Abstract
Creating new functional building blocks that expand the versatility of nanostructures depends on bottom-up self-assembly of amphiphilic biomolecules. Inspired by the unique physicochemical properties of hydrophobic perfluorocarbons, coupled with the powerful functions of nucleic acids, we herein report the synthesis of a series of diperfluorodecyl–DNA conjugates (PF–DNA) which can efficiently self-assemble into micelles in aqueous solution. On the basis of the micelle structure, both target binding affinity and enzymatic resistance of the DNA probe can be enhanced. In addition, based on the hydrophobic effect, the PF–DNA micelles (PFDM) can actively anchor onto the cell membrane, offering a promising tool for cell-surface engineering. Finally, the PFDM can enter cells, which is significant for designing carriers for intracellular delivery. The combined advantages of the DNA micelle structure and the unique physicochemical properties of perfluorocarbons make these PFDM promising for applications in bioimaging and biomedicine.
In nature, many exquisite supermolecular structures are built by bottom-up self-assembly of biomolecules, an essential step in implementing their ultimate biological functions. For example, the cell membrane, which serves as a gatekeeper to maintain cellular integrity and homeostasis, is constructed by self-assembly of amphipathic phospholipids.1 In addition, many artificial self-assembled nanostructures have been inspired by nature.2 Among them, micelle structures have attracted widespread attention because of their small nanoscale sizes (5–100 nm), uniform spherical shapes, and good biocompatibility.3 Creating amphipathic building blocks with biomolecules is an essential step toward constructing functional micelles for biological applications.4
Nucleic acids, as carriers of genetic information, are promising biomolecules for engineering functional nanostructures.5 Recently, micelle structures, composed of a hydrophobic polymer core and a hydrophilic DNA corona, have been developed and utilized for delivery of nucleic acid probes, hydrophobic drugs, and antisense DNA for biosensing, bioimaging, and cancer therapy.6–10 While the hydrophilic DNA segment has been well-explored, rare attention has been given to the development of the hydrophobic section, which is expected to broadly expand the application prospects of DNA micelles.
Perfluorocarbons (PFs) are chemically inert organic fluorine compounds composed only of carbon and fluorine. In recent years, the application of PFs in biomedical applications has gained considerable interest because of their unique physical and chemical properties.11 For example, owing to the low polarizability of fluorine, PFs exhibit low van der Waals and intermolecular interactions, making it easier for them to dissolve such gases as O2, CO2, NO, or N2.12–14 Meanwhile, because of their superior biostability in biological systems, PFs have been used as contrast agents for diagnostic ultrasound imaging.15–17 Moreover, because of high fluoride content, PFs can serve as a promising fluoromagnetic imaging tracers.16,18,19 Despite their potential, the applications of PF molecules in biomedical research have been hindered by their poor solubility. To address this issue, various nanomaterials, including inorganic nanoparticles,20–22 liposomes,23 and polymer nanoparticles,24,25 have been used to assist in the biological transport of PFs. While visible improvement has been achieved, most of these nanomaterials are limited by high toxicity and broad size distribution.17 As alternatives, amphipathic PF compounds were exploited by conjugating fluorocarbon chains with hydrophilic headgroups.26,27 For example, the conjugation of β-d-glucopyranosides with a single chain of PF allowed cellular delivery of pulmonary drugs and guidance synthesis of different glycolipids.28,29 To further promote the biological applications of PFs, introduction of functional biomolecules, including nucleic acids and peptides, as the hydrophilic section can be another promising direction.
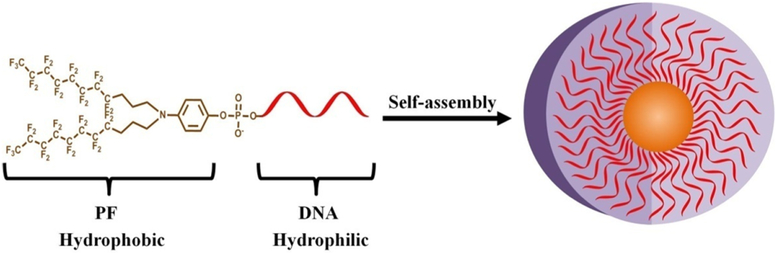
In this work, we have synthesized PF–DNA conjugates and used them as building blocks to construct PF–DNA micelles (PFDM) (Scheme 1). The micelle nanostructure was characterized by fluorescence spectrometry, native polyacrylamide gel electrophoresis (N-PAGE), dynamic light scattering (DLS), and atomic force microscopy (AFM). The DNA hybridization capability and DNA biostability of the PFDM were also investigated. In addition, the interaction of the PFDM with live cells and their cellular distribution were also studied using flow cytometry and confocal laser scanning microscopy (CLSM). Our experimental results show the high potential of this PFDM structure for biomedical research.
Scheme 1.
