Significance
The identification of genes that are differentially expressed provides a molecular foothold onto biological questions of interest. Whether some genes are more likely to be differentially expressed than others, and to what degree, has never been assessed on a global scale. Here, we reanalyze more than 600 studies and find that knowledge of a gene’s prior probability of differential expression (DE) allows for accurate prediction of DE hit lists, regardless of the biological question. This result suggests redundancy in transcriptomics experiments that both informs gene set interpretation and highlights room for growth within the field.
Keywords: transcriptomics, differential expression, metaanalysis, replicability, specificity
Abstract
Differential expression (DE) is commonly used to explore molecular mechanisms of biological conditions. While many studies report significant results between their groups of interest, the degree to which results are specific to the question at hand is not generally assessed, potentially leading to inaccurate interpretation. This could be particularly problematic for metaanalysis where replicability across datasets is taken as strong evidence for the existence of a specific, biologically relevant signal, but which instead may arise from recurrence of generic processes. To address this, we developed an approach to predict DE based on an analysis of over 600 studies. A predictor based on empirical prior probability of DE performs very well at this task (mean area under the receiver operating characteristic curve, ∼0.8), indicating that a large fraction of DE hit lists are nonspecific. In contrast, predictors based on attributes such as gene function, mutation rates, or network features perform poorly. Genes associated with sex, the extracellular matrix, the immune system, and stress responses are prominent within the “DE prior.” In a series of control studies, we show that these patterns reflect shared biology rather than technical artifacts or ascertainment biases. Finally, we demonstrate the application of the DE prior to data interpretation in three use cases: (i) breast cancer subtyping, (ii) single-cell genomics of pancreatic islet cells, and (iii) metaanalysis of lung adenocarcinoma and renal transplant rejection transcriptomics. In all cases, we find hallmarks of generic DE, highlighting the need for nuanced interpretation of gene phenotypic associations.
RNA-sequencing (RNA-seq) and microarray technology are valuable tools in the modern molecular biology toolkit, enabling large-scale analysis of the transcriptional changes associated with biological conditions of interest. Typically, expression levels for each gene are compared between sample groups, and the genes that pass certain statistical cutoffs are selected as the “hit list” for further interpretation and validation. This type of analysis has been used as the basis for key insights into the genes that drive physiological and disease mechanisms, for example, identifying novel circadian rhythm genes (1), characterizing transcriptional mechanisms of stem cell differentiation (2), and highlighting the critical role of cell proliferation-associated genes in predicting cancer metastasis and survival (3). In most cases, conclusions from differential expression (DE) studies are drawn using within-experiment data, which means that claims of specificity are relative to the control groups used for reference. In this work, we challenge this notion of specificity by probing whether certain gene expression profiles are generic, with a high probability of DE across a wide variety of biological conditions.
Our aim is related to previous efforts to identify genes that are preferentially variable within and across individuals, tissues, or genetic backgrounds (4–8). Each of these studies employed a similar strategy, controlling for all experimental variables to detect baseline transcriptional variation among well-matched samples. While each of these studies highlighted expression variability related to inflammation, hormone regulation, tissue composition, and stress responses, the identified gene lists were often surprisingly small, even with loosened statistical thresholds. Our present work starts where this previous research ends, with the hypothesis that many genes may show frequent DE, regardless of the specific biological question being addressed. If true, this would have a large impact on the interpretation of gene expression hit lists, where the appearance and reproducibility of these genes would now appear to be less surprising than we might naively expect.
Assessing studies to determine which genes are frequently differentially expressed creates a number of challenges, both technical and conceptual. To ensure uniform DE detection across studies, we had to reanalyze the data, rather than relying on reports based on publication-specific methods. This required the development of a rigorous pipeline for reanalysis of DE, including experimental annotation, quality control, batch modeling, and, finally, DE estimation. The Gemma database (https://gemma.msl.ubc.ca) contains thousands of curated and reanalyzed studies, and this resource allows us to make direct comparisons of DE hit lists, overcoming technical limitations (9). The major conceptual challenge is that studies vary in the number of differentially expressed genes that they observe, due to statistical power as well as biology. This means that naively creating a prior based on raw frequency of DE is unlikely to be optimal. Instead, we exploit a trick we have previously used in machine learning (10) to predict DE hit lists in the Gemma database, described in more detail below.
The basic premise is that we attempt to “cold read” the output of individual studies by calculating the ranked list of genes, which is mathematically optimal for predicting DE as measured by the area under the receiver operating characteristic curve (AUROC). This list, referred to as the “DE prior” or “global prior,” can be used to guess the hit list of any DE study without knowing anything about it. If the DE prior is highly predictive of most studies, then many genes—and specifically the ones the prior ranks highly—can be expected to arise frequently in DE hit lists. Within the same framework, we can evaluate any predictor, for example, ranking genes based on GC content, probability of mutation in the general population, or their expression level in adult tissues, and see how it compares.
Our work has three main contributions: (i) a quantitative definition of DE gene and hit list specificity, (ii) characterization of the biological processes that drive hit list overlaps, and (iii) a demonstration of how our approach can be used to interpret gene sets more broadly. In brief, we find that ranking genes by their contribution to DE allows us to predict hit lists with high performance (mean AUROC, >0.8), consistent with the existence of generic transcriptional patterns. The most common DE genes are enriched for important biological functions including sex, the extracellular matrix (ECM), as well as immune-related and stress responses. In three use cases, we show that the prior provides rich insight into gene set properties: validating that housekeeping genes are rarely DE, and highlighting the differential specificity of disease-associated genes and cell type markers. We expect that the simplicity, robustness, and general significance of the DE prior we have made available (11) will make it a valuable guide for interpreting and designing future transcriptomic studies.
Results
Data Processing and Quality Control.
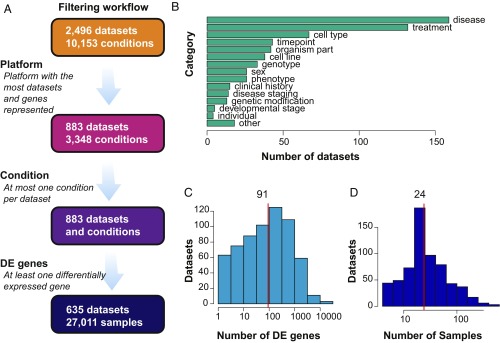
Gene expression microarray and RNA-seq technology have become increasingly common for untargeted, hypothesis-generating research into the genetic and transcriptional mechanisms underlying biological conditions of interest. In tandem, efforts to encourage data sharing have led to the deposition of raw data on public servers (12, 13), which may be thought of as a digital commons ready to be mined for scientific insight. However, there are major hurdles to the reuse of these public data, which include the need for consistent processing, as well as metadata extraction for downstream statistical analyses (14). The Gemma system was designed to overcome these issues by providing consistently processed and annotated expression data (9). For our assessment, DE was analyzed within Gemma, and then downloaded as flat files for further analysis in R. Details of the filtering and analysis process can be found in Methods and in Fig. 1. In brief, the database initially contained 2,496 human expression datasets comprising 10,153 individual experimental conditions across 105 platforms and platform combinations. To avoid complications due to platform-specific effects, we limited our analysis of Gemma to studies performed on the popular GPL570 (Affymetrix Human Genome U133 Plus 2.0 Array) platform as it allowed us to include the largest number of studies with the greatest transcriptome coverage (∼19,000 genes). All studies with at least one differentially expressed gene [absolute log2 fold change, >2; false-discovery rate (FDR), <0.05] were included, leaving us with a compendium of 635 datasets with a total of 27,011 samples and covering a wide range of biological conditions.
Fig. 1.
Data workflow and description. (A) Database filtering workflow. (B) Plot indicates the number of datasets per experimental category included in this assessment. Experiments ranged widely in their goals and design. (C) Histogram of the number of differentially expressed genes per dataset. Genes are considered to be differentially expressed if they have an absolute log2 fold change of >2 at FDR of <0.05. The red line indicates the median. (D) Histogram of the number of samples per dataset. The red line indicates the median.
