Significance
Marine phytoplankton are responsible for nearly half of the primary productivity on Earth, and the picocyanobacterium Synechococcus contributes up to 16% of this total. Synechococcus strains optimize their fitness by using different chromophores for photosynthetic light-harvesting in different light color niches. Many Synechococcus strains use chromatic acclimation to adjust chromophore composition to ambient blue to green light ratios. We determined that interplay between two chromophore attachment enzymes, MpeY and MpeZ, plays a role in chromatic acclimation at the level of gene expression. The unique ability of these two enzymes to attach different chromophores to the same residue makes them potentially useful for differential labeling of that residue for fluorescence-imaging technologies.
Keywords: light regulation, marine biology, photosynthesis, cyanobacteria, lyase
Abstract
Marine Synechococcus, a globally important group of cyanobacteria, thrives in various light niches in part due to its varied photosynthetic light-harvesting pigments. Many Synechococcus strains use a process known as chromatic acclimation to optimize the ratio of two chromophores, green-light–absorbing phycoerythrobilin (PEB) and blue-light–absorbing phycourobilin (PUB), within their light-harvesting complexes. A full mechanistic understanding of how Synechococcus cells tune their PEB to PUB ratio during chromatic acclimation has not yet been obtained. Here, we show that interplay between two enzymes named MpeY and MpeZ controls differential PEB and PUB covalent attachment to the same cysteine residue. MpeY attaches PEB to the light-harvesting protein MpeA in green light, while MpeZ attaches PUB to MpeA in blue light. We demonstrate that the ratio of mpeY to mpeZ mRNA determines if PEB or PUB is attached. Additionally, strains encoding only MpeY or MpeZ do not acclimate. Examination of strains of Synechococcus isolated from across the globe indicates that the interplay between MpeY and MpeZ uncovered here is a critical feature of chromatic acclimation for marine Synechococcus worldwide.
Picophytoplankton of the genus Synechococcus are distributed throughout the marine environment, constituting a global population of ∼7 × 1026 cells (1). The ubiquity of these organisms is partially due to the wide diversity of its photosynthetic pigments, which allows for niche occupancy of a large variety of light environments (2, 3). There are three main pigment types of Synechococcus strains adapted to distinct spectral niches in the marine environment: blue-light specialists, green-light specialists, and blue-green generalists (2, 4).
These three pigment types differ in the composition of their light-harvesting complexes, known as phycobilisomes (PBS). PBS are typically fan-shaped, with a central core that is appressed to the photosynthetic (thylakoid) membranes and multiple rods that extend from the core and provide increased surface area for photon capture. PBS consist of phycobiliproteins, apoproteins containing one to three covalently attached chromophores that absorb specific wavelengths of light (5), and linker proteins, which have structural and energy transfer roles (6). Although there are several kinds of phycobiliproteins, all are α/β heterodimers. In marine Synechococcus, PBS rods always contain the phycobiliprotein phycocyanin and usually contain one or two different forms of phycoerythrin called phycoerythrin-I (PE-I) and phycoerythrin-II (PE-II) (2, 3, 6). The α and β subunits of PE-I are encoded by the cpeBA operon and are called CpeA and CpeB, while the α and β subunits of PE-II are encoded by the mpeBA operon and are called MpeA and MpeB. These heterodimers are located in different regions of the rods, with CpeA and CpeB in the more core-proximal regions and MpeA and MpeB in the more core-distal regions.
Phycoerythrin in marine Synechococcus strains contains two different types of chromophores, blue-light–absorbing phycourobilin (PUB) [maximum absorbance (Absmax) ∼ 495 nm] and green-light–absorbing phycoerythrobilin (PEB) (Absmax ∼ 550 nm) (2, 7). Blue-light specialists have a high PUB:PEB ratio, while green-light specialists have a low PUB:PEB ratio. Blue-green generalists are capable of modifying their PUB:PEB ratio in response to the ambient blue-green ratio through a process known as type IV chromatic acclimation or CA4 (8, 9). In the CA4-capable strain Synechococcus sp. RS9916 (hereafter 9916), each CpeA/CpeB dimer contains four PEB and one PUB in green light and three PEB and two PUB in blue light. Each MpeA/MpeB dimer has four PEB and two PUB in green light and two PEB and four PUB in blue light (10).
Enzymes known as phycobilin lyases attach the light-harvesting chromophores to conserved cysteine residues in phycobiliproteins, forming thioether bonds (11, 12). Most lyases have high specificity for both particular chromophores and individual cysteine residues (12–14). While much is known about lyases capable of attaching phycocyanobilin (Absmax ∼ 660 nm) (11, 15–21), less is known about the enzymes involved in attaching PEB (22, 23) and PUB (10, 24, 25). Despite the rich chromophore diversity found in marine Synechococcus, only two phycobilin lyases, RpcG and MpeZ, have been characterized so far in this group (10, 24). RpcG attaches PUB to cysteine 83 (C83) of the phycocyanin α-subunit (24), while MpeZ attaches PUB to MpeA-C83 (10). These are dual function enzymes that have both lyase and isomerase activities, the latter allowing them to convert PEB into PUB. MpeU is required for attachment of PUB (25), but its precise function remains to be characterized. The attachment and isomerization of PEB to PUB to phycobiliproteins by lyase-isomerases is critical since these cyanobacteria have no known pathway that directly synthesizes PUB.
Blue-green generalists capable of CA4 comprise the most abundant pigment type, making up ∼40% of the global Synechococcus population of the oceans (26). By differentially producing PEB and PUB in response to changes in ambient light color, blue-green generalist strains can optimize light-harvesting in a variety of light color environments. Interestingly, CA4 has evolved twice, through acquisition of related, but distinct, genomic islands. CA4-A strains contain a genomic island encoding the PUB lyase-isomerase MpeZ, while CA4-B strains possess a genomic island encoding MpeW, an as yet uncharacterized member of the CpeY-MpeY-MpeZ family (7). Both CA4-A and CA4-B genomic islands also encode two master regulators, FciA and FciB. In the model CA4-A strain 9916, these proteins inversely control the expression of mpeZ and two other genes located in the same genomic island, which are more highly expressed in blue light than green light (4, 10). During CA4-A, MpeA-C83, MpeA-C140, and CpeA-C139 bind PUB in blue light and PEB in green light (10). Of these three changes, MpeZ catalyzes the attachment of PUB to MpeA-C83 in blue light. However, the lyase responsible for attaching PEB to MpeA-C83 in green light remained unknown.
