Abstract
A (GGGGCC) hexanucleotide repeat (HR) expansion in the C9ORF72 gene has been considered the major cause behind both frontotemporal dementia and amyotrophic lateral sclerosis, while a (GGGCCT) is associated with spinocerebellar ataxia 36. Recent experiments involving NMR, CD, optical melting and 1D 1H NMR spectroscopy, suggest that the r(GGGGCC) HR can adopt a hairpin structure with G-G mismatches in equilibrium with a Gquadruplex structure. G-Quadruplexes have also been identified for d(GGGGCC). As these experiments lack molecular resolution, we have used molecular dynamics microsecond simulations to obtain a structural characterization of the G-quadruplexes associated with both HRs. All DNA Gquadruplexes, parallel or antiparallel, with or without loops are stable, while only parallel and one antiparallel (stabilized by diagonal loops) RNA G-quadruplexes are stable. It is known that antiparallel G-quadruplexes require alternating guanines to be in a syn conformation that is hindered by the C3′-endo pucker preferred by RNA. Initial RNA antiparallel quadruplexes built with C2′-endo sugars evolve such that the transition (C2′-endo)-to-(C3′-endo) triggers unwinding and buckling of the flat G-tetrads, resulting in the unfolding of the RNA antiparallel quadruplex. Finally, a parallel G-quadruplex stabilizes an adjacent C-tetrad in both DNA and RNA (thus effectively becoming a mixed quadruplex of 5 layers). The C-tetrad is stabilized by the stacking interactions with the preceding G-tetrad, by cyclical hydrogen bonds C(N4)-(O2), and by an ion between the G-tetrad and the C-tetrad. In addition, antiparallel DNA G-quadruplexes also stabilize flat C-layers at the ends of the quadruplexes.
Keywords: Quadruplex, hexanucleotide repeats, DNA, RNA, G-quartets
Graphical Abstract
INTRODUCTION
Simple sequence repeats (SSRs) comprise all sequences with core motifs of 1–6 (and even 12) nucleotides that are repeated up to 30 times in the human genome, both in genetic and intergenic regions.1 SSRs exhibit “dynamic mutations” that do not follow Mendelian inheritance (which asserts that mutations in a single gene are stably transmitted between generations). Intergenerational expansion of SSRs is behind approximately 30 inherited neurological disorders known as “anticipation diseases”, where the age of the onset of the disease decreases and its severity increases with each successive generation.2–10 After a certain threshold in the length of the repeated sequence, the probability of further expansion and the severity of the disease increase with the length of the repeat. The expansion is believed to be primarily caused by some sort of slippage during DNA replication, repair, recombination or transcription.5,6,8–16
Recently, a (GGGGCC) hexanucleotide repeat (HR) expansion in the first intron of the C9ORF72 gene has been shown to be the major cause behind both frontotemporal dementia (FTD, here specifically indicated as C9FTD) and amyotrophic lateral sclerosis (ALS).17,18 While the unaffected population carries fewer than 20 repeats (generally no more than a couple), large expansions greater than 70 repeats and usually encompassing 250–1600 repeats have been found in C9FTD and ALS patients. C9FTD and ALS are two neurodegenerative diseases that can occur simultaneously, and are believed to be part of the same disease spectrum.19 ALS is characterized by a degeneration of motor neuronal cells in the brain and spinal chord leading to progressive muscle weakness and paralysis. C9FTD, on the other hand, is the most common cause behind early onset dementia due to a degeneration of the frontal and anterior temporal lobes. Another HR (GGGCCT), found in intron 1 of the NOP56 gene located on chromosome 20, has been associated with spinocerebellar ataxia 36 (SCA36).20 Normal alleles of the NOP56 gene are characterized by 5–14 HRs, while abnormal genes have repeats in the 800–2000 range. Autosomal-dominant spinocerebellar ataxias are a heterogeneous group of neurodegenerative disorders characterized by a progressive loss of balance, gait and limb ataxia. The specific form SCA36 is characterized by sensorineural hearing loss and late-onset motor neuron involvement with symptoms similar to ALS. Interestingly, in contrast to ALS and other SSR diseases, the severity of SCA36 does not seem to vary with the length of the HRs.20
SSR disorders can cause toxicity through different but nonexclusive mechanisms. First, the expansions originate in the DNA itself, and these expansions can alter the local chromatin structure, affecting RNA transcription and protein translation in the gene. Second, transcribed RNA can cause gain and/or loss of function. Third, translated repeats can also cause toxicity in the corresponding protein and its interaction partners, even when they reside in noncoding regions. For example, in ALS/ C9FTD patients, the transcribed introns contribute to neuropathology both through loss of function, as mRNA levels of C9ORF72 are decreased in C9FTD/ALS patients;17,18 and through gain of function, as RNA transcripts containing the (GGGGCC) HRs are accumulated in nuclear foci in the frontal cortex and spinal cord, leading to the sequestration of RNA-binding proteins.17 Antisense transcripts of the expansion, i.e., (CCCCGG) expanded repeats resulting from the bidirectional transcription of the DNA HR expansions, also form nuclear RNA foci.21,22 Moreover, the HR expansions in the noncoding region of the C9ORF72 gene can trigger protein translation in the absence of the start ATG codon, giving rise to the unconventional repeat-associated non-ATG (RAN) translation.22–25 RAN translation of the (GGGGCC) and (CCCCGG) expansions lead to Gly-Ala, Gly-Pro, Gly-Arg, Pro-Ala, and Pro-Arg poly dipeptide expansions, that have been detected in C9FTD/ALS patients.
