Significance
Hepatic injury is often accompanied by pulmonary inflammation. The underlying mechanism, however, remains unknown. Here we identify miR-122, released by injured liver cells in an exosome-independent manner, as the culprit driving liver injury-induced pulmonary dysfunction. Consistent with this, we show that mouse pulmonary inflammation is suppressed by depleting liver miR-122 or circulating miR-122, but initiated by directly injecting miR-122 via tail vein or respiratory trachea. Furthermore, circulating miR-122 is preferentially transported to lungs and taken up by alveolar macrophages, where it binds Toll-like receptor 7/8 (TLR7/8) to activate inflammatory responses. Our findings provide a mechanism for developing pulmonary inflammation/tissue damage and identify hepatic miR-122 and its activated TLR7/8 signal as potential therapeutic targets in controlling liver injury-induced pulmonary inflammation.
Keywords: liver injury, circulating miR-122, pulmonary inflammatory, macrophage, TLR7/8
Abstract
Hepatic injury is often accompanied by pulmonary inflammation and tissue damage, but the underlying mechanism is not fully elucidated. Here we identify hepatic miR-122 as a mediator of pulmonary inflammation induced by various liver injuries. Analyses of acute and chronic liver injury mouse models confirm that liver dysfunction can cause pulmonary inflammation and tissue damage. Injured livers release large amounts of miR-122 in an exosome-independent manner into the circulation compared with normal livers. Circulating miR-122 is then preferentially transported to mouse lungs and taken up by alveolar macrophages, in which it binds Toll-like receptor 7 (TLR7) and activates inflammatory responses. Depleting miR-122 in mouse liver or plasma largely abolishes liver injury-induced pulmonary inflammation and tissue damage. Furthermore, alveolar macrophage activation by miR-122 is blocked by mutating the TLR7-binding GU-rich sequence on miR-122 or knocking out macrophage TLR7. Our findings reveal a causative role of hepatic miR-122 in liver injury-induced pulmonary dysfunction.
The liver plays a central role in regulation of systematic organ inflammation. A diseased liver can have many deleterious effects on multiple organ systems, including the pulmonary system (1). Clinically, pulmonary abnormalities and symptoms in patients with chronic liver disease are common, and mild hypoxia is observed in approximately one-third of chronic liver disease patients. In fact, up to 70% of cirrhotic patients undergoing evaluation for liver transplantation complain of dyspnoea (2). Hepatopulmonary syndrome, characterized by intrapulmonary vascular dilatation, gas-exchange abnormalities, lung edema, and shortness of breath, is a major complication following liver ischemia and reperfusion subsequent to liver resection or transplantation (3, 4). Many factors, such as systemic inflammatory responses, are reportedly involved in hepatic injury-induced acute pulmonary inflammation and tissue damage. Lin et al. (5) observed that liver injury was associated with increased pulmonary inducible nitric oxide synthase (iNOS) expression, suggesting that iNOS expression might play a critical role in this phenomenon. In addition, massive cysteinyl leukotrienes (CysLTs)—including LTB4, LTC4, LTD4, and LTE4—were shown to be produced after hepatic injury and to play essential roles in lung edema development (6). Al-Amran et al. (7) also showed that LTs facilitate hemorrhagic shock-induced acute lung injury. However, although findings have accumulated characterizing the injurious processes occurring in the lung subsequent to liver injury, the molecular basis linking hepatic injury to acute pulmonary inflammation and tissue damage remains incompletely understood.
MicroRNAs (miRNAs), a class of noncoding RNAs ∼22 nucleotides in length, have been newly established as gene regulators in biological processes (8, 9). Previous studies by ourselves and others have shown that miRNAs are secreted by various types of cells through exosomes or protein-mediated pathways (10, 11). Furthermore, biological assays and functional studies have shown that extracellular miRNAs are highly stable and not only serve as biomarkers for various diseases, but also as novel signaling molecules mediating remote communication among various cell types and organs (12). Specifically, circulating miRNAs in exosomes can be delivered to recipient cells, where they function similar to endogenous miRNAs regulating multiple target genes or signaling events. For example, Zhang et al. (10) reported that miR-150 is selectively packaged into exosomes and actively secreted by human blood cells and cultured THP-1 cells. THP-1–derived exosomes can deliver miR-150 to human HMEC-1 cells, and elevated exogenous miR-150 effectively reduces c-Myb expression and enhances migration of HMEC-1 cells. On the other hand, certain miRNAs secreted in plasma are also undetectable in exosomes or other microvesicles (MVs) but associated with RNA-binding proteins, such as Argonaute 2 (AGO2) (11) and nucleophosmin 1 (NPM1) (13). However, elucidating the roles of MV-free circulating miRNAs in intercellular communication requires further attention.
In addition to translational repression or direct mRNA degradation via base-pairing complementary sites on target mRNAs, a conventional role of miRNAs in posttranscriptional gene regulation, miRNAs can also act directly as physiological ligands for certain RNA receptors. Among the Toll-like receptors (TLRs) identified to date, intracellular TLR7 in mice and TLR8 in humans can reportedly be recognized by exogenous GU-rich single-stranded RNA molecules, such as RN40 derived from HIV-1 (14). Recent studies showed that mature miRNAs with GU-rich sequences also contribute to immune stimulation by serving as physiological ligands for murine TLR7 and human TLR8 (15). For example, miR-21, miR-29a, miR-25-3p, and miR-92a-3p, all harboring GU-rich sequences, have been shown to interact with murine TLR7 and human TLR8 to activate the secretion of proinflammatory cytokines in macrophages (15–17). Consistent with this, Lehmann et al. (18) found that another GU-enriched miRNA, let-7b, acts as a potent activator of TLR7 signaling in neurons, which induces neurodegeneration. However, while miRNAs acting as TLR7/8 ligands suggests that they elicit bodily inflammatory responses by directly activating the TLR signaling pathway, whether miRNA binding to TLR plays a role in hepatic injury-induced acute pulmonary inflammation and tissue damage remains unknown.
In the present study, we provide evidence that released miR-122 by injured liver cells is a culprit driving hepatic injury-induced acute pulmonary inflammation and lung tissue damage. The plasma miR-122 level is strikingly increased under various liver injury conditions, and depleting miR-122 in mouse liver or mouse peripheral blood strongly attenuates acute pulmonary inflammation and lung tissue damage induced by liver injury. In situ labeling and qRT-PCR analysis show that circulating miR-122 released by injured mouse liver cells is delivered to lung tissue, particularly to alveolar macrophages. In macrophages, miR-122 activated TLR7 signaling, leading to macrophage M1 polarization and secretion of inflammatory cytokines, such as TNF-α and IL-6. Supporting the specific interaction between miR-122 and murine TLR7, activation of mouse alveolar macrophages by miR-122 was largely abolished by deleting TLR7 in mouse macrophages or mutating the TLR7-binding sequence within miR-122.
Results
Hepatic Injuries Result in Acute Pulmonary Inflammation and Tissue Damage.
