Abstract
The specific binding ability of DNA–lipid micelles (DLMs) can be increased by the introduction of an aptamer. However, supramolecular micellar structures based on self-assemblies of amphiphilic DLMs are expected to demonstrate low stability when interacting with cell membranes under certain conditions, which could lead to a reduction in selectivity for targeting cancer cells. We herein report a straightforward cross-linking strategy that relies on a methacrylamide branch to link aptamer and lipid segments. By an efficient photoinduced polymerization process, covalently linked aptamer–lipid units help stabilize the micelle structure and enhance aptamer probe stability, further improving the targeting ability of the resulting nanoassembly. Besides the development of a facile cross-linking method, this study clarifies the relationship between aptamer–lipid concentration and the corresponding binding ability.
Keywords: aptamer–lipid micelles, biostability, cross-linking, elf-assembly, specific cell recognition
Biomacromolecules combined with synthetic materials, such as peptide/protein–polymer conjugates[1] and DNA–block-copolymer (DBC) micelles,[2] have attracted much attention in bioanalysis and biomedicine. With versatile modifications, DBC micelles have been applied for drug delivery[3] and nanoreactors.[4] While controllable structures and precise nucleotide recognition properties make DBC micelles superior to many other biopolymers,[5] complicated and inefficient conjugations remain a challenge. For efficient synthesis and assembly, DNA–lipid micelles (DLMs), consisting of a single-stranded hydrophilic DNA head and a hydrophobic lipid tail, have been reported.[6] By solid-phase DNA synthesis, diacyllipid can be coupled onto the 5’-end of oligonucleotides.[7] Owing to the similarity between the diacyllipid in DLMs and the lipid bilayers in cell membranes, DLMs have shown low critical micelle concentration (CMC),[8] excellent biocompatibility,[7] and compelling membrane permeability.[9]
Previous studies have demonstrated the feasibility of using DLMs for intracellular imaging[7,10] and the inhibition of specific cellular biological functions[11] through interaction with target biomolecules. The introduction of aptamers into DLMs leads to even more rapid target-cell recognition.[12] At the same time, however, loss of DLM integrity and arbitrary insertion of disassembled lipid units could interfere with the recognition ability of the aptamer when interacting with cells.[6a,9] Therefore, a more stable micelle structure is required for target-cell recognition in practical applications. Various approaches have been investigated to stabilize copolymer micelles by employing covalent[13] or reducible linkages.[14] Moreover, both the Mirkin group[15] and the Rouge group[16] recently reported cross-linking methods for stabilizing DLMs, but they required long preparation times and additional cross-linkers. Thus, the challenge of constructing simple but specific and stable DLMs still remains.
Herein, we developed a cross-linked DNA–methacrylamide–lipid micelle (X-DLM) system (Scheme 1). This facile design incorporates a methacrylamide functionality between the hydrophilic and hydrophobic portions of DNA–lipid amphiphiles that can be cross-linked through free-radical polymerization under UV light after self-assembly in aqueous solution. In contrast to traditional non-cross-linked DLMs, well-defined X-DLMs offer better stability in a cellular environment, further providing excellent specificity for cell recognition when incorporated with targeting aptamers.
Scheme 1.
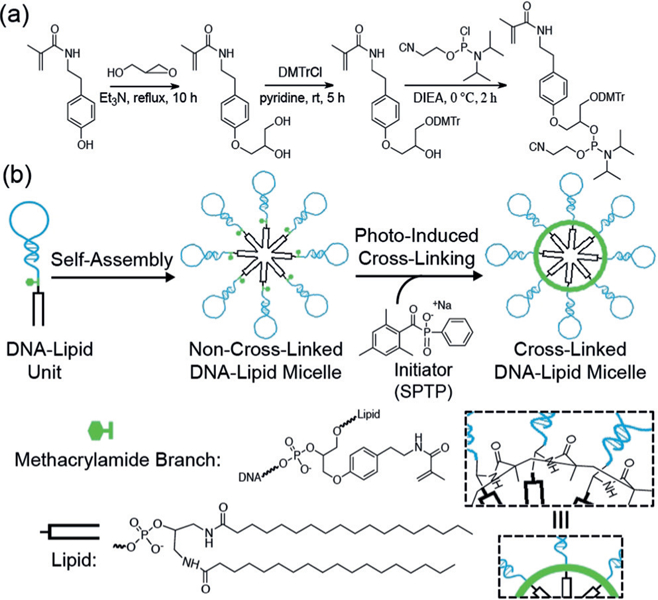
a) Synthesis of the DMT–methacrylamide phosphoramidite. b) Photoinduced cross-linking of self-assembled DNA–methacrylamide–lipid micelles. DIEA = diisopropylethylamine, DMTrCl = 4,4’-dimethoxytrityl chloride.
