Abstract
Cardiovascular diseases exist across all developed countries societies. Biomarkers that can predict or diagnose diseases early in their pathogeneses can reduce their morbidity and mortality in afflicted individuals. microRNAs are small regulatory RNAs that modulate translation and have been identified as potential fluid-based biomarkers across numerous maladies. We describe the current state of cardiovascular disease biomarkers across a range of diseases, including myocardial infarction, acute coronary syndrome, myocarditis, hypertension, heart failure, heart transplantation, aortic stenosis, diabetic cardiomyopathy, atrial fibrillation, and sepsis. We present the current understanding of microRNAs as possible biomarkers in these categories and where their best opportunities exist to enter clinical practice.
Keywords: microRNAs, biomarkers, cardiovascular disease, hypertension, sepsis, aortic stenosis
INTRODUCTION
In the field of cardiovascular disease, biomarkers are essential tools in a clinician’s armamentarium. The best of our biomarkers can aid in diagnosing an acute myocardial infarction (AMI), indicate long-standing heart failure, and predict the rejection of a transplanted heart. However, not all biomarkers are robust, and many fields within cardiovascular disease management await useful biomarkers to aid in diagnosis and assessing prognosis. Across all of these scenarios there is the opportunity for better biomarkers. It is with this appreciation that we critically evaluate microRNAs (miRNAs) as a new category of biomarkers for cardiovascular diseases. In this review, we analyze what the opportunities are for using miRNA biomarkers across particular disease states, specifically comparing miRNA biomarkers to what is currently used for the disease when appropriate. We also provide context for the intricacies of miRNA biomarker studies so each reader has the tools to critically evaluate any of those described herein or forthcoming miRNA studies for their potential clinical use in cardiovascular disease.
A PRIMER ON miRNAS AND BIOMARKERS IN CARDIOVASCULAR DISEASE
A Brief Description of miRNA Biogenesis
miRNAs are short (18–22 nucleotide) regulatory RNAs that bind mRNAs and decrease protein translation. miRNAs are generally cotranscribed with neighboring genes or co transcribed within a cluster of miRNAs (a polycistronic cluster). These primary miRNAs (pri-miRNAs) are then processed by Drosha into pre-miRNAs and cleaved by Dicer into mature miRNAs. One half of a mature miRNA (either the 5p or 3p sequence) is loaded into Argonaute 2 (Ago2) as part of RISC (the RNA-induced silencing complex). Although complicated, controversial, and with many exceptions to the rule, a six base pair seed sequence at the 5′ end binds to a complementary region along the 3′ untranslated region of an mRNA, thus enabling specific regulation of scores of mRNAs by a single miRNA (1, 2). This entire mechanism has been extensively described and reviewed elsewhere (3–5).
The Strict Nomenclature of miRNAs
miRNAs have a strict naming convention (6, 7). A miRNA name begins with a three-letter code for the species (e.g., hsa for Homo sapiens and cel for Caenorhabditis elegans). This is followed by miR and then a number of some sequential order. If miRNAs have the same seed sequence, indicating a shared family function, there may be a letter, in alphabetical order, after the number. Examples include hsa-miR-181a, hsa-miR-181b, and hsa-miR-181c. Each processed miRNA has two strands: a dominant strand (more abundant and loaded in RISC) and a passenger strand (degraded, less abundant), one from each of the 5p and 3p ends. These were denoted as the s (sense) and as (antisense) or * strands (star, denoting passenger) in earlier literature. A complete miRNA name would be hsa-miR-181a-3p or hsa-miR-181a-5p. The one exception to this naming convention is the let-7 family of miRNAs, which kept their original names. Once a species is established, the species code is not commented on further in most manuscripts. Equally, if the 5p or 3p designation is not stated, it is generally assumed that the dominant strand is being referenced.
The True Number of Human miRNAs is Unknown
The number of human miRNAs is controversial. miRBase (http://www.mirbase.org/) has been considered the central repository of all known miRNAs. Version 21 of miRBase listed 2,588 human miRNAs, a relatively stable number compared with prior versions of the database (6). However, since that version appeared, with the explosion in small RNA sequencing (RNA-seq) and generic novel miRNA discovery tools, thousands of additional human miRNAs have been proposed (8–11). Most of these have been collected into the new miRCarta database (https://mircarta.cs.uni-saarland.de/) that adds an additional 12,857 human miRNA precursors to our collection (12). Are all of these additional sequences truly in the miRNA oeuvre? Many groups suggest otherwise, indicating that miRBase is rife with non-miRNA species, and these newly minted novel miRNAs are mostly dead on arrival (13–17). In response to these concerns, MiRGeneDB (https://mircarta.cs.uni-saarland.de/) was developed as a hand-curated classification of miRNAs based on strict criteria, including genetic conservation and canonical development (14, 18). This database lists only 586 human miRNA genes. Clearly, the collection of small RNA–expressed species requires greater clarity and perhaps additional nomenclature to describe miRNA-like small RNAs (17). Regardless, for the purpose of utilizing miRNAs as biomarkers, any sequence that gives a consistent signal indicating the presence of disease can be useful.
Many miRNAs are Expressed in a Cell Type–Specific Fashion
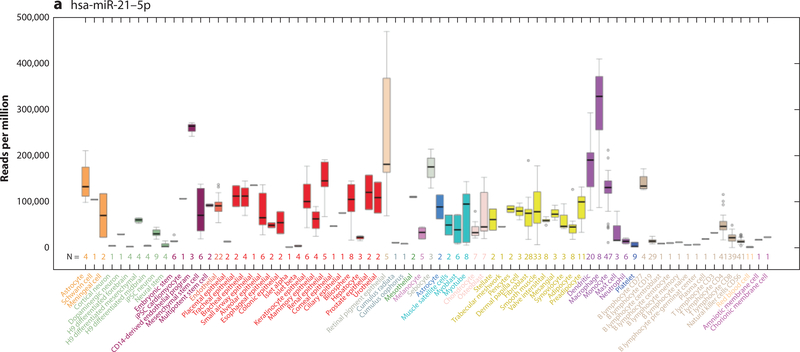
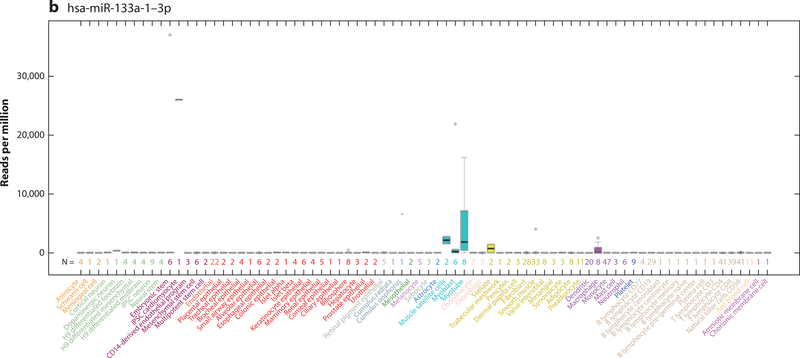
A factor in understanding the function and biomarker utility of miRNAs is the appreciation of the cells in which they are expressed. If a miRNA is expressed in a cardiomyocyte, it may have more relevance as a cardiovascular biomarker than one that also is expressed in a noncardiovascular cell (e.g., a hepatocyte). Early expression localization studies were performed in tissue (19–21). These limited our understanding of the contribution of the various cell types that are expressed across a variety of tissues—such as endothelial cells, red blood cells, and fibroblasts—to the changing miRNA levels in diseased tissue. This led to problems that plague the miRNA literature (7, 22, 23). Better characterization of miRNAs at the cellular level has been slow to develop. We have been actively engaged in the realization of miRNA cellular localization (24, 25), culminating in a cellular microRNAome that is based on miRNA data from 46 primary cell types and 42 cancer or immortalized cell lines (8). This work was complemented by two other papers in 2017 describing miRNA expression in many additional cell types (9, 26). These combined data are now accessible as a University of California Santa Cruz genome browser barChart that shows miRNA expression levels across 78 primary cell types and 51 cancer or immortalized cells (Figure 1) (17). For the first time, any investigator can quickly determine the cellular location of their miRNA of interest to determine its relevance to disease (Table 1). This information is vitally important to miRNA biomarker studies as seen below.
Figure 1.
University of California Santa Cruz Genome Browser barChart of the cellular expression patterns of microRNAs (miRNAs). (a) hsa-miR-21–5p expression is ubiquitous across most cell types, although the absolute expression level is variable. (b) hsa-miR-133a-1–3p shows cell type expression restricted to induced pluripotent stem cell cardiomyocytes, muscle satellite cells, myoblasts, myotubes, stellate cells, and macrophages at varying levels of expression.
Table 1.
Cellular localization of microRNAs described in biomarker studies in the review
| microRNAa | Cell-type enrichment |
|---|---|
| Let-7e | Ubiquitous |
| miR-19b | Ubiquitous |
| miR-21 | Ubiquitous |
| miR-28 | Ubiquitous |
| miR-29 | Ubiquitous |
| miR-30a | Nonhematologic cells |
| miR-30c | Ubiquitous |
| miR-99a | Ubiquitous (nonhematopoietic) |
| miR-122 | Hepatocytes |
| miR-126 | Endothelial cells, platelets |
| miR-134 | Platelets, iPSC neurons, islet β-cells |
| miR-143 | Smooth muscle cells, fibroblasts, cardiac muscle cells, skeletal muscle cells |
| miR-144 | Red blood cells |
| miR-145 | Smooth muscle cells, fibroblasts, cardiac muscle cells, skeletal muscle cells |
| miR-146a | Melanocytes, macrophages, lymphocytes, platelets |
| miR-146b | Dendritic cells, lymphocytes, macrophages |
| miR-150 | Lymphocytes |
| miR-155 | Lymphocytes |
| miR-182 | Epithelial cells |
| miR-192 | Epithelial cells |
| miR-197 | Neutrophils, macrophages, monocytes |
| miR-200a | Epithelial cells |
| miR-200b | Epithelial cells |
| miR-200c | Epithelial cells |
| miR-210 | Ubiquitous |
| miR-214 | Smooth muscle cells, fibroblasts, RPE cells, iPSC neurons |
| miR-215 | Epithelial cells |
| miR-221 | Ubiquitous |
| miR-223 | Macrophages, neutrophils |
| miR-296 | No strong cell signals |
| miR-323 | iPSC neurons |
| miR-328 | Platelets |
| miR-331 | Ubiquitous |
| miR-342 | Macrophages, lymphocytes |
| miR-361 | Lymphocytes |
| miR-375 | Islet β-cells (high expression), colonic epithelial cells (low expression) |
| miR-378a | Ubiquitous |
| miR-451 | Red blood cells |
| miR-517 | Embryonic stem cells, chorionic membrane cells |
| miR-637 | No strong cell signals |
| miR-652 | Platelets, macrophages, RPE cells |
| miR-664 | Lymphocytes (low expression) |
| miR-941 | Neutrophils, macrophages |
| miR-4516 | No strong cell signals |
Abbreviations: iPSC, induced pluripotent stem cell; RPE, retinal pigment epithelium.