Here, we investigate the role of a previously uncharacterized, putative PEB lyase named MpeY in CA4. We demonstrate that in 9916, MpeY attaches PEB to MpeA-C83 in green light. Additionally, we show that the expression ratio of mpeY and mpeZ mRNA determines if PEB or PUB is attached to MpeA-C83 in green and blue light. By analyzing environmental isolates from across the world’s oceans, we provide evidence that interplay between MpeY and MpeZ is globally conserved and that strains encoding only MpeY or MpeZ do not acclimate. Thus, the selective use of MpeY and MpeZ for the attachment of different chromophores at a single site within a phycobiliprotein is an evolutionary innovation that contributes to the process of CA4-A in marine Synechococcus.
Results
MpeY Controls Attachment of PEB in Green Light.
The 9916 genomic region implicated in MpeA and MpeB biosynthesis contains the mpeBA operon and genes encoding two putative phycobilin lyases, MpeY and MpeU (SI Appendix, Fig. S1A) (2, 25). While MpeU is not closely related to MpeZ, MpeY and MpeZ sequences from this strain share 47% amino acid sequence identity overall (SI Appendix, Fig. S1B), suggesting that these proteins are paralogs. Based on the genomic context of the mpeY gene and the relatedness of its encoded protein to MpeZ, we hypothesized that MpeY has a chromophore attachment role for either MpeA or MpeB.
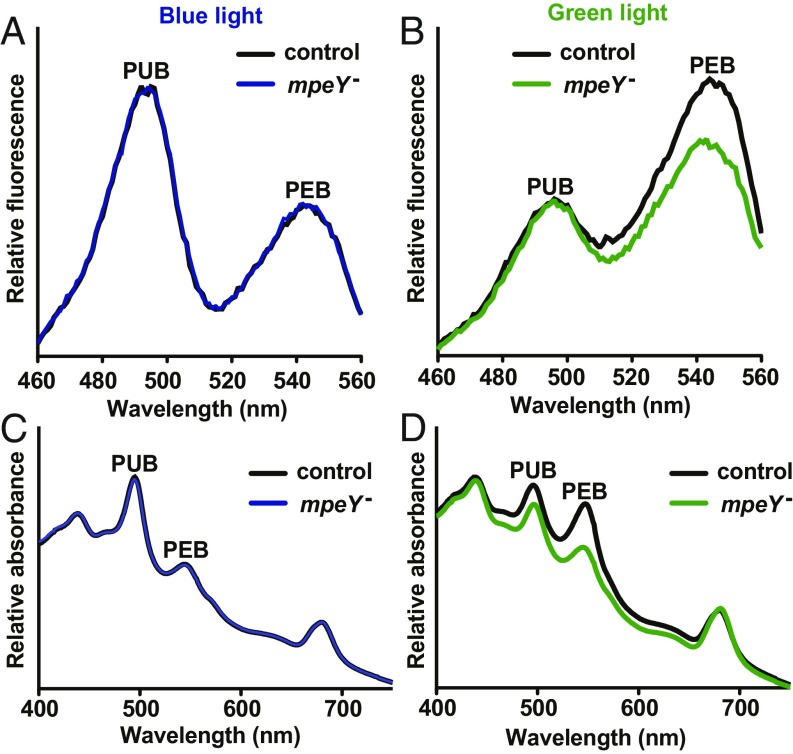
Fluorescence spectroscopy was used to determine the relative excitation at 495 and 550 nm, which provides an estimate of the ratio of PUB to PEB in an mpeY interruption mutant (SI Appendix, Fig. S1C) and control cells (Fig. 1 A and B). In blue light, the PUB:PEB ratio of the mpeY mutant was indistinguishable from control cells, but in green light, PEB fluorescence decreased, relative to PUB fluorescence, in mpeY mutant cells. Therefore, MpeY is likely required for PEB attachment in green light. The possibility of polar effects was tested by cloning the mpeY gene and its promoter into an autonomously replicating plasmid and transforming it into the mpeY mutant. In green light, this plasmid restored the normal PUB:PEB ratio to the mpeY mutant (SI Appendix, Fig. S2A), demonstrating that mpeY is necessary for PEB attachment in green light.
Fig. 1.
MpeY controls PEB attachment in green light. Fluorescence excitation spectra, with emission set at 580 nm and normalized to PUB at 495 nm, of control and mpeY mutant cells grown in (A) blue or (B) green light. Whole-cell absorbance spectra of control and mpeY mutant cells grown in (C) blue or (D) green light. Control cell results shown by black lines and mpeY mutant cell results shown by blue or green lines, which were all normalized to the chlorophyll absorbance peaks at 440 and 680 nm. Fluorescence and absorbance spectra are averages of three independent replicates.
Whole-cell absorbance spectroscopy showed that in blue light, mpeY mutant cells were indistinguishable from control cells, but that in green light, mpeY mutant cells had lower levels of both PUB and PEB absorbance relative to chlorophyll levels (Fig. 1 C and D). The effect on PEB was greater than the effect on PUB. Growth measurements demonstrated that mpeY mutant and control cells grew at the same rate in blue light, but that in green light, mpeY mutant cells grew more slowly (SI Appendix, Fig. S2B). This may be due to less efficient MpeA and MpeB biosynthesis in green light, resulting in a decrease in the number of complete PBS per cell and thus the overall amount of PUB and PEB in mpeY mutant cells. Together, these results demonstrate that MpeY is involved in PEB attachment in green light.
MpeY Attaches PEB to MpeA-C83.