An important breakthrough in the understanding of SSR diseases has been the recognition that stable atypical DNA secondary structure in the expanded repeats is “a common and causative factor for expansion in human disease”.7 G-rich sequences such as those in this study tend to form stable G-quadruplex structures, whose basic unit is a G-quartet (also named G-tetrad) that consists of a planar array of four Hoogsteen-bonded guanines residues. Consecutive quartets then stack to form the quadruplex stem, stabilized by monovalent cations lying in the central channel of the stem, in direct contact with the guanine carbonyl groups. G-Quadruplexes are well-known in the context of human telomeric DNA based on d(TTAGGG) repeats,26–31 where quadruplex formation helps maintain the length of the telomere. G-Quadruplexes are also associated with other SSRs, such as the CGG SSRs ssociated with the 5′-untranslated region of the Fragile X syndrome (FRAXA) gene FMR1,32,33 and the regulatory region of the insulin gene.33,34 Based on NMR and circular dichroism (CD) spectroscopy, DNA d(GGGGCC) oligomers with varying number of repeats have been found to adopt inter- and intramolecular G-quadruplex structures, in either parallel or antiparallel orientation.35 Using gel mobility shift assays and NMR and circular dichroism (CD) spectroscopy, RNA r(GGGGCC) expansions have been shown to adopt a stable, parallel G-quadruplex.36,37 However, recent experiments involving CD, optical melting and 1D 1H NMR spectroscopy, combined with chemical and enzymatic probing of a r(GGGGCC) repeat expansion point to a general scenario where the repeat expansion adopts a hairpin structure with G-G mismatches in equilibrium with a quadruplex structure.38,39 The equilibrium is temperature dependent, with T = 37° favoring hairpins and higher annealing temperatures favoring quadruplexes. The equilibrium is also controlled by the type of ions involved, and their ionic strength, with the larger K+ ions favoring G-quadruplexes and the smaller Na+ ions favoring hairpins.38 The decreased stability associated with decreasing ion size, i.e., K+ > Na+ > Li+, appears to be a characteristic of other G-quadruplexes as well. G-Quadruplexes have been extensively studied both experimentally and by using molecular dynamics (MD) simulations, and the reader is referred to refs 40–42 for reviews.
Given the clear connection between these human neurodegenerative diseases and the associated secondary structures, the focus of this paper is on understanding the structural and dynamical characteristics of the G-rich quadruplexes associated specifically with the (GGGGCC) and (GGGCCT) HRs. In order to do this, we carry out classical MD simulations to investigate the conformational space of various quadruplexes and associated dynamics; characterize the local conformations of the repeat mismatches and the ion distribution and binding; and when possible, discuss quadruplex relative stability. In previous work, we used MD to characterize the conformation and dynamics of the 12 homoduplexes that result from sense (GGGGCC) and antisense (CCCCGG) HRs under the three different reading frames for both DNA and RNA.43 We found that G-rich helices share common features. The inner G-G mismatches stay inside the helix in Gsyn–Ganti conformations and form two hydrogen bonds (HBs) between the Watson–Crick edge of Ganti and the Hoogsteen edge of Gsyn. In addition, Gsyn in RNA forms a base-phosphate HB. Inner G-G mismatches cause local unwinding of the helix. G-rich double helices are more stable than C-rich helices due to better stacking and HBs of G-G mismatches. C-rich helix conformations vary wildly. C mismatches flip out of the helix in DNA but not in RNA. Least (most) stable C-rich RNA and DNA helices have single (double) mismatches separated by two (four) Watson–Crick basepairs. The most stable DNA structure displays an “e-motif” where mismatched bases flip toward the minor groove and point in the 5′ direction.44 There are two RNA conformations, where the orientation and HB pattern of the mismatches is coupled to bending of the helix. Our previous work also includes conformational and dynamical characterization and free energy maps for (CAG)n and (GAC)n repeats45 and (CGG)n and (GCC)n repeats.46 All these trinucleotide repeats except for GAC are associated with a large number of late-onset progressive neurodegenerative diseases.47 A (GAC)5 repeat in the human gene for cartilage oligomeric matrix protein can only expand by one repeat (causing multiple epiphyseal dysplasia) or by two repeats (causing pseudoachondroplasia).48 The present work is thus part of our ongoing effort to study atypical secondary structures in SSRs.
RESULTS
Initial Structures.
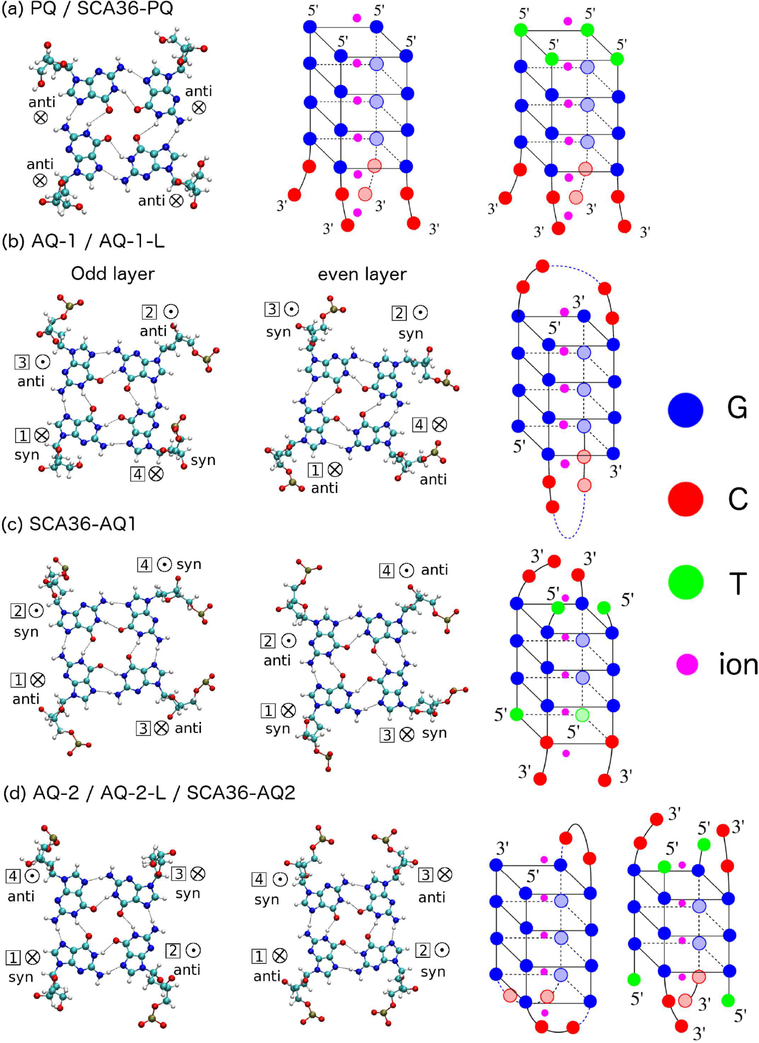
