Significance
Intracellular transport is driven by molecular motors that carry cargoes along cytoskeletal tracks. Molecular motors, such as kinesin-1, contain two motor domains so they can walk processively (take many steps per encounter) along a microtubule track. The two motor domains alternate their catalytic activities so that one of them is always attached to the track. Here we show that kinesins with one motor domain are not processive as individuals but can work collectively to drive continuous transport in cells. However, their transport is most efficient when the motor-to-cargo distance is short and the cargo imposes little load on the motor. These results lend insight into the minimal requirements for kinesin transport and the synergies gained through teamwork and coordination.
Keywords: molecular motor, kinesin, microtubule, intracellular transport, monomeric motor
Abstract
Kinesin motor proteins that drive intracellular transport share an overall architecture of two motor domain-containing subunits that dimerize through a coiled-coil stalk. Dimerization allows kinesins to be processive motors, taking many steps along the microtubule track before detaching. However, whether dimerization is required for intracellular transport remains unknown. Here, we address this issue using a combination of in vitro and cellular assays to directly compare dimeric motors across the kinesin-1, -2, and -3 families to their minimal monomeric forms. Surprisingly, we find that monomeric motors are able to work in teams to drive peroxisome dispersion in cells. However, peroxisome transport requires minimal force output, and we find that most monomeric motors are unable to disperse the Golgi complex, a high-load cargo. Strikingly, monomeric versions of the kinesin-2 family motors KIF3A and KIF3B are able to drive Golgi dispersion in cells, and teams of monomeric KIF3B motors can generate over 8 pN of force in an optical trap. We find that intracellular transport and force output by monomeric motors, but not dimeric motors, are significantly decreased by the addition of longer and more flexible motor-to-cargo linkers. Together, these results suggest that dimerization of kinesin motors is not required for intracellular transport; however, it enables motor-to-motor coordination and high force generation regardless of motor-to-cargo distance. Dimerization of kinesin motors is thus critical for cellular events that require an ability to generate or withstand high forces.
Cytoskeletal motor proteins drive the directional transport of cellular cargoes along actin or microtubule filaments. Defects in motor protein function result in impaired transport and are linked to numerous diseases, including neurodegeneration and cancer (1, 2). Transport kinesins such as kinesin-1, the founding member of the kinesin superfamily, generally dimerize through a coiled-coil stalk and thus have two motor domains for ATP hydrolysis and microtubule binding. The two motor domains undergo alternating (out-of-phase) ATPase cycles, thereby ensuring that one motor domain remains bound to the microtubule as the other takes a step forward (3). This enables individual dimeric kinesin-1 motors to be processive and maintain their interaction with the microtubule track for hundreds of catalytic cycles. Monomeric kinesin-1 motors generated by truncation of the coiled-coil stalk (4–8) or deletion of one of the motor domains (9) are not processive as single motors. Therefore, dimerization is required for processive motility of individual motors.
The relationship between dimerization, processivity, and a motor’s ability to drive cargo transport has been more difficult to establish. Leibler and Huse (10) provided a theoretical framework for how a motor’s interaction with the microtubule affects its ability to cooperate in teams. Processive motors such as kinesin-1 work as “porters” and can drive long-range transport alone or in small groups because the alternating ATPase cycles ensure that one motor remains bound to the track (high duty ratio). Nonprocessive motors, such as myosin-2 and axonemal dynein motors, must work in large ensembles because individual motors spend most of their time detached from the track (low duty ratio). Like “rowers” in a boat, individual nonprocessive motors interact only transiently with the track but collectively can generate force and large movements. Consistent with this framework, large ensembles of kinesin monomers can glide a microtubule or transport a bead, albeit at lower speeds and forces than their dimeric forms (4–8, 11, 12). In these studies, the rigid cargo enabled individual motors to communicate with each other and work collectively.
The ability of monomeric motors to collectively transport cargoes when attached to a lipid bilayer is less studied. Motors attached to a lipid bilayer are only weakly coupled to each other, but theoretical studies have predicted cooperative effects from unequal load sharing of Brownian motors transporting a vesicle (13, 14). Indeed, a recent study showed that single-headed versions of the kinesin-3 motor KIF1A attached to giant unilamellar vesicles are able to cluster at the leading edge of a tubule and drive its extraction from the vesicle (15). Theoretical work proposed that this motor’s unique ability to engage with the microtubule in a passive diffusive state enables it to cooperate effectively for transport of membrane-bound cargoes (16). Whether other kinesin motors can work in teams of monomers is unknown. Furthermore, the ability of monomeric kinesin motors to drive vesicle transport in a cellular environment has not been tested.
To determine whether kinesin monomers can work collectively to drive cargo transport in cells, we directly compared dimeric motors to artificial monomeric motors across the kinesin-1, -2, and -3 families in both in vitro and cellular assays. We find that surprisingly, the monomeric motors are able to drive the dispersion of peroxisomes to the cell periphery, indicating that dimerization and processive motility at the single-molecule level are not required for intracellular cargo transport by teams of motors. We further explore the mechanics of this cooperativity and find that in general, kinesin monomers are more efficient transporters if the motor-to-cargo distance is short and the cargo imposes minimal load on the motors. As the length of the stalk increases, monomers become less efficient, and dimerization becomes necessary to pull against load. Together, these results lend insight into the minimal requirements and mechanical modulators of collective kinesin cargo transport and synergies gained through teamwork and coordination. They may also shed light on why most kinesins evolved to function as dimers.
Results
Single-Headed KIF1A Motors Can Transport Membrane-Bound Cargo in Cells.
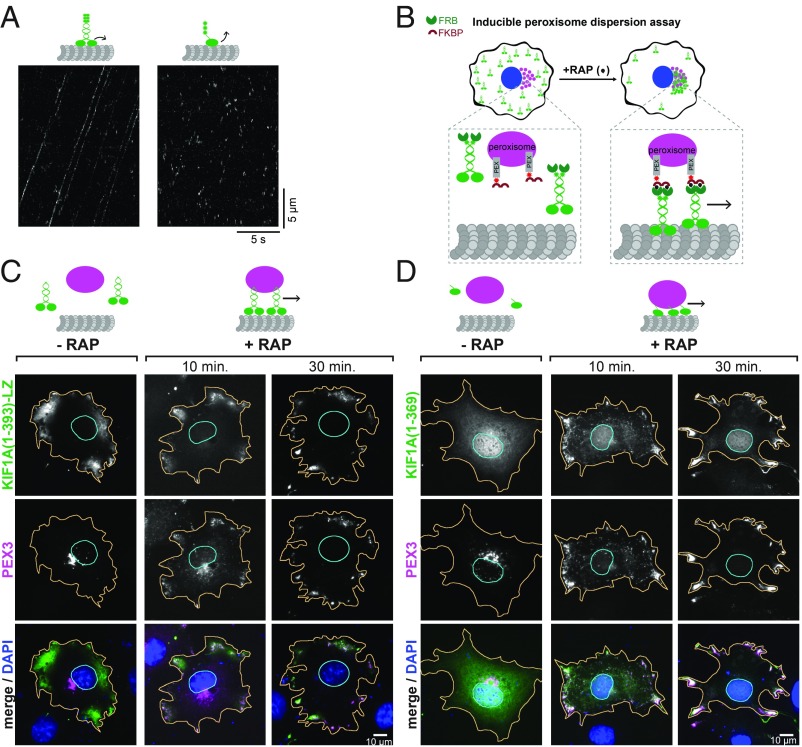
We first asked whether the kinesin-3 motor KIF1A, shown to work in teams to drive liposome transport in vitro (15), can work collectively to drive transport of a membrane-bound cargo in cells. We utilized a constitutively active, truncated version of KIF1A containing the motor domain, neck linker, neck coil, and the GCN4 leucine zipper [KIF1A(1-393)-LZ)] that is known to exist as a dimer, and we compared it to a monomeric version containing only the motor domain and neck linker [KIF1A(1-369)], based on our previous work (SI Appendix, Fig. S1) (17). We first verified the motility properties of individual motors by imaging 3xmCit-tagged motors by total internal reflection fluorescence (TIRF) microscopy. Dimeric KIF1A motors displayed long, processive, unidirectional runs at fast speeds (Fig. 1A, Left), consistent with previous studies (17), whereas monomeric KIF1A motors only had transient interactions with the microtubule and diffusive motion in both directions (Fig. 1A, Right), consistent with previous work (12, 18–20). Thus, dimerization is required for individual KIF1A motors to undergo robust processive motility along microtubules.
Fig. 1.
Single-headed KIF1A motors can cooperatively transport membrane-bound cargo in cells. (A) Single-molecule motility assays were carried out using TIRF microscopy to observe 3xmCit-tagged versions of the kinesin-3 motor KIF1A. Representative kymographs are shown for dimeric (Left) and monomeric (Right) KIF1A motors. Time is on the x axis and distance is on the y axis. [Scale bars, 5 s (x axis) and 5 μm (y axis).] (B) Schematic of the inducible peroxisome dispersion assay. A motor-mNG-FRB construct was coexpressed in COS-7 cells with PEX3-mRFP-FKBP. Motors were recruited to peroxisomes via rapamycin (RAP) addition and cells were fixed after 0, 10, or 30 min. Representative images at each time point after RAP addition are shown for (C) dimeric KIF1A and (D) monomeric KIF1A motors. (Scale bars, 10 μm.)
To test whether dimeric and monomeric KIF1A motors can work effectively in teams for cargo transport in cells, we utilized an artificial cargo trafficking assay in which the kinesin of interest is targeted to the peroxisome, and the subsequent redistribution of the peroxisome can be attributed to the transport capacity of the motor (Fig. 1B) (21). Peroxisomes are spherical membrane-bound organelles generally <250 nm in diameter and localized in the perinuclear region in COS-7 cells. Peroxisomes are relatively immotile (SI Appendix, Fig. S2A and Movie S1) (22, 23) and their dispersion requires <15-pN force generation by the recruited kinesin motors (24). This assay facilitates the analysis of motor behavior in a physiological environment where motors work in teams to transport membrane-enclosed cargoes.
