. (D) Roles of CTLA-4 and ADO in the DC-nTreg subset interplay. (D1) CD80 expression levels on iDCs cultured in the absence [control (Ctrl)] or presence of either IL-2/CD3/CD28–prestimulated nTreg subsets or αCTLA4 Ab 5 μg/mL. Mean ± SEM MFI values for CD80 are shown (n = 3). (D2) Schema of pericellular ATP metabolism. (D3) FACS analysis of ADA expression in nTreg subsets before and after stimulation with PMA/IONO; mean ± SEM percentage of ADA are shown (n = 3). (D4) ADO and inosine production by the three nTreg subsets incubated in the presence of exogenous ATP. Stimulated nTreg subsets were incubated with exogenous ATP (100 μM) for 120 min as described in SI Appendix, Materials and Methods. Mean ± SEM adenosine and inosine levels in supernatant, measured by ultra-high-performance liquid chromatography-coupled high-resolution mass spectrometry are shown. (E1) Suppressive activity of nTregs assessed in mDC-stimulated nTreg-Tconv CFSE cocultures. Mean ± SEM percentage of suppression are shown (n = 3). (E2) CD80 expression on iDCs induced to undergo maturation with LPS in the absence (Ctrl) or presence of either nTreg subsets or 5 μg/mL mAb αCTLA4 for 24 h. Mean ± SEM MFI values for CD80 are shown (n = 3). (E3) Amounts of IL-12 (Left, dark green) and IL-10 (Right, dark pink) in 2-d culture supernatants of mDCs stimulated with different concentrations of ADO measured by ELISA. Mean ± SEM of the cytokine concentrations are shown (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001.
This assumption is further experimentally supported by our findings that N1 and M1 subsets exhibit lower suppressive capacity when irradiated (Fig. 5 B3 and B4), and that cell contact nTreg suppressive function is reduced or lost when the nTreg suppressive assay is performed in the presence of target cells supplying higher doses of IL-2 (Tconv RO+ 25+) (SI Appendix, Fig. S5A and S5B). Of note, the cell–cell contact-suppressive activity of nTregs is neither cell type-specific nor HLA- restricted, as shown by the current polyclonal suppressive assay demonstrating that nTregs block the proliferation of CD8+ T or allogeneic CD4+ T cell targets (SI Appendix, Fig. S5C). Under inflammatory conditions, associated in vivo with adaptive reactions and including exogenous IL-2 production triggered by CD28 costimuli (signal 2) (26), the addition of anti-CD28 Ab to anti-CD3 stimulation in the current nTreg suppressive assay inhibited the cell contact-suppressive capacity of the three nTreg subsets in a dose-dependent manner (SI Appendix, Fig. S6 A1 and A2). Furthermore, when Transwell inserts separating nTreg subsets from preactivated CD4+ T cell targets were used (12), the N1 and M1 subsets, but not the M4 subset, exhibited suppressive capacity (SI Appendix, Fig. S6 B1 and B2). In effect, in an inflammatory context, while M4 subset cells undergo apoptosis (Fig. 2 C3 and C4), N1 and M1 subset cells proliferate, produce suppressive IL-10 cytokine, and, acting as Tr1-like cells, exert an IL-10–mediated inhibition of preactivated CD4 T cell targets (SI Appendix, Fig. S6B2), as has been previously reported for the whole nTreg population (12).
Ag-specific HLA-DR–restricted antigen-presenting cell stimulation prevailing in vivo.
