Significance
The plant hormone auxin coordinates almost all aspects of plant development. Throughout plant life, the expression of hundreds of genes involved in auxin regulation is orchestrated via several combinatorial and cell-specific auxin perception systems. An effective approach to dissect these complex pathways is the use of synthetic molecules that target specific processes of auxin activity. Here, we describe synthetic auxins, RubNeddins (RNs), which act as selective auxin agonists. The RN with the greatest potential for dissecting auxin perception was RN4, which we used to reveal a role for the chromatin remodeling ATPase BRAHMA in apical hook development. Therefore, the understanding of RN mode of action paves the way to dissecting specific molecular components involved in auxin-regulated developmental processes.
Keywords: auxin, chemical biology, selective agonist, prohormone, hormone perception
Abstract
Auxin phytohormones control most aspects of plant development through a complex and interconnected signaling network. In the presence of auxin, AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) transcriptional repressors are targeted for degradation by the SKP1-CULLIN1-F-BOX (SCF) ubiquitin-protein ligases containing TRANSPORT INHIBITOR RESISTANT 1/AUXIN SIGNALING F-BOX (TIR1/AFB). CULLIN1-neddylation is required for SCFTIR1/AFB functionality, as exemplified by mutants deficient in the NEDD8-activating enzyme subunit AUXIN-RESISTANT 1 (AXR1). Here, we report a chemical biology screen that identifies small molecules requiring AXR1 to modulate plant development. We selected four molecules of interest, RubNeddin 1 to 4 (RN1 to -4), among which RN3 and RN4 trigger selective auxin responses at transcriptional, biochemical, and morphological levels. This selective activity is explained by their ability to consistently promote the interaction between TIR1 and a specific subset of AUX/IAA proteins, stimulating the degradation of particular AUX/IAA combinations. Finally, we performed a genetic screen using RN4, the RN with the greatest potential for dissecting auxin perception, which revealed that the chromatin remodeling ATPase BRAHMA is implicated in auxin-mediated apical hook development. These results demonstrate the power of selective auxin agonists to dissect auxin perception for plant developmental functions, as well as offering opportunities to discover new molecular players involved in auxin responses.
The survival and reproductive success of all living organisms depend on their ability to perceive and integrate environmental and internal signals. As sessile organisms, plants have developed strategies to adapt to their surroundings, including an extensive developmental plasticity (1). Plant morphological changes are executed through regulation of hormone levels and signaling (2). The phytohormone auxin is involved in almost all aspects of plant development and adaptation. Auxin perception within the nucleus is mediated by the TRANSPORT INHIBITOR RESISTANT 1/AUXIN SIGNALING F-BOX (TIR1/AFB)–AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) (TIR1/AFB-AUX/IAA) coreceptor complex (3). The TIR1/AFB1-5 F-box proteins are subunits of the S-PHASE KINASE ASSOCIATED PROTEIN 1-CULLIN 1-F-BOX (SCF)-type E3 ligase and act as auxin receptors (4). Formation of the SCFTIR1/AFB–AUX/IAA-auxin complex leads to the ubiquitination of the AUX/IAA transcriptional repressors, targeting them for rapid degradation by the 26S proteasome (4). Removal of AUX/IAAs liberates the auxin response-activating AUXIN RESPONSE FACTOR (ARF) transcription factors from repression (4) and leads to the occurrence of an auxin-transcriptional response. There is significant variation in auxin-induced degradation rates among different AUX/IAA proteins, and at least some of this variation is attributable to the specificity in the interactions between the 29 AUX/IAAs and 6 TIR1/AFB F-box proteins in Arabidopsis (4–6). Amino acids within and outside the degron domain II (DII) of the AUX/IAA proteins determine the interaction strength of the coreceptor and specify AUX/IAA stability (5–7). The multiplicity of the potential coreceptor assembly is the first element mediating the complexity of the auxin response.
The ubiquitin-proteasome pathway plays an essential role in plant hormone signaling (8–10). Modification of the relevant components by the ubiquitin-like protein, RELATED TO UBIQUITIN/NEURAL PRECURSOR CELL EXPRESSED DEVELOPMENTALLY DOWN-REGULATED PROTEIN 8 (RUB/NEDD8), which is catalyzed by a cascade of enzymatic reactions analogous to ubiquitination, is critical for the full activity of the proteasome complex (11). In plants, the CULLINs (CUL1, CUL3, and CUL4) are NEDD8-modified proteins that form multimeric E3 ubiquitin ligase complexes (12). CUL1 acts as a scaffold within the SCF-type E3 ligases and neddylation states of CUL1 are essential for the ubiquitin ligase activity of the SCF complex (13). Loss of components of the neddylation pathway, such as the NEDD8-activating enzyme subunit AUXIN RESISTANT 1 (AXR1), reduces the response to several phytohormones, including auxin (14–17).
To understand how auxin perception mediates multiple aspects of plant development, we established an AXR1-dependent developmental defect-based chemical biology screen. Using this approach, we identified small synthetic molecules, RubNeddins (RNs), which selectively promote SCFTIR1/AFB–AUX/IAA coreceptor assembly, allowing local and precise modulation of auxin signaling pathways. Furthermore, these synthetic selective agonists possess the ability to identify and distinguish the molecular players involved in different aspects of auxin-regulated development, thereby dissecting the diversity of auxin action. We demonstrated this by employing these agonists to reveal different roles for specific AUX/IAA proteins during lateral root and apical hook development. In particular, the use of the selective auxin agonist RN4 revealed a role for the chromatin remodeling ATPase BRAHMA in apical hook development.
Results
The Rubylation/Neddylation Pathway Is Required for RNs to Alter Seedling Development.
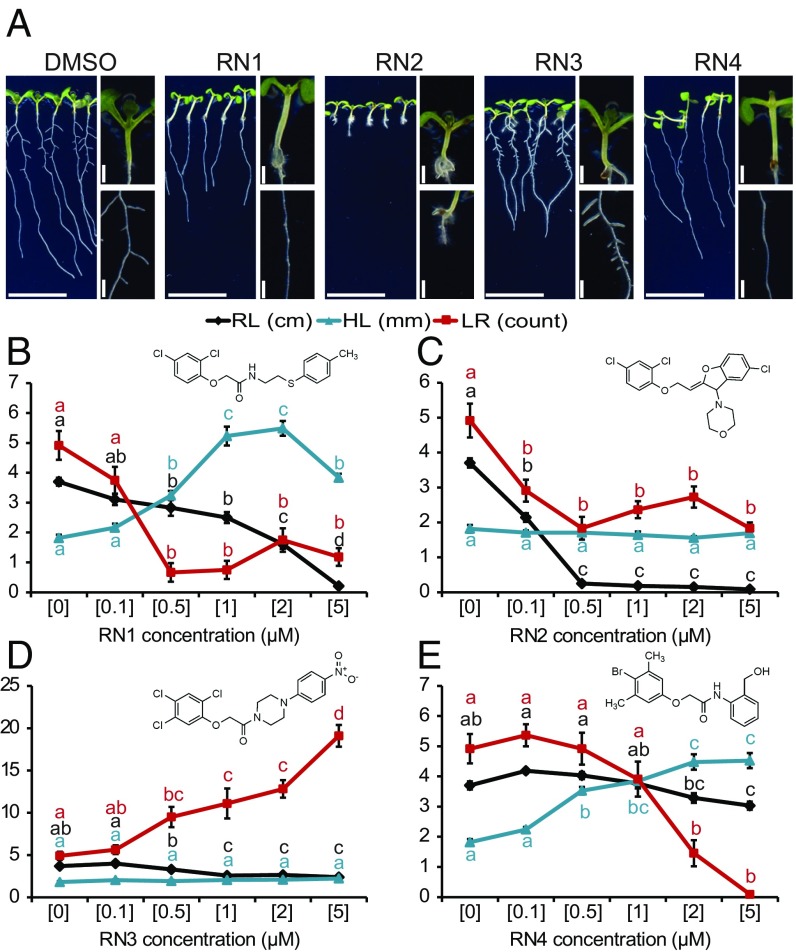
To address the complexity of auxin response, we established a chemical biology screen to isolate synthetic molecules targeting the NEDD8-mediated signaling pathway in Arabidopsis (SI Appendix, Fig. S1 A and B). We reasoned that some of these molecules might also target the auxin signaling pathway (SI Appendix, Fig. S1A) and we used 1-naphthaleneacetic acid (NAA) as control (SI Appendix, Fig. S1C). This strategy is complementary to previous ones aiming at isolating auxin-related small molecules (18, 19). Compounds affecting auxin-related developmental processes, such as primary root growth, hypocotyl elongation, and gravi- or photo-tropism responses in wild-type but not in axr1-30 seedlings, were selected (SI Appendix, Fig. S1B). This screening strategy, based on differential effects upon the two genetic backgrounds (Col-0 wild-type vs. axr1-30), was essential to filter out chemical activities with general impacts on seedling growth. We hypothesized that a small molecule for which activity was dependent on the AXR1 signaling machinery could be recognized by one or several TIR1/AFB–AUX/IAA coreceptor complexes. Of 8,000 diverse compounds (ChemBridge), we identified 34 small molecules (4.25‰) that selectively affected the growth of wild-type compared with axr1-30 seedlings. Four molecules, named RN1–4, were ultimately selected as they showed a dose-dependent activity and a high potency on wild-type seedling development in the micromolar range (SI Appendix, Fig. S1D). In detail, RN1 activity decreased lateral root number and primary root length, but increased hypocotyl length and adventitious root formation (Fig. 1 A and B and SI Appendix, Fig. S2A). RN2 application resulted in the inhibition of primary root growth and lateral root formation, without affecting hypocotyl length (Fig. 1 A and C). RN3 promoted the number of lateral roots (Fig. 1 A and D). RN4 activity increased hypocotyl elongation and inhibited lateral root formation (Fig. 1 A and E). Overall, these structurally similar compounds triggered specific morphological changes in wild-type, while axr1-30 was resistant to these effects, demonstrating that they require a functional RUB/NEDD8 signaling pathway.