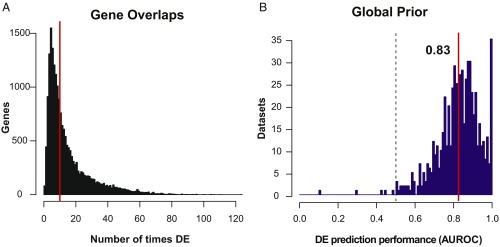
The objective of our analysis was to determine whether some genes are much more likely to be differentially expressed than others, and previous work in this area suggests that this is the case (e.g., ref. 4). We determined overlaps in DE results (DE hit lists) across the compendium. We find that nearly all genes are differentially expressed at least once, with most genes recurring in ∼10 datasets (Fig. 2A). We also find evidence of common DE, with 229 genes occurring in >10% of DE hit lists. The most extreme example is CXCL8, which is DE in nearly 20% of all datasets (124/635). CXCL8 (also known as IL8) is a chemokine known to be involved in attracting neutrophils toward sites of injury or infection (15), and it has been implicated in many disorders (16–18). Consistent with this, studies with DE of CXCL8 are not biased toward any particular area of research and include datasets comparing untreated (normal) cell lines and tissue types, as well as datasets comparing genetically or pharmacologically treated samples, and disease samples (Dataset S1). This suggests that overlaps in DE may reflect truly common biological or molecular processes, rather than biases in the composition of the compendium.
Fig. 2.
The global DE prior accurately predicts DE hit lists. (A) Recurrence of differentially expressed genes across datasets. The red line indicates the mean. On average, each gene is DE in 10 expression studies. However, the distribution has a long, right-sided tail, indicating a small number of genes that are frequently DE. (B) Distribution of AUROC scores using the global DE prior to predict hit lists across the 635 studies. The red line indicates the mean, and the dashed line indicates the null (0.5). On average, the prior has very high performance, distinguishing ∼80% of DE genes within each hit list, reflecting shared transcriptional features between studies.
In the following sections, we assess both the overlaps among studies, as well as their specificity, in more detail: first characterizing the degree to which we can predict DE using knowledge of overlaps, formalized as the DE prior; then characterizing the prior’s properties, documenting its robustness and superiority to even highly targeted priors; and finally demonstrating its application across three diverse use cases.
The Global DE Prior Predicts DE Results with High Accuracy.
Above, we described general patterns of overlapping DE between datasets. In this work, our goal is not only to find whether some genes are more commonly differentially expressed than others but also to determine whether differences in gene recurrence can be used to predict DE hit lists. This approach is useful because it provides a measure of specificity for each study: If an experiment’s hit list is well predicted by this generic ranking, it implies that many genes within that hit list are commonly DE in other studies. We can also use the same prediction approach to determine whether we can gain specificity in our predictions by creating subset-specific priors based on groups of studies that have common features (like those funded by the National Cancer Institute); or to investigate whether other gene properties, such as GC content, coding sequence length, or mutability in the population, might be predictive of DE, suggesting either biological or technical biases that contribute to common DE of genes.
To measure general redundancy in the compendium, we generate a global DE prior: This is the ranked list of genes that maximizes our ability to predict DE hit lists (Methods and Dataset S2). As described earlier, the global DE prior can be thought of as performing a “cold reading” of an experiment’s DE results. Just as a psychic will make predictions that are likely to be true based on general population statistics, we make predictions about genes based on the same principle; in our case, we take advantage of knowledge about a gene’s likelihood of DE before making a guess about a given experiment’s hit list. We determine how well we recover DE genes for each dataset using the ranking from the compendium-wide list (leaving out the study to be predicted) and report performance as the AUROC. This is roughly equivalent to the probability that we have correctly ranked a differentially expressed gene above a gene that is not differentially expressed within that experiment. Thus, 0.5 is random, 1.0 is perfect, and 0.7–0.8 is generally considered high performance.
We find that the global DE prior has remarkably high performance (mean AUROC, 0.83 ± 0.1; Fig. 2B; all scores, Dataset S1). In other words, given a random experiment, we could use the global prior to bet on which genes show up in the hit list and we would expect (on average) to accurately rank ∼80% of DE genes higher than other genes within each study. As a comparison, we tested the ability of any individual study to predict the results of all others. This is essentially a test of DE hit list overlap between all pairs of studies. Here, results were much closer to the null, with only 3 of the 635 studies showing appreciable performance (mean AUROC, >0.6), all of which were characterized by a large number of differentially expressed genes (median, ∼2,300, vs. all studies median, ∼90). Together, these results show that the prior contains an informative aggregate signal that is not recapitulated by any individual study.
Biological and Technical Features of the Prior.
Previous work has indicated that gene properties such as transcript length, the number of annotated functions, gene essentiality, or expression level in adult tissues can strongly predict the likelihood for that gene to be studied based on literature mining (19, 20). Other research has pointed to biases of length and GC content in expression analysis (21, 22), or gene multifunctionality effects on gene function prediction and network analysis (10). However, the degree to which generic gene properties may directly inform their probability of DE has not been assessed on a global scale.
To address this, we took the same approach that we took above, curating sets of gene properties, generating a prior for each, and reporting its ability to predict DE hit lists as the AUROC. Gene properties spanned four general categories: (i) technical features such as GC content and length (23); (ii) adult tissue expression levels sourced from GTEx (24) and lymphoblast cell lines from the Geuvadis project (25); (iii) functional properties such as mutability from ExAC (26), or node degree in aggregate coexpression networks (27); and (iv) annotation biases using multifunctionality in Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (10). None of these properties is nearly as predictive as the DE prior (mean across all categories, 0.55; Fig. 3). For example, a predictor based on prevalence in the GO (also known as gene multifunctionality) that predicts gene sets within the GO very well (mean AUROC, 0.83 ± 0.08) is close to useless at predicting DE (mean AUROC, 0.57 ± 0.03).
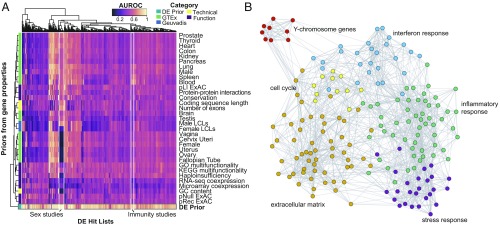
Fig. 3.
Multiple independent gene processes underlie the high performance of the DE prior. (A) Heatmap of AUROCs for each gene property-related prior (rows) and each study in the compendium (columns). Row colors indicate the gene property type. No single property performs as well as the DE prior. However, a few studies are clearly predicted by a number of features, including sex-related studies and immunity-related studies (both highlighted with white boxes). These groups are well predicted by expression level in males and expression level in blood, respectively. (B) Network visualization of genes in the top 1% of the DE prior. Each circle represents a gene, and edges connect all genes that are nearest neighbors based on their coexpression in an aggregate network. Genes are colored by cluster and labeled by GO enrichment. We find that genes in the top 1% of the prior show strong coexpression patterns, representing distinct functional modules that are commonly repurposed across many different conditions. ExAC, Exome Aggregation Consortium; GO, Gene Ontology; GTEx, Genotype-Tissue Expression project; KEGG, Kyoto Encyclopedia of Genes and Genomes; LCLs, lymphoblastoid cell lines; pLI, probability of intolerance to a single loss-of-function variant; pNull, probability of tolerance to loss-of-function variation; pRec, probability of intolerance of two loss-of-function variants.
Although no individual predictor performs as well as the DE prior across all studies, there are a small number of cases where gene properties perform quite well. For example, we find that predictors related to expression in females, GC content, and haploinsufficiency are strongly predictive of DE for the subset of hit lists deriving from comparisons of males and females (Fig. 3 and Dataset S1). Similarly, we find a small subset of studies that are preferentially predicted by gene expression levels in whole blood and in spleen, all of which are related to immune phenotypes.