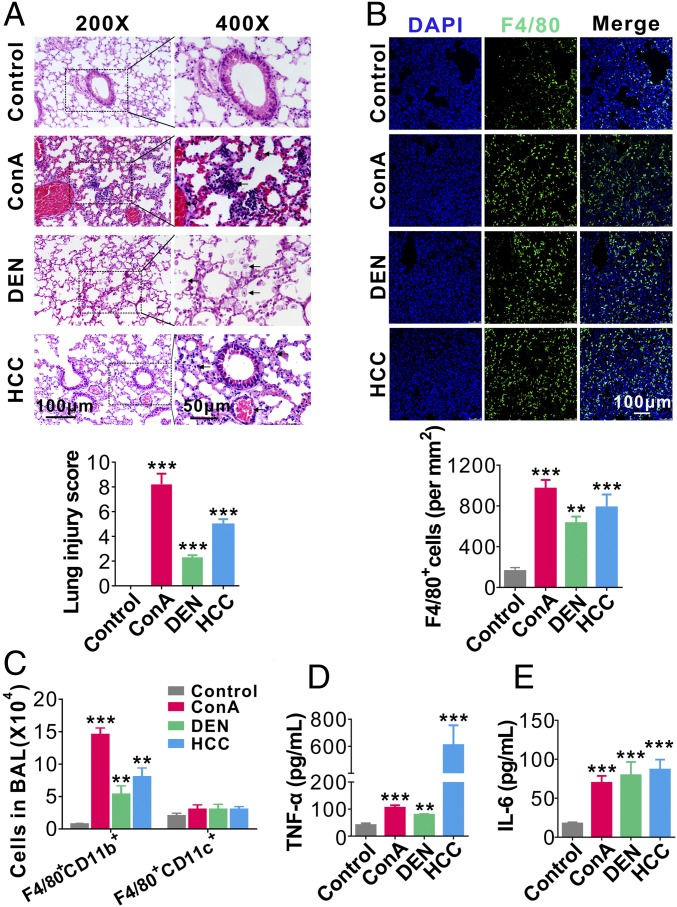
Clinic data strongly suggest a close association between hepatic injury and pulmonary abnormalities (2–5, 7). To test whether liver injuries can cause acute lung inflammation and tissue damage, we established three mouse models of liver injury: orthotopic transplanted hepatocellular carcinoma (HCC) (19), diethylnitrosamine (DEN)-induced chronic hepatitis (20), and Con A (ConA)-induced acute hepatitis (21, 22). As shown by H&E staining and biological analysis in SI Appendix, Fig. S1A, mice in three different liver injury models all displayed severe liver tissue damage and dysfunction. Interestingly, examining the tissue damage of different organs revealed that liver injuries in these mice were all accompanied by significant tissue damage in the lung, characterized by diffuse lesions with extensive interstitial inflammatory cell infiltration, alveolar edema, and hemorrhage (Fig. 1A, Upper). Quantifying the pulmonary inflammation and tissue damage based on the following parameters—pulmonary edema, inflammatory infiltration, hemorrhage, atelectasis, and hyaline membrane formation—we found significantly higher injury scores in liver-injured mice compared with those in the respective control groups (Fig. 1A, Lower). In contrast, only modest or little tissue damage was observed in the spleen, kidney, colon, and heart in mice suffering various liver diseases (SI Appendix, Fig. S1A). These results suggest that liver injury may be preferentially associated with pulmonary inflammation and tissue damage.
Fig. 1.
Lung inflammation and tissue damage caused by liver injuries. (A, Upper) Representative H&E staining of lung tissue sections in mice with or without ongoing ConA-induced acute hepatitis, DEN-induced chronic hepatitis, or orthotopic transplanted HCC. (Lower) Mouse lung injury scores analyzed by a double-blind examination for the following parameters: pulmonary edema, inflammatory infiltration, hemorrhage, atelectasis, and hyaline membrane formation (six mice per group, three to five images per mouse). The scores were assigned as follows: 0, no injury; 1, <25% injury; 2, 25–50% injury; 3, 50–75% injury; and 4 >75% injury. Eight to 10 high-magnitude fields from each slide were analyzed. (B, Upper) Representative images of F4/80+ cell infiltration into mouse lungs from mice with or without liver injuries. (Lower) Analysis of alveolar infiltration of F4/80+ macrophages (six mice oer group, three to five images per mouse). (C) FACS analysis of alveolar macrophage subpopulations (F4/80+CD11b+ or F4/80+CD11c+ cells) in BAL from mice with or without various liver injuries (six mice per group, n = 3). (D and E) Levels of TNF-α (D) and IL-6 (E) in BALF from mice with or without various liver injuries (six mice per group, n = 3). Data are show as means ± SEM. **P < 0.01, ***P < 0.001. One-way ANOVA followed by Bonferroni’s multiple comparisons test.
Consistent with the finding that liver injuries were associated with significant lung tissue damage, infiltration of inflammatory F4/80+ macrophages into mouse lungs was markedly increased in mice bearing HCC or treated with ConA or DEN compared with macrophage infiltration in control mice (Fig. 1B). We further isolated macrophages from mouse bronchoalveolar lavage (BAL) and analyzed their origin and cytokine secretion levels. As shown in Fig. 1C and SI Appendix, Fig. S1B, alveolar macrophages in mice with various liver injuries were CD11b+ but CD11c−, suggesting that they are mainly derived from myeloid lineage (23). Alveolar macrophages from mice with liver injuries also secreted significantly more proinflammatory cytokines, such as TNF-α (Fig. 1D) and IL-6 (Fig. 1E), than those from control mice.
Identification of Circulating Hepatic miR-122 as the Culprit Underlying Liver Injury-Induced Acute Lung Inflammation.
Given that different liver injuries caused similar acute pulmonary inflammation and tissue damage, we speculated that a common circulating factor might be involved. Because accumulating evidence has demonstrated that circulating miRNAs play a vital role in organ–organ communication (12), we next explored the potential role of these circulating miRNAs in the development of pulmonary inflammation and tissue damage in mice with ongoing liver injuries. In the experiment, we first profiled the levels of circulating miRNAs previously found to be associated with various liver dysfunctions in ConA-treated mice using qRT-PCR. As shown in SI Appendix, Fig. S2A, we found that miR-122 was the most significantly up-regulated miRNA in the plasma of mice subjected to ConA-induced liver injury. Screening the liver function-related miRNAs in mouse plasma samples also showed a markedly increased plasma miR-122 level from mice bearing HCC or treated with DEN (SI Appendix, Fig. S2B). Given that miRNAs can be secreted by cells through exosomes, particularly when cells are challenged by stimuli (12), we determined whether hepatic miR-122 in plasma is released from injured liver cells via exosomes. In this experiment, we separated plasma samples from mice with various liver injuries into exosome and exosome-free fractions (SI Appendix, Fig. S3 A and C). To our surprise, the majority of circulating miR-122 was detected in the exosome-free fraction (SI Appendix, Fig. S3D), suggesting that miR-122 might passively leak from injured liver cells. We also compared the miR-122 level in plasma samples collected from patients with active hepatitis (n = 10) or HCC (n = 10) with that from healthy donors (n = 10). As shown in SI Appendix, Fig. S3E, patients with hepatitis or HCC displayed a significantly higher plasma miR-122 level than healthy donors, which is consistent with previous reports that plasma or serum levels of miR-122 are elevated in patients with various liver injuries (24, 25). Consistent with our findings in mice, the majority of circulating miR-122 in the plasma from patients with ongoing hepatitis or HCC was also detected in the exosome-free fraction (SI Appendix, Fig. S3F).
To explore whether released miR-122 by liver cells is associated with cell death, such as apoptosis, necroptosis, and pyroptosis, the expression of protein markers of apoptosis (cleaved-caspase 3), necroptosis (RIP3), and pyroptosis (cleaved-caspase 1) were determined in liver tissue sections from mice bearing HCC or from mice whose liver injury was induced by ConA or DEN treatment. As shown in SI Appendix, Fig. S4A, various degrees of apoptosis, necroptosis, and pyroptosis were observed in these three liver injury mouse models. ConA treatment induced strong apoptosis in liver tissue. In contrast, DEN treatment caused more severe pyroptosis and mice bearing HCC displayed the highest level of necroptosis in liver. We also induced apoptosis, necroptosis, or pyroptosis in cultured primary hepatocytes (SI Appendix, Fig. S4B) and assayed miR-122 level in the culture supernatant (SI Appendix, Fig. S3 B and C). As shown in SI Appendix, Fig. S4C, liver cell apoptosis, necroptosis, or pyroptosis all significantly increased the level of miR-122 in culture medium, and in agreement with a previous observation, the majority of miR-122 in cell culture medium was in the exosome-free fraction.