The DNA–methacrylamide–lipid unit was synthesized on the basis of a two-step phosphoramidite coupling process (see the Supporting Information for details). First, a DMT– methacrylamide phosphoramidite (DMT= 4,4’-dimethoxytrityl) was synthesized as a basic building block to be attached to the 5’-end of DNA and then connected with a lipid (see the Supporting Information).[17] Then, after self-assembly in aqueous solution, radical polymerization facilitated the formation of C‒C covalent bonds between adjacent methacrylamide units, which served to link individual amphiphiles.[18] Water-soluble and biocompatible sodium phenyl-2,4,6-trimethylbenzoylphosphinate (SPTP) was chosen as the photoinitiator because of its rapid decomposition under UV irradiation (365 nm).[19] The rapid initiation rate provided complete cross-linking within 15 min at room temperature, thereby limiting overexposure of DNA to UV light. Therefore, as compared with previous micelle-stabilizing methods, the significance of our approach stems from the introduction of a DMT–methacrylamide phosphoramidite linking the DNA head and lipid tail to allow for efficient and simplified cross-linking with no need for cross-linkers.
In this study, sgc8 aptamer, which specifically recognizes the highly expressed membrane protein PTK7 on CCRF-CEM cells (T-cell acute lymphoblastic leukemia cell line), but not Ramos cells (B-cell human Burkitt lymphoma cell line),[20] was selected as the DNA segment to equip micelles with target-binding ability. To demonstrate the precise cross-linking of this approach, we used sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) to examine the mobility of carboxytetramethylrhodamine (TAMRA)-labeled micelles and the different building blocks (Figure 1 a). As compared to normal sgc8–methacrylamide–lipid micelles (Sgc8-LM) and all the constituent domains, cross-linked Sgc8-LM (X-Sgc8-LM) had the least mobility, most likely as a result of increased molecular weight of the cross-linked structures and better structural stability of the lipid core against disassembly by SDS. Because SDS disrupts non-covalent bonds, the hydrophobic interactions in Sgc8-LM were readily disintegrated in the presence of SDS, whereas the covalent bonds prevented X-Sgc8-LM from losing micellar integrity, even at a high concentration of SDS (0.8 % w/v; see Figure S8 in the Supporting Information).
Figure 1.
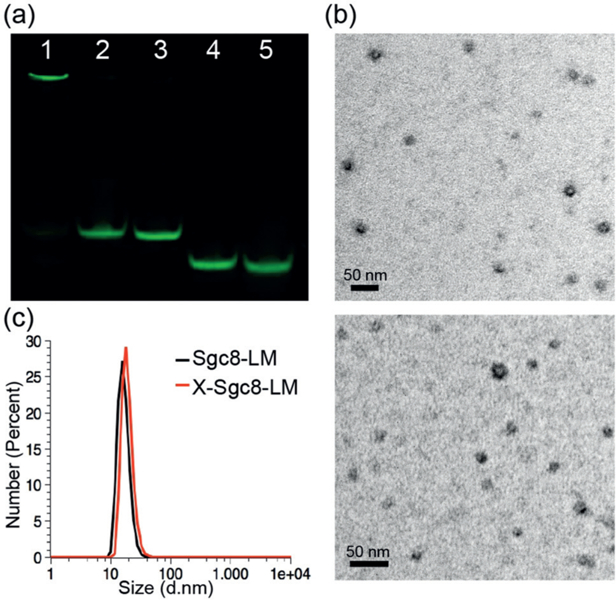
a) SDS-PAGE (12 %) analysis of different generations of TAMRA-labeled sgc8 micelles. Lanes: 1, X-Sgc8-LM; 2, Sgc8-LM; 3, Sgc8-Lipid; 4, Sgc8-Methacrylamide; 5, Sgc8. The gel was scanned in the TAMRA channel. b) TEM images showing the average sizes of Sgc8-LM (top) and X-Sgc8-LM (bottom). c) DLS measurements of micelles before (black) and after cross-linking (red).