This modularity, with certain properties predicting only subsets of studies, suggests that the DE prior is likely weighted toward genes from multiple functions, which would explain why individual features are poor predictors of DE hit lists. We thus aimed to characterize the biological properties of the DE prior in more detail. To do this, we focused on the genes that are in the top 1% of the prior (192 most commonly DE genes). We first asked whether they formed coexpressed gene modules in a high-quality coexpression network (27) (Fig. 3). Using a random-walk–based clustering approach (28), we find six clusters, ranging in size from 9 to 58 genes (SI Appendix, Table S1). To determine the robustness of the clusters, we performed a standard guilt-by-association analysis (29), which demonstrated that the clusters are strongly predictable from their coexpression patterns (mean AUROCs, 0.98 ± 0.02). In essence, all of the genes within each separate cluster are markers for the same process across their entire range of activity. GO analysis revealed that five of six clusters have significant functional enrichment at FDR of <0.05, with two clusters enriched for immune-related functions, one enriched for terms related to the ECM, one enriched for transcription factor activity and containing key stress response genes (FOS, JUN, ATF3, and EGR1/2/3), and one cluster enriched for cell cycle genes. The only cluster that was not attributed a GO function consists solely of Y-chromosome genes (EIF1AY, KDM5D, ZFY, NLGN4Y, DDX3Y, USP9Y, TXLNGY, TTTY14, and UTY), suggesting an obvious biological interpretation for their close association in the network. Altogether, these results support the notion that it is recurrence of biological processes across datasets, rather than any technical features, that allow the prior to accurately predict DE hit lists.
Predictions Are Not Improved by Increasing the Specificity of the Prior.
While we demonstrated that the DE prior has very good performance across studies, and that it is composed of commonly observed gene functional modules, it is possible that performance could be improved by taking a more biologically targeted approach. To address this, we did a series of computational experiments to try to maximally overfit to subsets of studies within the compendium and find predictors that outperform the DE prior. In brief, these consisted of (i) assessing studies funded by the same agency at the National Institutes of Health (NIH); (ii) clustering DE hit lists; and (iii) identifying the maximally predictive GO term for each hit list. Strikingly, across all of these experiments, and despite the extreme overfitting to potentially “beat” the global DE prior, we find only one biologically driven grouping where the global prior was outperformed by a more targeted prior. All others do not outperform the DE prior. We discuss these experiments in more detail below.
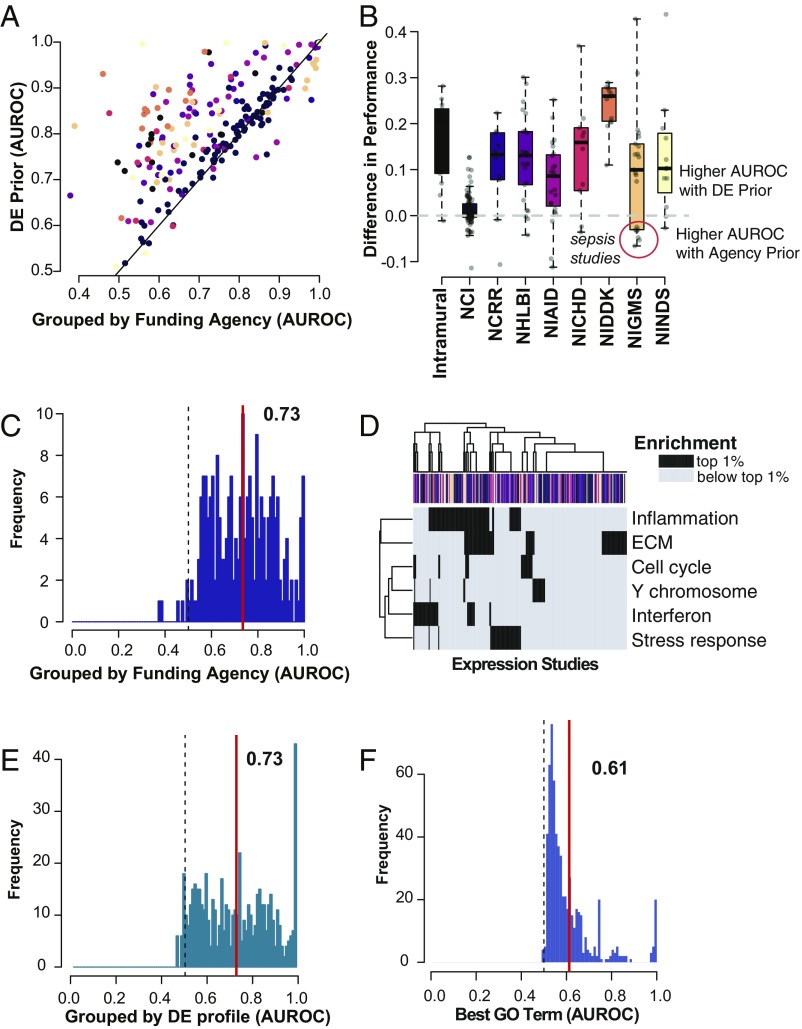
First, we used metadata to group studies based on their funding information, with the hypothesis that datasets funded by the same funding source (e.g., National Cancer Institute) are more likely to be biologically related than those across funding sources. If this is true, a prior generated from datasets funded by a single agency should outperform the global prior. Using data from PubMed, we found funding information for 307 of 635 datasets. We restricted our analyses to funders that supported 10 or more studies to ensure sufficient power for predictions, ultimately including 222 datasets from nine NIH institutes or other specific funding sources. To confirm whether the institutes tend to fund experiments targeted toward different end goals, we took advantage of the controlled vocabularies used to tag each experiment in Gemma, finding that studies funded by the National Cancer Institute are strongly enriched for the disease ontology term “organ system cancer” (DOID 0050686; P < 10−6) and that those funded by the National Institute of Allergy and Infectious Diseases are enriched for the term “disease by infectious agent” (DOID 0050117; P < 10−4), for example. Despite confirming research focus specificity, we find that the global DE prior outperforms almost all NIH institute-restricted priors (Fig. 4 A–C; mean global, 0.83 ± 0.1; mean funding agency, 0.73 ± 0.1). One exception is a subset within the group of studies funded by the National Institute of General Medical Sciences (NIGMS), all related to septic shock or traumatic injuries (Fig. 4B and Dataset S1). This is the sole biological grouping for which a more specific prior outperformed the global prior, and it can be attributed to an unusually high similarity in DE genes among these studies.
Fig. 4.
The global DE prior almost always outperforms targeted approaches to hit list prediction. (A) DE prior performance is plotted with respect to agency-specific prior performance. Each point represents an individual study, and studies are colored by their funding source, as in the boxplot in B. The identity line is shown in black. Hit lists are generally much better predicted by the global DE prior than by priors specified by funding source. (B) Difference in performance (global − agency). Boxplots show the quartiles, with whiskers extending to 1.5× the interquartile range. The majority of studies are better predicted by the DE prior, although there are a few exceptions among NIGMS funded studies (details in Dataset S1). (C) Distribution of AUROCs using funding agency-specific priors. The red line indicates the mean, and the dashed line shows the null. Mean performance is much lower than for the global prior. (D) Heatmap of enrichment for DE prior gene clusters (Fig. 3B). Rows indicate cluster labels, and columns are expression studies; colors indicate funding agency as in B. If the gene set is among the top 1% of enriched functions compared with all of the GO, the square is colored in black; otherwise, squares are gray. The majority of studies have enrichment for at least one of the clusters. Studies do not clearly group by funding agency. (E) Distribution of AUROCs using priors obtained after grouping hit lists into smaller subsets. Lines are as in C. (F) Distribution of AUROCs after selecting the maximally performing gene set from GO. Lines are as in C. NCI, National Cancer Institute; NCRR, National Center for Research Resources; NHLBI, National Heart, Lung and Blood Institute; NIAID, National Institute of Allergy and Infectious Diseases; NICHD, National Institute of Child Health and Human Development; NIDDK, National Institute of Diabetes and Digestive and Kidney Diseases; NIGMS, National Institute of General Medical Sciences; NINDS, National Institute of Neurological Disorders and Stroke.