We fused mNeonGreen (mNG) and the FK506 binding protein (FKBP) and rapamycin binding (FRB) domain to the C terminus of our dimeric and monomeric KIF1A motors. We coexpressed the tagged motors with a peroxisome-targeted mRFP-FKBP module (PEX3-mRFP-FKBP) in COS-7 cells. Addition of rapamycin induces the dimerization of FRB and FKBP, thereby rapidly recruiting the motor to the peroxisome surface (Fig. 1B). The cells were fixed at 0, 10, or 30 min of rapamycin treatment and the motor/cargo dispersion was observed by fluorescence microscopy (Fig. 1 C and D and SI Appendix, Fig. S2A). As expected, recruitment of dimeric KIF1A resulted in rapid redistribution of the peroxisomes to the cell periphery (Fig. 1C). To our surprise, we found that monomeric KIF1A motors were able to transport peroxisomes as well as the dimeric motor (Fig. 1D), despite lacking the single-motor processivity of the dimeric form. These results suggest that groups of monomeric KIF1A motors are able to work cooperatively while attached to a lipid bilayer to drive transport in a cellular environment. They also demonstrate that dimerization of kinesin motors is not required for cargo transport in cells.
Single-Headed Kinesin-1, -2, and -3 Motors Can Transport Membrane-Bound Cargo in Cells.
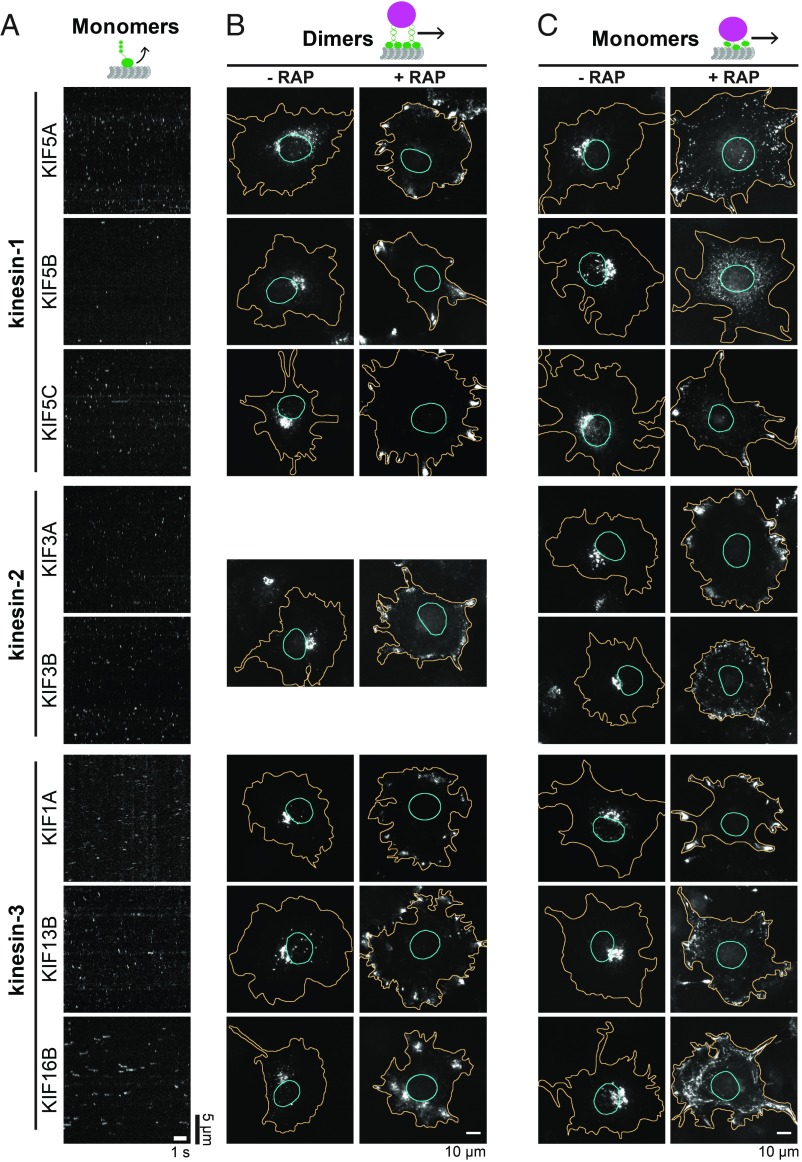
We next asked whether other kinesin motors can work as monomers to drive cargo transport in cells or whether this property is unique to KIF1A. A characteristic feature of kinesin-3 motors is the presence of a positively charged loop on the surface of the motor domain that enables short periods of diffusive motion along the negatively charged microtubule surface (Fig. 1A) (19, 20). Indeed, like KIF1A, truncated and monomeric versions of the kinesin-3 motors KIF13B and KIF16B displayed transient and diffusive interactions with microtubules (Fig. 2A). In contrast, artificial monomeric motors (SI Appendix, Fig. S1) of the kinesin-1 (KIF5A, KIF5B, KIF5C) and kinesin-2 (KIF3A, KIF3B, KIF17) families displayed rapid binding and unbinding events with little to no diffusive motion (Fig. 2A). Theoretical studies have suggested that the diffusive state provides an enhanced cooperativity that can generate collective forces required for cargo transport (16). We thus predicted that monomeric kinesin-3 motors, but not monomeric kinesin-1 or kinesin-2 motors, could work in teams to drive cargo transport in cells.
Fig. 2.
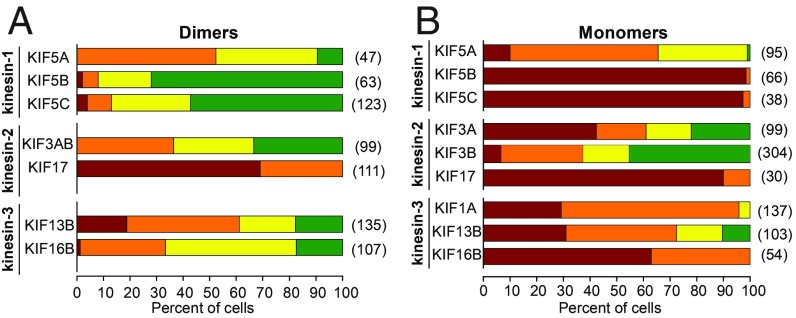
Single-headed kinesins can cooperate for peroxisome transport in cells. (A) Single-molecule motility assays of 3xmCit-tagged monomeric versions of motors from the kinesin-1, -2, and -3 families. Representative kymographs are shown. Time is on the x axis; distance is on the y axis. [Scale bars, 5 s (x axis) and 5 μm (y axis).] (B and C) Peroxisome dispersion assay. Representative images of COS-7 cells coexpressing PEX3-mRFP-FKBP and motor-mNG-FRB constructs of (B) dimeric or (C) monomeric versions of the indicated kinesin-1, kinesin-2, or kinesin-3 motors. Shown is the peroxisome distribution in the absence of RAP (Left) or 30 min after RAP addition (Right). See SI Appendix, Figs. S4–S6 for individual fluorescence channels. (Scale bars, 10 μm.)
To examine the ability of the dimeric and monomeric motors to drive peroxisome dispersion in cells, we tagged the dimeric and monomeric versions of each motor with mNG-FRB and coexpressed them in COS-7 cells with the PEX3-mRFP-FKBP construct. Motor recruitment to the peroxisome surface was acutely induced with rapamycin, and peroxisome dispersion was examined by fluorescence microscopy. Using the same components for chemically induced dimerization across all motors ensures that each motor is recruited to the peroxisome surface with the same affinity (SI Appendix, Fig. S3B). For the heterodimeric kinesin-2 motor KIF3AB, only the 3A subunit was tagged with mNG-FRB, and the recruitment of heterodimers was verified by colocalization of the KIF3B-TagBFP subunit. For simplicity and clarity, representative images of peroxisomes before and after 30-min rapamycin treatment are shown in Fig. 2 B and C, with individual fluorescence channels in SI Appendix, Figs. S4–S6.
As expected, dimeric versions of each kinesin motor were able to drive peroxisome dispersion (Fig. 2B and SI Appendix, Figs. S4–S6). Surprisingly, the monomeric motors were also able to drive peroxisome dispersion (Fig. 2C, SI Appendix, Figs. S4–S6, and Movie S2). These results indicate that the ability to diffuse along the microtubule is not required for motors in teams to drive cargo transport. Rather, the ability of kinesin motors to generate motion in cells requires only the catalytic core, which includes the force-generating elements [coverstrand and neck linker (25)].
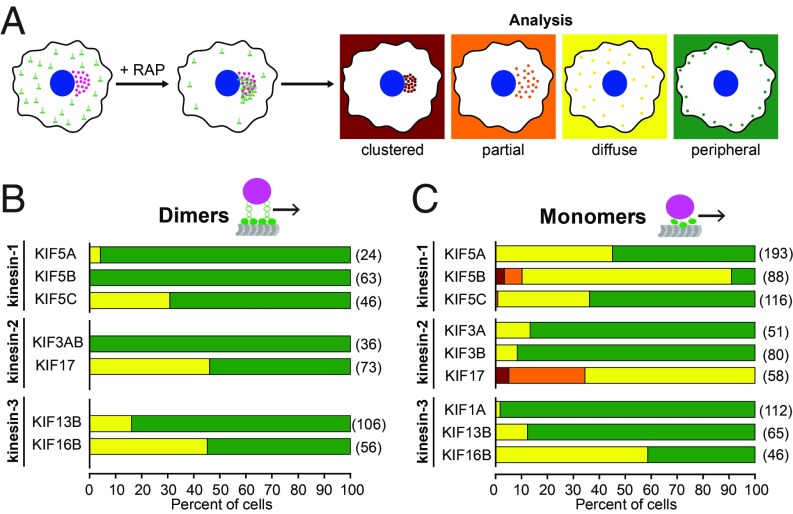
To quantify these results, we classified peroxisome localization into four categories: clustered, partially dispersed, diffuse, or peripherally dispersed (Fig. 3A). For the kinesin-1 motors, it is interesting that a dimeric KIF5B motor was most effective at full dispersion of the peroxisomes, whereas monomeric KIF5B was the least effective (Fig. 3 B and C). For the kinesin-2 motors, KIF17 was a less-effective motor than KIF3A/KIF3B in both the dimeric and monomeric states (Fig. 3 B and C). For the kinesin-3 motors, all motors were effective at peroxisome transport, although KIF16B appeared less effective in both the dimeric and monomeric states (Fig. 3 B and C). Together these results indicate that, despite the importance of dimerization for processive motion at the single-molecule level, dimerization is not required for kinesin motors to work effectively in teams to drive transport in cells.