Given that polyclonal TCR stimulation short circuits the antigen-presenting cell (APC) presentation required in vivo, we conducted an nTreg suppressive assay using APCs as stimulators to mimic the situation prevailing in vivo. Under steady state, when using autologous immature dendritic cells (iDCs) as stimulator cells, only mature memory M4 cells exercise a marked dose-dependent antiproliferative effect against preactivated CD4+ conventional targets (Fig. 5C). M4 cells exhibit a greater suppressive capacity than N1 and M1 in this setting, due chiefly to their greater expression of CTLA-4 and CD39 (Fig. 3B). The interaction of CTLA-4 and CD80 on iDCs inhibits DC maturation, resulting in attenuation of APC-induced effector T cell activation function (Fig. 5 D1). Furthermore, the presence of the ectonucleotidase CD39 together with the absence of CD26 and thus the ADA enables the M4 subset to convert extracellular ATP into immunosuppressive ADO and residual inosine (Fig. 5 D2 and D4). Under inflammation, when using allogeneic mature DCs as stimulator cells, the three subsets exert their suppressive activity by distinct regulatory mechanisms (Fig. 5 E1). Mature M4 cells activate the tolerization of TCR-stimulating DCs through their expression of CTLA-4–down-regulating B7 receptors (Fig. 5 E2) and the production of pericellular adenosine, which induces the secretion of high levels of IL-10 and low levels of IL-12 by the DCs (Fig. 5 E3). Immature M1 cells, which produce IL-10 under inflammation, can exert their suppressive activity as IL-10–secreting Tr1-like Tconvs (SI Appendix, Fig. S4), whereas naive precursor N1 cells proliferate and mature to M1 cells (Fig. 2A). In summary, during in vivo adaptive reaction-associated inflammation, all nTreg subsets, and particularly the M4 cells, trigger tolerogenic activation of the stimulating DCs. Such tolerogenic activation is characterized by inversion of the IL-12:IL-10 ratio (Fig. 5 E3). Under inflammation, TCR-stimulated N1 and M1 cells, which proliferate and produce IL-10, also could exert a Tr1-like suppressive activity. Finally, these experiments show that the regulatory capacity of nTregs varies according to both their stage of maturation and the microenvironmental context of their TCR stimulation.
Medical Implications.
The nTreg CD39/CD26 profile provides a blood biomarker for monitoring chronic inflammatory diseases and posthematopoietic stem cell transplantation acute myelogenous leukemia.
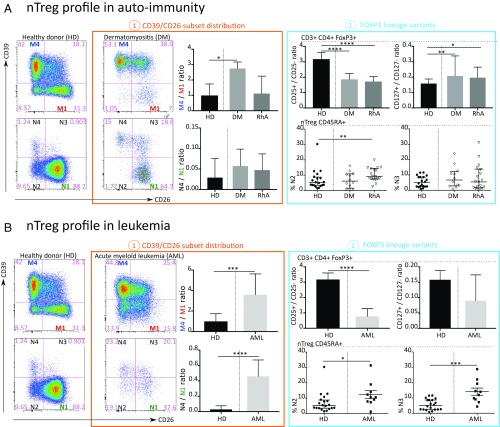
To investigate alterations in blood FOXP3 subpopulations in patients with chronic inflammatory diseases, cryopreserved PBMCs of 12 patients with untreated dermatomyositis (DM), 18 patients with rheumatoid arthritis (RhA) treated with immunosuppressive agents (representing autoimmune diseases associated with autoantibodies and T cell activation), and 10 patients with relapsed acute myelogenous leukemia (AML) after hematopoietic stem cell transplantation (HSCT) were compared with PBMCs from 20 healthy adults (SI Appendix, Table S3). The data are summarized in Fig. 6.
Fig. 6.
Distribution of blood FOXP3+ subpopulations is modified in autoimmunity and cancer. (A) Autoimmunity. (A1) Representative flow cytometry dot plots of CD39/CD26 major subsets and histograms of memory M4/M1 and naive N4/N1 frequency ratio. (A2) FOXP3 lineage variants in nTreg in healthy donors (HDs) compared with DM and RhA. (B) AML. (B1) Representative flow cytometry dot plots of CD39/CD26 major subsets and histograms of memory M4/M1 and naive N4/N1 frequency ratio. (B2) FOXP3 lineage variants in nTreg in HDs compared with AML. Data in histograms and scatterplots are presented as median (interquartile range) in HDs (n = 20) compared with patients with DM (n = 12), RhA (n = 18), and AML (n = 10). P values were calculated using the Wilcoxon Mann-Whitney U test. P values < 0.05 were considered significant. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. The initial CD3+CD4+ FOXP3+ was derived from a lymphocyte gate (defined on forward and side scatter) followed by single-cell discrimination, dead cell exclusion dye, and exclusion of iNKT and γδ T cells.