Ubiquitous indicates widespread expression, but not necessarily in all cell types.
MyomiRs
Between organ-specific data and cell-specific data, it has emerged that a group of miRNAs is found either exclusively or more abundantly in myocytes. These are termed myomiRs. As described in Table 2, some of these miRNAs are found within myosin heavy chain genes (27). Most of these myomiRs are also expressed in skeletal muscle, with miR-208 being the most specific of the myomiRs for cardiac muscle. This group of miRNAs is particularly important in cardiovascular disease. Because of their exclusivity to muscle cells, they should be found only in blood-based fluids (plasma and serum) if they are spilled from injured cells or perhaps be present if muscle cells use these miRNAs to signal to other cells (28). Of this group, miR-1 (miR-1–1 and miR-1–2 have identical sequences from different loci) has mainly been described in cardiac and skeletal muscle cells (8). However, it is also found in low levels in many organs (17, 29), suggesting that some noncardiac- and skeletal muscle cell expresses miR-1 in vivo. MyomiRs are an important group of miRNAs in cardiovascular biomarker studies, as discussed below.
Table 2.
Myocyte microRNAs (myomiRs) with increased or exclusive expression in skeletal and cardiac myocytes
| microRNA | Gene locus | Chromosome location | Cell type expression |
|---|---|---|---|
| miR-1–1 | NA | 20 | Cardiac, skeletal (and others at low levels?) |
| miR-1–2 | Intronic to MIB1 | 18 | Cardiac, skeletal (and others at low levels?) |
| miR-133a | Intronic to MIB1 | 18 | Cardiac, skeletal |
| miR-133b | Intronic to LINCMD1 | 6 | Skeletal |
| miR-206 | Downstream of LINCMD1 | 6 | Skeletal |
| miR-208a | Intronic to MYH6 | 14 | Cardiac |
| miR-208b | Intronic to MYH7 | 14 | Skeletal |
| miR-499a | Intronic to MYH7B | 20 | Cardiac, skeletal |
| miR-499b | Intronic to MYH7B | 20 | Cardiac, skeletal |
NA, not applicable.
Biomarkers
Biomarkers may be biochemical, molecular, histologic, radiographic, or physiologic. They may be obtained from whole blood, serum, plasma, body fluids, or tissues. For the purposes of this review, we discuss current biochemical and molecular biomarkers and newer, potential miRNA biomarkers. Biomarkers have multiple uses. They can be indicators of a pathologic or physiologic process; they can be used to monitor the progress of or provide the prognosis for a disease; they can be used to monitor the efficacy or toxicity of a therapeutic agent; or they may represent a therapeutic target (30). They may also be used to stratify patients for the purposes of a clinical trial. Thus, their usefulness has been well established.
However, they should be used only once they have been well validated for their intended purpose. They may also undergo considerable improvement in terms of sensitivity or specificity, or both. An excellent example of this is the refinement of the use of blood measurements of cardiac-specific troponin as an indicator of myocardial damage (31).
CURRENT BIOMARKERS OF CARDIOVASCULAR DISEASE
Myocardial Infarction
Of all the cardiovascular diseases, the diagnosis of an AMI, usually as the result of an acute coronary artery event, is of the most societal importance. Fortunately, robust science has produced biomarkers that rapidly detect an AMI. The measurement of cardiac troponins [either Troponin I (cTnI) or Troponin T (cTnT)] has emerged as our gold standard diagnostic test. Cardiac troponins are cardiomyocyte-specific proteins that are spilled into the circulation when a cardiomyocyte dies (32). They can be measured rapidly in clinical chemistry laboratories and as point-of-care testing (33). American Heart Association guidelines recommend taking two measurements, one at first assessment and a second 3–6 hours later (34). A test is considered positive if at least one value is >99th percentile of the upper reference limit. With the advent of a new high-sensitivity test (hs-cTnT), this biomarker is firmly established as a key diagnostic tool in confirming a myocardial event. However, troponins can be elevated in non-AMI settings, including myocarditis and end-stage renal disease, although they are generally at lower levels than following an AMI and have different kinetics over time due to their sustained release (also known as troponin leak) or accumulation. Any cardiac injury can cause a release of troponins, with the extent of the injury correlating roughly with the level of the troponin. Thus, it is a sensitive but not specific test for an AMI, and opportunities may exist to find biomarkers with more discrimination.
Hypertension
Hypertension, or high blood pressure, is a polygenic and multifactorial disease, and its course can be altered by many environmental factors. Hypertension is injurious to the body’s organs and is associated with increased incidences of stroke, myocardial infarction, and aortic aneurysm, among other problems. The diagnosis is easily made with the use of a sphygmomanometer to measure blood pressure, and the criteria for the diagnosis and treatment are well established (35, 36). Biomarkers beyond those obtained by the sphygmomanometer are clearly not needed for diagnosing hypertension; however, they may be of great value in helping to (a) stratify a patient for a therapeutic approach, (b) serve as prognostic indicators of hypertension-related sequelae or comorbidities, and (c) indicate a specific etiology of hypertension. It is in these roles that miRNA biomarkers may hold value.
Rejection of Orthotopic Heart Transplant
The gold standard for diagnosing heart transplant rejection is a pathologist’s review of endomyocardial biopsy material (37, 38). However, simpler blood-based methods have emerged. A gene expression profiling method called AlloMap (CareDx, Brisbane, CA) is used clinically to rule out cardiac rejection in patients (39). A second method, still in development, is a cell-free DNA assay that can detect increased levels of DNA from the donor heart in a patient’s blood, indicating heart injury (40). A third method utilizes brain natriuretic peptide (BNP) levels as indicators of rejection (41). The pros and cons of these methods as biomarkers are discussed in greater detail elsewhere, but no current blood-based test is useful at distinguishing between cellular and antibody-mediated rejection, among other limitations (42).
Myocarditis
Myocarditis can be difficult to diagnose, and having specific biomarkers could greatly improve our diagnostic abilities. Currently, endomyocardial biopsy remains the gold standard, as for heart transplantation (43). Since this is an invasive procedure and not widely offered, other methods are used as well. Even if myocarditis is present, the heterogeneous nature of the disease results in only ~50% of cases being diagnosed positively by biopsy (44). Other biomarker modalities that can be used include imaging studies and measurement of levels of troponin, BNP, and N-terminal pro-B-type natriuretic peptide (NT-proBNP). These tools have diagnostic overlap with myocardial infarction and other causes of heart failure, so there is not yet a robust biomarker specific to myocarditis.
Diabetic Cardiomyopathy
Diabetic cardiomyopathy is a well-recognized clinical entity that affects some patients with type 1 or type 2 diabetes mellitus. It is slowly progressive, and there are a number of risk factors, including a patient’s age, the duration of diabetes, and poor glycemic control. It associates with elevated serum creatinine and microalbuminuria, two nonspecific biomarkers of disease (45, 46). However, the discovery of specific biomarkers that could predict its presence before it becomes clinically manifest could lead to early and potentially specific treatments beyond simply controlling blood sugar and making diet and lifestyle modifications. Currently, no such biomarker exists.
Aortic Stenosis
Aortic stenosis is the most common valvular disorder in developed countries. Once a diagnosis of stenosis is made by auscultation of a murmur and follow-up echocardiogram, a patient is usually managed medically until an aortic valve replacement is needed. One of the major complications of aortic stenosis is left ventricular hypertrophy, with fibrosis leading to heart failure and death. The only blood-based biomarkers are BNP and NT-proBNP, which are nonspecific and usually indicative of decompensation, which occurs late in the management of the patient (47). Imaging modalities to identify myocardial fibrosis are being developed, but have not yet proven their value for aortic stenosis.
Heart Failure with Preserved Ejection Fraction
Heart failure with preserved ejection fraction (HFpEF) is, as the name implies, a cause of heart failure in which the ejection fraction is maintained, in contrast to the more common heart failure with reduced ejection fraction (HFrEF). HFpEF was previously called diastolic dysfunction or diastolic heart failure, and it is a catchall term for a variety of patients with diverse clinical phenotypes. It has remained a challenge to treat and discriminate across the different subsets of HFpEF. A number of typical cardiovascular and renal biomarkers are elevated in HFpEF (BNP, ST2, galectin 3, matrix metalloproteinases, osteopontin, cystatin C, troponins, and inflammatory cytokines) (48). None of these have much specificity for HFpEF, and diagnosing this entity remains a challenge. Although HFpEF and HFrEF can be distinguished by the determination of the ejection fraction, the differences between these entities remain an open scientific question.
Atrial Fibrillation
Atrial fibrillation can be readily diagnosed via electrocardiography (EKG) and can be designated as either paroxysmal or persistent. While patients can often be treated either electrically or pharmacologically, with reversion to a normal sinus rhythm, a percentage will revert to atrial fibrillation. In addition, atrial fibrillation is often the sequelae of underlying heart disease. Thus, while making the diagnosis of atrial fibrillation by EKG is established, a biomarker (or biomarkers) that could predict who is most prone to having atrial fibrillation for the first time or a recurrence could have a significant impact on the therapeutic strategies used for maintaining a normal sinus rhythm.
Sepsis
Although sepsis has long been characterized as a syndrome of disordered immune response to infection, a recent paradigm shift has altered the focus of sepsis research onto its cardiovascular dysfunction and resultant organ edema, ischemia, and failure. Further, despite numerous attempts to define its clinical features (49, 50), sepsis often presents nonspecifically and is frequently underrecognized early in its course. Thus, reliable diagnostic biomarkers could significantly improve sepsis outcomes, as poor recognition and delay in treatment is strongly associated with increased mortality (51). Procalcitonin, a precursor of the hormone calcitonin, has been demonstrated to have moderate discriminatory capability in the diagnosis of sepsis, and it has been incorporated sporadically into clinical practice (52). In order to gain acceptance by clinicians, miRNA-based diagnostic biomarkers would likely need to demonstrate superior test characteristics compared with procalcitonin.
A HISTORY OF miRNA BIOMARKER STUDIES
In 2008, Muneesh Tewari’s group (53) indicated that miRNAs were stable in blood fluids and could be useful as biomarkers of disease. This seminal paper initiated a new enterprise to identify miRNAs that could serve as biomarkers for all neoplastic and nonneoplastic diseases. Thousands of publications on a litany of diseases using a variety of miRNA biomarker strategies have since been published. While this canon is too extensive to describe fully, certain generalities apply to these data sets.
miRNA biomarkers are found in a variety of biological fluids. Most miRNA biomarker studies are performed using serum or plasma. Fewer studies have used urine, saliva, whole blood, or peripheral blood mononuclear cells (PBMCs). Within serum or plasma, miRNAs can be found in protein complexes, bound to Ago2, or located in exosomes derived from most human cell types in various and changing ratios.
miRNAs can be assayed by different methods. Most miRNA biomarker studies have relied on miRNA profiling using hybridization arrays or parallel quantitative polymerase chain reaction (qPCR) systems containing hundreds of miRNAs or directed qPCR approaches on a more limited set of miRNAs (54). Other methods include droplet digital PCR and small RNA-seq. Most of these methods rely on a normalization control, which may be an intrinsic RNA, an extrinsic (spike-in) RNA, or a global normalization method.