Fig. 3.
Quantification of peroxisome dispersion. (A) Schematic of classification system. The peroxisome distribution in individual cells was classified as clustered (red), partially dispersed (orange), diffuse (yellow), or peripheral (green). (B and C) Quantification of the proportion of cells in each peroxisome distribution category for cells expressing (B) dimeric or (C) monomeric versions of the indicated kinesin motors. The numbers in parentheses to the right of each bar indicate the number of cells counted across three independent experiments.
Monomeric Motors Are Impaired at High-Load Cargo Transport in Cells.
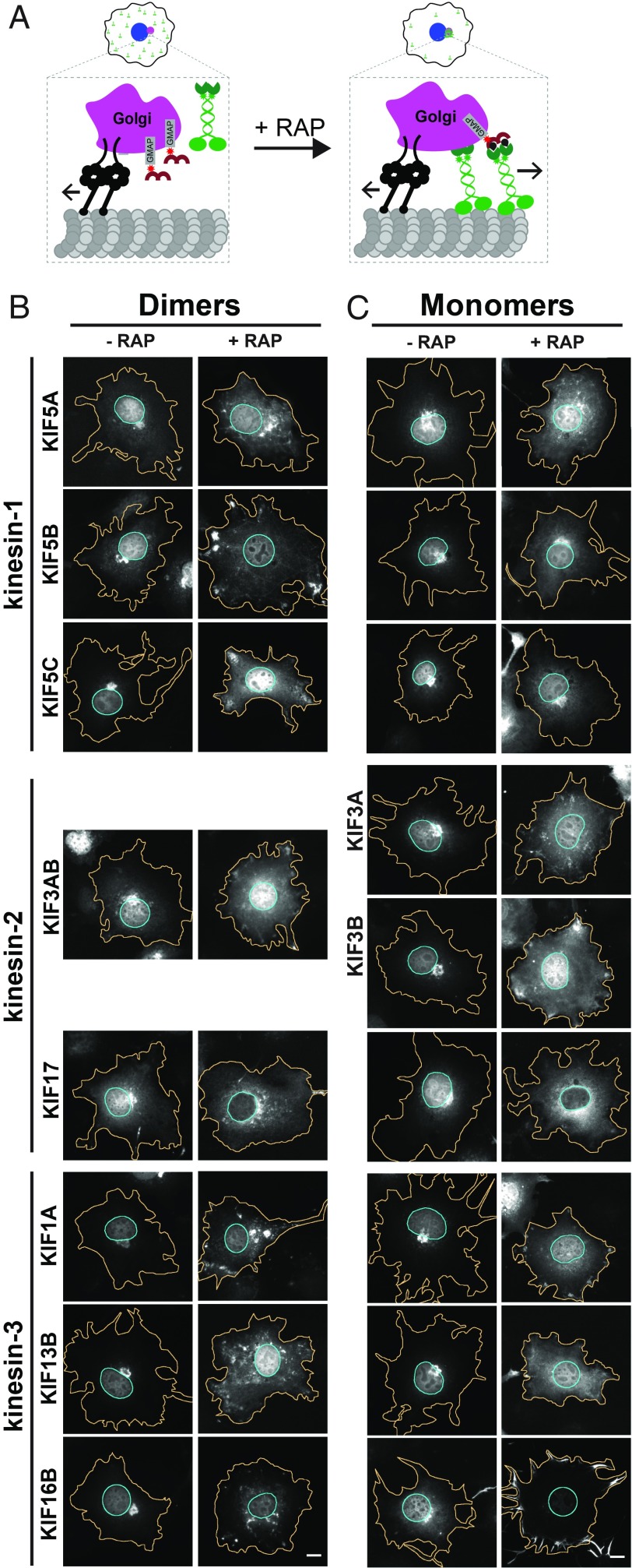
The fact that kinesin monomers are able to cooperatively transport peroxisomes in cells raises the question of why most kinesins exist as dimers. One potential advantage of being a dimer is that a dimeric motor can generate higher forces than monomeric motors working individually or in teams. We hypothesized that although monomer teams can generate sufficient force for transport of peroxisomes, these motor teams would be ineffective when challenged with a high-load cargo. To test this, we examined the ability of monomeric motors to drive dispersion of the Golgi complex in COS-7 cells. The Golgi is held in a tight cluster of stacked membranes in the perinuclear region of COS-7 cells by a combination of cytoplasmic dynein, myosin motors, and linker proteins (Movie S3) (26). Targeted kinesins must work against this opposing force to disperse the Golgi (Fig. 4A, SI Appendix, Fig. S2B, and Movie S4), with recent work suggesting that movement of the Golgi requires ∼200-pN force (27). We thus used the C-terminal region of GMAP210 to target the mRFP-FKBP module to the cis-Golgi membrane (28).
Fig. 4.
Motor dimerization facilitates transport of high-load cargoes in cells. (A) Schematic of the inducible Golgi dispersion assay. Targeted kinesins must work against the opposing force of endogenous proteins, for example, cytoplasmic dynein (black), that cluster the Golgi in the perinuclear region, making it a high-load cargo. Representative images of the Golgi distribution in COS-7 cells coexpressing GMAP210-mRFP-FKBP and motor-mNG-FRB constructs of (B) dimeric or (C) monomeric versions of the indicated kinesin-1, kinesin-2, or kinesin-3 motors. (Scale bars, 10 μm.) Individual fluorescence channels are displayed in SI Appendix, Figs. S7–S9.
Cells expressing a kinesin-mNG-FRB together with the Golgi-targeted GMAP210-mRFP-FKBP module were treated with rapamycin and then fixed and stained with an antibody against the Golgi marker Giantin to probe for dispersion of Golgi components (Fig. 4 B and C and SI Appendix, Figs. S7–S9). To directly compare Golgi and peroxisome dispersion, we classified Golgi dispersion using the same four phenotypes (Fig. 4A). Compared with peroxisome dispersion, there was much more variation both across and within families in terms of a motor’s ability to disperse the Golgi complex. In general, dimeric motors (Fig. 4B and SI Appendix, Figs. S7–S9) were better at dispersing the Golgi than their corresponding monomeric versions (Fig. 4C and SI Appendix, Figs. S7–S9). This supports the hypothesis that dimerization enables kinesin motors to generate higher forces necessary for the transport of high-load cargoes in a cellular context.
For kinesin-1 motors, we found that recruitment of dimeric KIF5B and KIF5C motors resulted in peripheral dispersion of the Golgi in the majority of cells (73% for KIF5B and 57% for KIF5C), whereas dimeric KIF5A motors were less effective (only 10% of cells achieved peripheral dispersion) (Fig. 5A). Interestingly, the monomeric motors showed the opposite trend; monomeric KIF5B and KIF5C could not drive Golgi dispersion (98% of cells contained clustered Golgi for KIF5B and 97% for KIF5C), whereas the monomeric KIF5A was relatively effective (Golgi remained clustered in only 11% of cells) (Fig. 5B).
Fig. 5.
Quantification of Golgi dispersion. The Golgi distribution in individual cells was classified as depicted in Fig. 3A: clustered (red), partially dispersed (orange), diffuse (yellow), and peripheral (green). Quantification of the proportion of cells in each Golgi distribution category for cells expressing (A) dimeric or (B) monomeric versions of the indicated kinesin motors. The numbers in parentheses to the right of each bar indicate the number of cells counted across three independent experiments.
For kinesin-2 motors, we found that the heterodimeric motor KIF3A/KIF3B was an effective motor for Golgi dispersion (32% of cells displayed peripheral Golgi dispersion), whereas dimeric KIF17 was completely ineffective (0% of cells had peripheral dispersion) (Fig. 5A). In fact, the majority of cells expressing KIF17 still had clustered Golgi despite the fact that the motor was well-targeted to the Golgi after rapamycin treatment (SI Appendix, Fig. S8). For the monomeric kinesin-2 motors, we were surprised to find that recruitment of either of the monomeric versions (KIF3A and KIF3B) of the heterodimeric motor resulted in Golgi dispersion in a significant population of the cells (23% of cells displayed peripheral dispersion for KIF3A and 42% for KIF3B) (Fig. 5B).
For kinesin-3 motors, all dimeric motors were capable of Golgi dispersion (peripheral dispersion in 18% cells of cells expressing KIF13B and 17% of cells expressing KIF16B) (Fig. 5A), whereas the monomeric versions were relatively inefficient at Golgi dispersion (30%, 30%, and 60% of cells still contained clustered Golgi for KIF1A, KIF13B, and KIF16B, respectively) (Fig. 5B). We had expected KIF1A to be effective as a monomer based on its ability to pull tubules out of lipid vesicles (15); however, KIF13B was the most effective kinesin-3 monomer in driving peripheral dispersion of the Golgi complex (10% of cells). Taken together, these results indicate that monomeric kinesin motors are generally ineffective at transport of a high-load cargo in a cellular environment. Thus, a benefit provided by dimerization is that the motor can generate sufficient forces required for transport of cellular cargoes.
The Length of KIF3B Monomers Modulates Their Cooperativity in Cells and in Vitro.