To obtain purified X-Sgc8-LM with the best cross-linking efficiency (ca. 60 %), we selected a 1 : 0.108 molar ratio of amphiphile unit to SPTP after comparing the intensities of product bands in the gel, followed by dialysis to remove unincorporated free units (see Figure S9). The possibility that the cross-linking resulted from linkages among DNA bases or lipid cores was also ruled out (see Figure S10), since none of the controls without methacrylamide formed cross-linked micelles under polymerization conditions. Therefore, cross-linking could only be carried out through methacrylamide polymerization while both DNA and lipid domain functionalities were maintained.
The homogeneous spherical structures of Sgc8-LM and X-Sgc8-LM were confirmed by negatively stained transmission electron microscopy (TEM) and dynamic light scattering (DLS; Figure 1). The change in particle size after cross-linking (from (17±2) nm, PDI=0.38 to (18±1) nm, PDI=0.27) was negligible, thus indicating that this cross-linking approach maintained a uniform architecture without affecting micelle size (Figure 1 b).
Besides undesirable micelle-structure disintegration, susceptibility of DNA to nuclease degradation also prevents current clinical translation of DNA micelle-based therapeutics.[21] Therefore, it is important to evaluate the stability of cross-linked micelles before their application in real cellular environments. Besides enhanced structural stability (Figure 1 a), X-Sgc8-LM also exhibited significantly increased resistance to nucleolytic degradation. Serum proteins can interact with lipids to prevent micelle formation and further cleave oligonucleotides.[22] In this study, we compared the biostability of X-Sgc8-LM, Sgc8-LM, and Sgc8 using serum and deoxyribonuclease I (DNase I). Since the Sgc8 aptamer was labeled with TAMRA dye in all groups, the dissociated aptamer–lipids and degraded aptamer pieces could be detected by the faster-moving bands through fluorescence (TAMRA channel scanning) on an agarose gel. Incubation of Sgc8 and Sgc8-LM in 10 % serum medium for 4 h resulted in (59±0.8) and (51±0.7)% cleavage of aptamer domains, respectively; whereas only 22±3% of the same sequences in X-Sgc8-LM were cleaved (Figure 2 a). A much clearer comparison was observed with DNase I (Figure 2 b). Sgc8 and Sgc8-LM were digested rapidly within 30 min, and finally only (10±3) and (14±7) %, respectively, of the aptamer remained intact in the presence of 100 mU DNase I. In contrast, cross-linked micelles maintained their probe stability, as evidenced by the presence of (58±0.9)% of the fluorescent bands remaining in the top well. The remarkably improved biostability of cross-linked micelles most probably resulted from the steric hindrance of the aptamer corona, as well as the efficient cross-linking that facilitated the rigidification of nanostructures. Consequently, this cross-linking approach could generate more robust and stable micellar assemblies, thus providing better opportunities for their application under physiological conditions.
Figure 2.