Illustration of a Diperfluorodecyl–DNA Micelle (PFDM) Nanostructure, Containing a Hydrophobic Perfluorocarbon Core and a Hydrophilic Oligonucleotide Corona
EXPERIMENTAL SECTION
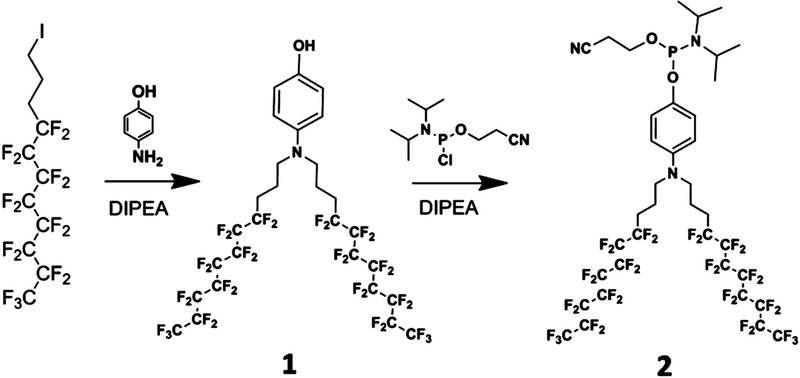
Synthesis of Diperfluorodecyl Phosphoramidite.
A 0.5 g sample of compound 1 (0.49 mmol) was dissolved in 20 mL of anhydrous dichloromethane, followed by addition of 0.19 g of N,N-diisopropylethylamine (DIPEA 1.47 mmol). After the mixture was cooled in an ice bath, 0.17 g of 2-cyanoethyl-N,N-diisopropylchlorophosphoramidite (0.73 mmol) was added. Then, the ice bath was removed, and the reaction was conducted at room temperature with stirring for an additional 2 h. The reaction mixture was successively washed with saturated NaHCO3, brine, and water. The organic compound was collected, dried over anhydrous Na2SO4, and concentrated under low pressure. After purification by a flash chromatographic column, compound 2 of ~0.4 g was obtained and identified by1H NMR (300 MHz, CDCl3) δ 6.95 (d, 2H), 6.71 (d, 2H), 3.94 (m, 2H), 3.73 (m, 2H), 3.22 (t, 4H), 2.65 (t, 2H), 2.14 (m, 4H), 1.82 (m, 4H), 1.14–1.25 (m, 12H),19F NMR (300 MHz, CDCl3) δ –80.82 (t, 6F), −113.92 (s, 4F), −121.73 (m, 12F), −122.75 (s, 4F), −123.45 (s, 4F), −126.15(s, 4F), and31P NMR (300 MHz, CDCl3) δ 147.10.
Solid-Phase Synthesis of Oligonucleotides.
All DNA synthesis reagents were purchased from Glen Research. Pyrene phosphoramidite (py) was synthesized in our lab according to a previous report.9 DNA synthesis, including conjugation of the diperfluorodecyl chain, was conducted via solid-phase phosphoramidite chemistry on a 12-column DNA/RNA synthesizer (Polygen) at 1.0 mM scale. For 5-carboxyfluorescein (FAM)-and 5-carboxytetramethylrhodamine (TAMRA)-labeled DNA, the corresponding controlled pore glass (CPG) was used. Detailed sequences are listed in Table S1. The obtained oligonucleotides were cleaved and deprotected from the CPG, followed by precipitation in cold salted ethanol solution at −20 °C overnight. The DNAs were dissolved in triethylammoniumacetate (TEAA, 100 mM, pH 7.5) after centrifugation to remove supernatant. After that, the DNAs were purified by reversed-phase high-performance liquid chromatography (HPLC) using a Bio Basic-4 column or a C18 reversed-phase column. The mobile phase was acetonitrile plus TEAA, and the elution program is listed in Table S2. For normal DNA molecules having no diperfluorodecyl chain, the 4,4′-dimethoxytrityl group was removed from DNA by adding 80% acetic acid aqueous solution, followed by another round of precipitation in cold ethanol. The resulting DNA products were dried with a rotary vacuum pump and subsequently quantified by measuring the absorbance at 260 nm.
Electrophoresis Characterization.
Self-assembly of the PFDM was characterized by N-PAGE (5 mL of 30% acrylamide, 0.11 mL of 10% ammonium persulfate, 3.0 mL of Tris–acetate–ethylenediaminetetraacetic acid (EDTA) buffer (TAE), 0.01 mL of N,N,N,N,tetramethylethylenediamine (TEMED), 6.9 mL of H2O). DNA molecules with and without diperfluorodecyl chain were dissolved in TAE/Mg2+ to give a final concentration of 6 μM. Ten microliters of each sample was mixed with 2 μL of 6× loading buffer and separated by 10% N-PAGE at 110 V for 90 min in 1× TAE (40 mM tris(hydroxymethyl)aminomethane, containing 1 mM EDTA and 20 mM acetic acid, pH 7.6).
Fluorescence Measurements.
All fluorescence measurements were performed on a Fluoromax-4 spectrofluorometer (HORIBA JobinYvon, Edison, NJ). The optical path length of the fluorescence cuvette was 1.0 cm. For pyrene (py), the samples were excited at 350 nm, and the emission spectra were collected from 360 to 650 nm. For FAM, the sample was excited at 488 nm, and the fluorescence intensity at 520 nm was used to evaluate the performance of the DNA micelles/probes.
Micelle Characterization.