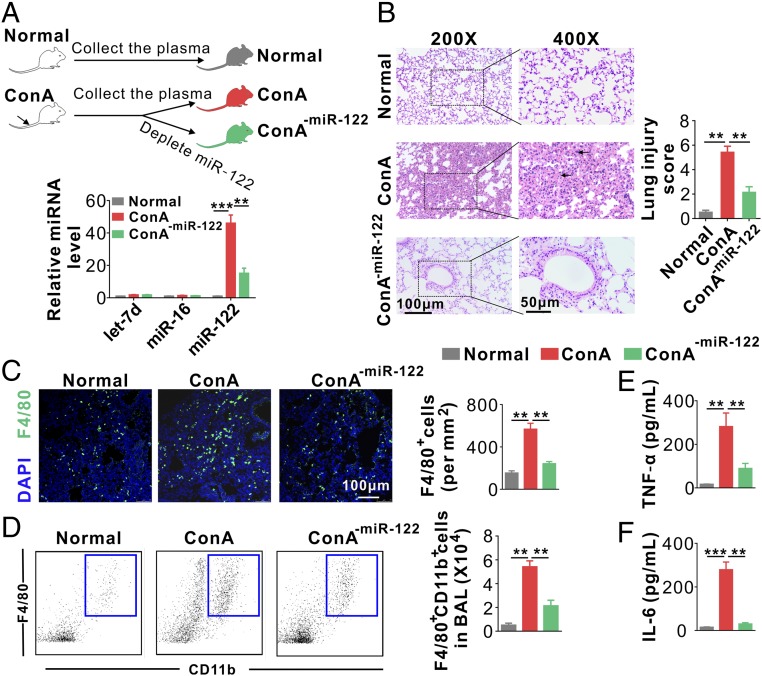
To test whether circulating miR-122 is a causative factor of liver injury-induced acute lung inflammation, we employed an adoptive transfer approach to test the effects of plasma isolated from ConA-treated mice on normal control mice with or without depleting plasma miR-122. Plasma samples were collected from both control and ConA-treated mice. To deplete plasma miR-122, ConA-treated mouse plasma samples were filtered through biotin–anti–miR-122 antisense oligonucleotides immobilized on streptavidin-conjugated Sepharose beads. qRT-PCR confirmed that miR-122 was effectively depleted from mouse plasma without affecting other miRNAs (Fig. 2A, Lower). We next injected mice with plasma from control (normal group) or ConA-treated mice with or without miR-122 depletion (ConA or ConA-miR-122 groups) via tail vein (Fig. 2A, Upper). H&E staining of mouse lungs revealed diffuse lesions with extensive interstitial hemorrhage, inflammatory cell infiltration, and alveolar edema in recipient mice with plasma from the ConA group but not the control group or miR-122–depleted ConA group (Fig. 2B, Left). A significantly higher score was observed in the ConA group compared with that in the other two groups (Fig. 2B, Right). Immunofluorescence (Fig. 2C) and flow cytometry (Fig. 2D) analyses of immune cell infiltration both indicated significantly higher infiltration of F4/80+CD11b+ macrophages into mouse lungs in the ConA group than in the other two groups. Consistent with this, the levels of inflammatory cytokines TNF-α and IL-6, in mouse BAL fluid (BALF), were significantly higher in mice given plasma from the ConA group than those from the control group, whereas TNF-α and IL-6 levels were strongly reduced by depleting plasma miR-122 (Fig. 2 E and F). Supporting the effect of hepatic miR-122 on activating pulmonary inflammation, detection of miR-122 level in the heart, spleen, lung, kidney, and colon from mice with ConA-induced acute hepatitis revealed that mouse lungs contained the highest level of miR-122 (SI Appendix, Fig. S5). To test whether liver injuries can cause a similar increase in miR-122 levels in patients’ lung tissues, we collected pulmonary effusion from 10 chronic hepatitis patients with pulmonary effusion and 10 severe pneumonia patients without liver disorder (SI Appendix, Table S2), and compared the miR-122 level in these pulmonary effusion samples. As shown in SI Appendix, Fig. S6, the average miR-122 level in pulmonary effusion samples from chronic hepatitis patients was increased fourfold compared with that from pneumonia patients without liver disorder. Taken together, these results suggest that miR-122, released by injured liver cells, may be a key causative factor in acute pulmonary inflammation and tissue damage induced by liver injury.
Fig. 2.
Hepatic miR-122 induces acute pulmonary inflammation and tissue damage. (A, Upper) Schematic of experimental design: 200 μL plasma samples from normal mice and ConA-treated mice with or without miR-122 depletion were separately injected into WT mice (six mice per group) every 6 h six times. (Lower) miR-122 was depleted from mouse plasma by miR-122 antisense oligonucleotide-conjugated Sepharose beads. (B) Mouse lung tissue damage analyzed by H&E staining (Left) and injury score (Right). (C) Representative images (Left) and analysis (Right) of F4/80+ macrophage infiltration into mouse lungs. Note that the infiltration of F4/80+ macrophages was significantly enhanced by plasma from ConA-treated mice, while depleting circulating miR-122 in ConA-treated mouse plasma markedly attenuated the infiltration of F4/80+ cells into alveoli (n = 6 mice per group, three to five images per mouse). (D) FACS analysis of F4/80+CD11b+ macrophages in BAL harvested from the recipient mice (six mice per group, n = 3). (E and F) Levels of TNF-α (E) and IL-6 (F) in BALF from the recipient mice (six mice per group, n = 3). Data are presented as mean ± SEM. **P < 0.01, ***P < 0.001. One-way ANOVA followed by Bonferroni’s multiple comparisons test.
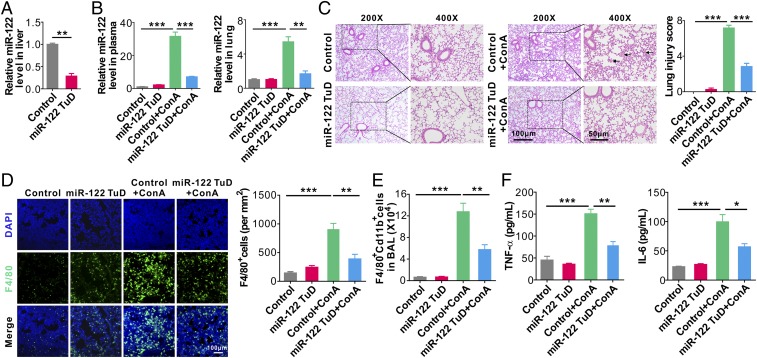
To further validate whether circulating miR-122 is a causative factor of liver injury-induced acute lung inflammation, we combined recombinant adeno-associated virus (rAAV) vectors with miR-122 “tough decoys” (TuDs) to specifically decrease miR-122 expression in liver (26). After 4 wk of AAV8 administration, qRT-PCR assay showed an ∼70% reduction of miR-122 in mice that received the miR-122 TuD vector (miR-122 TuD) compared with the mice that had not received miR-122 TuD (Control) (Fig. 3A). We then induced liver injury in the control mice or miR-122 TuD mice using ConA, and determined the levels of miR-122 in mouse plasma and lungs, respectively. As shown in Fig. 3B, the levels of miR-122 were strikingly increased in the control mice after inducing liver injury by ConA. However, ConA treatment did not show much increase of miR-122 in mouse plasma and lungs from mice receiving miR-122 TuD. H&E staining of mouse lungs revealed diffuse lesions with extensive interstitial hemorrhage, inflammatory cell infiltration, and alveolar edema in mice adoptively transferred with plasma from the Control+ConA group but not the mice in the miR-122 TuD group or miR-122 TuD+ConA group (Fig. 3C, Left). A significantly higher lung injury score was observed in the Control+ConA group compared with that in the other three groups (Control, miR-122 TuD, and miR-122 TuD+ConA) (Fig. 3C, Right). Immunofluorescence (Fig. 3D) and flow cytometry analyses (Fig. 3E) of immune cell infiltration both indicated significantly more infiltration of F4/80+CD11b+ macrophages into mouse lungs in the Control+ConA group than in the other three groups. Consistent with this, the levels of inflammatory cytokines TNF-α and IL-6, in mouse BALF, were significantly higher in the Control+ConA group than those from the other three groups (Fig. 3F).
Fig. 3.
Depleting mouse liver miR-122 greatly attenuates lung inflammation and tissue damage induced by injured liver. (A) Relative level of miR-122 in mouse liver following miR-122 TuD AAV8 administration. (B) Relative levels of miR-122 in mouse plasma (Left) and lungs (Right) in control mice (Control), miR-122 TuD mice (miR-122 TuD), control mice treated with ConA (Control+ConA), and miR-122 TuD mice treated with ConA (miR-122 TuD+ConA). (C) Mouse lung tissue damage analyzed by H&E staining (Left) and injury score (Right). (D) Representative images (Left) and analysis (Right) of F4/80+ macrophage infiltration into mouse lungs. (E) FACS analysis of F4/80+CD11b+ macrophages in BAL harvested from the recipient mice (six mice per group, n = 3). (F) Levels of TNF-α (Left) and IL-6 (Right) in BALF from the four group mice (six mice per group, n = 3). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. Student’s two-tailed, unpaired t-test (A), or one-way ANOVA followed by Bonferroni’s multiple comparisons test (B–F).