Quadruplexes are considerably easier to crystallize than duplexes: indeed the PDB contains several Gquadruplex structures but not a single duplex with 4 consecutives G’s that include mismatches. We therefore based our modeling on G-quadruplexes available in the PDB. We started with a parallel-stranded G-quadruplex DNA obtained in solution NMR (structure PDB ID 139D) with strand sequence TTGGGGT. We mutated this sequence so as to obtain the parallel quadruplex GGGGCC and TGGGCC, as shown in Figure 1a. We call this structure PQ and SCA36-PQ (PQ for parallel quadruplex). The geometries of two adjacent guanine residues within a quartet is anti-anti, as shown in Figure 1(a). We then constructed two antiparallel G-quadruplex structures for GGGGCC. One was built from the NMR structure 1JPQ which features two strands of (GGGG*BRU*TTTGGGG) arranged antiparallel with each other (BRU is bromodeoxyuridine), as shown in Figure 1b. By mutating the T/Bru bases and cutting the loops, we obtain a quadruplex structure made of two different strand types: 5′-GGGGCC-3′ and 5′-CCGGGG-3′. A quadruplex like this can possibly form from different parts of a long sequence or intermolecularly. However, if the C’s are linked similarly to the original 1JPQ NMR structure (as shown in Figure 1b), then it does not follow the HR pattern (the purpose of this simulation is to carry out comparative measures of stability). We call these structures AQ-1 and AQ-1-L, antiparallel quadruplex 1 without or with loops. The loops form a diagonal dimeric structure. Around the quartet, the guanine glycosidic geometries are anti-anti-syn-syn and syn-syn-anti-anti for adjacent quartets. The step patterns of adjacent guanine residues along the same strand are syn·anti, anti·syn, and syn· anti. A second antiparallel G-quadruplex was built starting from NMR structure with PDB ID 2N2D, and it is formed by a (GGGGCC)3-GGGG strand, which we used in two structures: with two independent strands (by cutting the loops), AQ-2, or with the original loops, AQ-2-L, as shown in Figure 1d.49 The loops form a chair-type monomeric model structure. Around the quartet, the guanine glycosidic geometries are anti-syn-anti-syn and syn-anti-syn-anti. The step patterns of adjacent guanine residues along the same strand are syn·anti, anti·syn, and syn·anti. Finally, for the TGGGCC sequence behind SCA36, we built two antiparallel quadruplexes. One (SCA36AQ-1) was built from the solution NMR structure 230D which features sequence of (GGGGTUTUGGGGTTTTGGGGUUUTTGGGI). The original structure 230D forms a basket-type monomeric structure. By mutating and cutting some residues, we obtain an antiparallel quadruplex structure with 4 strands of sequence ((TGGGCC)3-(TGGGC)), as shown in Figure 1c.The geometries of two adjacent guanine residues within a quartet are anti-syn, syn-syn, syn-anti, and anti-anti. For the G part of the quadruplex (with three G-quartets as opposed to four), there are two steps along the strand: anti·syn and syn·anti. The other antiparallel G-quadruplex of SCA36 was built from G-layers of 2N2D. We removed all C bases of 2N2D and extended the G-layers, and by cutting and mutating we generated SCA36-AQ-2, with anti·syn and syn·anti steps and anti–syn–anti–syn orientation within the same quartet layer (Figure 1d).49
Figure 1.
Quadruplex geometries. Left: Geometries of guanine residues within a quartet (odd layers in first column and even layers in second column). Right: full quadruplex configurations. (a) PQ and SCA36-PQ; (b) AQ-1/AQ-1-L; (c) SCA36-AQ1 (d) AQ-2/AQ-2-L/SCA36-AQ2. G’s have been labeled as blue, C’s are red, T’s are green, and ions are magenta. ⊗ indicates 5′ → 3′ strand direction pointing into the paper; ⊙indicates 5′ → 3′ strand direction pointing out of the paper.
In all cases, inner K+ ions were added to each quadruplex structure, as shown in Figure 1. Additional K+ ions were added to the solvent for neutralization. Finally, RNA structures were constructed by using LEAP to replace 2-deoxyribose by the alternative pentose sugar ribose. All the mutations were carried out with the Coot (Crystallographic Object-Oriented Toolkit) package.50 We have carried out two additional sets of simulations on DNA PQ, AQ-1, and AQ-2 models: one with all the K+ ions changed to Na+; the other one with all the initial ions K+ outside of the quadruplex. A summary of the quadruplexes used in our simulations is presented in Table 1.
Table 1.
Summary of the Quadruplexes Considered in This Study
| arrangement | model name | sequence | strands | form | ion type |
|---|---|---|---|---|---|
| parallel | PQ | 4 (GGGGCC) | 4 | DNA,RNA | K+ |
| PQ-Na | 4 (GGGGCC) | 4 | DNA | Na+ | |
| PQ-O | 4 (GGGGCC) | 4 | DNA | K+,outside | |
| SCA36-PQ | 4 (TGGGCC) | 4 | DNA,RNA(T→U) | K+ | |
| antiparallel | AQ-1 | 2 (GGGGCC), 2 (CCGGGG) | 4 | DNA,RNA | K+ |
| AQ-1-Na | 2 (GGGGCC), 2 (CCGGGG) | 4 | DNA | Na+ | |
| AQ-1-O | 2 (GGGGCC), 2 (CCGGGG) | 4 | DNA | K+,outside | |
| AQ-2 | 3 (GGGGCC), 1 (GGGG) | 4 | DNA,RNA | K+ | |
| AQ-2-Na | 3 (GGGGCC), 1 (GGGG) | 4 | DNA | Na+ | |
| AQ-2-O | 3 (GGGGCC), 1 (GGGG) | 4 | DNA | K+,outside | |
| AQ-1-L | 2 (GGGGCCCCGGGG) | 2 looped | DNA,RNA | K+ | |
| AQ-2-L | (GGGGCC)3-GGGG | 1 looped | DNA,RNA | K+ | |
| SCA36-AQ1 | 3 (TGGGCC), 1 (TGGGC) | 4 | DNA,RNA(T→U) | K+ | |
| SCA36-AQ2 | 4 (TGGGCC) | 4 | DNA,RNA(T→U) | K+ |
G-Rich Quadruplex.