Different PF–DNA conjugates were diluted with Dulbecco’s phosphate-buffered saline (DPBS) buffer (5 mM Mg2+) to 2 μM and left at room temperature for about 2 h. Ten microliters of this solution was deposited on a freshly cleaved mica surface and allowed to adsorb for 5 min. The mica surface was washed twice with a further 20 μL of Millipore water and dried by nitrogen gas. Then, AFM was performed on a Multimode 8 (Bruker/U.S.A.). In addition, 100 μL of this solution was analyzed with DLS on a Malvern Zetasizer Nano ZS90 (Malvern Instruments, Ltd., Worcestershire, U.K.).
Melting Temperature Measurement.
To compare the melting temperatures of PFDM with free DNA molecules, the FAM-labeled PFDM or FAM-labeled DNA molecules (500 nM) were mixed with equivalent DABCYL-labeled cDNA in 1× PBS. Fifty microliters of the mixture was transferred to a qPCR tube and analyzed with a qPCR instrument. The temperature was increased from 20 to 80 °C at a rate of 1 °C/min. FAM fluorescence intensity was measured over time.
Nuclease Digestion Assay.
FAM-labeled PFDM or FAM-labeled DNA molecules (DNA concentration fixed at 500 nM) were mixed with DABCYL-labeled cDNA (1000 nM) in buffer (10 mM Tris, 2.5 mM MgCl2, and 0.5 mM CaCl2, pH = 7.8). To facilitate DNA hybridization, the mixture was heated at 95 °C for 1 h, and then slowly cooled to room temperature in a thermally insulated container. The entire cooling process lasted about 5 h. After dilution to 150 nM with buffer, the resulting mixture was transferred to a 96-well plate (100 μL per well), and DNase I was added (5 units/mL). The kinetic fluorescence change at 520 nm was detected using a Synergy H4 hybrid reader (BioTek) with excitation at 488 nm.
Cell Culture.
CEM (a T lymphoblast-like cell line), 3T3-L1 (a mouse embryonic cell line), and HeLa (a cervical adenocarcinoma cell line) were cultured in RPMI 1640 medium containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA, U.S.A.) and 0.5 mg/mL penicillin–streptomycin (KeyGEN Biotech, Nanjing, China) in 5% CO2 at 37 °C.
Cell Binding Affinity Assay.
CEM cells (2 × 105) were treated with 200 nM PF–T15–TAMRA or T15–TAMRA in incubation buffer (DPBS containing 5 mM Mg2+, 4.5 mg/mL glucose) at room temperature for 30 min. Subsequently, the cells were washed three times with DPBS (Gibco). After resuspension in incubation buffer, the cells were analyzed by flow cytometry (BD FACSVerse) and confocal laser scanning microscopy (FV1000-X81, Olympus).
Confocal Fluorescence Microscopy Imaging.
HeLa cells were plated in a confocal dish and grown for 24 h. Then, the cells were incubated with 500 nM TAMRA-labeled PFDM in incubation buffer at 37 °C for 2 h. After washing with DPBS three times, the cells were treated with nucleic indicator Hoechst 33342 (0.2 μg/mL), lysosomal indicator LysoTracker green (50 nM), or plasma membrane tracker (30 nM) for 30 min, respectively. After washing, the cells were imaged with confocal laser scanning microscopy.
Cytotoxicity Assay.
Cytotoxicity was determined using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. Briefly, HeLa cells and 3T3-L1 cells (6 × 103 cells per well) were plated in 96-well plates and grown for 24 h before treatment, followed by incubation with compound 2 or PFDM of different concentrations (0.5–10 μM) for 48 h. For CEM cells, 3 × 104 cells per well were directly treated with compound 2 or PFDM of different concentrations (0.5–10 μM) for 48 h. After removal of cell medium, CellTiter reagent (20 μL) diluted with fresh medium (100 μL) was added to each well and incubated for 1–2 h. The absorbance (490 nm) of the resulting solution was measured with a Synergy H4 hybrid reader (BioTek).
RESULTS AND DISCUSSION
Successful Construction of PFDM.
Precise control over the components, sizes, and shapes of nanostructures is essential for manipulating their functionality. To generate monodisperse micelle structures, we focused on synthesizing uniform amphipathic building blocks. Conventional DNA–polymer conjugation conducted in homogeneous systems has been limited by low product yield, poor molecular uniformity, and difficult purification.30,31 Recently, solid-phase synthesis has significantly improved the efficiency of DNA/polymer conjugation.6,32 Here, a heptadecafluorodecyl iodide was used as a primary monomer. To improve the hydrophobicity of the PF chain, diperfluorodecyl, a dimer of heptadecafluorodecyl, was synthesized via nucleophilic reaction.11 Subsequently, a phosphoramidite moiety was coupled onto one terminus of the diperfluorodecyl compound (Scheme 2). As such, the diperfluorodecyl chain could be conjugated with the DNA segment using the solid-phase synthesis strategy. The successful synthesis of compounds 1 and 2 was characterized by1H NMR,19F NMR, and31P NMR (Figures S1–S5). The diperfluorodecyl phosphoramidite was directly conjugated onto the 5′-end of the DNA on a fully automated DNA synthesizer. The resulting PF–DNA conjugates were purified by HPLC with a product yield of about 57% (Figure S6). The successful PF–DNA conjugation was further confirmed by mass spectrometry (Figure S7).
Scheme 2.