The role of circulating miR-122 in eliciting acute lung inflammation and tissue damage was also validated by directly injecting mice with synthetic miR-122 through either tail vein or respiratory trachea (SI Appendix, Fig. S7A). As shown, direct miR-122 injection through both routes strongly increased miR-122 expression in mouse lungs (SI Appendix, Fig. S7B, Left), whereas only tail vein injection increased the plasma level of miR-122 (SI Appendix, Fig. S7B, Right). Mice injected with miR-122 via their tail vein or respiratory trachea displayed significant lung injuries compared with those injected with the vehicle control as determined by H&E staining and injury scoring system (SI Appendix, Fig. S7C). Immunofluorescence labeling of mouse lung tissues indicated that significantly more inflammatory F4/80+ macrophages had infiltrated into mouse lungs following miR-122 injection via tail vein or respiratory trachea (SI Appendix, Fig. S7D). Flow cytometry analysis of isolated cells in mouse BAL confirmed the enhanced infiltration of inflammatory macrophages in mouse lungs after miR-122 injection (SI Appendix, Fig. S7E). Moreover, the level of TNF-α and IL-6 were significantly higher in BALF from mice administered with miR-122 than those from mice treated with vehicle control (SI Appendix, Fig. S7F). Given that secretion of inflammatory cytokines is a main feature of inflammatory M1 phenotypic activation (27), this result suggested that miR-122 injection had skewed alveolar macrophages toward M1 polarization. Consistent with this, qRT-PCR data showed that alveolar macrophages isolated from miR-122–injected mice expressed significantly higher levels of M1 polarized gene transcripts, such as iNOS and TNF-α (SI Appendix, Fig. S7G).
Hepatic miR-122 Enters Alveolar Macrophages and Activates Intracellular TLR7 Signaling.
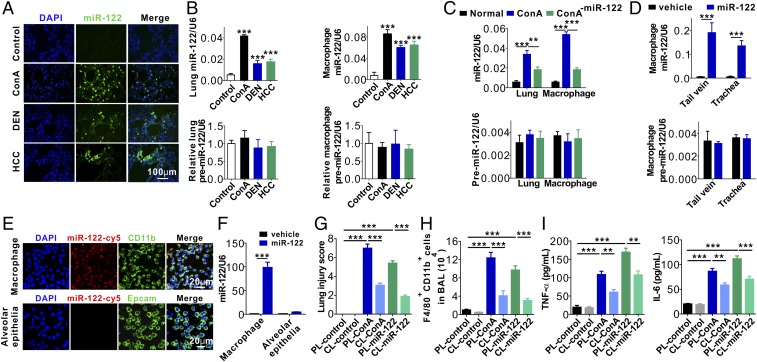
To further characterize the role of miR-122 in activating alveolar macrophages and eliciting acute pulmonary inflammation, we examined whether miR-122 released by injured liver cells could enter alveolar macrophages. First, an in situ hybridization assay was performed to determine miR-122 distribution patterns in lung tissues from mice with and without ongoing liver injuries. As shown in Fig. 4A, substantial amounts of miR-122 entered mouse lung tissues after the development of various hepatic injuries. The levels of mature miR-122, but not premiR-122, determined by qRT-PCR, were markedly increased in lung tissues and alveolar macrophages from mice bearing HCC, as well as mice treated with ConA or DEN (Fig. 4B). The results suggest that miR-122 in mouse lung tissues and alveolar macrophage is likely derived not from de novo synthesis but from the delivery of exogenous miR-122 under liver injury conditions.
Fig. 4.
Uptake of hepatic miR-122 by mouse alveolar macrophages. (A) In situ hybridization of miR-122 in mouse lungs following various liver injuries. (B) Levels of miR-122 or premiR-122 in mouse lungs and alveolar macrophages isolated from mice with various liver injuries. (C) Levels of miR-122 (Upper) and premiR-122 (Lower) in lungs and alveolar macrophages isolated from mice after adoptive transfer of ConA-treated mouse plasma with or without miR-122 depletion. (D) Levels of miR-122 (Upper) and premiR-122 (Lower) in alveolar macrophages isolated from mice with or without direct injection of miR-122 via tail vein or respiratory trachea. (E and F) Uptake of synthetic miR-122-cy5 by mouse primary macrophages and mouse alveolar epithelial cells assayed by immunofluorescence (E) and qRT-PCR (F). (G) Mouse lung tissue damage analyzed by injury score in mice of control group (PL-control), macrophage-depletion group (CL-control), control mice injected with ConA group (PL-ConA), control mice injected with synthetic miR-122 group (PL–miR-122), macrophage-depletion mice injected with ConA group (CL-ConA) and macrophage depletion mice injected with synthetic miR-122 group (CL–miR-122). (H) FACS analysis of F4/80+CD11b+ macrophages in BAL harvested from the recipient mice (six mice per group, n = 3). (I) Levels of TNF-α (Left) and IL-6 (Right) in BALF from the six group mice (six mice per group, n = 3). Data are presented as mean ± SEM. **P < 0.01, ***P < 0.001. One-way ANOVA followed by Bonferroni’s multiple comparisons test (B, C, and G–I) or student’s two-tailed, unpaired t test (D and F).
We next determined the levels of miR-122 and premiR-122 in mouse lungs and alveolar macrophages following the adoptive transfer of ConA-treated mouse plasma with or without miR-122 depletion. As shown in Fig. 4C, the levels of miR-122, but not premiR-122, were strikingly increased after the adoptive transfer of ConA-treated mouse plasma. Depleting miR-122 from ConA-treated mouse plasma significantly reduced the level of miR-122 in mouse lungs and alveolar macrophages. In line with the notion that alveolar macrophages are the major recipient cells of circulating miR-122 in mouse lungs, direct injection of miR-122 through the tail vein or respiratory trachea both strongly increased miR-122 levels, but not premiR-122 levels, in alveolar macrophages (Fig. 4D). To validate whether exosome-free miR-122 enters mouse alveolar macrophages, we incubated mouse alveolar macrophages and primary alveolar epithelial cells with synthetic miR-122-Cy5. We found that miR-122–Cy5 rapidly entered mouse alveolar macrophages but not primary alveolar epithelial cells (Fig. 4E). After 2 h of incubation at 37 °C, miR-122 expression in macrophages was increased nearly 100-fold, while only a two- to threefold increase was observed in alveolar epithelial cells (Fig. 4F).
To further demonstrate that macrophages are critical for liver injury-induced pulmonary inflammation in vivo, we depleted macrophages using clodronate liposomes (CL) (28) before injection with ConA or synthetic miR-122. Mice treated with PBS liposomes (PL) served as controls. H&E staining of mouse lung tissue sections displayed diffuse lesions with extensive interstitial hemorrhage, inflammatory cell infiltration, and alveolar edema in control mice treated with ConA or synthetic miR-122 (PL-ConA group and PL–miR-122 group), whereas macrophage-depleted mice injected with ConA or synthetic miR-122 (CL-ConA group and CL–miR-122 group) had no apparent diffuse lesions, inflammatory cell infiltration, and alveolar edema (SI Appendix, Fig. S8A). A significantly higher lung injury score was registered in PL-ConA and PL–miR-122 groups compared with that in CL-ConA and CL–miR-122 groups (Fig. 4G). Immunofluorescence (SI Appendix, Fig. S8B) and flow cytometry (Fig. 4H) analyses of immune cell infiltration both showed significantly more infiltration of F4/80+CD11b+ macrophages into mouse lungs in the PL-ConA and PL–miR-122 groups compared with that in the CL-ConA and CL–miR-122 groups. Consistent with this, the levels of inflammatory cytokines TNF-α and IL-6 in mouse BALF were strongly reduced in macrophage-depleted mice injected with ConA or synthetic miR-122 (CL-ConA group and CL–miR-122 group) compared with those in the PL-ConA and PL–miR-122 groups (Fig. 4I).
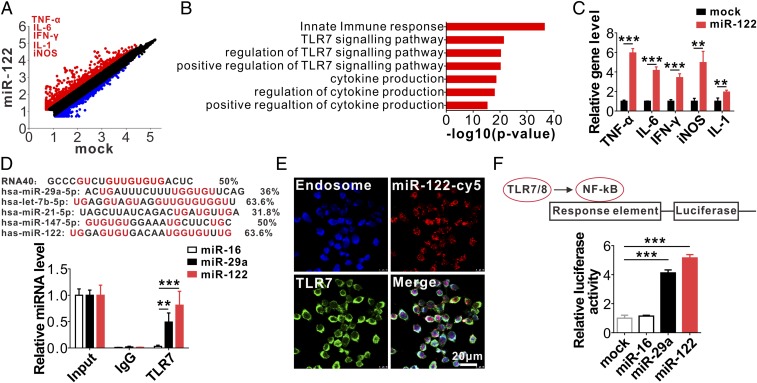
To explore the molecular basis underlying the effect of hepatic miR-122 on alveolar macrophage inflammatory activation, we performed transcription profiling in macrophages treated with or without miR-122. As shown in Fig. 5A, incubation with miR-122 markedly altered the gene-expression profile in macrophages (29). Among 29,280 genes detected, 1,089 were up-regulated (Fig. 5A, red) and 849 were down-regulated (Fig. 5A, blue) (fold-change > 2) in miR-122–treated macrophages compared with untreated macrophages. The expression levels of macrophage M1 polarization-related genes, such as Tnf, Il6, Ifng, Il1, and Nos2, were significantly elevated. Gene ontology analysis indicated that several top-ranked pathways enriched in macrophages incubated with miR-122 were related to the innate immune response, especially the TLR7 signaling pathway (Fig. 5B). qRT-PCR assay further confirmed the up-regulation of TLR7 signaling pathway-associated inflammatory molecules Tnf, Il6, Ifng, Il1, and Nos2 in macrophages treated with miR-122 compared with those in untreated macrophages (Fig. 5C). These results collectively suggest that miR-122 may activate the macrophage intracellular TLR7 signaling pathway, leading to macrophage M1 polarization and enhanced Tnf, Il6, Ifng, Il1, and Nos2 expression.