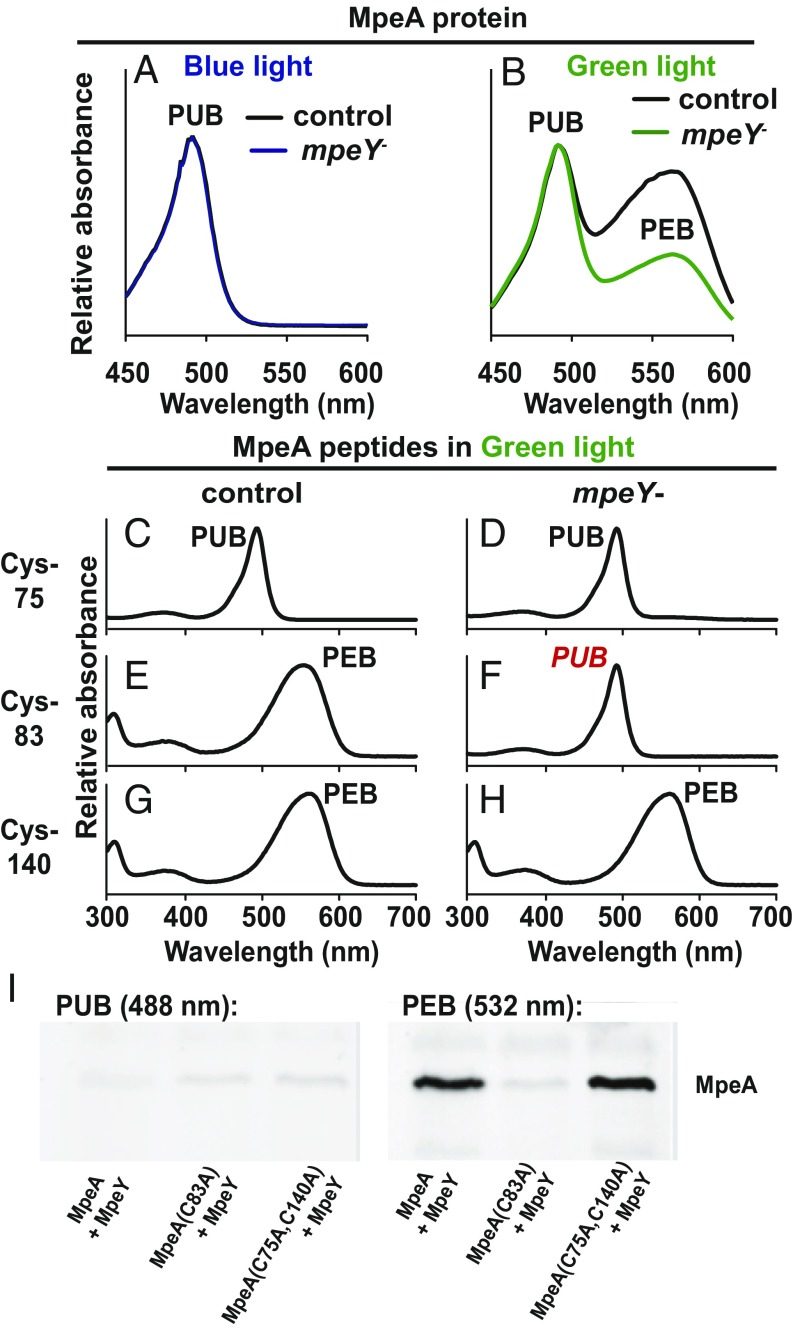
The function of MpeY was further examined by using HPLC to separate the phycoerythrin subunits MpeA, MpeB, CpeA, and CpeB from PBS that had been purified from mpeY mutant cells grown in blue or green light. UV-visible (VIS) absorption spectroscopy of these proteins revealed that MpeA isolated from blue-light–grown mpeY mutant and control cells had identical profiles, while the PUB:PEB absorbance ratio for MpeA was higher in the mpeY mutant than in control cells after growth in green light (Fig. 2 A and B). In addition, the absorption profiles of MpeB, CpeA, and CpeB isolated from mpeY mutant cells were indistinguishable from control cells after growth in either blue or green light (SI Appendix, Fig. S3 A–F). We therefore conclude that among the four PE subunits and two light color conditions, MpeY is specifically required for PEB attachment to MpeA in green light.
Fig. 2.
MpeY attaches PEB to MpeA-C83. Absorbance spectra of purified MpeA protein from control (black lines) and mpeY mutant cells (blue or green lines) grown in (A) blue or (B) green light. MpeA peptides containing C75 (C and D), C83 (E and F), or C140 (G and H) from control (C, E, and G) or mpeY mutant (D, F, and H) cells grown in green light. Red italic, chromophore difference between control and mpeY mutant. Spectra are representative of three independent replicates. (I) Zinc-enhanced fluorescent gel showing PEB or PUB covalently bound to HT-MpeA from E. coli under various coexpression conditions (shown on bottom). Representative of three replicates.
In 9916, chromophores are attached to MpeA at C75, C83, and C140. To determine which of these residues were targeted by MpeY, we used tandem mass spectrometry combined with UV-VIS absorption spectroscopy to examine peptides containing these residues from mpeY mutant and control cells (SI Appendix, Fig. S4). Cells grown in blue light had PUB attached to all three residues from both mpeY mutant and control cells (SI Appendix, Fig. S5). In addition, for cells grown in green light, PUB was attached to C75 (Fig. 2 C and D), and PEB was attached to C140 (Fig. 2 G and H) from both cell types. The only observable difference in chromophorylation was at MpeA-C83 from cells grown in green light, where PEB was attached in control cells (Fig. 2E), and PUB was present in mpeY mutant cells (Fig. 2F). Collectively, these data show that out of the 11 cysteines that are chromophorylated in the four PE subunits (10), MpeY is required specifically for PEB attachment to MpeA-C83, and this attachment can only be detected in green light.
The lyase activity of recombinant MpeY was examined using a heterologous plasmid coexpression system in Escherichia coli. Three different hexahistidine-tagged (HT) versions of MpeA were used as substrates: wild type, HT-MpeA; a mutant with C83 replaced by alanine (A), HT-MpeA (C83A); and a mutant with C75 and C140 each replaced with A, HT-MpeA (C75A, C140A). We also expressed NusA-tagged (Nus) MpeY along with enzymes required for PEB synthesis in E. coli. After coexpression and cell lysis, HT-MpeA and mutant derivatives were purified using Ni-NTA affinity chromatography and analyzed using SDS/PAGE (Fig. 2I). Zinc-enhanced fluorescence was used to show that MpeY is sufficient to covalently attach PEB to C83 of MpeA (Fig. 2I) and thus is a PEB lyase.
mpeY− Suppressor Mutations Increase mpeZ Expression.
