Significance
Lipoprotein lipase (LPL) plays a central role in plasma lipid metabolism, hydrolyzing the triglycerides in lipoproteins and releasing fatty acid nutrients for vital tissues. LPL is synthesized by adipocytes and myocytes and secreted into the interstitial spaces, whereupon it is captured by an endothelial cell protein, GPIHBP1, and transported to its site of action within the capillary lumen. For decades, LPL has been assumed to be a homodimer, with dimerization thought to be required for both secretion and catalytic activity. In the present study, we revisited the notion that LPL is a homodimer. Our findings indicate that LPL and GPIHBP1-bound LPL are active in a monomeric state.
Keywords: lipase, triglycerides, lipolysis
Abstract
Lipoprotein lipase (LPL), the enzyme that hydrolyzes triglycerides in plasma lipoproteins, is assumed to be active only as a homodimer. In support of this idea, several groups have reported that the size of LPL, as measured by density gradient ultracentrifugation, is ∼110 kDa, twice the size of LPL monomers (∼55 kDa). Of note, however, in those studies the LPL had been incubated with heparin, a polyanionic substance that binds and stabilizes LPL. Here we revisited the assumption that LPL is active only as a homodimer. When freshly secreted human LPL (or purified preparations of LPL) was subjected to density gradient ultracentrifugation (in the absence of heparin), LPL mass and activity peaks exhibited the size expected of monomers (near the 66-kDa albumin standard). GPIHBP1-bound LPL also exhibited the size expected for a monomer. In the presence of heparin, LPL size increased, overlapping with a 97.2-kDa standard. We also used density gradient ultracentrifugation to characterize the LPL within the high-salt and low-salt peaks from a heparin-Sepharose column. The catalytically active LPL within the high-salt peak exhibited the size of monomers, whereas most of the inactive LPL in the low-salt peak was at the bottom of the tube (in aggregates). Consistent with those findings, the LPL in the low-salt peak, but not that in the high-salt peak, was easily detectable with single mAb sandwich ELISAs, in which LPL is captured and detected with the same antibody. We conclude that catalytically active LPL can exist in a monomeric state.
Lipoprotein lipase (LPL) is arguably the central molecule in plasma lipid metabolism, hydrolyzing the triglycerides in lipoproteins and releasing fatty acid nutrients for use by vital tissues (1). LPL is synthesized by parenchymal cells, mainly adipocytes and myocytes, and secreted into the interstitial spaces, where it is captured by an endothelial cell protein, GPIHBP1, and transported to the capillary lumen (2). LPL is a member of a lipase family that includes pancreatic lipase, hepatic triglyceride lipase, and endothelial lipase. Each of these enzymes has an amino-terminal α/β-hydrolase domain harboring a catalytic triad and a carboxyl-terminal β-barrel domain that interacts with lipids. In the case of LPL, the carboxyl-terminal domain mediates GPIHBP1 binding, as well as binding of lipoprotein particles (3–6).
Mature human LPL contains 448 amino acids and two N-linked glycans. By SDS/PAGE, LPL has a molecular mass of ∼55 kDa (7), but the functional (catalytically active) unit of LPL is widely assumed to be a homodimer, with a size of ∼110 kDa (8–14). Several observations have been interpreted as supporting the idea that LPL is a homodimer. One of these relates to interactions of LPL with heparin-Sepharose beads. LPL bound to heparin-Sepharose can be eluted with a linear NaCl gradient in two peaks: a low-salt (∼0.5 M NaCl) peak containing inactive (or minimally active) LPL and a high-salt (∼1.2 M NaCl) peak containing catalytically active LPL (8–10). The low-salt peak is thought to contain catalytically inactive LPL monomers, while the high-salt peak is thought to contain active homodimers (8–10). By virtue of having two LPL molecules, homodimers have been thought to bind more avidly to heparin-Sepharose, explaining why higher NaCl concentrations are required to elute the active form (10, 15).
Another observation favoring LPL homodimers is the size of LPL in sucrose density gradient ultracentrifugation studies. Density gradient ultracentrifugation studies by several laboratories have indicated that the size of heparin-stabilized LPL is ∼110 kDa, corresponding to the expected size of homodimers (15–17). Immunochemical studies have also been interpreted as favoring the existence of LPL homodimers. For example, several groups have shown that the LPL in postheparin plasma can be detected with a “single mAb” sandwich ELISA (i.e., an ELISA in which the LPL is both captured and detected with the same mAb) (11, 18). Each of these lines of evidence is subject to caveats, however. For example, one could argue that the LPL in the high-salt peak from a heparin-Sepharose column is monomeric, and that both the avid heparin binding and the robust catalytic activity simply reflect proper folding of LPL monomers. Moreover, one could also argue that the ∼110-kDa size of heparin-stabilized LPL in density gradient ultracentrifugation studies is related either to the binding of heparin or to heparin-induced LPL dimers/multimers. The immunochemical observations are also subject to caveats. Of note, sandwich ELISAs that use two different LPL-specific mAbs (capturing LPL with one mAb and detecting the bound LPL with another mAb) are far more sensitive than single-mAb sandwich ELISAs (18). One potential explanation for the reduced sensitivity of single-mAb sandwich ELISAs is that most LPL is in the form of monomers (undetectable by a single-antibody sandwich ELISA), and that only a subpopulation is in the form of dimers (detectable by a single-antibody sandwich ELISA).
Recently, Birrane et al. (19) solved the structure of a lipid-free LPL:GPIHBP1 complex by X-ray crystallography. Two LPL molecules were present in the crystallographic unit, and they interacted in a reciprocal fashion at a single site, between the hydrophobic Trp-rich motif in the carboxyl-terminal domain of one LPL molecule (sequences that mediate lipoprotein binding) (5, 6) and the hydrophobic catalytic pocket in the amino-terminal domain of the other LPL molecule (sequences that hydrolyze triglycerides). The same orientation was observed for LPL in solution by small-angle X-ray scattering (SAXS) analyses (19). The conformation observed by X-ray crystallography and SAXS would appear to support the long-held assumption that LPL is a homodimer. Again, however, there are caveats. One caveat is that the LPL homodimers observed by X-ray crystallography and SAXS occurred in the setting of high concentrations of LPL (0.7–15 mg/mL). It is likely that high protein concentrations favor homodimer formation. Another important caveat is that the conformation observed by X-ray crystallography differs substantially from what would be expected for a catalytically active homodimer, in that the intercalation of the Trp-rich motif of one LPL molecule into the catalytic pocket of the other LPL molecule seemingly would preclude lipoprotein binding and triglyceride hydrolysis (19). If catalytically active homodimers exist physiologically, then it seems likely that they would exhibit a conformation distinct from that observed in the crystal structure.
In the present study, we revisited the notion that LPL is active only as a homodimer, motivated in part by the realization that the evidence favoring LPL homodimers is subject to caveats. In addition, the fact that pancreatic lipase is active as a monomer (20, 21) makes it reasonable to suspect that other members of the same family (e.g., LPL) might also be active as monomers. In addition, we reasoned that the availability of new research tools (e.g., recombinant LPL, recombinant GPIHBP1, LPL-specific mAbs) might allow us to gain fresh insight into the properties of catalytically active LPL.
Results
Assessing LPL Size by Density Gradient Ultracentrifugation.
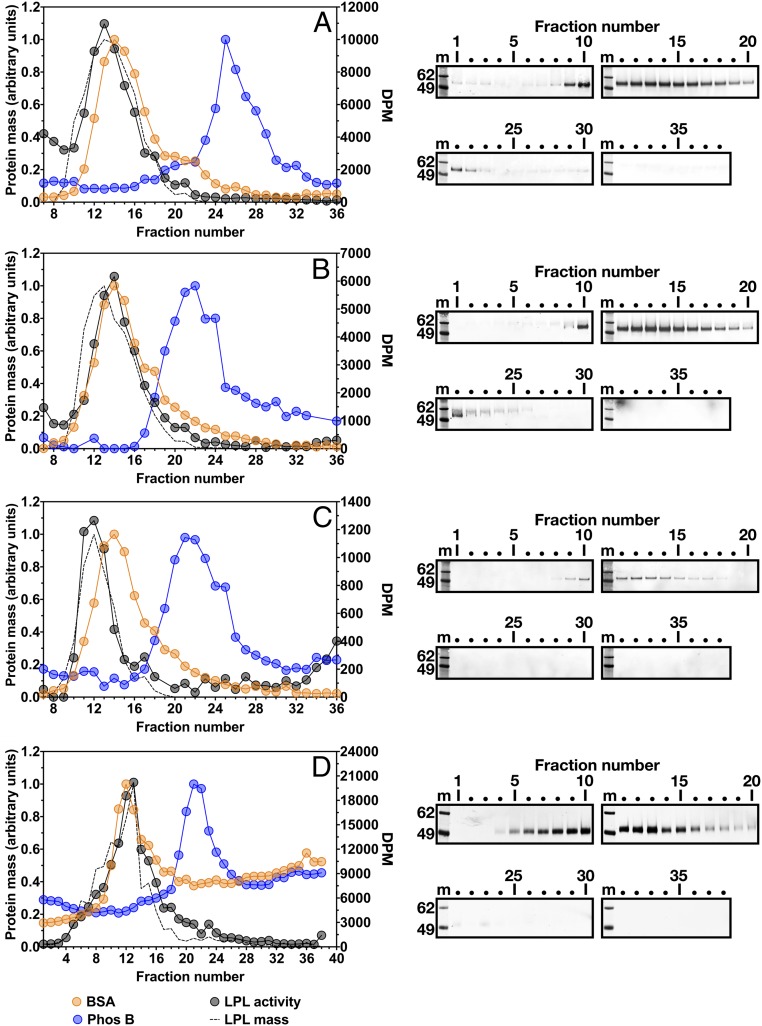
We used density gradient ultracentrifugation to assess the size of purified preparations of human and bovine LPL (Fig. 1 A–C), as well as LPL in the conditioned medium of stably transfected CHO cells (Fig. 1D). LPL activity in each density fraction was measured with a [3H]triolein substrate, and LPL mass was assessed by Western blot analysis. With purified human or bovine LPL, almost all the mass and activity eluted in density fractions 10–18, slightly before fractions containing a BSA standard (66 kDa). Small amounts of LPL protein were detected in fractions 19–23 [partially overlapping with fractions containing the phosphorylase b (Phos B) standard (97.2 kDa)], but catalytic activity in the latter fractions was quite low (Fig. 1 A–C). When medium from human LPL-expressing CHO cells was subjected to density gradient ultracentrifugation, most of the LPL activity and mass appeared in the same fractions as BSA, with only small amounts located in fractions containing the Phos B standard (Fig. 1D).