Fig. 1.
Four RN chemicals trigger different morphological changes. (A) Col-0 seedlings were grown on RN-supplemented media for 8 d. DMSO was used as control. Images display the effects of the RN at a representative concentration: RN1: 2 μM; RN2: 0.5 μM; RN3: 2 μM; RN4: 5 μM. (Scale bars, 1 cm.) (B–E) RN1 (B), RN2 (C), RN3 (D), and RN4 (E) selectively affected primary root length (RL), hypocotyl length (HL), and the number of lateral roots (LR). For each graph, the RN structure is reported. Statistics were performed using ANOVA and Tukey’s test. Means ± SEM are shown, n = 10 seedlings for each concentration of the dose–response; different letters indicate significant differences at P < 0.05. Concentrations in micromolars are indicated in brackets (B–E).
The RNs Act as Developmental Regulators in Several Land Plants.
We then analyzed RN effects on Populus (poplar) and Physcomitrella patens (moss). RN1, which induced hypocotyl elongation and promoted adventitious root formation in Arabidopsis, and RN3, which increased lateral root number in Arabidopsis, were applied to three different lines of poplar explants (SI Appendix, Fig. S2 B–D). The poplar lines were selected for their different rooting abilities; T89 is an easy rooting hybrid while SwAsp19 and -35 have a low rooting capacity even when treated with indole-3-butyric acid, an auxin commonly used as a rooting agent. Interestingly, both RN1 and RN3 promoted adventitious root formation preferentially in the SwAsp lines. Next, the effects of the RNs were investigated in moss and compared with those of IAA (SI Appendix, Fig. S3). Similar to IAA, most of the RNs inhibited caulonemal colony outgrowth (SI Appendix, Fig. S3A). The RN-induced effects on shoots were more diverse. At the tested concentrations, while no effect of RN1 was observed, application of RN2 caused a clear increase in shoot length, RN3 treatment resulted in thinner leaves, and RN4 slightly reduced shoot size (SI Appendix, Fig. S3B). At low concentration, IAA increased the number of buds/shoots per colony after 1 wk (SI Appendix, Fig. S3C), while it reduced bud/shoot formation after 2 wk regardless of the concentrations tested (SI Appendix, Fig. S3D). This dual effect of IAA was mimicked by RN4. RN1 and RN3 treatment resulted mainly in an increase of the bud/shoot number per colony after 1 wk and RN2 only reduced bud/shoot formation after 2 wk. These results demonstrate that the activities of the RNs are mediated by pathways present in several species.
The RNs Partly Function as Prohormones.
RN1, RN3, and RN4 share structural similarities with previously described prohormones (19, 20). Because prohormones are hydrolyzed in vivo to release the active hormone moieties (21), we examined the potential metabolism of the RN compounds in liquid treatment media and in planta (SI Appendix, Fig. S4). In RN-supplemented MS media without plants, negligible concentrations of free acids were detected at the 0 h time point, except for 2,4-dichlorophenoxyacetic acid (2,4-D) originating from RN2 and 2,4,5-trichloroacetic acid (2,4,5-T) from RN3 (SI Appendix, Fig. S4D). Importantly, in these plant-free media, no obvious degradation of RN compounds was observed 24 h after treatment. However, in the presence of seedlings, higher levels of the corresponding free acids, 2,4-D, 2,4,5-T, and RN4-1, were found after 24 h in the media treated with RN1, RN3, and RN4, respectively, although the level of 2,4-D in RN2-treated media was not changed (SI Appendix, Fig. S4D). As expected, in Arabidopsis seedlings treated by the RNs for 24 h, all free acids were detected in the range from 0.4 to 2% relative to the levels of the corresponding RNs (SI Appendix, Fig. S4E).
These results imply that even though the RN compounds are fairly stable in liquid media, their biological activities might result from their metabolism in planta to the free acids 2,4-D (RN1 and RN2) and 2,4,5-T (RN3), which are known to possess auxinic activity and RN4-1 (RN4), which contains a bromo group, an electron-withdrawing substituent that can give rise to a high auxinic activity (22). To address this possibility, we first determined the appropriate treatment concentrations of 2,4-D, 2,4,5-T, and RN4-1 that lead to their accumulation within roots to similar levels as found after treatments with RN1, RN3, and RN4, respectively (SI Appendix, Fig. S5 A, C, and E). Then, using these determined treatment concentrations, we investigated the effects of 2,4-D on primary root length in 5-d-old seedlings (SI Appendix, Fig. S5B) and of 2,4,5-T and RN4-1 on lateral root density in 8-d-old seedlings (SI Appendix, Fig. S5 D and F). The results revealed that 2,4-D, at an in planta concentration intermediate to that resulting from treatments with 0.5 and 2 µM RN1, had an effect on primary root length that was correspondingly intermediate between these two concentrations of RN1 (SI Appendix, Fig. S5B). This suggests that the effect of RN1 on primary root length is likely to be due to the release of 2,4-D. However, in the case of lateral root density, a much weaker effect for 2,4,5-T, or no effect at all for RN4-1, compared with the relevant RN compound was found (SI Appendix, Fig. S5 D and F). These results show that the effects of RN3 and RN4 on lateral root density are only partially, or not at all, due to their degradation to the free acids 2,4,5-T or RN4-1, respectively.
We next performed a structure activity relationship (SAR) analysis by comparing the effects of various RN analogs, 2,4-D, 2,4,5-T, and RN4-1 on plant development and on the expression pattern of the auxin-responsive promoter DR5 in seedlings of pDR5::GUS (23) (SI Appendix, Fig. S6). The SAR analysis indicated that the absence of chlorine at position C2 in the 2,4-D substructure of RN1 (analog RN1-1) or the complete loss of the 2,4-D moiety (analog RN1-2) significantly reduced the effects of RN1 on plant development (SI Appendix, Fig. S6 A and E), implying that the 2,4-D substructure is important for RN1 activity. Modification of the 2,4-D core structure in RN2 (analog RN2-2) abolished its potency, whereas analogs displaying a side-chain modification (RN2-1 or RN2-3) were as potent as RN2 (SI Appendix, Fig. S6 B and F), indicating that the activity of RN2 is most probably attributable to the release of 2,4-D in the growing media. Like RN2, none of the RN2 analogs visibly altered the pDR5::GUS expression pattern compared with the DMSO control. RN3 mainly promoted lateral root number, while its effect on primary root elongation was mild (Fig. 1D). Analogs RN3-2 and RN3-3, with modifications on the phenylpiperazine side chain, behaved similarly to RN3 (SI Appendix, Fig. S6 C, G, and H). However, removal of the whole side chain from RN3, generating 2,4,5-T, abolished its positive effect on lateral root number and introduced a strong inhibitory effect on primary root length (SI Appendix, Fig. S6H), suggesting a difference in potency between the two compounds. Moreover, the activity of RN3 was significantly compromised by disruption of the substructure of 2,4,5-T (analog RN3-1) via loss of the three chlorines (SI Appendix, Fig. S6 C, G, and H). These results suggest that the 2,4,5-T substructure is critical for RN3’s potency. Further comparisons using analogs only differing in the number of chlorines on the 2,4,5-T substructure, such as between RN3-2, RN3-4, and RN3-6, or between RN3-3, RN3-5, and RN3-7, indicated that C5 chlorination of the 2,4,5-T moiety is crucial for RN3’s selective activity. Intriguingly, while RN3 did not alter the pDR5::GUS expression pattern compared with the DMSO control, fluorination of the phenyl in RN3 induced pDR5::GUS expression in some cases (analog RN3-3 compared with RN3-2), while reducing it in other cases (analogs RN3-5 and RN3-7 compared with RN3-4 and RN3-6, respectively) (SI Appendix, Fig. S6C). These results reinforce the importance of C5 chlorination of the 2,4,5-T moiety for the selective activity of RN3.
We showed that RN4 releases the free acid RN4-1 in planta (SI Appendix, Figs. S4 D and E and S5E), possibly by hydrolysis. As expected, considering the presence of a bromo group, this compound strongly induced pDR5::GUS expression, in contrast to RN4 itself (SI Appendix, Fig. S6D). While RN4-1 significantly enhanced hypocotyl elongation, it was not as potent in this regard as RN4 (SI Appendix, Fig. S6 D and I). Comparison of the effects of modifications of the RN4-1 substructure (analog RN4-2) and of the hydroxymethylphenylamine substructure (analog RN4-10) of RN4 indicate that while the intact auxinic RN4-1 moiety is indispensable for RN4’s effect on the hypocotyl, the nonauxinic side chain is also required to induce maximal hypocotyl elongation (SI Appendix, Fig. S6 D and I). Further comparison between RN4-2 and RN4, as well as their free acids (RN4-3 and RN4-1, respectively), highlight the key contribution of the bromophenoxy methylation to the selective activity of RN4 on hypocotyl rather than primary root (SI Appendix, Fig. S6 D, I, and J). Consistent with the SAR results, even though RN4-2 shows a bipartite structure, it was still able to induce pDR5::GUS expression (SI Appendix, Fig. S6D). RN4-10, in which the nonauxinic moiety of RN4 is modified, induced pDR5::GUS expression slightly more than RN4 (SI Appendix, Fig. S6D). We also designed RN4 analogs with predicted low hydrolysis capacity (RN4-4, RN4-8, RN4-9, and RN4-11). As expected, none of these analogs could induce hypocotyl growth (SI Appendix, Fig. S6 D and I), indicating that the typical bipartite prohormone structure of RN4 is important for its effect on hypocotyl elongation and that hydrolysis is required to liberate this activity. Moreover, except for RN4-9, these compounds could not induce pDR5::GUS. Interestingly, the analog RN4-11, generated by methylation of RN4 on the amide bond, inhibited primary root elongation without affecting hypocotyl length (SI Appendix, Fig. S6J). Because the predicted corresponding free acid RN4-1 did not reduce primary root length, this result indicates that the full, nonhydrolyzed RN4 structure possesses additional auxin-like activity.