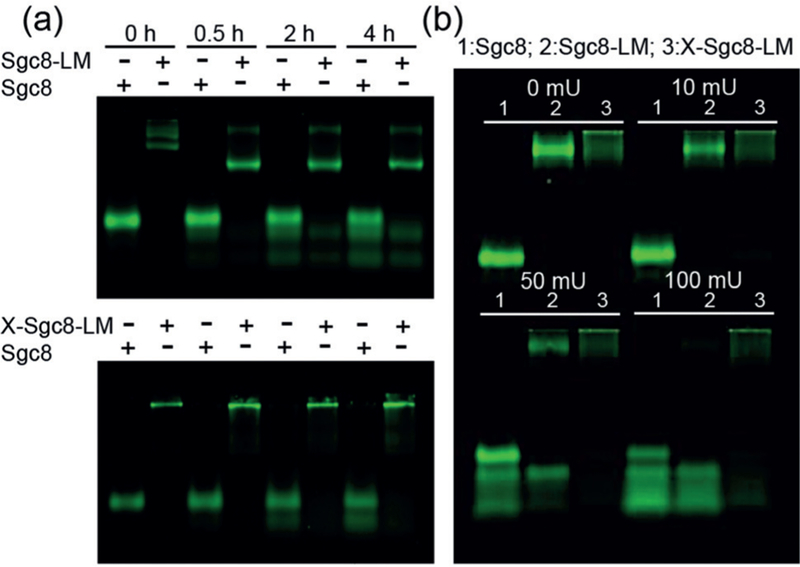
Agarose gel (4 %) showing a) serum stability and b) DNase I resistance of TAMRA-labeled Sgc8, Sgc8-LM, and X-Sgc8-LM. a) Samples were incubated with RPMI cell culture medium containing 10 % FBS at 37°C for 0, 0.5, 2, and 4 h. The + symbols indicate which species was mounted in the respective lane. b) Samples were incubated with DNase I of different concentrations (0, 10, 50, and 100 mU) at room temperature for 30 min.
Having demonstrated the superior stability of cross-linked micelles, we next examined their targeting abilities. Until now, lipidic domains have been considered as nonspecific “glue” that could partition into any hydrophobic cellular membranes.[8] Thus, after a certain incubation time (e.g., over 30 min), we hypothesized that aptamer–lipid micelles would not be able to discriminate between target and control cells, as the appended lipid chains could dynamically disassemble and insert into random cell membranes. However, as shown in flow cytometry histograms (Figure 3 a, top), TAMRA-labeled Sgc8-Lipid exhibited good selectivity for target CEM cells at very low concentration (25 nm) after incubation for 30 min at 4°C, as compared with library DNA–lipid (Lib-Lipid) micelles. Interestingly, when incubated with negative Ramos cells, both Sgc8-and Lib-Lipid showed gradually increasing binding shifts as the amphiphile concentration increased (Figure 3 a, bottom), thus indicating dose-dependent binding ability to Ramos. The competition between specific targeting ability of the aptamer and low off-rates of the lipid on cell membranes could account for these unanticipated results.[12b] When the aptamer–lipid concentration is low, the selective binding property of Sgc8 is the dominant factor that enables rapid cell-type discrimination. However, as the concentration increases, more aptamer–lipid units anchor onto the cell-membrane surface, and could not readily diffuse away because of the preferred thermal stability, thereby compromising the recognition specificity of the aptamer.
Figure 3.
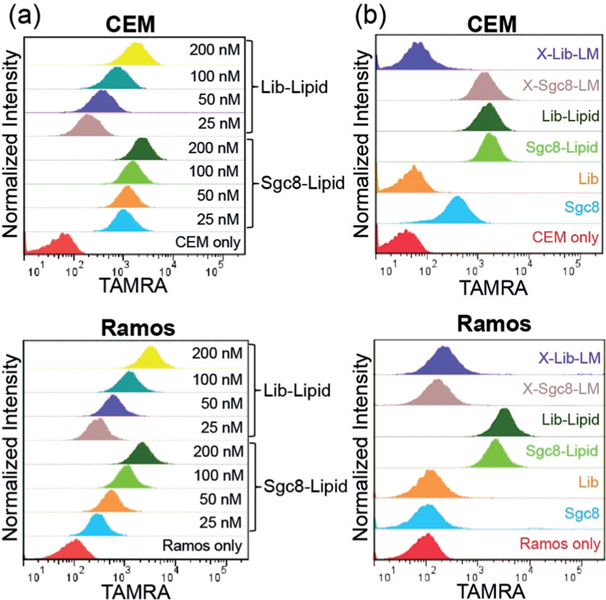
Flow cytometry histograms showing a) different binding abilities of Sgc8-Lipid and Lib-Lipid to CEM (top) and Ramos cells (bottom) at different concentrations, and b) the selective recognition of target CEM cells by X-Sgc8-LM with a high DNA concentration (200 nm; top) as opposed to nontarget Ramos cells (bottom). Lib: library DNA.