Fig. 1.
Analyzing human and bovine LPL by density gradient ultracentrifugation. Samples were loaded onto density gradients and centrifuged in an SW41 rotor (Beckman Coulter) for 23 h at 39,000 rpm. Gradients were unloaded in 38 fractions. Each fraction (70 μL) was tested for LPL activity with a [3H]triolein substrate [plotted as disintegrations per minute (DPM); y-axis on the right]. The black circles show LPL activity in the density fractions. LPL mass was assessed by Western blot analysis (blots shown on the right). The Western blots were scanned and quantified with an infrared scanner. The dotted black line depicts the intensity of the LPL band in each lane, normalized to the lane with the highest-intensity band. m, molecular mass standards. Two size markers were examined: BSA (66 kDa) and phosphorylase b (Phos B; 97.2 kDa). For each marker, protein concentration was quantified and normalized to the fraction with the highest protein concentration (protein mass is depicted on the y-axis on the left). (A and B) Size of human LPL, as judged by density gradient ultracentrifugation. Here 26 μg of purified human LPL was loaded onto a 10–30% glycerol gradient (A) or a 5–20% sucrose gradient (B). Western blot analysis was performed under nonreducing conditions with the human LPL-specific mAb 4-1a. (C) Size of bovine LPL by density gradient ultracentrifugation (5–20% sucrose gradient). Here 20 μg of purified bovine LPL was loaded onto the gradient, and Western blot analysis was performed with mAb 5D2. (D) Size of freshly secreted, catalytically active human LPL, as judged by density gradient ultracentrifugation. CHO cells stably expressing human LPL were grown in suspension culture for 2 h at 37 °C. The conditioned medium (225 μL) was loaded onto a 10–30% glycerol gradient. Western blot analysis was performed under nonreducing conditions with a rabbit polyclonal antibody against human LPL. Sucrose and glycerol density gradient studies yielded similar findings, but glycerol gradients resulted in somewhat improved preservation of LPL activity and improved separation of the BSA and Phos B markers. In D, the uneven baseline for protein standards reflects the use of an old batch of glycerol (known to interfere with the protein assay). Comparing density gradients from purified LPL and freshly secreted LPL, two differences are apparent. The freshly secreted LPL appears to be slightly larger than the purified LPL. In addition, there was no LPL activity or mass in the early fractions (fractions 1–5) with freshly secreted LPL. However, lipase activity was evident in fractions 1–5 with purified LPL, likely due to a contaminating, exogenous lipase, because human LPL was either undetectable or present in only very small amounts on the Western blot.
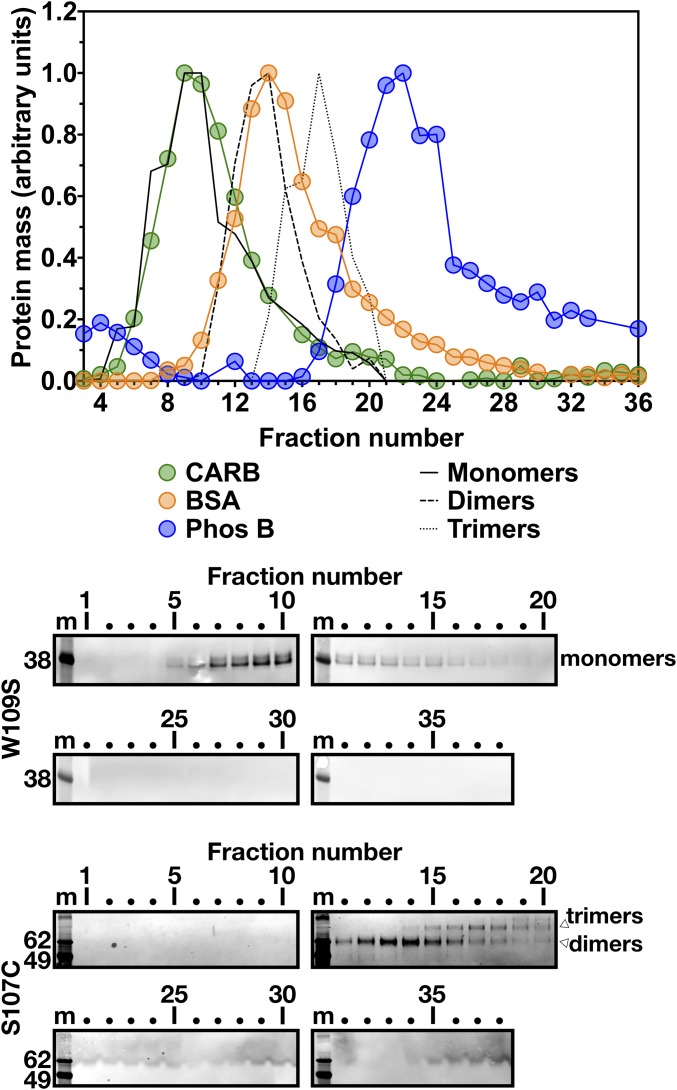
In parallel, we used density gradient ultracentrifugation to assess the size of uPAR-tagged, soluble versions of human GPIHBP1. We examined GPIHBP1-W109S (which folds normally and has little propensity to form disulfide-linked dimers) and GPIHBP1-S107C (which contains an unpaired cysteine and folds improperly, resulting in disulfide-linked dimers and multimers) (22). In density gradient studies, the GPIHBP1-W109S monomers (30.3 kDa) peaked in fractions 9 and 10, overlapping with the carbonic anhydrase (CARB) standard (29 kDa) (Fig. 2). GPIHBP1-S107C dimers (60.6 kDa) overlapped with the BSA standard, and GPIHBP1-S107C trimers (90.9 kDa) peaked between the BSA and Phos B standards (Fig. 2).
Fig. 2.
Size of human GPIHBP1 as measured by density gradient ultracentrifugation (5–20% sucrose gradient). Here 800 μL of medium from Drosophila S2 cells stably expressing GPIHBP1-W109S or GPIHBP1-S107C (both containing an amino-terminal uPAR tag and a carboxyl-terminal 11A12 epitope tag) was loaded onto density gradients and centrifuged in an SW41 rotor (Beckman Coulter) for 23 h at 39,000 rpm. GPIHBP1 in the 38 fractions was assessed by Western blot analysis with mAb 11A12 (blots shown below). For each GPIHBP1 species (monomers, solid black line; dimers, dashed black line; trimers, dotted black line), band intensity was normalized to the highest-intensity band, and the distribution of each GPIHBP1 species was plotted. m, molecular weight standards. GPIHBP1 monomers (30.3 kDa) were quantified from the GPIHBP1-W109S Western blots, and GPIHBP1 dimers (60.6 kDa) and trimers (90.9 kDa) were quantified from the GPIHBP1-S107C Western blots. The three size markers— carbonic anhydrase (CARB; 29 kDa), BSA (66 kDa), and phosphorylase b (Phos B; 97.2 kDa)—were examined and plotted as described in Fig. 1.