Overall, we showed that RN1, RN3, and RN4 function as prohormones, being metabolized in planta to release more potent auxin agonists, while the effects of RN2 are most likely due to its degradation to 2,4-D. However, our SAR results also suggest that the nonhydrolyzed forms of RN1, RN3, and RN4 display additional auxin-like effects and therefore might themselves act as selective auxin agonists.
The RNs Act as Selective Auxin Agonists.
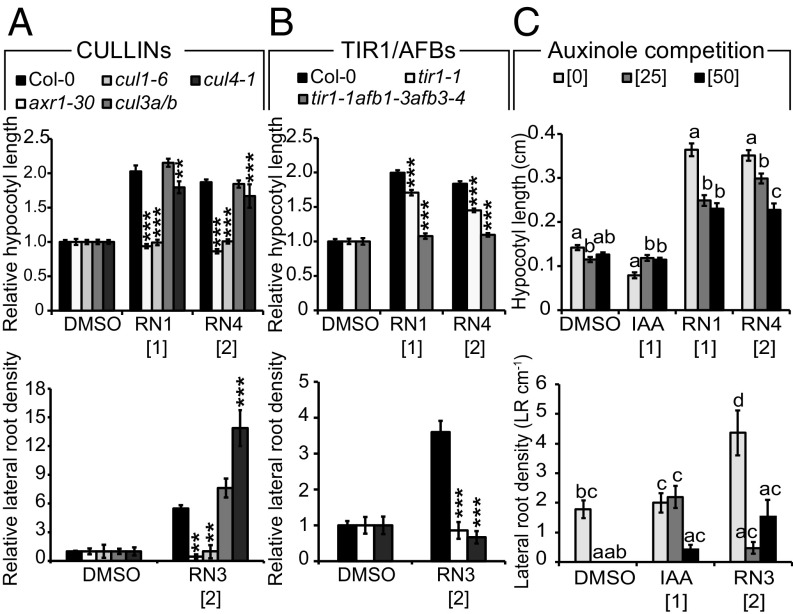
AXR1 is a component of the neddylation pathway targeting, among others, the CUL proteins (11). To determine which CUL proteins might be involved in mediating the effects of each RN, we tested their potency on the loss-of-function cul1-6, cul3a/b, and cul4-1 mutants. We limited these tests to RN1, RN3, and RN4 as we showed that RN2 activity is most probably due to its in vitro cleavage into 2,4-D, an already well-described synthetic auxin. All three tested RNs had a lesser effect on the cul1-6 mutant than on other CUL mutant lines (Fig. 2A), indicating that they function at the level of or upstream of CUL1. Given that signaling pathways mediated by AXR1 and CUL1 converge at the SCF complex, and that the chemical structures and activities of the three RNs are related to auxin, we hypothesized that auxin receptor F-box proteins might also be required for RN activities. To test this, we examined tir1 single and tir1/afb multiple mutants and found that the RN-induced phenotypes were strongly reduced when the compounds were applied on tir1-1 and tir1-1afb1-3afb3-4 (24, 25) (Fig. 2B). Thus, a functional SCFTIR1/AFB complex is essential for the effects of the RNs. To further confirm this result, we tested the effect of cotreatment of the compound auxinole (26), an auxin antagonist specific for SCFTIR1/AFB, together with each of the three RNs or the endogenous auxin IAA in the wild-type. The RN-induced phenotypes were inhibited by auxinole (Fig. 2C), demonstrating that auxin coreceptor complex formation is essential for RN activities.
Fig. 2.
RN-induced phenotypes require the formation of a functional auxin–SCFTIR1/AFB complex. Relative (treated/DMSO) (A and B) or absolute (C) hypocotyl length (Upper charts) and lateral root density (Lower charts) were measured for wild-type (Col-0) and mutant seedlings grown on media supplemented with RN compounds for 7 d. DMSO was used as control. (A) axr1-30, cul1-6, cul3a/b, and cul4-1. (B) tir1-1 and tir1-1afb1-3afb3-4. (C) Auxinole competition assay on Col-0. Statistics were performed using ANOVA and Tukey’s test. Means ± SEM are shown, n = 30 seedlings across three independent replicates, **P < 0.01, ***P < 0.001 (A and B) or different letters indicate significant differences at P < 0.05 (C). Concentrations in micromolars are indicated in brackets.
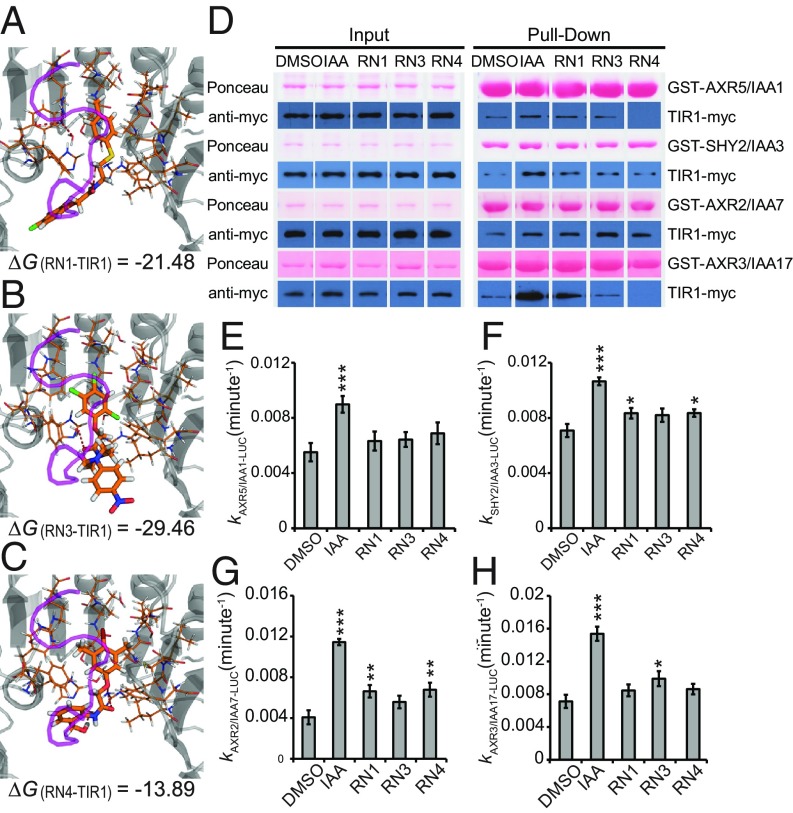
Next, we employed a molecular modeling strategy to explore the possible interactions of the RNs with the DII degron of AUX/IAA7 in the auxin-binding pocket of TIR1. Docking experiments validated that the physical property of the auxin-binding pocket was promiscuous enough to accommodate the potential steric hindrance of RN1, RN3, or RN4 (Fig. 3 A–C and Movie S1). The calculated free energies (ΔG) of binding also revealed thermodynamic stability for the three RNs inside the auxin pocket of TIR1 (Fig. 3 A–C and SI Appendix, Fig. S7A). The positive control IAA was able to bind TIR1 with a ΔG(IAA-TIR1) of −11.68, whereas the negative control Tryptophan (Trp) was not, with a ΔG(Trp-TIR1) of 63.34 (SI Appendix, Fig. S7A). Among the RN analogs, RN4-1 and RN4-2 showed stronger thermodynamic stability compared with IAA. RN2 and the inactive analog RN4-8 could not dock inside the auxin-binding site to stabilize TIR1 (SI Appendix, Fig. S7A). This last result confirmed once again that RN2 activity is most likely due to its cleavage into 2,4-D.
Fig. 3.
RN3 and RN4 act as selective agonists of auxin. (A–C) The RNs showed different thermodynamic stabilities from the calculated free energies (ΔG). RN1 (A), RN3 (B), and RN4 (C) were sterically favorable for the binding of the AUX/IAA7 DII degron. TIR1 is presented in gray and the AUX/IAA7 DII degron, which was included afterward to observe any conflict with the RNs, is in purple. Thermodynamic stability was computed within the TIR1 auxin binding pocket and the most stable conformations are represented. (D) The potential of the RNs (at 50 μM) to promote the formation of the coreceptor complex was performed using in vitro translated TIR1-myc and recombinant GST-AUX/IAAs. Depending on the GST-AUX/IAA translational fusion used for the in vitro GST pull-down, the RNs selectively increased the recovery of TIR1-myc. (E–H) AUX/IAA degradation was assayed in planta using Arabidopsis lines constitutively expressing different AUX/IAA-LUCs in the presence of RNs at 50 μM. Effects of the RNs on the in vivo degradation rate k of AXR5/IAA1-LUC (E), SHY2/IAA3-LUC (F), AXR2/IAA7-LUC (G), or AXR3/IAA17-LUC (H) translational fusions. Statistical analyses were performed using the Student’s t test. Means ± SEM are shown, n = 30 seedlings across five independent replicates, *P < 0.05, **P < 0.01, ***P < 0.001.