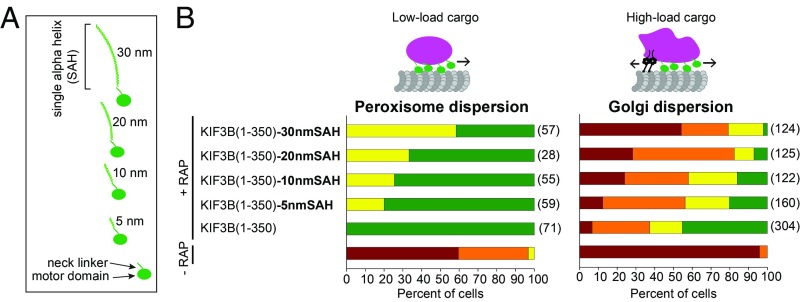
Kinesins are generally elongated molecules containing extensive nonmotor segments that contribute to oligomerization, cargo binding, and regulation of motor activity (29, 30). We thus asked how the addition of nonmotor elements would affect the ability of kinesin monomers to cooperate in intracellular cargo transport. We speculated that teams of monomers may only be able to “row” along the microtubule and generate force if there is a short distance between the motor and the cargo. To test this, we took advantage of the unique ability of KIF3B to function in Golgi dispersion and appended single α-helix (SAH) domains of varying length (5, 10, 20, or 30 nm) after the neck linker of the KIF3B monomer (Fig. 6A). SAHs are stable helical structures found in numerous proteins, including myosin motors, and their defined length and mechanical properties make them ideal for protein engineering applications (31). We tested the functionality of the extended motors in the peroxisome and Golgi dispersion assays. With both cargoes, we observed a robust trend: as the motor-to-cargo distance increased, the monomeric KIF3B motors became less efficient at working in teams to transport both peroxisomes and Golgi in cells (Fig. 6B).
Fig. 6.
Increasing the motor-to-cargo distance reduces transport by KIF3B monomers in a length-dependent manner. (A) Schematic of constructs. SAH sequences of known length (5, 10, 20, or 30 nm) were fused in-frame after the neck linker of the monomeric KIF3B(1-350) construct. (B) KIF3B motors with extended motor-to-cargo linkers were tested in the peroxisome (Left) and Golgi (Right) dispersion assays. The distribution of each organelle in the absence of rapamycin (−RAP) is shown in the bottom bar of each graph. The numbers in parentheses to the right of each bar indicate the number of cells counted across three independent experiments.
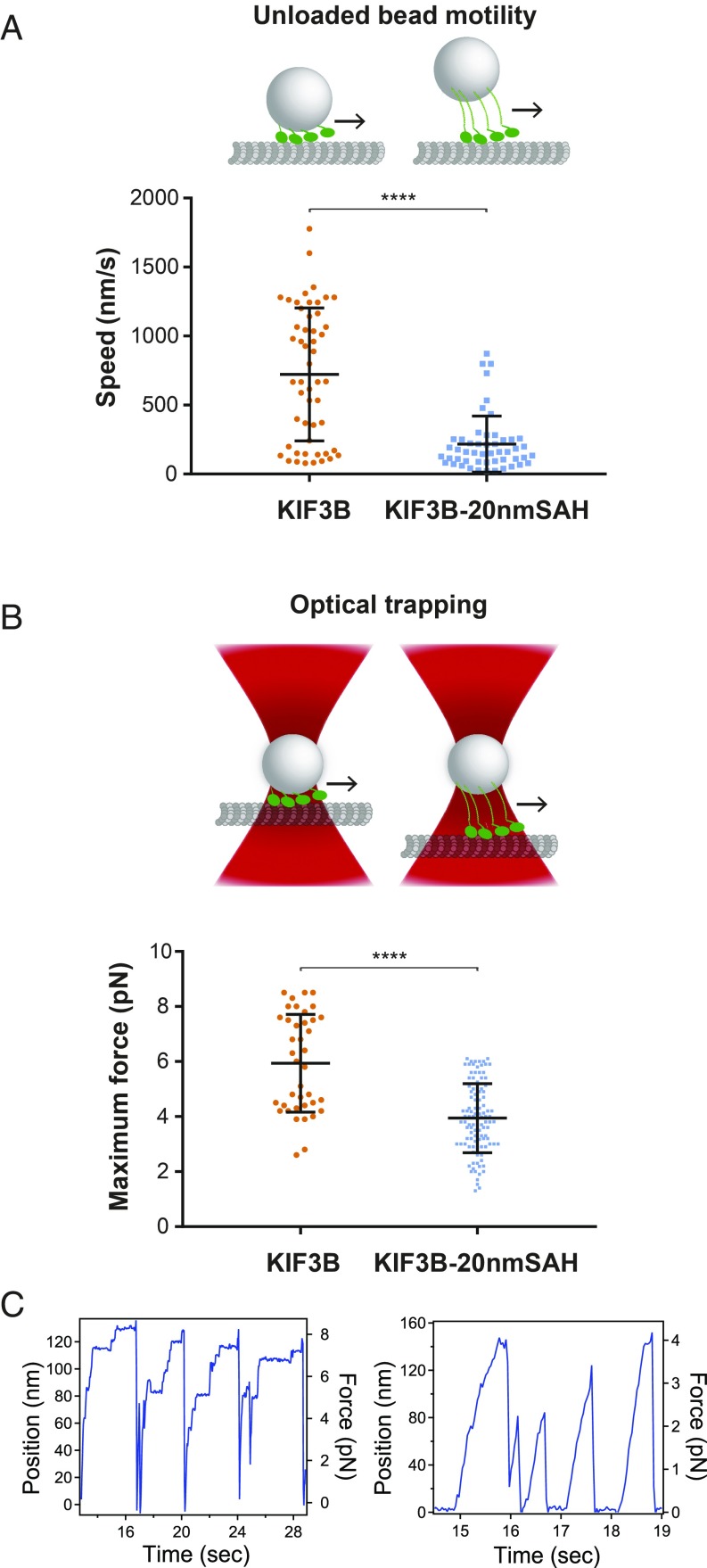
To gain a mechanistic understanding of the ability of short versus elongated monomers to cooperate, we bound monomeric, biotinylated KIF3B motors to streptavidin-coated beads and measured their speed (Fig. 7A) and force generation (Fig. 7 B and C) in a multimotor context. For these assays, we compared the minimal KIF3B monomer (no SAH) to the monomer with a 20-nm SAH (KIF3B-20nmSAH) because these showed significant differences in the peroxisome and Golgi dispersion assays (Fig. 6). Beads coated with multiple KIF3B and KIF3B-20nmSAH monomers showed consistent unidirectional motility along microtubules. The short KIF3B monomers drove bead motility with faster speeds (722.8 ± 482.1 nm/s) than the longer KIF3B-20nmSAH monomers (217.4 ± 203.9 nm/s) (Fig. 7A). To measure the force production of the KIF3B monomers, we used optical trapping and found that beads coated with short KIF3B monomers moved quickly out of the trap and stalled at 4–8 pN of force before returning to the center of the trap, whereas beads coated with longer KIF3B-20nmSAH monomers moved more slowly out of the trap and detached at lower forces before returning to the trap center (Fig. 7C). Strikingly, in some events, the short KIF3B monomers were able to withstand over 8 pN of force. Quantification of multiple events demonstrated that short KIF3B monomers detached from the microtubule track at an average of 6 pN, whereas longer KIF3B-20nmSAH monomers detached from the microtubule at an average of 4 pN (Fig. 7B). These results indicate that the impaired ability of the longer KIF3B-20nmSAH monomers to drive cargo transport in cells may be due, at least in part, to their reduced speed and force generation in a multimotor context.
Fig. 7.
A short motor-to-cargo linker enables KIF3B monomers to drive faster unloaded bead motility and generate higher forces. Monomeric KIF3B(1-350) motors containing no extension (KIF3B) or a 20-nm SAH extension (KIF3B-20nmSAH) were biotinylated, bound to 0.4-μm streptavidin beads, and tested for their ability to drive transport and generate force. (A, Upper) Schematic of unloaded bead motility assays. Fluorescent beads with bound biotinylated motors were imaged by TIRF microscopy and their speeds were measured by kymograph. (Lower) KIF3B monomers: average bead motility 723 ± 482 nm/s (SD), n = 52 events over three independent experiments; KIF3B-20nmSAH monomers, average bead motility 217 ± 204 nm/s (SD), n = 51 over three independent experiments. ****P < 0.0001 (two-sample Kolmogorov–Smirnov test). (B, Upper) Schematic of optical trapping assay. (Lower) KIF3B monomers average stall forces 5.9 ± 1.8 pN (SD), n = 39 events over three independent experiments; KIF3B-20nmSAH monomers 3.9 ± 1.3 pN (SD), n = 108 events over three independent experiments. ****P < 0.0001 (two-sample Kolmogorov–Smirnov test). (C) Example traces of optically trapped beads coated with KIF3B monomers (Left) or KIF3B-20nmSAH monomers (Right).
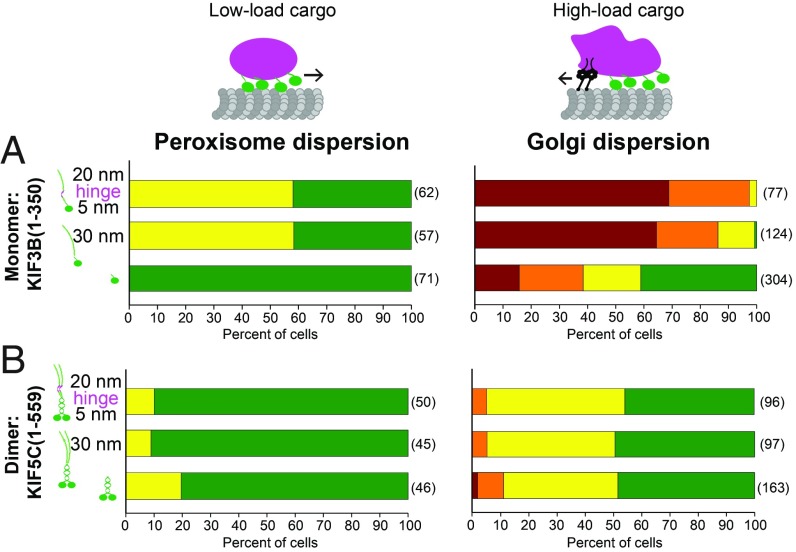
In our experiments, the SAH domains are assumed to act as inert spacers of increasing length; however, the longer SAH domains may additionally affect compliance as they are longer than the persistence length of 15 nm (31). To examine how the flexibility of the motor-to-cargo linker impacts the ability of motors in teams to cooperate during cellular transport, we linked two shorter SAHs via a flexible hinge segment (5nmSAH–hinge–20nmSAH) while maintaining the approximate length of the longest SAH (30nmSAH). This architecture mimics that of many kinesin motors that contain flexible hinge regions separating rigid coiled-coil segments. For the monomeric KIF3B motor, appendage of the 5nmSAH–hinge–20nmSAH segment resulted in a decrease in transport efficiency similar to that observed upon appendage of the uninterrupted 30nmSAH to the motor. This effect was observed in both the peroxisome and Golgi dispersion assays (Fig. 8A) and suggests that the ability of monomeric motors to cooperate effectively is primarily influenced by the distance from the cargo. This result is in contrast to the dimeric KIF5C motor where appendage of either the longer 30nmSAH or the longer and more flexible 5nmSAH–hinge–20nmSAH had no significant impact on the ability of the motor to drive peroxisome or Golgi dispersion in a cellular context (Fig. 8B).