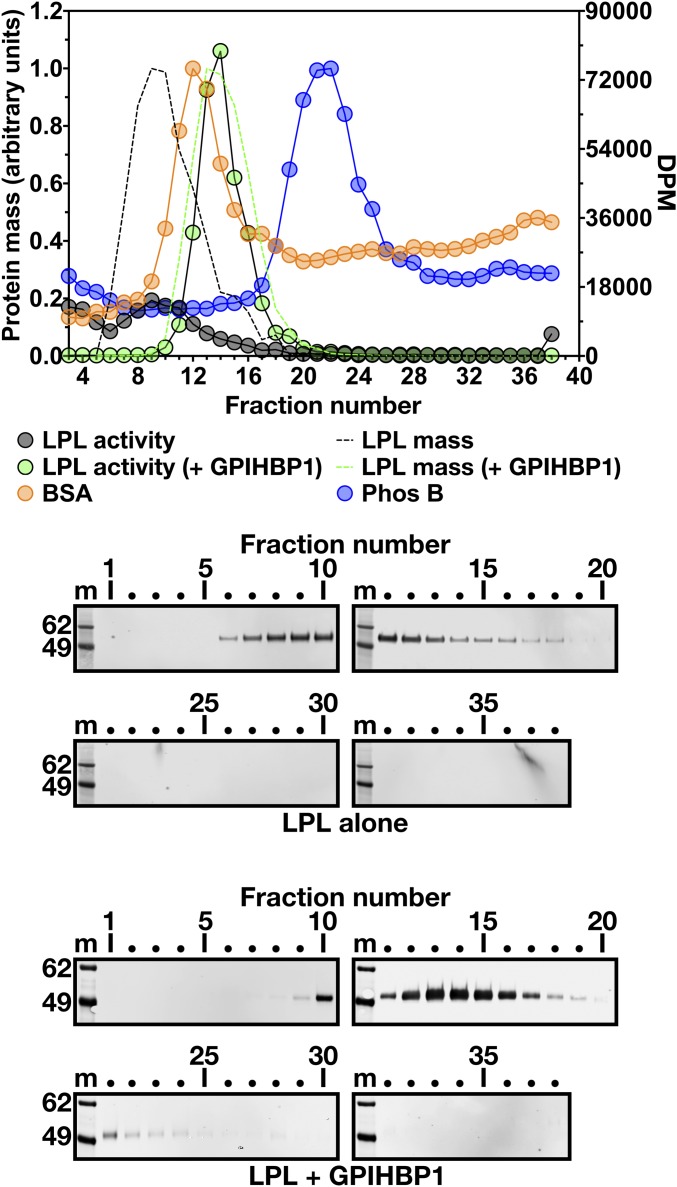
We also examined, by density gradient ultracentrifugation, the size of LPL complexed to wild-type soluble GPIHBP1 containing a uPAR-tag (Fig. 3). In that setting, LPL activity and mass peaked in density gradient fractions 13–14 (Fig. 3), overlapping with the BSA standard but with minimal overlap with Phos B. The mass and activity of LPL alone (in the absence of GPIHBP1 binding) peaked in fractions 8–11, several fractions before the BSA standard (Fig. 3). The catalytic activity in density fractions containing GPIHBP1-bound LPL was higher than in density fractions containing LPL alone (Fig. 3), consistent with GPIHBP1’s ability to prevent unfolding of the LPL hydrolase domain (23).
Fig. 3.
Size of GPIHBP1-bound LPL measured by density gradient ultracentrifugation. Purified human LPL (26 μg) was preincubated on ice for 10 min alone or in combination with a uPAR-tagged wild-type human GPIHBP1 (LPL + GPIHBP1) (23.7 μg) and then loaded on 10–30% glycerol gradients and centrifuged in an SW41 rotor (Beckman Coulter) for 23 h at 39,000 rpm. Two size markers were examined, BSA (66 kDa) and phosphorylase b (Phos B; 97.2 kDa), as described in Fig. 1. The uneven baseline for protein standards reflects the use of an old batch of glycerol (known to interfere with the protein assay). 70 μL of the “LPL alone” density fractions (black circles) and 0.5 μL of fractions for LPL + GPIHBP1 (green circles) were tested for LPL activity with a [3H]triolein substrate (plotted as DPM on the y-axis on the right). LPL mass is also shown, based on quantification of LPL bands on the Western blots shown below. Western blots were performed under nonreducing conditions with a rabbit polyclonal antibody against human LPL. Quantification of the Western blot data was performed as described in Fig. 1. The black dotted line represents data for LPL alone, and the green dotted line represents data for LPL + GPIHBP1. m, molecular weight standards.
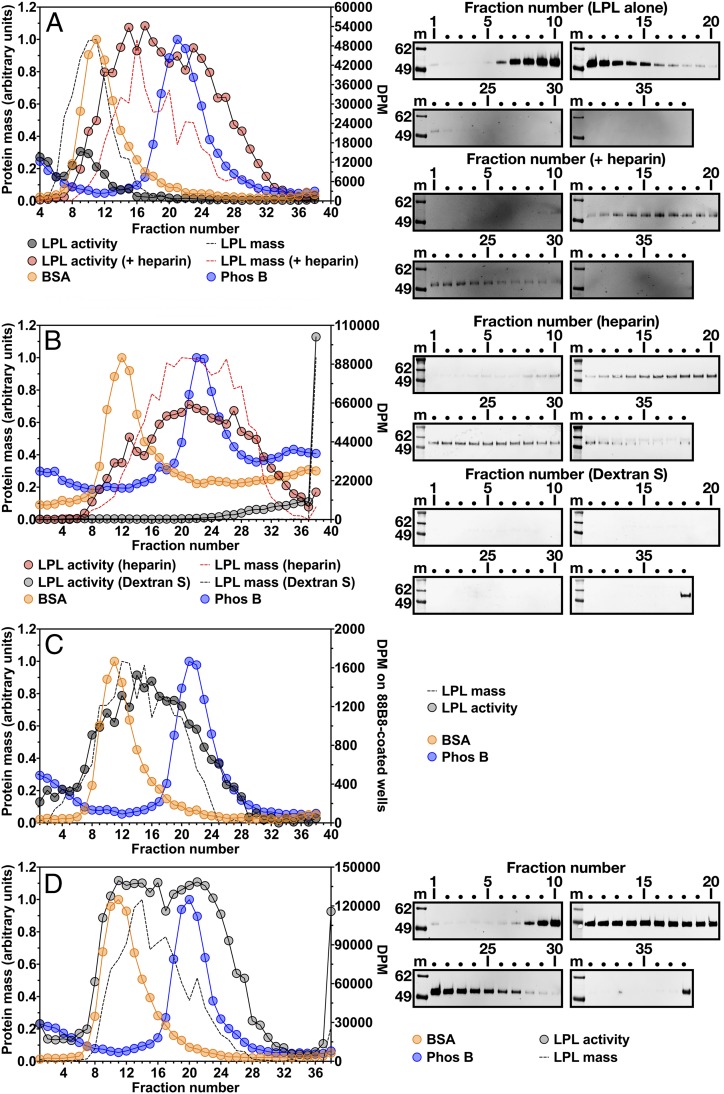
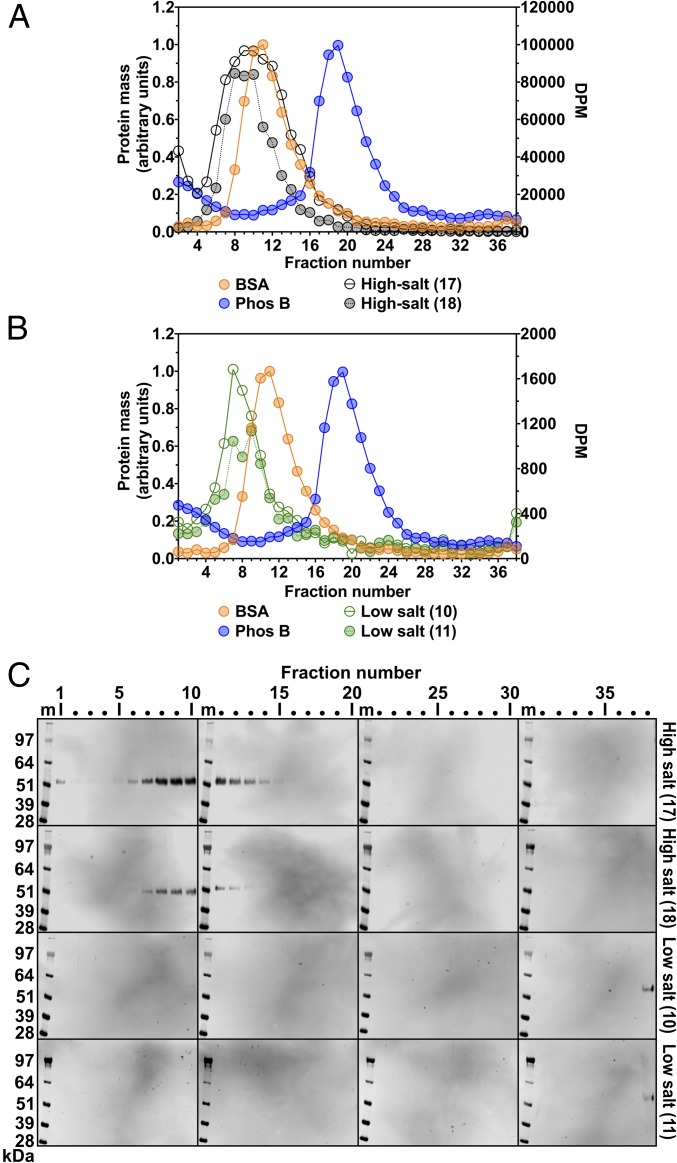
In previous sucrose density gradient ultracentrifugation studies by other groups (15–17), heparin-stabilized LPL exhibited the expected size of homodimers (∼110 kDa). To explore the possibility that heparin influences the size of LPL, we used density gradient ultracentrifugation studies to examine the size of purified preparations of human LPL before and after the addition of heparin (10 U/mL, molecular mass ∼15 kDa). Heparin increased the size of LPL, such that there was only partial overlap with the BSA standard and substantial overlap with Phos B (Fig. 4A). We observed similar findings when we assessed the size of LPL produced by stably transfected CHO cells that had been grown in the presence of heparin (Fig. 4B). Moreover, density gradient ultracentrifugation studies on postheparin human plasma revealed that LPL mass and activity were broadly distributed, overlapping with both BSA and Phos B standards (Fig. 4C).