Synthesis Route of Diperfluorodecyl Phosphoramidite
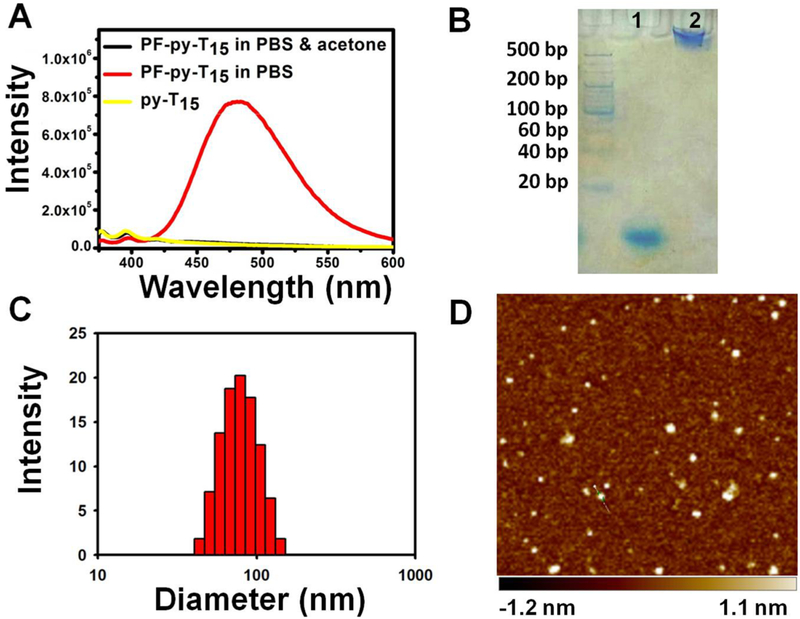
To establish a fluorescence signal that indicates the formation and dissociation of the micelle structure, a spatially sensitive dye, pyrene (py), was incorporated between the DNA headgroup and the PF chain. In the monomeric state, pyrene emits an ensemble of fluorescence peaks in the low-wavelength range. Upon self-assembly into a micelle structure, multiple pyrene molecules aggregate and generate a strong excimer fluorescence peak with large Stokes shift.33 The structural change of PF–py–DNA was studied with fluorescence spectrometry. As shown in Figure 1A, PF–py–DNA conjugates (PF–py–T15) spontaneously self-assembled into a micelle structure in phosphate-buffered saline (PBS), as indicated by broad excimer fluorescence emission peaking at 475 nm under excitation of a 350 nm laser. However, without hydrophobic interaction of the PF chains, py–DNA (py–T15) remained in a monodisperse state in PBS, and only a weak monomeric fluorescence band ranging from 375 to 391 nm was detected. Moreover, with the addition of acetone (v/v = 1:1), the micelle structure (PF–py–T15) was dissociated, accompanied by the disappearance of the excimer fluorescence.
Figure 1.
(A) Fluorescence spectra of PF–py–DNA and py–DNA dissolved in PBS and PF–py–DNA dissolved in a mixture of PBS and acetone (v/v = 1:1). (B) 10% N-PAGE analysis of T15 (1) and PF–T15 (2). (C) Dynamic light scattering (DLS) result of the self-assembled PF–T15 micelles. (D) AFM topography image of the self-assembled PF–T15 micelles deposited on a mica surface.
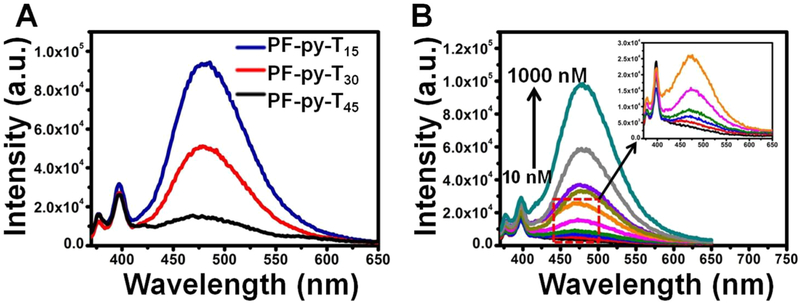
Formation of the PFDM was further confirmed with 10% N-PAGE. As shown in Figure 1B, PFDM migrated more slowly than the corresponding DNA without a PF chain, suggesting the successful formation of larger nanostructures. The hydrodynamic diameter of PFDM in PBS was 57.8 nm with a particle dispersion index (PDI) of 0.382 (Figure 1C), as measured by DLS. In addition, the morphology of PFDM was visualized with AFM. Figure 1D shows a layer of uniform spherical particles with diameter of 22.9 ± 3.4 nm. Notably, the vertical forces of the tapping mode and the strong electrostatic interaction between the soft micelle material and the hard mica sheet34 resulted in compressing the height of PFDM to about 1.8 ± 0.3 nm. These results demonstrate that PF–DNA conjugates can efficiently and spontaneously self-assemble into uniform micelle structures in aqueous solution. To assess the impact of DNA length on micelle formation, three PF–py–DNA conjugates with different DNA lengths (15T, 30T, and 45T) were synthesized. As shown in Figure 2A, the pyrene excimer fluorescence gradually decreased as the DNA length was extended from 15 to 45 mer, indicating a size-dependent assembly of the PFDM structure. The hydrodynamic diameters in PBS for PF–py–T30 and PF–py–T45 were 105.7 nm (PDI, 0.492) and 164.1 nm (PDI, 0.566), respectively (Figure S8). To obtain a relatively stable micelle structure, DNA probes of no more than 15 mer were used in the following studies.