Although Sgc8-Lipid micelles showed good target binding at low concentrations, we could not overlook the nonspecific shifts of Lib-Lipid micelles towards CEM cells and the shifts of the two micelles towards Ramos cells under the same conditions. Importantly, in terms of drug delivery and other bioapplications for targeting cancer cells, loss of micelle specificity at high concentrations would be an issue. However, the methacrylamide cross-linking approach addressed these issues (Figure 3b; see also Figure S11). X-Sgc8-LM at both high and low aptamer–lipid concentrations (200 and 50 nm) displayed clear shifts towards CEM cells, but not Ramos cells, whereas the cross-linked library DNA–methacrylamide–lipid micelles (X-Lib-LM) exhibited negligible shifts towards both cell types, thus indicating the excellent recognition capability of X-Sgc8-LM towards target cells. Moreover, the excellent specificity of X-Sgc8-LM was still maintained in the presence of serum proteins (see Figure S12). In contrast to the barely satisfactory performance of unmodified Sgc8 and the arbitrary binding of non-cross-linked micelles, the significant improvement of X-Sgc8-LM in selectivity demonstrated that strengthened structure and probe stability could help enhance target-cell-recognition ability of aptamer-functionalized cross-linked micelles.
The target specificity of X-Sgc8-LM was also evidenced by confocal microscopy (Figure 4). At a high concentration (200 nm), disassembled Sgc8-LM freely inserted into cell membranes of both CEM and Ramos without specificity after incubation at 37°C for 2 h. Conversely, the red fluorescence of X-Sgc8-LM was observed only after incubation with CEM under the same conditions. Cross-linking is, therefore, able to overcome the disassembly of the lipid core to give better target-recognition ability. Moreover, as compared with the strong binding of Lib-LM towards both positive and negative cells, X-Lib-LM recognized neither CEM nor Ramos, thus suggesting the loss of nonselective binding ability after cross-linking (see Figure S13). Thus, considering the selective targeting ability, lipid-core loading capacity, and excellent biocompatibility (see Figure S14) of the cross-linked aptamer-lipid micelles reported herein, we envision that future studies will focus on applications in targeted bioanalysis and cancer theranostics.
Figure 4.
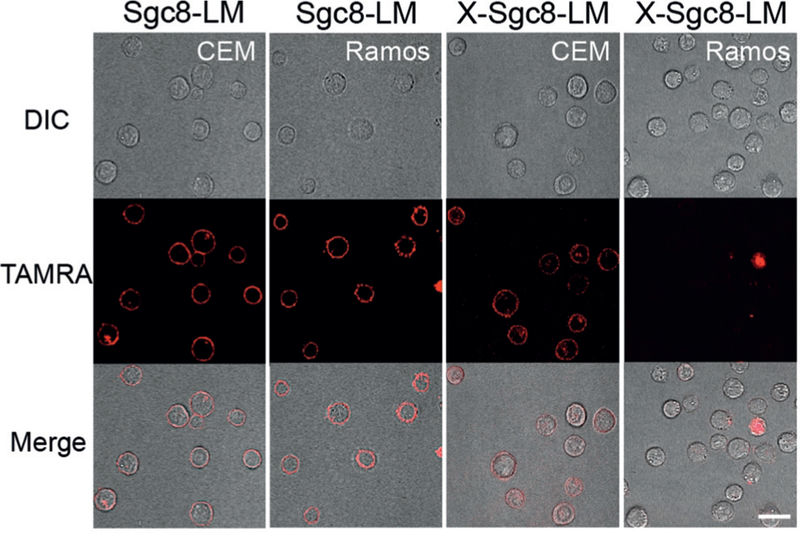
Confocal microscopy images of CEM and Ramos cells treated with Sgc8-LM and X-Sgc8-LM. X-Sgc8-LM (200 nm) targeted only CEM cells, whereas non-cross-linked Sgc8-LM recognized both CEM and Ramos cells after incubation for 2 h. Scale bar: 20 μm.