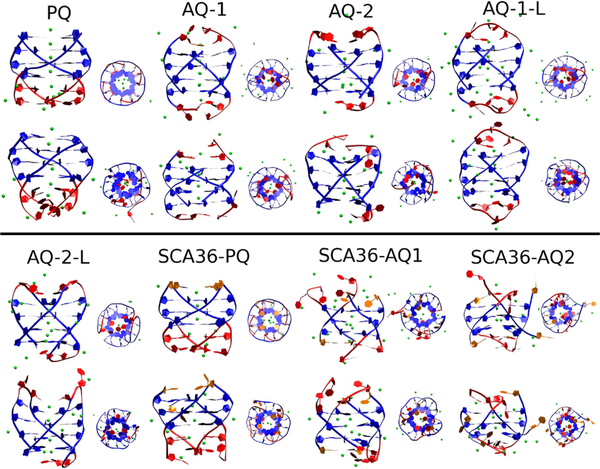
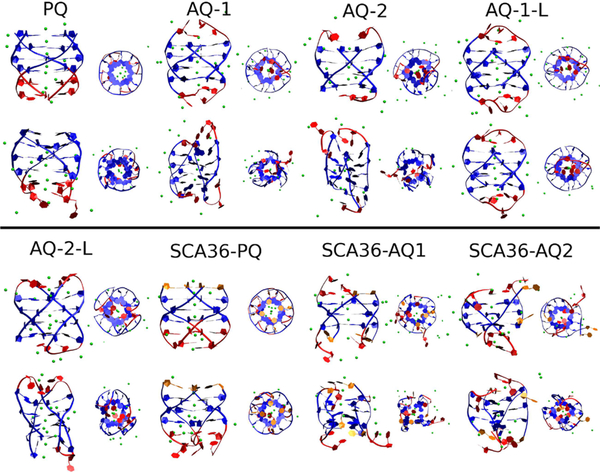
Figures 2 and 3 show a snapshot of the initial structures (after a few nanoseconds of equilibration) and the corresponding structures at 1 μs for DNA and RNA, respectively. The main results of these simulations are that all DNA quadruplexes, both parallel and antiparallel, are stable, at least up to 1 μs, even without stabilizing loops. On the other hand, as the RNA snapshots qualitatively show, the parallel arrangement PQ and SCA36-PQ RNA are preserved by the 1 μs simulation, but the good initial quadruplex conformations are lost for all but AQ-1-L (that does not correspond exactly to the HR sequence, see Figure 1).
Figure 2.
Snapshot of initial conformation (odd rows) and final conformation (even rows) of DNA quadruplexes.
Figure 3.
Snapshot of initial conformation (odd rows) and final conformation (even rows) of RNA quadruplexes.
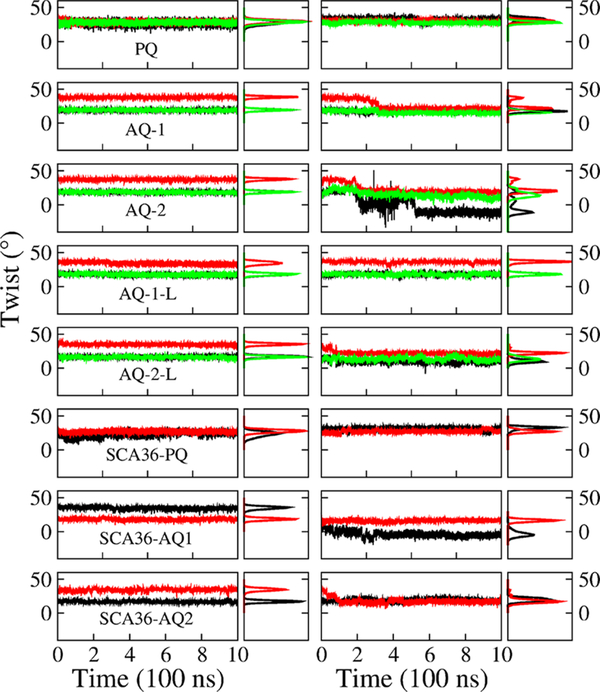
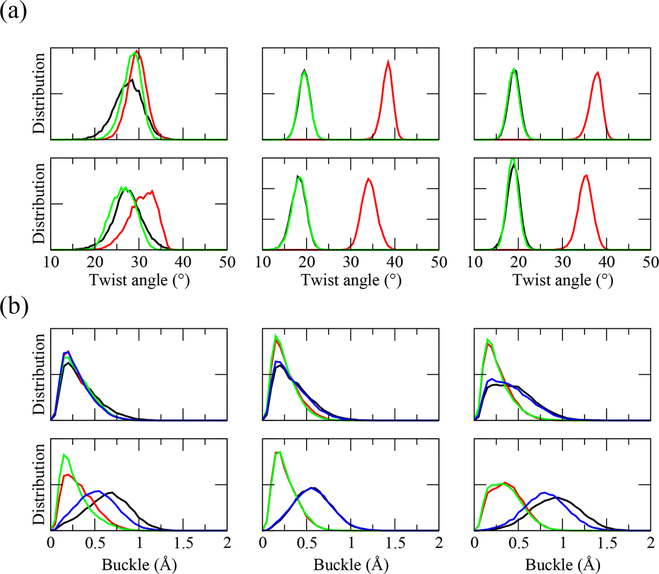
We quantify the stability of the quadruplexes by tracking the value of their twist angle and buckle displacement as a function of time. Figure 4 shows the twist angle for various steps (see SI for definition) as a function of time (odd columns) and the corresponding distribution (even columns) for each Gquadruplex in DNA and RNA form. Parallel DNA model PQ favors 29° for the twist, while the antiparallel models AQ-1, AQ-2, AQ-1-L and AQ-2-L present two values at 20° and 37° (AQ-2-L has a lower value at 18° and 35°). The two values are due to the anti· syn step patterns characteristic of the AQ quadruplexes, instead of anti· anti favored in parallel PQ models. In the RNA quadruplexes, PQ displays a stable quadruplex and the twist angles favor 30°. The antiparallel models, on the other hand, tend to unwind with a consequent decay in the twist angles. The twist angle of AQ-1 drops below 18°, with the third layer unwinding dramatically from 40° to 22°. The twist angle of AQ-2 drops from 40° to 20° in the third layer, and from 20° to −20° in the first layer. The presence of loops confers more stability to the quadruplexes. Thus, the looped model AQ-1-L maintains a relative stable bimodal distribution at peaks 19° and 38°. The other looped model AQ-2-L also unwinds from its initial values, but not much. Finally, SCA36 models behave similarly to ALS models. Parallel DNA SCA36-PQ favors 28.5°, while the antiparallel models SCA36-AQ1 and SCA36-AQ2 favor two values at 18.5°–35.5° and 17°–34.5°, respectively. For the RNA quadruplexes, SCA36-PQ favors a stable twist angle of 26.5° while the antiparallel models tend to unwind with a consequent decay in the twist angles. The twist angle of the first step in SCA36-AQ1 drops to −4.5°, and for the second step it drops to 15.5°. The twist angle of the second step of SCA36-AQ2 drops to 17°.
Figure 4.
Quadruplex twist angle. Twist angle as a function of time (odd columns) and the corresponding distribution (even columns) of step 1 (top, between Q-layer 1 and Q-layer 2, black), step 2 (middle, red), and step 3 (bottom, green) in quadruplex models in DNA (left) and RNA (right) form.