Figure 2.
(A) Fluorescence spectra of PF–py–T15, PF–py–T30, and PF–py–T45 in PBS with concentration of 1 μM. (B) Fluorescence spectra of py–T15 at 1000 nM (black line) and PF–py–T15 at different concentrations (10, 50, 75, 100, 200, 300, 500, 700, 1000 nM) in PBS.
The critical micelle concentration (cmc) of PF–py–T15 was determined by measuring the fluorescence spectrometry of PF–py–T15 at a series of concentrations. The cmc is defined as the threshold concentration of amphipathic molecules above which micelle structures start to form. As illustrated in Figure 2B, the excited state fluorescence of py–T15 did not appear, even at a high concentration (1 μM), while excimer fluorescence intensity of PF–py–T15 increased in a dose-dependent manner and showed a low cmc of 10 nM, indicating stable formation of the PFDM.
Enhanced Target Binding Affinity.
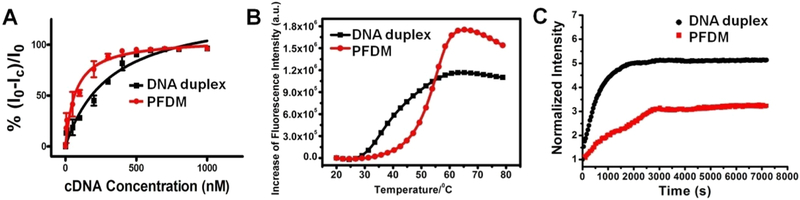
Having demonstrated the successful formation of the PFDM structure, we proceeded to test its performance in aqueous solution. To compare the target recognition capability of PFDM with free corresponding DNA probe, the 3′-end of the DNA was labeled with a FAM fluorophore, and the 5′-end of its fully complementary DNA sequence was labeled with a DABCYL moiety. Upon hybridization, the FAM fluorophore and the DABCYL quencher were in close proximity, resulting in fluorescence being quenched. By using fluorescence titration assay, the fluorescence intensity of both PFDM and free DNA showed a dose-dependent decrease in response to cDNA with the concentration ranging from 5 to 1000 nM, while PFDM showed a sharper transition (Figure S9). The dissociation constants (Kd) were calculated according to the fluorescence variation value. The Kd of the PFDM/cDNA complex was 71.4 nM, which was 4.5 times lower than that of the DNA/cDNA duplex (Kd = 317.8 nM), indicating a stronger DNA binding affinity of PFDM (Figure 3A). The hybridization thermodynamics of PFDM and free DNA probe were also compared using a real-time polymerase chain reaction (qPCR). To perform this experiment, equivalent PFDM/cDNA complex and free DNA/cDNA duplex were heated from 20 to 80 °C at a rate of 1 °C/min, and the fluorescence intensity was read for each 2 °C increment. Because of DNA dehybridization resulting from increased temperature, the quenched FAM fluorescence was gradually restored. Figure 3B shows a higher melting temperature (Tm) of PFDM (53.3 °C) with a sharper transition in comparison to that of DNA (40.2 °C). These results demonstrate that formation of the micelle structure can improve the target recognition capability of the DNA probes, a phenomenon which can be attributed to the synergistic effect of oligonucleotide hybridization resulting from the highly accumulated DNA probe on the micelle surface.
Figure 3.
(A) Comparison of target binding affinity between PFDM and free DNA. FAM fluorescence variation between PFDM and DNA with addition of cDNA at different concentrations. The concentration of PF–DNA–FAM and DNA–FAM was fixed at 1 μM, and the concentration of cDNA ranged from 5 nM to 1 μM. (B) Melting transition of PFDM/cDNA complex and DNA/cDNA duplex. The concentrations of PF–DNA–FAM, DNA–FAM, and cDNA were fixed at 500 nM. (C) The kinetic fluorescence assays of PFDM/cDNA complex and DNA/cDNA duplex with addition of DNAase I (5 units/mL).
While various DNA-based biosensing and drug delivery systems have been developed, clinical translation has lagged behind because of low biostability. It was reported that polyvalent DNA nanoparticles can, to some extent, protect DNA molecules from enzymatic digestion.35 To test whether our micelle nanostructure could improve DNA biostability, the PFDM/cDNA complex and the DNA/cDNA duplex were incubated with endonuclease DNase I (5 units/mL). It is well-known that DNase I can cleave double-stranded DNA (dsDNA) in a nonspecific manner. If DNA duplex is cleaved, then the FAM fluorophore on the DNA probe will be separated from the quencher on the cDNA, leading to fluorescence restoration. Thus, the kinetics of fluorescence restoration can serve as a marker of enzymatic digestion of the DNA probe. As shown in Figure 3C, the fluorescence signal of the PFDM/cDNA complex increased more slowly than that of the DNA/cDNA duplex, indicating that the formation of the micelle structure could, indeed, improve the stability of DNA molecules.