In summary, we have developed a facile and effective cross-linking strategy to equip aptamer–lipid micelles with enhanced stability and improved specificity to target cancer cells. Micelles exhibited specificity at a low concentration (25 nm), but lost targeting ability at high concentration (200 nm), thus indicating the necessity of stabilizing micellar structures. The novel incorporation of biocompatible methacrylamide units[23] provides a straightforward approach with no extra cross-linker needed. The simplified free-radical cross-linking of aptamer–lipid micelles promotes cancer-cell recognition, while significantly mitigating nonspecific binding. Continued research on building stimuli-responsive micelle structures will further lead to feasible drug-release systems. Thus, given the potential for versatile modification of the lipid core and aptamer, this cross-linked nanoassembly is promising for applications in targeted cellular imaging, gene therapy, and drug delivery.
Supplementary Material
Acknowledgements
We thank Dr. K. R. Williams for manuscript review. This work is supported by NIH GM R35 127130 and NSF 1645215, and by NSFC grants (NSFC 21521063).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Xiaowei Li, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiology and Functional Genomics Health Cancer Center, UF Genetics Institute and McKnight Brain Institute, University of Florida Gainesville, FL 32611 (USA).
C. Adrian Figg, George and Josephine Butler Polymer Research Laboratory Center for Macromolecular Science and Engineering Department of Chemistry, University of Florida Gainesville, FL 32611-7200 (USA).
Ruowen Wang, Molecular Science and Biomedicine Laboratory (MBL), State Key Laboratory of Chemo/Bio-Sensing and Chemometrics, College of Life Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha, Hunan, 410082 (China), Institute of Molecular Medicine, Renji Hospital, Shanghai Jiao Tong University School of Medicine, College of Chemistry and Chemical Engineering, Shanghai, 200240 (China).
Ying Jiang, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA), Molecular Science and Biomedicine Laboratory (MBL), State Key LaboratoryofChemo/Bio-SensingandChemometrics,CollegeofLife Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha,Hunan, 410082 (China).
Yifan Lyu, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA), Molecular Science and Biomedicine Laboratory (MBL), State Key LaboratoryofChemo/Bio-SensingandChemometrics,CollegeofLife Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha,Hunan, 410082 (China).
Hao Sun, George and Josephine Butler PolymerResearch Laboratory Center for Macromolecular Science and Engineering Department of Chemistry,University of Florida Gainesville, FL 32611-7200 (USA).
Yuan Liu, Molecular Science and Biomedicine Laboratory (MBL), State Key LaboratoryofChemo/Bio-SensingandChemometrics,CollegeofLife Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha,Hunan, 410082 (China).
Yanyue Wang, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA).
I-Ting Teng, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA).
Weijia Hou, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA).
Ren Cai, Molecular Science and Biomedicine Laboratory (MBL), State Key LaboratoryofChemo/Bio-SensingandChemometrics,CollegeofLife Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha,Hunan, 410082 (China).
Cheng Cui, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA), Molecular Science and Biomedicine Laboratory (MBL), State Key LaboratoryofChemo/Bio-SensingandChemometrics,CollegeofLife Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha,Hunan, 410082 (China).
Long Li, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA).
Xiaoshu Pan, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA).
Brent S. Sumerlin, George and Josephine Butler PolymerResearch Laboratory Center for Macromolecular Science and Engineering Department of Chemistry,University of Florida Gainesville, FL 32611-7200 (USA)
Weihong Tan, Center for Research at the Bio/Nano Interface, Department of Chemistry and Department of Physiologyand Functional Genomics Health Cancer Center,UFGenetics Institute and McKnightBrain Institute, University of Florida Gainesville, FL 32611 (USA), Molecular Science and Biomedicine Laboratory (MBL), State Key LaboratoryofChemo/Bio-SensingandChemometrics,CollegeofLife Sciences and College of Chemistry and Chemical Engineering Aptamer Engineering, Center of Hunan Province, Hunan University Changsha,Hunan, 410082 (China), Institute of Molecular Medicine,Renji Hospital,Shanghai Jiao Tong UniversitySchool of Medicine, College of Chemistry and Chemical Engineering, Shanghai, 200240 (China).