Robust miRNA biomarker studies tend to have two stages. Generally, in the first stage, an agnostic, array-based approach is used on a moderate-size population (20–50 samples) to identify several potential individual targets or a small collection of miRNAs that in combination predict a certain disease or outcome. Then a second, larger population is assayed for only this subset of miRNAs, usually by qPCR. Another common approach is to use literature searches and prior similar biomarker studies to prioritize a more focused list of miRNAs, which are then assayed directly in one large study.
Practical Considerations for Using miRNAs as Biomarkers
The overwhelming excitement about miRNA biomarkers (and the huge number of patent filings) quickly gave way to some underappreciated complexities of assaying for miRNAs. Perhaps the initial publication warning of potential confounders was again written by Tewari’s group (55). They demonstrated that many of the miRNAs identified as being associated with various neoplasms were, in fact, hematologic-specific miRNAs that correlated with blood cell levels. For example, miR-150 is found exclusively in lymphocytes and correlates with a patient’s absolute lymphocyte count, which can vary between healthy and diseased patients. Thus, many of these miRNA biomarkers merely indicated what a complete blood count would have shown. This lack of knowledge of miRNA expression patterns has hampered a significant number of miRNA biomarker studies, in which miRNAs unrelated to the disease process are described.
We followed up Tewari and colleagues’ work by investigating nonneoplastic miRNAs and focusing on whether a miRNA that was claimed as a biomarker for a particular disease was biologically plausible as a specific marker for that disease (25). Unfortunately, the vast majority of miRNAs failed to show usefulness based on these criteria. Six miRNAs claimed as specific biomarkers were biomarkers for nine or more diseases, which makes them hard to claim as specific. We did note that myomiRs were frequently upregulated in a variety of cardiac diseases that injure muscle, such as myocardial infarction and myocarditis. So what causes the difficulties described in these publications?
Technical Causes of Variability in miRNA Biomarker Studies
The technical causes of variation in miRNA levels are well known (Table 3). The major cause of variation is likely sample preparation. As seen in one example from plasma, the extent of centrifugation can change the level of some miRNAs by >1,000-fold, but it may change others by <4-fold (56).
Table 3.
Technical and biological factors affecting fluid-based studies of microRNA biomarkers
| Technical factors | Biological factors |
|---|---|
| Extent of centrifugation | Fluid source (serum, plasma, PBMCs, exosome) |
| Spike-in control | Sex |
| Platform | Time of day |
| RNA isolation method | Hemolysis |
| Storage method | Genetic variation |
| Analysis method | Patient phenotype |
| Fluid collection method | Age |
| Alignment method (for small RNA sequencing) | |
| RNA storage (stability) | |
| Lower level of expression limitation |
Abbreviation: PBMCs, peripheral blood mononuclear cells.
Another important source of variation is the normalization approach used. In tissue, the small nuclear U6 RNA (known as RNU6B or U6) can be used as a housekeeping gene to normalize miRNA levels. It has also been used for biomarker studies of plasma and serum. Unfortunately, RNU6B is not native to plasma or serum, being released only after the death of cells; thus, it should not be used as a normalizing control for this type of sample (25, 54), and studies based on U6 normalization should be questioned.
Different miRNA platforms using a variety of methods (hybridization, PCR) have their own biases that can affect miRNA levels. This issue was explored nicely by the miRQC (microRNA quality control) consortium (57). Finally, the statistical methods used for normalization—global methods or specific controls—can strongly influence biomarker discovery (58). One common flaw seen across many biomarker studies is the lack of a basal threshold for expression. Thus, groups find strong signals in the noise part of the data. Many of these purported miRNA biomarkers (often miRNAs with four-number names, such as miR-3168 or miR-1915) have little to no expression (although absolute expression is often not commented on) and are the result of spurious signals. These findings are rarely, if ever, replicated between studies (7).
Biological Causes of Variability in miRNA Biomarker Studies
In addition to technical causes of variation, biological factors also are worthy of consideration (Table 3). A major decision point in a biomarker study is the fluid source used for the miRNAs. Plasma and serum have both been used, although historically this decision was likely based on what samples were already present in a freezer, rather than planned in advance. miRNAs are known to vary between these fluids (54). For plasma, the choice of different preparations, such as platelet-poor plasma or platelet-rich plasma, can contribute to miRNA differences. miRNAs have also been reported to vary by sex (59, 60), patient’s age (61, 62), and time of day of collection (63). The last of these suggests there are fluctuations in the half-life of circulating miRNAs, which is a potential problem when trying to determine a temporal relationship with the disease of interest. Single nucleotide polymorphisms (SNPs) also exist in a small number of mature miRNAs. Even if the SNPs do not change the levels of the miRNAs, they can impact hybridization to miRNA-specific probes and qPCR primer binding or affect miRNA counting in alignment tools (64). A number of publications have nicely addressed certain pitfalls and approaches to consider when developing and critically reviewing a miRNA biomarker study (7, 25, 65–67).
STUDIES OF miRNA BIOMARKERS IN CARDIOVASCULAR DISEASE
miRNA Biomarkers in Acute Myocardial Infarction and Acute Coronary Syndrome
Of all the cardiovascular miRNA biomarker opportunities, none has received as much attention as AMI and acute coronary syndrome (ACS). This is likely due to the high societal demand, clear phenotype, and potential financial reward. However, compared with many other cardiovascular diseases, the barrier for entry of a new, clinically useful biomarker is significantly higher due to the strength of troponin biomarkers in the current marketplace. Thus, to prove their usefulness, miRNA biomarkers for AMI and ACS must have better test statistics or other attributes that will make them more powerful than the troponins.
More than 100 manuscripts have evaluated miRNAs as biomarkers for this category, and it is beyond the scope of this review to cover them all. One of the first AMI biomarker studies was the work of Ai et al. (68) in 2010 that explored miR-1 and miR-133, finding that miR-1, but not miR-133, was elevated in participants with AMI. Many additional manuscripts explored these and other myomiRs, essentially reporting finding consistent elevations in cases of AMI and ACS (Table 2) (69). This is logical, as the death of cardiomyocytes will allow cellular components (i.e., miRNAs, genes, proteins) to spill into the blood. In the absence of myocardial or skeletal injury, the levels of these myomiRs in blood should be low to undetectable. Thus, myomiRs are essentially equivalent to troponins.
How do myomiRs compare with troponins? Here the data are more variable. Wang et al. (70) investigated the miRNAs miR-1, miR-133a, miR-499, and miR-208a in a population of 66 participants and showed that each miRNA was elevated in the setting of an AMI. Most importantly, they compared receiver operating characteristic (ROC) curves with cTnI and found that, at best, miR-208a was equivalent to the troponin. However, miR-208a is a expressed at relatively low levels and can be difficult to detect in blood-based fluids. Li et al. (71) studied plasma levels of these same four myomiRs in 99 participants. They also reported elevations in each myomiR, but all were inferior indicators compared with cTnT. Oerlemans et al. (72) performed a similar study in an emergency room setting by investigating 332 patients with suspected ACS. A combination of three miRNAs—miR-1, miR-499, and miR-21—had better test statistics (area under the curve; AUC) than hs-cTnT alone (AUC, 0.96 versus 0.86). In one of the strongest evaluations of myomiRs as biomarkers for AMI, the Wagner group (73) compared miR-208 and miR-499 with hs-cTnT in 510 participants. This paper was important in showing that miR-499 was superior to miR-208 and had some favorable test characteristics compared with hs-cTnT. Specifically, 93% of patients presenting within the first 3 h of having an AMI were positive for elevated miR-499, while only 88% were positive by hs-cTnT. It was suggested that a single miR-499 level might be superior to serial testing of hs-cTnT. Yet overall, miR-499 and hs-cTnT were essentially equivalent. Therefore, while several robust studies have been performed to specifically address whether myomiRs are superior to troponins, it seems that, at best, they behave equivalently and, at worst, are slightly inferior to troponins as biomarkers of AMI and ACS.
A number of non-myomiRs have also been evaluated in AMI and ACS. These include miR-941 (74), miR-323–3p (75), miR-652 (75), miR-221 (76), miR-328 (77), and miR-134 (77). Due to the large number of AMI studies that performed global surveys of miRNAs and failed to detect these miRNAs, it is challenging to think that these individual studies may be replicated and that measuring these miRNAs may be superior to measuring troponins. However, a secondary value of these miRNAs may be able to predict other facets of AMI and ACS and could be worthy of exploration.
One important study of the ability to predict a myocardial infarction is worth discussing. The Mayr group (78) robustly investigated 19 candidate miRNAs in 820 participants to assess their ability to predict incident myocardial infarction. They found elevated miR-126 was positively associated with risk, while miR-223 and miR-197 were inversely associated. miR-126 is modestly expressed in platelets, highly expressed in endothelial cells, and has well-known functions in both, thus a potential rationale for this finding exists (79). However, miR-126 has been claimed as a specific biomarker for a variety of diseases by the same group, including diabetes (25, 80). Rather than being a biomarker for a specific disease, it may be an indicator of abnormal platelet or endothelial cell function.
miRNA Biomarkers in the Rejection of Heart Transplants
As described above, three blood-based biomarkers of cardiac rejection exist (i.e., the gene expression panel, cell-free DNA, and NT-proBNP). Of these, the gene-expression panel is used clinically, although its test statistics indicate there is room for improvement, particularly in discriminating between cellular and antibody-mediated rejection. An evaluation of the differential expression of miRNAs in endomyocardial biopsy tissue from rejecting and nonrejecting hearts identified nine modulated miRNAs that were all inflammatory cell–enriched miRNAs, essentially demonstrating that more inflammatory cells were present in the rejecting tissue (as expected) (81). Three studies of miRNA biomarkers of acute cellular rejection from two groups yielded 13 circulating miRNAs, of which only miR-142–3p was shared by both groups (82–84). The larger study, by Duong Van Huyen et al. (82), evaluated 60 participants in a test cohort and 53 in a validation cohort. Despite the rigor of these three studies, one group identified miR-144 and the other group identified miR-451 as biomarkers of transplant rejection. These two miRNAs, in a bicistronic cluster, are expressed only in red blood cells and likely reflect hematocrit levels. A single study by Neumann et al. (85) investigated 40 patients with variable levels of transplant vasculopathy using miRNA profiling. They found miR-628 had the best test statistics (using the AUC) and could serve as a biomarker for the progression of coronary artery vasculopathy. While opportunities exist, more work is needed to settle on suitable miRNAs that can serve as biomarkers of rejection and transplant vasculopathy.
miRNA Biomarkers in Myocarditis
As seen above, myocarditis can be challenging to diagnose and confirm with our current set of biomarkers. Therefore, there is an opportunity to identify helpful miRNA biomarkers. Although there have been many studies showing miRNA dysregulation in myocarditis in human samples and animal models, the number of studies using blood-based biomarkers of myocarditis have been fewer. Corsten et al. (69), in a study of several heart diseases, investigated 14 acute myocarditis plasma samples for several myomiRs, miR-223, miR-146a, miR-146b, and miR-155. They found modest but significant increases in miR-208b and miR-499, but no change in inflammation-related miRNAs. Although cTnT values were known, no direct head-to-head comparisons of superiority were reported. More recently, Wang et al. (86) studied 119 children with myocarditis and 120 age-matched controls, comparing miR-1, miR-146b, and other cardiovascular biomarkers. Based on ROC curves, these miRNAs were inferior to cTnI, interleukin (IL)-18, and tumor necrosis factor (TNF)-α.
miRNA Biomarkers in Hypertension
As stated above, miRNA biomarkers are not needed to diagnose hypertension, but they may be useful in stratifying patients for various therapeutic approaches, prognosticating comorbidities, or identifying an underlying etiology.
