Significance
Acetogenic bacteria are the most primordial living organisms. They can live solely on inorganic compounds by using carbon monoxide or molecular hydrogen as energy sources to fix carbon dioxide to acetate and rely on a chemiosmotic mechanism for energy conservation. Most microorganisms possess a complex respiratory chain that is composed of many different components to establish the chemiosmotic gradient. Here, we dissect the bioenergetics in a chemolithoautotrophic thermophilic acetogenic bacterium. This living fossil uses a simple respiration comprising only a hydrogenase and an ATP synthase for energy conservation. This two-module respiration system is sufficient to sustain primordial microbial life.
Keywords: acetogenesis, bioenergetics, energy-converting hydrogenase, respiratory mechanism, chemiosmosis
Abstract
The ancient reductive acetyl-CoA pathway is employed by acetogenic bacteria to form acetate from inorganic energy sources. Since the central pathway does not gain net ATP by substrate-level phosphorylation, chemolithoautotrophic growth relies on the additional formation of ATP via a chemiosmotic mechanism. Genome analyses indicated that some acetogens only have an energy-converting, ion-translocating hydrogenase (Ech) as a potential respiratory enzyme. Although the Ech-encoding genes are widely distributed in nature, the proposed function of Ech as an ion-translocating chemiosmotic coupling site has neither been demonstrated in bacteria nor has it been demonstrated that it can be the only energetic coupling sites in microorganisms that depend on a chemiosmotic mechanism for energy conservation. Here, we show that the Ech complex of the thermophilic acetogenic bacterium Thermoanaerobacter kivui is indeed a respiratory enzyme. Experiments with resting cells prepared from T. kivui cultures grown on carbon monoxide (CO) revealed CO oxidation coupled to H2 formation and the generation of a transmembrane electrochemical ion gradient (). Inverted membrane vesicles (IMVs) prepared from CO-grown cells also produced H2 and ATP from CO (via a loosely attached CO dehydrogenase) or a chemical reductant. Finally, we show that Ech activity led to the translocation of both H+ and Na+ across the membrane of the IMVs. The H+ gradient was then used by the ATP synthase for energy conservation. These data demonstrate that the energy-converting hydrogenase in concert with an ATP synthase may be the simplest form of respiration; it combines carbon dioxide fixation with the synthesis of ATP in an ancient pathway.
The pioneer organism in a primordial world was probably a chemolithoautotrophic thermophilic anaerobe that employed the reductive acetyl-CoA pathway (1, 2). This pathway is also called Wood–Ljungdahl pathway (WLP), in dedication to its discoverers, and encompasses CO2 fixation to acetyl CoA, CO2 assimilation into biomass, and energy conservation (3, 4). The WLP has prevailed in three groups of strictly anaerobic organisms: methanogenic archaea, sulfate reducing bacteria, and acetogenic bacteria.
Acetogenic bacteria are ubiquitous in nature and occupy an increasingly important role in biotechnological and industrial applications (5–8). They can grow heterotrophically on, for example, sugars, alcohols, and aldehydes but also chemolithoautotrophically on hydrogen plus carbon dioxide or on carbon monoxide (3, 9–11). Lithotrophic metabolism involves reduction of two mol CO2 to acetate with electrons derived from H2 according to:
| [1] |
One molecule of CO2 is reduced to formate, the formate is then bound in an ATP-dependent reaction to formyl-tetrahydrofolate (THF), followed by a reduction of the formyl group to a methenyl group via methenyl-THF and methylene-THF (12–15). A second molecule of CO2 is reduced by the CO dehydrogenase/acetyl CoA synthase (CODH/Acs) to enzyme-bound CO that combines with the methyl group and CoA on the CODH/Acs to acetyl-CoA (12, 16). Acetyl-CoA is then converted via acetyl phosphate to acetate and ATP.
CO metabolism follows the same principle but involves oxidation of CO to CO2 according to:
| [2] |
In sum, CO is oxidized to acetate according to:
| [3] |
The net yield of ATP by substrate level phosphorylation (SLP) is zero, but since the bacteria do grow under lithotrophic conditions, additional ATP synthesis must occur by a chemiosmotic mechanism (11).
How the WLP can be coupled to the synthesis of ATP has been solved only recently for a mesophilic acetogen. Acetobacterium woodii, which grows optimally at 30 °C, has a simple respiratory chain consisting of a ferredoxin-oxidizing, NAD-reducing respiratory enzyme encoded by the rnf genes and an ATP synthase that are connected by a sodium ion circuit across the cytoplasmic membrane (17, 18): The Rnf complex uses redox energy to generate a transmembrane electrochemical Na+ gradient across the cytoplasmic membrane that then drives ATP synthesis by a Na+ F1FO ATP synthase. rnf genes are not present in every acetogen and, thus, a second mode of energy conservation must exist in acetogenic bacteria. Interestingly, inspection of genome sequences led to the detection of genes encoding energy-converting, possibly ion-translocating, membrane-bound hydrogenases (Ech) in Rnf-free acetogens, leading to the postulate that in these acetogens Rnf is replaced by Ech (15).