Autoimmunity.
Although there was no difference in the frequency of CD4+ T cells and FOXP3+ cells within CD4+ T cells (SI Appendix, Fig. S7A), we observed great changes within the FOXP3+ population. We questioned whether the major CD39/CD26 subset distribution of blood nTregs represents a novel pathogenic biomarker for monitoring patients with chronic inflammatory diseases. In the patients with DM, we observed an elevation of the memory M4/M1 ratio (Fig. 6 A1) in relation to accumulation of the M4-differentiated nTreg population. Regarding FOXP3+ lineage variants, the analysis revealed a decreased CD25+/CD25− ratio (Fig. 6 A2) and a significantly increased CD127+/CD127− ratio (Fig. 6 A2). In the patients with RhA, we noted a marked increase in the naive CD39−CD26− N2 subpopulation (Fig. 6 A2).
AML.
We observed significant decreases in CD4+ T cells and of CD45RA+ cells in both Tconvs and nTregs after HSCT, as described previously (34). There was no difference in the frequency of FOXP3+ cells within CD4+ T cells (SI Appendix, Fig. S7B).
Concerning the CD39/CD26 subset distribution, patients with AML after HSCT exhibited highly elevated memory M4/M1 and naive N4/N1 ratios (Fig. 6 B1) in relation to an accumulation of the M4-differentiated nTreg population and abnormal CD39+ N4 naive Tregs, respectively (Fig. 6 B2). Regarding FOXP3+ Treg variants, our analysis revealed a decreased CD25+/CD25− ratio (Fig. 6 B2) indicating elevation of a CD25− abnormal variant. We also observed an accumulation of the naive CD39−CD26− N2 and CD39+CD26+ N3 subpopulations (Fig. 6 B2).
In summary, our blood CD39/CD26 profile biomarkers study illustrates that within the blood FOXP3+ T cell population, chronic inflammatory diseases (DM and AML) are associated with skewing toward high expression of CD39+ markers (mainly subset M4). We also observed an abnormal accumulation of FOXP3+CD25− cells and FOXP3+CD127+ cells. Moreover, AML and RhA were associated with elevations of abnormal naive CD39/CD26 FOXP3 subsets.
Microenvironmental factors govern the transdetermination of TH0 naive CD4+ T cells into FOXP3+ iTregs.
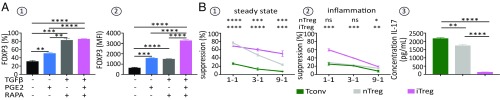
Chen et al. (19) have reported that TGFβ, in combination with TCR stimulation and IL-2, transdifferentiates naive TH0 CD4+ T cells into iTregs. To optimize the generation and expansion of iTregs, we isolated naive CD4+ T cells from human cord blood mononuclear cells and stimulated them in presence of TGFβ and IL-2, together with rapa or rapa plus PGE2. Cells treated with IL-2 only or with IL-2 and PGE2 served as controls. While the two polarizing media induced equally high fractions of FOXP3-expressing cells (Fig. 7 A1), the FOXP3 MFI was highest when PGE2 was added in addition to rapa in the culture media (Fig. 7 A2). Importantly, we further showed that (i) iTregs, generated and expanded for 21 d in the presence of rapa and PGE2, display greater suppressive activity than fresh nTregs (Fig. 7 B1), and (ii) these iTregs still maintained their suppressive activity when the functional suppressive assays are performed in presence of a highly inflammatory medium containing IL-2, IL-1β, IL-6, IL-21, and IL-23 cytokines, while fresh nTregs lose their regulatory capacity under these culture conditions of stimulation (Fig. 7 B2). This effect can be accounted for by the fact that, in contrast to iTregs, TCR-stimulated fresh nTregs in the presence of highly inflammatory medium secrete IL-17 (Fig. 7 B3).
Fig. 7.