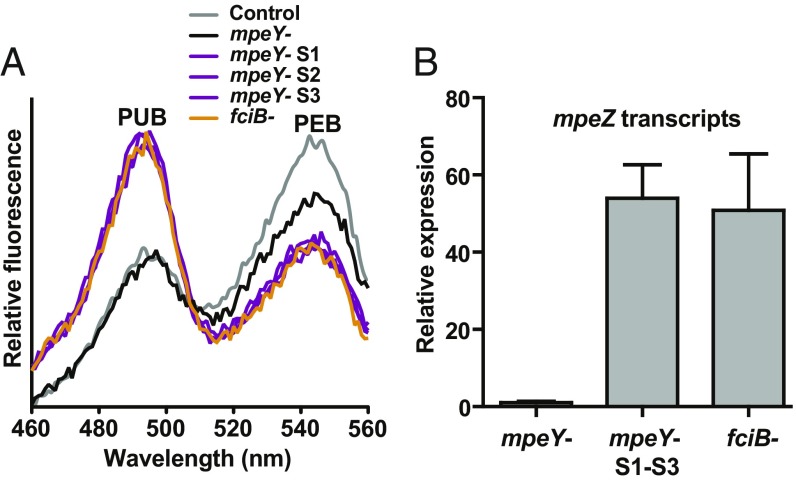
MpeY attaches PEB to MpeA-C83 in green but not blue light, indicating that it plays a role in the chromophore changes at that residue during CA4. The mechanism through which MpeY operates was further characterized by selecting for spontaneous suppressor mutants with higher growth rates than the mpeY mutant. Passaging of three independent mpeY mutant cultures for ∼100 generations selected for apparent suppressor mutations in the CA4 genomic island (SI Appendix, Fig. S6A). These mutations led to increased growth rates relative to the mpeY mutant (SI Appendix, Fig. S6B) yet maintained their initial interruptions of mpeY (SI Appendix, Fig. S6C). Fluorescence spectroscopy analysis of each putative suppressor mutant revealed that all had an increased PUB:PEB ratio during growth in white light, compared with the original mpeY mutant and control cells (Fig. 3A) and were indistinguishable from one another, suggesting they could be caused by similar mutations.
Fig. 3.
mpeY- suppressor mutations increase mpeZ expression. (A) Fluorescence excitation spectra, with emission set at 580 nm, of control (gray line), mpeY mutant (black line), mpeY mutant suppressor mutants S1-S3 (purple lines), and fciB mutant (orange lines) cells grown in white light, in which control cells have the same pigmentation phenotype as when grown in green light. Fluorescence spectra are an average of three independent replicates. (B) Mean mpeZ transcript levels from mpeY mutant, mpeY− suppressor mutants S1-S3, and fciB mutant cells grown in white light. Values from mpeY mutant cells were set to 1 after rRNA normalization. Transcript levels for the mpeY and fciB mutants are averages of three independent replicates with error bars showing the SEM. Transcript levels for the mpeY mutant suppressor mutants S1-S3 are the combined average of one independent replicate each of S1, S2, and S3.
A similarly high PUB:PEB ratio had been previously observed for an interruption mutant of fciB in all light conditions examined (4), and the suppressor mutants grew at a rate that was very similar to the fciB mutant (SI Appendix, Fig. S6B). In addition, all of the suppressor mutants had a higher level of PUB absorbance than the original mpeY mutant and were phenotypically indistinguishable from the fciB mutant (SI Appendix, Fig. S6D). These data collectively suggested that the suppressor mutations interrupted the normal expression of fciB. Sequencing revealed a unique fciB mutation site in each of the three suppressor mutants examined, while no fciB mutation was present in control cells that were also passaged for 100 generations (SI Appendix, Fig. S6A). Two of the suppressor mutants contained frameshift mutations in the genomic region encoding the N-terminal portion of FciB. One was due to a 5′ TC 3′ insertion (Suppressor 1), and the other was due to a 5′ TC 3′ deletion (Suppressor 3) in a region containing six 5′ TC 3′ repeats. Suppressor 2 contained a glycine to arginine missense mutation at residue 251 in the C-terminal portion of FciB, which is predicted to encode a helix-turn-helix DNA-binding motif (7). Fluorescence excitation spectroscopy showed that the original mpeY mutant phenotype was restored after transformation of each suppressor strain with fciA and fciB, which previously complemented an fciB interruption mutant (4) (SI Appendix, Fig. S7A), demonstrating that these fciB mutations caused the suppressor phenotypes.
Based on previous results showing that mpeZ transcript levels were constitutively high in both blue and green light in an fciB mutant (4), we tested the mpeY− suppressor mutants for elevated mpeZ transcript levels during growth in white light that produces the same phenotype as green light. RNA-Seq data demonstrated that absolute mpeZ RNA abundance was much higher than mpeY RNA abundance in blue light, but much lower in green light, and that neither was ever completely absent (SI Appendix, Fig. S7B). RNA blot analyses showed that mpeY mutant cells had low levels of mpeZ transcripts, while both mpeY− suppressor and fciB mutant cells had ∼50-fold higher mpeZ transcript levels (Fig. 3B). Therefore, the mpeY− suppressor mutants appear to modify the mpeY mutant growth phenotype in white light by inactivating fciB, leading to the elevation of the expression of mpeZ. Because MpeZ is known to attach PUB to MpeA-C83 (10), the phenotypes of these suppressor mutants strongly suggest that the relative expression of mpeY and mpeZ controls the attachment of PEB versus PUB.
Interplay Between MpeZ and MpeY Controls Chromophorylation of the Same Cysteine During CA4.