Ech belongs to the group 4 [NiFe] hydrogenases (respiratory hydrogenases), which comprise a very diverse and still largely enigmatic group of H2-evolving, possibly ion-translocating, energy-conserving membrane-associated hydrogenases (19, 20). The first described complex of this kind was the formate-hydrogen lyase (FHL), discovered in the most common microbial model organism Escherichia coli (21). To date, however, FHL complexes have never been demonstrated to translocate ions across the cytoplasmic membrane in bacteria. More complex Ech complexes with 6 to 14 subunits are present in archaea, and evidence has been presented for ion transport for the enzymes from Methanosarcina, Pyrococcus, and Thermococcus (22–24). The six-subunit complex as found in the methanogen Methanosarcina mazei is H+-translocating and the first enzyme in a chain of respiratory enzymes. The 14-subunit complex of Pyrococcus was described as proton translocating, but the recent high-resolution cryo-EM structure revealed a row of potential Na+/H+ antiporter modules leading to the assumption that the enzyme translocates both H+ and Na+ (25). Indeed, simultaneous transport of both H+ and Na+ has been experimentally demonstrated for the related enzyme from Thermococcus (24). However, in none of these examples is Ech the only coupling site or not even the predominant mode of energy conservation.
The thermophilic acetogenic bacterium Thermoanaerobacter kivui does not have rnf genes but two ech gene clusters and no other possible respiratory enzyme (26). It can grow lithotrophically on H2+CO2 (27) and has been adapted to grow on CO or CO+CO2 or syngas (CO+H2+CO2) (28). The scope of this work was to analyze whether Ech is indeed the respiratory enzyme of T. kivui.
Results
Energetics of CO Oxidation.
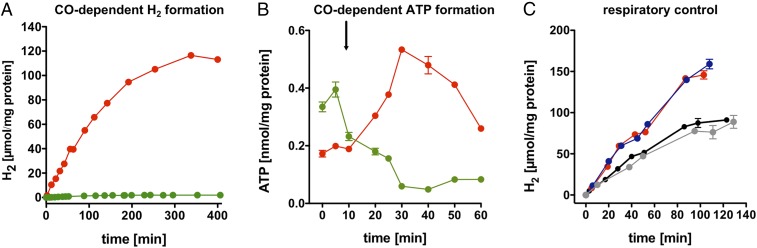
To address whether CO oxidation is coupled to the formation of H2 and ATP, T. kivui cells were grown on CO or glucose, and resting cells were prepared. After addition of CO, glucose-grown cells produced only little H2, whereas CO-grown cells produced H2 at a rate of 584 ± 3 nmol·min−1·mg−1 protein (n = 2, SD) (Fig. 1A), indicating that the ability to oxidize CO was induced during growth on CO. Concomitant with H2 formation, the intracellular ATP content increased (Fig. 1B). ATP synthesis depended on an energized membrane as evident from the inhibition by the protonophore 3,3′,4′,5-tetrachlorosalicylanilide (TCS) or sodium ionophore N,N,N′,N′-tetracyclohexyl-1,2-phenylenedioxydiacetamide (ETH2120) and NaCl (SI Appendix, Fig. S1). To address whether CO oxidation is coupled to the generation of an electrical field across the membrane (due to ion translocation), we tested the effect of ionophores on electron transport (H2 formation). The protonophore TCS or the sodium ionophore ETH2120 and NaCl (Fig. 1C) both stimulated H2 formation [84 ± 30% or 69 ± 2% (n = 2, SD), respectively]. This is reminiscent of “respiratory control” as observed in mitochondria: Charge translocation leads to the build-up of an electrical field that slows down further electron transport; disruption of the electrical field then stimulates electron transport. Thus, the experiments described above are consistent with the hypothesis that H2 formation from CO in T. kivui is coupled to the generation of a transmembrane electrochemical ion gradient.
Fig. 1.
Energetics of CO oxidation in T. kivui. (A) Resting cells (1 mg/mL) pregrown on CO (red) produced H2 from CO but not glucose-pregrown cells (green). (B) Resting cells (1 mg/mL) pregrown on CO (red) produced ATP from CO but not glucose-pregrown cells (green). (C) H2 evolution from CO by resting cells (0.5 mg/mL) pregrown on CO was stimulated when preincubated with 30 μM protonophore TCS (blue) or 30 μM sodium ionophore ETH2120 and 5 mM NaCl (red), but not in the absence of ionophore (black) or in the presence of ethanolic solvent alone (gray).
To investigate whether Na+ might be the chemiosmotic coupling ion, we analyzed the effect of Na+ on H2 evolution from CO. H2 formation from CO was already quite high at the lowest (contaminating) Na+ concentration (90 µM), and addition of NaCl (0.25–10 mM) only slightly increased H2 formation (SI Appendix, Fig. S2). KCl did not stimulate H2 formation. Na+ stimulation was highest at the lowest pH (SI Appendix, Fig. S3). This coincides with the increasing H2 evolution rate from CO at decreasing pH (due to higher substrate availability; SI Appendix, Fig. S4).
Preparation of Intact Inverted Membrane Vesicles of T. kivui.