Fig. 5.
MiR-122 specifically binds to mouse macrophage endosomal TLR7. (A–C) Macrophage activation by internalized synthetic miR-122 detected by microarray gene-expression profiling (A and B) and qRT-PCR (C). (D, Upper) Comparison of GU enrichment in miR-122 with other TLR7/8-binding miRNAs; (Lower) Detection of miR-122 and miR-29a but not miR-16 in anti-TLR7 antibody-immunoprecipitated complex by qRT-PCR. (E) Colocalization of miR-122-Cy5 and TLR7 in endosomal structures of mouse macrophages. (F, Upper) schematic of NF-κB luciferase reporter assay of miR-122 binding human TLR8 in HEK293T cells. (Lower) Enhancement of NF-κB activity in HEK293T cells by miR-122 and miR-29a via specific binding to human TLR8. Data are presented as mean ± SEM (n = 3). **P < 0.01, ***P < 0.001. Student’s two-tailed, unpaired t test (C) or one-way ANOVA followed by Bonferroni’s multiple comparisons test (D and F).
Given that synthetic mature miRNA has a low affinity for AGO2, an important component of the RNA-induced silencing complex (RISC) required for miRNA function, we postulated that the single-strain mature miR-122 may execute its role in activating macrophage TLR7 (murine) or TLR8 (human) through a RISC-independent nonconventional mechanism. Although base-pairing with complementary sequences on target mRNAs is considered the conventional mechanism by which miRNAs posttranscriptionally regulate target gene expression, recent studies reported that miRNAs with GU-rich sequences can directly act as physiological ligands for TLR7/8 (15–18, 30, 31). Because miR-122 is also GU-enriched, with its GU percentage being higher than or similar to that of other miRNAs known to interact with TLR7/8 (Fig. 5D, Upper), we postulated that exosome-free miR-122 promotes macrophage inflammatory responses by activating the TLR7 signaling pathway. To verify this hypothesis, we first performed coimmunoprecipitation assays to assess binding between miR-122 and mouse macrophage TLR7. In this experiment, TLR7 was first immunoprecipitated from macrophages by an anti-TLR7 antibody and then incubated with miR-122, miR-29a, or miR-16. The levels of TLR-bound miRNAs were assessed by qRT-PCR. In this experiment, miR-29a (GU-rich) or miR-16 (not GU-rich) served as the positive or negative controls. As shown in Fig. 5D, Lower, miR-122 and miR-29a, but not miR-16, were enriched in the complex immunoprecipitated by the anti-TLR7 antibody. Intracellular localization of fluorescent miR-122 also supported the association of internalized miR-122 with endosomal TLR7 in murine macrophages (Fig. 5E). In this experiment, primary mouse macrophages were incubated with Cy5-conjugated miR-122 for 2 h before labeling the endogenous TLR7 with anti-TLR7 antibody and the endosomes with LysoTracker Blue. The colocalization of Cy5-conjugated miR-122 (Fig. 5E, red) and TLR7 (Fig. 5E, green) in endosomes was detected in macrophages, suggesting that exogenous miR-122 can bind to endosomal TLR7.
Because NF-κB, a transcription factor modulating the gene expression of various inflammatory cytokines, is activated by TLR7/8 (14, 15), we next tested whether miR-122 can affect NF-κB activity in human HEK-293T cells via activating TLR7/8. In this experiment, we first constructed an NF-κB reporter system (Fig. 5F, Upper) and then transfected HEK-293T cells with an NF-κB reporter construct to establish an NF-κB reporter system in HEK-293T cells. Cells were then incubated with N-[1-(2,3-Dioleoyloxy)propyl]-N,N,Ntrimethylammoniummethylsulfate (DOTAP) (14, 15) alone or DOTAP formulations containing miR-16, miR-29a, or miR-122. NF-κB activity was assessed with a luciferase assay as previously described (14). The result clearly revealed that NF-κB could be activated in TLR8-expressing HEK-293 cells by miR-122 or miR-29a, but not by miR-16 (Fig. 5F, Lower).
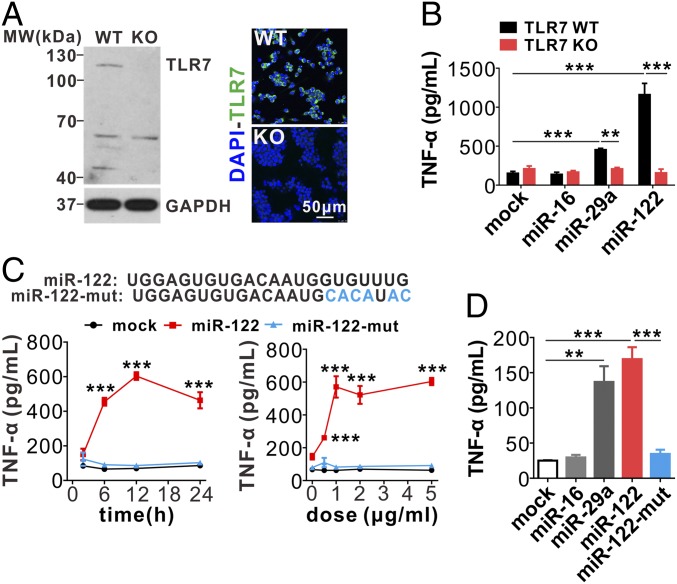
Alveolar Macrophage Activation by miR-122 Is Abolished by Deleting Macrophage TLR7 or Mutating TLR7-Binding Sequence on miR-122.
Given that TLR7 activation will lead to the production of inflammatory cytokines in mouse macrophages, such as TNF-α, we next determined whether the effect of miR-122 on promoting TNF-α secretion in mouse macrophages is mediated through activating mouse TLR7 signaling. First, we knocked out Tlr7 in RAW264.7 macrophages to generate stable Tlr7-knockout (Tlr7-KO) cells using CRISPR-Cas9 and a specific guide RNA (Fig. 6A). Both Western blot and immunofluorescence label showed TLR7-KO in RAW264.7 macrophages. Tlr7-WT and Tlr7-KO RAW264.7 macrophages were then treated with synthetic mature miR-122, miR-16, or miR-29a and subjected to TNF-α expression detection. As shown in Fig. 6B, miR-122 and miR-29a both significantly increased TNF-α production in Tlr7-WT macrophages but not in Tlr7-KO macrophages. Serving as a negative control, miR-16 did not induce TNF-α production in both macrophages.
Fig. 6.
MiR-122 elicits mouse macrophages inflammatory responses via activating Tlr7 signal pathway. (A) Tlr7 knockout in mouse macrophages by the CRISPR-cas9 technique. (B) Tlr7 KO abolishes the effect of miR-122 on mouse macrophage inflammatory activation. (C, Upper) Mutation of GU-rich sequences in miR-122. (Lower) miR-122 but not miR-122 mutant stimulates RAW246.1 cells to secrete TNF-α in a time- and dose-dependent manner. (D) TNF-α secretion by mouse alveolar macrophages treated with various miRNAs, including miR-16, miR-29a, miR-122, and miR-122 mutant. Data are presented as mean ± SEM. **P < 0.01, ***P < 0.001. Two-way ANOVA followed by Bonferroni’s multiple comparisons test (B), or student’s two-tailed, unpaired t test (C), or one-way ANOVA followed by Bonferroni’s multiple comparisons test (D).