In all other cases, the strong performance of the DE prior across funding agencies can readily be understood by visualizing study-specific enrichment of genes in our six clusters (Fig. 4D). We see that studies from multiple NIH institutes are enriched for the six biological processes that comprise the top 1% of the prior, and that study results can often be enriched for more than one of these processes. Because prior construction is always performed via cross-validation (leaving out the study to be predicted), the robust cross-institute signal captured by the global DE prior allows it to outperform the less well-powered average from studies funded by the same NIH institute.
For our next experiment, we aimed to see whether we could improve performance by clustering DE hit lists to find groups of studies that are highly similar to one another. Our outcome measure is the same as in the previous, determining whether cluster-specific priors outperform the global DE prior. Because each cluster-specific prior is created from a set of experiments chosen specifically to be more similar than the average, at worst, the cluster-priors should perform as well as the global prior since it can simply be reused in each cluster. If their performance is not substantially higher than the DE prior, then the groupings have not successfully discovered distinct patterns. In fact, we obtain much lower performance for the cluster-specific priors than we do for the global prior (mean AUROC, 0.73 ± 0.2; Fig. 4E). This means that the clusters are not sufficiently distinct. Similar to what was observed for the NIH institute groupings, this suggests that the use of the smaller number of experiments for each cluster-specific prior simply results in a less robust version of the global prior.
As a final attempt to overfit priors for our 635 datasets, we tested every GO group for its ability to predict DE hit lists. In this case, each GO group is individually tested for its overlaps with the DE hit lists of each experiment, and we report the GO group with the highest performance (maximal overlap) for each study. Despite overfitting to each dataset by picking only the GO group with the post hoc highest performance, the mean of the maximal AUROCs is only 0.61 (Fig. 4F; all scores in Dataset S1), significantly lower than the average performance of the DE prior (AUROC, 0.83).
Together, these results demonstrate the very high performance of the DE prior for predicting hit lists. It is apparently challenging to improve on its performance, even with the intention of overfitting. Of course, predicting DE genes well on average does not mean that we know everything there is to know about the individual experiments, and some DE genes are unexplained by global features, potentially reflecting distinctive biology. The improvement of the NIGMS prior over the global for prediction of sepsis studies is a good example of this, and it indicates that the inclusion of significantly more data may eventually allow for improved performance of cluster-specific priors. However, only comparison with the global prior can establish that specificity has been achieved. More broadly, the high performance and robustness of the global prior is useful in its own right, allowing for the reinterpretation of established and novel gene sets, explored in the three use cases below.
Use Case 1: Reexamining PAM50 Genes and Housekeeping Genes with the DE Prior.
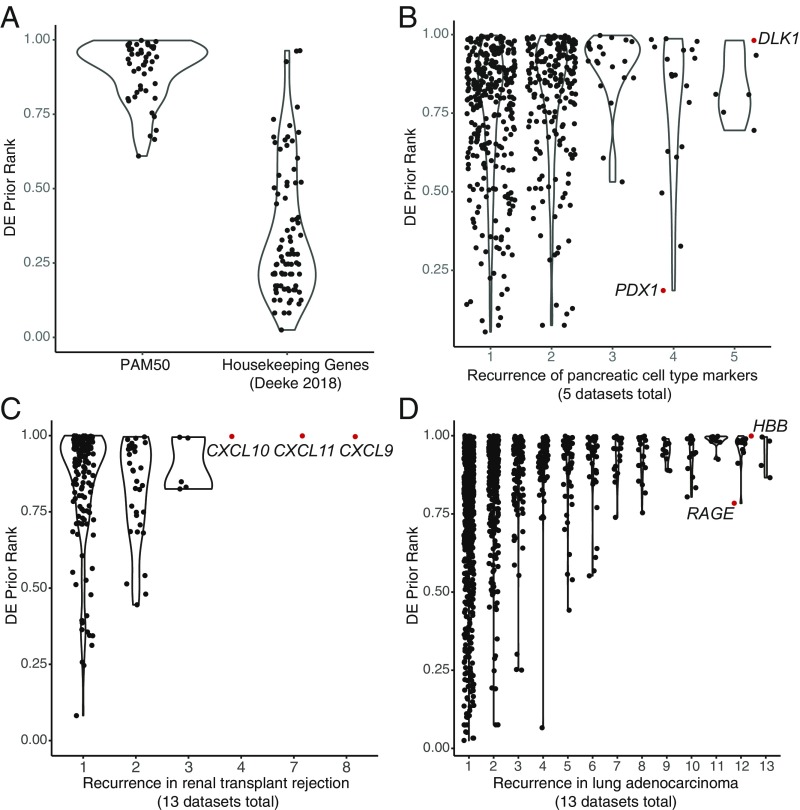
One prominent goal of human expression analysis has been to classify disease samples. Almost two decades ago, it was observed that breast cancer subtypes were distinguishable by their mRNA expression patterns (30). Since then, the set of genes required for classification was increasingly refined (31, 32), and now a set of 50 genes called “PAM50” for “Prediction Analysis of Microarray 50,” are available as a commercial kit for clinical use (33). While our DE prior cannot determine the relationship between the genes and the outcome (i.e., cancer subtype and, by extension, prognosis), it can shed light onto the general likelihood of these genes to be differentially expressed across many different studies. We plot each gene with respect to its prior probability, defined by the gene’s rank in the minimum-rank prior (Methods; 0 = not commonly DE; 1 = commonly DE). Notably, we find that the PAM50 genes are very commonly DE, ranking in the top 10% of the prior on average (mean rank, 0.90 ± 0.1, for the 48 of 50 genes assessed on GPL570; MIA and PTTG1 are not assayed). This result does not invalidate the usefulness of PAM50 DE for prognosis when a breast cancer diagnosis is known. However, it indicates that DE of these genes is not limited to breast cancer, which is consistent with the recent finding that the same signature can be employed to predict prostate cancer subtypes (34).
A contrasting goal in expression analysis has been to identify genes that are stably expressed across a broad range of conditions [sometimes referred to as housekeeping genes (35, 36)], with the idea that these genes may be used for data normalization (37, 38). Recent work in this area has turned to single-cell RNA-seq data to identify stably expressed genes (39, 40). If these genes are stable across conditions, we would expect them to show infrequent DE. Indeed, we find that all housekeeping sets tested have low average ranks in the DE prior (Eisenberg microarray, 0.41 ± 0.3; Eisenberg RNA-seq, 0.38 ± 0.2; Lin, 0.42 ± 0.2; Deeke, 0.35 ± 0.2; Deeke displayed in Fig. 5A; all others in SI Appendix, Fig. S1). This is very far into the tail of gene set predictabilities within our DE distribution (bottom 1%). However, there is quite a bit of variability in the prior probability of DE among these putative housekeepers, which is perhaps to be expected given previous evaluations (39). In line with this, very few genes overlap among all of the housekeeping sets (6 of 4,513 total), although we find an encouraging trend toward decreasing prior probability of DE as the replicability of the gene’s status as a housekeeper increases (SI Appendix, Fig. S1). One striking exception to this trend is RPS24, a gene that encodes a component of the 40S ribosome and is included in all four lists. We find that this gene is very commonly differentially expressed (DE prior rank, 0.96), which undermines its utility for normalization. This may be a consequence of the distinct biology that some ribosomal proteins exhibit [e.g., response to stress (41)] and reflected by RPS24’s disease association to Diamond–Blackfan anemia (42).