To assess ion translocation coupled to H2 evolution on a subcellular level, we established a procedure to prepare energetically intact inverted membrane vesicles (IMVs) from T. kivui. IMVs were prepared under strictly anoxic conditions from CO-grown cells by a gentle cell disruption in a French Press cell. To test the membrane integrity of the IMVs, an artificial H+ gradient was established across the membrane of the IMVs and it was analyzed whether IMVs could hold such an artificial pH gradient. Therefore, they were loaded with ammonium chloride and subsequently diluted in a fluorescence cell containing ammonium-free buffer and the fluorescent dye 9-amino-6-chloro-2-methoxyacridine (ACMA). The dilution leads to the dissociation of NH4Cl into NH3 and H+. The ammonia diffuses across the membrane, whereas H+ are retained, leading to an acidification within the vesicle (∆pH) that is detected as a quenching of the fluorescence of ACMA. The quenching was indeed observed, and the pH gradient was stable for 5 min (SI Appendix, Fig. S5A). The pH gradient could be partially dissipated by the addition of protonophore TCS (SI Appendix, Fig. S5B) and fully dissipated upon addition of the membrane-perforating 1-butanol (SI Appendix, Fig. S5C). Thus, the procedure led to energetically intact IMVs.
IMVs Exhibit Ech Activity.
First, we had to determine whether IMVs retained the respiratory enzyme during preparation. To ensure that soluble proteins (particularly soluble hydrogenases) were removed from the vesicles, one-time washed IMVs (= washed IMVs) were used for all measurements, unless indicated otherwise. Washed IMVs were incubated in vesicle buffer and CO was added. Indeed, hydrogen was produced, demonstrating the presence of Ech at the membrane (SI Appendix, Fig. S6). Since ferredoxin may be used as an electron carrier between CO dehydrogenase and Ech, we analyzed the effect of ferredoxin (0–50 µM; purified from Clostridium pasteurianum as described in ref. 29) on the H2 formation rate and the amount of H2 produced. Hydrogen formation rate and the amount of hydrogen produced increased with increasing ferredoxin concentrations and were highest at 50 µM (rate: 100 ± 5%, final H2 evolved: 100%) and lowest without additional ferredoxin (rate: 37 ± 3%, final H2 evolved: 31%). Hence, assays analyzing H2 evolution from CO were routinely supplemented with 30 µM ferredoxin. With this assay, washed IMVs usually evolved H2 at 100–700 nmol·min−1·mg−1 protein from CO, depending on the preparation. The chemical reductants sodium dithionite (NaDt) or titanium (III) citrate (Ti3+) could replace CO as reductant. IMVs (200 µg/mL) incubated in buffer supplemented with NaDt (1.5 mM) as reductant evolved H2 at 301 ± 16 nmol·min−1·mg−1 protein (n = 2, SD). The H2 evolution rate increased threefold with additional ferredoxin (30 µM) to 889 ± 127 nmol·min−1·mg−1 protein (final H2 evolved without or with ferredoxin 4.5 or 12 µmol/mg protein). Ferredoxin did not stimulate H2 evolution with Ti3+ as reductant. Hence, electron transfer from CO or NaDt to H+ proceeds via ferredoxin, a redox reaction characteristic for the Ech complex.
To consolidate that the H2-forming activity is indeed anchored in the membrane as anticipated for Ech, we washed the vesicles multiple times and measured the total H2 formation activity (Utot). Protein preparations (200 µg/mL) were incubated in assay buffer containing ferredoxin (30 µM) and energized with CO (flushing) or NaDt (1.5 mM). The unwashed IMVs possessed a Utot of 2.3 ± 0.3 and 6.0 ± 0.3 Utot with NaDt and CO as reductant, respectively. IMVs washed once resulted in a loss of Utot with CO as reductant (but not NaDt). IMVs washed twice still possessed 95 ± 3% Utot, but only 10 ± 1% Utot with NaDt and CO as reductant, respectively. The supernatant fractions did not exhibit H2 formation. These experiments demonstrated a rather stable anchorage of the H2-evolving Ech complex and an only loose attachment of the CODH to the membrane.
Ech Activity Leads to the Establishment of a Chemiosmotic Gradient Composed of both H+ and Na+.