Cell Membrane Anchoring and Cellular Internalization.
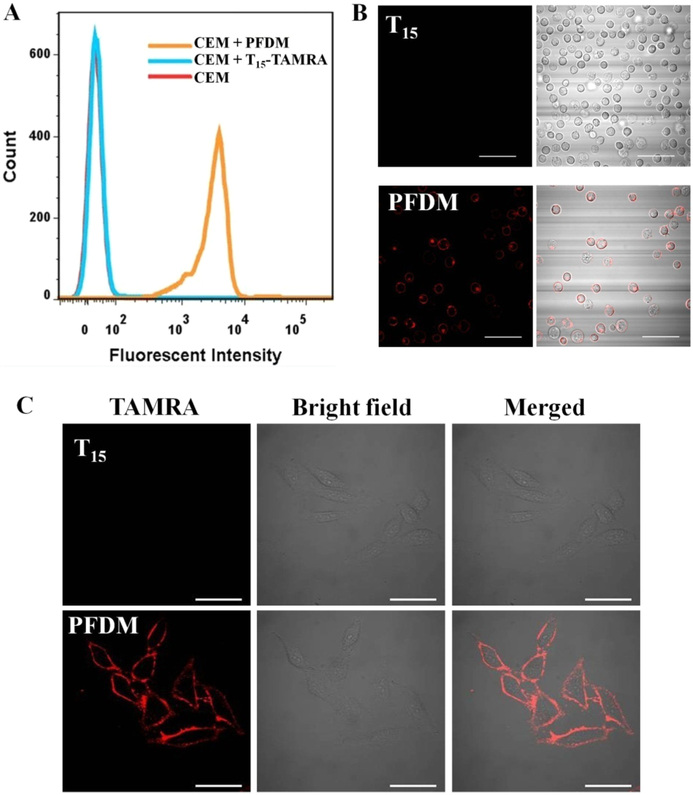
Having confirmed the enhanced biostability of PFDM, we next investigated its interaction with live cells. Previous reports demonstrated that oligonucleotides conjugated with hydrophobic alkyl chains can anchor on the cell membrane by hydrophobic interaction between alkyl chains and hydrophobic layer of the cell membrane.6 Therefore, we investigated the cell membrane anchoring of PFDM. To avoid the interference of fluorescence signal from altered pH values in different cellular compartments, the DNA probes were labeled with a pH-insensitive rhodamine dye (TAMRA). A T lymphoblast-like cell line, CCRF-CEM, was used as our model. CEM cells were incubated with TAMRA-labeled PFDM or TAMRA-labeled DNA for 0.5 h. After removal of unbound probes, the cells were examined by flow cytometry. Figure 4A shows a significant TAMRA fluorescence shift in the cells treated with PFDM, while only minimal fluorescence signal was observed in the cells treated with free DNA probes. The mean TAMRA fluorescence intensity from the cells treated with PFDM was about 136.6 times higher than that from the cells treated with the control probe, indicating that introduction of a PF tail can greatly facilitate the interaction of DNA macromolecules with live cells. The cellular location of PFDM was also visualized with confocal microscopy. After treatment with PFDM, a clear TAMRA signal was localized on the cell membrane (Figure 4B). Apart from the suspension cell line, TAMRA-labeled PFDM can anchor onto the membrane of HeLa cells, an adherent cell line with well-stretched cellular structure (Figure 4C). These results were correlated well with our previous work,4 demonstrating that the DNA micelle structure tends to dissociate and spontaneously insert into the plasma membrane based on hydrophobic interaction. This membrane-anchoring feature makes our PFDM a potentially useful tool for cell membrane engineering.
Figure 4.
Interaction of PFDM with cells. CEM cells were treated with TAMRA-labeled PFDM (200 nM) or control TAMRA-labeled DNA for 30 min and subsequently examined by flow cytometry (A) and confocal microscopy (B). (C) HeLa cells were treated with TAMRA-labeled PFDM (200 nM) or control TAMRA-labeled DNA for 30 min and subsequently examined by confocal microscopy. The scale bar represents 50 μm.