The tolerogenic microenvironment dictates the ex vivo induction of FOXP3 iTregs by CD4-naive TH0 cells transdetermination. (A) Analysis of FOXP3+ expression in iTregs generated ex vivo from polyclonally stimulated naive CD4+ T cells with different nTreg polarizing media. Naive CD4+ T cells were stimulated for 12 d with plate-bound anti-CD3 (4 µg/mL) in the presence of IL-2 (100 IU/mL). Where indicated, TGFβ (5 ng/mL), rapa (10 nM), and PGE2 (1 µM) were added. (A1 and A2) Frequency (A1) and expression level (evaluated by MFI) (A2) of FOXP3 in CD4+ T cell culture. (B) Ex vivo suppressive capacity of human Tregs generated with the polarizing medium containing TGFβ (5 ng/mL), rapa (10 nM), and PGE2 (1 µM). (B1 and B2) The suppressive capacity of ex vivo-generated Tregs was evaluated in quiescent (B1) and inflammatory (B2) conditions with the standard polyclonal nTreg assay. CFSE-labeled Tconvs were cocultured with ex vivo-generated Tregs at different ratios. The percent inhibition of TconvCFSE proliferation by Tregs is depicted. Fresh nTregs and Tconvs served as controls. (B3) IL-17 production by ex vivo-generated iTregs measured in supernatant culture by ELISA. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Discussion
Although there have been more than 50,000 publications on nTregs since their discovery in 1995 (2), our basic knowledge of these cells remains ill-defined and even confusing. Given their medical implication in autoimmune diseases (35) and their administration by adoptive transfer to treat these diseases or GVHD (21, 36), a better understanding of these cells is an urgent need. The present study has attempted to clarify some critical issues concerning nTregs.
The first question is which Tregs belong to the nTregs lineage. These cells, initially defined by their CD4+CD25+ phenotype (2), their natural thymic developmental origin, and their necessary but not sufficient FOXP3 transcript expression, were further structurally well identified at a resting stage in human PBMCs by their demethylated FOXP3-TSDR region (30) and lack of IL-2 production (31). These typical nTregs are to be distinguished from activated memory T cells originating from different CD4+ or CD8+ subtypes, which in a tolerogenic microenvironment transdifferentiate to express the regulatory FOXP3 transcript (15).
In our phenotypic study based on CD39/CD26 profile, we found that the nTreg population was heterogeneous and composed of five major subsets (N1, M1, M4, and two transient subsets, M2 and M3) (Fig. 1). nTreg heterogeneity was further confirmed by RNA sequencing data (Fig. 4). These subsets represent different stages of FOXP3 Treg maturation, as revealed by in vitro experiments performed on separate subsets stimulated under different culture media, critically including IL-2 and/or PGE2 and/or TGFβ (Fig. 2). The parental maturation relationship between nTreg subsets was further confirmed by structural data observed in (i) naive precursor N1 cells expressing early life cell cycle markers (Fig. 3A) and a relatively lower level of FOXP3-TSDR demethylation and (ii) memory mature M4 cells expressing high levels of activation markers (including CD25 and HLA-DR) (Fig. 3B) together with high levels of both FOXP3 regulatory transcript and TSDR demethylation. Interestingly, the nTreg cell cycle maturation is also associated with a parallel increased expression of regulatory markers from N1 to M1 to M4 (SI Appendix, Fig. S2). Finally, the supervised analysis of mRNA expression levels of markers implicated in the different phases of nTreg life (cell activation, proliferation, functional regulatory differentiation, and senescence) reflects the N1 to M1 to M4 subset parental maturation (SI Appendix, Fig. S3).