Fig. 4.
The size of LPL, as judged by density gradient ultracentrifugation, is increased by heparin or dextran sulfate. Samples were loaded onto 10–30% glycerol gradients and centrifuged in an SW41 rotor (Beckman Coulter) for 23 h at 39,000 rpm. LPL activity in each fraction was tested with a [3H]triolein substrate (DPM on the y-axis on the right). For A, B, and D, LPL mass in each fraction was assessed with Western blot analysis of SDS-polyacrylamide gels (nonreduced samples). Western blots (shown on the right) were scanned and quantified, and the data were plotted as described in Fig. 1. m, molecular weight standards. Two protein standards, BSA (66 kDa) and phosphorylase b (Phos B; 97.2 kDa), were examined, as described in Fig. 1. (A) Size of purified human LPL in the presence and absence of heparin. Purified human LPL (20 µg) or purified human LPL (5 µg) that had been incubated with heparin (10 U/mL) was loaded onto density gradients. Density fractions (70 µL for LPL alone fractions, 25 µL for LPL + heparin fractions) were tested for LPL activity, and the data are plotted as gray and red circles, respectively. Western blot analysis was performed with a rabbit polyclonal antibody against human LPL; LPL mass data from the LPL alone and LPL + heparin Western blots are plotted as black and red dotted lines, respectively. (B) The size of freshly secreted LPL is influenced by the inclusion of heparin or dextran sulfate in the culture medium. Human LPL-expressing cells were grown in suspension culture (8 × 106 cells/mL) for 2 h at 37 °C in medium containing either heparin (50 U/mL) or dextran sulfate (molecular mass >500 kDa; 1 g/L). Conditioned medium (225 μL) was loaded onto gradients. The distribution of LPL catalytic activity in density fractions (70 μL) is plotted. The red circles represent LPL from medium containing heparin; gray circles, LPL from medium containing dextran sulfate. Western blots for LPL were performed with mAb 4-1a, and the band intensities were quantified. The red dotted line represents the distribution of LPL mass when cells were grown in medium containing heparin; the black dotted line, the distribution of LPL mass when cells were grown in medium containing dextran sulfate. The uneven baseline for protein standards reflects the use of an old batch of glycerol (known to interfere with the protein assay). (C) Size of LPL in postheparin plasma by density gradient ultracentrifugation. Pooled human postheparin plasma (200 μL) was loaded onto the gradient. To assess LPL activity, each fraction (150 μL) was loaded onto mAb 88B8-coated wells, and the triglyceride hydrolase activity of the captured LPL was measured with a [3H]triolein substrate. The gray circles represent LPL activity in the density fractions. Relative amounts of LPL mass were measured by adding 50 μL of each fraction to mAb 88B8-coated wells and then detecting bound LPL with HRP-labeled mAb 5D2. The OD450 value of each fraction, reflecting relative amounts of LPL mass, was normalized to the fraction with the highest OD450 value (fraction 12). LPL mass is plotted as a dotted black line. (D) Size of human LPL purified by ion-exchange chromatography in the presence of dextran sulfate (5 kDa). Here 20 µg of the LPL was loaded onto the density gradient. Each fraction (25 µL) was tested for LPL activity, and the data are plotted as gray circles. Western blot analysis was performed with a rabbit polyclonal antibody against human LPL; the LPL mass data are plotted as a black dotted line.
We also assessed, by density gradient ultracentrifugation, the size of human LPL produced by stably transfected CHO cells grown in the presence of dextran sulfate (molecular mass 500 kDa), which binds LPL and stabilizes its catalytic activity (24). The presence of dextran sulfate in the cell culture medium markedly increased the size of freshly secreted LPL, such that nearly all the LPL mass and activity was at the bottom of the tube (fraction 38) (Fig. 4B). When we evaluated the size of human LPL that had been purified in the presence of dextran sulfate (5 kDa), some of the LPL activity and mass eluted in fractions overlapping with the BSA and Phos B standards, but large amounts were found at the bottom of the tube (fraction 38) (Fig. 4D).
We also investigated the behavior of LPL that had been incubated with 1 M or 6 M guanidine hydrochloride. As expected, guanidine hydrochloride eliminated the catalytically active high-salt peak, as measured by heparin-Sepharose chromatography (SI Appendix, Fig. S1A). Of note, the size of LPL, as measured by density gradient ultracentrifugation, was unaffected by guanidine hydrochloride treatment; that is, the size of untreated LPL and guanidine hydrochloride-treated LPL appeared in fractions before and overlapping with the BSA standard (SI Appendix, Fig. S1 B and C).
LPL in the Low-Salt and High-Salt Peaks from a Heparin-Sepharose Column.
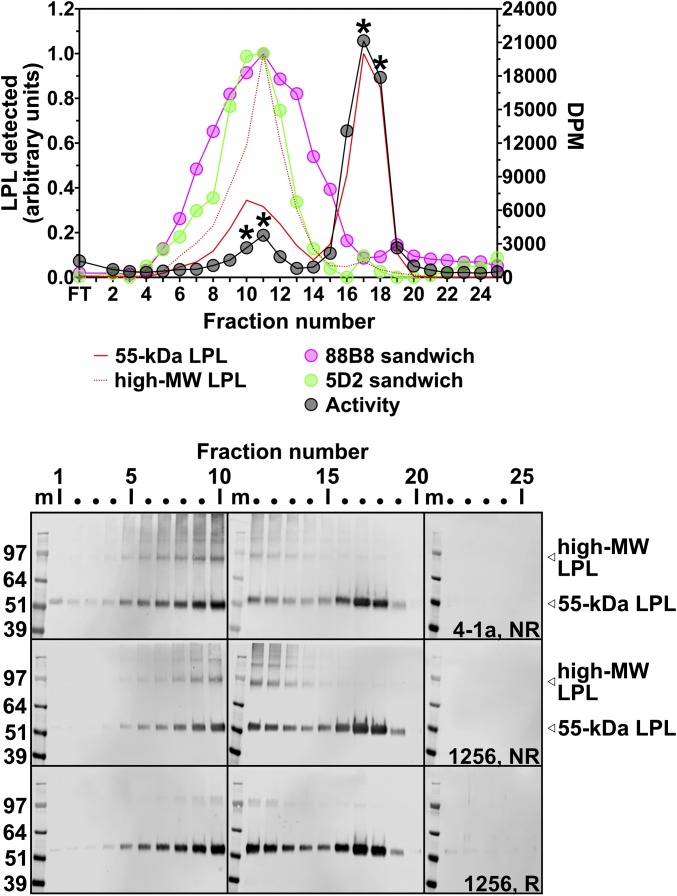
Conditioned medium from human LPL-expressing CHO cells was loaded onto a heparin-Sepharose column, and the LPL was eluted with a 0.4–2 M NaCl gradient. As expected, most of the catalytic activity was found in the high-salt peak (peaking in fractions 17–18), but a small amount was located in the low-salt peak (peaking in fractions 10–11) (Fig. 5). LPL mass in each fraction was assessed by SDS-polyacrylamide gel electrophoresis under nonreducing conditions, followed by Western blot analysis with an LPL-specific antibody. As expected, the 55-kDa LPL bands peaked in fractions 10 and 17, corresponding to the low-salt and high-salt peaks, respectively (Fig. 5). Under nonreducing conditions, higher molecular mass, SDS-resistant LPL species were present in fractions spanning the low-salt peak (fractions 5–13) (Fig. 5). When those fractions were electrophoresed under reducing conditions, the intensity of the higher molecular mass LPL bands decreased, and the intensity of the 55-kDa LPL bands increased (Fig. 5).
Fig. 5.
Elution of human LPL from a heparin-Sepharose column with a linear NaCl gradient. CHO cells expressing human LPL were grown in suspension culture for 16 h at 37 °C in medium containing protease inhibitors. Then 40 mL of medium was loaded onto a 3-mL heparin-Sepharose column, and the LPL was eluted with a NaCl gradient (0.4–2 M). For LPL activity measurements, 10 μL of each fraction was added to 88B8-coated wells, and triglyceride hydrolase activity was assessed with a [3H]triolein substrate (plotted as DPM on the y-axis on the right). The relative amount of LPL mass in the different fractions was assessed by Western blot analysis of SDS-polyacrylamide gels using mAb 4-1a or a rabbit polyclonal antibody against human LPL (antibody 1256). Samples were electrophoresed under reducing (R) and nonreducing (NR) conditions. A 55-kDa LPL band and a high molecular mass band (∼100 kDa) from the Western blots were quantified with an infrared scanner, normalized to the lane with the highest-intensity band, and averaged. The Western blot band intensity data (solid red line, 55-kDa LPL; dotted red line, high molecular weight LPL) were plotted along with the enzymatic activity data (black circles). LPL was characterized in each fraction (10 μL) with two different single-mAb sandwich ELISAs (an 88B8-88B8 sandwich ELISA and a 5D2-5D2 sandwich ELISA). The OD450 readings, after normalization to the well with the highest OD450 value, are plotted in pink for the 88B8-88B8 ELISA and in green for the 5D2-5D2 ELISA. *Denotes fractions that were subjected to density gradient ultracentrifugation (Fig. 6). FT, flow-through; m, SDS/PAGE protein size markers; MW, molecular weight.