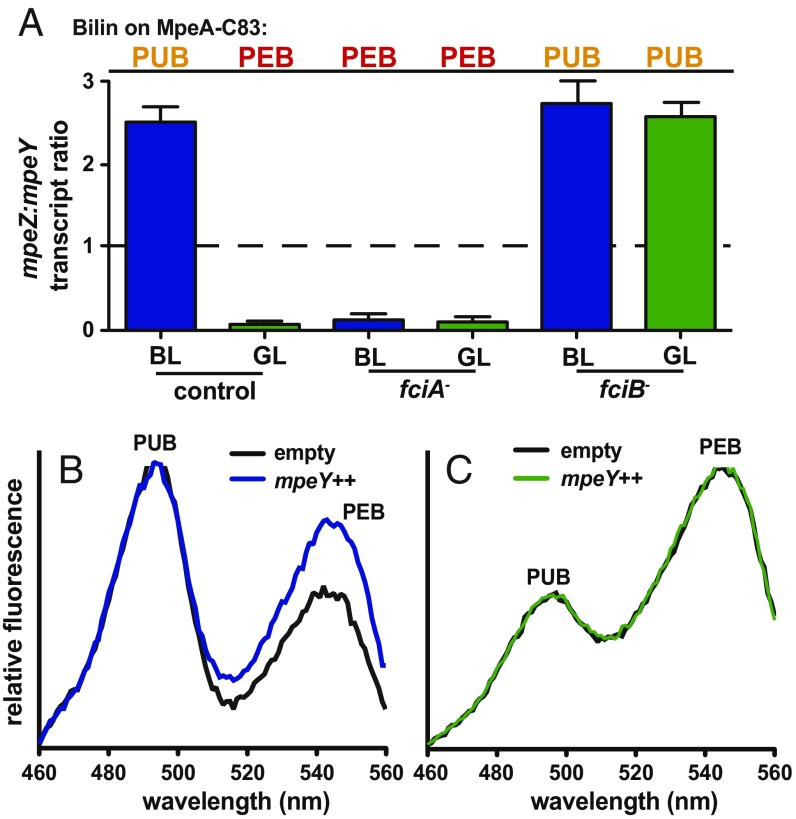
Our discovery that mpeY suppressor mutants increased mpeZ expression led us to hypothesize that the MpeZ:MpeY ratio, driven by the relative expression levels of mpeZ and mpeY, controls whether PUB or PEB is attached at MpeA-C83 during CA4. We tested this by examining existing RNA-Seq data from fciA and fciB interruption mutants. As previously shown (4), the former is locked in CA4 green-light phenotype and always has PEB at MpeA-C83, while the latter is locked in the CA4 blue-light phenotype and always has PUB at MpeA-C83 (Fig. 4A). In addition, the mpeZ:mpeY expression ratio differed from the control in the fciA and fciB mutants, being high in the fciB mutant and low in the fciA mutant in both blue and green light (Fig. 4A). Thus, the mpeZ:mpeY mRNA expression ratio is correlated with the type of chromophore attached to MpeA-C83, suggesting that this ratio has a major role in determining whether PUB or PEB is attached to MpeA-C83 during CA4.
Fig. 4.
The ratio of transcripts encoding MpeY and MpeZ determines the chromophorylation state of MpeA-C83. (A) Summary of bilins attached to MpeA-C83 in different light colors and genetic backgrounds (4). Transcript-level ratios from RNA-sequencing of mpeZ and mpeY in control, fciA mutant, and fciB mutant cells grown in blue (blue bars) or green (green bars) light. Error bars show the SEM of three independent replicates for each strain and light condition. Fluorescence excitation spectra, with emission set at 580 nm and normalized to PUB at 495 nm, of wild-type cells with either empty vector (black lines) or a vector overexpressing mpeY (indicated by mpeY++ with blue or green lines) grown in (B) blue or (C) green light. Fluorescence spectra are averages of three independent replicates. BL, blue light; GL, green light.
This was directly tested by introducing an autonomously replicating plasmid expressing mpeY from its native promoter into wild-type cells. The mpeY RNA levels in this transformed line were eightfold higher in green light and 14-fold higher in blue light than in wild-type cells transformed with the empty vector (SI Appendix, Fig. S7 C and D). Fluorescence emission spectra obtained from green-light–grown cells showed that PUB and PEB fluorescence spectra for the mpeY overexpression strain were indistinguishable from the empty vector control strain. However, for cells grown in blue light the mpeY overexpression strain exhibited a higher PEB to PUB fluorescence ratio than the empty vector control strain (Fig. 4 B and C). The simplest and most reasonable explanation for this result is that overproduced MpeY was outperforming MpeZ in blue light, leading to the addition of PEB instead of PUB at MpeA-C83. Therefore, we conclude that interplay between MpeZ and MpeY controls which of these two chromophores are attached at this residue during CA4.
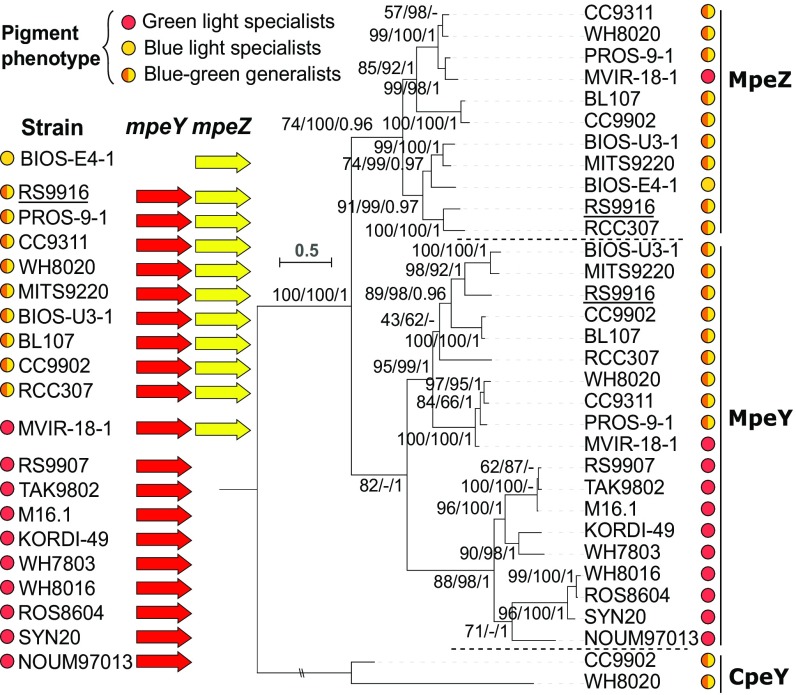
Interplay Between MpeY and MpeZ Is Widespread, Suggesting a Paradigm for Pigment Diversification in the Marine Environment.