Fig. 8.
Monomeric but not dimeric motors are sensitive to the length and flexibility of the motor-to-cargo linker. A 30nmSAH or 5nmSAH–hinge–20nmSAH motor-to-cargo linker was fused in-frame after the neck linker of (A) the monomeric KIF3B(1-350) construct or (B) the dimeric KIF5BC(1-559) construct and the motors were tested in the peroxisome (Left) and Golgi (Right) dispersion assays. The distribution of each organelle after 30 min of rapamycin (+RAP) treatment is shown in the bar graphs. The numbers in parentheses to the right of each bar indicate the number of cells counted across three independent experiments.
These results demonstrate that the ability of monomeric kinesins to work effectively in teams and transport cargo depends on the distance between the motor domain and the cargo. For kinesins known to be involved in transport of membrane-bound cargoes in cells, the presence of nonmotor segments for oligomerization and motor regulation results in an increased motor-to-cargo distance and thereby imposes a constraint on motor function that dimerization appears to resolve.
Discussion
It has long been thought that the ability of kinesin motors to dimerize for processive motility is critical for their ability to transport vesicles and other cargoes in cells. Here we demonstrate that kinesin monomers that are not processive as individual motors can carry out efficient, long-range transport in cells when grouped on the same cargo. While other studies have demonstrated that monomeric kinesins can generate processive motion when present at high densities on rigid surfaces, this study demonstrates continuous cargo transport by motors attached to a fluid membrane and pulling against a load in a cellular environment. We find that transport driven by monomeric kinesins is most efficient (i) under low-load conditions and (ii) when the motor-to-cargo distance is short.
Intracellular Transport Driven by Diverse Kinesin Monomers.
Monomeric versions of the kinesin-3 motor KIF1A can undergo processive motion as individual motors by a biased diffusion mechanism and can cooperate at high densities to drive the motion of rigid cargoes, such as a bead or glass surface (11, 12, 18, 19). The first hints that monomeric KIF1A motors could work effectively in teams when attached to a fluid lipid layer came from studies with giant unilamellar vesicles (15). Our work extends these findings to show that monomeric KIF1A can drive the dispersion of membrane-bound vesicles and organelles in a cellular environment (Fig. 1).
Furthermore, our work demonstrates that the ability of a monomeric kinesin to drive membrane transport in cells is not restricted to KIF1A. Indeed, we were surprised to find that all kinesin monomers that we tested could transport peroxisomes in cells, despite being nonprocessive as individual motors (Fig. 2). These results indicate that the ability of kinesins to transport a fluid cargo in cells requires only a catalytic motor domain that includes the force-generating elements of the coverstrand and neck linker. These minimal elements enable the motors to work as rowers where each member of the team briefly binds to the track, generates an impulse of force, and then releases from the track. Strikingly, the ability to drive transport does not appear to require specific properties of the catalytic motor domain, because diverse monomers across the kinesin-1, -2, and -3 families were all able to transport peroxisomes effectively.
Previous theoretical and experimental work on KIF1A proposed that the ability of monomeric motors to cooperate efficiently for transport of membrane-bound cargoes requires the motor to be able to interact with the microtubule in a diffusive state (13–16). Our data rule out this possibility as monomeric kinesin-1 and kinesin-2 motors that do not diffuse on the microtubule lattice are still capable of working in teams to drive peroxisome dispersion (Fig. 2). Rather, our data suggest that a weak and transient interaction with the microtubule is sufficient for nonprocessive motors to carry out long-range transport in teams. This is consistent with previous work showing that weak motor–microtubule interactions are sufficient for nonprocessive axonemal dynein motors to move microtubules (32). Furthermore, some dimeric motors display little to no processivity as individual motors but can generate continuous motion when working in teams (33–36). Our results extend these findings and suggest that nonprocessive motors, whether monomeric or dimeric, can drive continuous cargo transport as long as high-force generation is not required.
One feature that influences the ability of monomeric motors to work collectively in cells is the distance of the motor–cargo linkage. We found that as the motor-to-cargo linker length increased, the ability of the KIF3B monomer to transport a cargo decreased, particularly in high-force situations (Figs. 6 and 8). Monomeric motors with a short motor-to-cargo distance are not known to exist in the kinesin superfamily but can be found in the myosin superfamily where myosin I motors generate collective force and motility when attached to a lipid bilayer (37, 38). A short motor-to-cargo distance enables the motors to work in a confined environment where their spatial proximity to the track allows them to quickly rebind and engage in productive rowing. A shorter and less compliant motor-to-cargo linkage also more effectively couples motors by communicating their force and position to the cargo and other team members.
High-Load Transport Driven by Monomeric KIF3A and KIF3B Motors.
Although all monomers could transport peroxisomes, only a few were capable of transporting the Golgi, a high-load cargo. The unique ability of KIF3A and KIF3B (kinesin-2) monomers to drive transport of a high-load cargo in cells was striking (Figs. 4 and 5). That KIF3A and KIF3B are uniquely well-suited to transport high-load cargoes even as monomers is particularly interesting given their unique assembly as a heterodimeric KIF3A/KIF3B motor in the native molecule. Insights about these monomeric motors could inform studies seeking to explain the functional significance of the heterodimeric KIF3A/KIF3B architecture.
We were also surprised at the magnitude of force production by KIF3B when attached to beads and manipulated in an optical trap in vitro. We found that multiple KIF3B monomers could withstand up to 8 pN of force (Fig. 7). In contrast, multiple kinesin-1 KIF5B or kinesin-3 Unc-104 monomers are capable of withstanding only 2–3 pN of force (7, 11, 12) and we find that monomeric kinesin-1 and kinesin-3 motors are unable to transport high-load cargo in cells (Figs. 4 and 5). The molecular mechanisms by which teams of KIF3A and KIF3B monomers are able to generate such high forces are presently unclear and require further study.
Recent work on dimeric kinesin-2 motors (native KIF3A/KIF3B or engineered KIF3A/KIF3A or KIF3B/KIF3B) has revealed molecular properties that may shed light on how their monomeric versions are uniquely suited to work in teams and drive high-load transport in cells. The dimeric motors were found to rapidly unbind from the microtubule track in response to opposing force (39–41). Similarly, we find that monomeric versions of KIF3B detach rather than stall at limiting forces (Fig. 7C), a property that could reduce frictional interference between microtubule-bound motors. The dimeric kinesin-2 motors were also found to have faster reattachment rates compared with kinesin-1 (42), a property that may help to maintain a minimum number of engaged motors (43, 44). Further work is need to determine whether these molecular properties can explain the ability of the monomeric KIF3A and KIF3B motors to drive high-load transport in cells.
The Advantages of Being a Dimer.
We have shown that although dimerization is required for processive motion of individual kinesin motors, it is not required for teams of motors to drive continuous movement of membrane-bound cargoes in cells. Why then do most kinesins exist as dimers? Dimerization permits tight motor-to-motor coupling, which has several advantages for a molecular motor. First, tight mechanical coupling enables two motor domains to coordinate their kinetic cycles and ensures that one motor domain is attached to the microtubule track during force generation (45–50). For monomeric motors, a tight motor-to-motor coupling requires mechanical coupling through a rigid cargo (4–8, 11, 12) or perhaps a membrane with decreased fluidity (51, 52).
Tight motor-to-motor coupling also permits an increase in the motor-to-cargo distance while maintaining the ability to generate force and undergo processive motility. Our data demonstrate that monomeric motors become less-efficient transporters as the motor-to-cargo distance increases, especially under high-load conditions (Figs. 6 and 8), whereas dimeric motors are not influenced by motor-to-cargo distance (Fig. 8). These results agree with recent work demonstrating that an increased linker length has minimal effect on transport by dimeric kinesin-1 or kinesin-7 motors attached to beads (53) but contrast with previous work carried out with dimeric motors attached to a glass surface in a microtubule gliding assay (54, 55). We thus suggest that kinesin motors largely evolved a dimeric state to incorporate nonmotor domains for oligomerization and regulation while maintaining the ability to drive continuous, high-load cargo transport via motor-to-motor coupling.
Dimerization is also advantageous for motor proteins in that it provides a built-in partner and thus permits motors to work effectively at limiting concentrations. In contrast, monomeric motors require high concentrations to drive transport (15). Although collective transport by both dimeric and monomeric motors could be enhanced by generating locally high concentrations of motors on membranes, for example by dynamic clustering in response to load (13, 14, 56, 57) or by sorting into membrane microdomains (58), we note that close proximity of monomeric motors on the cargo surface can drive dimerization and thereby generate processive motors for cargo transport. Indeed, a cargo-induced monomer-to-dimer transition has been suggested to regulate members of both the kinesin and myosin superfamilies. For kinesin motors, cargo-induced dimerization was first suggested for the kinesin-3 family members murine KIF1A and its Caenorhabditis elegans homolog Unc-104 (12, 59–61) and has subsequently been demonstrated for additional members of the kinesin-3 family (17, 62–67). For myosin motors, cargo-mediated dimerization has been demonstrated to regulate the motility of members of the myosin-6, myosin-7, and myosin-10 families (68–74).
Surprising Features of Multimotor Transport by Dimeric Kinesins in Cells.