Regarding the physiological characteristics of nTreg subsets, the present study shows that cytokine expression levels are governed chiefly by the microenvironmental context of their stimulation, although subtle differences in activities were detected among the major subsets (SI Appendix, Fig. S4). Under a steady state, TCR-stimulated FOXP3 Tregs from all of the subsets remain anergic (Fig. 1 D5), whereas in the presence of IL-2 or in contact with autoreactive conventional T cells supplying the IL-2 growth factor, the anergic status is abrogated and TCR-stimulated nTregs proliferate, but in N1 and M1 cells more than in M4 cells (Fig. 2 C1 and C2). In an inflammation and particularly an overinflammation/danger context, composed of inflammatory complement proteins, nTregs (chiefly the M1 subset) release IL-10 and behave as Tr1-like cells (SI Appendix, Fig. S4), whereas the majority of M4 mature cells undergo rapid apoptosis (Fig. 2 C3 and C4). In contrast, in a proinflammatory context, including IL-2, IL-1β, IL-6, and IL-23, cultured memory nTregs, which release IL-17 and behave as Th-17–like cells, are not suppressive under these conditions. Of particular importance for adoptive nTreg-based therapy, we found that ex vivo naive Tconv-induced iTregs in such an inflammatory microenvironment do not exert Th-17–like activity, and that their function remains suppressive independent of the context (Fig. 7B).
Focusing on the nTreg suppressive function investigated in the present study, following polyclonal TCR stimulation exclusively prevailing ex vivo, we confirmed that under steady state, all the subsets inhibit proliferation of immune-activated cell targets by cell-cell contact. This regulatory function is neither antigen-specific nor HLA-restricted (SI Appendix, Fig. S5), and furthermore is inhibited under inflammation (SI Appendix, Fig. S6). Under inflammation, nTregs, particularly M1 ones, which produce IL-10 (SI Appendix, Fig. S4), act as Tr1-like cells via humoral factors as observed in a Transwell coculture suppressive assay for the total nTreg population (12). Conversely, following a specific APC stimulation as required in vivo, under steady-state TCR stimulated by immature autologous DC, nTreg subsets and particularly the mature M4 cells, which express CD39 and CTLA-4 at a resting stage (Fig. 3B), suppress by cell-cell contact preactivated autologous conventional T CD4+ targets experimentally, accounting for self-tolerance (Fig. 5C). However, under inflammation, if nTreg cell-cell contact suppressive capacity is inhibited (SI Appendix, Fig. S6) (12), mature M4 cells could tolerogenize their TCR-stimulating DCs (Fig. 5E), while immature M1 cells could still exert a Tr1-like suppressive activity by producing IL-10 (SI Appendix, Fig. S4) and precursor N1 cells mature to M1 (Fig. 2).
Thus, in vitro experiments mimicking the Ag-specific HLA-DR–restricted conditions of TCR stimulation prevailing in vivo, strongly suggest that the reported mechanisms (6) by which FOXP3 Tregs achieve their suppressive activities are used in a nonredundant way but are conditioned chiefly by the microenvironmental context of their stimulation. Of note, the distinctive regulatory signaling pathways ultimately lead to IL-2 deprivation of the activated target cell proliferation (Fig. 5B and SI Appendix, Fig. S5A). Indeed, IL-2 starvation results either from juxtacrine IL-2 consumption by Tregs under steady state (Fig. 5B) or from an increased ADO concentration (Fig. 5 D4), which is known to inhibit IL-2 synthesis of target cells via activation of cAMP (24). As to the increased ADO concentration, our culture experiments performed on nTreg-Tconv cocultures in the presence of stimulating DCs (Fig. 5 E1) or following stimulation of isolated DCs (Fig. 5 E2 and E3) enabled us to identify CTLA-4, CD39, and the absence of CD26 (ADA-binding receptor) as factors triggering tolerogenic activation of DCs (Fig. 5D). Fig. 5 E3 further shows that the tolerogenic DC-nTreg interplay triggers the release of IL-10 associated with an inhibition of IL-12 production; DC-induced release of IL-10 could inhibit ADA expression on activated T cell targets, thereby enhancing ADO concentration, as reported previously (37).
Most importantly, this basic study focusing on the effects of microenvironmental factors on nTreg structural heterogeneity and functional plasticity has relevant medical implications for both clinical diagnostics and therapeutics. At the clinical level, the finding of a stable CD39/CD26 profile in healthy individuals over time but a variable interindividual profile prompted us to evaluate whether this profile is a novel biomarker for use in monitoring nTreg dysfunction in chronic inflammatory diseases. Initial phenotypic analysis carried out on blood T cells from patients with an autoimmune disease from DM and RhA as well as from AML patients after HSCT, not only showed disease-specific CD39/CD26 profiles but also abnormal expression of minor nTreg subsets including naive N2-4 subsets, CD25− variant and FOXP3+ CD127+ Tregs as illustrated in Fig. 6. Finally, the present study suggests that, given their inability to produce IL-17, ex vivo generated and expanded FOXP3 Tregs represent an optimal source of Tregs to be administered without any risk of inflammatory complications in nTreg-based adoptive therapies to treat autoimmune pathologies and GVHD (21).
