Abstract
Persistence of latent HIV-1 in long-lived resting memory CD4+ T cells is a major barrier to curing HIV-1 infection, and thus a biomarker for latently infected cells would be of great scientific and clinical importance.1,2,3,4,5 Through an elegant discovery-based approach, Descours et al. reported that CD32a, an Fcγ receptor not normally expressed on T cells, is a potential biomarker for latently infected cells.6 Using the quantitative viral outgrowth assay, we show that CD32+ CD4+ T cells do not harbor the majority of intact proviruses in the latent reservoir and that the enrichment found by Descours et al. may in part reflect the use of an ultrasensitive ELISA for HIV-1 p24 antigen that does not predict exponential viral outgrowth. Our studies show that CD32 is not a biomarker for the major population of latently infected CD4+ T cells.
If CD32a is a specific biomarker for latent HIV-1 infection in CD4+ T cells and is never expressed on CD4+ T cells in the absence of HIV-1 infection, then there a difference in the frequency of CD4+ T cells that express CD32 in HIV-1-infected individuals relative to healthy donors is expected. We isolated total CD4+ T cells from infected and uninfected donors by negative selection, stained them for CD32 and CD4, and analyzed expression of these proteins by flow cytometry. In healthy donors, we found that an average of 0.019% of CD4+ T cells were also CD32+ (Fig. 1A). This value is not significantly different from values we found in HIV-1-infected individuals (Fig. 1A, average 0.011%, p=0.1143) nor is it different from the values previously reported by Descours et al. in HIV-1-infected individuals (0.016%, p = 0.66). Thus CD32 does not appear to be a specific biomarker of latently infected CD4+ T cells.
Fig. 1.
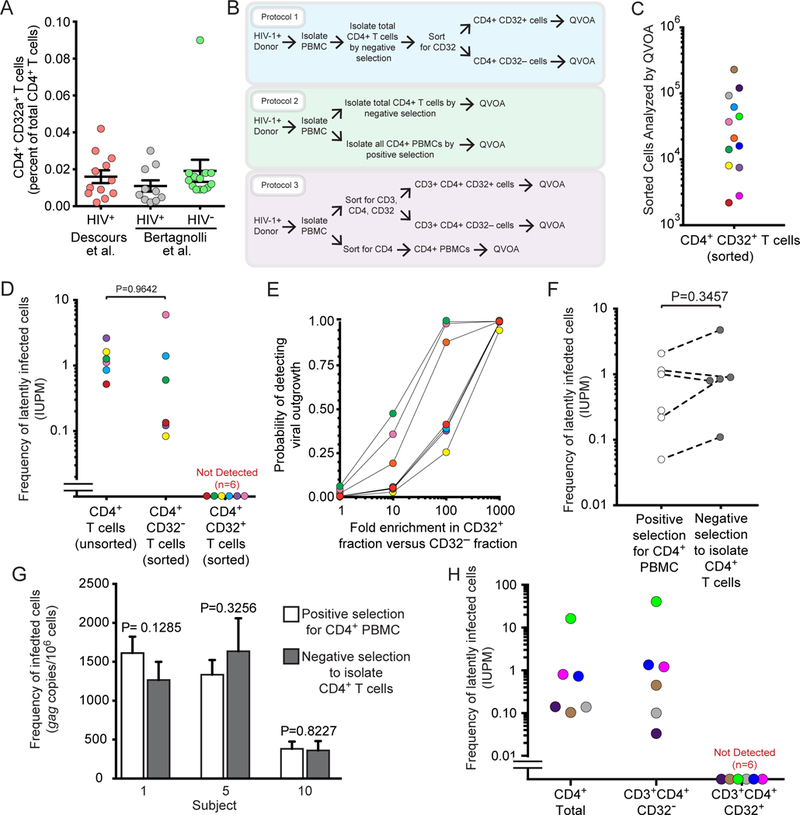
A) Percentage of CD4+CD32hi T cells relative to total CD4+ T cells in healthy donors and HIV-1 infected donors. Infected donor values reported by Descours et al. were obtained from Supplementary Table 4 of reference 6. B) Schematic depicting 3 strategies used to obtain different populations of CD4+ T cells plated in QVOAs. C) Numbers of sorted CD4+CD32hi and CD3+CD4+CD32hi T cells from each subject plated in QVOAs. D) Frequencies of latently infected cells among CD4+CD32hi T cells and CD4+CD32neg T cells and among total CD4+ T cells from the same subjects previously measured in separate experiments. Cells were isolated using protocol 1 (colors coordinate with subject values from Fig 1c). E) Probability of detecting outgrowth based on measured frequencies of latently infected cells among the CD4+CD32neg fraction and number of CD4+CD32hi cells plated assuming various degrees of enrichment of HIV-1 in CD32hi cells. F) Frequencies of latently infected cells measured in QVOAs using positive or negative selection to obtain total CD4+ cells (protocol 2; positive selection was accomplished using either sorting or CD4 microbead strategies, with similar results). G) Comparison of proviral DNA measurements obtained with Qpcr on total CD4+ cells purified using positive or negative selection (protocol 2). H) Frequencies of latently infected cells among total CD4 cells, and CD3+CD4+CD32neg and CD3+CD4+CD32hi populations. Cells were isolated using protocol 3 (colors coordinate with subject values from Fig 1c).