One recommendation for treating patients with hypertension is to encourage lifestyle modifications, specifically having them restrict salt intake. However, this is not always effective, and some patients have been classified as inverse salt sensitive. Thus, there is an opportunity to identify a biomarker that could predict this class of hypertensive patient. In a small study (10 participants), Gildea et al. (87) measured miRNAs in urinary exosomes and characterized them based on a patient’s salt sensitivity status. miR-4516, intronic to the PKD1 gene locus, was the only miRNA that was higher in salt-sensitive patients and lower in inverse salt-sensitive patients compared with controls. Qi et al. (88) used whole blood and qPCR to screen potential miRNAs as biomarkers for salt sensitivity in 91 participants. Their strongest signal was for miR-361–5p, which was associated with the risk of salt sensitivity. These studies suggest an opportunity for using urine or blood to categorize a patient’s sensitivity to salt and provide the appropriate dietary advice.
Another opportunity for biomarkers is in cases of white-coat hypertension, in which a patient has elevated blood pressure readings in a medical center, but is otherwise normotensive. Several studies have screened for potential blood miRNA biomarkers to look for associations with white-coat hypertension. Huang et al. (89) found that three miRNAs—miR-30a, miR-29, and miR-133 (normalizedtocel-miR-54)—have potential as screening tools for white-coat hypertension. Cengiz et al. (90) also measured 10 plasma miRNAs (normalized to U6) in 90 participants (30 with white-coat hypertension, 30 hypertensive, and 30 normotensive). Of these miRNAs, miR-21, miR-122, miR-637, and let-7e expression levels were significantly upregulated in the hypertensive group compared with the normotensive group. miR-296–5p levels were significantly downregulated in the hypertensive patients and upregulated in the patients with white-coat hypertension compared with the normotensive patients. miR-122 is exclusively expressed in hepatocytes, suggesting that its elevation is related to low-level damage or microvesicle release from the liver.
Changes in vascular smooth muscle and arterial vessel wall thickness are common sequelae of hypertension, and noninvasive serial monitoring of these changes could alter how aggressively a hypertensive patient is treated. Cengiz et al. (91) measured plasma miR-21 in a group of 28 hypertensive patients and 28 matched controls. They found miR-21 to be significantly increased both in the hypertensive patients and individuals with greater carotid intimal thickening.
In another interesting study, Karolina et al. (92) attempted to identify circulating miRNA profiles of metabolic syndrome by comparing different groups. They performed miRNA profiling of 46 healthy controls and 50 patients with metabolic syndrome, 30 with hypertension, 89 with hypercholesterolemia, and 50 with type 2 diabetes. miRNA profiles were determined from whole blood and exosomes, with comparable results for both. A cluster of three miRNAs—miR-130a, miR-195, and miR-92a—distinguished hypertensive patients from metabolic syndrome patients.
Kontaraki et al. (93) studied PBMCs in hypertensive men (n = 60) and healthy normotensive controls (n = 29). They measured the levels of five miRNAs (normalized to U6) that were purported to have effects on vascular smooth muscle phenotypic expression. Compared with the healthy controls, in hypertensive patients, the miRNAs miR-143, miR-145, and miR-133 were all significantly decreased, while miR-21 and miR-1 were significantly higher. Interestingly, the levels of miR-143, miR-145, and miR-21 were negatively correlated with 24-h ambulatory diastolic blood pressures, while miR-133 was positively correlated. Although these findings will not supersede the use of the sphygmomanometer in diagnosing hypertension, they may suggest pathways modulated in hypertension.
Some patterns of miRNA expression are beginning to emerge from these studies, raising the possibility that miRNA biomarkers will be useful for diagnostic, prognostic, or etiologic characterization of the hypertensive patient. For a more comprehensive review of miRNAs, their SNPs, and blood pressure regulation, the reader is referred to Marques & Charchar (94).
miRNA Biomarkers in Diabetic Cardiomyopathy
Diabetic cardiomyopathy is an important complication in a subset of diabetic individuals. There may be an opportunity to identify early miRNA biomarkers that can predict disease. De Gonzalo-Calvo et al. (95) studied myomiRs in serum from 72 participants with type 2 diabetes and myocardial steatosis, a feature of diabetic cardiomyopathy. miRNAs were normalized to a cel-miR-39 control. De Gonzalo-Calvo et al. (95) found a modest but significant association between miR-1 and miR-133a and myocardial steatosis, and they further showed that these miRNAs enhanced their ROC predictive model that was based on clinical factors. Then they investigated mice fed a high-fat diet, replicating the increased serum miR-1 and miR-133 levels. As described for other cardiovascular diseases, the serum increase of these two myomiRs likely reflects myocardial injury and the leakage of these myomiRs into the circulation. No other studies of blood-based biomarkers exist for diabetic cardiomyopathy.
A study of right atrial appendage and left ventricle tissues from 28 patients with type 2 diabetes and 38 nondiabetic patients explored myomiRs and miR-126 relative to U6 (96). In heart tissue from diabetic patients, researchers found lower levels of miR-1, miR-133, miR-126, and miR-499, and higher levels of miR-208 compared with tissue from the nondiabetic patients. That discrepancy between most myomiRs and miR-208 is difficult to reconcile, but it could suggest miR-208 dysregulation in cardiac remodeling. The general overall reduction in the myomiRs in diabetic tissues could be the result of a changing cell ratio in which more fibroblasts and thus fewer myocytes are present.
Many additional miRNAs have been implicated functionally in diabetic cardiomyopathy based on animal and cellular studies. They are beyond the scope of this review, but the reader is referred to additional manuscripts for further insights (97–102).
miRNA Biomarkers in Aortic Stenosis
Multiple studies have investigated aspects of miRNAs serving as biomarkers of aortic stenosis, primarily related to myocardial complications. Garcia et al. (103) investigated plasma miR-133a in 74 participants with aortic stenosis. Interestingly, Garcia et al. (103) demonstrated that elevated preoperative miR-133a predicted improved normalization of left ventricular mass after surgery. Also of importance, they sampled blood from the coronary sinus (venous return from the heart) and right atrium (systemic venous return) and noted significantly higher values of miR-133a from the sinus (140.4 versus 72.3 relative expression units normalized to cel-miR-39), establishing the heart as the source of miR-133a in blood.
Chen et al. (104) investigated miR-1, miR-133a, and miR-378a in plasma from 152 participants with and without aortic stenosis. When normalized to U6, they noted that these miRNAs were lower in patients with aortic stenosis and that a low level of miR-378a alone could distinguish between patients with and without left ventricular hypertrophy. Beaumont et al. (105) investigated serum levels of seven miRNAs from 57 participants with or without aortic stenosis, showing there were reduced levels of miR-133a and miR-19b (normalized to cel-miR-39) in patients with aortic stenosis. The sum of these studies is a developing understanding of the relationship of miR-133a with aortic stenosis outcome.
Other studies have investigated other miRNA biomarkers for aortic stenosis. Fabiani et al. (106) investigated plasma miR-21 as a potential biomarker of myocardial fibrosis in severe aortic stenosis. They correlated elevated miR-21 with biopsy-based collagen levels in 23 patients. This confirmed the findings of Villar et al. (107), who showed that elevated miR-21 (normalized to cel-miR-39) correlated with the severity of aortic stenosis in both plasma and tissue biopsy samples from >100 participants. Coffey et al. (47) attempted a broad miRNA profiling study in 51 individuals with or without aortic stenosis. Although they found four miRNAs that varied by disease state (including miR-451a), these markers failed to be validated, and the authors concluded that no real biomarkers were discovered in the study. Finally, the Omland group (108) found that elevated miR-210 serum levels correlated with aortic stenosis in a study of 67 participants.
miRNA Biomarkers in Heart Failure with Preserved Ejection Fraction
In back-to-back manuscripts, Wong et al. (109) and Watson et al. (110) explored miRNAs in plasma from participants with HFpEF versus those with HFrEF. In Wong et al. (109), 39 HFrEF, 19 HFpEF, and 28 control participants were compared in a miRNA array platform. Numerous miRNAs varied across these three groups, with seven miRNAs being discriminatory for HFpEF. The second study investigated plasma from 270 participants with HFpEF, HFrEF, or no heart failure (110). After initial miRNA profiling and subsequent validation in the wider cohort, five miRNAs (miR-375, miR-146a, miR-30c, miR-328, and miR-221) were found to vary in heart failure. While none of these miRNAs outperformed BNP in predicting heart failure, the AUC test statistic was optimized when they were added to the log BNP value.
miRNA Biomarkers in Atrial Fibrillation
A variety of studies of miRNA biomarkers for atrial fibrillation have described a number of potential biomarkers. One of the more interesting miRNAs in this group is miR-328. Lu et al. (111) used a canine model of atrial fibrillation to identify four miRNAs (miR-223, miR-328, miR-664, and miR-517) that were elevated in atrial myocardium, with miR-328 having the greatest increase. Elevation of miR-328 was also identified at the time of open-heart surgery in the right atrium of patients with atrial fibrillation (n = 12) compared with those without atrial fibrillation (n = 10). In additional studies in a transgenic mouse model, they further demonstrated that overexpression of miR-328 increased vulnerability to atrial fibrillation, potentially through the regulation of the target genes CACNA1C and CACNB1, both of which encode for L-type calcium channels (111).
More support for a role of miR-328 in atrial fibrillation comes from the Framingham Heart Study (112). miRNA profiling was performed on the whole blood of 2,467 participants. Four miRNAs—miR-328, miR-150, miR-331, and miR-28—were negatively nominally associated with prevalent atrial fibrillation. However, only lower miR-328 was significant after adjustments for age, sex, and technical factors. Thus, while miR-328 elevations appear to occur in myocardium involved in atrial fibrillation, lower miR-328 is seen in blood.
Liu et al. (113) used small RNA-seq to obtain a plasma miRNA expression profile in patients with paroxysmal atrial fibrillation (n = 5), persistent atrial fibrillation (n = 5), and healthy controls (n = 5). They found differences in miRNAs between these groups. Of note, miR-328 was not variable. They followed up four of these miRNAs in a separate population of patients with paroxysmal atrial fibrillation (n = 30), persistent atrial fibrillation (n = 30), and controls (n = 30). Only elevated miR-150 (in contrast to the Framingham Heart Study’s reduction) was replicated in this larger cohort.