Fig. 5.
Reinterpreting gene sets with the DE prior. (A) PAM50 genes and housekeepers are plotted with respect to their rank in the DE prior, and each point represents a single gene (0, never differentially expressed; 1, frequently differentially expressed). As expected, PAM50 genes have high DE prior ranks (mean, 0.9), whereas housekeepers are not frequently DE and have low DE prior ranks (mean, 0.35). (B) Alpha and beta cell markers derived from single-cell RNA-seq are plotted with respect to their DE prior rank, and their recurrence among five independent hit lists. Increasing recurrence is associated with higher DE ranks. Differences in prior ranks between highly recurrent genes suggests differential condition specificity. (C) DE genes from metaanalysis of 13 renal transplant rejection studies are plotted with respect to their DE prior rank and their recurrence. The only genes that recur among most studies are all very frequently DE (high DE prior ranks), and thus unlikely to be specifically associated with renal transplant rejection. (D) DE genes from metaanalysis of 13 lung adenocarcinoma studies are plotted with respect to their DE prior rank and their recurrence. RAGE, a known biomarker, has a relatively low DE prior rank.
Use Case 2: Interpreting Marker Genes from Single-Cell RNA-Seq.
Ideally, the DE prior should inform the interpretation of differentially expressed genes. One potential limitation is that biases in the composition of the database, which comprises many disease-associated studies, might mean that the prior will not be capable of characterizing studies of “normal” variability, such as those of DE between cell types. We evaluated this by looking at the performance for a type of experiment not represented in Gemma, specifically DE from five single-cell RNA-seq experiments between alpha and beta cells of the pancreas (43–47). As previously, we define DE genes within each experiment as those with FDR of <0.05 and log2 fold change >2 (details in Methods). We find that the prior has high performance in predicting DE between alpha and beta cells, with a mean AUROC of 0.78 ± 0.03 (SI Appendix, Fig. S2), comparable to the average performance across our original compendium. Along with our previous assessments of data subsetting, both by funding source and by DE profiles, this confirms that the prior is not unduly biased toward disease-associated studies.
We next looked at the prior probability for each putative marker gene. At the low end of prior probability are a number of interesting genes. For example, among the 19 genes differentially expressed in four of five studies, the gene with the lowest prior probability of DE is PDX1 (DE prior rank, 0.19). PDX1 is a transcriptional activator required for pancreatic beta cell function, with highly restricted expression in adult tissues (48). For this reason, PDX1 is likely to be a more tissue-specific marker than other genes such as DLK1 (five of five studies; prior, 0.98), which is a good beta cell marker but also has functions outside of the pancreas, notably in adipogenesis (49).
Use Case 3: Interpreting DE Hit Lists from Metaanalysis.
In our final use case, we used the prior to inform the interpretation of DE hit lists from metaanalysis, with the hypothesis that metaanalysis will preferentially identify generic DE genes because of differential prior probability. We reanalyzed previously published lung adenocarcinoma (50–62) and renal transplant rejection datasets (63–75), comprising 13 experiments for each, and over 2,600 samples (SI Appendix, Table S2). Previous metaanalyses of these datasets identified expression changes related to immune activation and angiogenesis (68, 76), hinting that they would be well predicted by the DE prior. Accordingly, we found evidence supporting our hypothesis that highly recurrent genes tend to have high global DE prior probability (Fig. 5 C and D).
We noted that there were very few recurrent DE genes among the transplant rejection studies, even though we reduced our fold change threshold for DE gene calling (fold >1.3 rather than 2). Consistent with the lack of signal associated with transplant rejection, the three genes found in four or more datasets are all very commonly differentially expressed (DE prior rank, >0.99); even a group of studies without any specific overlap in condition (i.e., a random sample of studies) could expect to see recurrence of such high-prior genes. Notably, these genes are all ligands of the CXCR3 receptor and are induced by interferon signaling to recruit T cells to sites of injury or infection (77), and all are included in our interferon and chemokine signaling related cluster. The association of these genes with renal transplant rejection is thus highly probable, but their specificity to the phenotype is not.
In the lung adenocarcinoma metaanalysis, we also see a relationship between recurrence and prior probability, with most genes that recur in more than one-half of the DE hit lists having a DE prior probability of >0.9. One notable exception is the RAGE receptor gene (also known as AGER), a known biomarker of lung cancer (78), which is recurrently differentially expressed in 12 of 13 datasets but has lower global prior probability of DE (DE prior rank, 0.78). These results suggest that RAGE should be interpreted differently than other genes of similar recurrence: it should be considered more likely to be specific to the phenotype than, for example, the beta-hemoglobin gene (HBB), which also recurs in 12 datasets but has a global DE prior rank of >0.999.
Discussion
In expression analysis, it is common to claim a condition-specific association for a gene, for example, to define a novel tissue-specific marker, or to associate a gene with a particular perturbation. However, these claims are limited by the specificity of the comparisons made within each study. In this work, we reexamined condition specificity by assessing gene overlaps between more than 600 expression studies. We discover that human DE experiments are highly predictable, suggesting limited specificity of a major fraction of DE hit lists. Underlying their predictability is the fact that the same gene modules are consistently repurposed across conditions. These modules include the immune response and inflammation, the ECM, cell cycling, stress responses, and sex. Our results have major implications for interpreting DE hit lists from metaanalysis, which are very likely to report generic (high-DE prior) genes, as well as for other well-characterized gene sets, such as disease biomarkers, or housekeeping genes.
The characterization of previously published housekeeping gene sets is a useful and revealing test case for the global DE prior. The four gene sets we tested were initially identified by the stability of their expression levels, meaning that all four were designed to act as references for data normalization. In line with this, we found that their average ranks within the global prior are extremely low, particularly with respect to the distribution for our 635 DE hit lists. This confirmed that most of these possible housekeeping genes are rarely differentially expressed and supports their potential for data normalization. RPS24 is a clear outlier with respect to the other housekeepers that are common to all sets, and we would predict that it would be dangerous to use it for normalization (79). This example demonstrates how the prior can provide valuable context for the interpretation of gene candidates, in this case by interrogating the assumption of gene stability.
As described, the housekeepers provide insight into the interpretation of low-ranking genes in the DE prior. Housekeepers have low ranks due to their stable expression across conditions, but a gene’s low rank may also arise due to its lack of expression, or because it is DE in rarely targeted conditions. How are we to interpret high-ranking genes? We believe these are usefully understood through the lens of our two targeted metaanalyses. In the case of renal transplant rejection, very few genes were found to recur across hit lists, and the top candidates from this metaanalysis are all frequently DE. The small number of hits we observe, even after lowering the effect size threshold, suggests that these studies are too heterogeneous to converge on specific, shared DE genes. Upon combining them, only generic signals emerge. In contrast, our lung adenocarcinoma metaanalysis was much more successful, identifying many specific (low-DE prior) and plausible candidates. Our interpretation is that lung adenocarcinoma is a more homogeneous category than kidney rejection (at least for the datasets we analyzed), allowing for greater ascertainment of specific hits both within and across studies. In lung adenocarcinoma, the recurrent DE genes varied with respect to their prior probability of DE, suggesting differences in the degree to which they may be considered specific to the phenotype. A gene like RAGE, which is highly recurrent but of lower prior probability than other recurrent genes, is more likely to be specifically related to the disorder than genes that are DE in many more conditions.
We emphasize that, despite being generically differentially expressed, high-prior genes are biologically important, with roles in many critical processes. Their high rank simply indicates the poor degree to which they can specifically be associated with a particular phenotype. The high ranks of the PAM50 genes are useful to consider here. These genes have proven clinical and prognostic significance for breast cancer, yet they are clearly not only of significance for this condition. This is true of all high-ranking genes. Moreover, in keeping with their biological significance, the high-ranking genes will be familiar to many biologists, and their presence in the DE prior will come as no surprise. The value of our DE prior is its ability to meaningfully calibrate a degree of surprise. Knowing which genes are of high prior empowers rapid and objective prioritization of gene hit lists for further study, whether that might mean an emphasis on well-studied pathways or the evaluation of more unusual gene associations.