Our data show that in 9916, a strain isolated from the Gulf of Aqaba in the Red Sea, MpeY and MpeZ both act on the same residue as part of CA4. To explore if interplay between MpeY and MpeZ is widespread, we examined 20 strains isolated from seven regions around the globe that encode MpeY and/or MpeZ (SI Appendix, Table S1), analyzing the phylogenetic relatedness of these enzymes (Fig. 5, Right). Nine of these isolates encoded only MpeY, one isolate encoded only MpeZ, and 10 isolates encoded both MpeY and MpeZ (Fig. 5, Left). While the single isolate encoding only MpeZ, BIOS-E4-1, was a blue-light specialist (PUB-rich), all nine isolates that only encode MpeY were green-light specialists (PEB-rich). Nine of the ten strains, isolated from six different oceanic regions, encoded both MpeY and MpeZ and were blue-green generalists capable of CA4. The single exception, MVIR-18-1, was previously found to lack significant blue-green regulation of its mpeZ gene (7). From an evolutionary point of view, sequence homologies between MpeY and MpeZ and with CpeY, characterized as a PEB lyase that acts on CpeA-C82 in Fremyella diplosiphon (23), clearly indicate that these three proteins share a common ancestor. The fact that CpeY is found in all phycoerythrin-containing cyanobacteria, while both MpeY and MpeZ are present only in marine Synechococcus, indicates that this common ancestor was more closely related to CpeY (2, 7). Phylogenetic analysis showed that sequences of MpeZ, MpeY from green-light specialists, and MpeY from chromatic acclimaters formed three independent clusters (Fig. 5, Right). However, the order of appearance of these three groups remains unclear because, although the tree topology shown in Fig. 5 was found to be the most probable, the high P values (>0.05) obtained using an approximately unbiased (AU) test (27, 28) did not allow us to reject the two alternative tree topologies (SI Appendix, Table S1). In any case, the most probable evolutionary scenario is that after an initial duplication event leading to the appearance of the MpeYZ precursor from a CpeY-related ancestor, this precursor likely gained site specificity for MpeA-C83 while conserving its lyase function. The MpeZ precursor most likely diverged afterward to further gain an isomerase function. Our findings support the proposal that two paralogous enzymes, the lyase MpeY and the lyase-isomerase MpeZ, can act on the same residue either independently (for example, in green-light specialists) or in a concerted fashion (in CA4-capable strains), thus contributing to pigment diversity in marine Synechococcus.
Fig. 5.
Interplay between MpeY and MpeZ is conserved throughout Synechococcus isolates capable of CA4. (Left) Phycobilisome pigment phenotype of 20 Synechococcus isolates and corresponding presence of mpeY (red) and/or mpeZ (yellow) in their genomes. (Right) Maximum Likelihood (ML) analysis of MpeY and MpeZ using the distantly related phycobilin lyase CpeY as a root (23). Colored circles indicate the pigment phenotype (See key in upper left corner). Series of three numbers at nodes correspond to bootstrap values for ML and neighbor-joining analyses and Bayesian posterior probabilities (PP, ranging between 0 and 1). Only values higher than 60% for bootstrap values and 0.60 for PP are shown on the phylogenetic tree. Underlining denotes Synechococcus sp. RS9916, the strain used throughout this study.
Discussion
The successful expansion of marine Synechococcus throughout the photic zone of the world’s oceans likely relied in part on the diversification of its photosynthetic light-harvesting abilities. Blue-light specialist, green-light specialist, and blue-green generalist strains have evolved to incorporate an optimal combination of chromophores in their PBS to efficiently utilize solar energy in all light color niches encountered in the underwater environment. Specific energy absorption and transfer characteristics of each type of PBS are determined by which chromophore is added at each cysteine attachment site throughout the light-harvesting structure. These additions are catalyzed by phycobilin lyases and lyase-isomerases, making this a pivotal group of enzymes for successful light color adaptation of Synechococcus in the marine environment. Marine Synechococcus strains have up to 14 genes per genome that are predicted to encode different phycobilin lyases that attach chromophores to distinct cysteine residues within the apoproteins of the PBS, although it is yet to be determined how specificity is achieved between each lyase and its cognate residue (10, 13, 14).
The discovery that two enzymes, MpeY and MpeZ, independently chromophorylate one PBS residue, MpeA-C83, in 9916 provides insights into the mechanism of CA4. First, the fact that mpeZ expression is up-regulated in blue light (4, 10) and is in the CA4 genomic island (4, 7, 10) suggests that the ancestor of all CA4-A strains was a green-light specialist with sufficient mpeY expression to always have PEB at MpeA-C83. Subsequent acquisition of the CA4 genomic island likely led to high-level expression of mpeZ in blue light, since two CA4 master regulators encoded in the island, FciA and FciB, impart this light color control. Thus, the high MpeZ:MpeY ratio in blue light results in PUB attachment at MpeA-C83, while the low MpeZ:MpeY ratio in green light leads to PEB attachment. This mechanism is supported by the MpeY overexpression results shown in Fig. 4B. It must also be true in wild-type cells since MpeA-C83 binds PUB in the green-light–grown mpeY mutant (Fig. 2F), demonstrating that MpeZ is present under these conditions, but that its activity must be overcome by the activity of MpeY in wild-type cells. Furthermore, PEB is attached to MpeA-C83 in blue-light–grown mpeZ mutant cells (10), showing that in blue-light–grown wild-type cells, MpeY is present, but that its activity must be overcome by the activity of the more abundant MpeZ. Our current model of CA4 is further strengthened by the observation that MpeZ and MpeY are independently sufficient to attach PUB (10) and PEB (Fig. 2) to MpeA-C83 in a heterologous expression system.
The relationship between mpeY and/or mpeZ presence and their pigment type in many marine Synechococcus strains (Fig. 5) provides even further support for our model that the interplay between MpeY and MpeZ contributes to CA4. The almost perfect correlation between mpeY and/or mpeZ presence and pigment type suggests that our discovery has implications for other strains of marine Synechococcus. Interestingly, the only exception is also consistent with our current understanding of CA4. Isolate MVIR-18-1, which has both mpeY and mpeZ but is not a blue-green specialist, shows little to no blue-green–light regulation of mpeZ gene expression and thus is unable to carry out normal CA4 (7). Also, BIOS-E4-1, a blue-light specialist and the only strain that has mpeZ but not mpeY, lacks a light color-regulated mpeZ gene (7). It was recently proposed that BIOS-E4-1 previously had both mpeZ and mpeY, then lost mpeY and the light color regulation of mpeZ, leading to a blue-light specialist phenotype (26). Interestingly, the BIOS-E4-1 genotype is similar to the mutants that arose in our mpeY− suppressor screen, where inactivating mpeY led to more rapidly growing mutants (SI Appendix, Fig. S6) that lost CA4 by expressing mpeZ at blue-light levels even in green-light conditions (Fig. 3B). BIOS-E4-1 also lacks fciA and fciB, which is consistent with its inability to undergo CA4. The loss of these CA4 regulators might have been the next evolutionary step after the inactivation of mpeY and the generation of an fciB− suppressor mutant. Thus, our inactivation of mpeY may have initiated a series of genetic changes that parallel an evolutionary process that is occurring in the oceans. In the natural environment, BIOS-E4-1–like genotypes appear to predominate in some permanently iron-poor areas such as the tropical South Pacific Ocean (26).