We also analyzed the fractions from the heparin-Sepharose column with two different single-mAb sandwich ELISAs, one in which the LPL was captured and detected with mAb 5D2 and a second in which the LPL was bound and detected with mAb 88B8 (Fig. 5). The vast majority of the LPL detectable by the single-mAb ELISAs was found in the low-salt fractions. Neither single-mAb sandwich ELISA detected significant amounts of LPL in the high-salt peak, even though the high-salt peak contained most of the LPL mass and activity (Fig. 5).
We next examined individual fractions from the heparin-Sepharose column (specifically, fractions from within the low-salt and high-salt peaks) by density gradient ultracentrifugation. Nearly all the LPL activity in two fractions from the high-salt peak (fractions 17 and 18; Fig. 5) appeared in density gradient fractions 5–15 (slightly before and overlapping with the BSA standard) (Fig. 6A). LPL mass was also located in density fractions 5–15, as detected by Western blot analysis of SDS-polyacrylamide gels under nonreducing conditions (Fig. 6C and SI Appendix, Fig. S2).
Fig. 6.
Density gradient ultracentrifugation studies to assess the size of the LPL in fractions from the low- and high-salt peaks from a heparin-Sepharose column. Here 800 μL of two fractions from the low-salt peak (fractions 10 and 11) and 500 μL of two fractions from the high-salt peak (fractions 17 and 18) of the heparin-Sepharose column (shown in Fig. 5) were loaded onto 10–30% glycerol gradients and centrifuged in an SW41 rotor (Beckman Coulter) for 30 h at 39,000 rpm. Triglyceride hydrolase activity in 30 μL of each density fraction was assessed with a [3H]triolein substrate. (A) LPL activity (plotted as DPM on the y-axis on the right) for density gradient fractions derived from “high-salt peak fractions” 17 and 18. (B) LPL activity (plotted as DPM on the y-axis on the right) for density gradient fractions derived from “low-salt peak” fractions 10 and 11. Note the different scales on the right y-axis for A and B. (C) Assessing LPL mass in density gradient fractions by Western blot analysis with a rabbit polyclonal antibody against human LPL. Density gradient fractions derived from the high-salt peak fractions 17 and 18 were size-fractioned by SDS/PAGE under nonreducing conditions, while the density gradient fractions derived from the low-salt peak fractions 10 and 11 were electrophoresed under reducing conditions. m, SDS/PAGE protein size markers. Western blot band intensities for the high-salt fractions in C are quantified and plotted in SI Appendix, Fig. S2. SI Appendix, Fig. S3 depicts the distribution of LPL in density gradient fractions derived from the low-salt peak fractions 10 and 11, as measured by 88B8-88B8 and 5D2-5D2 single-mAb ELISA.
When fractions from the low-salt peak (fractions 10 and 11; Fig. 5) were examined by density gradient ultracentrifugation, only small amounts of LPL activity were detectable in the density gradient fractions, peaking in fractions 7–9, before and overlapping with the BSA standard (Fig. 6B; note the marked difference in scale compared with Fig. 6A). Most of the LPL mass was found at the bottom of the tube (fraction 38), as shown in Western blot analysis (Fig. 6C). That LPL, almost certainly in the form of aggregates, was readily detected by single-mAb ELISAs (SI Appendix, Fig. S3).
Testing Whether CHO Cells That Express Two Differentially Tagged LPL Proteins Produce Mixed LPL Species Containing Both Epitope Tags.
The density gradient ultracentrifugation studies revealed that freshly secreted, catalytically active LPL exhibits the size of a monomer (∼55 kDa). To further examine the properties of freshly secreted LPL, we cotransfected CHO cells with expression vectors for two differentially tagged LPLs, FLAG-tagged human LPL and S-protein–tagged human LPL, with the goal of determining whether the cotransfected cells produce LPL species containing both FLAG and S-protein tags (i.e., “mixed LPL species”). In one set of cotransfection experiments, both epitope tags were placed at the amino terminus of LPL (FLAGwt + Swt; Fig. 7 A–C); in another set of cotransfection studies, the FLAG tag was placed at the amino terminus of LPL while the S-protein tag was placed at the carboxyl terminus (FLAGwt + wS; Fig. 7 D–F).
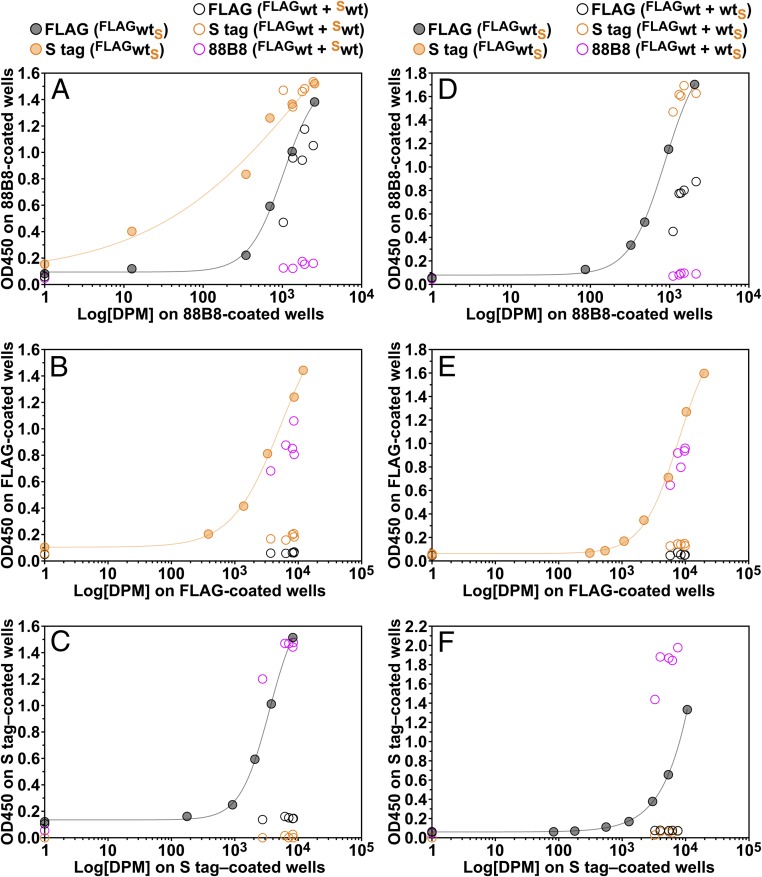
Fig. 7.
Testing the capacity of ELISAs to detect mixed LPL species in the medium of CHO cells that were cotransfected with two differentially tagged wild-type (wt) human LPL expression vectors (FLAG, S-protein). In one set of cotransfection experiments (A–C), both epitope tags were at the amino terminus of LPL (FLAGwt + Swt); in another set of cotransfection experiments (D–F), the FLAG tag was at the amino terminus, while the S-protein tag was at the carboxyl terminus (FLAGwt + wtS). As a control, we examined medium from cells that expressed a doubly tagged LPL (FLAGwtS). All of the tagged LPL constructs expressed catalytically active LPL. At 24 h after transfection, the medium was replaced, and the cells were incubated for 2 h at 37 °C in fresh medium containing 0.1% FBS. Samples of the conditioned medium were collected and applied to duplicate 96-well plates that had been coated with mAb 88B8 (A and D), a FLAG mAb (B and E), or an S-protein antibody (C and F). One set of 96-well plates was used solely for measuring LPL activity in samples of conditioned medium. Aliquots of medium from the cotransfected cells (either FLAGwt + Sw or FLAGwt + wtS) or dilutions of medium from cells expressing doubly tagged LPL (FLAGwtS) were added to 96-well plates, and the activity of antibody-captured LPL was assessed with a [3H]triolein substrate. As expected, all the samples of medium contained LPL activity (>103 DPM) (A–F). The second set of 96-well plates was used for ELISAs. The antibody-captured LPL was detected with HRP-labeled versions of mAb 88B8, FLAG antibody, or S-protein antibody. (The HRP antibody used is shown in the legend at the top of A and D.) The LPL activity was plotted on the x-axis, and the OD450 of the ELISA (reflecting relative amounts of LPL mass) was plotted on the y-axis. When captured with mAb 88B8 (A and D), the tagged LPLs were readily detected with the HRP-labeled epitope tag antibodies but not with HRP–mAb 88B8. Similarly, when the LPL was captured with the FLAG antibody (B and E), the LPL could be easily detected with HRP-mAb 88B8 but not with HRP-labeled FLAG antibody. When the LPL was captured with the S-protein antibody (C and F), the LPL could be easily detected with HRP-mAb 88B8 but not with HRP-labeled S-protein antibody. In addition, the LPL captured by the FLAG antibody (B and E) yielded a very low signal with the HRP-labeled S-protein antibody, and the LPL captured by the S-protein antibody (C and F) yielded a very low signal with the HRP-labeled FLAG antibody. In contrast, at similar activity levels, the LPL produced by FLAGwtS-transfected cells yielded a very robust signal in both ELISAs (B, C, E, and F). Considered together, these findings indicate that there were very low amounts of mixed LPL species [i.e., LPL species containing both FLAGwt and Swt (B and C) or both FLAGwt and wtS (E and F)] in the medium of the cotransfected cells. Of note, as shown in Fig. 8, the very low amounts of mixed LPL species present in the medium did not contribute to the catalytic activity found in the medium of the cotransfected cells.