Natsume et al. (114) performed miRNA profiling of the serum of 10 patients with atrial fibrillation and 5 healthy controls to identify predictors of atrial fibrillation. Four miRNAs—miR-99a–5p, miR-192–5p, miR-214–3p, and miR-342–5p—were elevated in atrial fibrillation patients relative to controls, and the latter two had the highest accuracy based on ROC curves.
Dawson et al. (115) measured miRNAs in plasma and the right atrial appendages of patients with atrial fibrillation with (n = 16) or without congestive heart failure (n = 17) and controls (n = 30). Plasma levels of miR-29b and miR-21 were significantly decreased in patients with atrial fibrillation. Interestingly, in both human and canine right atrial appendages, levels of miR-29b were also significantly decreased by atrial fibrillation. The authors further went on to demonstrate a potential role for miR-29b in cardiac fibrosis.
McManus et al. (116) studied plasma miRNA biomarkers of prevalent atrial fibrillation in 112 patients with and 99 patients without atrial fibrillation. Atrial tissue was also obtained from patients undergoing cardiac surgery (n = 31). The plasma miRNAs miR-21 and miR-150 were both twofold lower in patients with atrial fibrillation compared with patients without. Both miRNAs increased threefold 1 month after ablation. Atrial levels of miR-21 but not miR-150 were lower in patients with atrial fibrillation compared with patients who did not have atrial fibrillation.
miRNA Biomarkers in Sepsis
A number of investigators have examined the role of circulating miRNAs for use as diagnostic biomarkers in sepsis (117–119). While most miRNAs have not been validated in multiple studies, a few miRNAs have been repeatedly identified across studies as being associated with sepsis, including miR-150, miR-223, and miR-146a (120–127). Several of these studies are confounded by methodologic flaws, however, and at present none of these miRNAs are used as biomarkers in clinical practice.
miR-150 has been investigated as both a diagnostic and a prognostic biomarker. Ma et al. (122) used small RNA-seq to compare miRNA expression in whole blood between cohorts of septic patients and nonseptic but critically ill patients. They observed that miR-150 expression was higher in sepsis, although the expression levels were not normalized (122). Several investigators have demonstrated that plasma miR-150 levels are correlated with disease severity and prognosis. Vasilescu and colleagues (120) examined the expression of miR-150 in the white blood cells of septic patients and found that miR-150 levels were inversely correlated with disease severity and levels of IL-10, IL-18, and TNFα. Similarly, other investigators have shown that serum and plasma levels of miR-150 are lower in patients with higher Sequential Organ Failure Assessment scores and in nonsurvivors of sepsis (121, 128). However, because miR-150 has been identified as a lymphocyte-specific miRNA, these findings may simply be a function of the circulating lymphocyte count and may not add value above a complete blood count with differential.
Similarly, investigators have examined the diagnostic and prognostic abilities of miR-223, a miRNA that is specific to neutrophils and macrophages. Initial studies were promising, as plasma miR-223 levels were found to be significantly lower in septic patients compared with nonseptic patients undergoing cardiac surgery (123), and miR-223 plasma expression was significantly lower among nonsurvivors of sepsis independent of illness severity and other relevant clinical variables (124). The prognostic potential of miR-223 in sepsis was further supported by a separate study demonstrating that plasma miR-223 levels are reduced in more severe sepsis compared with mild disease (125). A more recent analysis in a larger cohort demonstrated mixed results for the utility of miR-223 as a sepsis biomarker. Benz et al. (129) measured serum miR-223 levels in 221 critically ill patients (septic = 137, nonseptic = 84). There was no difference in miR-223 expression between the two groups, suggesting that it may have limited value as a diagnostic biomarker. However, reduced miR-223 levels were correlated with mortality in the intensive care unit, suggesting that miR-223 may have prognostic capability (129). It is unclear whether miR-223 expression outperforms or has additive value to more traditional and widely used systems for scoring the severity of illness.
Although the association between miR-146a and sepsis has been examined several times, the results are inconsistent and contradictory. An early study examining miR-146a expression in sepsis was performed by Wang et al. (123), who found that (similar to miR-223) plasma miR-146a levels were reduced in patients with sepsis compared with levels in nonseptic acutely ill patients. Because miR-146a is known to regulate Toll-like receptor 4 signaling, this observation may have a plausible biological rationale (130). Subsequent studies, however, have inconsistently corroborated these findings, with one group finding no correlation between plasma miR-146a levels and sepsis among patients presenting to the emergency room (128), while another group observed increased expression of plasma miR-146a in pediatric sepsis (126). Thus, the clinical utility of miR-146a as a diagnostic biomarker of sepsis is uncertain.
RECOMMENDATIONS FOR BEST PRACTICES
As is evident from this collection of manuscripts, the technical approaches used to measure miRNA levels have varied widely (Table 4). It is clear that to generate robust data about miRNA biomarkers, the next generation of studies needs to adhere to stricter guidelines and follow recommendations so that results can be replicated (7, 65, 131–133). Some technically driven best practices, based on these and other reviews of miRNA biomarker studies, are to ensure the inclusion of large, well-characterized populations; steady normalization signals (e.g., spike-in RNAs); and consistent preparation of appropriate blood-based fluids; to avoid using as biomarkers miRNAs that have only low expression; and to use robust statistical analyses of the signal. Biologically based best practices include ensuring that individuals are well-matched between groups, using consistent fluid collection practicesand investigating biologically relevant targets.
Table 4.
Fluid-based studies of microRNA biomarkers of cardiovascular diseases
| microRNAs investigateda,b | Source | Method | Study size | Type of study | Reference |
|---|---|---|---|---|---|
| Acute myocardial infarction or acute coronary syndrome | |||||
| miR−1; −133 | Plasma | qPCR | 93 AMI; 66 controls | D | 68 |
| miR−1; −133a; −208b; −499 | Plasma | qPCR | 67 AMI; 32 controls | D | 71 |
| miR−1; −208a; −499; −21; −146a | Serum | qPCR | 332 patients with chest pain | D | 72 |
| miR−208b; −499 | Plasma | qPCR | 510 AMI; 87 controls | D | 73 |
| Multiple microRNAs;b miR−941 | PBMC | ELOSA QC assay; qPCR | 72 patients with chest pain | D | 74 |
| Multiple microRNAs; miR−323; −27b; −652; −103 | Plasma | Exiqon qPCR panel; qPCR | 240 ACS; 120 controls | D | 75 |
| Multiple microRNAs | Plasma | Qiagen miRNome PCR array | 27 patients with chest pain; 16 controls | D | 76 |
| miR−328; −134 | Plasma | qPCR | 359 AMI; 30 controls | D | 77 |
| Multiple microRNAs; miR−24; −126; −140; −150; −197; −223; −486 | Plasma | TaqMan Array A and B Cards; qPCR | 820 Bruneck study participants | P | 78 |
| Heart transplantation | |||||
| miR−326; −142; −101; −144; −27a; −424; −339 |
Serum | qPCR | 10 patients at 3 time points | D | 84 |
| miR−142; −101; −424; −27a; −144; −339; −326 |
Serum | qPCR | 63 patients | D | 83 |
| miR−10a; −21; −31; −92a; −142; −155; −451; −126; −221; −296; −208; −181a; −181b; −182 |
Plasma | qPCR | 30 patients with rejection; 30 patients without rejection; 53 validation participants | D | 82 |
| Multiple microRNAs; miR−628; −155; −34a; −98; −204 | Plasma | TaqMan Array A and B Cards; qPCR | 40 patients; samples used twice | D | 85 |
| Myocarditis | |||||
| miR−1; −133a, −146a; −146b; −155; −208b; −23; −499 | Plasma | qPCR | 14 patients with acute myocarditis; 20 with remote myocarditis; 20 controls | D | 69 |
| miR−1; −146b | Serum | qPCR | 119 pediatric patients with myocarditis; 120 controls | D | 86 |
| Hypertension | |||||
| Multiple microRNAs; miR−1268b; −5002; 4516; −3183; −3940; −4649; −320a | Urinary exosomes | Microarray; qPCR | 10 hypertensives | D | 87 |
| Multiple microRNAs; miR−15b; −19a; −382; −26b; −361; −423; −210; −361 | Whole blood | Small RNA sequencing; qPCR | 56 salt-sensitive hypertensives; 56 salt-resistant hypertensives | D | 88 |
| miR−30a; −29; −133 | Plasma | qPCR | 35 white-coat hypertensives; 35 hypertensives; 35 normotensives | D | 89 |
| miR−21; −122; −125a; −126; −130a; −155; −195; −296; −637; let−7e | Plasma | qPCR | 30 white-coat hypertensives; 30 hypertensives; 30 normotensives | D | 90 |
| miR−21 | Plasma | qPCR | 28 hypertensives; 28 normotensives | D | 91 |
| Multiple microRNAs; miR−103; −17; −183; −197; −23a; −509; −584; −652; −130a; −195; −92a; −150; −192; −27a; −320a | Whole blood and exosomes | miRCURY LNA microRNA Array; qPCR | 50 patients with metabolic syndrome; 30 hypertensives; 50 with type 2 diabetes; 89 with hypercholesterolemia; 46 controls | D | 92 |
| miR−143; −145; −133, −21; −1 | PBMCs | qPCR | 60 hypertensives; 29 normotensives | D | 93 |
| Diabetic cardiomyopathy | |||||
| miR−1; −133a; −133b; −208a; −208b; −499 | Serum and exosomes | qPCR | 78 with type 2 diabetes from the PIRAMID study | D | 95 |
| Aortic stenosis | |||||
| miR−133a | Plasma | qPCR | 74 patients | P | 103 |
| miR−1; −133a; −378a | Plasma | qPCR | 112 patients; 40 controls | D | 104 |
| miR−19b; −133a; −21; −29; −1; −208a; −499 | Serum | qPCR | 28 patients; 29 controls | Y | 105 |
| miR−21 | Whole blood | qPCR | 23 patients undergoing surgery | P | 106 |
| miR−21 | Plasma | qPCR | 75 patients; 32 surgical controls; 25 controls | D | 107 |
| Multiple microRNAs; miR−22; −23; −382; −451a; −21 | Plasma | Affymetrix GeneChip miRNA 2.0 Array; qPCR | 24 patients; 27 controls | D | 47 |
| miR−210; −22; −425 | Serum | qPCR | 57 patients; 13 controls | D | 108 |
| Heart failure with preserved ejection fraction | |||||
| Multiple microRNAs; miR−1225; −1233; −125a; −1299; −130a; −1322; −145; −17; −1825; −183; −186; −190a; −193b–3p; −193–5p; −204; −211; −301a; −320d; −326; −361; −423; −431; −485; −494; −509; −545; −550a; −625; −629; −638; −671; −92b | Whole blood and plasma | miRCURY LNA microRNA Array; qPCR | 118 patients with heart failure; 58 controls from the SHOP and SLAS studies | D | 109 |
| Multiple microRNAs; miR−30c; −146a; −221; −328; −375 | Serum | TaqMan Array A and B Cards; qPCR | 90 patients with HFpEF; 90 with HFrEF; 90 controls from the STOP-HF Study | D | 110 |
| Atrial fibrillation | |||||