To experimentally confirm the binding of the RNs within the auxin coreceptor complex, we tested their ability to promote the interactions between TIR1 and AUX/IAA proteins using in vitro pull-down assays. First, TIR1-myc protein purified from wheat germ extract and four different GST-AUX/IAA proteins were used (27–29). IAA stimulated the interaction of TIR1-myc with all AUX/IAAs tested (Fig. 3D and SI Appendix, Fig. S7B). All three RNs stimulated the recovery of TIR1-myc in complex with GST-SHY2/IAA3 or GST-AXR2/IAA7 to a similar extent (Fig. 3D and SI Appendix, Fig. S7B). In the case of GST-AXR5/IAA1, RN1 stimulated the interaction with TIR1-myc, while RN3 had little effect and surprisingly, RN4 decreased the basal interaction (Fig. 3D and SI Appendix, Fig. S7B). When GST-AXR3/IAA17 was used as bait, RN1 strongly promoted the interaction with TIR1-myc, while RN3 had little effect and again, RN4 reduced the basal interaction (Fig. 3D and SI Appendix, Fig. S7B). These data imply that RN3 and RN4 are able to selectively promote the interactions between specific TIR1 and AUX/IAA protein combinations in this system, while RN1 and IAA promoted each interaction, as shown previously for IAA (27–29).
To test that these effects on TIR–AUX/IAA complex formation were not dependent on metabolism of the RN compounds in the wheat germ extract, we next performed a complementary pull-down experiment using insect cell-expressed TIR1 (as a His-MBP-FLAG-TIR1 fusion protein) with bacterially expressed GST-AXR2/IAA7 or GST-AXR3/IAA17 in the presence of the RNs or the RN4 degradation product RN4-1 (SI Appendix, Fig. S7 C and D). In this system, the RNs again promoted selective interactions between TIR1 and AXR2/IAA7 or AXR3/IAA17, this time in the absence of potential plant hydrolases (in insect cells). Importantly, the promotion and inhibition of TIR1 interaction with AXR2/IAA7 and AXR3/IAA17, respectively, by RN3 and RN4 were identical in the two in vitro systems. Moreover, the degradation product RN4-1 behaved differently from RN4, by not promoting the interaction between TIR1 and AXR2/IAA7 and slightly promoting the interaction between TIR1 and AXR3/IAA17, which might explain these compounds’ different activities in vivo. In fact, we were able to confirm that the observed TIR1–AXR/IAA interactions in this system were induced or repressed specifically by the RNs and not by their free-acid degradation products, as no 2,4-D, 2,4,5-T, or RN4-1 could be detected at relevant time points in the pull-down reactions treated with RN1, RN3, or RN4, respectively (SI Appendix, Fig. S7E). These data demonstrate that RN3 and RN4 are able to selectively promote the interactions between TIR1 and certain AUX/IAA proteins. Hence, our results suggest that RN3 and RN4 are not just prohormones, but also act consistently as selective auxin agonists in two different in vitro experimental conditions and their effects on plant development may therefore be attributable to selective auxin agonistic activity.
To test whether the RNs might also act as selective auxin agonists in planta, we assayed their potency in promoting the in vivo degradation of the AUX/IAA proteins. In a 1-h time course, IAA significantly increased the degradation rate of the four tested AUX/IAA-LUCIFERASE (LUC) proteins, while the RNs had different potency depending on the AUX/IAA proteins used (Fig. 3 E–H and SI Appendix, Fig. S7F). Therefore, the RN molecules act as selective auxin agonists both in vitro and in vivo, but the specificity of the interactions seems to be dependent on the experimental conditions, as the predicted behavior of AUX/IAA proteins based on their sensitivity to RN3 and RN4 in our in planta LUC assays did not always match that in our in vitro pull-down assays. While the conditions tested in vivo reflect RN capacity to enhance the interactions of the different SCFTIR1/AFB–AUX/IAA coreceptors within a complex molecular surrounding, those tested in vitro reflect the interactions in much simpler conditions. Nonetheless, our results imply that altering interaction affinity within each coreceptor complex with selective auxin agonists might modulate a multitude of specific plant development aspects.
RN3 and RN4 Induce Selective Early Transcriptional Responses.
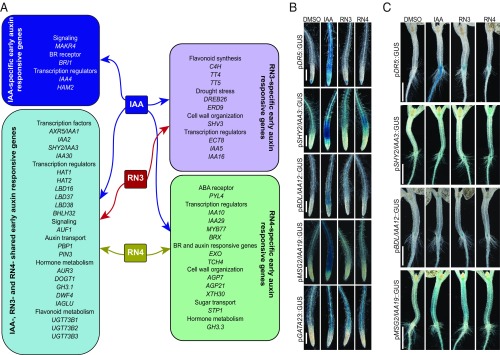
The in vitro assays indicated that RN3 and RN4 are the most selective auxin agonists, showing different effects on different AUX/IAA proteins. Moreover, RN3 and RN4 induced distinct developmental processes, particularly on lateral root development. While RN3 enhanced the density of lateral roots without affecting primary root length in the wild-type, RN4 inhibited lateral root development (Fig. 1). Because these RNs promoted fast degradation of AUX/IAA proteins fused to LUC, we investigated how their activities fine-tuned events downstream of coreceptor complex formation. To this end, we performed transcriptome-wide expression profiling of Arabidopsis cell suspension cultures treated with IAA, RN3, and RN4, to characterize the early transcriptional responses induced by these compounds (Dataset S1). The data have been deposited at the European Nucleotide Archive (www.ebi.ac.uk/ena) under the accession number PRJEB31496 (30). Analysis of the differentially expressed genes (DEGs) revealed subsets that were up- or down-regulated specifically by one, two, or all three chemical treatments (SI Appendix, Fig. S8A and Table S1). Among the early auxin-responsive genes identified, AXR5/IAA1, IAA2, SHORT HYPOCOTYL 2 (SHY2)/IAA3, and IAA30 were significantly up-regulated by IAA, RN3, and RN4 (Fig. 4A and SI Appendix, Table S1). IAA5 and IAA16 expressions were induced specifically by IAA and RN3, while IAA10 and IAA29 expressions were up-regulated selectively by IAA and RN4, revealing some differences between RN3 and RN4 in their capacity to induce early-responsive AUX/IAA genes. In total, 121 genes were differentially up-regulated by IAA, RN3, and RN4, such as LATERAL ORGAN BOUNDARIES-DOMAIN 16 (LBD16), BASIC HELIX–LOOP–HELIX 32 (BHLH32), PINOID-BINDING PROTEIN 1 (PBP1), and PIN-FORMED 3 (PIN3) (31–34) (Fig. 4A), confirming the potential of the RNs to modulate auxin-related developmental processes. The genes CINNAMATE 4 HYDROXYGENASE (C4H), TRANSPARENT TESTA 4 (TT4), TT5, DEHYDRATION RESPONSE ELEMENT-BINDING PROTEIN 26 (DREB26), and EARLY-RESPONSIVE TO DEHYDRATION 9 (ERD9) were commonly up-regulated by IAA and RN3 but not by RN4. These five genes are known to be tightly regulated in a tissue-specific and auxin-dependent manner to modulate lateral root density and architecture (35–39). Among the genes commonly regulated by IAA and RN4 but not RN3, we identified MYELOBLASTOSIS 77 (MYB77) and BREVIX RADIX (BRX) transcription factors, which have been shown to control lateral root formation in an auxin-dependent manner (40, 41). These results correlate with the differential effects of RN3 and RN4 on lateral root development. Taken together, these data demonstrate the potential of RN3 and RN4 to specifically identify auxin-responsive genes involved in defined developmental processes, such as lateral root formation. Overall, we showed that RN molecules are able to selectively trigger specific auxin perception machinery, inducing expression of specific sets of genes, and resulting in distinct developmental traits.
Fig. 4.
RN3 and RN4 activate independent auxin responses. (A) Selected sets of up-regulated genes in cell culture representing: IAA-specific induced genes (dark blue); IAA-, RN3-, and RN4-induced genes (light blue); IAA- and RN3-specific induced genes (lilac); and IAA- and RN4-specific induced genes (green) (see Dataset S1 for the complete list of genes and SI Appendix, Table S1 for fold induction values of the selected genes). (B and C) Five-day-old seedlings expressing pDR5::GUS, pSHY2/IAA3::GUS, pBDL/IAA12::GUS, pMSG2/IAA19::GUS, or pGATA23::GUS transcriptional fusions treated with IAA, RN3, and RN4 at 10 μM for 16 h. DMSO was used as control. (B) Representative primary roots after GUS staining. (C) Representative hypocotyl-root junctions after GUS staining. (Scale bars, 100 μm in B and 1 mm in C.)
RN3 and RN4 Induce Specific Subsets of Auxin Responsive Promoters.