Materials and Methods
The study was approved by the respective institutional review boards of UCL and AP-HP and conducted in accordance with current ethical and legal frameworks. Blood samples were obtained after informed consent. Detailed descriptions of (i) the cellular material originating from healthy individuals or patients with autoimmune diseases or AML; (ii) the reagents and methods for cell purification, culture, RNA-sequencing and DNA methylation experiments, adenosine and inosine measurements, flow cytometry analysis, and functional assays; and (iii) the statistical methods used in the experiments and for RNA-sequencing data processing and analysis are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Béatrice Drouet for skillful scientific help; Dominique Bron (Université Libre de Bruxelles) for helpful discussions; Victor Renault and Emmanuel Tubacher (Laboratory for Bioinformatics), Fondation Jean Dausset Centre d'Etude du Polymorphisme Humain for bioinformatics support; and Fabienne Georges and Laure Finet (biobank of CHU UCL Namur) for ensuring high quality samples. This work was supported by Neovacs and by a Televie grant (to M.B. and C.C.).
Footnotes
Conflict of interest statement: D.Z. is a cofounder of Neovacs, and R.C. is an employee of eBioscience.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812471116/-/DCSupplemental.
References
- 1.Murphy KM, Weaver CT. Janeway’s Immunobiology. 9th Ed Garland Science; New York: 2016. [Google Scholar]
- 2.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25): Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. [PubMed] [Google Scholar]
- 3.Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol. 2005;6:345–352. doi: 10.1038/ni1178. [DOI] [PubMed] [Google Scholar]
- 4.Gershon RK, Cohen P, Hencin R, Liebhaber SA. Suppressor T cells. J Immunol. 1972;108:586–590. [PubMed] [Google Scholar]
- 5.Groux H, et al. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 6.Vignali DAA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maizels RM, Smith KA. Regulatory T cells in infection. Adv Immunol. 2011;112:73–136. doi: 10.1016/B978-0-12-387827-4.00003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeuchi Y, Nishikawa H. Roles of regulatory T cells in cancer immunity. Int Immunol. 2016;28:401–409. doi: 10.1093/intimm/dxw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Noval Rivas M, Chatila TA. Regulatory T cells in allergic diseases. J Allergy Clin Immunol. 2016;138:639–652. doi: 10.1016/j.jaci.2016.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beres AJ, Drobyski WR. The role of regulatory T cells in the biology of graft-versus-host disease. Front Immunol. 2013;4:163. doi: 10.3389/fimmu.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Garra A, Vieira PL, Vieira P, Goldfeld AE. IL-10–producing and naturally occurring CD4+ Tregs: Limiting collateral damage. J Clin Invest. 2004;114:1372–1378. doi: 10.1172/JCI23215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Buanec H, et al. IFN-α and CD46 stimulation are associated with active lupus and skew natural T regulatory cell differentiation to type 1 regulatory T (Tr1) cells. Proc Natl Acad Sci USA. 2011;108:18995–19000. doi: 10.1073/pnas.1113301108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarantopoulos S, Lu L, Cantor H. Qa-1 restriction of CD8+ suppressor T cells. J Clin Invest. 2004;114:1218–1221. doi: 10.1172/JCI23152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1062. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Ioan-Facsinay A, van der Voort EIH, Huizinga TWJ, Toes REM. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 16.Kmieciak M, et al. Human T cells express CD25 and Foxp3 upon activation and exhibit effector/memory phenotypes without any regulatory/suppressor function. J Transl Med. 2009;7:89. doi: 10.1186/1479-5876-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayyoub M, et al. Human memory FOXP3+ Tregs secrete IL-17 ex vivo and constitutively express the T(H)17 lineage-specific transcription factor RORgamma t. Proc Natl Acad Sci USA. 2009;106:8635–8640. doi: 10.1073/pnas.0900621106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beriou G, et al. IL-17–producing human peripheral regulatory T cells retain suppressive function. Blood. 2009;113:4240–4249. doi: 10.1182/blood-2008-10-183251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA. IL-2 is essential for TGF-beta to convert naive CD4+CD25− cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J Immunol. 2007;178:2018–2027. doi: 10.4049/jimmunol.178.4.2018. [DOI] [PubMed] [Google Scholar]