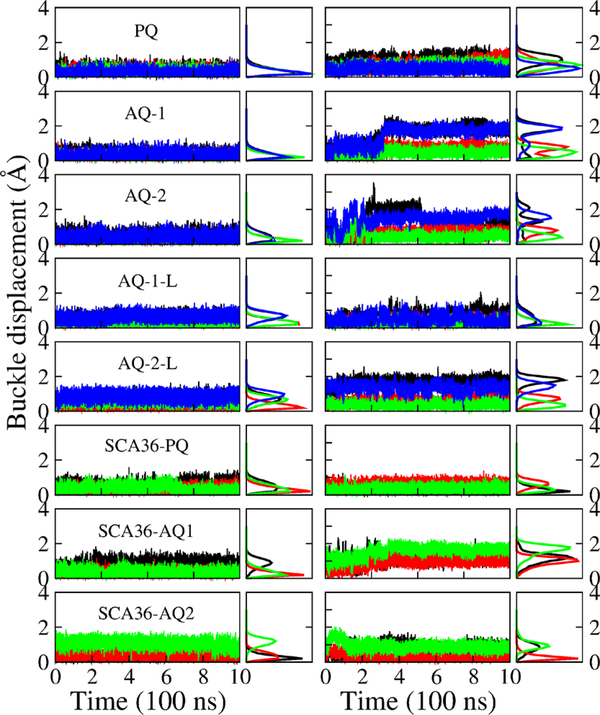
Figure 5 shows the buckle displacement (see the Supporting Information (SI) for definition) as a function of time (odd columns) and the corresponding distribution (even columns) for each G-quadruplex layer in DNA and RNA form. The buckle displacement of DNA models PQ, AQ-1, and AQ-2 favors 0.2 Å, which corresponds to stable flat quartet layers. AQ-1-L has a stable buckle displacement distribution at 0.2 Å for middle layers but a higher value (0.8 Å) for the edge G layers. Only the second G layer in AQ-2-L favors 0.2 Å, the other layers have higher values of buckle displacement, from 0.8 to 1.1 Å. RNA exhibit less stable quadruplexes than DNA, as manifested by the buckle displacement: PQ favors buckle values from 0.5 to 1.1 Å; and AQ-1 and AQ-2 only favor a lower buckle displacement (less than 1 Å) in the middle layers, the edge layers experience larger buckle displacement up to 2 Å, corresponding a “disordered” G-quadruplex. In agreement with the twist angle, the buckle displacement of AQ-1-L is small (0.2 Å) and therefore signals a stable G-quadruplex. The buckle displacement of AQ-2-L favors less than 1 Å for middle layers but larger values (up to 2 Å) for edge layers. Similar trends are observed for the SCA36 quadruplexes. For the edge G layers, the buckle displacement of DNA models SCA36-PQ, SCA36-AQ1 and SCA36-AQ2 favors 0.2 Å, 0.9 and 1.2 Å, respectively. In RNA models, SCA36-PQ favors 0.2 Å for the edge G layers and 0.6 Å for the middle layers. SCA36-AQ1 and SCA36-AQ2 favor 1.2 and 0.2 Å for middle layers, but higher values of 1.8 and 0.9 Å for edge layers.
Figure 5.
Quadruplex buckle displacement. Buckle displacement as a function of time (odd columns) and the corresponding distribution (even columns) of G-layer 1 (black), 2 (red), 3 (green), and 4 (blue) (from top to bottom) in quadruplex models in DNA (left) and RNA (right) form.
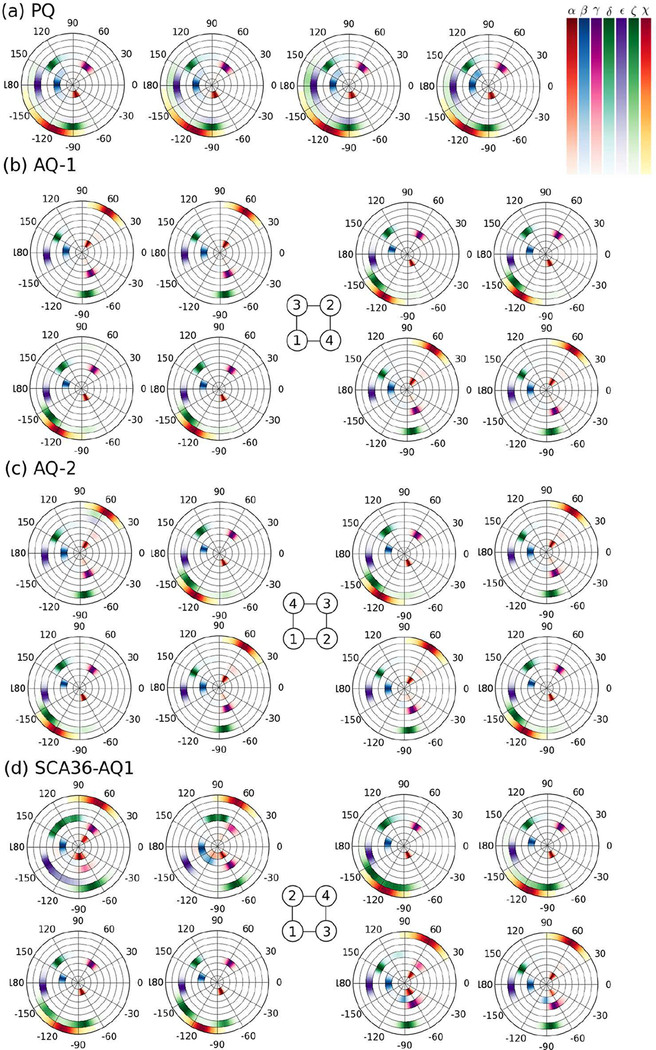
Examples of the density distribution of backbone and glycosidic torsion angles for DNA are presented in Figure 6 and Table 2, that also lists results for RNA. The distributions are obtained for the last 500 ns of the simulations. In Figure 6, distributions of backbone torsion angles α, β, γ, δ, ϵ, ζ and glycosidic torsion angle χ are shown from the inner to the outer circular rings for the particular layers indicated in the figure caption. The table is only an attempt to sum up the results of the full distributions for all layers and all models in DNA and RNA, which are quite complex and are presented in full in the SI. Table 2 lists the maximum value of the distribution; two or more values corresponds to a multimodal distribution, while numbers in square brackets correspond to a range of values without a clear maximum. For DNA PQ, AQ-1, AQ-2, and the three SCA36 models; and for RNA PQ and SCA36-PQ, the backbone torsion angles α and γ satisfy α + γ = C, with C in the range (–16°,0°). There are strong correlations between the anti/syn states and the backbone torsion angles for DNA AQ-1 and AQ-2: when G basis are in anti conformation (χ = −120° or −110°), they have α = −60°, β = 166°, γ = 55°, δ = 130°, ζ = −138°; when they are in syn conformation (χ = 60°), they have the other set of values α = 60°, β = 180°, γ = −60° or −70°, δ = 150°, ζ ≃ – 84°. In both cases, ϵ = −178°. The presence of loops in DNA forces values of backbone angles that do not follow these correlations. On the other hand, RNA antiparallel models do not exhibit correlated torsion angles as DNA does, another indication that the antiparallel G-quadruplexes in RNA are not stable.