Moving forward, we anticipate that the prior will be most usefully exploited to design experiments that address the role of generic functions: either controlling for the processes that are of high prior probability explicitly or targeting them in more detail. Because of their high recurrence, simply reporting that these processes are associated with a phenotype should not be considered particularly novel or informative. Instead, it will be important to understand the detailed interplay of high prior functions within systems of interest. Studies of single-cell transcriptomics are perhaps the most obvious method for doing this, as they allow for expression to be dissected into cell type-specific signals, already helping to deconvolve immune responses within the tumor microenvironment, for example (80). Building on our knowledge from many experiments offers a route toward progressive refinement of expression studies, improving our understanding of molecular mechanisms in both health and disease.
Methods
Data Availability.
The DE prior is available for download from GitHub (https://github.com/maggiecrow/DEprior) (11) and may also be found in Dataset S2. Dataset S1 contains all study metadata from Gemma, and associated prior AUROCs supporting Figs. 1, 2, and 4. Additional supplementary materials may be found on GitHub.
Gemma, Gene Sets, and R.
Data preprocessing and DE analyses were performed using the Gemma web server (9). In brief, Gemma imports data series from Gene Expression Omnibus (GEO) (GSE*) along with sample annotations if they are available (GDS*). Annotations are supplemented with manual curation of both samples and experiments using ontologies with fixed vocabularies to assist with data retrieval (e.g., Disease Ontology, Cell Line Ontology). To facilitate cross-platform comparisons, probe sets from each expression platform (GPL*) are reannotated at the sequence level as described (81). Quality control checks are performed to remove outliers and adjust for possible batch effects, and DE analysis is computed using linear modeling approaches followed by multiple hypothesis test correction as described (82, 83). R (84) was used for all other analyses. Means ± SDs are reported throughout, unless otherwise specified.
Human GO and gene annotations were downloaded from the GO Consortium in February 2017. For platform-specific analyses, GO was subset to genes assayed within a given platform. For all tests, only GO terms with 20–1,000 annotated genes were used. Other gene functional annotations were curated as in ref. 85 and are available on GitHub (https://github.com/sarbal/EffectSize). PAM50 genes were sourced from ref. 86. Housekeeping genes from bulk microarray (35), and the two single-cell gene sets (39, 40), were downloaded from the personal website of Johann Gagnon-Bartsch, Department of Statistics, University of Michigan, Ann Arbor, MI, in December 2018 (www-personal.umich.edu/∼johanngb/ruv/). Housekeeping genes from bulk RNA-seq (36) were downloaded in December 2018 from https://www.tau.ac.il/∼elieis/HKG/.
DE Prior.
Priors were calculated as described (10) using the calculate_multifunc function in the EGAD package (29). In brief, this involves scoring each gene as a function of both the number of gene sets it appears in (e.g., DE hit lists), as well as the size of each gene set. This can be expressed mathematically as follows:
where each gene set has an Npos (number of genes within it) and an Nneg (number of genes outside it). A given gene’s score is then calculated as the sum, over all sets of which that gene is a member, of the reciprocal of the product of Npos and Nneg for each gene set. Genes are then ranked by their score. We specifically call this “multifunctionality” when the gene sets are derived from the GO as in ref. 10 and use the term “DE prior” for gene sets empirically derived from expression data. We use leave-one-out cross-validation to avoid overfitting DE priors, repeating the calculation for each DE hit list to be predicted. To generate a stable ranked list for characterization, we assign each gene its minimum rank from the cross-validated priors. This “minimum-rank” prior has slightly higher performance on average than the fully cross-validated version (AUROC 0.87 vs. 0.83; SI Appendix, Fig. S3). As noted, it is used for the evaluation of external gene sets, such as PAM50, and is the basis of the clustering and enrichment analysis described in Fig. 3.
Filtering Gemma to Define the Compendium.
In Gemma, experiments commonly have multiple conditions in their study design. For example, a single GEO series may contain multiple condition comparisons, such as age, sex, treatment, or tissue type, and these would all be assessed separately as parameters in a single linear model. To avoid weighting the DE prior toward studies with many experimental factors, we subset the database to include only a single condition contrast per GEO accession, chosen based on the maximum total number of samples. Where there were ties, the first listed condition was taken to be the exemplar. For each experiment, DE results were thresholded to only include genes with an absolute log2 fold change >2 and those with FDR of <0.05. This left us with a vector of 0s and 1s that indicated whether or not a gene was differentially expressed within each study, and these were used as our “gene sets” for calculating the DE prior, as defined above. These strict inclusion criteria were imposed to minimize the inclusion of false positives among DE hit lists, as it has previously been shown that, at the same FDR threshold, genes of larger effect size are more replicable across studies (87). Reducing the threshold to include all genes with log2 fold change of >|1| at FDR < 0.05 would have increased the total number of datasets in the compendium from 635 to 719 while slightly decreasing the performance of the prior (AUROC, 0.79 ± 0.1).
Gene Properties as Priors.
To explore properties of genes that contribute to high performance more broadly, we took advantage of previous efforts from the J.G. laboratory to compile functional annotation data from a wide variety of sources (85) and used these as priors for predicting DE hit lists. This consisted of ranking each annotation vector using tied ranks, and then using these as the “optimallist” for the auc_multifunc function from the EGAD package (29) to calculate AUROCs for each DE hit list. AUROCs for each prior and study may be found on GitHub (https://github.com/maggiecrow/DEPrior).
Characterizing Top Genes.
To determine whether top 1% genes might be regulated as modules, we assessed their coexpression in a large aggregate network generated from 75 expression studies as described previously (27). In brief, the network is a dense weighted network in which each individual study is rank-standardized across all gene–gene correlations and then those matrices are averaged across all studies. Node degrees are calculated conventionally as the summation of weights for a gene to all other gene pairs. The aggregate network (30,491 × 30,491 genes) was subset to genes that make up the top 1% of the prior (186 of 192 could be identified), and a nearest-neighbor graph was built using the nng function in the cccd package (88), using 1-coexpression as the distance metric and setting k = 10. The nearest-neighbor graph was then clustered using the Walktrap algorithm (28) as implemented in the igraph package (89). Cluster robustness was assessed using the neighbor_voting function within the EGAD package (29), performing threefold cross-validation on the subnetwork (186 × 186 genes) and specifying AUROC as the output metric. Finally, clusters were evaluated for GO enrichment using the hypergeometric test.
Data Subsetting by Funding Information and Clustering.
To find funding information for studies, we used the RISmed package (90), querying for Agency based on PubMed identifiers included in Gemma. Funding sources that had supported 10 or more experiments were included for further analysis (222 of 635). Priors for studies funded by the same source were defined using leave-one-out cross-validation, as described. To directly compare with global prior performance (Fig. 4 A and B), we recalculated the DE prior using only the subset of studies included for the funding agency analysis. To plot the heatmap in Fig. 4D, we performed functional enrichment for all 222 studies using GO terms with 20–1,000 genes (∼4,000 terms) as well as our six clusters. Clusters were marked as enriched within studies where they ranked in the top 1% of all gene sets by P value.
Clusters of similar DE hit lists were defined by first performing hierarchical clustering of Euclidean distances with average linkage, then using the cutreeDynamic method (22) with the following parameter settings: deepsplit = 2, minClusterSize = 10, and pamRespectsDendro = FALSE. This resulted in 155 clusters, 88 of which contained two or more studies and were used for further analysis. Cluster-specific priors were defined using leave-one-out cross-validation as described.
Reanalysis of Single-Cell Pancreas RNA-Seq, Kidney Transplant Rejection, and Lung Adenocarcinoma Data.
Five single-cell datasets from human pancreas (43–47) were downloaded from the laboratory website of M. Hemberg, Wellcome Sanger Institute, Hinxton, UK (https://hemberg-lab.github.io/scRNA.seq.datasets/human/pancreas/), in July 2016. DE was performed between samples labeled as alpha cells and beta cells using the Wilcoxon rank sum test in R (wilcox.test), and fold changes were calculated as log2(mean(alpha) + 1) − log2(mean(beta) + 1) on standardized counts. Since only one gene met the thresholds in the Baron dataset, we included all genes with the minimum P value in the DE hit list.
Processed and parsed kidney transplant rejection data and lung adenocarcinoma data were kindly provided by Purvesh Khatri, Stanford University, Stanford, CA, and Timothy Sweeney, Inflammatix, Burlingame, CA, as RDS files (87), and limma (91) was used for DE analysis. We defined DE genes within each experiment as those with FDR of <0.05 and log2 fold change of >2 in the case of lung adenocarcinoma and >1.3 in the case of transplant rejection. Six of these experiments were included in our original compendium, so in addition to plotting the DE prior ranks for all genes (Fig. 5), we also recalculated priors after excluding the overlapping studies for each phenotype (SI Appendix, Fig. S4), which yielded comparable results.
Supplementary Material
Acknowledgments
We thank Dr. Purvesh Khatri and Dr. Timothy Sweeney for providing the lung adenocarcinoma and transplant rejection data, and Manthan Shah for assistance with ontology parsing. We are grateful to the anonymous reviewers for their thoughtful comments. M.C. was supported by NIH Grant F32MH114501 and a National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Grant from the Brain & Behavior Research Foundation. S.B. and J.G. were supported by NIH Grants R01MH113005 and R01LM012736. N.L. and P.P. were supported by NIH Grant R01MH111099 and the Natural Sciences and Engineering Research Council of Canada. N.L. was also supported by a Four Year Doctoral Fellowship from the University of British Columbia.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The DE prior is available for download from GitHub (https://github.com/maggiecrow/DEprior).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1802973116/-/DCSupplemental.
References
- 1.McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ren J, Jin P, Wang E, Marincola FM, Stroncek DF. MicroRNA and gene expression patterns in the differentiation of human embryonic stem cells. J Transl Med. 2009;7:20. doi: 10.1186/1479-5876-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van ’t Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 4.Pritchard CC, Hsu L, Delrow J, Nelson PS. Project normal: Defining normal variance in mouse gene expression. Proc Natl Acad Sci USA. 2001;98:13266–13271. doi: 10.1073/pnas.221465998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pritchard C, Coil D, Hawley S, Hsu L, Nelson PS. The contributions of normal variation and genetic background to mammalian gene expression. Genome Biol. 2006;7:R26. doi: 10.1186/gb-2006-7-3-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vedell PT, Svenson KL, Churchill GA. Stochastic variation of transcript abundance in C57BL/6J mice. BMC Genomics. 2011;12:167. doi: 10.1186/1471-2164-12-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng WC, et al. Intra- and inter-individual variance of gene expression in clinical studies. PLoS One. 2012;7:e38650. doi: 10.1371/journal.pone.0038650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCall MN, Illei PB, Halushka MK. Complex sources of variation in tissue expression data: Analysis of the GTEx lung transcriptome. Am J Hum Genet. 2016;99:624–635. doi: 10.1016/j.ajhg.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoubarev A, et al. Gemma: A resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics. 2012;28:2272–2273. doi: 10.1093/bioinformatics/bts430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillis J, Pavlidis P. The impact of multifunctional genes on “guilt by association” analysis. PLoS One. 2011;6:e17258. doi: 10.1371/journal.pone.0017258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow M. 2018 DEprior. Available at https://github.com/maggiecrow/DEprior. Deposited December 13, 2018.
- 12.Brazma A, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 13.Ball CA, et al. Submission of microarray data to public repositories. PLoS Biol. 2004;2:E317. doi: 10.1371/journal.pbio.0020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 15.Baggiolini M, Walz A, Kunkel SL. Neutrophil-activating peptide-1/interleukin 8, a novel cytokine that activates neutrophils. J Clin Invest. 1989;84:1045–1049. doi: 10.1172/JCI114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mukaida N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am J Physiol Lung Cell Mol Physiol. 2003;284:L566–L577. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- 17.Bartosik-Psujek H, Stelmasiak Z. The levels of chemokines CXCL8, CCL2 and CCL5 in multiple sclerosis patients are linked to the activity of the disease. Eur J Neurol. 2005;12:49–54. doi: 10.1111/j.1468-1331.2004.00951.x. [DOI] [PubMed] [Google Scholar]
- 18.Arican O, Aral M, Sasmaz S, Ciragil P. Serum levels of TNF-α, IFN-γ, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005;2005:273–279. doi: 10.1155/MI.2005.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandey AK, Lu L, Wang X, Homayouni R, Williams RW. Functionally enigmatic genes: A case study of the brain ignorome. PLoS One. 2014;9:e88889. doi: 10.1371/journal.pone.0088889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoeger T, Gerlach M, Morimoto RI, Nunes Amaral LA. Large-scale investigation of the reasons why potentially important genes are ignored. PLoS Biol. 2018;16:e2006643. doi: 10.1371/journal.pbio.2006643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo WP, Jenssen T-K, Butte AJ, Ohno-Machado L, Kohane IS. Analysis of matched mRNA measurements from two different microarray technologies. Bioinformatics. 2002;18:405–412. doi: 10.1093/bioinformatics/18.3.405. [DOI] [PubMed] [Google Scholar]
- 22.Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casper J, et al. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 2018;46:D762–D769. doi: 10.1093/nar/gkx1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium G. GTEx Consortium Human genomics. The genotype-tissue expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lappalainen T, et al. Geuvadis Consortium Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballouz S, Verleyen W, Gillis J. Guidance for RNA-seq co-expression network construction and analysis: Safety in numbers. Bioinformatics. 2015;31:2123–2130. doi: 10.1093/bioinformatics/btv118. [DOI] [PubMed] [Google Scholar]
- 28.Pons P, Latapy M. International Symposium on Computer and Information Sciences. Springer Nature Switzerland; Cham, Switzerland: 2005. Computing communities in large networks using random walks; pp. 284–293. [Google Scholar]
- 29.Ballouz S, Weber M, Pavlidis P, Gillis J. EGAD: Ultra-fast functional analysis of gene networks. Bioinformatics. 2017;33:612–614. doi: 10.1093/bioinformatics/btw695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perou CM, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 31.Sorlie T, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris LN, et al. American Society of Clinical Oncology Use of biomarkers to guide decisions on adjuvant systemic therapy for women with early-stage invasive breast cancer: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2016;34:1134–1150. doi: 10.1200/JCO.2015.65.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao SG, et al. Associations of luminal and basal subtyping of prostate cancer with prognosis and response to androgen deprivation therapy. JAMA Oncol. 2017;3:1663–1672. doi: 10.1001/jamaoncol.2017.0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–365. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 36.Eisenberg E, Levanon EY. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 37.Lippa KA, Duewer DL, Salit ML, Game L, Causton HC. Exploring the use of internal and externalcontrols for assessing microarray technical performance. BMC Res Notes. 2010;3:349. doi: 10.1186/1756-0500-3-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gagnon-Bartsch JA, Speed TP. Using control genes to correct for unwanted variation in microarray data. Biostatistics. 2012;13:539–552. doi: 10.1093/biostatistics/kxr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deeke JM, Gagnon-Bartsch JA. 2018. Stably expressed genes in single-cell RNA-sequencing. bioRxiv:10.1101/475426. Preprint, posted November 21, 2018.
- 40.Lin Y, et al. 2018. Evaluating stably expressed genes in single cells. bioRxiv:10.1101/229815. Preprint, posted November 22, 2018.
- 41.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulirsch JC, et al. The genetic landscape of Diamond–Blackfan anemia. Am J Hum Genet. 2018;103:930–947. doi: 10.1016/j.ajhg.2018.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baron M, et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang YJ, et al. Single-cell transcriptomics of the human endocrine pancreas. Diabetes. 2016;65:3028–3038. doi: 10.2337/db16-0405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muraro MJ, et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 2016;3:385–394.e3. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segerstolpe Å, et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 2016;24:593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xin Y, et al. RNA sequencing of single human islet cells reveals type 2 diabetes genes. Cell Metab. 2016;24:608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 48.Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251–4259. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falix FA, Aronson DC, Lamers WH, Gaemers IC. Possible roles of DLK1 in the Notch pathway during development and disease. Biochim Biophys Acta. 2012;1822:988–995. doi: 10.1016/j.bbadis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Feng L, et al. Gene expression profiling in human lung development: An abundant resource for lung adenocarcinoma prognosis. PLoS One. 2014;9:e105639. doi: 10.1371/journal.pone.0105639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hou J, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabbout M, et al. ETS2 mediated tumor suppressive function and MET oncogene inhibition in human non-small cell lung cancer. Clin Cancer Res. 2013;19:3383–3395. doi: 10.1158/1078-0432.CCR-13-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Landi MT, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3:e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lo FY, et al. The database of chromosome imbalance regions and genes resided in lung cancer from Asian and Caucasian identified by array-comparative genomic hybridization. BMC Cancer. 2012;12:235. doi: 10.1186/1471-2407-12-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okayama H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 56.Robles AI, et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J Thorac Oncol. 2015;10:1037–1048. doi: 10.1097/JTO.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rousseaux S, et al. Ectopic activation of germline and placental genes identifies aggressive metastasis-prone lung cancers. Sci Transl Med. 2013;5:186ra66. doi: 10.1126/scitranslmed.3005723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Selamat SA, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–1211. doi: 10.1101/gr.132662.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stearman RS, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Su LJ, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei TY, et al. Protein arginine methyltransferase 5 is a potential oncoprotein that upregulates G1 cyclins/cyclin-dependent kinases and the phosphoinositide 3-kinase/AKT signaling cascade. Cancer Sci. 2012;103:1640–1650. doi: 10.1111/j.1349-7006.2012.02367.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xi L, et al. Whole genome exon arrays identify differential expression of alternatively spliced, cancer-related genes in lung cancer. Nucleic Acids Res. 2008;36:6535–6547. doi: 10.1093/nar/gkn697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dean PG, Park WD, Cornell LD, Gloor JM, Stegall MD. Intragraft gene expression in positive crossmatch kidney allografts: Ongoing inflammation mediates chronic antibody-mediated injury. Am J Transplant. 2012;12:1551–1563. doi: 10.1111/j.1600-6143.2011.03964.x. [DOI] [PubMed] [Google Scholar]
- 64.Einecke G, et al. A molecular classifier for predicting future graft loss in late kidney transplant biopsies. J Clin Invest. 2010;120:1862–1872. doi: 10.1172/JCI41789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flechner SM, et al. Kidney transplant rejection and tissue injury by gene profiling of biopsies and peripheral blood lymphocytes. Am J Transplant. 2004;4:1475–1489. doi: 10.1111/j.1600-6143.2004.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halloran PF, et al. Potential impact of microarray diagnosis of T cell-mediated rejection in kidney transplants: The INTERCOM study. Am J Transplant. 2013;13:2352–2363. doi: 10.1111/ajt.12387. [DOI] [PubMed] [Google Scholar]
- 67.Hayde N, et al. The clinical and genomic significance of donor-specific antibody-positive/C4d-negative and donor-specific antibody-negative/C4d-negative transplant glomerulopathy. Clin J Am Soc Nephrol. 2013;8:2141–2148. doi: 10.2215/CJN.04240413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khatri P, et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J Exp Med. 2013;210:2205–2221. doi: 10.1084/jem.20122709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maluf DG, et al. Evaluation of molecular profiles in calcineurin inhibitor toxicity post-kidney transplant: Input to chronic allograft dysfunction. Am J Transplant. 2014;14:1152–1163. doi: 10.1111/ajt.12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010;21:1987–1997. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reeve J, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013;13:645–655. doi: 10.1111/ajt.12079. [DOI] [PubMed] [Google Scholar]
- 72.Rekers NV, et al. Increased metallothionein expression reflects steroid resistance in renal allograft recipients. Am J Transplant. 2013;13:2106–2118. doi: 10.1111/ajt.12314. [DOI] [PubMed] [Google Scholar]
- 73.Saint-Mezard P, et al. Analysis of independent microarray datasets of renal biopsies identifies a robust transcript signature of acute allograft rejection. Transpl Int. 2009;22:293–302. doi: 10.1111/j.1432-2277.2008.00790.x. [DOI] [PubMed] [Google Scholar]
- 74.Toki D, et al. The role of macrophages in the development of human renal allograft fibrosis in the first year after transplantation. Am J Transplant. 2014;14:2126–2136. doi: 10.1111/ajt.12803. [DOI] [PubMed] [Google Scholar]
- 75.Ó Broin P, et al. A pathogenesis-based transcript signature in donor-specific antibody-positive kidney transplant patients with normal biopsies. Genom Data. 2014;2:357–360. doi: 10.1016/j.gdata.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen R, et al. A meta-analysis of lung cancer gene expression identifies PTK7 as a survival gene in lung adenocarcinoma. Cancer Res. 2014;74:2892–2902. doi: 10.1158/0008-5472.CAN-13-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Groom JR, Luster AD. CXCR3 ligands: Redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–215. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jing R, Cui M, Wang J, Wang H. Receptor for advanced glycation end products (RAGE) soluble form (sRAGE): A new biomarker for lung cancer. Neoplasma. 2010;57:55–61. doi: 10.4149/neo_2010_01_055. [DOI] [PubMed] [Google Scholar]
- 79.Jaffe AE, et al. Practical impacts of genomic data “cleaning” on biological discovery using surrogate variable analysis. BMC Bioinformatics. 2015;16:372. doi: 10.1186/s12859-015-0808-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pavlidis P, Noble WS. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001;2:RESEARCH0042. doi: 10.1186/gb-2001-2-10-research0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pavlidis P. Using ANOVA for gene selection from microarray studies of the nervous system. Methods. 2003;31:282–289. doi: 10.1016/s1046-2023(03)00157-9. [DOI] [PubMed] [Google Scholar]
- 84.R Core Team 2018. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
- 85.Ballouz S, Gillis J. Strength of functional signature correlates with effect size in autism. Genome Med. 2017;9:64. doi: 10.1186/s13073-017-0455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bastien RRL, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics. 2012;5:44. doi: 10.1186/1755-8794-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sweeney TE, Haynes WA, Vallania F, Ioannidis JP, Khatri P. Methods to increase reproducibility in differential gene expression via meta-analysis. Nucleic Acids Res. 2017;45:e1. doi: 10.1093/nar/gkw797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marchette DJ. Random Graphs for Statistical Pattern Recognition. Wiley; Hoboken, NJ: 2005. [Google Scholar]
- 89.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Systems. 2006;2006:1695. [Google Scholar]
- 90.Kovalchik S. 2017 RISmed: Download content from NCBI databases. R Package, Version 2.1.7. Available at https://cran.r-project.org/web/packages/RISmed/RISmed.pdf. Accessed May 9, 2018.
- 91.Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. Limma: Linear models for microarray data; pp. 397–420. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DE prior is available for download from GitHub (https://github.com/maggiecrow/DEprior) (11) and may also be found in Dataset S2. Dataset S1 contains all study metadata from Gemma, and associated prior AUROCs supporting Figs. 1, 2, and 4. Additional supplementary materials may be found on GitHub.