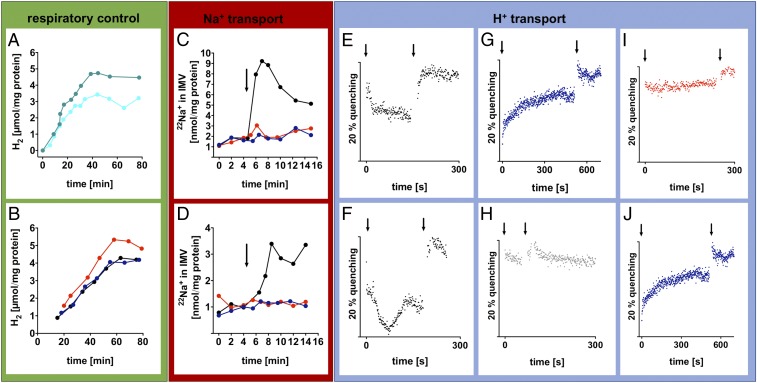
To test whether Ech activity is coupled to the generation of an electrical potential across the vesicle membrane, washed IMVs were incubated in assay buffer in the presence of the potassium ionophore valinomycin and KCl that disrupt electrical fields. Ech activity was stimulated by 43% with a H2 evolution rate from CO of 139.9 ± 9.8 compared with 96.8 ± 6.5 nmol·min−1·mg−1 protein in a solvent control assay (Fig. 2A), demonstrating respiratory control. The sodium ionophore ETH2120 stimulated by 30%, whereas the protonophore TCS had no significant stimulatory effect (Fig. 2B; H2 evolution rates for the assay containing sodium ionophore, protonophore, or no ionophore were 92.5 ± 3.3, 74.0 ± 3.8, or 71.5 ± 2.6 nmol·min−1·mg−1 protein). This is in contrast to the experiments described above for whole cells; the reason remains unclear but may result from a weaker energetic coupling in IMVs compared with whole cells. In addition, in whole cells, acetate formation is inhibited by the protonophore and, thus, more H2 can be formed from CO (30). The experiments with uncoupling agents clearly demonstrate that, also on a subcellular level, Ech activity is coupled to ion translocation and subsequent generation of an electrochemical ion gradient.
Fig. 2.
Ech establishes a chemisomotic gradient composed of Na+ and H+. (A and B) Ech activity is coupled to the electrical field. Washed IMVs (60 µg/mL) catalyzed H2 formation from CO, and this activity was stimulated in assays preincubated with 100 mM KCl and 20 μM K+ ionophore valinomycin (green) but not the solvent DMSO alone (light blue), or 10 μM Na+ ionophore ETH2120 (red), but not 10 μM protonophore TCS (blue) or the solvent ethanol alone (black). (C and D) Ech activity leads to Na+ transport. Washed IMVs (500 μg/mL) preincubated with 1.0 µCi/mL 22NaCl (and 30 μM ferredoxin in D) accumulated 22Na+ in the vesicle lumen when energized (arrow) with 1 mM Ti3+ (C) or CO flushing (D) (black). Preincubation with either 20 μM ETH2120 (red) or 20 μM TCS (blue) abolished 22Na+ transport in both assays. (E–J) Ech activity leads to H+ transport. A ∆pH was detected upon addition (arrow) of reduced ferredoxin (20 µM) to three times washed IMVs (500 μg/mL) by measuring the fluorescence quench of ACMA (4 µM) at 40 °C (E) or 60 °C (F). Reduced ferredoxin was (re)generated by supplemented purified CODH (30 μg from A. woodii) in a 100% CO atmosphere. The quench was abolished by addition (second arrow) of 10 μL of 90% (NH4)2SO4. (G) IMVs were omitted. (H) Ferredoxin was omitted. (I and J) Assays were preincubated with 20 μM ETH2120 and 150 mM NaCl in I or 20 μM TCS in J. Shown is one representative of two biologically independent experiments.
Finally, to prove that Ech activity indeed is a chemiosmotic coupling site, we performed translocation experiments with the radioisotope 22Na+ or the fluorescence dye ACMA. In the first instance, washed IMVs were incubated in assay buffer containing 1.0 µCi/mL 22NaCl. Upon addition of Ti3+, up to 9 nmol/mg protein 22Na+ accumulated in the lumen of the IMVs (Fig. 2C). Either sodium ionophore ETH2120 or protonophore TCS abolished 22Na+ accumulation (a control assay containing the ethanolic solvent did not show this effect). However, since Ti3+ is a strong artificial reductant and H2 evolution from Ti3+ was not ferredoxin-dependent, the experiment was repeated under more physiological conditions with ferredoxin and CO as reductant. In this assay, 3.5 nmol/mg protein 22Na+ also accumulated in the lumen of the IMVs (Fig. 2D). Again, either ionophore (but not the solvent alone) abolished 22Na+ translocation. Thus, this experiment clearly demonstrates that Ech establishes a Na+ gradient across the cytoplasmic membrane in T. kivui.
These data clearly demonstrated Na+ transport coupled to Ech activity. To test for a possible H+ transport alongside Na+ transport, the generation of a ΔpH was studied by ACMA quenching as described above. Neither NaDt or Ti3+ were suitable as reductants since they eradicated the quenching ability of ACMA nor the usual start of the reaction via flushing with CO was possible due to the small reaction volume of the fluorescence cell. Therefore, IMVs were washed three times to detach the attached CODH from the Ech and enable a distinct start of the redox reaction. Three times washed IMVs were incubated in a fluorescence cell containing IMV assay buffer, ACMA, and purified CODH (30 µg, from A. woodii; ref. 31) in a 100% CO atmosphere. First experiments were conducted at lower temperatures (40 °C) to ensure functionality of the supplemented CODH from the mesophilic A. woodii, and stability of the vesicles, which is of particular importance when measuring H+ transport (due to the small atomic radius). The reaction was started by addition of ferredoxin, which was immediately reduced by the CODH. This led to a continuous decrease in fluorescence at 40 °C (Fig. 2E) and 60 °C (Fig. 2F), demonstrating a ∆pH formation due to H+ transport into the vesicle lumen. Thus, the system was intact even at 60 °C. The quench was larger at 60 °C but was not sustained continuously. The fluorescence increased again to some extent (= dequench), which is most certainly a result of H+ efflux due to high(er) kinetic energies and concomitant higher membrane permeability to H+. The fluorescence was however only fully restored in both assays (Fig. 2 E and F) upon addition of (NH4)2SO4, which dissipates the H+ gradient. The control assays that contained no IMVs (Fig. 2G) or no ferredoxin (Fig. 2H) showed neither a decrease in fluorescence upon ferredoxin addition nor an impact on fluorescence upon addition of (NH4)2SO4. Preincubation of IMVs with sodium ionophore ETH2120 (Fig. 2I) or protonophore TCS (Fig. 2J) did not lead to quenching or dequenching. In summary, the experiment clearly showed that Ech activity also leads to the establishment of a +.