Figure 6.
Density distribution of backbone and glycosidic torsion angles for DNA. Distributions of backbone torsion angles (α, β, γ, δ, ϵ, ζ) and glycosidic torsion angle χ are shown from the inner to the outer circular rings for each base at the middle G-layers during the last 500 ns of the simulations. (a) PQ (second G-layer) (b) AQ-1 (second (left) and third (right) G-layers) (c) AQ-2 (second (left) and third (right) G-layers) (d) SCA36-AQ1 (first (left) and second (right) G-layers). For the antiparallel models, the strands’ orientation–as sketched in Figure 1– is shown in the central schemes.
Table 2.
Backbone and Glycosidic Torsion Anglesa
| DNA | |||||
| ALS | PQ | AQ-1 | AQ-2 | AQ-1-L | AQ-2-L |
| α | −70 | −60 (60) | −60 (60) | −70, 78 | −60 (60) |
| β | −174 | 166 (180) | 166 (180) | −162, 170 | 166 (180) |
| γ | 54 | 55 (−60) | 54 (−70) | −66, 58 | −170, ± 60 |
| δ | 138 | 130 (150) | 130 (150) | [86–154] | 130 (150) |
| ϵ | −178 | −178 | −178 | −178 | −170 |
| ζ | −90 | −138 (−82) | −138 (−86) | −174, −86 | −82, −140 |
| χ | [[−118]–[−98]] | −120 (60) | −110 (60) | −110 (58) | −110 (60) |
| DNA | |||||
| SCA36 | SCA36-PQ | SCA36-AQ1 | SCA36-AQ2 | ||
| α | −70 | ±60 | ±60 | ||
| β | [166–180] | [166–180] | [166–180] | ||
| γ | 54 | ±54 | ±60 | ||
| δ | 102, 134 | 78, [130–150] | [130–150] | ||
| ϵ | −178 | [[−180]–[−138]] | −178 | ||
| ζ | −90 | −140, −86, −66 | −142, −86, 82 | ||
| χ | [[−120]–[−100]] | −110 (65) | −110 (65) | ||
| RNA | |||||
| PQ | AQ-1 | AQ-1-L | SCA36-PQ | ||
| α | −74 | −78,86 | −75 (54) | −74 | |
| β | −178,170 | [[−180]–[−160]] | [170–180] | [165–180] | |
| γ | 60 | 60 | 70 (−60) | 60 | |
| δ | 80,140 | 80, 150 | 78,140 | [60–90], [130–150] | |
| ϵ | [[−180]–[−150]] | −70, [[–170]–[−150]] | [[−180]–[−150]] | [[−180]–[−150]] | |
| ζ | [[−100]–[−60]] | [[−90]–[−70]], 94 | −140, –90 | [[−110]–[−60]] | |
| χ | [[−180]–[−150]],−120 | [[−120]–[−100]] (60) | [[−120]–[−90]] ([60–80]) | −110, −150 | |
This table lists the maximum value of each angle distribution. Numbers separated by commas correspond to main maxima in multimodal distributions. Numbers is square brackets separated by a dash indicate a range of values without a clear maximum. Numbers not surrounded by parentheses correspond to anti conformations, while number in parentheses correspond to syn conformations (PQ models are all anti). In antiparallel quadruplexes, if there is only one or several entries but none in parentheses for a given angle in a column, it simply means that both syn and anti conformations share those maxima or range of the distributions.
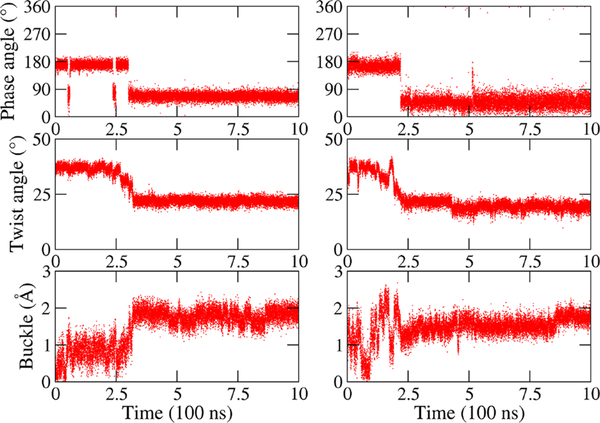
In order to determine the role of sugar puckering in RNA, we track the evolution of the sugar rings. Initially, the RNA quadruplexes were assembled in the same sugar conformations as DNA, mainly C2′-endo with a few exceptions (original to the PDB structures) and allowed to evolve without restraints. Sugar puckering is a relatively fast degree of freedom that can be sampled by regular MD; examples for the pseudorotation phase angle for RNA AQ-1 and AQ-1-L are given in SI Figures S3 and S4. We found that as the RNA Gsyn bases recover their preferred sugar conformation of C3′-endo, the reaccommodation triggers the instability of the antiparallel quadruplex, as shown for example in Figure 7. This figure shows the strong correlation between the (C2′-endo) → (C3′-endo) transition and the decay of twist and increase of buckle displacement. Some other Gsyn bases undergo intermediate states of puckering rearrangement. Figures S3 and S4 show a general trend for the antiparallel RNA models: quadruplexes without loop undergo many more puckering transitions than quadruplexes with loops. In particular, the pseudorotation phase angle undergoes nonnegligible transitions in only four Gsyn bases in RNA AQ-1-L, but it undergoes non-negligible transitions in both syn and anti bases in most (except four) bases in RNA AQ-1. Indeed, the G bases in AQ-1-L are C2′-endo except for four ending bases— two of them connected to Cs, and other two free ending—that transit between C3′-endo and C2′-endo frequently.
Figure 7.
Example of the coupling between sugar puckering, twist and buckle displacement in RNA. Pseudo rotation phase angle (top row), twist angle (middle), and buckle displacement (bottom) for a chosen Gsyn and its quartet layer in RNA AQ-1 (left) and RNA AQ-2 (right).