We further investigated the abilities of RN3 and RN4 to selectively induce later auxin responses using various auxin-responsive reporter lines after 45 min, 5 h, or 16 h of RN treatment. We found that neither the auxin-responsive reporter pDR5::GUS nor the indicator of nuclear auxin perception p35S::DII-Venus (42) showed any response to RN treatment in the primary root (Fig. 4B and SI Appendix, Fig. S8 B and D). However, in the root–hypocotyl junction, the expression of pDR5::GUS was promoted by either longer treatment (24 h) or higher concentration (50 µM) of RN3 or RN4 (Fig. 4C and SI Appendix, Fig. S8C). To determine whether these effects were specific to the RNs or rather due to their free-acid degradation products, we first determined the appropriate treatment concentrations of 2,4,5-T and RN4-1 that lead to their accumulation within the roots to similar levels as found after 16-h treatments with RN3 and RN4, respectively (SI Appendix, Fig. S9 A and B). While treatment with 2,4,5-T, similar to RN3, had no effect on pDR5::GUS expression in the root (SI Appendix, Fig. S9C), treatment with RN4-1, in contrast to RN4, induced pDR5::GUS expression in the root (SI Appendix, Fig. S9D). For other auxin-responsive reporter lines tested, RN3 and RN4 induced expression patterns that partially overlapped with those induced by IAA (Fig. 4 B and C). In the primary root, the RN compounds induced pSHY2/IAA3::GUS and pBODENLOS(BDL)/IAA12::GUS expression with different patterns compared with that induced by IAA, but did not stimulate pMASSUGU2(MSG2)/IAA19::GUS expression (Fig. 4B). Both compounds also promoted the expression of pGATA23::GUS, a marker of lateral root founder cell identity (43). RN4 additionally induced pSHY2/IAA3::GUS expression in the hypocotyl and the shoot apical meristem (Fig. 4C). In contrast to the primary root, RN3 and RN4 induced pMSG2/IAA19::GUS expression in the hypocotyl (Fig. 4C), although only RN4 induced hypocotyl elongation (Fig. 1B). Treatment of these auxin-responsive reporter lines with 2,4,5-T induced similar expression patterns in the primary root as treatment with RN3 (SI Appendix, Fig. S9C), suggesting that the observed effects of RN3 may in fact be due to 2,4,5-T activity. However, as found for the DR5 promoter, RN4-1 induced the expression of most of the other promoters tested more strongly than RN4 in the primary root (SI Appendix, Fig. S9D), suggesting that these two compounds affect auxin-responsive promoter expression rather differently. Despite the release of RN4-1 during RN4 treatment, the effects of RN4 appear to be prominent as this compound did not induce pDR5::GUS despite the presence of RN4-1. Our data indicate that RN3 and RN4 may be able to induce specific auxin-regulated promoters, which might be responsible for their selective activities on plant development. Indeed, these RNs activate some but not all modules of the auxin signaling pathway within the same tissue, confirming their selective auxin agonist activities.
A summary of the results obtained for the four RNs is presented in SI Appendix, Table S2. In particular, RN3 and RN4 behave as auxin agonists, which selectively promote or inhibit AUX/IAA degradation in a reproducible manner, leading to specific transcriptional regulation and developmental outputs.
AUX/IAA Sensitivity to RN3 and RN4 in Planta.
We hypothesized that as the RN molecules show selectivity toward the auxin coreceptor complex, they might help to dissect specific functions of individual AUX/IAAs in distinct developmental processes. One approach to achieve this could be to investigate the responses of AUX/IAA gain-of-function mutants to auxin treatment; however, such a genetic approach could prove problematic due to high redundancy among the AUX/IAAs. As a potentially more effective alternative, we challenged such mutants with the specific auxin analogs RN3 and RN4.
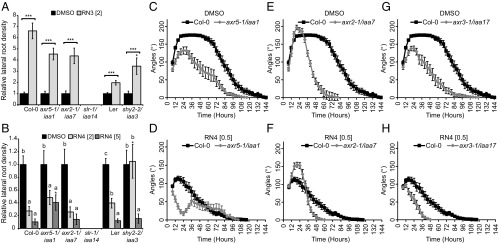
We first focused on lateral root development as RN3 and RN4 had opposite effects on this process (Fig. 1 D and E). Furthermore, based on our transcriptomic analysis, RN3 and RN4 induce different sets of IAA-responsive genes that are known to be involved in the regulation of lateral root development (Fig. 4A). We therefore investigated the sensitivities of 8-d-old seedlings of AUX/IAA gain-of-function mutants axr5-1/iaa1 (28), axr2-1/iaa7 (44), shy2-2/iaa3 (45, 46), and solitary root (slr-1)/iaa14 (47) to treatments of RN3 and RN4 with regards to lateral root development. We tested the sensitivities of these gain-of-function mutants to RN3, which increases lateral root density in Col-0 and Ler, with the Col-0 accession interestingly showing much higher sensitivity to this effect (Fig. 5A). We found that most of the mutants were also sensitive to this effect, with the exception of slr-1/iaa14 (Fig. 5A). The mutant shy2-2/iaa3 was more sensitive to this effect of RN3 than the wild-type (Fig. 5A); however, it is important to note that in this mutant, this compound mainly induced the slight emergence of lateral root primordia rather than the emergence of well-developed lateral roots. These data suggest that apart from SLR/IAA14, the AUX/IAAs we tested are not required for the stimulatory activity of RN3 on lateral root density. We next aimed to characterize RN4 activity on lateral root development in these mutants. RN4 reduced lateral root density in Col-0 and Ler (Fig. 5B). Compared with Col-0, axr5-1/iaa1 was resistant to this effect of RN4 at 5 µM, while axr2-1/iaa7 was sensitive at both tested RN4 concentrations (Fig. 5B). Interestingly, shy2-2/iaa3 was sensitive to RN4 at 5 µM, but resistant at 2 µM, compared with Ler (Fig. 5B). Our results suggest that AXR5/IAA1 and SHY2/IAA3 might be degraded by RN4 to reduce lateral root density.
Fig. 5.
RN-induced phenotypes require the degradation of specific AUX/IAAs. (A and B) Relative lateral root density (treated/DMSO) was measured for gain-of-function mutants axr5-1/iaa1, axr2-1/iaa7, slr-1/iaa14, and shy2-2/iaa3 and their respective wild-type grown on media supplemented with RN3 (A) and RN4 (B) for 8 d. DMSO was used as control. Statistical analyses were performed using Student’s t test (A), or ANOVA and Tukey’s test (B) to compare the effect of RN3 (A) or RN4 (B) relative to the DMSO control for each genotype, as indicated with triple asterisks and square brackets (A) or different letters (B). The Student’s t test was used to compare the relative effect of RN3 (A) or RN4 (B) on the mutants to that on the relevant wild-type, as indicated with single asterisks. (C–H) Gain-of-function mutants axr5-1/iaa1 (C and D), axr2-1/iaa7 (E and F), and axr3-1/iaa17 (G and H) were grown in the dark on DMSO (C, E, and G) and RN4 (D, F, and H) -supplemented media for 6 d. Measurement of apical hook angle was performed every 3 h. Means ± SEM are shown, n > 20 seedlings across three independent replicates; *P < 0.05, **P < 0.01, ***P < 0.001, different letters indicate significant differences at P < 0.05. Concentrations in micromolars are indicated in brackets.
By using the RN molecules, we revealed potential contributions of specific AUX/IAAs to the complicated process of lateral root development. However, the sensitivities of the aux/iaa gain-of-function mutants to the RNs in terms of lateral root development did not exactly match the RN-induced AUX/IAA degradation/stabilization results found with our binding affinity assays. Lateral root development is a complicated process that requires the formation of a new meristem and emergence through several root layers, suggesting that the specific tissue context may affect RN activity and selectivity. We therefore decided to switch our focus to apical hook development in etiolated seedlings, a rather simpler process than lateral rooting, but one also regulated by auxin (48). Apical hook development is characterized by differential growth between the two sides of the apical hypocotyl and comprises the formation, maintenance, and opening phases (49, 50). We first tested the effects of RN3 and RN4 on apical hook development in the wild-type (SI Appendix, Fig. S10A). While 2 µM RN3 did not affect apical hook development, RN4 completely abolished hook formation in a dose-dependent manner (SI Appendix, Fig. S10 A and B).
We decided to exploit RN4 to understand whether selected AUX/IAAs play specific roles during apical hook development. We tested the effects of 0.5 µM RN4 on hook development in the gain-of-function mutants axr5-1/iaa1, axr2-1/iaa7, and axr3-1/iaa17 for 6 d in the dark. All three mutants showed altered apical hook development compared with the wild-type in control conditions (Fig. 5 C, E, and G). A detailed analysis of these results indicates that AXR5/IAA1 and AXR3/IAA17 need to be degraded for a proper apical hook to develop, while AXR2/IAA7 is likely stabilized during the formation phase and degraded during the maintenance phase. Similar to the wild-type, axr5-1/iaa1 showed sensitivity to RN4 during the formation phase, with no hook being present at 24 h; however, by 36 h the mutant had attained a slight hook curvature of 50°, which then started opening directly (Fig. 5D). The mutant axr2-1/iaa7 was resistant to RN4 in the formation phase (Fig. 5F) and axr3-1/iaa17 was sensitive to RN4 (Fig. 5H). Taken together, these results indicate that all three AUX/IAAs tested here play a role during apical hook development. In particular, our results suggest that AXR2/IAA7 is stabilized during apical hook formation while AXR5/IAA1 stabilization occurs during the maintenance phase.
The effects of 0.5 µM RN4 on AUX/IAA mutants during the first 24 h of apical hook development (Fig. 5 D, F, and H) correlate strikingly with our in vitro pull-down assay results (Fig. 3D). AXR2/IAA7 proteins strongly interacted with TIR1 in the presence of RN4 (Fig. 3 D and G and SI Appendix, Fig. S7B), suggesting that a stabilized version of this AUX/IAA should confer resistance to the RN4 auxin agonist, which is indeed what we found with the axr2-1/iaa7 gain-of-function mutant (Fig. 5F). In contrast, AXR5/IAA1 and AXR3/IAA17 did not interact with TIR1 when RN4 was present in the pull-down assay (Fig. 3 D, E, and H and SI Appendix, Fig. S7B) and the corresponding gain-of-function mutants were sensitive to the effects of RN4 on hook development (Fig. 5 D and H).