The H+ Gradient Drives ATP Synthesis.
That Ech activity leads to Na+ transport was initially surprising, since we did not encounter Na+ dependence for growth (SI Appendix, Fig. S7) (26), and the ATP synthase does not contain a conserved Na+ binding motif (26). To complete the elucidation of the chemiosmotic mechanism in T. kivui, we thus carried out biochemical analyses to assess the ion specificity of the gradient-consuming F1FO ATP synthase. As expected, we observed a ΔpH formation at IMVs (SI Appendix, Fig. S8) and not a pNa in response to ATP hydrolysis. Thus, the ATP synthase consumes the H+ gradient, which is established by the Ech complex.
Both ech Clusters Are Transcriptionally Up-Regulated During Autotrophic Growth.
The Ech complex in T. kivui is the product of either one or both of the two ech gene clusters present in the genome. Both gene clusters were highly up-regulated during autotrophic growth, especially with CO. The relative transcript level of ech1 and ech2 increased 6- and 16-fold in cells grown on H2 and 31- and 43-fold in cells grown on CO, normalized to cells grown on glucose. This up-regulation is most likely also a crucial reason for the successful adaptation of T. kivui to grow on CO (28).
Discussion
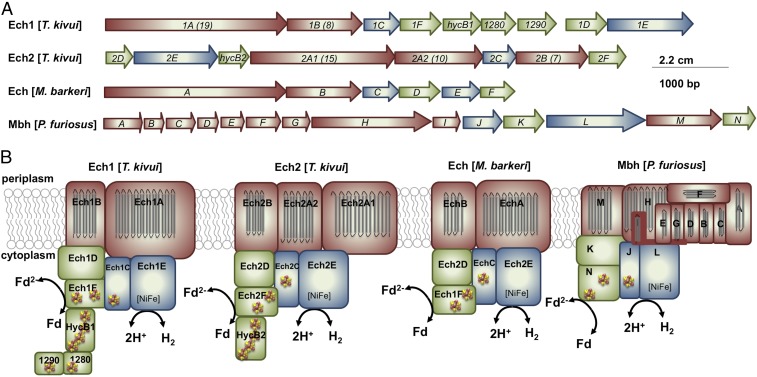
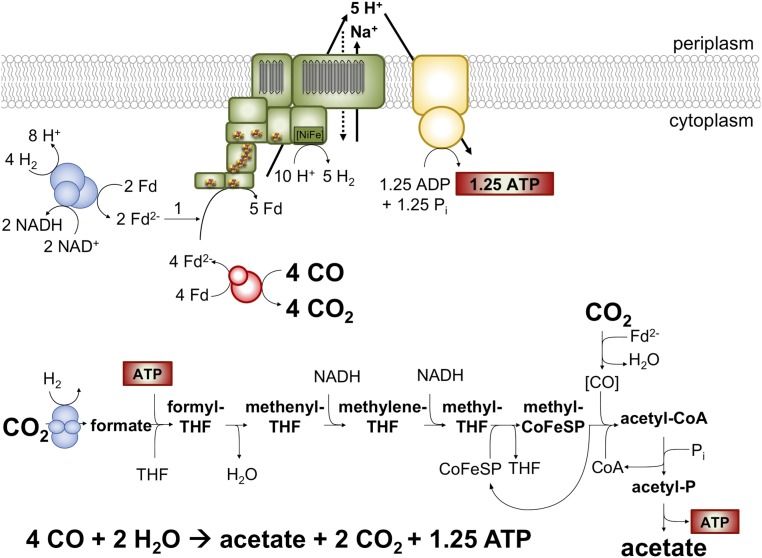
The occurrence of two (or more) clusters encoding group 4 [NiFe] hydrogenases within an organism might be a commonality, as it applies also to the other Ech-containing acetogens Moorella thermoacetica, Thermacetogenium phaeum, several archaea (32), and even E. coli (33). The number of genes and genetic arrangement of these clusters is very diverse (19). The ech1 and ech2 clusters in T. kivui contain 9 and 8 genes, whereas the most simple ech cluster as found in Methanosarcina barkeri contains 6 genes (34) and the mbh cluster in P. furiosus comprises 14 genes (35) (Fig. 3A). The derived protein complexes have three modules: the electron input module, an electron output module comprising the [NiFe] hydrogenase, and a membrane integral domain for ion translocation and anchorage (Fig. 3B). Bioinformatic analyses suggested the presence of Na+/H+ antiporter modules in the membrane arm of the Mbh from Pyrococcus or Thermococcus (32, 36) and, thus, the idea arose that these enzymes can translocate both H+ and Na+. Experiments with whole cells and IMVs of Thermococcus onnurineus revealed that protons are extruded first and sodium ions second by action of a Na+/H+ antiport mechanism (24). The secondary Na+ gradient then drives ATP synthesis via a Na+ A1AO ATP synthase (37). This idea is supported by the recent high-resolution structure of the 14-subunit Mbh from P. furiosus (25). The membrane arm can be divided into a H+ and a Na+ translocation unit, the latter resembles an Mrp-type Na+/H+ antiporter. Based on this model it was suggested that as in T. onnurineus, protons are extruded first and Na+ second. Again, the latter drives the synthesis of ATP by a Na+ A1AO ATP synthase. Similarly, Ech activity in T. kivui also established a Na+ and H+ gradient. A simultaneous transport of both ions could, for example, be explained by a promiscuous enzyme as described for the ATP synthase in Methanosarcina acetivorans (38), a simultaneous action of one H+ and one Na+ specific Ech complex (Ech1/2), or an Na+/H+ antiport. Although mrp genes encoding the Na+/H+ antiport in P. furiosus are missing in T. kivui, the membrane-integral subunits of both Ech complexes (Fig. 3B) share highest sequence similarities with Na+/H+ antiporters (SI Appendix, Table S1). There are no