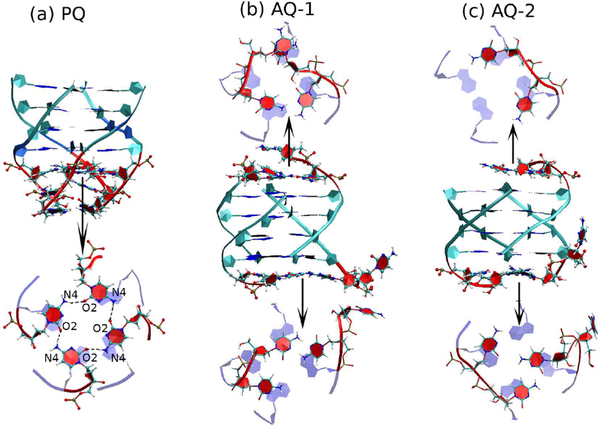
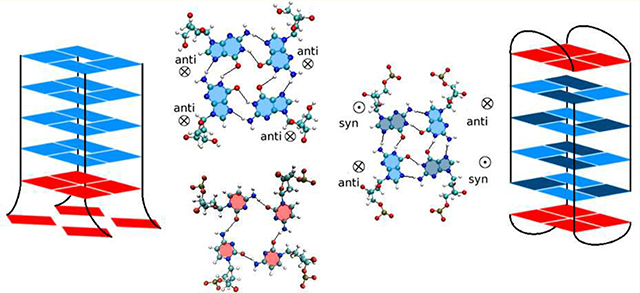
Although quartets formed by four unmodified cytosine bases are very rare, we find that parallel G-quadruplexes do stabilize the adjacent C bases into a quartet in both DNA and RNA (thus effectively becoming a mixed quadruplex of 5 layers). A typical conformation is shown in Figure 8 for DNA. As the figure clearly shows, the C-quartet is stabilized by the stacking interactions with the G bases in the preceding quartet, and by hydrogen bonds C(N4)–C(O2), whose population is ≃88% for both DNA and RNA PQ models. In addition, there is an ion between the G-quartets and the C-quartet. The end C layer, on the other hand, is unstable, with a total hydrogen bond population of ≃40% for both DNA and RNA. Ordered C bases are also present in the antiparallel DNA models AQ-1 and AQ-2 (but not in antiparallel RNA). In model AQ-1 there are two strands of two linked Cs across the diagonal in each end of the duplex (Figure 1), and via bending of the backbone these bases attempt to form a quartet, with three bases managing a distinct layer and a fourth one flipping freely, as shown in Figure 8. Even model AQ-2 (with two two-base strands on one end, but only one two-base strand on the other) favors the formation of ordered layers. This stabilizing of the C layers is driven entirely by the stacking with the guanines in the G-quartets.
Figure 8.
C-Layers coupled to the G-quadruplexes in DNA. This figure shows snapshots of DNA conformations where the C bases at the end of the quadruplexes form flat layers. Top projections are depicted in the 5′→3′ direction while bottom ones are shown in the 3′→5′ direction. (a) PQ model with a symmetric C-tetrad; (b) AQ-1; (c) AQ-2.
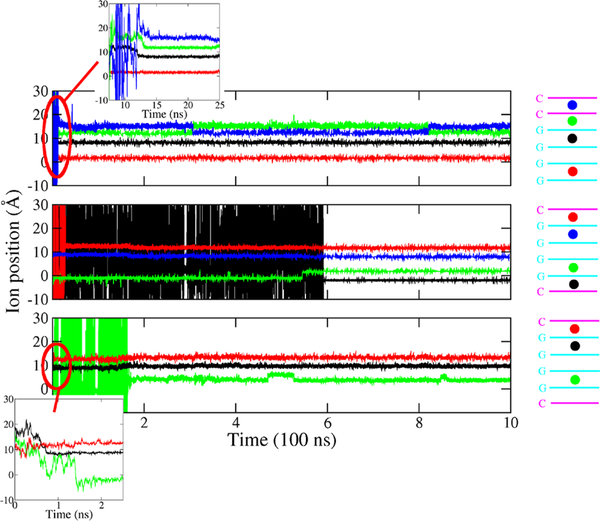
Finally, we present results concerning the stability of the quadruplexes as informed by the ion species. To compare the effect of the K+ and Na+ ions on the stability of G-quadruplexes, we carried out an additional set of 1 μs simulations for PQ-1, AQ-1, and AQ-2 in DNA form. For the G-layers steps, Figure 9 shows the twist angle and the buckle displacement distributions with either with K+ or Na+ as neutralizing ion. Both K+ and Na+ ions favor a stable twist distribution, with Na+ yielding in general slightly smaller twist angles, especially for the edge steps of PQ and the middle steps of AQ models. On the other hand, the models with Na+ as neutralizing ion have larger buckle displacement (>0.5 Å) than K+ models (0.25 Å) for edge layers, i.e., quadruplex stabilization by K+ is better than by Na+. In addition, we carried out another set of simulations of PQ, AQ-1, and AQ-2 in DNA form, where the initial neutralization ions K+ are completely outside the helix. Interestingly, the ions diffused into the central quadruplex channel within the first few nanoseconds. Figure 10 shows the ions diffusing through the inner quadruplex channel as a function of time. We found that the ions entered through the nearest channel opening, moved one or two layers, and remained there, blocking a middle step in the tunnel over our simulation time scales.
Figure 9.
Twist angle and buckle displacement distributions as a function of ion type. (a) Twist angle distribution of step 1 (black), 2 (red) and 3 (green) in quadruplex models PQ (left), AQ-1 (middle) and AQ-2 (right) in DNA form with K+ (top) and Na+ (bottom) as neutralizing ion. (b) Buckle displacement distribution of G-layer 1 (black), 2 (red), 3 (green), and 4 (blue) in quadruplex models PQ (left), AQ-1 (middle), and AQ-2 (right) in DNA form with K+ (top) and Na+ (bottom) as neutralizing ion.
Figure 10.
Ion diffusion through the quadruplex channel. Ion position (Å) (left) through the inner interbase channel of the quadruplex for PQ (top), AQ-1 (middle), and AQ-2 (bottom). The origin corresponds the middle of first G layer, shown close to the bottom of each cartoon. In the three cases, only an inner step is left unoccupied.