Overall, our study of the effects of RN4 in particular on the AUX/IAA gain-of-function mutants, distinguishes the involvement of specific AUX/IAAs in lateral root and apical hook development. Thus, we demonstrated the potential of such selective auxin agonists in dissecting auxin perception controlling specific developmental processes in vivo.
Mutation in the ATPase Domain of AtBRM Confers Resistance to RN4.
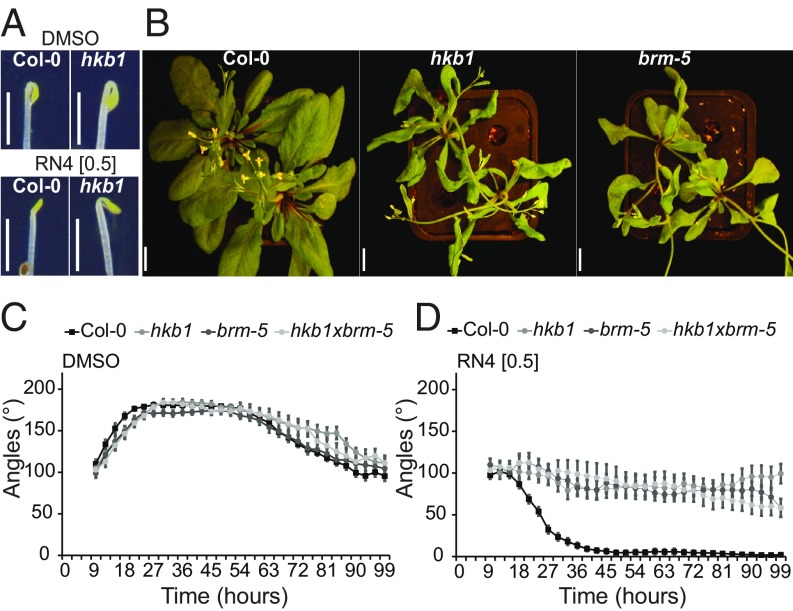
RN4 represents a useful tool to investigate the role of auxin during early stages of skotomorphogenesis. To identify new molecular players involved in apical hook development, we performed a forward genetic screen of sensitivity to RN4, using an ethyl methanesulfonate-mutagenized Col-0 population and selected those mutants that were able to form an apical hook in the presence of 0.5 µM RN4 in the dark, which we named hookback (hkb) mutants. We then further selected only those of the mutants that were sensitive to the effects of 75 nM 2,4-D on seedling phenotype in the light (SI Appendix, Fig. S10C). Using this strategy, we could exclude known auxin resistant mutants that might appear in the screen. Several independent hkb lines, each carrying a single recessive mutation, were isolated from the screen and we focused on characterizing one of these, hkb1. In contrast to Col-0, hkb1 had formed well-curved apical hooks in the presence of RN4 24 h after germination, while under mock-treated conditions there were no major differences between the two genotypes (Fig. 6A). Whole-genome sequencing of hkb1 revealed the presence of one nonsynonymous ethyl methanesulfonate-like mutation (C-to-T nucleotide substitution) in the coding region of the AT2G46020 gene that encodes for the SWItch/Sucrose Non-Fermentable (SWI/SNF) chromatin remodeling ATPase BRAHMA (BRM). The data have been deposited at the European Nucleotide Archive (www.ebi.ac.uk/ena) under the accession number PRJEB21529 (51). To confirm that the mutation in BRM is responsible for the resistance of hkb1 against the negative effect of RN4 on apical hook formation, we carried out several analyses. First, we checked the phenotypes of available T-DNA mutants for BRM, including brm-1, brm-2, brm-4, and brm-5 (ectopic expression of seed storage proteins3, essp3) (52, 53). However, we focused our investigations on brm-5 because both hkb1 and brm-5 contain a mutation in the ATPase domain (54) and 4-wk-old plants of the two mutants showed similar phenotypes, including twisted leaves and less siliques than wild-type (Fig. 6B). Importantly, brm-5 showed similar resistance to the effect of 0.5 µM RN4 on apical hook formation to that shown by hkb1 (Fig. 6 C and D). These results strongly suggest that the mutation in the ATPase domain of BRM in hkb1 is responsible for the resistance of this mutant to RN4. Next, we crossed hkb1 with brm-5 and the F2 generation was analyzed. The hkb1xbrm-5 mutant showed the same apical hook phenotype and similar RN4 resistance as the single hkb1 and brm-5 mutants (Fig. 6 C and D), confirming that the mutation that confers resistance against RN4 in hkb1 is in the BRM gene.
Fig. 6.
The hkb1 mutant is resistant to the RN4 effect on apical hook development and carries a mutation on BRM. (A) Comparison of apical hook phenotype in Col-0 and hkb1 seedlings 24 h after germination in the dark. The seedlings were grown on media supplemented with DMSO (Upper) or RN4 (Lower). (Scale bars, 2 mm.) (B) Four-week-old Col-0, hkb1, and brm-5 grown in long-day greenhouse conditions. (Scale bars, 1 cm.) (C and D) Apical hook angle in Col-0, hkb1, brm-5, and hkb1xbrm-5 grown on DMSO (C) and 0.5 µM RN4 (D) supplemented media for 6 d in the dark. Measurement of apical hook angle was performed every 3 h. Means ± SEM are shown, n > 18 seedlings across two independent replicates. Concentrations in micromolars are indicated in brackets.
Our results suggest that BRM may function as a negative regulator of apical hook formation. Considering the resistance of both the axr2/iaa7 gain-of-function mutant and hkb1/brm-5 to the effect of RN4 on apical hook formation, we hypothesize that AXR2/IAA7 might negatively regulate BRM-induced gene transcription. We suggest that RN4 induces degradation of AXR2/IAA7, which may lead to BRM-mediated promotion of transcription of genes negatively regulating apical hook formation, potentially through chromatin remodeling.
Overall, our results show that selective auxin agonists can enable us to dissect the roles of specific AUX/IAAs in developmental processes, leading to the dissection of the molecular mechanisms of these processes.
Discussion
Complicated auxin perception modules translate auxin signals into a multitude of developmental responses (55, 56). Several studies have demonstrated that IAA displays different affinities for different SCFTIR1/AFB–AUX/IAA coreceptor complex combinations (6, 57) and specific auxin perception modules have even been shown to act sequentially during development (58). In this work, we isolated the RNs as selective auxin agonists and revealed their potential to dissect the complex and redundant mechanisms of auxin perception machinery that control specific aspects of plant development. We employed RN4 in particular as a tool to characterize specific auxin perception modules and their potential targets. Remarkably, we even found variability of RN sensitivity between different accessions in both Arabidopsis and poplar, pointing to future challenges toward developing the most suitable auxin agonists for specific species and accessions. However, it is important to emphasize that we identified degradation products released from all four RNs in planta, which in some cases also induced plant responses. This finding highlights that it is essential to investigate the stability of any such identified auxin agonists and take into account any degradation products released.
Auxin behaves like molecular glue within the SCFTIR1/AFB–AUX/IAA complex (55) by fitting into a space between the TIR1/AFB receptor and AUX/IAA coreceptor and extending the hydrophobic protein interaction surface. It has long been known that the auxin-binding pocket of SCFTIR1/AFB is promiscuous, a feature that was heavily investigated during the early years of auxin research in the 1940s (59, 60). During this time, several auxinic compounds were discovered, including NAA, 2,4-D, and picolinate auxins, such as picloram (61), which are widely used today for basic research and agricultural applications. The 2,4-D and NAA modes of action are similar to that of IAA, as they also enhance the binding affinity between TIR1 and the AUX/IAAs. Their affinity to the coreceptor complex is lower than that of IAA, but they are more stable metabolically, which explains their robust activity. Although the full details of the mode of action of these synthetic auxins are not yet known, they have been instrumental in the discoveries of crucial auxin signaling components, such as AXR1, AXR3/IAA17, AXR5/IAA1, AFB4, and AFB5 (62–66). Thus, synthetic compounds with auxin-like activities hold the potential to dissect the convoluted mechanisms of auxin signaling. Moreover, our isolation and characterization of RN4 revealed different activity and selectivity compared with most of the currently available synthetic auxins, and thus open up new possibilities to identify novel actors in auxin biological responses.
Here, we have shown the selective capacity of RN3 and RN4 to promote the interaction of TIR1 with specific AUX/IAA coreceptors, highlighting a strong potential for such auxin agonists in defining AUX/IAA involvement in specific transcriptional responses and developmental traits. This potential was strongly supported by our genetic approach, showing that different AUX/IAA gain-of-function mutants display defined sensitivities to RN3 and RN4 in terms of lateral root development. Importantly, we uncoupled the effects of RN3 and RN4 on TIR1-AUX/IAA interactions and lateral root development from their free acid degradation products, thus confirming the usefulness of these RN compounds as selective auxin agonists. Multiple AUX/IAA-ARF modules act sequentially over time and space to orchestrate lateral root development (58, 67). Our data indicate that RN3 may promote development of lateral roots through SLR/IAA14 degradation and the stabilization of SHY2/IAA3, but we cannot yet conclude whether degradation of additional AUX/IAAs is also required for this effect. On the other hand, the resistance of the axr5-1/iaa1 mutant to high concentrations of RN4 revealed a role for AXR5/IAA1 as a positive regulator of lateral root development.