other apparent gene products identified as potential Na+/H+ antiporters, neither in the genome of T. kivui, nor in the genome of the closely related acetogen M. thermoacetica. Intriguingly, the existence of an electrogenic Na+/H+ antiport has been demonstrated in M. thermoacetica (39). However, the sequence of translocation events may be different. A primary transport of Na+ was indicated by the stimulatory effect of relieving thermodynamic backup pressure at IMVs with sodium ionophore, but not protonophore (Fig. 2B) and the prevented ΔpH formation across IMVs in the presence of sodium ionophore (Fig. 2I). This would argue for Na+ first and H+ second, which is also supported by the fact that the ATP synthase of T. kivui is proton coupled. However, Na+ transport into IMVs was inhibited instead of stimulated in the presence of protonophore (Fig. 2 C and D). This could be explained by a Na+/H+ antiport rate that exceeds the primary transport, as described for an antiport activity in the Ech-containing methanogen M. barkeri (40). To resolve the sequence and mechanism of ion translocation, a genetic approach or analyses of purified Ech complex would be required. Anyway, the use of an antiporter module connected to the primary ion pump is of decisive importance for growth of microorganisms at the thermodynamic limit of life: A Na+/H+ antiport with nonintegral stoichiometry would allow the translocation of less than one ion per redox reaction, a prerequisite for growth at ΔG values that do not allow for the translocation of even one ion (24, 41). By demonstrating that Ech forms a functional respiratory enzyme in T. kivui, a second mode of energy conservation in acetogenic bacteria was experimentally verified. Ech is a more ancient energy conserving system, as reflected by simple iron sulfur cofactors as opposed to complex organic flavin cofactors in the other chemiosmotic coupling site of acetogens, the Rnf complex (20, 42, 43). Finally, we propose the following chemolithoautotrophic metabolism in T. kivui: 4 mol CO are converted to 1 mol acetate, yielding 1.25 mol of ATP. The Ech as well as the electron-bifurcating hydrogenase of T. kivui are essential for the process; a model is given in Fig. 4.
Fig. 3.
Genetic arrangement of ech and mbh clusters and cartoon models of protein complexes of Ech(s) or Mbh in T. kivui, M. barkeri, and P. furiosus. Gene clusters are depicted as deposited for T. kivui DSM 2030, M. barkeri Fusaro DSM 804, and P. furiosus COM1 on IMG/M ER (44). (A) Genes containing TMHs are depicted in red, genes encoding soluble proteins are depicted in green, and genes encoding the large and small subunit of the [NiFe] hydrogenase are depicted in blue. (B) The same color code applies to the corresponding protein subunits. TMHs (gray bars), iron-sulfur clusters (cubes), and the [NiFe] active site were predicted with InterPro (45).
Fig. 4.
A redox-balanced model for CO metabolism in T. kivui. The Ech complex (green) is constituted of either one or both Ech1 and Ech2. It catalyzes Fd2− oxidation and H2 evolution coupled to the transport of H+ and Na+. The F1FO ATP synthase (yellow) has an assumed H+/ATP stoichiometry of 12/3. The electron bifurcating hydrogenase and hydrogen-dependent CO2 reductase are depicted in blue. Numbers are rounded.
In conclusion, this work demonstrated that (i) an Ech hydrogenase is a respiratory enzyme in a bacterium, (ii) a respiratory hydrogenase and an ATP synthase are sufficient to enable microbial life at the thermodynamic limit of life, and (iii) there is a second mode of energy conservation in acetogenic bacteria.