The second mechanistic insight provided by our data regarding CA4 is that, since this process involves PUB/PEB changes at MpeA-C83, MpeA-C140, and CpeA-C139 in 9916 (4, 10), there must be one or more additional processes responsible for these changes at MpeA-C140 and CpeA-C139. If all CA4 control and mechanistic processes involve components encoded within the CA4 genomic island, the best candidate genes are the two additional genes on the island that are highly up-regulated in blue light, fciC and unk10. These both encode small proteins with limited or no similarity to known proteins, making it challenging to discover their function.
A final challenge to understanding CA4 in 9916 will be to determine how this organism is sensing light color to control CA4. The CA4 master regulators FciA and FciB are both putative AraC transcription factor family members, leaving this question open. While our hypothesis is that FciA and FciB control CA4 through transcriptional regulation, this hypothesis has yet to be formally tested. Additional genetic tools such as reporter gene systems will need to be developed for marine Synechococcus in order for this question to be answered.
Overall, our results show that the interplay between MpeY and MpeZ is important for CA4 in 9916. We hypothesize that other forms of such interplay between other paralogous lyase/lyase-isomerase pairs are likely to occur within the marine Synechococcus. In addition to furthering our basic understanding of a globally important marine picophytoplankton, the previously unidentified ability of two enzymes to vie for attachment of spectrally different chromophores to the same cysteine residue will provide tools for further development of multicolor fluorescence-labeling technology.
Materials and Methods
Strains and Growth Conditions.
Growth conditions were as previously described (10). Details are provided in SI Appendix.
In Vivo Heterologous Expression and Purification of Recombinant Proteins.
Plasmid constructs are provided in SI Appendix. Recombinant proteins were expressed and purified from E. coli BL21 (DE3) as previously described (19, 23). Details are provided in SI Appendix. Cell lysis, centrifugation, and protein purification were conducted as previously described (23).
Protein Analysis by Spectroscopy and Gel-Electrophoresis.
Polypeptide samples were resolved by polyacrylamide gel electrophoresis (PAGE, 15% wt/vol) in the presence of SDS and visualized as previously described (23).
mpeY Disruption/Complementation/Overexpression.
Transformation of 9916 by conjugation was performed as previously described (4, 10, 29). Details are provided in SI Appendix.
HPLC and MS-MS Analysis of Phycobiliproteins.
Phycobilisomes were purified as described (10, 30). HPLC was used to separate each phycobiliprotein subunit, and samples were digested with trypsin as described previously (10, 23). HPLC-separated and trypsin-digested phycobiliprotein subunits were analyzed by UV-VIS absorption spectroscopy and MS-MS as described (10).
Suppressor Analyses.
To select for mpeY mutant suppressors, three independent mpeY mutant cell lines were grown in white light for ∼100 generations. Details are provided in SI Appendix.
RNA Analyses.
RNA-sequencing data were further analyzed from previous experiments (4), except that instead of values for the blue-light control being set to 100 and values normalized across each replicate separately, the raw values were averaged. RNA blots were performed as previously described (4, 31). Values were normalized as previously described (4, 31). Details are provided in SI Appendix.
Comparative Genomics and Phylogenetic Analyses.
Details on retrieval of mpeY and mpeZ gene content are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the D.M.K. laboratory, especially Lisa Wiltbank, as well as Andrian Gutu, Jeff Palmer, and Jake McKinlay for helpful discussions and comments and thank Hidetoshi Shimodaira and Theophile Grébert for their kind hints in running the AU test. This research was supported by National Institutes of Health Training Grant T32-GM007757 (to J.E.S.), National Science Foundation Grants MCB-1029414 and MCB-1818187 (to D.M.K.) and MCB-1244339 (to W.M.S.), by the Office of the Vice Provost for Research at Indiana University, Bloomington, through its Bridge Funding Program (D.M.K.) and by the French “Agence Nationale de la Recherche” Program CINNAMON (ANR-17-CE2-0014-01) (for F.P. and L.G.). Scientific exchanges were made possible by PICS (Projet International de Coopération Scientifique) ChromaCya.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810491116/-/DCSupplemental.