To examine whether infectious proviruses were present in CD4+CD32hi T cells, total CD4+ T cells were isolated by negative selection from 6 HIV-1+ individuals on suppressive antiretroviral therapy (ART) for at least 6 months (see Table S1 for patient characteristics). The cells were stained and sorted to obtain CD4+CD32hi and CD4+CD32neg populations which were subsequently plated in quantitative viral outgrowth assays (QVOAs) as described in Laird et al.7 (Fig. 1b, Protocol 1). The number of CD4+CD32hi cells assayed by QVOA for each subject is shown in Fig. 1c. On day 14, viral outgrowth was measured using a standard ELISA for HIV-1 p24 antigen. CD4+CD32hi wells from all subjects were negative for p24 on day 14 and remained negative after an additional week of culture. Conversely, viral outgrowth was observed in CD4+CD32neg wells from all subjects on both days 14 and 21. The mean infected cell frequency, 1.37 infectious units per million (IUPM), was comparable to values previously measured in resting CD4+ T cells in several studies (0.03–3 IUPM8; 0.97 IUPM in chronic patients9) and to values previously measured in the same subjects (mean 1.33 IUPM) (Fig. 1d; Table S2). If the enrichment of proviruses in CD32+ cells reported by Descours et al. was characteristic of replication-competent proviruses, then it is highly probable that outgrowth would have been seen in cultures with CD4+CD32hi T cells (Fig 1e).
These results indicate that a substantial fraction of latently infected CD4+ T cells do not express CD32. One possible explanation for the discrepancy between our results and those of Descours et al. is that some latent HIV-1 may be present in a unique and previously undescribed population of CD4+ T cells that express CD32 together with other non-T cell lineage markers. If this were the case, such cells would be removed during the standard negative selection approaches used to isolate CD4+ T cells. We tested whether a CD4+ T cell enrichment based on negative selection could be removing any CD4+ cell subpopulations harboring latent HIV-1 (e.g. CD4+CD32hi T cells). We isolated total CD4+ cells from infected donors on suppressive ART using two different methods: negative selection to remove other lineages, leaving untouched CD4+ T cells and positive selection for cells expressing CD4 (Fig. 1b, Protocol 2). Both CD4+ populations were tested for the presence of infectious virus by QVOA. Importantly, no statistically significant difference was observed in frequencies of latently infected cells between the two purified cells populations (Fig. 1f). Furthermore, no significant difference in levels of proviral DNA was observed between the purified cell populations (Fig. 1g). As CD4 is required for HIV-1 entry, cell populations obtained via positive selection for CD4 should include every latently infected CD4+ T cell. Given that neither infected cell frequencies nor proviral DNA levels differed between the purified cell populations, we conclude that no additional sizable population of latently infected cells was recovered by positive CD4 selection.
In further studies, we used a cell sorting strategy identical to that described by Descours et al. on samples from 6 HIV-1 positive subjects. Peripheral blood mononuclear cells (PBMCs) from HIV-1+ subjects were stained and sorted to obtain CD3+CD4+CD32hi and CD3+CD4+CD32neg populations and the resulting cell populations were tested for latently infected cells using the QVOA. The numbers of CD3+CD4+CD32hi cells assayed by QVOA for each subject are shown in Fig. 1c and Table S3. In addition, total CD4+ cells were obtained by staining PBMCs for CD4 and sorting for CD4+ cells (Fig. 1b, Protocol 3). QVOA results showed that both the CD3+CD4+CD32neg and the total CD4+ T cell populations had the same infected cell frequencies that were comparable to infected cell frequencies measured in other studies10. However, we observed no outgrowth in CD3+CD4+CD32hi cultures (Fig. 1h; Table S2).