Our results highlight the complex interplay of features that determines the efficiency of motors working in teams in a cellular environment. We note that motors that are strong individually are not always the most effective motors under high-load conditions. For example, we were surprised to find that the kinesin-2 motor KIF17 could not drive Golgi dispersion in teams. It is unclear why KIF17 is ineffective in this context because individual KIF17 motors are relatively fast and processive motors and can step against a 6-pN hindering load (75, 76). We also note that the motors most effective as dimers are not the most effective as monomers. For example, we were surprised to find that although kinesin-1 dimers, the canonical porters, are able to drive Golgi dispersion better than kinesin-2 dimers, the kinesin-1 monomers (KIF5A, KIF5B, KIF5C) are less effective at Golgi dispersion than kinesin-2 monomers (KIF3A, KIF3B). This suggests that the stalk and other nonmotor domains play important and possibly deterministic roles in the force-generating capability of motors. Similarly, from a dynamics perspective, the kinetic signature of each motor domain likely impacts its ability to function in teams. Further work investigating the motility of motor–cargo ensembles in live cells is required to unravel the contributions of force generation, motor number, and motor kinetics to transport by teams of motors on membrane-bound cargoes.
Materials and Methods
Plasmids.
The amino acid sequences of the dimeric and monomeric kinesins and their design are described in SI Appendix, Fig. S1. Motors were tagged with three tandem mCitrine (mCit) fluorescent proteins for single-molecule imaging, with tandem mNG and FRB sequences for the cargo dispersion assays in cells, or with the AviTag (aa sequence GLNDIFEAQKIEWHE) for biotinylation and attachment to beads in motility and optical trap assays. Plasmids encoding FKBP and FRB were obtained from ARIAD Pharmaceuticals and are now available from Takara Bio as DmrA and DmrC, respectively. The plasmid encoding bacterial biotin ligase BirA tagged with HA (HA-BirA) and the peroxisome-targeting construct PEX3-mRFP-2xFKBP were gifts from C. Hoogenraad, Utrecht University, The Netherlands (21). Plasmids encoding mNG were obtained from Allele Biotechnology. DNA fragments encoding SAH domains were amplified by PCR cloning to obtain the 5-nm helix from Homo sapiens translation initiation factor IF-2, 10-nm helix from Sus scrofa myosin VI medial tail, 20-nm helix from S. cerevisiae mannosyltransferase MNN4, and 30-nm helix from Trichomonas vaginalis Kelch-motif family protein (77). For the 5nmSAH–hinge–20nmSAH construct, the SAH sequences were separated by the hinge 1 from KIF3B (amino acid sequence SIGRRKRREKRREGGGSGGGGEEEEEEGEEGEEDGDDKD). All fusion proteins were expressed under the control of the cytomegalovirus promoter in the EGFP-N1 vector (Takara Bio); this vector also contains an SV40 origin for replication in mammalian cells and a kanamycin-resistance cassette for amplification in Escherichia coli. All plasmids were verified by DNA sequencing.
Cell Culture and Extract Preparation.
COS-7 cells (African green monkey kidney fibroblasts; American Type Culture Collection) were cultured in Dulbecco’s Modified Eagle Medium (Gibco) + 10% (vol/vol) Fetal Clone III (HyClone) + 2 mM GlutaMAX (l-alanyl-l-glutamine dipeptide in 0.85% NaCl; Gibco) at 37 °C with 5% CO2. Cells were seeded and transfected 24 h later at a density of ∼60–80% confluent with plasmids encoding motor-3xmCit, TransIT-LT1 transfection reagent (Mirus), and Opti-MEM Reduced Serum Medium (Gibco). Twenty-four hours after transfection, cells were harvested with 0.05% Trypsin-EDTA (Gibco) and centrifuged 3 min at 3,000 × g at 4 °C. The pellet was washed once with PBS, centrifuged 3 min at 3,000 × g at 4 °C, and resuspended in ice-cold lysis buffer [25 mM Hepes/KOH, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, and 1% (vol/vol) Triton X-100, pH 7.4] freshly supplemented with 1 mM ATP, 1 mM phenylmethylsulfonyl fluoride, and 1% (vol/vol) protease inhibitor mixture (P8340; Sigma-Aldrich). The lysate was clarified by centrifuging for 10 min at 20,000 × g at 4 °C and aliquots of motor-containing supernatant were snap-frozen in liquid nitrogen and stored at −80 °C. Cell lysates containing biotinylated motors tagged with the AviTag were obtained the same way except cells were cotransfected with a plasmid encoding HA-BirA, 0.05% digitonin was substituted for 1% Triton X-100, and the samples were incubated 5 min on ice before centrifugation.
Inducible Cargo Dispersion Assays.
COS-7 cells were seeded onto glass coverslips and cotransfected with plasmids encoding motor-mNG-FRB and GMAP210-mRFP-2xFKBP for Golgi dispersion assays or with motor–mNG-FRB and PEX3-mRFP-2xFKBP for peroxisome dispersion assays. Approximately 14 h after transfection, rapamycin (Calbiochem) was added to each well at a final concentration of 44 nM to induce motor recruitment to the Golgi or peroxisome surface. Addition of an equivalent volume of ethanol vehicle was used as a control. After incubation for 10 or 30 min, cells were washed with PBS and then fixed with 3.7% formaldehyde in PBS, quenched with 50 mM NH4Cl in PBS, permeabilized with 0.2% Triton X-100 in PBS, and blocked with blocking buffer (0.2% fish skin gelatin in PBS). Primary and secondary antibodies were applied in blocking buffer for 1 h at room temperature in the dark. Commercial antibodies used were polyclonal antibody against the cis-Golgi marker giantin (1:1,200 PRB-114C; Covance) and goat anti-rabbit Alexa680-labeled secondary antibody (1:500; Jackson ImmunoResearch). Nuclei were stained with DAPI (final concentration 10.9 µM) and cover glasses were mounted in ProlongGold (Invitrogen).
Images were collected on an inverted epifluorescence microscope (Nikon TE2000E) with a 40×, 0.75 NA objective and Photometrics CoolSnapHQ camera. Only cells expressing low to medium levels of the motor-mNG-FRB construct were selected for imaging and included in the quantification (SI Appendix, Fig. S3A). No dispersion of either peroxisomes or Golgi was observed during control experiments performed in the absence of rapamycin or absence of motor expression (SI Appendix, Fig. S2C). We also verified that the ability of monomeric motors to drive cargo transport in cells was unrelated to the tandem FKBP as identical dispersion phenotypes were obtained for cells expressing motor-mNG-FRB + PEX3-mRFP-FKBP as for motor-mNG-FRB + PEX3-mRFP-2xFKBP. The dispersion phenotype in each cell was scored as clustered, partial dispersion, diffuse, or peripheral dispersion based on the location of the colocalized motor and PEX3 signals (for peroxisome dispersion) or the motor, GMAP210, and giantin signals (for the Golgi dispersion). The scoring system was found to correspond nearly completely to an unbiased distance-based analysis using a custom ImageJ plugin and thus the scoring system was used across all experiments to compare across motor constructs and conditions.
To quantify motor and PEX3 expression levels per cell (SI Appendix, Fig. S3 A and B) and per cargo (SI Appendix, Fig. S3 C–F), custom analysis pipelines were developed using CellProfiler (https://cellprofiler.org) (78). At least 30 cells were analyzed for each motor in the absence (SI Appendix, Fig. S3 A and B) and presence (SI Appendix, Fig. S3 C–F) of rapamycin. All imaging was carried out using identical conditions to ensure fair comparison of fluorescence intensities. Briefly, fluorescence channels were aligned and background signal was subtracted separately for each channel. Nuclei were identified and used as seeds to identify cell peripheries. Each cell was manually checked and the outline was adjusted. Cells were masked to measure mean pixel intensity in each fluorescence channel within the cell boundary. Peroxisome objects were identified using three-class, adaptive, Otsu thresholding and then the output was manually checked to add or remove objects. Mean and integrated pixel intensities were measured for all objects in the motor and PEX3 channels and grouped by parent cell. Normalized distance was calculated as the distance from the cell centroid divided by the distance from the cell centroid to the cell edge for each cargo.
Single-Molecule Motility Assays.
HiLyte647-labeled microtubules were polymerized from unlabeled and HiLyte647-labeled tubulins (Cytoskeleton) in BRB80 (80 mM Pipes/KOH, 1 mM MgCl2, 1 mM EGTA, pH 6.8) supplemented with MgCl2 and GTP at 37 °C for 60 min. Microtubules were stabilized by adding 2 µM taxol in BRB80 and incubating another 60 min. Taxol-stabilized microtubules were stored at room temperature in the dark. A flow cell (∼10-μL volume) was assembled by attaching a clean #1.5 coverslip (Thermo Fisher Scientific) to a glass slide (Thermo Fisher Scientific) with two stripes of double-sided tape. Polymerized microtubules were diluted in BRB80 buffer containing 10 μM taxol and then were infused into flow cells and incubated for 5 min at room temperature for nonspecific adsorption to the coverslips. Sequentially, the flow cells were incubated with: (i) blocking buffer [15 mg/mL BSA in P12 (12 mM Pipes/KOH, 2 mM MgCl2, 1 mM EGTA, pH 6.8) with 10 μM taxol] and then (ii) 3xmCit-tagged motors in the motility mixture (0.5–6 μL cell lysate, 2 mM ATP, 9 mg/mL BSA, 0.4 mg/mL casein, 10 μM taxol, 1 mM MgCl2, 1 mM DTT, 10 mM glucose, 2 mg/mL glucose oxidase, and 80 µg/mL catalase in P12). The flow cells were sealed with molten paraffin wax and imaged on an inverted TIRF microscope Ti-E/B (Nikon) equipped with 100× 1.49 NA oil immersion TIRF objective (Nikon), 488-, 561-, and 640-nm lasers, and an electron-multiplying charge-coupled device detector (iXon X3DU897; Andor). Image acquisition was controlled with Elements software (Nikon). One image of HiLyte647-labeled microtubules was acquired and then images were acquired in the 488-nm channel at an acquisition rate of 10–20 Hz for 30 s. Monomers were imaged at twice the rate as dimers to capture any diffusive motion. Maximum-intensity projections were generated, and kymographs were produced by drawing along these tracks using Elements software or ImageJ. The velocity was defined as the distance on the y axis of the kymograph divided by the time on the x axis of the kymograph. All in vitro assays were performed at room temperature.