References
- [1] (a).Gauthier MA, Klok HA, Chem. Commun 2008, 2591–2611; [DOI] [PubMed] [Google Scholar]; (b) Dutta K, Hu D, Zhao B, Ribbe AE, Zhuang J, Thayumanavan S, J. Am. Chem. Soc 2017, 139, 5676–5679; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Figg CA, Bartley AN, Kubo T, Tucker BS, Castellano RK, Sumerlin BS, Polym. Chem 2017, 8, 2457 – 2461. [Google Scholar]
- [2] (a).Schnitzler T, Herrmann A, Acc. Chem. Res 2012, 45, 1419–1430; [DOI] [PubMed] [Google Scholar]
- (b).Yang CJ, Pinto M, Schanze K, Tan W, Angew. Chem. Int. Ed 2005, 44, 2572–2576; Angew. Chem. 2005, 117, 2628–2632; [DOI] [PubMed] [Google Scholar]; (c) Jeong JH, Park TG, Bioconjugate Chem 2001, 12, 917–923. [DOI] [PubMed] [Google Scholar]
- [3] (a).Oh SS, Lee BF, Leibfarth FA, Eisenstein M, Robb MJ, Lynd NA, Hawker CJ, Soh HT, J. Am. Chem. Soc 2014, 136, 15010–15015; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Tan XY, Lu XG, Jia F, Liu XF, Sun YH, Logan JK, Zhang K, J. Am. Chem. Soc 2016, 138, 10834–10837; [DOI] [PubMed] [Google Scholar]; (c) Jeong JH, Kim SW, Park TG, Bioconjugate Chem 2003, 14, 473–479. [DOI] [PubMed] [Google Scholar]
- [4] (a).Trinh T, Chidchob P, Bazzi HS, Sleiman HF, Chem. Commun 2016, 52, 10914–10917; [DOI] [PubMed] [Google Scholar]; (b) Alemdaroglu FE, Ding K, Berger R, Herrmann A, Angew. Chem. Int. Ed 2006, 45, 4206–4210; Angew. Chem. 2006, 118, 4313 – 4317. [DOI] [PubMed] [Google Scholar]
- [5] (a).Kim CJ, Hu X, Park SJ, J. Am. Chem. Soc 2016, 138, 14941–14947; [DOI] [PubMed] [Google Scholar]; (b) Li Z, Zhang Y, Fullhart P, Mirkin CA, Nano Lett 2004, 4, 1055–1058. [Google Scholar]
- [6] (a).Liu HP, Zhu Z, Kang HZ, Wu YR, Sefan K, Tan WH, Chem. Eur. J 2010, 16, 3791–3797; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pokholenko O, Gissot A, Vialet B, Bathany K, Thiery A, Barthelemy P, J. Mater. Chem. B 2013, 1, 5329–5334; [DOI] [PubMed] [Google Scholar]; (c) Edwardson TGW, Carneiro KMM, McLaughlin CK, Serpell CJ, Sleiman HF, Nat. Chem 2013, 5, 868–875; [DOI] [PubMed] [Google Scholar]; (d) Wang YY, Wu CC, Chen T, Sun H, Cansiz S, Zhang LQ, Cui C, Hou WJ, Wu Y, Wan S, Cai R, Liu Y, Sumerlin BS, Zhang XB, Tan WH, Chem. Sci 2016, 7, 6041–6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You MX, Han D, Zhang XB, Tan WH, Angew. Chem. Int. Ed 2013, 52, 2012–2016; Angew. Chem. 2013, 125, 2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Anaya M, Kwak M, Musser AJ, Mgllen K, Herrmann A, Chem. Eur. J 2010, 16, 12852–12859. [DOI] [PubMed] [Google Scholar]
- [9].Patwa A, Gissot A, Bestel I, Barthelemy P, Chem. Soc. Rev 2011, 40, 5844–5854. [DOI] [PubMed] [Google Scholar]
- [10].Wu CC, Chen T, Han D, You MX, Peng L, Cansiz S, Zhu GZ, Li CM, Xiong XL, Jimenez E, Yang CJ, Tan WH, ACS Nano 2013, 7, 5724–5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] (a).Rush AM, Nelles DA, Blum AP, Barnhill SA, Tatro ET, Yeo GW, Gianneschi NC, J. Am. Chem. Soc 2014, 136, 7615–7618; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Godeau G, Staedel C, Barthelemy P, J. Med. Chem 2008, 51, 4374 – 4376. [DOI] [PubMed] [Google Scholar]