We also analyzed CD3+CD4+CD32hi and CD3+CD4+CD32neg cells isolated by the method of Descours et al. for the presence of proviral DNA by qPCR. We found 89 copies of gag per million CD3+CD4+CD32neg cells, which is similar to prior measurements in total CD4+ T cells11. However, no proviral DNA was detected after DNA extraction from 39,000 CD3+CD4+CD32hi cells and subsequent qPCR analysis (data not shown). This finding makes it highly unlikely that this cell population is enriched for HIV-1 to a level of more than one provirus copy per cell as previously reported by Descours et al. We caution that normalization of very low-level HIV-1 DNA measurements from qPCR reactions with a low number of input cell equivalents could artificially produce apparent enrichments in HIV-1 DNA.
In a further attempt to explain the discordant QVOA results obtained in our studies and those of Descours et al., we tested whether the use of the ultra-sensitive p24 digital ELISA12 and the low cell input can affect IUPM calculations, leading to erroneous overestimation of latent infection. QVOA culture supernatants were assayed for HIV-1 p24 using the ultrasensitive SIMOA p24 2.0 assay (Quanterix, MA) on days 5, 9, 14, and 21. Using the lower limit of quantification (LLOQ, 0.01 pg/ml) as the cut off, we found that wells from 2 out of 3 QVOAs containing CD4+CD32hi cells tested positive for p24 by this assay even though the same wells were negative by standard ELISA, which is several orders of magnitude less sensitive (Fig 2a). Exponential outgrowth is the hallmark of replication-competent viruses. In QVOA cultures of CD4+CD32neg cells, only a fraction of the wells that were positive by SIMOA showed exponential outgrowth as determined by the standard ELISA on day 21 (Fig. 2b). Importantly, CD4+CD32hi culture wells that tested positive by SIMOA p24 assay showed no exponential outgrowth and had significantly lower levels of p24 (Fig 2c). It is possible that low positive SIMOA values could reflect an assay artifact or the presence of defective proviruses still capable of producing low levels of Gag13. A further concern is that IUPM calculations are based on cell input, fold dilutions, and the number of technical replicates14, and thus QVOAs performed with very small numbers of sorted CD4+CD32hi cells can dramatically skew the frequency of cells harboring replication-competent proviruses (5-fold dilutions from 800 to 1 cell in Descours et al.). When we used the results obtained with SIMOA p24 assay, IUPM values ranged from zero to 3134 and 554 (patients 4 and 5, respectively; Fig. 2d). As a consequence, when we calculated the “fold enrichment” of IUPM in the CD4+CD32hi cells over CD4+CD32neg cells, we observed a mean fold enrichment of 665 (range 152–1179, from the two patients with positive p24 using SIMOA), similar to what was reported by Descours et al. (Fig 2e), even though no outgrowth was observed from these cultures.
Fig. 2.
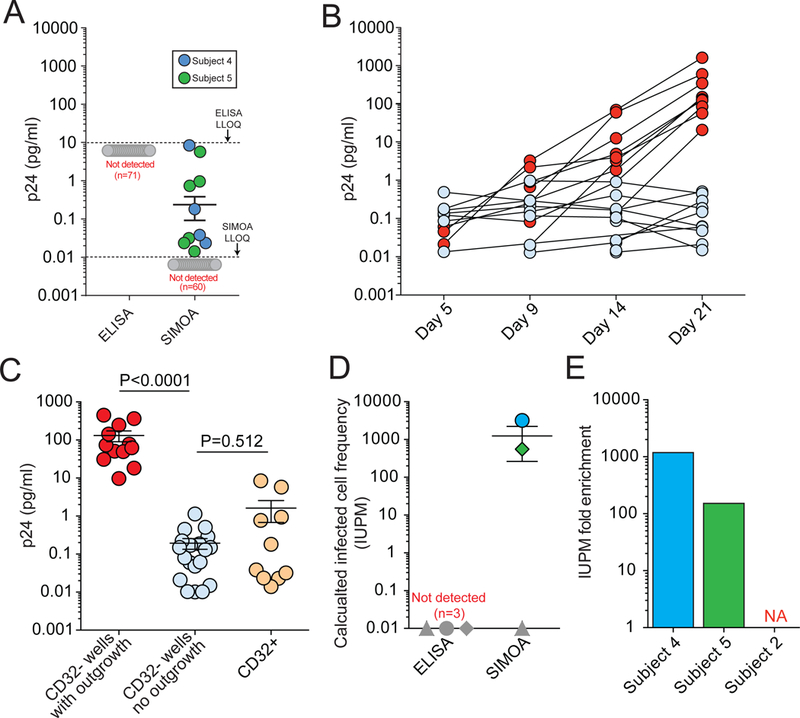
Ultrasensitive p24 measurements as a possible factor in overestimation of latent infection. A) p24 values from CD32+ culture wells measured with ELISA versus SIMOA (Lower limit of quantification (LLOQ): 5–10 pg/mL and 0.01 pg/mL, respectively) (data collected from three subjects, for a total of 71 wells). B) Longitudinal p24 values measured with SIMOA in individual culture wells in the QVOA for CD32neg cells from subject 5, showing wells with and without viral outgrowth (red and blue circles, respectively). C) p24 values in QVOA wells of CD32hi and CD32neg cultures. Day 21 values are shown for CD32neg culture wells with outgrowth as detected by ELISA (red), CD32neg culture wells with no outgrowth by ELISA but with positive SIMOA values (blue), and CD32hi culture wells with no outgrowth by ELISA but with positive SIMOA values (orange). Date were collected from subjects 2, 4, and 5. p values were calculated with a non-parametric t-test. D) IUPM calculation based on ELISA versus SIMOA. Symbols in dark grey represent values below the limit of detection. E) Fold enrichment of IUPM in CD32+ cells (subject ID shown on the horizontal axis).