Materials and Methods
Experimental procedures for cultivation of the organism; preparation and experiments with resting cells; determination of H2, CO, and acetate concentrations; preparation of IMVs; measurement of Ech activity at IMVs; measurement of 22Na+ or H+ translocation; and determination of relative transcript levels are described in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818580116/-/DCSupplemental.
References
- 1.Wächtershäuser G. On the chemistry and evolution of the pioneer organism. Chem Biodivers. 2007;4:584–602. doi: 10.1002/cbdv.200790052. [DOI] [PubMed] [Google Scholar]
- 2.Martin W, Russell MJ. On the origin of biochemistry at an alkaline hydrothermal vent. Philos Trans R Soc Lond B Biol Sci. 2007;362:1887–1925. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drake HL, Gössner AS, Daniel SL. Old acetogens, new light. Ann N Y Acad Sci. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- 4.Ljungdahl L, Wood HG. Incorporation of C14 from carbon dioxide into sugar phosphates, carboxylic acids, and amino acids by Clostridium thermoaceticum. J Bacteriol. 1965;89:1055–1064. doi: 10.1128/jb.89.4.1055-1064.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller V, Frerichs J. 2013. Entry for “Acetogenic bacteria.” Encyclopedia of Life Science (John Wiley & Sons Ltd, Chichester, UK)
- 6.Köpke M, Noack S, Dürre P. 2011. The past, present, and future of biofuels–Biobutanol as promising alternative. Biofuel Production–Recent Developments and Prospects (InTech, Rijeka, Croatia), pp 451–486.
- 7.Liew F, et al. Gas fermentation-a flexible platform for commercial scale production of low-carbon-fuels and chemicals from waste and renewable feedstocks. Front Microbiol. 2016;7:694. doi: 10.3389/fmicb.2016.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bertsch J, Müller V. Bioenergetic constraints for conversion of syngas to biofuels in acetogenic bacteria. Biotechnol Biofuels. 2015;8:210. doi: 10.1186/s13068-015-0393-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bache R, Pfennig N. Selective isolation of Acetobacterium woodii on methoxylated aromatic acids and determination of growth yields. Arch Microbiol. 1981;130:255–261. [Google Scholar]
- 10.Eichler B, Schink B. Oxidation of primary aliphatic alcohols by Acetobacterium carbinolicum sp. nov., a homoacetogenic anaerobe. Arch Microbiol. 1984;140:147–152. [Google Scholar]
- 11.Müller V. Energy conservation in acetogenic bacteria. Appl Environ Microbiol. 2003;69:6345–6353. doi: 10.1128/AEM.69.11.6345-6353.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ragsdale SW. Enzymology of the wood-Ljungdahl pathway of acetogenesis. Ann N Y Acad Sci. 2008;1125:129–136. doi: 10.1196/annals.1419.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ljungdahl LG. 1994. The acetyl-CoA pathway and the chemiosmotic generation of ATP during acetogenesis. Acetogenesis (Chapman & Hall, New York), pp 63–87.
- 14.Wood HG, Ragsdale SW, Pezacka E. The acetyl-CoA pathway of autotrophic growth. FEMS Microbiol Rev. 1986;39:345–362. [Google Scholar]
- 15.Schuchmann K, Müller V. Autotrophy at the thermodynamic limit of life: A model for energy conservation in acetogenic bacteria. Nat Rev Microbiol. 2014;12:809–821. doi: 10.1038/nrmicro3365. [DOI] [PubMed] [Google Scholar]
- 16.Pezacka E, Wood HG. Role of carbon monoxide dehydrogenase in the autotrophic pathway used by acetogenic bacteria. Proc Natl Acad Sci USA. 1984;81:6261–6265. doi: 10.1073/pnas.81.20.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biegel E, Müller V. Bacterial Na+-translocating ferredoxin:NAD+ oxidoreductase. Proc Natl Acad Sci USA. 2010;107:18138–18142. doi: 10.1073/pnas.1010318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritz M, Müller V. An intermediate step in the evolution of ATPases–The F1F0-ATPase from Acetobacterium woodii contains F-type and V-type rotor subunits and is capable of ATP synthesis. FEBS J. 2007;274:3421–3428. doi: 10.1111/j.1742-4658.2007.05874.x. [DOI] [PubMed] [Google Scholar]
- 19.Vignais PM, Billoud B. Occurrence, classification, and biological function of hydrogenases: An overview. Chem Rev. 2007;107:4206–4272. doi: 10.1021/cr050196r. [DOI] [PubMed] [Google Scholar]
- 20.Schut GJ, et al. The role of geochemistry and energetics in the evolution of modern respiratory complexes from a proton-reducing ancestor. Biochim Biophys Acta. 2016;1857:958–970. doi: 10.1016/j.bbabio.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 21.Böhm R, Sauter M, Böck A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 22.Welte C, Krätzer C, Deppenmeier U. Involvement of Ech hydrogenase in energy conservation of Methanosarcina mazei. FEBS J. 2010;277:3396–3403. doi: 10.1111/j.1742-4658.2010.07744.x. [DOI] [PubMed] [Google Scholar]