| Multiple microRNAs | Whole blood | Array | 153 with prevalent AF; 107 with incident AF; 2,185 without AF (controls) | P | 112 |
| Multiple microRNAs; miR−19a; −146a; −150; −375 | Plasma | Small RNA sequencing; qPCR | 35 with persistent AF; 35 with paroxysmal AF; 35 controls | D | 113 |
| Multiple microRNAs; miR−27b; −30a; −328; −99a, −192; −214, −342; −125b; −130b; −362; −424 | Serum | TaqMan Array A and B Cards; qPCR | 30 with paroxysmal AF; 30 with chronic AF; 55 controls | D | 114 |
| miR−29b; −21; −133; −15 | Plasma | qPCR | 33 with persistent AF; 32 with congestive heart failure; 30 controls | D | 115 |
| 86 microRNAs | Plasma | qPCR | 112 with AF; 99 controls from the miRhythm Study | D | 116 |
| Sepsis | |||||
| miR−150; −182; −342–5p, −486 | PBMCs | Agilent hybridized microarray | 24 with sepsis; 32 healthy controls | D | 120 |
| miR−146a; −223; −126; −132; −155; let−7i | Serum | qPCR | 50 with sepsis; 30 controls with SIRS; 20 healthy controls | D | 123 |
| miR−574–5p; −297 | Serum | Affymetrix microarray | 78 sepsis survivors; 64 sepsis nonsurvivors | P | 134 |
| Multiple microRNAs | Serum | Solexa sequencing with qPCR validation | 117 sepsis survivors; 97 sepsis nonsurvivors | P | 124 |
| miR−15a, −16 | Serum | qPCR | 166 with sepsis; 32 with SIRS; 24 healthy controls | D | 135 |
| miR−223; −15b; −483; −499; −122; −193b | Plasma | qPCR | 166 with sepsis; 24 healthy controls | D, P | 125 |
| miR−181b | Plasma | qPCR | 36 sepsis; 17 controls in intensive care unit | D | 136 |
| miR−146a | Plasma | qPCR | 14 with sepsis; 14 with SIRS | D | 127 |
| Multiple microRNAs | Plasma | qPCR | 22 with sepsis; 22 with SIRS; 17 healthy controls | D | 122 |
| miR−122; −193b; −483; −574 | Serum | Affymetrix microarray | 232 with sepsis; 24 healthy controls | P | 137 |
| miR−21; −125b; −132; −146a; −155; −223 | Plasma | qPCR | 40 pediatric patients with sepsis; 20 pediatric patients with SIRS; 15 healthy pediatric controls | D | 126 |
| miR−223 | Serum | qPCR | 137 with sepsis; 84 without sepsis; 75 healthy controls | D, P | 129 |
| miR−150; −146a; −223 | Plasma | qPCR | 69 with sepsis; 24 healthy controls | D, P | 128 |
| let−7a; miR−150; −1249; −199b | PBMCs | Geniom Biochip microarray | 22 with sepsis; 20 healthy controls | D | 138 |
| miR−15a; −27a; −34a | Plasma | qPCR | 62 with sepsis; 32 healthy controls | P | 139 |
| miR−30d; −30a; −192; −26a; −23a; −191–5p | Plasma | qPCR | 29 with sepsis; 44 with SIRS; 16 controls | D | 140 |
Abbreviations: ACS, acute coronary syndrome; AF, atrial fibrillation; AMI, acute myocardial infarction; D, diagnostic biomarker study; ELOSA, enzyme-linked oligosorbent assay; HFpEF, heart failure with preserved ejection fraction; HFrEF, heart failure with reduced ejection fraction; P, prognostic biomarker study; PBMCs, peripheral blood mononuclear cells; QC, quality control; qPCR, quantitative polymerase chain reaction; SIRS, systemic inflammatory response syndrome; Y, pathophysiologic study.
The microRNAs listed reflect those assayed, which were not necessarily found to have significant associations with disease.
Multiple microRNAs implies the use of a multiplexed array system or small RNA sequencing.
CONCLUSIONS AND FUTURE CONSIDERATIONS
The outlook for using miRNA biomarkers for cardiovascular disease is complex. In the areas in which miRNA biomarker studies have made the most progress (e.g., AMI and ACS), other robust biomarkers exist. Where there is more opportunity and need for cardiovascular diagnostics, miRNA studies are inconsistent and less robust. Those who are working in this field would be well advised to learn from first-generation miRNA studies, focus on opportunities where there is a lower barrier of entry for miRNA-based diagnostics, and then perform robust and reproducible studies using biologically applicable samples. Only then will miRNAs succeed as a class of biomarkers for cardiovascular disease.
ACKNOWLEDGMENTS
M.K.H. was supported by the US National Institutes of Health (grant no. 1R01HL137811) and the American Heart Association (Grant-in-Aid no. 17GRNT33670405). P.V.H. was supported by the US National Institutes of Health (grant nos. UL1TR001450 and 1R01GM113995). A.J.G. was supported by the US National Institutes of Health (grant nos. 1K23HL135263-01A and 1R01GM113995).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Lewis BP, Burge CB, Bartel DP. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20 [DOI] [PubMed] [Google Scholar]
- 2.Pinzon N, Li B, Martinez L, Sergeeva A, Presumey J, et al. 2017. microRNA target prediction programs predict many false positives. Genome Res. 27:234–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai Y, Yu X, Hu S, Yu J. 2009. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform 7:147–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauersachs J, Thum T. 2011. Biogenesis and regulation of cardiovascular microRNAs. Circ. Res 109:334–47 [DOI] [PubMed] [Google Scholar]
- 5.Kim VN. 2005. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol 6:376–85 [DOI] [PubMed] [Google Scholar]
- 6.Kozomara A, Griffiths-Jones S. 2014. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witwer KW, Halushka MK. 2016. Toward the promise of microRNAs—enhancing reproducibility and rigor in microRNA research. RNA Biol. 13:1103–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCall MN, Kim MS, Adil M, Patil AH, Lu Y, et al. 2017. Toward the human cellular microRNAome. Genome Res. 27:1769–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Rie D, Abugessaisa I, Alam T, Arner E, Arner P, et al. 2017. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol 35:872–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Londin E, Loher P, Telonis AG, Quann K, Clark P, et al. 2015. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. PNAS 112: E1106–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backes C, Meder B, Hart M, Ludwig N, Leidinger P, et al. 2016. Prioritizing and selecting likely novel miRNAs from NGS data. Nucleic Acids Res. 44:e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backes C, Fehlmann T, Kern F, Kehl T, Lenhof HP, et al. 2018. miRCarta: a central repository for collecting miRNA candidates. Nucleic Acids Res. 46:D160–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludwig N, Becker M, Schumann T, Speer T, Fehlmann T, et al. 2017. Bias in recent miRBase annotations potentially associated with RNA quality issues. Sci. Rep 7:5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, et al. 2015. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu. Rev. Genet 49:213–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang HR, Schoenfeld LW, Ruby JG, Auyeung VC, Spies N, et al. 2010. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 24:992–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Liu XS. 2011. Systematic curation of miRBase annotation using integrated small RNA high-throughput sequencing data for C. elegans and Drosophila. Front. Genet 2:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halushka MK, Fromm B, Peterson KJ, McCall MN. 2018. Big strides in cellular microRNA expression. Trends Genet. 34:165–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fromm B, Domanska D, Hackenberg M, Mathelier A, Hoye E, et al. 2018. MirGeneDB2.0: the curated microRNA gene database. bioRxiv 258749 [DOI] [Google Scholar]
- 19.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol 12:735–39 [DOI] [PubMed] [Google Scholar]
- 20.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, et al. 2007. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129:1401–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, et al. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. PNAS 101:9740–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halushka MK. 2016. MicroRNA-144 is unlikely to play a role in bronchiolitis obliterans syndrome. J. Heart Lung Transplant 35:543. [DOI] [PubMed] [Google Scholar]
- 23.Kent OA, McCall MN, Cornish TC, Halushka MK. 2014. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 42:7528–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCall MN, Kent OA, Yu J, Fox-Talbot K, Zaiman AL, Halushka MK. 2011. MicroRNA profiling of diverse endothelial cell types. BMC Med. Genom 4:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. 2014. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLOS ONE 9:e89565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juzenas S, Venkatesh G, Hubenthal M, Hoeppner MP, Du ZG, et al. 2017. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 45:9290–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Rooij E, Quiat D, Johnson BA, Sutherland LB, Qi X, et al. 2009. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Dev. Cell 17:662–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das S, Halushka MK. 2015. Extracellular vesicle microRNA transfer in cardiovascular disease. Cardiovasc. Pathol 24:199–206 [DOI] [PubMed] [Google Scholar]
- 29.Kumar B, Rosenberg AZ, Choi SM, Fox-Talbot K, de Marzo AM, et al. 2018. Cell-type specific expression of oncogenic and tumor suppressive microRNAs in the human prostate and prostate cancer. Sci. Rep 8:7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern AD, Alexander BM, Chandra A. 2018. Innovation incentives and biomarkers. Clin. Pharmacol. Ther 103:34–36 [DOI] [PubMed] [Google Scholar]
- 31.Antman EM. 2018. The introduction and clinical use of cardiac-specific troponin assays. Clin. Pharmacol. Ther 103:31–33 [DOI] [PubMed] [Google Scholar]
- 32.Streng AS, Jacobs LH, Schwenk RW, Cardinaels EP, Meex SJ, et al. 2014. Cardiac troponin in ischemic cardiomyocytes: intracellular decrease before onset of cell death. Exp. Mol. Pathol 96:339–45 [DOI] [PubMed] [Google Scholar]
- 33.Bingisser R, Cairns C, Christ M, Hausfater P, Lindahl B, et al. 2012. Cardiac troponin: a critical review of the case for point-of-care testing in the ED. Am. J. Emerg. Med 30:1639–49 [DOI] [PubMed] [Google Scholar]
- 34.White H, Thygesen K, Alpert JS, Jaffe A. 2014. Universal MI definition update for cardiovascular disease. Curr. Cardiol. Rep 16:492. [DOI] [PubMed] [Google Scholar]
- 35.Carey RM, Whelton PK. 2018. Prevention, detection, evaluation, and management of high blood pressure in adults: synopsis of the 2017 American College of Cardiology/American Heart Association hypertension guideline. Ann. Intern. Med 168:351–58 [DOI] [PubMed] [Google Scholar]
- 36.Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, et al. 2018. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 71:e13–115 [DOI] [PubMed] [Google Scholar]
- 37.Stewart S, Winters GL, Fishbein MC, Tazelaar HD, Kobashigawa J, et al. 2005. Revision of the 1990 working formulation for the standardization of nomenclature in the diagnosis of heart rejection. J. Heart Lung Transplant 24:1710–20 [DOI] [PubMed] [Google Scholar]