Moreover, we used the RN with the greatest potential, RN4, as a tool to identify which of several AUX/IAA proteins are directly involved in apical hook development and revealed the implication of auxin-signaling components, such as the SWI/SNF chromatin remodeling ATPase BRM. Remarkably, BRM has already been shown to be involved in auxin-dependent floral fate acquisition (68). In the inflorescence, when MONOPTEROS (MP)/ARF5 is free from AUX/IAA repression, it recruits BRM or its homolog SPLAYED (SYD) to remodel chromatin and thus promote gene transcription. Interestingly, in a yeast three-hybrid assay, AXR3/IAA17 and BDL/IAA12 have been shown to prevent the association of MP to BRM (68). According to these results and our data showing the resistance of axr2-1/iaa7 and hkb1/brm-5 to RN4-mediated suppression of apical hook formation, we hypothesize that BRM, by associating with an unknown ARF transcription factor, might promote transcription of genes negatively regulating hook formation. We also hypothesize that AXR2/IAA7 might prevent the association of the ARF to BRM. Application of RN4 prompts the degradation of AXR2/IAA7, which may facilitate the association of the ARF to BRM, promoting transcription of downstream genes negatively regulating apical hook formation, potentially through chromatin remodeling. However, the hypothesis that stabilization of AXR2/IAA7 during apical hook formation blocks BRM activity raises the question of whether MP plays a role during hook development or whether BRM is recruited by other ARFs.
The different affinities of AUX/IAA proteins for IAA, RN3, and RN4 might lie in differences in residues within the DII domain. Our study thus brings us a step closer to a better quantitative understanding of the TIR1–AUX/IAA interaction system of auxin perception in a tissue-specific manner. Besides IAA, several other phytohormones including jasmonate-isoleucine, gibberellin, brassinosteroids, and abscisic acid (ABA), also function by modulating the protein–protein interactions of their coreceptors (69). Isolation of novel molecules modulating such interactions could therefore also be useful in uncovering the signaling components of these phytohormones.
Auxins have many uses in agriculture, horticulture, forestry, and plant tissue culture (59). The selective auxin agonists described here may also find niche applications in these fields. RN activities in the low micromolar range and conservation of their specific developmental effects in land plants enforces this possibility. Moreover, the availability of models for ligand-bound coreceptors may allow rational design of a wider array of auxin agonists using RN structures, in particular RN4, as a starting point. Indeed, a rational design approach has already paved the way for developing agrochemicals interacting specifically with a subset of ABA receptors (70). Such an approach might also have the potential to overcome the limitations of some of the RNs, for example by enhancing stability to eliminate the release of degradation products.
Overall, the isolation and characterization of chemical modulators of plant hormone signaling is an effective way to better understand the specificity of hormonal receptors. Because of the availability of genetic and genomic methods, most chemical biology approaches are performed in model species, such as Arabidopsis. However, chemicals that induce well-characterized effects in Arabidopsis can be applied to nonmodel species to improve crop and tree value in agriculture and forestry, respectively. The complexity of the genomes of such nonmodel species may also be unraveled by the use of chemicals for which target proteins or pathways are known, giving a better understanding of evolutionary mechanisms.
Materials and Methods
See SI Appendix for detailed experimental procedures.
Arabidopsis thaliana seedlings were grown on 1/2 MS medium supplemented with 0.05% Mes, 1% sucrose, and 0.7% agar at pH 5.6. Stock solutions of all compounds were dissolved in DMSO, which was also used in equal volume as a solvent control. Docking experiments were performed using SwissDock (71, 72) with the ZINC ID of the RNs and 2P1Q crystal structure of TIR1 with the DII domain of AXR2/IAA7 (60). The best conformation was chosen according to the FullFitness (kcal/mol). The corresponding binding energies for every conformation of each ligand were calculated using Hybrid-DFT-D3. In vitro pull-down assays, with epitope-tagged TIR1 expressed with TnT-T7 coupled wheat germ extract (Promega), were performed as described previously (29, 73). For the luciferase assay, 7-d-old seedlings were incubated in Bright-Glo luciferase assay system (Promega) luciferine solution (LS) for 30 min before treatment with 50-µM compounds dissolved in LS. Light emission was recorded for 5 min using a LAS-3000 (Fujifilm) and the natural log of the normalized relative light unit was calculated as described previously (74). The degradation rate k (min−1) was used to compare treatments. The transcriptomic responses induced by the RNs were investigated by RNA sequencingeq, using A. thaliana ecotype Col-0 cell suspension culture (75) treated with 50 µM RN3, RN4, or IAA for 30 min. Total RNA was extracted from filtered cells using the RNeasy Plant Mini Kit (Qiagen) and sent to the SNP&SEQ Technology Platform in Uppsala University for sequencing. Genes were considered significantly differentially expressed if the adjusted P values after false discovery rate) correction for multiple testing were lower than 0.05. For GUS assays, seedlings were fixed in 80% acetone, washed with 0.1 M phosphate buffer and transferred to 2 mM X-GlcA (Duchefa Biochemie) in GUS buffer (0.1% triton X-100; 10 mM EDTA; 0.5 mM potassium ferrocyanide; 0.5 mM potassium ferricyanide) in the dark at 37 °C before stopping the reaction with 70% ethanol.
Supplementary Material
Acknowledgments
We thank the many researchers who kindly provided us with published Arabidopsis lines; the Arabidopsis Biological Research Center and Nottingham Arabidopsis Stock Centre for distributing seeds; the Swedish Metabolomics Centre (www.swedishmetabolomicscentre.se/) for access to instrumentation; O. Keech for critical reading of the manuscript; M. Quareshy, V. Uzunova, and R. Napier (University of Warwick) for assistance with insect cell expression of epitope-tagged TIR1; L. Bako, for providing us with Arabidopsis cell culture; Pardeep Singh for sharing powder of 2,4,5-T and RN4-1 with us; and R. Bhalerao for sharing the system for time-lapse imaging of dark grown seedlings. J.C. thanks J. Brown for technical assistance, and K. Dreher and J. Gilkerson for helpful discussion and for transgenic line characterization. Sequencing was performed by the SNP&SEQ Technology Platform, Science for Life Laboratory at Uppsala University, a national infrastructure supported by the Swedish Research Council and the Knut and Alice Wallenberg Foundation. RNA sequencing data analysis was in part performed by Bioinformatics Infrastructure for Life Sciences. Amplification of in vitro poplar SwAsp lines was performed by the Poplar Transgenics Facility, Umeå Plant Science Centre. Whole-genome resequencing was performed by Novogene and the results were analyzed with the support of Nicolas Delhomme and Iryna Shutava, Umeå Plant Science Centre Bioinformatics Platform. This work was supported by Vetenskapsrådet VR 2013-4632 and VINNOVA (to T.V., M.Ł., S.M.D., P.-A.E., and S. Robert); Vetenskapsrådet VR 2016-00768 (to Q.M.); The Knut and Alice Wallenberg Foundation (A.R.); The Knut and Alice Wallenberg Foundation ShapeSystems Grant 2012-0050 (to S.M.D., S. Robert, Q.M., and K.L.); the Olle Engkvist Byggmästare Foundation (S. Raggi); Kempestiftelserna (Q.M., D.K.B., and P.-A.E.); the Carl Tryggers Foundation (Q.M. and M.T.); SweTree Technologies (S.M.D.); an EMBO short-term fellowship (to T.V.); an Seth M. Kempe short-term fellowship (to T.V.); a travel grant from the Bröderna Edlunds Foundation (to T.V.); National Science Foundation MCB–0929100 (to J.C.); National Institutes of Health Grant NIH GM43644 (to M.E.); the Biotechnology and Biological Sciences Research Council BB/L010623/1 (to S.K.); and the Ministry of Education, Youth and Sports of the Czech Republic (European Regional Development Fund-Project “Plants as a tool for sustainable global development” no. CZ.02.1.01/0.0/0.0/16_019/0000827) (to B.P. and O.N.). L.P. was funded by a postdoctoral scholarship and research Grant 1507013N of the Research Foundation Flanders. Chemical Biology Consortium Sweden is primarily funded by the Swedish Research Council (P.-A.E.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The whole genome sequencing data of hkb1 has been deposited at the European Nucleotide Archive, https://www.ebi.ac.uk/ena (accession number: PRJEB21529). The RNA sequencing data of Arabidopsis cell suspension culture treated with IAA, RN3, and RN4 have been deposited at the European Nucleotide Archive, http://www.ebi.ac.uk/ena (accession number: PRJEB31496).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809037116/-/DCSupplemental.