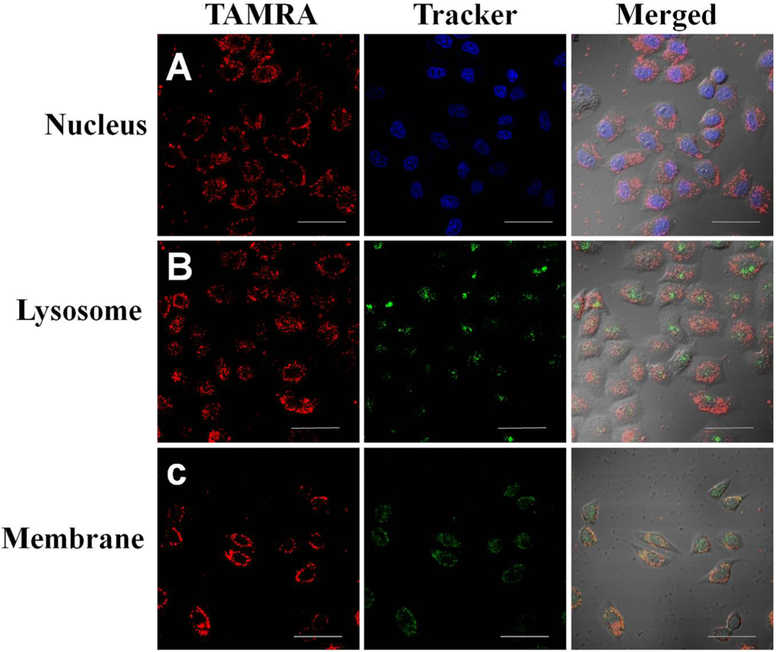
The cytotoxicities of compound 2 and PFDM were tested with MTS assay. Neither of them showed observable impact on the cell viability of 3T3-L1 cells (a mouse embryonic cell line), CEM cells, and HeLa cells even at a high concentration of 10 μM (Figure S10), suggesting an excellent biocompatibility of both compound 2 and PFDM. After that, the cell internalization capability and the cellular distribution of PFDM were studied; HeLa cells was used as our model. HeLa cells were incubated with PFDM at 37 °C for 2 h and subsequently stained with different fluorescence dyes to indicate cellular organelles. Cell membrane was stained with membrane tracker. As shown in Figure 5, the TAMRA signal was not only observed on the cell membrane, but also in the intracellular compartment, indicating the cellular internalization capability of PFDM. Meanwhile, the nucleic location of PFDM was excluded by negligible colocalization between the nucleic indicator signal, Hoechst 33342, and the TAMRA signal. Interestingly, only minimal TAMRA fluorescence signal was observed in the lysosome, as indicated by LysoTracker green, suggesting the ability of PF–DNA molecules to escape from the lysosomal compartment. This result agrees with a previous work which reported that fluorinated chains could facilitate lysosomal escape of nanoparticles.36 It is well-known that lysosomes contain various enzymes which can rapidly digest biomolecules, seriously limiting the effective delivery of DNA/RNA molecules to their target destination, such as cytoplasm and nucleus. Therefore, by taking advantage of lysosomal escape, PFDM has vast potential for applications in bioimaging and gene therapy. The DNA probe could efficiently anchor onto the cell membrane and, more importantly, enter the cells in a lysosome-escape pathway, suggesting cytoplasmic delivery of nucleic acids for bioimaging and gene therapy. Moreover, by combining the versatile capacity of DNA molecules with the unique physicochemical properties of PF, PFDM provide potentially powerful tools for biological and biomedical research.
Figure 5.
CLSM images of HeLa cells incubated with TAMRA-labeled PFDM (500 nM) at 37 °C for 2 h, and then treated with Hoechst 33342 (A), LysoTracker green (B), or membrane tracker (C). The scale bar represents 50 μm.
CONCLUSIONS
In summary, we have successfully synthesized a diperfluorodecyl phosphoramidite and efficiently conjugated it with a DNA segment through a solid-phase synthesis strategy. These PF–DNA conjugates could spontaneously self-assemble into micelle structures. Our experiments demonstrated that this micelle nanostructure could enhance the target binding affinity, thermostability, and enzymatic resistance of the DNA probe. Meanwhile, with introduction of a PF chain, the DNA probe could efficiently anchor onto the cell membrane and, more importantly, enter the cells in a lysosome-escape pathway, suggesting cytoplasmic delivery of nucleic acids for bioimaging and gene therapy. Consequently, by combining the versatile capacity of DNA molecules with the unique physicochemical properties of PF, these PFDM provide potentially powerful tools for biological and biomedical research.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Foundation for Innovative Research Groups of NSFC (21521063) and NSFC Grants (91753109, 21505039, 21325520). This work is also supported by NIH GM127130 and NSF 1645215.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.8b01005.
Materials, synthesis of compound 1, NMR characterization of compounds 1 and 2, high-performance liquid chromatography profile of PF–T15–TAMRA, ESI-MS analysis of PF–T15–TAMRA, dynamic light scattering data, comparison of target binding affinity between PFDM and free DNA, cell viability of compound 2 and PFDM, DNA sequences designed in this work, and the elution program for HPLC purification of lipidconjugated oligonucleotides (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Zhang S Nat. Biotechnol 2003, 21, 1171–1178. [DOI] [PubMed] [Google Scholar]
- (2).Mai Y; Eisenberg A Chem. Soc. Rev 2012, 41, 5969–5985. [DOI] [PubMed] [Google Scholar]
- (3).Blanazs A; Armes SP; Ryan A Macromol. Rapid Commun 2009, 30, 267–277. [DOI] [PubMed] [Google Scholar]
- (4).Cavalli S; Albericio F; Kros A Chem. Soc. Rev 2010, 39, 241–263. [DOI] [PubMed] [Google Scholar]