To test for the presence of mixed LPL species containing both epitope tags, we used two-antibody sandwich ELISAs, capturing LPL with one epitope tag antibody and detecting the LPL with an HRP-labeled antibody against the other epitope tag. To confirm that the sandwich ELISAs were capable of detecting LPL species harboring two different epitope tags, we tested the ability of the ELISA to detect LPL in the medium of cells that had been transfected with an LPL construct containing an amino-terminal FLAG tag as well as a carboxyl-terminal S-protein tag (FLAGwtS). The vast majority of cells that had been cotransfected with two differentially tagged LPLs expressed both proteins (SI Appendix, Fig. S4 A and B). The conditioned medium from cells that had been cotransfected with two differentially tagged LPLs as well as medium from FLAGwtS-expressing cells contained catalytically active LPL (DPM plotted on the x-axis in Fig. 7). The LPL in the conditioned medium from the cotransfected cells was analyzed with a series of sandwich ELISAs in which the LPL was captured with mAb 88B8 (Fig. 7 A and D), a FLAG antibody (Fig. 7 B and E), or an S-protein antibody (Fig. 7 C and F). Two-antibody sandwich ELISAs using any combination of an epitope tag antibody and mAb 88B8 (Fig. 7) readily detected LPL in aliquots of the cell culture medium. In contrast, an “88B8-88B8” single-mAb sandwich ELISA (using mAb 88B8 to capture and detect LPL) detected only very small amounts of LPL in the same samples (Fig. 7 A and D). Similarly, only very small amounts of LPL were detected with single-antibody ELISAs using either of the epitope tag antibodies (Fig. 7 B, C, E, and F). Finally, we used two-antibody ELISAs to test for the presence of LPL species containing both epitope tags (i.e., ELISAs in which the LPL was captured with one epitope tag antibody and detected with the other epitope tag antibody). Only very small amounts of mixed LPL species were detected (Fig. 7 B, C, E, and F). Using the same ELISA, large amounts of doubly tagged LPL were detected in the medium of FLAGwtS-expressing cells (Fig. 7 B, C, E, and F). Similar findings were observed in an independent series of experiments (SI Appendix, Fig. S4 C–E).
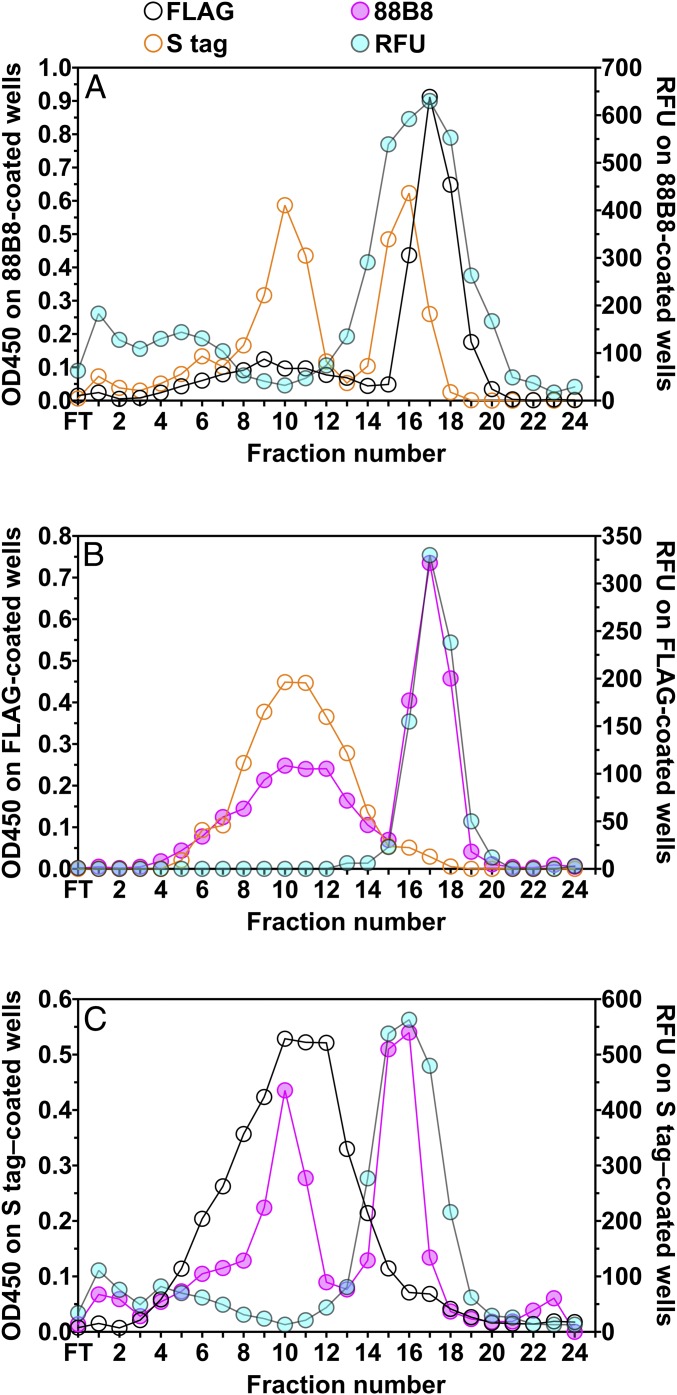
The experiments shown in Figs. 5 and 6 revealed that some of the LPL in the conditioned medium from LPL-expressing cells was in the form of aggregates (found in the low-salt peak from a heparin-Sepharose column). Unsurprisingly, those aggregates were readily detectable with single-mAb sandwich ELISAs using mAb 5D2 or mAb 88B8. In cells coexpressing two differentially tagged LPLs, we observed substantial amounts of LPL mass and activity in the conditioned medium but only very small amounts of mixed LPL species harboring both epitope tags (Fig. 7 B, C, E, and F and SI Appendix, Fig. S4 C–E). We predicted that the low amounts of mixed LPL species containing both epitope tags would be inactive and would be detected in the low-salt peak by heparin-Sepharose chromatography. Indeed, this was the case. Analysis of medium from cells coexpressing FLAG-tagged LPL and S-protein–tagged LPL by heparin-Sepharose chromatography revealed high catalytic activity in the high-salt peak but little activity in the low-salt peak (Fig. 8). Of note, the mixed LPL species harboring both FLAG and S-protein tags were largely confined to the low-salt peak and were nearly absent from the high-salt peak (Fig. 8 B and C).
Fig. 8.
Heparin-Sepharose chromatography analysis of properties of mixed LPL species in the medium of cells coexpressing two differentially tagged LPL constructs, one for LPL with an amino-terminal FLAG tag and the other for LPL containing a carboxyl-terminal S-protein tag (FLAGwt and wtS). Conditioned medium (40 mL) from the cotransfected cells was loaded onto a heparin-Sepharose column, and the LPL was eluted with a NaCl gradient into 25 fractions. Samples of each fraction were collected and applied to duplicate sets of 96-well plates that had been coated with mAb 88B8 (A), a FLAG mAb (B), or an S-protein antibody (C). To assess LPL activity, 175 μL from each fraction was added to one set of 96-well plates, and the esterase activity of antibody-captured LPL was measured with the DGGR substrate (plotted as relative fluorescent units; y-axis on the right). LPL mass in the samples was assessed with ELISAs by adding fractions (1–10 μL) onto the second set of 96-well plates. The antibody-captured LPL was then detected with HRP-labeled FLAG antibody, S-protein antibody, or mAb 88B8. The OD450 value, reflecting relative amounts of LPL mass in each fraction, is plotted on the y-axis on the left. With mAb 88B8-coated plates (A), LPL catalytic activity was found in fractions from the high-salt peak (fractions 14–21) but was nearly absent in fractions from the low-salt peak (fractions 7–13). However, the two differentially tagged LPLs were detected in both the low-salt and high-salt peaks, as determined by ELISAs using HRP-labeled FLAG or S-protein antibodies. In plates coated with the FLAG antibody (B), LPL activity was confined to fractions of the high-salt peak, but the FLAG-tagged LPL was found in fractions from both the low-salt and high-salt peaks, as determined by an ELISA using HRP-labeled mAb 88B8. Of note, the mixed LPL species (i.e., FLAG antibody-captured LPL that could be detected with the HRP-labeled S-protein antibody) were confined to fractions of the catalytically inactive low-salt peak. Similarly, in plates coated with the S-protein antibody (C), LPL activity was confined to fractions of the high-salt peak, but the S-protein-tagged LPL was found in fractions of both the low-salt and high-salt peaks, as determined by ELISA using an HRP-labeled mAb 88B8. However, mixed LPL species (S-protein antibody–captured LPL that could be detected with HRP-labeled FLAG antibody) were confined primarily to fractions from the inactive low-salt peak. FT, flow-through.