- 23.Sapra R, Bagramyan K, Adams MWW. A simple energy-conserving system: Proton reduction coupled to proton translocation. Proc Natl Acad Sci USA. 2003;100:7545–7550. doi: 10.1073/pnas.1331436100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim JK, Mayer F, Kang SG, Müller V. Energy conservation by oxidation of formate to carbon dioxide and hydrogen via a sodium ion current in a hyperthermophilic archaeon. Proc Natl Acad Sci USA. 2014;111:11497–11502. doi: 10.1073/pnas.1407056111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, et al. Structure of an ancient respiratory system. Cell. 2018;173:1636–1649.e16. doi: 10.1016/j.cell.2018.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hess V, Poehlein A, Weghoff MC, Daniel R, Müller V. A genome-guided analysis of energy conservation in the thermophilic, cytochrome-free acetogenic bacterium Thermoanaerobacter kivui. BMC Genomics. 2014;15:1139. doi: 10.1186/1471-2164-15-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daniel SL, Hsu T, Dean SI, Drake HL. Characterization of the H2- and CO-dependent chemolithotrophic potentials of the acetogens Clostridium thermoaceticum and Acetogenium kivui. J Bacteriol. 1990;172:4464–4471. doi: 10.1128/jb.172.8.4464-4471.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weghoff MC, Müller V. CO metabolism in the thermophilic acetogen Thermoanaerobacter kivui. Appl Environ Microbiol. 2016;82:2312–2319. doi: 10.1128/AEM.00122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schönheit P, Wäscher C, Thauer RK. A rapid procedure for the purification of ferredoxin from Clostridia using polyethyleneimine. FEBS Lett. 1978;89:219–222. doi: 10.1016/0014-5793(78)80221-x. [DOI] [PubMed] [Google Scholar]
- 30.Kottenhahn P, Schuchmann K, Müller V. Efficient whole cell biocatalyst for formate-based hydrogen production. Biotechnol Biofuels. 2018;11:93. doi: 10.1186/s13068-018-1082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess V, Schuchmann K, Müller V. The ferredoxin:NAD+ oxidoreductase (Rnf) from the acetogen Acetobacterium woodii requires Na+ and is reversibly coupled to the membrane potential. J Biol Chem. 2013;288:31496–31502. doi: 10.1074/jbc.M113.510255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim JK, Kang SG, Lebedinsky AV, Lee JH, Lee HS. Identification of a novel class of membrane-bound [NiFe]-hydrogenases in Thermococcus onnurineus NA1 by in silico analysis. Appl Environ Microbiol. 2010;76:6286–6289. doi: 10.1128/AEM.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sargent F. The model [NiFe]-hydrogenases of Escherichia coli. Adv Microb Physiol. 2016;68:433–507. doi: 10.1016/bs.ampbs.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 34.Künkel A, Vorholt JA, Thauer RK, Hedderich R. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur J Biochem. 1998;252:467–476. doi: 10.1046/j.1432-1327.1998.2520467.x. [DOI] [PubMed] [Google Scholar]
- 35.Silva PJ, et al. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur J Biochem. 2000;267:6541–6551. doi: 10.1046/j.1432-1327.2000.01745.x. [DOI] [PubMed] [Google Scholar]
- 36.Schut GJ, Boyd ES, Peters JW, Adams MW. The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol Rev. 2013;37:182–203. doi: 10.1111/j.1574-6976.2012.00346.x. [DOI] [PubMed] [Google Scholar]
- 37.Mayer F, Lim JK, Langer JD, Kang SG, Müller V. Na+ transport by the A1AO-ATP synthase purified from Thermococcus onnurineus and reconstituted into liposomes. J Biol Chem. 2015;290:6994–7002. doi: 10.1074/jbc.M114.616862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlegel K, Leone V, Faraldo-Gómez JD, Müller V. Promiscuous archaeal ATP synthase concurrently coupled to Na+ and H+ translocation. Proc Natl Acad Sci USA. 2012;109:947–952. doi: 10.1073/pnas.1115796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terracciano JS, Schreurs WJ, Kashket ER, Membrane H. Membrane H conductance of Clostridium thermoaceticum and Clostridium acetobutylicum: Evidence for electrogenic Na/H antiport in Clostridium thermoaceticum. Appl Environ Microbiol. 1987;53:782–786. doi: 10.1128/aem.53.4.782-786.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Müller V, Blaut M, Gottschalk G. Generation of a transmembrane gradient of Na+ in Methanosarcina barkeri. Eur J Biochem. 1987;162:461–466. doi: 10.1111/j.1432-1033.1987.tb10624.x. [DOI] [PubMed] [Google Scholar]
- 41.Müller V, Hess V. The minimum biological energy quantum. Front Microbiol. 2017;8:2019. doi: 10.3389/fmicb.2017.02019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedrich T, Scheide D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5. doi: 10.1016/s0014-5793(00)01867-6. [DOI] [PubMed] [Google Scholar]
- 43.Backiel J, et al. Covalent binding of flavins to RnfG and RnfD in the Rnf complex from Vibrio cholerae. Biochemistry. 2008;47:11273–11284. doi: 10.1021/bi800920j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markowitz VM, et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 2014;42:D560–D567. doi: 10.1093/nar/gkt963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn RD, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45:D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.