In summary, we find no evidence that CD32 expression indicates the presence of latent HIV-1 and demonstrate that at least a substantial fraction of the HIV-1 latent reservoir is in CD3+CD4+CD32neg T cells. While no outgrowth could be found in cultures containing CD4+CD32hi T cells, viral outgrowth comparable to historical measurements was found in cultures containing CD4+CD32neg T cells. We have shown that the use of an ultrasensitive p24 ELISA assay may account for the apparent enrichment observed by culture experiments by Descours et al. In short, our results have demonstrated that CD32 does not define the HIV-1 reservoir and that future research is still needed to identify biomarkers for latently infected cells.
Methods
QVOAs isolated CD4+ T cells using negative depletion and sorted for CD32a+ cells (Fig. 1b, Protocol 1). To test whether negative depletion was causing a loss of CD32+ CD4+ T cells, outgrowth and proviral DNA was compared from QVOAs where CD4+ T cells were isolated using positive selection to measurements using negative depletion. Outgrowth measurements and proviral DNA were also measured using the same methods described by Descours et al. Proviral DNA measurements were performed using qPCR15. HIV-1 p24 values were measured using both a standard ELISA for p24 antigen (Perkin Elmer, MA) and SIMOA (Quanterix, MA). Details are provided in Supplementary Methods.
Supplementary Material
Acknowledgements
We would like to thank the study participants without whom this research would not be possible. Funding was provided by the by the NIH Martin Delaney I4C, Beat-HIV and DARE Collaboratories by the Johns Hopkins Center for AIDS Research (P30AI094189), by NIH grant 43222, and by the Howard Hughes Medical Institute and the Bill and Melinda Gates Foundation.
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Finzi D et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278, 1295–1300 (1997). [DOI] [PubMed] [Google Scholar]
- 2.Chun TW et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U.S.A 94, 13193–13197 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong JK et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278, 1291–1295 (1997). [DOI] [PubMed] [Google Scholar]
- 4.Richman DD et al. The challenge of finding a cure for HIV infection. Science 323, 1304–7 (2009) [DOI] [PubMed] [Google Scholar]
- 5.Deeks SG et al. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12, 607–14 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Descours B et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 543, 564–567 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Laird GM et al. Measuring the Frequency of Latent HIV-1 in Resting CD4+ T Cells Using a Limiting Dilution Coculture Assay. Methods Mol Biol 1354, 239–53 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Siliciano JD et al. Long-tern follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 6, 727–8 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Eriksson S et al. Comparative analysis of measure of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crooks AM et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis 212, 1361–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besson GJ et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 59, 1312–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passaes CP et al. HIV cure research: advances and prospects. Virology 454–455, 340–52 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Pollack RA et al. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host Microbe 21, 494–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosenbloom DI et al. Designing and Interpreting Limiting Dilution Assays: General Principles and Applications to the Latent Reservoir for Human Immunodeficiency Virus- 1. Open Forum Infect Dis 2, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massanella M et al. Quantification of Total and 2-LTR (Long terminal repeat) HIV DNA, HIV RNA and Herpesvirus DNA in PBMCs. Bio Protoc 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