Since the LPL in tissues is normally bound to GPIHBP1 on capillaries, we tested whether coexpression of V5-tagged LPL-S159G (a catalytically inactive LPL in which the serine of the catalytic triad is replaced with glycine) and FLAG-tagged LPL-C445Y [a catalytically active LPL mutant that lacks the capacity to bind to GPIHBP1 (25)] would result in the secretion of mixed LPL species that are catalytically active and capable of binding GPIHBP1. Cells were transfected with these LPL constructs alone or together, and the medium from the cotransfected cells was added to GPIHBP1-coated (SI Appendix, Fig. S5A) or FLAG antibody-coated wells (SI Appendix, Fig. S5B). Relative amounts of LPL captured on the wells were assessed with HRP-labeled FLAG, V5, or 5D2 antibodies, and the catalytic activity of the captured LPL was assessed with a DGGR substrate. FLAG-tagged LPL-C445Y, when expressed alone, failed to bind to GPIHBP1 (SI Appendix, Fig. S5A) but did bind to the FLAG antibody and was catalytically active (SI Appendix, Fig. S5B). V5-tagged LPL-S159G, when expressed alone, readily bound to GPIHBP1 and could be detected with 5D2 and V5 antibodies, but there was no catalytic activity (SI Appendix, Fig. S5A). Examination of the medium from cells that had been cotransfected with FLAG-tagged LPL-C445Y and V5-tagged LPL-S159G revealed binding of the V5-tagged LPL-S159G to GPIHBP1; however, there was little or no association with FLAG-tagged LPL-C445Y and no catalytic activity (SI Appendix, Fig. S5A). Similarly, FLAG-tagged LPL-C445Y could be captured from the medium with a FLAG antibody, as shown by the binding of 5D2 or by catalytic activity assays, but there was little or no association with V5-tagged LPL-S159G (SI Appendix, Fig. S5B). Thus, we found little evidence that the cotransfected cells produce significant amounts of mixed LPL species containing both LPL-C445Y and LPL-S159G.
Discussion
The concept that LPL is catalytically active only as a homodimer has been widely accepted (8–13) and has guided both the formulation of new hypotheses and the interpretation of new experimental data (26–28). In the present study, we provide evidence that LPL is active in the monomeric state. Freshly secreted LPL, when subjected to density gradient ultracentrifugation, exhibited the size expected of a monomer, with both activity and mass peaks appearing in fractions overlapping with or slightly before the 66-kDa BSA standard. Only low amounts of LPL activity were observed in fractions corresponding to the expected size of homodimers. The activity and mass peaks for GPIHBP1-bound LPL also exhibited the size of monomers, with substantial overlap with the BSA standard and little overlap with the 97.2-kDa Phos B standard. LPL within the high-salt peak from a heparin-Sepharose column, long thought to represent LPL homodimers, also exhibited the size of monomers by density gradient ultracentrifugation. When LPL was denatured with guanidine hydrochloride, enzymatic activity was lost, but there was little or no effect on LPL size by density gradient ultracentrifugation. The LPL in the low-salt peak, long thought to be in the form of monomers (10, 29, 30), was largely in the form of aggregates at the bottom of the ultracentrifugation tube in the case of the low-salt peak isolated from fresh cell culture medium (Fig. 6). The LPL in the low-salt peak, but not the LPL in the high-salt peak, yielded a robust signal in single-mAb sandwich ELISAs (i.e., ELISAs that capture and detect the LPL with the same mAb). Taken together, these findings suggest that the LPL in the low-salt peak is misfolded, accounting for the propensity for aggregation, the negligible levels of catalytic activity, and the lower avidity for heparin-Sepharose. Conversely, we suspect that LPL monomers in the high-salt peak are properly folded, explaining both the robust catalytic activity and the avid binding to heparin-Sepharose.
Studies of CHO cells coexpressing differentially tagged LPL proteins provided further support for the notion that active LPL is monomeric. When CHO cells were cotransfected with expression vectors for two differentially tagged LPL proteins, both LPLs were secreted into the medium and were catalytically active, but the medium contained only small amounts of mixed LPL species (i.e., LPL species harboring both epitope tags). On heparin-Sepharose chromatography, these mixed LPL species were largely confined to the catalytically inactive low-salt peak. Moreover, when two different mutant LPLs (one mutant with a catalytic triad mutation but capable of binding GPIHBP1 and a second mutant that is catalytically active but unable to bind GPIHBP1) were coexpressed in CHO cells, we observed little evidence for catalytically active mixed LPL species (i.e., LPL species that were both catalytically active and capable of binding GPIHBP1) in the cell culture medium.
Our density gradient ultracentrifugation studies and LPL coexpression studies strongly favor the concept that LPL is active as a monomer; however, it is important to place our experiments in context. We emphasize that the concentrations of LPL in our ELISAs and density gradient fractions were low: no more than ∼3 μg/mL in the ELISAs and no more than 10 μg/mL in individual density gradient fractions. We believe that LPL–LPL interactions can and do occur in the setting of high concentrations of purified LPL. Indeed, examination of higher concentrations of LPL (∼0.7 mg/mL) by SAXS revealed homodimers (19). In addition, the generation of crystals for the LPL:GPIHBP1 complex involved high concentrations of LPL (∼15 mg/mL) (19). The crystal structure revealed two LPL molecules interacting in a head-to-tail orientation, with the hydrophobic Trp-rich motif in the carboxyl-terminal domain of one LPL monomer buried within the hydrophobic catalytic pocket in the amino-terminal domain of the partner monomer (19). As noted earlier, we are skeptical that the LPL–LPL interactions observed in the crystal structure are compatible with lipoprotein binding or triglyceride hydrolysis; nevertheless, it remains possible that LPL–LPL interactions, including those uncovered in the crystal structure, occur transiently in physiological settings in which triglyceride-rich lipoproteins are absent (e.g., the subendothelial spaces). We also emphasize that all the present experiments, as well as the earlier SAXS and crystallographic analyses (19), were performed in the absence of lipids or lipoproteins. The conformation of LPL when associated with lipoproteins is unknown. Finally, we point out that all our experiments were performed with LPL in solution, but the vast majority of LPL in mammalian tissues is tethered to GPIHBP1 on capillary endothelial cells (2, 31–33). GPI-anchored proteins are known to form clusters on the plasma membrane (34), presumably resulting in clustering of LPL as well. Whether that sort of clustering would promote transient LPL–LPL interactions is unknown.
As noted earlier, previous density gradient ultracentrifugation studies found that LPL exhibits the size of a homodimer (15–17), but these studies were performed with heparin-stabilized LPL. Our studies show that adding heparin to purified preparations of LPL—or including heparin in the cell culture medium—increases the size of LPL in density gradient ultracentrifugation experiments, such that a substantial fraction of the LPL exhibits the size expected for LPL homodimers. Dextran sulfate also increases the size of LPL in density gradient ultracentrifugation studies.
Single-mAb sandwich ELISAs have been proposed as a methodology to detect catalytically active homodimers (11). In our studies, single-mAb ELISAs using mAb 5D2 or mAb 88B8 readily detected LPL in the low-salt peak from a heparin-Sepharose column (i.e., LPL with minimal catalytic activity), but the signal was low in the high-salt peak, where the vast majority of the catalytically active LPL is found. Thus, we are highly skeptical that single-mAb sandwich ELISAs are specific for catalytically active LPL homodimers—at least in settings where the LPL is free of heparin or dextran sulfate. In earlier studies (11, 18), single-mAb sandwich ELISA successfully detected active LPL in postheparin plasma. In that setting, it seems possible that the single-mAb ELISA detected heparin-induced LPL dimers or multimers, or perhaps LPL associated with lipoproteins.
Our present study showing that LPL is active as a monomer could prompt additional studies of the functions of key proteins in LPL biology. The assumption that LPL is active only as a homodimer has influenced notions about other key molecules in plasma triglyceride metabolism; for example, acceptance of the “LPL homodimer paradigm” was evident in formulating ideas about the role of lipase maturation factor 1 (LMF1) in LPL secretion (26, 27). In wild-type cells, LPL is promptly secreted, but in LMF1-deficient cells, LPL forms aggregates within the endoplasmic reticulum and is degraded (26, 27). How LMF1 promotes LPL secretion has not been definitively established, but it has been proposed that the function of LMF1 is to assemble inactive LPL monomers into catalytically active, secretion-competent homodimers (26, 27). Given that freshly secreted, catalytically active LPL exhibits the size of monomers, we suspect that the prevailing ideas regarding LMF1 function may warrant further scrutiny. An alternative hypothesis for LMF1 function would be that LMF1, along with other chaperones, facilitates LPL secretion by ensuring proper folding of LPL monomers. Another example of the influence of the LPL homodimer paradigm is evident in a consideration of the function of ANGPTL4 in regulating LPL activity. Sukonina et al. (28) observed that treating LPL with ANGPTL4 inactivates LPL catalytic activity and increases the low-salt peak by heparin-Sepharose chromatography, prompting the conclusion that ANGPTL4 functions by converting active LPL homodimers into inactive monomers. Recent hydrogen-deuterium exchange/mass spectrometry studies (35) revealed that ANGPTL4 catalyzes the unfolding of LPL’s amino-terminal hydrolase domain. In light of those findings and the observation that LPL is active as a monomer, we suspect that ANGPTL4 likely inactivates LPL by promoting the unfolding of active LPL monomers rather than by converting catalytically active homodimers into inactive monomers. Further analyses, including additional SAXS studies, are needed to address this issue.
Materials and Methods
Human LPL was expressed in CHO cells (19), and human GPIHBP1 was expressed in Drosophila S2 cells (23, 35). Human LPL proteins containing epitope tags were expressed as described previously (19). Mutations were introduced into the LPL constructs with the QuikChange Lightning kit (Agilent Technologies). For solid-phase immunoassays, wells of 96-well ELISA plates were coated with antibodies or GPIHBP1 (23). Samples containing LPL were added to the wells and incubated overnight at 4 °C. After washing, HRP-labeled antibodies against LPL or epitope tags (100 μL) were added to the wells, followed by incubation for 2 h at 4 °C. After washing, 1-step Ultra TMB substrate (50 μL; Thermo Fisher Scientific) was added to the wells, followed by incubation on ice. Reactions were stopped with 2 M sulfuric acid (50 μL), and the OD450 values were recorded.