- [12] (a).Huang FJ, You MX, Chen T, Zhu GZ, Liang HJ, Tan WH, Chem. Commun 2014, 50, 3103–3105; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wu YR, Sefah K, Liu HP, Wang RW, Tan WH, Proc. Natl. Acad. Sci. USA 2010, 107, 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13] (a).Sun XK, Rossin R, Turner JL, Becker ML, Joralemon MJ, Welch MJ, Wooley KL, Biomacromolecules 2005, 6, 2541–2554; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McHale R, Patterson JP, Zetterlund PB, O’Reilly RK, Nat. Chem 2012, 4, 491–497; [DOI] [PubMed] [Google Scholar]; (c) Joralemon MJ, O’Reilly RK, Hawker CJ, Wooley KL, J. Am. Chem. Soc 2005, 127, 16892–16899; [DOI] [PubMed] [Google Scholar]; (d) Kwak M, Musser AJ, Lee J, Herrmann A, Chem. Commun 2010, 46, 4935–4937. [DOI] [PubMed] [Google Scholar]
- [14] (a).Li YP, Xiao K, Luo JT, Xiao WW, Lee JS, Gonik AM, Kato J, Dong TA, Lam KS, Biomaterials 2011, 32, 6633–6645; [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Lee S-Y, Tyler JY, Kim S, Park K, Cheng J-X, Mol. Pharm 2013, 10, 3497–3506; [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Bapat AP, Ray JG, Savin DA, Sumerlin BS, Macromolecules 2013, 46, 2188–2198; [Google Scholar]; (b) Bapat AP, Ray JG, Savin DA, Hoff EA, Patton DL, Sumerlin BS, Polym. Chem 2012, 3, 3112–3120. [Google Scholar]
- [15].Banga RJ, Meckes B, Narayan SP, Sprangers AJ, Nguyen ST, Mirkin CA, J. Am. Chem. Soc 2017, 139, 4278–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Awino JK, Gudipati S, Hartmann AK, Santiana JJ, Cairns-Gibson DF, Gomez N, Rouge JL, J. Am. Chem. Soc 2017, 139, 6278–6281. [DOI] [PubMed] [Google Scholar]
- [17].Yang L, Meng L, Zhang X, Chen Y, Zhu G, Liu H, Xiong X, Sefah K, Tan W, J. Am. Chem. Soc 2011, 133, 13380–13386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Giz A, Catalgil-Giz H, Alb A, Brousseau JL, Reed WF, Macromolecules 2001, 34, 1180–1191. [Google Scholar]
- [19] (a).Benedikt S, Wang J, Markovic M, Moszner N, Dietliker K, Ovsianikov A, Grützmacher H, Liska R, J. Polym. Sci. Part A 2016, 54, 473–479; [Google Scholar]; (b) Tan JB, Sun H, Yu MG, Sumerlin BS, Zhang L, ACS Macro Lett 2015, 4, 1249–1253. [DOI] [PubMed] [Google Scholar]
- [20].Shangguan D, Li Y, Tang ZW, Cao ZHC, Chen HW, Mallikaratchy P, Sefah K, Yang CYJ, Tan WH, Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shi J, Kantoff PW, Wooster R, Farokhzad OC, Nat. Rev. Cancer 2017, 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22] (a).Kastantin M, Missirlis D, Black M, Ananthanarayanan B, Peters D, Tirrell M, J. Phys. Chem. B 2010, 114, 12632–12640; [DOI] [PubMed] [Google Scholar]; (b) Wilner SE, Sparks SE, Cowburn D, Girvin ME, Levy M, J. Am. Chem. Soc 2015, 137, 2171–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Duncan R, Adv. Drug Delivery Rev 2009, 61, 1131–1148. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