- 38.Berry GJ, Burke MM, Andersen C, Bruneval P, Fedrigo M, et al. 2013. The 2013 International Society for Heart and Lung Transplantation Working Formulation for the standardization of nomenclature in the pathologic diagnosis of antibody-mediated rejection in heart transplantation. J. Heart Lung Transplant 32:1147–62 [DOI] [PubMed] [Google Scholar]
- 39.Pham MX, Teuteberg JJ, Kfoury AG, Starling RC, Deng MC, et al. 2010. Gene-expression profiling for rejection surveillance after cardiac transplantation. N. Engl. J. Med 362:1890–900 [DOI] [PubMed] [Google Scholar]
- 40.De Vlaminck I, Valantine HA, Snyder TM, Strehl C, Cohen G, et al. 2014. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci. Transl. Med 6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittleson MM, Skojec DV, Wittstein IS, Champion HC, Judge DP, et al. 2009. The change in B-type natriuretic peptide levels over time predicts significant rejection in cardiac transplant recipients. J. Heart Lung Transplant. 28:704–9 [DOI] [PubMed] [Google Scholar]
- 42.Halushka MK, Mitchell RN, Padera RF. 2016. Heart failure therapies: new strategies for old treatments and new treatments for old strategies. Cardiovasc. Pathol 25:503–11 [DOI] [PubMed] [Google Scholar]
- 43.Pollack A, Kontorovich AR, Fuster V, Dec GW. 2015. Viral myocarditis—diagnosis, treatment options, and current controversies. Nat. Rev. Cardiol 12:670–80 [DOI] [PubMed] [Google Scholar]
- 44.Chow LH, Radio SJ, Sears TD, McManus BM. 1989. Insensitivity of right ventricular endomyocardial biopsy in the diagnosis of myocarditis. J. Am. Coll. Cardiol 14:915–20 [DOI] [PubMed] [Google Scholar]
- 45.Nichols GA, Hillier TA, Erbey JR, Brown JB. 2001. Congestive heart failure in type 2 diabetes: prevalence, incidence, and risk factors. Diabetes Care 24:1614–19 [DOI] [PubMed] [Google Scholar]
- 46.Carr AA, Kowey PR, Devereux RB, Brenner BM, Dahlof B, et al. 2005. Hospitalizations for new heart failure among subjects with diabetes mellitus in the RENAAL and LIFE studies. Am. J. Cardiol 96:1530–36 [DOI] [PubMed] [Google Scholar]
- 47.Coffey S, Williams MJ, Phillips LV, Jones GT. 2015. Circulating microRNA profiling needs further refinement before clinical use in patients with aortic stenosis. J. Am. Heart Assoc 4:e002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma K, Kass DA. 2014. Heart failure with preserved ejection fraction: mechanisms, clinical features, and therapies. Circ. Res 115:79–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bone RC, Balk RA, Cerra FB, Dellinger RP, Fein AM, et al. 1992. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. Chest 101:1644–55 [DOI] [PubMed] [Google Scholar]
- 50.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, et al. 2016. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 315:801–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, et al. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit. Care Med 34:1589–96 [DOI] [PubMed] [Google Scholar]
- 52.Tang BM, Eslick GD, Craig JC, McLean AS. 2007. Accuracy of procalcitonin for sepsis diagnosis in critically ill patients: systematic review and meta-analysis. Lancet Infect. Dis 7:210–17 [DOI] [PubMed] [Google Scholar]
- 53.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. 2008. Circulating microRNAs as stable blood-based markers for cancer detection. PNAS 105:10513–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. 2012. Comparing the microRNA spectrum between serum and plasma. PLOS ONE 7:e41561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, et al. 2012. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res 5:492–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheng HH, Yi HS, Kim Y, Kroh EM, Chien JW, et al. 2013. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLOS ONE 8:e64795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mestdagh P, Hartmann N, Baeriswyl L, Andreasen D, Bernard N, et al. 2014. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 11:809–15 [DOI] [PubMed] [Google Scholar]
- 58.Schwarzenbach H, da Silva AM, Calin G, Pantel K. 2015. Data normalization strategies for microRNA quantification. Clin. Chem 61:1333–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon LM, Edelstein LC, Nagalla S, Woodley AB, Chen ES, et al. 2014. Human platelet microRNA–mRNA networks associated with age and gender revealed by integrated plateletomics. Blood 123:e37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo L, Liang T, Yu J, Zou Q. 2016. A comprehensive analysis of miRNA/isomiR expression with gender difference. PLOS ONE 11:e0154955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olivieri F, Spazzafumo L, Santini G, Lazzarini R, Albertini MC, et al. 2012. Age-related differences in the expression of circulating microRNAs: miR-21 as a new circulating marker of inflammaging. Mech. Ageing Dev 133:675–85 [DOI] [PubMed] [Google Scholar]
- 62.Sredni ST, Gadd S, Jafari N, Huang CC. 2011. A parallel study of mRNA and microRNA profiling of peripheral blood in young adult women. Front. Genet 2:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heegaard NH, Carlsen AL, Lilje B, Ng KL, Ronne ME, et al. 2016. Diurnal variations of human circulating cell-free micro-RNA. PLOS ONE 11:e0160577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baras AS, Mitchell CJ, Myers JR, Gupta S, Weng LC, et al. 2015. miRge—a multiplexed method of processing small RNA-seq data to determine microRNA entropy. PLOS ONE 10:e0143066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Witwer KW. 2015. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin.Chem 61:56–63 [DOI] [PubMed] [Google Scholar]
- 66.Becker N, Lockwood CM. 2013. Pre-analytical variables in miRNA analysis. Clin. Biochem 46:861–68 [DOI] [PubMed] [Google Scholar]
- 67.Khan J, Lieberman JA, Lockwood CM. 2017. Variability in, variability out: best practice recommendations to standardize pre-analytical variables in the detection of circulating and tissue microRNAs. Clin. Chem. Lab. Med 55:608–21 [DOI] [PubMed] [Google Scholar]
- 68.Ai J, Zhang R, Li Y, Pu J, Lu Y, et al. 2010. Circulating microRNA-1 as a potential novel biomarker for acute myocardial infarction. Biochem. Biophys. Res. Commun 391:73–77 [DOI] [PubMed] [Google Scholar]
- 69.Corsten MF, Dennert R, Jochems S, Kuznetsova T, Devaux Y, et al. 2010. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet 3:499–506 [DOI] [PubMed] [Google Scholar]
- 70.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, et al. 2010. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur. Heart J 31:659–66 [DOI] [PubMed] [Google Scholar]
- 71.Li YQ, Zhang MF, Wen HY, Hu CL, Liu R, et al. 2013. Comparing the diagnostic values of circulating microRNAs and cardiac troponin T in patients with acute myocardial infarction. Clinics 68:75–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oerlemans MI, Mosterd A, Dekker MS, de Vrey EA, van Mil A, et al. 2012. Early assessment of acute coronary syndromes in the emergency department: the potential diagnostic value of circulating microRNAs. EMBO Mol. Med 4:1176–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Devaux Y, Vausort M, Goretti E, Nazarov PV, Azuaje F, et al. 2012. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin. Chem 58:559–67 [DOI] [PubMed] [Google Scholar]
- 74.Bai R, Yang Q, Xi R, Li L, Shi D, Chen K. 2017. miR-941 as a promising biomarker for acute coronary syndrome. BMC Cardiovasc. Disord 17:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pilbrow AP, Cordeddu L, Cameron VA, Frampton CM, Troughton RW, et al. 2014. Circulating miR-323–3p and miR-652: candidate markers for the presence and progression of acute coronary syndromes. Int. J. Cardiol 176:375–85 [DOI] [PubMed] [Google Scholar]
- 76.Coskunpinar E, Cakmak HA, Kalkan AK, Tiryakioglu NO, Erturk M, Ongen Z. 2016. Circulating miR-221–3p as a novel marker for early prediction of acute myocardial infarction. Gene 591:90–96 [DOI] [PubMed] [Google Scholar]
- 77.He F, Lv P, Zhao X, Wang X, Ma X, et al. 2014. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol. Cell. Biochem 394:137–44 [DOI] [PubMed] [Google Scholar]
- 78.Zampetaki A, Willeit P, Tilling L, Drozdov I, Prokopi M, et al. 2012. Prospective study on circulating microRNAs and risk of myocardial infarction. J. Am. Coll. Cardiol 60:290–99 [DOI] [PubMed] [Google Scholar]
- 79.Chistiakov DA, Orekhov AN, Bobryshev YV. 2016. The role of miR-126 in embryonic angiogenesis, adult vascular homeostasis, and vascular repair and its alterations in atherosclerotic disease. J. Mol. Cell. Cardiol 97:47–55 [DOI] [PubMed] [Google Scholar]
- 80.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, et al. 2010. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ. Res 107:810–17 [DOI] [PubMed] [Google Scholar]
- 81.Van Aelst LN, Summer G, Li S, Gupta SK, Heggermont W, et al. 2016. RNA profiling in human and murine transplanted hearts: identification and validation of therapeutic targets for acute cardiac and renal allograft rejection. Am. J. Transplant 16:99–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duong Van Huyen JP, Tible M, Gay A, Guillemain R, Aubert O, et al. 2014. MicroRNAs as non-invasive biomarkers of heart transplant rejection. Eur. Heart J 35:3194–202 [DOI] [PubMed] [Google Scholar]
- 83.Sukma Dewi I, Hollander Z, Lam KK, McManus JW, Tebbutt SJ, et al. 2017. Association of serum miR-142–3p and miR-101–3p levels with acute cellular rejection after heart transplantation. PLOS ONE 12:e0170842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sukma Dewi I, Torngren K, Gidlof O, Kornhall B, Ohman J. 2013. Altered serum miRNA profiles during acute rejection after heart transplantation: potential for non-invasive allograft surveillance. J. Heart Lung Transplant 32:463–66 [DOI] [PubMed] [Google Scholar]
- 85.Neumann A, Napp LC, Kleeberger JA, Benecke N, Pfanne A, et al. 2017. MicroRNA 628–5p as a novel biomarker for cardiac allograft vasculopathy. Transplantation 101:e26–33 [DOI] [PubMed] [Google Scholar]
- 86.Wang D, Li T, Cui H, Zhang Y. 2016. Analysis of the indicating value of cardiac troponin I, tumor necrosis factor-α, interleukin-18, miR-1 and miR-146b for viral myocarditis among children. Cell. Physiol. Biochem 40:1325–33 [DOI] [PubMed] [Google Scholar]
- 87.Gildea JJ, Carlson JM, Schoeffel CD, Carey RM, Felder RA. 2013. Urinary exosome miRNome analysis and its applications to salt sensitivity of blood pressure. Clin. Biochem 46:1131–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Qi H, Liu Z, Liu B, Cao H, Sun W, et al. 2017. micro-RNA screening and prediction model construction for diagnosis of salt-sensitive essential hypertension. Medicine 96:e6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huang YQ, Huang C, Chen JY, Li J, Feng YQ. 2016. The association of circulating miR-30a, miR-29 and miR-133 with white-coat hypertension. Biomark. Med 10:1231–39 [DOI] [PubMed] [Google Scholar]