To examine the specificity of interaction between miR-122 and TLR7, we removed the TLR7 “binding sequence” in miR-122 by mutating GU bases in the 3′-terminus of miR-122 to AT (Fig. 6C, Upper); the miR-122 mutant (miR-122-mut) completely lost the capacity to activate RAW264.7 macrophage TNF-α expression in a time- and concentration-dependent manner (Fig. 6C, Lower). We also treated mouse primary alveolar macrophages with miR-122, miR-122-mut, miR-29a, and miR-16, and assessed TNF-α production. As shown in Fig. 6D, miR-122 and miR-29a, but not miR-122-mut or miR-16, induced TNF-α production in primary alveolar macrophages.
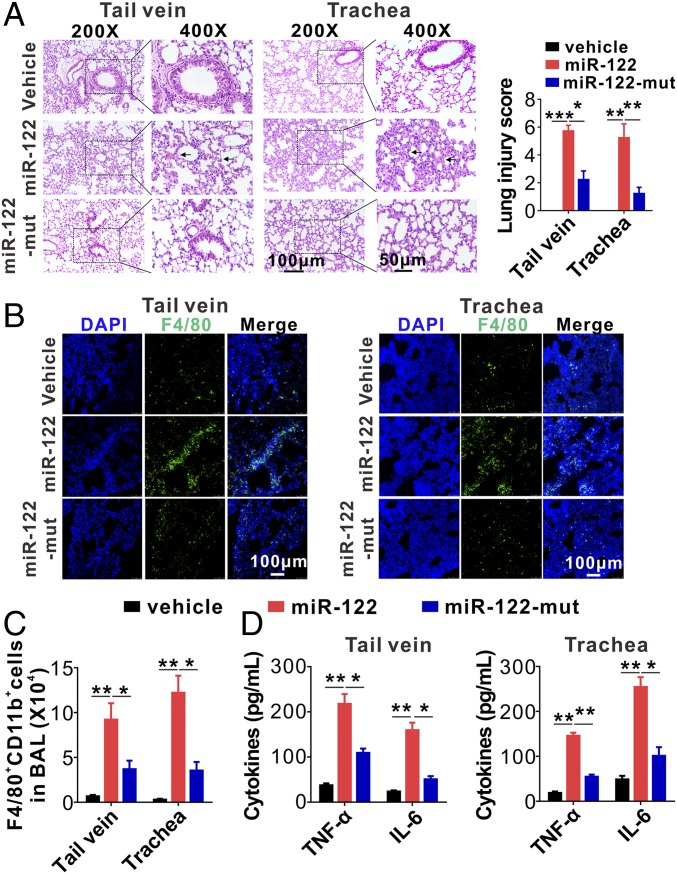
To further validate the role of miR-122 in eliciting mouse acute pulmonary inflammation by activating alveolar macrophage TLR7 signaling, we directly injected saline (vehicle), synthetic miR-122, or miR-122-mut into mice via tail vein or respiratory trachea. At 6-h postinjection, the mice were killed and the miR-122 level in mouse peripheral blood, lungs, and alveolar macrophages, as well as the degree of lung-tissue injury, infiltration of inflammatory macrophages, and cytokine production, were monitored. Although the levels of both miR-122 and miR122-mut were both significantly increased in mouse lung tissue following injection via tail vein or respiratory trachea (SI Appendix, Fig. S9), miR-122 induced a much stronger lung injury than miR-122-mut and vehicle control (Fig. 7A). Immunofluorescence labeling of mouse lung tissue sections clearly showed that injection of miR-122 via tail vein (Fig. 7B, Left) and respiratory trachea (Fig. 7B, Right) induced stronger lung infiltration of inflammatory F4/80+ macrophages (Fig. 7B, green) than injection of miR-122-mut and vehicle control. We next isolated macrophages from mouse BAL and assessed the cell populations and cytokine release via flow cytometry and ELISA, respectively. As shown in Fig. 7C, injection of miR-122 via tail vein or respiratory trachea both induced significantly more infiltration of F4/80+CD11b+ macrophages into mouse lungs than injection of miR-122-mut and vehicle control. Consistent with this result, alveolar macrophages isolated from mice injected with miR-122 via tail vein (Fig. 7D, Left) and respiratory trachea (Fig. 7D, Right) displayed significantly higher TNF-α and IL-6 levels than those isolated from mice injected with miR-122-mut and vehicle control. These results collectively suggest that hepatic miR-122 elicits mouse alveolar macrophage inflammatory responses via activating the Tlr7 signaling pathway.
Fig. 7.
MiR-122-mut fails to elicit mouse acute lung inflammation and tissue damage. (A) Mouse pulmonary inflammation and lung tissue damage quantitated by H&E staining (Left) and lung injury scoring (Right) (six mice per group, three to four images for each mouse). (B) Alveolar infiltration of F4/80+ macrophages in mice with injection of vehicle control, miR-122, or miR-122 mutant via tail vein or respiratory trachea, respectively. Macrophage infiltration was detected by immunofluorescence. (C) FACS analysis of F4/80+CD11b+ macrophages in BALF harvested from the mice injected with miR-122 or miR-122 mutant (six mice per group, n = 3). (D) Levels of inflammatory cytokines in BALF harvested from recipient mice following injection of miR-122 or miR-122 mutant via tail vein (Left) or respiratory trachea (Right) (six mice per group, n = 3). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA followed by Bonferroni’s multiple comparisons test.
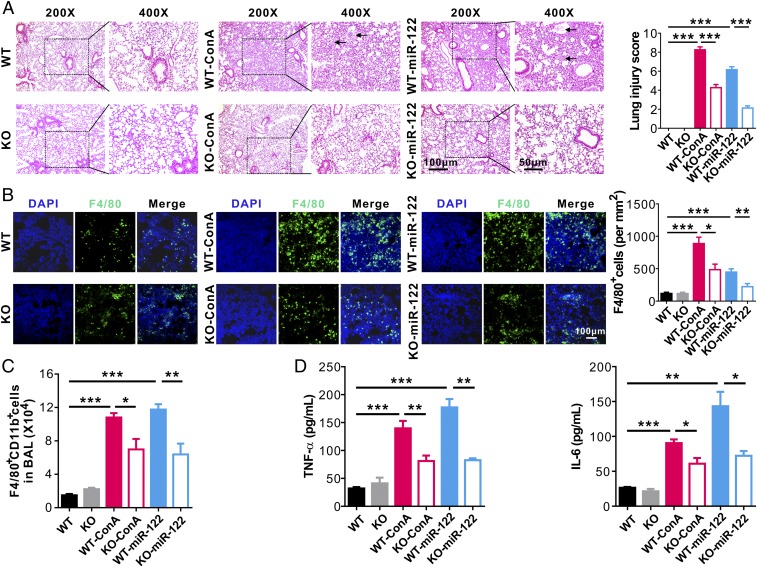
Finally, to determine the effects of Tlr7 signaling on miR-122–induced pulmonary inflammation and lung injury, WT and Tlr7-KO mice (32) were subjected to ConA or synthetic miR-122 treatment. As shown in Fig. 8A, Left, WT mice treated with ConA or synthetic miR-122 (WT-ConA or WT–miR-122 group) displayed more diffuse lesions with extensive interstitial hemorrhage, inflammatory cell infiltration, and alveolar edema compared with Tlr7-KO mice subjected to the same treatment (KO-ConA and KO–miR-122 groups). A significantly higher lung injury score was observed in the WT-ConA group and WT–miR-122 group compared with that in the other four groups (Fig. 8A, Right). Immunofluorescence (Fig. 8B) and flow cytometry (Fig. 8C) analysis of immune cell infiltration both indicated more infiltration of F4/80+CD11b+ macrophages into mouse lungs in the WT-ConA group and WT–miR-122 group compared with that in the Tlr7-KO groups. Consistent with this, the levels of inflammatory cytokines TNF-α and IL-6 in mouse BALF were significantly higher in WT mice injected with ConA or synthetic miR-122 (WT-ConA group and WT–miR-122 group) than those from the Tlr7-KO groups subjected to the same treatment (KO-ConA group and KO–miR-122) (Fig. 8D).
Fig. 8.
Lung inflammation and tissue damage induced by miR-122 is abolished by deleting Tlr7. (A) Mouse lung tissue damage analyzed by H&E staining (Left) and lung injury scoring (Right) in mice of control group (WT), Tlr7 depletion group (KO), control mice injected with ConA group (WT-ConA), control mice injected with synthetic miR-122 group (WT–miR-122), Tlr7-depletion mice injected with ConA group (KO-ConA), and Tlr7-depletion mice injected with synthetic miR-122 (KO–miR-122) (six mice per group, three to four images for each mouse). (B) Alveolar infiltration of F4/80+ macrophages in six group mice. Macrophage infiltration was detected by immunofluorescence. (C) FACS analysis of F4/80+CD11b+ macrophages in BAL harvested from the six group mice (six mice per group, n = 3). (D) Levels of TNF-α (Left) and IL-6 (Right) in BALF from the six group mice (six mice per group, n = 3). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. One-way ANOVA followed by Bonferroni’s multiple comparisons test.