- (5).Meng HM; Liu H; Kuai H; Peng R; Mo L; Zhang XB Chem. Soc. Rev 2016, 45, 2583–2602. [DOI] [PubMed] [Google Scholar]
- (6).Liu H; Zhu Z; Kang H; Wu Y; Sefan K; Tan W Chem. -Eur. J 2010, 16, 3791–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wu Y; Sefah K; Liu H; Wang R; Tan W Proc. Natl. Acad. Sci. U. S. A 2010, 107, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Chen T; Wu CS; Jimenez E; Zhu Z; Dajac JG; You M; Han D; Zhang X; Tan W Angew. Chem 2013, 125, 2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Jin C; Liu X; Bai H; Wang R; Tan J; Peng X; Tan W ACS Nano 2017, 11, 12087–12093. [DOI] [PubMed] [Google Scholar]
- (10).Huang F; You M; Chen T; Zhu G; Liang H; Tan W Chem. Commun 2014, 50, 3103–3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sletten EM; Swager TM J. Am. Chem. Soc 2014, 136, 13574–13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Riess JG Artif. Cells. Blood. Sub 2005, 33, 47–63. [DOI] [PubMed] [Google Scholar]
- (13).Cheng Y; Cheng H; Jiang C; Qiu X; Wang K; Huan W; Yuan A; Wu J; Hu Y Nat. Commun 2015, 6, 8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Song X; Feng L; Liang C; Yang K; Liu Z Nano Lett. 2016, 16, 6145–6153. [DOI] [PubMed] [Google Scholar]
- (15).Matsunaga TO; Sheeran PS; Luois S; Streeter JE; Mullin LB; Banerjee B; Dayton PA Theranostics 2012, 2, 1185–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Rapoport N Wires. Nanomed. Nanobi 2012, 4, 492–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Huang Y; Vezeridis AM; Wang J; Wang Z; Thompson M; Mattrey RF; Gianneschi NC J. Am. Chem. Soc 2017, 139, 15–18. [DOI] [PubMed] [Google Scholar]
- (18).Ahrens ET; Flores R; Xu H; Morel PA Nat. Biotechnol 2005, 23, 983–987. [DOI] [PubMed] [Google Scholar]
- (19).Janjic JM; Ahrens ET Wires. Nanomed. Nanobi 2009, 1, 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Niu D; Wang X; Li Y; Zheng Y; Li F; Chen H; Gu J; Zhao W; Shi J Adv. Mater 2013, 25, 2686–2692. [DOI] [PubMed] [Google Scholar]
- (21).Wang X; Chen H; Chen Y; Ma M; Zhang K; Li F; Zheng Y; Zeng D; Wang Q; Shi J Adv. Mater 2012, 24, 785–791. [DOI] [PubMed] [Google Scholar]
- (22).Jia X; Cai X; Chen Y; Wang S; Xu H; Zhang K; Ma M; Wu H; Shi J; Chen H ACS Appl. Mater. Interfaces 2015, 7, 4579–4588. [DOI] [PubMed] [Google Scholar]
- (23).Huynh E; Leung BY; Helfield BL; Shakiba M; Gandier J-A; Jin CS; Master ER; Wilson BC; Goertz DE; Zheng G Nat. Nanotechnol 2015, 10, 325–332. [DOI] [PubMed] [Google Scholar]
- (24).Wilson K; Homan K; Emelianov S Nat. Commun 2012, 3, 618. [DOI] [PubMed] [Google Scholar]
- (25).Rapoport N; Nam K-H; Gupta R; Gao Z; Mohan P; Payne A; Todd N; Liu X; Kim T; Shea J; Scaife C; Parker DL; Jeong E-K; Kennedy AM J. Controlled Release 2011, 153, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Shepherd FH; Holzenburg A Anal. Biochem 1995, 224, 21–27. [DOI] [PubMed] [Google Scholar]
- (27).Frotscher E; Danielczak B; Vargas C; Meister A; Durand G; Keller S Angew. Chem., Int. Ed 2015, 54, 5069–5073. [DOI] [PubMed] [Google Scholar]
- (28).Li X; Turanek J; Knötigová P; Kudláčková H; Mašek J; Pennington DB; Rankin SE; Knutson BL; Lehmler H-J New J. Chem 2008, 32, 2169–2179. [Google Scholar]
- (29).Kasuya MCZ; Tojino M; Mizuno M; Hatanaka KJ J. Fluorine Chem 2010, 131, 655–659. [Google Scholar]
- (30).Li Z; Zhang Y; Fullhart P; Mirkin CA Nano Lett. 2004, 4, 1055–1058. [Google Scholar]
- (31).Alemdaroglu FE; Ding K; Berger R; Herrmann A Angew. Chem., Int. Ed 2006, 45, 4206–4210. [DOI] [PubMed] [Google Scholar]
- (32).Storhoff JJ; Elghanian R; Mucic RC; Mirkin CA; Letsinger RL J. Am. Chem. Soc 1998, 120, 1959–1964. [Google Scholar]
- (33).Conlon P; Yang CJ; Wu Y; Chen Y; Martinez K; Kim Y; Stevens N; Marti AA; Jockusch S; Turro NJ; Tan WJ Am. Chem. Soc 2008, 130, 336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Liang X; Mao G; Ng KY S. Colloids Surf., B 2004, 34, 41–51. [DOI] [PubMed] [Google Scholar]
- (35).Liu H; Moynihan KD; Zheng Y; Szeto GL; Li AV; Huang B; Van Egeren DS; Park C; Irvine DJ Nature 2014, 507, 519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Li L; Song L; Liu X; Yang X; Li X; He T; Wang N; Yang S; Yu C; Yin T; Wen Y; He Z; Wei X; Su W; Wu Q; Yao S; Gong C; Wei Y ACS Nano 2017, 11, 95–111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.