References
- 1.Flombaum P, et al. Present and future global distributions of the marine cyanobacteria Prochlorococcus and Synechococcus. Proc Natl Acad Sci USA. 2013;110:9824–9829. doi: 10.1073/pnas.1307701110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Six C, et al. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: A comparative genomics study. Genome Biol. 2007;8:R259. doi: 10.1186/gb-2007-8-12-r259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ong LJ, Glazer AN, Waterbury JB. An unusual phycoerythrin from a marine cyanobacterium. Science. 1984;224:80–83. doi: 10.1126/science.224.4644.80. [DOI] [PubMed] [Google Scholar]
- 4.Sanfilippo JE, et al. Self-regulating genomic island encoding tandem regulators confers chromatic acclimation to marine Synechococcus. Proc Natl Acad Sci USA. 2016;113:6077–6082. doi: 10.1073/pnas.1600625113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glazer AN. Light guides. Directional energy transfer in a photosynthetic antenna. J Biol Chem. 1989;264:1–4. [PubMed] [Google Scholar]
- 6.Ong LJ, Glazer AN. Phycoerythrins of marine unicellular cyanobacteria. I. Bilin types and locations and energy transfer pathways in Synechococcus spp. phycoerythrins. J Biol Chem. 1991;266:9515–9527. [PubMed] [Google Scholar]
- 7.Humily F, et al. A gene island with two possible configurations is involved in chromatic acclimation in marine Synechococcus. PLoS One. 2013;8:e84459. doi: 10.1371/journal.pone.0084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palenik B. Chromatic adaptation in marine Synechococcus strains. Appl Environ Microbiol. 2001;67:991–994. doi: 10.1128/AEM.67.2.991-994.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everroad C, et al. Biochemical bases of type IV chromatic adaptation in marine Synechococcus spp. J Bacteriol. 2006;188:3345–3356. doi: 10.1128/JB.188.9.3345-3356.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla A, et al. Phycoerythrin-specific bilin lyase-isomerase controls blue-green chromatic acclimation in marine Synechococcus. Proc Natl Acad Sci USA. 2012;109:20136–20141. doi: 10.1073/pnas.1211777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairchild CD, et al. Phycocyanin alpha-subunit phycocyanobilin lyase. Proc Natl Acad Sci USA. 1992;89:7017–7021. doi: 10.1073/pnas.89.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheer H, Zhao KH. Biliprotein maturation: The chromophore attachment. Mol Microbiol. 2008;68:263–276. doi: 10.1111/j.1365-2958.2008.06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schluchter WM, et al. Phycobiliprotein biosynthesis in cyanobacteria: Structure and function of enzymes involved in post-translational modification. In: Hallenbeck PC, editor. Advances in Experimental Medicine and Biology. Springer; New York: 2010. pp. 211–228. [DOI] [PubMed] [Google Scholar]
- 14.Bretaudeau A, et al. CyanoLyase: A database of phycobilin lyase sequences, motifs and functions. Nucleic Acids Res. 2013;41:D396–D401. doi: 10.1093/nar/gks1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen G, Schluchter WM, Bryant DA. Biogenesis of phycobiliproteins: I. cpcS-I and cpcU mutants of the cyanobacterium Synechococcus sp. PCC 7002 define a heterodimeric phyococyanobilin lyase specific for beta-phycocyanin and allophycocyanin subunits. J Biol Chem. 2008;283:7503–7512. doi: 10.1074/jbc.M708164200. [DOI] [PubMed] [Google Scholar]
- 16.Saunée NA, Williams SR, Bryant DA, Schluchter WM. Biogenesis of phycobiliproteins: II. CpcS-I and CpcU comprise the heterodimeric bilin lyase that attaches phycocyanobilin to CYS-82 of beta-phycocyanin and CYS-81 of allophycocyanin subunits in Synechococcus sp. PCC 7002. J Biol Chem. 2008;283:7513–7522. doi: 10.1074/jbc.M708165200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao KH, et al. Lyase activities of CpcS- and CpcT-like proteins from Nostoc PCC7120 and sequential reconstitution of binding sites of phycoerythrocyanin and phycocyanin beta-subunits. J Biol Chem. 2007;282:34093–34103. doi: 10.1074/jbc.M703038200. [DOI] [PubMed] [Google Scholar]
- 18.Zhao KH, et al. Phycobilin:cystein-84 biliprotein lyase, a near-universal lyase for cysteine-84-binding sites in cyanobacterial phycobiliproteins. Proc Natl Acad Sci USA. 2007;104:14300–14305. doi: 10.1073/pnas.0706209104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen G, et al. Identification and characterization of a new class of bilin lyase: The cpcT gene encodes a bilin lyase responsible for attachment of phycocyanobilin to Cys-153 on the beta-subunit of phycocyanin in Synechococcus sp. PCC 7002. J Biol Chem. 2006;281:17768–17778. doi: 10.1074/jbc.M602563200. [DOI] [PubMed] [Google Scholar]
- 20.Fairchild CD, Glazer AN. Oligomeric structure, enzyme kinetics, and substrate specificity of the phycocyanin α subunit phycocyanobilin lyase. J Biol Chem. 1994;269:8686–8694. [PubMed] [Google Scholar]
- 21.Zhou J, Gasparich GE, Stirewalt VL, de Lorimier R, Bryant DA. The cpcE and cpcF genes of Synechococcus sp. PCC 7002. Construction and phenotypic characterization of interposon mutants. J Biol Chem. 1992;267:16138–16145. [PubMed] [Google Scholar]
- 22.Wiethaus J, et al. CpeS is a lyase specific for attachment of 3Z-PEB to Cys82 of beta-phycoerythrin from Prochlorococcus marinus MED4. J Biol Chem. 2010;285:37561–37569. doi: 10.1074/jbc.M110.172619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas A, et al. Characterization of the activities of the CpeY, CpeZ, and CpeS bilin lyases in phycoerythrin biosynthesis in Fremyella diplosiphon strain UTEX 481. J Biol Chem. 2011;286:35509–35521. doi: 10.1074/jbc.M111.284281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blot N, et al. Phycourobilin in trichromatic phycocyanin from oceanic cyanobacteria is formed post-translationally by a phycoerythrobilin lyase-isomerase. J Biol Chem. 2009;284:9290–9298. doi: 10.1074/jbc.M809784200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahmoud RM, et al. Adaptation to blue light in marine Synechococcus requires MpeU, an enzyme with similarity to phycoerythrobilin lyase isomerases. Front Microbiol. 2017;8:243. doi: 10.3389/fmicb.2017.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grébert T, et al. Light color acclimation is a key process in the global ocean distribution of Synechococcus cyanobacteria. Proc Natl Acad Sci USA. 2018;115:E2010–E2019. doi: 10.1073/pnas.1717069115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 28.Everroad RC, Wood AM. Phycoerythrin evolution and diversification of spectral phenotype in marine Synechococcus and related picocyanobacteria. Mol Phylogenet Evol. 2012;64:381–392. doi: 10.1016/j.ympev.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 29.Brahamsha B. A genetic manipulation system for oceanic cyanobacteria of the genus Synechococcus. Appl Environ Microbiol. 1996;62:1747–1751. doi: 10.1128/aem.62.5.1747-1751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collier JL, Grossman AR. Chlorosis induced by nutrient deprivation in Synechococcus sp. strain PCC 7942: Not all bleaching is the same. J Bacteriol. 1992;174:4718–4726. doi: 10.1128/jb.174.14.4718-4726.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seib LO, Kehoe DM. A turquoise mutant genetically separates expression of genes encoding phycoerythrin and its associated linker peptides. J Bacteriol. 2002;184:962–970. doi: 10.1128/jb.184.4.962-970.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.