- 21.Gliwiński M, Iwaszkiewicz-Grześ D, Trzonkowski P. Cell-based therapies with T regulatory cells. BioDrugs. 2017;31:335–347. doi: 10.1007/s40259-017-0228-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baecher-Allan C, Brown JA, Freeman GJ, Hafler DA. CD4+CD25high regulatory cells in human peripheral blood. J Immunol. 2001;167:1245–1253. doi: 10.4049/jimmunol.167.3.1245. [DOI] [PubMed] [Google Scholar]
- 23.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohta A, Sitkovsky M. Extracellular adenosine-mediated modulation of regulatory T cells. Front Immunol. 2014;5:304. doi: 10.3389/fimmu.2014.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong RP, et al. Characterization of adenosine deaminase binding to human CD26 on T cells and its biologic role in immune response. J Immunol. 1996;156:1349–1355. [PubMed] [Google Scholar]
- 26.Thompson CB, et al. CD28 activation pathway regulates the production of multiple T-cell-derived lymphokines/cytokines. Proc Natl Acad Sci USA. 1989;86:1333–1337. doi: 10.1073/pnas.86.4.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acosta-Rodriguez EV, Napolitani G, Lanzavecchia A, Sallusto F. Interleukins-1β and -6 but not transforming growth factor-β are essential for the differentiation of interleukin-17–producing human T helper cells. Nat Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 28.Kemper C, et al. Activation of human CD4+ cells with CD3 and CD46 induces a T-regulatory cell 1 phenotype. Nature. 2003;421:388–392. doi: 10.1038/nature01315. [DOI] [PubMed] [Google Scholar]
- 29.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 30.Baron U, et al. DNA demethylation in the human FOXP3 locus discriminates regulatory T cells from activated FOXP3(+) conventional T cells. Eur J Immunol. 2007;37:2378–2389. doi: 10.1002/eji.200737594. [DOI] [PubMed] [Google Scholar]
- 31.Jonuleit H, et al. Identification and functional characterization of human CD4(+)CD25(+) T cells with regulatory properties isolated from peripheral blood. J Exp Med. 2001;193:1285–1294. doi: 10.1084/jem.193.11.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferraro A, et al. Interindividual variation in human T regulatory cells. Proc Natl Acad Sci USA. 2014;111:E1111–E1120. doi: 10.1073/pnas.1401343111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santegoets SJAM, et al. Monitoring regulatory T cells in clinical samples: Consensus on an essential marker set and gating strategy for regulatory T cell analysis by flow cytometry. Cancer Immunol Immunother. 2015;64:1271–1286. doi: 10.1007/s00262-015-1729-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogonek J, et al. Immune reconstitution after allogeneic hematopoietic stem cell transplantation. Front Immunol. 2016;7:507. doi: 10.3389/fimmu.2016.00507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paust S, Cantor H. Regulatory T cells and autoimmune disease. Immunol Rev. 2005;204:195–207. doi: 10.1111/j.0105-2896.2005.00247.x. [DOI] [PubMed] [Google Scholar]
- 36.Trzonkowski P, et al. First-in-man clinical results of the treatment of patients with graft versus host disease with human ex vivo expanded CD4+CD25+CD127− T regulatory cells. Clin Immunol. 2009;133:22–26. doi: 10.1016/j.clim.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 37.Mandapathil M, et al. Generation and accumulation of immunosuppressive adenosine by human CD4+CD25highFOXP3+ regulatory T cells. J Biol Chem. 2010;285:7176–7186. doi: 10.1074/jbc.M109.047423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.