Bead Motility Assays.
The amount of biotinylated motors in cell lysates was quantified using a dot blot by spotting serial dilutions of cell lysates and a known control biotinylated protein on a nitrocellulose membrane. The membrane was blocked with 5% BSA in TBST, probed with streptavidin-Alexa 680, and imaged with an Azure imager. Spots were quantified using ImageJ with background subtraction from mock-transfected cells. Various amounts of biotinylated motors were bound to beads by incubating 5-µL sonicated beads, 2 µL P12 buffer (+10 mM ATP, 5 mM MgCl2, 5 mM DTT), and cell lysate on ice for 30 min with periodic mixing. A motor concentration of 35 nM was found to produce reliable, unidirectional bead motility for both KIF3B and KIF3B-20nmSAH monomers. Streptavidin-coated fluorescent yellow beads (SVFP-0552-5, 0.4-µm diameter; Spherotech) were sonicated at 40% for 2 min, the surface was blocked for 10 min with 5 mg/mL casein in P12 with 10 μM taxol, and then incubated with cell lysate in P12. The motor/bead mixture was added to assay buffer prepared immediately beforehand containing 2 mM ATP, 1 mg/mL casein, 10 μM taxol, 1 mM MgCl2, 1 mM DTT, 10 mM glucose, 2 mg/mL glucose oxidase, and 80 µg/mL catalase in P12. A flow cell was assembled and polymerized microtubules were flowed in and allowed to adsorb to the coverslip for 20 min in a humidified chamber to promote sticking. The flow cells were sealed with molten paraffin wax and imaged on an inverted TIRF microscope as described above. Images were acquired at an acquisition rate of 10 Hz for 30 s in the 561-nm channel to simultaneously view microtubules and fluorescent yellow beads. Kymographs were generated and analyzed as above.
Optical Trapping Assays.
Identical cell lysates and bead preparations were used in the bead motility and optical trapping assays. Microtubules were prepared as described previously (79). Purified tubulin from PurSolutions (bovine, 1001) was reconstituted in the supplied polymerization buffer. Thirteen microliters PEM104 buffer (104 mM Pipes, 1.3 mM EGTA, 6.3 mM MgCl2, pH adjusted to 6.9 with KOH), 2.2 µL 10 mM GTP, and 2.2 µL DMSO were mixed. 4.8 µL of 10 mg/mL tubulin were added to the mixture and allowed to incubate for 40 min at 37 °C. Subsequently, 2 µL of stabilization solution [STAB, 38.6 µL P12, 0.5 µL 100 mM GTP, 4.7 µL 65 g/L NaN3 (Sigma S-8032), 1.2 µL 10 mM Taxol (Cytoskeleton TXD01), 5 µL DMSO (Cytoskeleton)] was added to the stock microtubule solution at room temperature.
Optical trapping assays with motor-coated beads were performed as described previously (79). A flow cell that holds a volume of ∼15 µL was assembled using a microscope slide, etched coverslips, and double-sided tape. Before assembly, etched coverslips were incubated in a solution of 100 µL poly-l-lysine (PLL, Sigma P8920) in 30 mL ethanol for 15 min. The coverslip was then dried with a filtered air line. After flow cell assembly, microtubules were diluted 150 times from the stock in a solution of PemTax (P12 with 20 µM taxol). The diluted microtubules were added to the flow cell and allowed to adhere to the PLL surface for 10 min. Unbound microtubules were then washed out with 20 µL PemTax. A solution of casein (Blotting-Grade Blocker, Bio-Rad 1706404) diluted in PemTax (1:8 mixture) was then added to the flow cell and allowed to incubate for 10 min to block the remainder of the surface to prevent nonspecific binding. After the incubation, the flow cell was washed with 50 µL PemTax and 80 µL assay buffer (AB). Various amounts of biotinylated motors were tested and a motor concentration of 7 nM was found to produce reliable trapping of motor-coated beads for both KIF3B and KIF3B-20nmSAH monomers. Ten microliters of the motor/bead incubation were added to fresh assay buffer as described for bead motility assays and then diluted further by 50 µL of P12 buffer to avoid overcrowding of the flow cell with beads.
Optical trapping measurements were obtained using a custom-built instrument with separate trapping and detection systems. The instrument set-up and calibration procedures have been described previously (80). Briefly, beads were trapped with a 1,064-nm laser that was coupled to an inverted microscope with a 100×/1.3 NA oil-immersion objective. Bead displacements from the trap center were recorded at 3 kHz and further antialias-filtered at 1.5 kHz. Position calibration and trap stiffness measurements were obtained using custom LabVIEW programs.
Supplementary Material
Acknowledgments
We thank Andy Poulos for help in establishing the inducible Golgi dispersion assay; and members of the K.J.V. laboratory for discussions and support. This work was supported by National Institutes of Health Grant GMR01070862 (to K.J.V.) and National Science Foundation Grant 1330792 (to M.J.L.). B.G.B. was supported by a Graduate Research Fellowship from the National Science Foundation (DGE 1256260) and by the Cellular and Molecular Biology Training Grant T32-GM007315 from the National Institutes of Health. D.N.R. was supported by the Graduate Research Fellowship Program under Grant 1445197 from the National Science Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817924116/-/DCSupplemental.
References
- 1.Lucanus AJ, Yip GW. Kinesin superfamily: Roles in breast cancer, patient prognosis and therapeutics. Oncogene. 2018;37:833–838. doi: 10.1038/onc.2017.406. [DOI] [PubMed] [Google Scholar]
- 2.Brady ST, Morfini GA. Regulation of motor proteins, axonal transport deficits and adult-onset neurodegenerative diseases. Neurobiol Dis. 2017;105:273–282. doi: 10.1016/j.nbd.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hancock WO. The kinesin-1 chemomechanical cycle: Stepping toward a consensus. Biophys J. 2016;110:1216–1225. doi: 10.1016/j.bpj.2016.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berliner E, Young EC, Anderson K, Mahtani HK, Gelles J. Failure of a single-headed kinesin to track parallel to microtubule protofilaments. Nature. 1995;373:718–721. doi: 10.1038/373718a0. [DOI] [PubMed] [Google Scholar]
- 5.Diehl MR, Zhang K, Lee HJ, Tirrell DA. Engineering cooperativity in biomotor-protein assemblies. Science. 2006;311:1468–1471. doi: 10.1126/science.1122125. [DOI] [PubMed] [Google Scholar]
- 6.Inoue Y, et al. Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc Natl Acad Sci USA. 1997;94:7275–7280. doi: 10.1073/pnas.94.14.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamei T, Kakuta S, Higuchi H. Biased binding of single molecules and continuous movement of multiple molecules of truncated single-headed kinesin. Biophys J. 2005;88:2068–2077. doi: 10.1529/biophysj.104.049759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young EC, Mahtani HK, Gelles J. One-headed kinesin derivatives move by a nonprocessive, low-duty ratio mechanism unlike that of two-headed kinesin. Biochemistry. 1998;37:3467–3479. doi: 10.1021/bi972172n. [DOI] [PubMed] [Google Scholar]
- 9.Hancock WO, Howard J. Processivity of the motor protein kinesin requires two heads. J Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leibler S, Huse DA. Porters versus rowers: A unified stochastic model of motor proteins. J Cell Biol. 1993;121:1357–1368. doi: 10.1083/jcb.121.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okada Y, Higuchi H, Hirokawa N. Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature. 2003;424:574–577. doi: 10.1038/nature01804. [DOI] [PubMed] [Google Scholar]
- 12.Tomishige M, Klopfenstein DR, Vale RD. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 2002;297:2263–2267. doi: 10.1126/science.1073386. [DOI] [PubMed] [Google Scholar]
- 13.Brugués J, Casademunt J. Self-organization and cooperativity of weakly coupled molecular motors under unequal loading. Phys Rev Lett. 2009;102:118104. doi: 10.1103/PhysRevLett.102.118104. [DOI] [PubMed] [Google Scholar]
- 14.Orlandi JG, Blanch-Mercader C, Brugués J, Casademunt J. Cooperativity of self-organized Brownian motors pulling on soft cargoes. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82:061903. doi: 10.1103/PhysRevE.82.061903. [DOI] [PubMed] [Google Scholar]
- 15.Oriola D, Roth S, Dogterom M, Casademunt J. Formation of helical membrane tubes around microtubules by single-headed kinesin KIF1A. Nat Commun. 2015;6:8025. doi: 10.1038/ncomms9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oriola D, Casademunt J. Cooperative force generation of KIF1A Brownian motors. Phys Rev Lett. 2013;111:048103. doi: 10.1103/PhysRevLett.111.048103. [DOI] [PubMed] [Google Scholar]
- 17.Soppina V, et al. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okada Y, Hirokawa N. A processive single-headed motor: Kinesin superfamily protein KIF1A. Science. 1999;283:1152–1157. doi: 10.1126/science.283.5405.1152. [DOI] [PubMed] [Google Scholar]
- 19.Okada Y, Hirokawa N. Mechanism of the single-headed processivity: Diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA. 2000;97:640–645. doi: 10.1073/pnas.97.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Soppina V, Verhey KJ. The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors. Mol Biol Cell. 2014;25:2161–2170. doi: 10.1091/mbc.E14-01-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapitein LC, et al. Probing intracellular motor protein activity using an inducible cargo trafficking assay. Biophys J. 2010;99:2143–2152. doi: 10.1016/j.bpj.2010.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapp S, et al. Microtubule-based peroxisome movement. J Cell Sci. 1996;109:837–849. doi: 10.1242/jcs.109.4.837. [DOI] [PubMed] [Google Scholar]
- 23.Wiemer EA, Wenzel T, Deerinck TJ, Ellisman MH, Subramani S. Visualization of the peroxisomal compartment in living mammalian cells: Dynamic behavior and association with microtubules. J Cell Biol. 1997;136:71–80. doi: 10.1083/jcb.136.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Efremov AK, et al. Delineating cooperative responses of processive motors in living cells. Proc Natl Acad Sci USA. 2014;111:E334–E343. doi: 10.1073/pnas.1313569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang W, Lang MJ, Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Brownhill K, Wood L, Allan V. Molecular motors and the Golgi complex: Staying put and moving through. Semin Cell Dev Biol. 2009;20:784–792. doi: 10.1016/j.semcdb.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Guet D, et al. Mechanical role of actin dynamics in the rheology of the Golgi complex and in Golgi-associated trafficking events. Curr Biol. 2014;24:1700–1711. doi: 10.1016/j.cub.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 28.Engelke MF, et al. Engineered kinesin motor proteins amenable to small-molecule inhibition. Nat Commun. 2016;7:11159. doi: 10.1038/ncomms11159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 30.Verhey KJ, Hammond JW. Traffic control: Regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 31.Swanson CJ, Sivaramakrishnan S. Harnessing the unique structural properties of isolated α-helices. J Biol Chem. 2014;289:25460–25467. doi: 10.1074/jbc.R114.583906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vale RD, Soll DR, Gibbons IR. One-dimensional diffusion of microtubules bound to flagellar dynein. Cell. 1989;59:915–925. doi: 10.1016/0092-8674(89)90614-4. [DOI] [PubMed] [Google Scholar]