Discussion
The present study provides evidence that hepatic miR-122 released from injured liver cells is a major causative factor of acute pulmonary inflammation and tissue damage induced by liver dysfunction. In this circulating miR-122-based mechanism, injured liver cells first release substantial amounts of miR-122 into the circulation in an exosome-independent pathway, and then the free-circulating miR-122 were preferentially transported into lung tissue, particularly alveolar macrophages. Inside alveolar macrophages, hepatic miR-122 activates the endosomal TLR7 signaling pathway and elicits macrophage inflammatory responses (SI Appendix, Fig. S10).
In agreement with previous reports (2, 3, 6), our results—derived from three different mouse liver injury models—showed that various liver diseases could cause pulmonary inflammation and tissue damage. This finding suggests that a common factor or signaling pathway may be responsible for liver injury-induced pulmonary inflammation and tissue damage. Although different factors—including reactive oxygen species or cytokines produced in liver ischemia and reperfusion (33, 34), CysLTs (6), and LTs (35)—have been identified to play a critical role in the different models, they are not the common factors mediating the pulmonary inflammation induced by various liver injuries. However, as a substantial amount of miR-122 was found to be released by injured liver cells in different liver injury mouse models, and circulating miR-122 can enter alveolar macrophages and activate the macrophage TLR7 signal pathway, miR-122 released by injured livers may serve as the common factor for eliciting pulmonary inflammation under various liver diseases.
Several pieces of evidence support that circulating miR-122 released by injured liver cells is a primary culprit of liver injury-induced pulmonary inflammation and tissue damage. First, miR-122 is the most enriched miRNA in livers, as it normally comprises nearly 70% of total liver miRNAs (36). Moreover, it has been well known that miR-122 can be released by hepatic cells through both active and passive mechanisms. Like other secreted miRNAs in circulation, miR-122 is stable and can be delivered to other organs and cells. Although the mechanism remains unclear, assessing miR-122 distribution by qRT-PCR showed that the liver-released miR-122 was preferentially delivered into mouse lung tissues (SI Appendix, Fig. S5), which implicates the involvement of hepatic miR-122 in acute pulmonary inflammation and tissue damage induced by various liver injuries. Second, both in vivo and in vitro assays showed that miR-122 was rapidly internalized into alveolar macrophages (Fig. 4). Third, combining biological and functional analyses, we demonstrated that, following macrophage internalization, miR-122 could specifically bind macrophage endosomal TLR7 (murine) and TLR8 (human) via a GU-rich TLR-binding sequence, leading to NF-κB activation and proinflammatory cytokine secretion. Both mutating the GU-rich sequence and directly deleting mouse macrophage endosomal Tlr7 completely abolished the effect of miR-122 on inflammatory activation of mouse macrophages, confirming the role of miR-122–TLR7 interaction in liver injury-induced pulmonary acute inflammation and tissue damage. Finally, miR-122 is an effective tumor suppressor, and its expression is significantly decreased in HCC. Therefore, increasing miR-122 levels in tumor cells has become a promising antitumor strategy (37–39). However, inflammatory “side-effects” of miR-122 are often observed upon its injection into animals. Interestingly, by modifying a GU motif in miR-122, Peacock et al. (40) found that immune stimulation induced by miR-122 could be controlled. These studies strongly support our finding that miR-122 binds to macrophage endosomal TLR7/8 via GU-rich sequences, leading to TLR7/8-mediated macrophage inflammatory responses.
Systemic measurement of miR-122 levels in various tissues found that, following liver injury (such as ConA-induced acute hepatitis), the level of miR-122 in mouse lungs was significantly higher than other tissues, except the spleen (SI Appendix, Fig. S5), suggesting that levels of circulating miR-122 are higher in the portal circulation compared with the systemic circulation. This notion is supported by the effect of hepatic miR-122 on activating tissue damage mainly in mouse lungs but not other tissues (SI Appendix, Fig. S1). It is also intriguing that injured liver-released miR-122 in the exosome-free fraction is quite stable. Because we identified the role of miR-150 released by stimulated monocytes in modulating endothelial cell function (10), we think that extracellular miRNAs are mainly packed into exosomes and secreted to serve as signal molecules in mediating cell–cell communication. However, qRT-PCR assays of both human and mouse samples clearly showed that plasma miR-122 was in the exosome-free fractions (SI Appendix, Fig. S3). To address the question how exosome-free miR-122 escapes the degradation of RNases, which are abundant in plasma, we performed protein analysis using LC-MS/MS after immunoprecipitating plasma miR-122 from hepatitis patients via biotin-conjugated miR-122 antisense oligonucleotide (41). As shown in SI Appendix, Table S4, a panel of proteins—particularly α-2-macroglobulin (A2M), α-2 globin chain (HBA2), apolipoprotein A-I (APOA1), haptoglobin (HP), hemoglobin-β (HBβ), and others—were found to be associated with miR-122. The binding of serum proteins to plasma miRNA was previously reported by Arroyo et al. (11). It is possible that these proteins bound to miR-122 play a role in protecting miRNA against degradation by RNases. Interestingly, HBA2 and HBβ are subunit of hemoglobin, and HP (42) has been previously reported to be able to bind to hemoglobin. Given that hemoglobin plays an essential role in oxygen-exchange in alveolar space, binding of miR-122 to hemoglobin or the proteins with high affinity to hemoglobin may also facilitate the preferential delivery of miR-122 into lungs.
Although our results show that hepatic miR-122 plays an important role in inducing acute pulmonary inflammation and lung tissue damage, other factors, such as high-mobility group box 1 (HMGB1) released by injured cells (43), may also contribute to the pulmonary inflammation and lung tissue injury. Huebener et al. (43) demonstrated that epithelial HMGB1, through interacting with receptor for advanced glycation end products, triggers neutrophil-mediated injury amplification following necrosis. Given that damaged hepatocytes can release HMGB1 into the circulation, it is possible that these circulating HMGB1 initiate alveolar macrophage inflammatory responses. However, because our LC-MS/MS analysis of miR-122–bound proteins did not show HMGB1 (SI Appendix, Table S4), the circulating HMGB1 may not play a major role in lung acute inflammation induced by an injured liver.
Elucidating the role of miR-122 in linking acute pulmonary inflammation and tissue damage to various liver injuries not only validates the concept that secreted miRNAs in circulation are essential signaling molecules that mediate communication among nonadjacent cells and organs (10, 12, 44, 45), but also provides evidence that liver dysfunctions can have deleterious effects on other organ systems. Consistent with the observation that miR-122 released by injured mouse liver cells was delivered to lung tissues (Fig. 4), chronic hepatitis patients with pulmonary effusion also displayed markedly higher miR-122 levels in pulmonary effusion patients than pneumonia patients without liver disorder (SI Appendix, Fig. S6). Our results suggest that human lung dysfunction can also be derived from liver injury and that hepatic miR-122 released from injured liver cells can activate alveolar macrophage inflammatory responses and elicit acute pulmonary inflammation and tissue damage. Clinically, many patients experience acute pulmonary inflammation and lung injury with no identifiable infectious sources and are resistant to antibacterial treatment. Given that liver injury, specifically miR-122 released from injured liver cells, potentially underlies acute pulmonary inflammation and tissue damage, patients with these symptoms should be examined for liver injury and dysfunction.
In conclusion, the present study demonstrates evidence that miR-122 released from injured liver cells can elicit acute pulmonary inflammation responses via activating alveolar macrophage TLR7/8. By identifying this unconventional role of hepatic miR-122 in stimulating lung immune injury, our findings provide a mechanism by which pulmonary inflammation and tissue damage can be caused, and identify hepatic miR-122 and its activated TLR7/8 signaling pathway as potential therapeutic targets in controlling liver injury-induced pulmonary inflammation.
Materials and Methods
Human Samples, Patient Characteristics, and Clinical Features.