DISCUSSION
In this work, we have examined a series of both parallel and antiparallel DNA and RNA basic G-quadruplexes. G-quadruplexes have been well characterized in the literature.40–42 They display right-handed helicity and result from the hydrophobic stacking of two or more G-quartets. A G-quartet consists of a planar array of four guanines, linked by a cyclic array of hydrogen bonds from the Watson–Crick and the Hoogsteen faces. Consecutive quartets then stack to form the quadruplex stem, stabilized by monovalent cations lying in the central channel of the stem, in direct contact with the guanine carbonyl groups. G-quadruplexes generally have loops, and there is literature reporting the role of loop length, type, and sequence in the stability of G-quadruplexes. However, for SSRs that can contain hundreds of repeats, the stems of the quadruplexes are presumed relatively large, with the loop playing a lesser role in the stability of the noncanonical motif. Thus, in this initial work we have mainly examined the “stem” of the quadruplexes, as shown in Figure 1.
Parallel G-quadruplexes have all the four strands parallel to each other and their guanine residues in anti conformations,51 resulting in very similar structures. When antiparallel strands are added, more polymorphism appears.51 In particular, the AQ-2 model, where each strand has two adjacent nearest neighbors in the opposite direction, is a fully antiparallel model (the strands across the diagonal, or second neighbors, are parallel). Two additional hybrid models can be constructed. In the AQ-1 type shown in Figure 1, each strand has one adjacent neighbor in the same direction, and the other in the opposite direction. A second hybrid topology has three strands run in the same direction and the fourth strand in the opposite direction. Both hybrid models are usually labeled antiparallel and produce two parallel and two antiparallel nearest-neighbor strands. (AQ-1 has antiparallel diagonal strands, and the third model has one pair of diagonal strands parallel and the other antiparallel). In this initial work, we have concentrated in the fully parallel model, and antiparallel models with two pairs of strands in the opposite direction. In the AQ-1 type, adjacent Gquartets display the syn-syn-anti-anti and anti-anti-syn-syn topologies. In the AQ-2 type, adjacent G-quartets display the anti-syn-anti-syn and syn-anti-syn-anti topologies. In all types of G-quadruplexes, strands that have the same direction have the same glycosidic conformation (and strands in opposite direction have the opposite glycosidic orientation). In addition, syn and anti glycosidic conformations alternate for adjacent guanines along the same strand.52,53 Experimentally, it has been observed that G-quadruplexes formed by two or four Gquartets favor a 5′-syn·anti(·syn·anti)-3′ conformation along the same strand.54–58 This trend has been explained in a study that involves MD and free energy calculations.59 These calculations indicate that along a given strand, the relative stability of the steps is syn·anti > anti·anti > anti·syn > syn·syn. Thus, in G-quadruplexes formed by an even number of layers, a 5′-syn· anti starting conformation ensures the number of syn· anti steps is greater than the number of anti·syn steps by one (obviously this effect waters down for long quadruplexes). For G-quadruplexes formed by an odd number of layers, the number of anti· syn and of syn· anti steps is the same, independently of the initial guanine conformation.
Based on NMR and circular dichroism (CD) spectroscopy, DNA d(GGGGCC) oligomers with varying number of repeats have been found to adopt inter- and intramolecular Gquadruplex structures, in either parallel or antiparallel orientation.35 Using gel mobility shift assays and NMR and circular dichroism (CD) spectroscopy, RNA r(GGGGCC) expansions have been shown to adopt a stable, parallel Gquadruplex.36,37 However, recent experiments involving CD, optical melting and 1D 1H NMR spectroscopy, combined with chemical and enzymatic probing of a r(GGGGCC) repeat expansion point to a general scenario where the repeat expansion adopts a hairpin structure with G-G mismatches in equilibrium with a quadruplex structure.38,39 These experiments lack molecular resolution, and in this work we have aimed at a structural characterization of the G-quadruplexes associated with the hexanucleotide repeats.
All in all, we have tested 22 different G-quadruplex models for 1 μs each (see Table 1). We find that all DNA models, parallel or antiparallel, with or without loops are stable, while only parallel RNA quadruplexes (models PQ for ALS and SCA36-PQ) are stable, with the only exception of the antiparallel model AQ-1-L, where the presence of the two diagonal loops help stabilize the quadruplex (AQ-1-L does not exactly correspond to an HR sequence). The stability or unfolding of the quadruplexes is qualitatively shown in Figures 2 and 3, and has been quantified by quadruplex twist and quadruplex buckle displacement, as well as by the distribution of the backbone and glycosidic torsion angles. For a stable structure, twist has a single value for a parallel quadruplex (with guanosines all in anti conformations) or two values for antiparallel quadruplexes (corresponding to anti/syn conformations). The twist values stay constant for the RNA parallel models but quickly decay for the antiparallel models, signaling unwinding. Buckle displacement has low values for DNA (around 0.2 Å), especially for models PQ, AQ-1, AQ-2, signaling stable flat quartet layers. In RNA models, on the other hand, there is a considerable increase of buckle displacement, up to 2 Å in outer quadruplex layers. SCA36 models in general are expected to exhibit less stability, as they are formed only by three G-quartets as opposed to four. In particular, antiparallel DNA SCA36-AQ1 and SCA36-AQ2 models seem less stable than parallel DNA SCA36-PQ, as measured by buckle displacement (but twist is preserved). Backbone angles also reveal the conformational stability of the quadruplexes. For DNA AQ-1 and AQ-2, each conformation of the glycosidic torsion angle, anti (χ = −120° or −110°) or syn (χ = 60°) is associated with a different set of backbone torsion angles. On the other hand, RNA antiparallel models do not exhibit correlated torsion angles as DNA does, another indication that the antiparallel G-quadruplexes in RNA are not stable.
It has been argued that RNA favors parallel quadruplexes because of its nucleoside sugar most favorable conformation, C3′-endo (it is generally C2′-endo in DNA). The anti conformation can be formed whatever the sugar conformation is (C2′- or C3′-endo), whereas the syn position is unfavorable in the case of C3′-endo sugar conformation because of the steric hindrance between O3′ and C5′.42 However, more recent data shows that RNA guanines can adopt the syn conformation without “external” aids, such as chemical modifications, special loops, protein interactions, etc. In addition to the case of the left-handed Z-RNA helix,60–62 this has been described in G-G mismatches in tri- and hexanucleotide repeats in helical conformation (We note that DNA also forms a similar lefthanded Z-form helix63). In previous work, we used MD to characterize the conformation and dynamics of the 12 homoduplexes that result from sense (GGGGCC) and antisense (CCCCGG) HRs under the three different reading frames for both DNA and RNA.43 We found that G-rich helices share common features. The inner G-G mismatches stay inside the helix in Gsyn-Ganti conformations and form two hydrogen bonds (HBs) between the Watson–Crick edge of Ganti and the Hoogsteen edge of Gsyn. In addition, