Significance
γ-Secretase cleaves multiple transmembrane proteins, but little is known about how it controls its substrate specificity. γ-Secretase activating protein (GSAP) has been reported to differentially activate γ-secretase for APP and Notch cleavages. The mechanism by which GSAP regulates γ-secretase specificity is elusive. Here, we demonstrate that GSAP directly regulates γ-secretase activity and specificity. Furthermore, GSAP functions as a switch between two forms of γ-secretase that have different activities for APP and Notch substrates, leading to different specificities. These findings open a new avenue for drug development through targeting the specificity of modifying proteins. This work also suggests that the association of GSAP with aging, Alzheimer’s disease, and Down syndrome could be attributed to the function of GSAP in the regulation of γ-secretase.
Keywords: Alzheimer’s disease, γ-secretase modulation, GSAP, active-site labeling, conformational change
Abstract
The mechanism by which γ-secretase activating protein (GSAP) regulates γ-secretase activity has not yet been elucidated. Here, we show that knockout of GSAP in cultured cells directly reduces γ-secretase activity for Aβ production, but not for Notch1 cleavage, suggesting that GSAP may induce a conformational change contributing to the specificity of γ-secretase. Furthermore, using an active-site–directed photoprobe with double cross-linking moieties, we demonstrate that GSAP modifies the orientation and/or distance of the PS1 N-terminal fragment and the PS1 C-terminal fragment, a region containing the active site of γ-secretase. This work offers insight into how GSAP regulates γ-secretase specificity.
The γ-secretase protease is a large intramembrane protein complex comprising four essential components: presenilin 1 (PS1), nicastrin (Nct), anterior pharynx-defective 1 (Aph1), and presenilin enhancer 2 (Pen2) (1, 2). PS1 is the catalytic core of the complex (3–5) and requires an activation step by endoproteolysis (6) that is dependent on Pen2 (5, 7, 8). In addition to these mandatory subunits, γ-secretase is also regulated by nonessential proteins such as CD147, TPM21, the γ-secretase activating protein (GSAP), and Hif-1α (9–12). GSAP is an ∼98-kDa holoprotein that undergoes extensive processing, resulting in an ∼16-kDa C-terminal fragment (12). This fragment was found to form a ternary complex with γ-secretase and amyloid precursor protein (APP)-C99 and to regulate the cleavage at γ sites, but not ε sites, controlling the generation of amyloid beta (Aβ) and APP intracellular domain, respectively.
Decreasing GSAP expression in cells significantly reduced Aβ levels (12, 13) without affecting the cleavage of other substrates, such as Notch (12). However, the role of GSAP in the regulation of γ-secretase has been challenged (13) due to lack of direct mechanistic evidence. Nevertheless, GSAP RNAi mice crossed with double transgenic APPsweXPS1ΔE9 Alzheimer’s disease (AD) model mice have reduced Aβ burden (12), indicating that GSAP is a novel therapeutic target for the treatment of AD. Furthermore, genetic studies have shown that GSAP is linked with aging, AD, and Down syndrome (14–16). Therefore, investigating the function of GSAP in Aβ production and in γ-secretase activity and specificity is critical for developing an in-depth understanding of γ-secretase modulation and effective AD therapeutics. However, such studies have been hindered due to lack of suitable technologies.
In this study, we use cell-free assays with recombinant substrates to show that knocking out GSAP reduces γ-secretase activity for Aβ production without interfering with Notch1 cleavage, providing strong evidence that GSAP directly regulates γ-secretase activity and specificity. Furthermore, using an active-site–directed photoprobe, we show that γ-secretase has different conformations in the presence and absence of GSAP, which have different catalytic activities for APP and Notch substrates. This work offers insight into how GSAP regulates γ-secretase specificity.
Results
GSAP KO Reduces Aβ Secretion and γ-Secretase Activity.
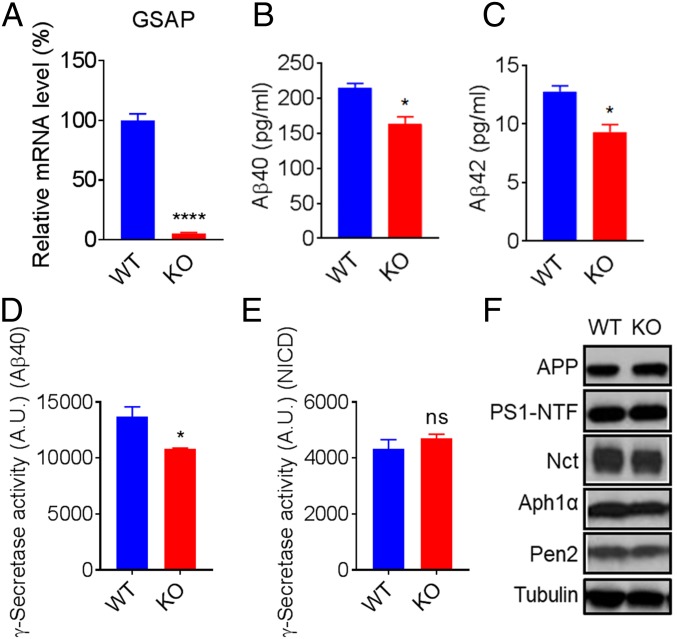
Previous studies examined the function of GSAP using siRNA knockdown methods (12, 13). To further investigate the role of GSAP in γ-secretase regulation, we knocked out GSAP with CRISPR-Cas9 technology in HEK293 cells that stably express APP (HEK-APP). Knockout (KO) of GSAP was confirmed by genomic sequencing (SI Appendix, Fig. S1). mRNA levels of GSAP in GSAP-KO cells were not detectable (Fig. 1A), whereas the expression of APP mRNA was not changed (SI Appendix, Fig. S1). Secretion of Aβ40 and Aβ42 in the GSAP-KO cells was only 75% and 73% of HEK-APP WT cells, respectively (Fig. 1 B and C).
Fig. 1.
HEK-APP GSAP-KO reduces Aβ secretion and γ-secretase activity for APP without changing γ-secretase components. (A) GSAP mRNA levels in HEK-APP WT versus HEK-APP GSAP-KO cells. (B and C) Secretion of Aβ40 (B) and Aβ42 (C) in HEK-APP GSAP WT versus HEK-APP GSAP-KO cells showing significant reduction in KO cells. Data in A–C represent means ± SEM; n = 3. (D) γ-Secretase activity levels toward recombinant APP are decreased in HEK-APP GSAP-KO compared with WT counterpart. (E) γ-Secretase activity levels toward recombinant Notch remain the same in HEK-APP GSAP-KO compared with WT counterpart. NICD, Notch1 intracellular domain. Data in D and E represent means ± SEM; n = 6. (F) Western blot analysis of APP and total γ-secretase subunits PS1-NTF, Nct, Aph1a, and Pen2 in HEK-APP WT and KO. *P < 0.05, ****P < 0.0001; ns, not significant.
To directly measure the effect of GSAP on γ-secretase activity, we performed exo-cell assays (17) using recombinant APP or Notch substrate (18), which allows for the immediate and real-time analysis of γ-secretase activity for both substrates. HEK-APP WT and GSAP-KO cells were seeded in a 96-well plate overnight. The recombinant substrates were then added to the cells in the presence of 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO) and incubated for 2.5 h to measure γ-secretase cleavage. The cleaved products were detected with an AlphaLISA assay (18). γ-Secretase activity was calculated by normalizing to protein concentration. HEK-APP GSAP-KO cells have only 71% γ-secretase activity for Aβ40 production compared with WT (Fig. 1C) but have the same level of Notch1 cleavage (Fig. 1D), indicating that GSAP solely impacts the processing of APP without affecting Notch processing. Next, we tested whether the reduced γ-secretase activity and Aβ secretion were a result of changes in expression levels of the γ-secretase subunits. The protein levels of APP, PS1 N-terminal fragment (PS1-NTF), Nct, Aph1a, and Pen2 remained unchanged in HEK-APP KO compared with WT (Fig. 1E), indicating that GSAP KO directly affects γ-secretase activity without altering overall steady-state levels of γ-secretase subunits. In addition, we generated GSAP-KO SH-5YSY cells and found that GSAP KO reduces γ-secretase activity for Aβ40, but not for Notch cleavage (SI Appendix, Fig. S2).
Overexpression of hGSAP in GSAP-KO Cells Rescues γ-Secretase Activity and Aβ Secretion.
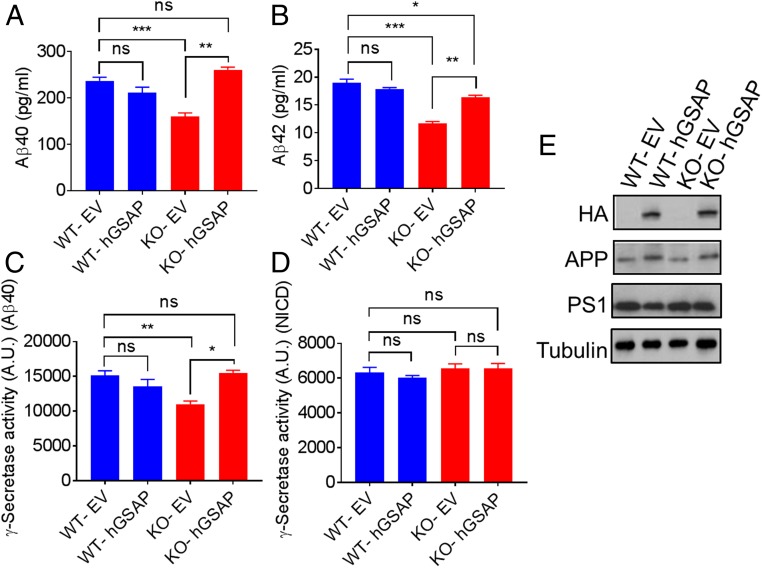
To determine whether the reduction of γ-secretase activity in the GSAP-KO cells directly results from elimination of GSAP, we performed rescue studies by overexpressing GSAP in the KO cells. Empty vector (EV) or the full-length human GSAP with a C-terminal HA tag (hGSAP) construct was transfected into the HEK-APP WT or GSAP-KO cells, and secreted Aβ species were measured 48 h posttransfection. We found that overexpression of hGSAP in HEK-APP KO cells can fully or partially restore secreted Aβ40 and Aβ42 (Fig. 2 A and B). However, overexpression of hGSAP in HEK-APP WT cells had no effect on Aβ secretion, indicating that the endogenous level of GSAP is sufficient for γ-secretase activation for the processing of APP. Next, we measured γ-secretase activity using cell membranes prepared 48 h posttransfection. In agreement with the Aβ production data, expression of hGSAP in KO cells rescued γ-secretase activity for APP, but the γ-secretase activity remained unchanged in the WT membrane (Fig. 2C). Remarkably, γ-secretase activity for cleavage of recombinant Notch substrate was not changed with GSAP overexpression in any of the cell lines (Fig. 2D). Expression of hGSAP, migrating as expected at ∼98-kDa protein, was confirmed by Western blot using anti-HA antibodies (Fig. 2E). Moreover, the overexpression of GSAP did not alter the expression of APP or PS1 (Fig. 2E). Similar results were obtained rescuing the γ-secretase activity and Aβ secretion with SH-5YSY GSAP-KO cells (SI Appendix, Fig. S3). The expression of GSAP on the GSAP-KO background can rescue γ-secretase activity for the processing of APP, demonstrating that the GSAP-KO effect on γ-secretase activity comes from the abolition of GSAP.
Fig. 2.
Overexpression of hGSAP in GSAP KO cells rescues γ-secretase activity and Aβ secretion. EV or hGSAP was transfected into the HEK-APP GSAP WT or KO cells. (A and B) Aβ secretion was measured 48 h posttransfection using Meso Scale Discovery Aβ detection for Aβ40 (A) and Aβ42 (B). (C and D) Membrane fractions prepared from 48-h posttransfection of EV and hGSAP in HEK-APP WT or KO cells were assayed for γ-secretase activity using recombinant APP (C) or Notch (D). NICD, Notch1 intracellular domain. All data represent means ± SEM; n = 3. (E) Western blot analysis of HA (hGSAP), APP, and PS1-NTF in HEK-APP WT and KO cells transfected with either EV or hGSAP. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant.
GSAP Modifies γ-Secretase Catalytic Efficiency for APP, but Not for Notch.
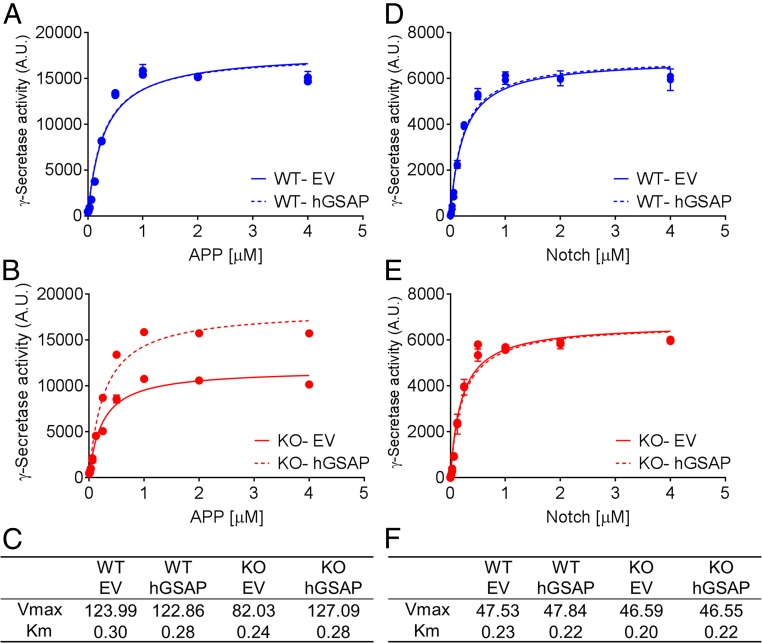
To better understand the effect of GSAP on γ-secretase, we measured the kinetics of γ-secretase in membrane fractions prepared from four cell lines: HEK-APP GSAP WT and GSAP-KO cells transfected with EV or hGSAP. First, we found that Km and Vmax values for APP of HEK-APP WT are 0.29 µM and 123.99 a.u.⋅µg−1⋅min−1, respectively, and for GSAP-KO, the values are 0.24 µM and 82.03 a.u.⋅µg−1⋅min−1, respectively (Fig. 3 A and B). Second, transfection of hGSAP in the KO cells increased Vmax (AlphaLISA arbitrary unit) without modifying the Km value (µM) (Fig. 3 B and C) but had no effect on HEK-APP GSAP WT γ-secretase. Finally, γ-secretase from all four cell lines had similar Km and Vmax values for Notch substrate regardless of WT or KO and hGSAP transfection (Fig. 3 D–F). These results indicate that GSAP does not change the binding of γ-secretase to APP substrate, but rather alters the Vmax. Moreover, GSAP specifically modulates γ-secretase for Aβ production, but not Notch1 cleavage.
Fig. 3.
GSAP modifies γ-secretase catalytic efficiency for APP, but not for Notch. Kinetic curves fitted to Michaelis–Menten model of γ-secretase activity processing recombinant APP (A and B) or Notch (D and E) measured from membrane fraction of either HEK-APP GSAP WT (A and D) or GSAP-KO (B and E) cells transfected with EV or hGSAP. Vmax (a.u.⋅µg−1⋅min−1) and Km (µM) values calculated for APP (C) or Notch (F) substrate. The data are representative of three independent experiments.
GSAP Modifies the Active-Site Conformation of γ-Secretase.
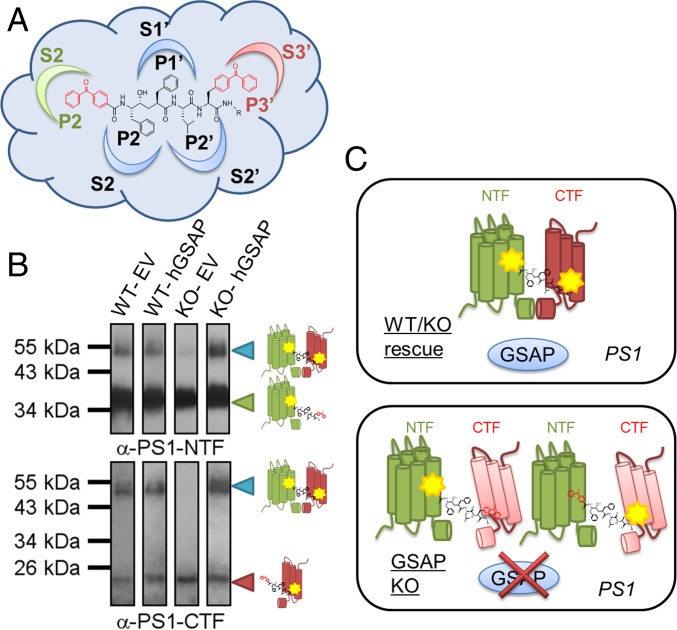
Kinetic analysis suggests that GSAP might alter the active site of γ-secretase, leading to different catalytic efficiencies for APP substrates. To detect the changes in the active site in the presence and absence of GSAP, we used active-site–directed γ-secretase inhibitors that directly interact with PS1-NTF and PS1-CTF (C-terminal fragment) (4, 18). Moreover, these active-site–directed inhibitors only label the active form of γ-secretase, not full-length PS1 (4). L631 contains two benzoylphenylalanine (BPA) groups at its P2 and P3′ positions, allowing for the photolabeling of γ-secretase (Fig. 4A) (19). L631 has been demonstrated to label PS1-NTF, PS1-CTF, and cross-linked PS1-NTF and PS1-CTF together (19). The photolabeling efficiency depends on direct contact between the residues and the corresponding subpockets (S2 and S3′) in the active site (Fig. 4A). Any conformational changes induced by GSAP, which alter the distance or orientation between a subpocket and the photoprobe, may lead to different cross-linking efficiencies (18). Four cell type membranes (WT-EV, WT-hGSAP, KO-EV, and KO-hGSAP) were photolabeled with L631. Labeled species were isolated by streptavidin beads and analyzed by PS1-NTF and PS1-CTF antibodies. L631 photolabels both PS1-NTF (Fig. 4B, Upper, ∼34-kDa band) and PS1-CTF (Fig. 4B, Lower, ∼20-kDa band) in membranes from all four cell lines. One striking difference is that L631 does not cross-link PS1-NTF and PS1-CTF together (a ∼55-kDa band) in GSAP-deficient cells (KO-EV) (Fig. 4B), and the labeling of the 55-kDa band can be restored by the reexpression of GSAP (KO-hGSAP). In the presence of GSAP, we propose that PS1-NTF and PS1-CTF align in a specific conformation in which L631 can cross-link the two fragments, resulting in higher γ-secretase activity for Aβ production. However, when GSAP is absent, the conformation of the active site is transformed into a different form, which does not allow for L631 to cross-link the two fragments (Fig. 4C) and results in lower γ-secretase activity for Aβ production, but not for Notch1 cleavage.
Fig. 4.
GSAP modifies the active-site conformation of PS1. (A) Structure of photoprobe L631 with two BPA groups embedded in P2 and P3′ that are photoactivatable and can detect conformational changes corresponding to the γ-secretase active-site subpockets S2 and S3′. (B) Western blot analysis with antibodies for PS1-NTF (Upper) and PS1-CTF (Lower) of the photolabeling efficiency by L631 in HEK-APP WT or KO cells transfected with EV or hGSAP. (C) Schematic representation of GSAP modification of PS1: In the presence of GSAP (Upper), PS1-NTF and PS1-CTF are aligned in a specific confirmation whereby the L631 probe can photolabel and cross-link both together, but when GSAP is absent (Lower), this active-site confirmation is modified and the L631 cross-linking is absent.
Discussion
γ-Secretase cleaves an array of substrates (20), including Notch proteins and key molecules that regulate many biological processes ranging from neuronal development to tumorigenesis (21, 22). Therefore, understanding how γ-secretase activity and specificity are regulated has been a critical question that remains unanswered. It has been suggested that besides mandatory subunits, γ-secretase is regulated by modulatory proteins such as CD147, TPM21, GSAP, and Hif-1α (9–12). While the essential subunits are ubiquitously expressed, only small portion of γ-secretase complexes are catalytically active (23–26). GSAP has emerged as a promising γ-secretase regulator to be targeted in AD because of its unique interference with APP processing without modifying Notch cleavage (12). Although the effect of GSAP on the production of Aβ has been established, its role in the activation and specificity of γ-secretase has been controversial (13, 27, 28) due to lack of practical and suitable approaches to investigate its mechanism.
The failure to detect changes in γ-secretase activity and Aβ secretion simply by overexpressing GSAP has led previous studies to question whether GSAP can modulate γ-secretase (13, 28). However, our data show that the level of endogenous GSAP in WT cells is sufficient to regulate γ-secretase activity and that overexpression of exogenous GSAP cannot further enhance γ-secretase cleavage in WT cells. This indicates that the endogenous GSAP is sufficient for the regulation of γ-secretase in these cells. Interestingly, another γ-secretase modulatory protein, CD147, was shown to have no significant effect on Aβ production when overexpressed in WT cells (10) and may have a different mechanism in the regulation of γ-secretase.
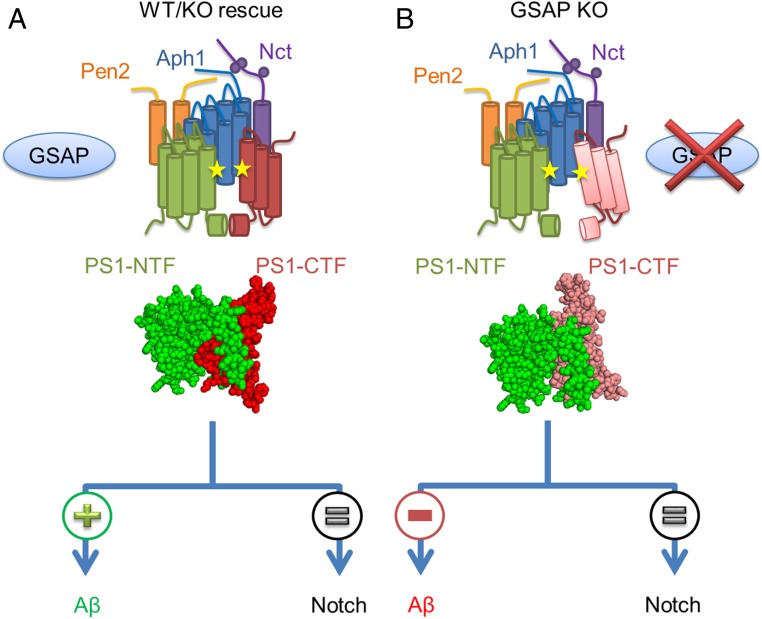
Even though an atomic structure of γ-secretase has been reported (29), there is little information about the active site of γ-secretase and conformational changes. Activity-based probes designed from transition-state inhibitors have been used broadly to study the active γ-secretase complex because they do not bind to the inactive complex (4, 25, 26). Subsequently, a “photophore walking” approach (18) has been developed to detect conformational changes in the γ-secretase active site. In this technique, a transition-state inhibitor that directly interacts with the active site is modified by incorporating photoactivatable groups into different side chains along the probe and, therefore, can be cross-linked to different subpockets within the active site. Since the efficiency of photolabeling depends on the contact region and proximity to residues within the active site, any conformational changes that occur alter the orientation or distance between a subpocket and lead to different cross-linking efficiencies. Using an L458 derivative with two BPA groups incorporated in P2 and P3′ (L631) (19), we were able to detect the changes in the γ-secretase active site in the presence and absence of GSAP, a mechanism which was not shown previously. Furthermore, our photolabeling studies indicate that γ-secretase has at least two conformations modulated through GSAP. Switching between these two forms can affect γ-secretase activity for Aβ production, but not Notch1 cleavage. These findings are also consistent with our previous report that GSAP does not affect the ε-site cleavage, similar to the Notch cleavage site (12). When GSAP is present in the cells, like in the WT or KO cells transfected with GSAP (Fig. 5A), PS1 adopts a native conformation allowing for the normal processing of APP and Notch. In this native conformation, PS1-NTF and PS1-CTF are in a specific orientation allowing for the L631 photoprobe to cross-link the two fragments. However, KO of GSAP results in a different PS1 conformation, associated with a reduction of γ-secretase activity for APP, leading to reduction in Aβ secretion (Fig. 5B). Our studies show that GSAP directly affects the conformation of the active site, providing proof that GSAP directly regulates γ-secretase activity and specificity. Our findings provide a route to the investigation of γ-secretase modulation and, ultimately, to the development of therapeutic agents for AD. In addition, abnormal activation of γ-secretase by GSAP that is associated with aging, AD, and Down syndrome (14–16) may lead to AD.
Fig. 5.
Proposed model of GSAP modulation of γ-secretase activity. (A and B) γ-Secretase complex presented as transmembrane rods containing PS1-NTF (green), PS1-CTF (red), Nct (purple), Aph1 (blue), and Pen2 (orange) in the presence (A) of GSAP (blue sphere) in WT or in GSAP rescue with induced PS1 conformation, which leads to γ-secretase activity for both APP and Notch. When GSAP is absent (B) PS1 adopts a different conformation, which leads to a decrease in APP processing and a reduction in Aβ secretion, but not in Notch processing.
Materials and Methods
Cell Culture.
HEK-APP cell lines were cultured in DMEM supplemented with 10% FBS and 1% penicillin and streptomycin. Human neuroblastoma SH-5YSY cell lines were grown in MEM/F-12 supplemented with 10% FBS and 1% penicillin. Transfection was done using Lipofectamine LTX with Plus Reagent according to manufacturer’s instructions.
CRISPR-Cas9 GSAP-KO Generation and Isolation.
Human GSAP CRISPR-Cas9 plasmid with gRNA targeting exon 16 (CATTGCCCTTTACAGTCATT) was design and cloned into PX459 by the Memorial Sloan Kettering Cancer Center (MSKCC) RNAi core facility. HEK-APP or SH-5YSY cells were transfected and selected with 2 µg/mL puromycin. Single clones were isolated and analyzed by DNA sequencing of GSAP exon 16. Both HEK-APP and SH-5YSY hGSAP-KO clones contain a single-nucleotide deletion, which creates early termination.
RNA Isolation and Real-Time RT-PCR.
Total RNA was isolated with the QIAGEN RNeasy Mini Kit according to the manufacturer’s protocols. RNA (1 µg) was reversely transcribed to cDNA using the SuperScript III First-Strand Synthesis System (Invitrogen). qRT-PCR analysis was performed with designated cDNA samples using TaqMan Gene Expression Assay (Applied Biosystems). All real-time qPCR was performed in triplicate on the Fast 7500 Real-Time PCR System (Applied Biosystems). TaqMan primers were hGSAP (Hs01383759_m1) and ribosomal 18S (Hs03003631_g1) from Applied Biosystems. Relative quantitation between samples was analyzed using the ΔΔCT method.
Meso Scale Discovery.
Secreted human Aβ species were detected using Meso Scale Discovery multiplex (6E10) from cell culture media 48 h posttransfection according to the manufacturer’s instructions.
Western Blot and Antibodies.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris, pH8.0, 150 nM NaCl, 0.1% vol/vol Nonidet P-40, and 0.5% wt/vol deoxycholic acid) containing protease inhibitor mixture. Protein concentration was determined by the DC Protein Assay Kit (Bio-Rad). Antibodies used for Western blot are as follows: PS1-NTF and Nct (from our laboratory), PS1-CTF (MAB5232; Millipore), Aph1a (38-3600; Invitrogen), Pen2 (18189; Abcam), APP (MABN10; Millipore), and HA (18181; Abcam).
γ-Secretase Activity Assays.
The exo-cell assay was performed as previously described (17). Briefly, cells were seeded in 96-well culture plates for 24 h and were washed with PBS after removing media. Next, Sb4 substrate (1 µM) or NTM2 substrate (0.4 µM) was added and incubated in 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (Pipes) buffer (50 mM Pipes, pH 7.0, 150 mM KCl, 5 mM CaCl2, 5 mM MgCl2) and 0.25% CHAPSO detergent at 37 °C for 2.5 h. γ-Secretase products were detected by AlphaLISA methods using G2-10 or SM320 antibodies for Aβ40 or Notch1 intracellular domain, respectively (18). Activity readout was expressed as arbitrary AlphaLISA units. Specific activity was normalized to protein concentration. Cell membrane preparation and γ-secretase assays were described previously (18, 30, 31)
Activity-Based Photoaffinity Labeling.
Active-site–based photoaffinity labeling was performed with cell lines in a 12-well tissue culture dish with 10 nM L631 in PBS (pH 7.4) and 0.25% CHAPSO. Photolabeling experiments were carried out as described previously (4, 19). Reaction mixtures were solubilized with RIPA buffer for 1 h at room temperature, and streptavidin beads were added to capture labeled PS1 at 4 °C overnight. Beads were washed and eluted by boiling in 2× Laemmli sample buffer. Ensuing samples were resolved on SDS/PAGE followed by Western blotting with PS1-NTF or PS1-CTF antibody.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants R01NS096275 and RF1AG057593 (to Y.-M.L.) and R01AG047781 (to P.G.); the Fisher Center for Alzheimer’s Research (P.G.); and the JPB Foundation (Y.-M.L. and P.G.). G.P.L. is supported by the Institutional Training Grant 5T32GM073546. We also acknowledge the Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748), Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research, the Experimental Therapeutics Center of MSKCC, and the William Randolph Hearst Fund in Experimental Therapeutics.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1820160116/-/DCSupplemental.
References
- 1.De Strooper B. Aph-1, Pen-2, and nicastrin with presenilin generate an active gamma-secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- 2.Sun L, et al. Structural basis of human γ-secretase assembly. Proc Natl Acad Sci USA. 2015;112:6003–6008. doi: 10.1073/pnas.1506242112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esler WP, et al. Transition-state analogue inhibitors of gamma-secretase bind directly to presenilin-1. Nat Cell Biol. 2000;2:428–434. doi: 10.1038/35017062. [DOI] [PubMed] [Google Scholar]
- 4.Li YM, et al. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature. 2000;405:689–694. doi: 10.1038/35015085. [DOI] [PubMed] [Google Scholar]
- 5.Ahn K, et al. Activation and intrinsic γ-secretase activity of presenilin 1. Proc Natl Acad Sci USA. 2010;107:21435–21440. doi: 10.1073/pnas.1013246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thinakaran G, et al. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 7.Takasugi N, et al. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–441. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 8.Luo WJ, et al. PEN-2 and APH-1 coordinately regulate proteolytic processing of presenilin 1. J Biol Chem. 2003;278:7850–7854. doi: 10.1074/jbc.C200648200. [DOI] [PubMed] [Google Scholar]
- 9.Chen F, et al. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- 10.Zhou S, Zhou H, Walian PJ, Jap BK. CD147 is a regulatory subunit of the gamma-secretase complex in Alzheimer’s disease amyloid beta-peptide production. Proc Natl Acad Sci USA. 2005;102:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villa JC, et al. Nontranscriptional role of Hif-1α in activation of γ-secretase and Notch signaling in breast cancer. Cell Rep. 2014;8:1077–1092. doi: 10.1016/j.celrep.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He G, et al. Gamma-secretase activating protein is a therapeutic target for Alzheimer’s disease. Nature. 2010;467:95–98. doi: 10.1038/nature09325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hussain I, et al. The role of γ-secretase activating protein (GSAP) and imatinib in the regulation of γ-secretase activity and amyloid-β generation. J Biol Chem. 2013;288:2521–2531. doi: 10.1074/jbc.M112.370924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu M, et al. Common GSAP promoter variant contributes to Alzheimer’s disease liability. Neurobiol Aging. 2014;35:2656 e1–2656 e7. doi: 10.1016/j.neurobiolaging.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Chu J, Wisniewski T, Praticò D. GATA1-mediated transcriptional regulation of the γ-secretase activating protein increases Aβ formation in Down syndrome. Ann Neurol. 2016;79:138–143. doi: 10.1002/ana.24540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez SE, Nadeem M, Malek-Ahmadi MH, He B, Mufson EJ. Frontal cortex and hippocampal γ-secretase activating protein levels in prodromal Alzheimer disease. Neurodegener Dis. 2017;17:235–241. doi: 10.1159/000477937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shelton CC, Tian Y, Frattini MG, Li YM. An exo-cell assay for examining real-time gamma-secretase activity and inhibition. Mol Neurodegener. 2009;4:22. doi: 10.1186/1750-1326-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chau DM, Crump CJ, Villa JC, Scheinberg DA, Li YM. Familial Alzheimer disease presenilin-1 mutations alter the active site conformation of γ-secretase. J Biol Chem. 2012;287:17288–17296. doi: 10.1074/jbc.M111.300483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu M, et al. Gamma-secretase: Characterization and implication for Alzheimer disease therapy. Neurobiol Aging. 2002;23:1023–1030. doi: 10.1016/s0197-4580(02)00126-4. [DOI] [PubMed] [Google Scholar]
- 20.Kopan R, Ilagan MX. Gamma-secretase: Proteasome of the membrane? Nat Rev Mol Cell Biol. 2004;5:499–504. doi: 10.1038/nrm1406. [DOI] [PubMed] [Google Scholar]
- 21.Lathia JD, Mattson MP, Cheng A. Notch: From neural development to neurological disorders. J Neurochem. 2008;107:1471–1481. doi: 10.1111/j.1471-4159.2008.05715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu Y, et al. The presenilin proteins are components of multiple membrane-bound complexes that have different biological activities. J Biol Chem. 2004;279:31329–31336. doi: 10.1074/jbc.M401548200. [DOI] [PubMed] [Google Scholar]
- 24.Beher D, et al. In vitro characterization of the presenilin-dependent gamma-secretase complex using a novel affinity ligand. Biochemistry. 2003;42:8133–8142. doi: 10.1021/bi034045z. [DOI] [PubMed] [Google Scholar]
- 25.Placanica L, et al. Pen2 and presenilin-1 modulate the dynamic equilibrium of presenilin-1 and presenilin-2 gamma-secretase complexes. J Biol Chem. 2009;284:2967–2977. doi: 10.1074/jbc.M807269200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai MT, et al. Presenilin-1 and presenilin-2 exhibit distinct yet overlapping gamma-secretase activities. J Biol Chem. 2003;278:22475–22481. doi: 10.1074/jbc.M300974200. [DOI] [PubMed] [Google Scholar]
- 27.Alzforum 2014 GSAP revisited: Does it really play a role in processing Aβ? Available at https://www.alzforum.org/news/research-news/gsap-revisited-does-it-really-play-role-processing-av. Accessed January 3, 2014.
- 28.Deatherage CL, Hadziselimovic A, Sanders CR. Purification and characterization of the human γ-secretase activating protein. Biochemistry. 2012;51:5153–5159. doi: 10.1021/bi300605u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bai XC, et al. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li YM, et al. Presenilin 1 is linked with gamma-secretase activity in the detergent solubilized state. Proc Natl Acad Sci USA. 2000;97:6138–6143. doi: 10.1073/pnas.110126897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian Y, Bassit B, Chau D, Li YM. An APP inhibitory domain containing the Flemish mutation residue modulates gamma-secretase activity for Abeta production. Nat Struct Mol Biol. 2010;17:151–158. doi: 10.1038/nsmb.1743. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.