- 33.Furuta K, et al. Measuring collective transport by defined numbers of processive and nonprocessive kinesin motors. Proc Natl Acad Sci USA. 2013;110:501–506. doi: 10.1073/pnas.1201390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jonsson E, Yamada M, Vale RD, Goshima G. Clustering of a kinesin-14 motor enables processive retrograde microtubule-based transport in plants. Nat Plants. 2015;1:15087. doi: 10.1038/nplants.2015.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hutterer A, Glotzer M, Mishima M. Clustering of centralspindlin is essential for its accumulation to the central spindle and the midbody. Curr Biol. 2009;19:2043–2049. doi: 10.1016/j.cub.2009.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norris SR, et al. Microtubule minus-end aster organization is driven by processive HSET-tubulin clusters. Nat Commun. 2018;9:2659. doi: 10.1038/s41467-018-04991-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyrpassopoulos S, et al. Force generation by membrane-associated myosin-I. Sci Rep. 2016;6:25524. doi: 10.1038/srep25524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McIntosh BB, Pyrpassopoulos S, Holzbaur ELF, Ostap EM. Opposing kinesin and myosin-I motors drive membrane deformation and tubulation along engineered cytoskeletal networks. Curr Biol. 2018;28:236–248.e5. doi: 10.1016/j.cub.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andreasson JO, Shastry S, Hancock WO, Block SM. The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr Biol. 2015;25:1166–1175. doi: 10.1016/j.cub.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arpağ G, Shastry S, Hancock WO, Tüzel E. Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys J. 2014;107:1896–1904. doi: 10.1016/j.bpj.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroeder HW, 3rd, et al. Force-dependent detachment of kinesin-2 biases track switching at cytoskeletal filament intersections. Biophys J. 2012;103:48–58. doi: 10.1016/j.bpj.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Q, Mickolajczyk KJ, Chen GY, Hancock WO. Motor reattachment kinetics play a dominant role in multimotor-driven cargo transport. Biophys J. 2018;114:400–409. doi: 10.1016/j.bpj.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campàs O, et al. Coordination of kinesin motors pulling on fluid membranes. Biophys J. 2008;94:5009–5017. doi: 10.1529/biophysj.107.118554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leduc C, et al. Cooperative extraction of membrane nanotubes by molecular motors. Proc Natl Acad Sci USA. 2004;101:17096–17101. doi: 10.1073/pnas.0406598101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andreasson JO, et al. Examining kinesin processivity within a general gating framework. eLife. 2015;4:e07403. doi: 10.7554/eLife.07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clancy BE, Behnke-Parks WM, Andreasson JO, Rosenfeld SS, Block SM. A universal pathway for kinesin stepping. Nat Struct Mol Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dogan MY, Can S, Cleary FB, Purde V, Yildiz A. Kinesin’s front head is gated by the backward orientation of its neck linker. Cell Rep. 2015;10:1967–1973. doi: 10.1016/j.celrep.2015.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mickolajczyk KJ, Hancock WO. Kinesin processivity is determined by a kinetic race from a vulnerable one-head-bound state. Biophys J. 2017;112:2615–2623. doi: 10.1016/j.bpj.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shastry S, Hancock WO. Interhead tension determines processivity across diverse N-terminal kinesins. Proc Natl Acad Sci USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yildiz A, Tomishige M, Gennerich A, Vale RD. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson SR, Trybus KM, Warshaw DM. Motor coupling through lipid membranes enhances transport velocities for ensembles of myosin Va. Proc Natl Acad Sci USA. 2014;111:E3986–E3995. doi: 10.1073/pnas.1406535111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grover R, et al. Transport efficiency of membrane-anchored kinesin-1 motors depends on motor density and diffusivity. Proc Natl Acad Sci USA. 2016;113:E7185–E7193. doi: 10.1073/pnas.1611398113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gudimchuk N, et al. Probing mitotic CENP-E kinesin with the tethered cargo motion assay and laser tweezers. Biophys J. 2018;114:2640–2652. doi: 10.1016/j.bpj.2018.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bieling P, Telley IA, Piehler J, Surrey T. Processive kinesins require loose mechanical coupling for efficient collective motility. EMBO Rep. 2008;9:1121–1127. doi: 10.1038/embor.2008.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crevenna AH, et al. Secondary structure and compliance of a predicted flexible domain in kinesin-1 necessary for cooperation of motors. Biophys J. 2008;95:5216–5227. doi: 10.1529/biophysj.108.132449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chowdary PD, Kaplan L, Che DL, Cui B. Dynamic clustering of dyneins on axonal endosomes: Evidence from high-speed darkfield imaging. Biophys J. 2018;115:230–241. doi: 10.1016/j.bpj.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shaklee PM, et al. Bidirectional membrane tube dynamics driven by nonprocessive motors. Proc Natl Acad Sci USA. 2008;105:7993–7997. doi: 10.1073/pnas.0709677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rai A, et al. Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell. 2016;164:722–734. doi: 10.1016/j.cell.2015.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Al-Bassam J, et al. Distinct conformations of the kinesin Unc104 neck regulate a monomer to dimer motor transition. J Cell Biol. 2003;163:743–753. doi: 10.1083/jcb.200308020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klopfenstein DR, Tomishige M, Stuurman N, Vale RD. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 2002;109:347–358. doi: 10.1016/s0092-8674(02)00708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Klopfenstein DR, Vale RD. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol Biol Cell. 2004;15:3729–3739. doi: 10.1091/mbc.E04-04-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hammond JW, et al. Mammalian kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol. 2009;7:e72. doi: 10.1371/journal.pbio.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huo L, et al. The CC1-FHA tandem as a central hub for controlling the dimerization and activation of kinesin-3 KIF1A. Structure. 2012;20:1550–1561. doi: 10.1016/j.str.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 64.Huckaba TM, Gennerich A, Wilhelm JE, Chishti AH, Vale RD. Kinesin-73 is a processive motor that localizes to Rab5-containing organelles. J Biol Chem. 2011;286:7457–7467. doi: 10.1074/jbc.M110.167023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee JR, et al. An intramolecular interaction between the FHA domain and a coiled coil negatively regulates the kinesin motor KIF1A. EMBO J. 2004;23:1506–1515. doi: 10.1038/sj.emboj.7600164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren J, et al. Coiled-coil 1-mediated fastening of the neck and motor domains for kinesin-3 autoinhibition. Proc Natl Acad Sci USA. 2018;115:E11933–E11942. doi: 10.1073/pnas.1811209115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamada KH, Hanada T, Chishti AH. The effector domain of human Dlg tumor suppressor acts as a switch that relieves autoinhibition of kinesin-3 motor GAKIN/KIF13B. Biochemistry. 2007;46:10039–10045. doi: 10.1021/bi701169w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park H, et al. Full-length myosin VI dimerizes and moves processively along actin filaments upon monomer clustering. Mol Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 69.Phichith D, et al. Cargo binding induces dimerization of myosin VI. Proc Natl Acad Sci USA. 2009;106:17320–17324. doi: 10.1073/pnas.0909748106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakai T, Umeki N, Ikebe R, Ikebe M. Cargo binding activates myosin VIIA motor function in cells. Proc Natl Acad Sci USA. 2011;108:7028–7033. doi: 10.1073/pnas.1009188108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Spudich G, et al. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to disabled-2 and PtdIns(4,5)P2. Nat Cell Biol. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Umeki N, et al. Phospholipid-dependent regulation of the motor activity of myosin X. Nat Struct Mol Biol. 2011;18:783–788. doi: 10.1038/nsmb.2065. [DOI] [PubMed] [Google Scholar]
- 73.Yang Y, et al. Dimerized Drosophila myosin VIIa: A processive motor. Proc Natl Acad Sci USA. 2006;103:5746–5751. doi: 10.1073/pnas.0509935103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu C, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138:537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 75.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Milic B, Andreasson JOL, Hogan DW, Block SM. Intraflagellar transport velocity is governed by the number of active KIF17 and KIF3AB motors and their motility properties under load. Proc Natl Acad Sci USA. 2017;114:E6830–E6838. doi: 10.1073/pnas.1708157114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Norris SR, et al. A method for multiprotein assembly in cells reveals independent action of kinesins in complex. J Cell Biol. 2014;207:393–406. doi: 10.1083/jcb.201407086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carpenter AE, et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Reinemann DN, et al. Collective force regulation in anti-parallel microtubule gliding by dimeric Kif15 kinesin motors. Curr Biol. 2017;27:2810–2820.e6. doi: 10.1016/j.cub.2017.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Khalil AS, et al. Kinesin’s cover-neck bundle folds forward to generate force. Proc Natl Acad Sci USA. 2008;105:19247–19252. doi: 10.1073/pnas.0805147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.