References
- 1.Teichmann T, Muhr M. Shaping plant architecture. Front Plant Sci. 2015;6:233. doi: 10.3389/fpls.2015.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolters H, Jürgens G. Survival of the flexible: Hormonal growth control and adaptation in plant development. Nat Rev Genet. 2009;10:305–317. doi: 10.1038/nrg2558. [DOI] [PubMed] [Google Scholar]
- 3.Calderon-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception—Structural insights. Cold Spring Harb Perspect Biol. 2010;2:a005546. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weijers D, Wagner D. Transcriptional responses to the auxin hormone. Annu Rev Plant Biol. 2016;67:539–574. doi: 10.1146/annurev-arplant-043015-112122. [DOI] [PubMed] [Google Scholar]
- 5.Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calderón Villalobos LIA, et al. A combinatorial TIR1/AFB-Aux/IAA co-receptor system for differential sensing of auxin. Nat Chem Biol. 2012;8:477–485. doi: 10.1038/nchembio.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss BL, et al. Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol. 2015;169:803–813. doi: 10.1104/pp.15.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downes B, Vierstra RD. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans. 2005;33:393–399. doi: 10.1042/BST0330393. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelley DR, Estelle M. Ubiquitin-mediated control of plant hormone signaling. Plant Physiol. 2012;160:47–55. doi: 10.1104/pp.112.200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mergner J, Schwechheimer C. The NEDD8 modification pathway in plants. Front Plant Sci. 2014;5:103. doi: 10.3389/fpls.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 13.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 14.Leyser HM, et al. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature. 1993;364:161–164. doi: 10.1038/364161a0. [DOI] [PubMed] [Google Scholar]
- 15.Timpte C, Lincoln C, Pickett FB, Turner J, Estelle M. The AXR1 and AUX1 genes of Arabidopsis function in separate auxin-response pathways. Plant J. 1995;8:561–569. doi: 10.1046/j.1365-313x.1995.8040561.x. [DOI] [PubMed] [Google Scholar]
- 16.Schwechheimer C, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292:1379–1382. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 17.Tiryaki I, Staswick PE. An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol. 2002;130:887–894. doi: 10.1104/pp.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J. SIR1, an upstream component in auxin signaling identified by chemical genetics. Science. 2003;301:1107–1110. doi: 10.1126/science.1084161. [DOI] [PubMed] [Google Scholar]
- 19.Christian M, Hannah WB, Lüthen H, Jones AM. Identification of auxins by a chemical genomics approach. J Exp Bot. 2008;59:2757–2767. doi: 10.1093/jxb/ern133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savaldi-Goldstein S, et al. New auxin analogs with growth-promoting effects in intact plants reveal a chemical strategy to improve hormone delivery. Proc Natl Acad Sci USA. 2008;105:15190–15195. doi: 10.1073/pnas.0806324105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ettmayer P, Amidon GL, Clement B, Testa B. Lessons learned from marketed and investigational prodrugs. J Med Chem. 2004;47:2393–2404. doi: 10.1021/jm0303812. [DOI] [PubMed] [Google Scholar]
- 22.Aibibuli Z, Wang Y, Tu H, Huang X, Zhang A. Facile synthesis and herbicidal evaluation of 4H-3,1-benzoxazin-4-ones and 3H-quinazolin-4-ones with 2-phenoxymethyl substituents. Molecules. 2012;17:3181–3201. doi: 10.3390/molecules17033181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 25.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi K, et al. Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol. 2012;7:590–598. doi: 10.1021/cb200404c. [DOI] [PubMed] [Google Scholar]
- 27.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, et al. The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1) Plant J. 2004;40:772–782. doi: 10.1111/j.1365-313X.2004.02254.x. [DOI] [PubMed] [Google Scholar]
- 29.Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vain T, et al. 2019 RNAseq analysis of Arabidopsis cell suspension culture treated with IAA, RN3 and RN4. European Nucleotide Archive. Available at www.ebi.ac.uk/ena/data/view/PRJEB31496. Deposited March 1, 2019.
- 31.Goh T, Joi S, Mimura T, Fukaki H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development. 2012;139:883–893. doi: 10.1242/dev.071928. [DOI] [PubMed] [Google Scholar]
- 32.De Rybel B, et al. Plant development. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 33.Vilches-Barro A, Maizel A. Talking through walls: Mechanisms of lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol. 2015;23:31–38. doi: 10.1016/j.pbi.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 34.Benjamins R, Ampudia CS, Hooykaas PJ, Offringa R. PINOID-mediated signaling involves calcium-binding proteins. Plant Physiol. 2003;132:1623–1630. doi: 10.1104/pp.103.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buer CS, Djordjevic MA. Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot. 2009;60:751–763. doi: 10.1093/jxb/ern323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis DR, et al. Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 2011;156:144–164. doi: 10.1104/pp.111.172502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krishnaswamy S, Verma S, Rahman MH, Kav NNV. Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol. 2011;75:107–127. doi: 10.1007/s11103-010-9711-7. [DOI] [PubMed] [Google Scholar]
- 38.Chen J-H, et al. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2012;158:340–351. doi: 10.1104/pp.111.181875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang H-W, et al. A glutathione S-transferase regulated by light and hormones participates in the modulation of Arabidopsis seedling development. Plant Physiol. 2010;154:1646–1658. doi: 10.1104/pp.110.159152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin R, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, et al. BREVIS RADIX is involved in cytokinin-mediated inhibition of lateral root initiation in Arabidopsis. Planta. 2009;229:593–603. doi: 10.1007/s00425-008-0854-6. [DOI] [PubMed] [Google Scholar]
- 42.Brunoud G, et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 43.De Rybel B, et al. A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr Biol. 2010;20:1697–1706. doi: 10.1016/j.cub.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 44.Nagpal P, et al. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 2000;123:563–574. doi: 10.1104/pp.123.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian Q, Reed JW. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development. 1999;126:711–721. doi: 10.1242/dev.126.4.711. [DOI] [PubMed] [Google Scholar]
- 46.Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H. Multiple AUX/IAA-ARF modules regulate lateral root formation: The role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci. 2012;367:1461–1468. doi: 10.1098/rstb.2011.0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 48.Abbas M, Alabadí D, Blázquez MA. Differential growth at the apical hook: All roads lead to auxin. Front Plant Sci. 2013;4:441. doi: 10.3389/fpls.2013.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Raz V, Ecker JR. Regulation of differential growth in the apical hook of Arabidopsis. Development. 1999;126:3661–3668. doi: 10.1242/dev.126.16.3661. [DOI] [PubMed] [Google Scholar]
- 50.Zádníková P, et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development. 2010;137:607–617. doi: 10.1242/dev.041277. [DOI] [PubMed] [Google Scholar]
- 51.Vain T, et al. 2019 DNA sequencing for RN4-resistant Arabidopsis thaliana EMS mutant hookback1 (hkb1). European Nucleotide Archive. Available at www.ebi.ac.uk/ena/data/view/PRJEB21529. Deposited June 27, 2017.
- 52.Hurtado L, Farrona S, Reyes JC. The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol Biol. 2006;62:291–304. doi: 10.1007/s11103-006-9021-2. [DOI] [PubMed] [Google Scholar]
- 53.Tang X, et al. The Arabidopsis BRAHMA chromatin-remodeling ATPase is involved in repression of seed maturation genes in leaves. Plant Physiol. 2008;147:1143–1157. doi: 10.1104/pp.108.121996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farrona S, Hurtado L, Bowman JL, Reyes JC. The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development. 2004;131:4965–4975. doi: 10.1242/dev.01363. [DOI] [PubMed] [Google Scholar]
- 55.Wang R, Estelle M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salehin M, Bagchi R, Estelle M. SCFTIR1/AFB-based auxin perception: Mechanism and role in plant growth and development. Plant Cell. 2015;27:9–19. doi: 10.1105/tpc.114.133744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Havens KA, et al. A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol. 2012;160:135–142. doi: 10.1104/pp.112.202184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lavenus J, et al. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 59.Jönsson Å. Chemical structure and growth activity of auxins and antiauxins. In: Ruhland W, editor. Encyclopedia of Plant Physiology. Springer; Berlin: 1961. pp. 959–1006. [Google Scholar]
- 60.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 61.Ma Q, Grones P, Robert S. Auxin signaling: A big question to be addressed by small molecules. J Exp Bot. 2018;69:313–328. doi: 10.1093/jxb/erx375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maher EP, Martindale SJB. Mutants of Arabidopsis thaliana with altered responses to auxins and gravity. Biochem Genet. 1980;18:1041–1053. doi: 10.1007/BF00484337. [DOI] [PubMed] [Google Scholar]
- 63.Estelle MA, Somerville C. Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. MGG Mol Gen Genet. 1987;206:200–206. [Google Scholar]
- 64.Woodward AW, Bartel B. Auxin: Regulation, action, and interaction. Ann Bot. 2005;95:707–735. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walsh TA, et al. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 2006;142:542–552. doi: 10.1104/pp.106.085969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Prigge MJ, et al. The Arabidopsis auxin receptor F-Box proteins AFB4 and AFB5 are required for response to the synthetic auxin picloram. G3 (Bethesda) 2016;6:1383–1390. doi: 10.1534/g3.115.025585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.De Smet I, et al. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:2705–2710. doi: 10.1073/pnas.0915001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu MF, et al. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. eLife. 2015;4:e09269. doi: 10.7554/eLife.09269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rigal A, Ma Q, Robert S. Unraveling plant hormone signaling through the use of small molecules. Front Plant Sci. 2014;5:373. doi: 10.3389/fpls.2014.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park S-Y, et al. Agrochemical control of plant water use using engineered abscisic acid receptors. Nature. 2015;520:545–548. doi: 10.1038/nature14123. [DOI] [PubMed] [Google Scholar]
- 71.Grosdidier A, Zoete V, Michielin O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 2011;39:W270–W277. doi: 10.1093/nar/gkr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grosdidier A, Zoete V, Michielin O. Fast docking using the CHARMM force field with EADock DSS. J Comput Chem. 2011;32:2149–2159. doi: 10.1002/jcc.21797. [DOI] [PubMed] [Google Scholar]
- 73.Yu H, et al. Mutations in the TIR1 auxin receptor that increase affinity for auxin/indole-3-acetic acid proteins result in auxin hypersensitivity. Plant Physiol. 2013;162:295–303. doi: 10.1104/pp.113.215582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gilkerson J, et al. Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics. 2009;181:945–963. doi: 10.1534/genetics.108.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fülöp K, et al. The Medicago CDKC;1-CYCLINT;1 kinase complex phosphorylates the carboxy-terminal domain of RNA polymerase II and promotes transcription. Plant J. 2005;42:810–820. doi: 10.1111/j.1365-313X.2005.02421.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.