Deidentified archived samples of human postheparin plasma (stored at −80 °C) from a study by two of the authors (P.J.H. and K.L.S.) (36) were tested for LPL mass and activity. The human studies were approved by the Institutional Review Board at the University of California, Davis, and informed consent was obtained from all subjects in the study.
Triglyceride hydrolase activity in plasma samples and in cell culture medium was measured with a [3H]triolein substrate (31) using rat serum as a source of apo-CII, and esterase activity was measured with a DGGR (1,2-di-O-lauryl-rac-glycero-3-glutaric acid 6′-methylresorufin ester) substrate. In many cases, activity was measured on LPL that had been captured on wells of 96-well plates.
The binding of LPL to a heparin-Sepharose chromatography column and the elution of LPL with a NaCl gradient were performed as described previously (31). Sucrose or glycerol density-gradient ultracentrifugation studies were performed according to standard techniques. Western blot analyses of untagged LPL and immunocytochemistry studies on LPL were performed as described previously (3). More details are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. André Bensadoun (Division of Nutritional Science, Cornell University) for providing the bovine LPL used in this study. This work was supported by Grants HL090553, HL087228, and HL125335 from the National Heart, Lung, and Blood Institute and Transatlantic Network Grant 12CVD04 from the Leducq Foundation.
Footnotes
Conflict of interest statement: M. Meiyappan and J.E.G. are employees of Shire, now part of Takeda, and Takeda stock owners. K.M. is an employee of Immunobiologic Laboratories and holds stock in that company. K.N. holds stock in Immunobiologic Laboratories and serves as a consultant for Skylight and Sysmex.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900983116/-/DCSupplemental.
References
- 1.Fong LG, et al. GPIHBP1 and plasma triglyceride metabolism. Trends Endocrinol Metab. 2016;27:455–469. doi: 10.1016/j.tem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies BS, et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. doi: 10.1016/j.cmet.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beigneux AP, et al. Assessing the role of the glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J Biol Chem. 2011;286:19735–19743. doi: 10.1074/jbc.M111.242024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gin P, et al. Binding preferences for GPIHBP1, a glycosylphosphatidylinositol-anchored protein of capillary endothelial cells. Arterioscler Thromb Vasc Biol. 2011;31:176–182. doi: 10.1161/ATVBAHA.110.214718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lookene A, Groot NB, Kastelein JJ, Olivecrona G, Bruin T. Mutation of tryptophan residues in lipoprotein lipase: Effects on stability, immunoreactivity, and catalytic properties. J Biol Chem. 1997;272:766–772. doi: 10.1074/jbc.272.2.766. [DOI] [PubMed] [Google Scholar]
- 6.Goulbourne CN, et al. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 2014;19:849–860. doi: 10.1016/j.cmet.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bensadoun A, et al. A new monoclonal antibody, 4-1a, that binds to the amino terminus of human lipoprotein lipase. Biochim Biophys Acta. 2014;1841:970–976. doi: 10.1016/j.bbalip.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hata A, et al. Binding of lipoprotein lipase to heparin: Identification of five critical residues in two distinct segments of the amino-terminal domain. J Biol Chem. 1993;268:8447–8457. [PubMed] [Google Scholar]
- 9.Bengtsson G, Olivecrona T, Höök M, Riesenfeld J, Lindahl U. Interaction of lipoprotein lipase with native and modified heparin-like polysaccharides. Biochem J. 1980;189:625–633. doi: 10.1042/bj1890625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lookene A, Zhang L, Hultin M, Olivecrona G. Rapid subunit exchange in dimeric lipoprotein lipase and properties of the inactive monomer. J Biol Chem. 2004;279:49964–49972. doi: 10.1074/jbc.M407419200. [DOI] [PubMed] [Google Scholar]
- 11.Peterson J, et al. Structural and functional consequences of missense mutations in exon 5 of the lipoprotein lipase gene. J Lipid Res. 2002;43:398–406. [PubMed] [Google Scholar]
- 12.Zambon A, Schmidt I, Beisiegel U, Brunzell JD. Dimeric lipoprotein lipase is bound to triglyceride-rich plasma lipoproteins. J Lipid Res. 1996;37:2394–2404. [PubMed] [Google Scholar]
- 13.Wang H, Eckel RH. Lipoprotein lipase: From gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 14.Iverius PH, Ostlund-Lindqvist AM. Lipoprotein lipase from bovine milk: Isolation procedure, chemical characterization, and molecular weight analysis. J Biol Chem. 1976;251:7791–7795. [PubMed] [Google Scholar]
- 15.Zhang L, Lookene A, Wu G, Olivecrona G. Calcium triggers folding of lipoprotein lipase into active dimers. J Biol Chem. 2005;280:42580–42591. doi: 10.1074/jbc.M507252200. [DOI] [PubMed] [Google Scholar]
- 16.Wong H, et al. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc Natl Acad Sci USA. 1997;94:5594–5598. doi: 10.1073/pnas.94.11.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ben-Zeev O, Doolittle MH. Determining lipase subunit structure by sucrose gradient centrifugation. Methods Mol Biol. 1999;109:257–266. doi: 10.1385/1-59259-581-2:257. [DOI] [PubMed] [Google Scholar]
- 18.Sato K, et al. The majority of lipoprotein lipase in plasma is bound to remnant lipoproteins: A new definition of remnant lipoproteins. Clin Chim Acta. 2016;461:114–125. doi: 10.1016/j.cca.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Birrane G, et al. Structure of the lipoprotein lipase-GPIHBP1 complex that mediates plasma triglyceride hydrolysis. Proc Natl Acad Sci USA. 2019;116:1723–1732. doi: 10.1073/pnas.1817984116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler FK, D’Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 21.van Tilbeurgh H, Sarda L, Verger R, Cambillau C. Structure of the pancreatic lipase-procolipase complex. Nature. 1992;359:159–162. doi: 10.1038/359159a0. [DOI] [PubMed] [Google Scholar]
- 22.Beigneux AP, et al. GPIHBP1 missense mutations often cause multimerization of GPIHBP1 and thereby prevent lipoprotein lipase binding. Circ Res. 2015;116:624–632. doi: 10.1161/CIRCRESAHA.116.305085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mysling S, et al. The acidic domain of the endothelial membrane protein GPIHBP1 stabilizes lipoprotein lipase activity by preventing unfolding of its catalytic domain. eLife. 2016;5:e12095. doi: 10.7554/eLife.12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuoka N, Saito Y, Yoshida S. Effects of dextran sulfate on stabilization of milk lipoprotein lipase and VLDL triglyceride hydrolysis in vitro. Tohoku J Exp Med. 1986;149:61–66. doi: 10.1620/tjem.149.61. [DOI] [PubMed] [Google Scholar]
- 25.Voss CV, et al. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci USA. 2011;108:7980–7984. doi: 10.1073/pnas.1100992108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doolittle MH, Ehrhardt N, Péterfy M. Lipase maturation factor 1: Structure and role in lipase folding and assembly. Curr Opin Lipidol. 2010;21:198–203. doi: 10.1097/MOL.0b013e32833854c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doolittle MH, Péterfy M. Mechanisms of lipase maturation. Clin Lipidol. 2010;5:71–85. doi: 10.2217/clp.09.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci USA. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osborne JC, Jr, Bengtsson-Olivecrona G, Lee NS, Olivecrona T. Studies on inactivation of lipoprotein lipase: Role of the dimer to monomer dissociation. Biochemistry. 1985;24:5606–5611. doi: 10.1021/bi00341a048. [DOI] [PubMed] [Google Scholar]
- 30.van Tilbeurgh H, Roussel A, Lalouel JM, Cambillau C. Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: Consequences for heparin binding and catalysis. J Biol Chem. 1994;269:4626–4633. [PubMed] [Google Scholar]
- 31.Beigneux AP, et al. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allan CM, et al. Mobility of “HSPG-bound” LPL explains how LPL is able to reach GPIHBP1 on capillaries. J Lipid Res. 2017;58:216–225. doi: 10.1194/jlr.M072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristensen KK, et al. A disordered acidic domain in GPIHBP1 harboring a sulfated tyrosine regulates lipoprotein lipase. Proc Natl Acad Sci USA. 2018;115:E6020–E6029. doi: 10.1073/pnas.1806774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha S, Anilkumar AA, Mayor S. GPI-anchored protein organization and dynamics at the cell surface. J Lipid Res. 2016;57:159–175. doi: 10.1194/jlr.R062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mysling S, et al. The angiopoietin-like protein ANGPTL4 catalyzes unfolding of the hydrolase domain in lipoprotein lipase and the endothelial membrane protein GPIHBP1 counteracts this unfolding. eLife. 2016;5:e20958. doi: 10.7554/eLife.20958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shirakawa T, et al. Comparison of the effect of post-heparin and pre-heparin lipoprotein lipase and hepatic triglyceride lipase on remnant lipoprotein metabolism. Clin Chim Acta. 2015;440:193–200. doi: 10.1016/j.cca.2014.07.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.