- 90.Cengiz M, Karatas OF, Koparir E, Yavuzer S, Ali C, et al. 2015. Differential expression of hypertension-associated microRNAs in the plasma of patients with white coat hypertension. Medicine 94:e693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cengiz M, Yavuzer S, Kiliçkiran Avci B, Yürüyen M, Yavuzer H, et al. 2015. Circulating miR-21 and eNOS in subclinical atherosclerosis in patients with hypertension. Clin. Exp. Hypertens 37:643–49 [DOI] [PubMed] [Google Scholar]
- 92.Karolina DS, Tavintharan S, Armugam A, Sepramaniam S, Pek SL, et al. 2012. Circulating miRNA profiles in patients with metabolic syndrome. J. Clin. Endocrinol. Metab 97:E2271–76 [DOI] [PubMed] [Google Scholar]
- 93.Kontaraki JE, Marketou ME, Zacharis EA, Parthenakis FI, Vardas PE. 2014. Differential expression of vascular smooth muscle–modulating microRNAs in human peripheral blood mononuclear cells: novel targets in essential hypertension. J. Hum. Hypertens 28:510–16 [DOI] [PubMed] [Google Scholar]
- 94.Marques FZ, Charchar FJ. 2015. microRNAs in essential hypertension and blood pressure regulation. Adv. Exp. Med. Biol 888:215–35 [DOI] [PubMed] [Google Scholar]
- 95.de Gonzalo-Calvo D, van der Meer RW, Rijzewijk LJ, Smit JW, Revuelta-Lopez E, et al. 2017. Serum microRNA-1 and microRNA-133a levels reflect myocardial steatosis in uncomplicated type 2 diabetes. Sci. Rep 7:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rawal S, Ram TP, Coffey S, Williams MJ, Saxena P, et al. 2016. Differential expression pattern of cardiovascular microRNAs in the human type-2 diabetic heart with normal ejection fraction. Int. J. Cardiol 202:40–43 [DOI] [PubMed] [Google Scholar]
- 97.Leon LE, Rani S, Fernandez M, Larico M, Calligaris SD. 2016. Subclinical detection of diabetic cardiomyopathy with microRNAs: challenges and perspectives. J. Diabetes Res 2016:6143129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu H, Buchan RJ, Cook SA. 2010. MicroRNA-223 regulates Glut4 expression and cardiomyocyte glucose metabolism. Cardiovasc. Res 86:410–20 [DOI] [PubMed] [Google Scholar]
- 99.Guo R, Nair S. 2017. Role of microRNA in diabetic cardiomyopathy: from mechanism to intervention. Biochim. Biophys. Acta 1863:2070–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X, Liu S. 2017. Role of microRNAs in the pathogenesis of diabetic cardiomyopathy. Biomed. Rep 6:140–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gilca GE, Stefanescu G, Badulescu O, Tanase DM, Bararu I, Ciocoiu M. 2017. Diabetic cardiomyopathy: current approach and potential diagnostic and therapeutic targets. J. Diabetes Res 2017:1310265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Copier CU, Leon L, Fernandez M, Contador D, Calligaris SD. 2017. Circulating miR-19b and miR-181b are potential biomarkers for diabetic cardiomyopathy. Sci. Rep 7:13514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garcia R, Villar AV, Cobo M, Llano M, Martin-Duran R, et al. 2013. Circulating levels of miR-133a predict the regression potential of left ventricular hypertrophy after valve replacement surgery in patients with aortic stenosis. J. Am. Heart Assoc 2:e000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chen Z, Li C, Xu Y, Li Y, Yang H, Rao L. 2014. Circulating level of miR-378 predicts left ventricular hypertrophy in patients with aortic stenosis. PLOS ONE 9:e105702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Beaumont J, Lopez B, Ravassa S, Hermida N, Jose GS, et al. 2017. MicroRNA-19b is a potential biomarker of increased myocardial collagen cross-linking in patients with aortic stenosis and heart failure. Sci. Rep 7:40696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fabiani I, Scatena C, Mazzanti CM, Conte L, Pugliese NR, et al. 2016. Micro-RNA-21 (biomarker) and global longitudinal strain (functional marker) in detection of myocardial fibrotic burden in severe aortic valve stenosis: a pilot study. J. Transl. Med 14:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Villar AV, Garcia R, Merino D, Llano M, Cobo M, et al. 2013. Myocardial and circulating levels of microRNA-21 reflect left ventricular fibrosis in aortic stenosis patients. Int. J. Cardiol 167:2875–81 [DOI] [PubMed] [Google Scholar]
- 108.Røsjø H, Dahl MB, Bye A, Andreassen J, Jørgensen M, et al. 2014. Prognostic value of circulating microRNA-210 levels in patients with moderate to severe aortic stenosis. PLOS ONE 9:e91812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wong LL, Armugam A, Sepramaniam S, Karolina DS, Lim KY, et al. 2015. Circulating microRNAs in heart failure with reduced and preserved left ventricular ejection fraction. Eur. J. Heart Fail 17:393–404 [DOI] [PubMed] [Google Scholar]
- 110.Watson CJ, Gupta SK, O’ Connell E, Thum S, Glezeva N, et al. 2015. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur. J. Heart Fail 17:405–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, et al. 2010. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation 122:2378–87 [DOI] [PubMed] [Google Scholar]
- 112.McManus DD, Lin H, Tanriverdi K, Quercio M, Yin X, et al. 2014. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Heart Rhythm 11:663–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu Z, Zhou C, Liu Y, Wang S, Ye P, et al. 2012. The expression levels of plasma microRNAs in atrial fibrillation patients. PLOS ONE 7:e44906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Natsume Y, Oaku K, Takahashi K, Nakamura W, Oono A, et al. 2018. Combined analysis of human and experimental murine samples identified novel circulating microRNAs as biomarkers foratrial fibrillation. Circ. J 82:965–73 [DOI] [PubMed] [Google Scholar]
- 115.Dawson K, Wakili R, Ordog B, Clauss S, Chen Y, et al. 2013. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation 127:1466–75 [DOI] [PubMed] [Google Scholar]
- 116.McManus DD, Tanriverdi K, Lin H, Esa N, Kinno M, et al. 2015. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study). Heart Rhythm 12:3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Benz F, Roy S, Trautwein C, Roderburg C, Luedde T. 2016. Circulating microRNAs as biomarkers for sepsis. Int. J. Mol. Sci 17:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dumache R, Rogobete AF, Bedreag OH, Sarandan M, Cradigati AC, et al. 2015. Use of miRNAs as biomarkers in sepsis. Anal. Cell. Pathol 2015:186716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kingsley SMK, Bhat BV. 2017. Role of microRNAs in sepsis. Inflamm. Res 66:553–69 [DOI] [PubMed] [Google Scholar]
- 120.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, et al. 2009. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLOS ONE 4:e7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roderburg C, Luedde M, Vargas Cardenas D, Vucur M, Scholten D, et al. 2013. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLOS ONE 8:e54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ma Y, Vilanova D, Atalar K, Delfour O, Edgeworth J, et al. 2013. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLOS ONE 8:e75918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, et al. 2010. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem. Biophys. Res. Commun 394:184–88 [DOI] [PubMed] [Google Scholar]
- 124.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie L. 2012. Serum microRNA signatures identified by Solexa sequencing predict sepsis patients’ mortality: a prospective observational study. PLOS ONE 7:e38885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wang HJ, Zhang PJ, Chen WJ, Feng D, Jia YH, Xie LX. 2012. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J. Trauma Acute Care Surg. 73:850–54 [DOI] [PubMed] [Google Scholar]
- 126.Wu Y, Li C, He Y, Li Q, Wang G, et al. 2014. [Relationship between expression of microRNA and inflammatory cytokines plasma level in pediatric patients with sepsis]. Zhonghua Er Ke Za Zhi 52:28–33 (In Chinese) [PubMed] [Google Scholar]
- 127.Wang L, Wang HC, Chen C, Zeng J, Wang Q, et al. 2013. Differential expression of plasma miR-146a in sepsis patients compared with non-sepsis-SIRS patients. Exp. Ther. Med 5:1101–04 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Puskarich MA, Nandi U, Shapiro NI, Trzeciak S, Kline JA, Jones AE. 2015. Detection of microRNAs in patients with sepsis. J. Acute Dis 4:101–6 [Google Scholar]
- 129.Benz F, Tacke F, Luedde M, Trautwein C, Luedde T, et al. 2015. Circulating microRNA-223 serum levels do not predict sepsis or survival in patients with critical illness. Dis. Markers 2015:384208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Taganov KD, Boldin MP, Chang KJ, Baltimore D. 2006. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. PNAS 103:12481–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Witwer KW, Soekmadji C, Hill AF, Wauben MH, Buzas EI, et al. 2017. Updating the MISEV minimal requirements for extracellular vesicle studies: building bridges to reproducibility. J. Extracell. Vesicles 6:1396823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chevillet JR, Lee I, Briggs HA, He Y, Wang K. 2014. Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules 19:6080–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sunderland N, Skroblin P, Barwari T, Huntley RP, Lu R, et al. 2017. MicroRNA biomarkers and platelet reactivity: the clot thickens. Circ. Res 120:418–35 [DOI] [PubMed] [Google Scholar]
- 134.Wang H, Meng K, Chen W, Feng D, Jia Y, Xie L. 2012. Serum miR-574–5p: a prognostic predictor ofsepsis patients. Shock 37:263–67 [DOI] [PubMed] [Google Scholar]
- 135.Wang H, Zhang P, Chen W, Feng D, Jia Y, Xie LX. 2012. Evidence for serum miR-15a and miR-16 levels as biomarkers that distinguish sepsis from systemic inflammatory response syndrome in human subjects. Clin. Chem. Lab. Med 50:1423–28 [DOI] [PubMed] [Google Scholar]
- 136.Sun X, Icli B, Wara AK, Belkin N, He S, et al. 2012. MicroRNA-181b regulates NF-κB-mediated vascular inflammation. J. Clin. Investig 122:1973–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang H, Yu B, Deng J, Jin Y, Xie L. 2014. Serum miR-122 correlates with short-term mortality in sepsis patients. Crit. Care 18:704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.How CK, Hou SK, Shih HC, Huang MS, Chiou SH, et al. 2015. Expression profile of microRNAs in Gram-negative bacterial sepsis. Shock 43:121–27 [DOI] [PubMed] [Google Scholar]
- 139.Goodwin AJ, Guo C, Cook JA, Wolf B, Halushka PV, Fan H. 2015. Plasma levels of microRNA are altered with the development of shock in human sepsis: an observational study. Crit. Care 19:440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Caserta S, Kern F, Cohen J, Drage S, Newbury SF, Llewelyn MJ. 2016. Circulating plasma microRNAs can differentiate human sepsis and systemic inflammatory response syndrome (SIRS). Sci. Rep 6:28006. [DOI] [PMC free article] [PubMed] [Google Scholar]