Study protocols involved human sample collection and treatment were approved by the Institutional Review Board at Drum Tower Hospital Affiliated to Medical School of Nanjing University (Nanjing, China). Written informed consent was obtained from each individual before enrolment. Peripheral blood (15 mL each) was drawn from 10 healthy donors, 10 patients with ongoing hepatitis, and 10 patients with HCC admitted to Nanjing Drum Tower Hospital, respectively. The clinical features of the patients are detailed in SI Appendix, Table S1. Pulmonary effusion samples were collected from 10 chronic hepatitis patients with pulmonary effusion and 10 patients with severe pneumonia but without liver disorder. After centrifugation at 1,500 × g for 10 min, the supernatants of pulmonary effusion were collected and stored at −80 °C until analysis. The clinical features of the patients are detailed in SI Appendix, Table S2.
Mouse Studies.
C57Bl/6J and SCID (severe combined immune deficiency) mice were purchased from the Model Animal Research Center, Nanjing University (Nanjing, China). Tlr7-KO mice were obtained from The Jackson Laboratory (Bar Harbor, Maine) and all the KO mice were backcrossed with C57BL/6 mice over 10 generations (32). All mouse protocols followed the National Institutes of Health guidelines for the care and use of mice and were approved by the Institutional Animal Care and Use Committee of Nanjing University (Nanjing, China).
Supplementary Material
Acknowledgments
We thank Dr. Jill Leslie Littrell (Georgia State University) for critical reading and constructive discussion of the manuscript. This work was supported by Ministry of Science and Technology of China Grant 2018YFA0507100; National Natural Science Foundation of China Grants 31670917, 91640103, 31770981, and 31741075; Natural Science Foundation of Jiangsu Province Grant BK20170076; Six talent peaks project of Jiangsu Province (YY-012) and the Fundamental Research Funds for the Central Universities Grants 020814380039 and 020814380082; National Science Foundation for Young Scientists of China Grant 81401895; The National Postdoctoral Program for Innovative Talents Grant BX20180139; and the Scientific Research Foundation of Graduate School of Nanjing University Grant 2016CL08.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.R.B. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE124360).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814139116/-/DCSupplemental.
References
- 1.Huffmyer JL, Nemergut EC. Respiratory dysfunction and pulmonary disease in cirrhosis and other hepatic disorders. Respir Care. 2007;52:1030–1036. [PubMed] [Google Scholar]
- 2.Fallon MB, Abrams GA. Pulmonary dysfunction in chronic liver disease. Hepatology. 2000;32:859–865. doi: 10.1053/jhep.2000.7519. [DOI] [PubMed] [Google Scholar]
- 3.Rodriguez-Roisin R, Krowka MJ. Hepatopulmonary syndrome—A liver-induced lung vascular disorder. N Engl J Med. 2008;358:2378–2387. doi: 10.1056/NEJMra0707185. [DOI] [PubMed] [Google Scholar]
- 4.Lange PA, Stoller JK. The hepatopulmonary syndrome. Ann Intern Med. 1995;122:521–529. doi: 10.7326/0003-4819-122-7-199504010-00008. [DOI] [PubMed] [Google Scholar]
- 5.Lin HI, et al. Reperfusion liver injury induces down-regulation of eNOS and up-regulation of iNOS in lung tissues. Transplant Proc. 2006;38:2203–2206. doi: 10.1016/j.transproceed.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Takamatsu Y, et al. Role of leukotrienes on hepatic ischemia/reperfusion injury in rats. J Surg Res. 2004;119:14–20. doi: 10.1016/j.jss.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Al-Amran FG, Hadi NR, Hashim AM. Leukotriene biosynthesis inhibition ameliorates acute lung injury following hemorrhagic shock in rats. J Cardiothorac Surg. 2011;6:81. doi: 10.1186/1749-8090-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. In: Abbas AK, Galli SJ, Howley PM, editors. Annual Review of Pathology: Mechanisms of Disease. Vol 9. Annual Reviews; Palo Alto, CA: 2014. pp. 287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen X, Liang H, Zhang J, Zen K, Zhang C-Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 15.Fabbri M, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci USA. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He WA, et al. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. Proc Natl Acad Sci USA. 2014;111:4525–4529. doi: 10.1073/pnas.1402714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casadei L, et al. Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res. 2017;77:3846–3856. doi: 10.1158/0008-5472.CAN-16-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehmann SM, et al. An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 19.Jin F, et al. MiR-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017;8:e2540. doi: 10.1038/cddis.2016.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S, Kamata H, Luo JL, Leffert H, Karin M. IKKbeta couples hepatocyte death to cytokine-driven compensatory proliferation that promotes chemical hepatocarcinogenesis. Cell. 2005;121:977–990. doi: 10.1016/j.cell.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 21.Diao W, et al. The protective role of myeloid-derived suppressor cells in concanavalin A-induced hepatic injury. Protein Cell. 2014;5:714–724. doi: 10.1007/s13238-014-0069-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gantner F, Leist M, Lohse AW, Germann PG, Tiegs G. Concanavalin A-induced T-cell-mediated hepatic injury in mice: The role of tumor necrosis factor. Hepatology. 1995;21:190–198. doi: 10.1016/0270-9139(95)90428-x. [DOI] [PubMed] [Google Scholar]
- 23.Hussell T, Bell TJ. Alveolar macrophages: Plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 24.Wang K, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L-M, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798–9807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 26.Xie J, et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat Methods. 2012;9:403–409. doi: 10.1038/nmeth.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cai X, et al. Re-polarization of tumor-associated macrophages to pro-inflammatory M1 macrophages by microRNA-155. J Mol Cell Biol. 2012;4:341–343. doi: 10.1093/jmcb/mjs044. [DOI] [PubMed] [Google Scholar]
- 28.van Rooijen N, van Kesteren-Hendrikx E. “In vivo” depletion of macrophages by liposome-mediated “suicide”. Methods Enzymol. 2003;373:3–16. doi: 10.1016/s0076-6879(03)73001-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, et al. 2019 RAW246.1 cells activation by internalized synthetic miR-122. Gene Expression Omnibus. Available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE124360. Deposited December 26, 2018.
- 30.Challagundla KB, et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J Natl Cancer Inst. 2015;107:djv135. doi: 10.1093/jnci/djv135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabbri M. TLRs as miRNA receptors. Cancer Res. 2012;72:6333–6337. doi: 10.1158/0008-5472.CAN-12-3229. [DOI] [PubMed] [Google Scholar]
- 32.Fu Y-J, et al. Effects of different principles of traditional Chinese medicine treatment on TLR7/NF-κB signaling pathway in influenza virus infected mice. Chin Med. 2018;13:42. doi: 10.1186/s13020-018-0199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiu MH, et al. Protective effect of melatonin on liver ischemia-reperfusion induced pulmonary microvascular injury in rats. Transplant Proc. 2012;44:962–965. doi: 10.1016/j.transproceed.2012.01.097. [DOI] [PubMed] [Google Scholar]
- 34.Yeh DYW, Yang YC, Wang JJ. Hepatic warm ischemia-reperfusion-induced increase in pulmonary capillary filtration is ameliorated by administration of a multidrug resistance-associated protein 1 inhibitor and leukotriene D4 antagonist (MK-571) through reducing neutrophil infiltration and pulmonary inflammation and oxidative stress in rats. Transplant Proc. 2015;47:1087–1091. doi: 10.1016/j.transproceed.2014.10.061. [DOI] [PubMed] [Google Scholar]
- 35.Al-Amran FG, Hadi NR, Hashim AM. Cysteinyl leukotriene receptor antagonist montelukast ameliorates acute lung injury following haemorrhagic shock in rats. Eur J Cardiothorac Surg. 2013;43:421–427. doi: 10.1093/ejcts/ezs312. [DOI] [PubMed] [Google Scholar]
- 36.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from HCR mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 37.Hsu SH, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122:2871–2883. doi: 10.1172/JCI63539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsai W-C, et al. MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis. J Clin Invest. 2012;122:2884–2897. doi: 10.1172/JCI63455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122—A key factor and therapeutic target in liver disease. J Hepatol. 2015;62:448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 40.Peacock H, et al. Nucleobase and ribose modifications control immunostimulation by a microRNA-122-mimetic RNA. J Am Chem Soc. 2011;133:9200–9203. doi: 10.1021/ja202492e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang R, et al. Mouse miRNA-709 directly regulates miRNA-15a/16-1 biogenesis at the posttranscriptional level in the nucleus: Evidence for a microRNA hierarchy system. Cell Res. 2012;22:504–515. doi: 10.1038/cr.2011.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wassell J. Haptoglobin: Function and polymorphism. Clin Lab. 2000;46:547–552. [PubMed] [Google Scholar]
- 43.Huebener P, et al. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J Clin Invest. 2015;125:539–550. doi: 10.1172/JCI76887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ying W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Yin Y, et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014;24:1164–1180. doi: 10.1038/cr.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.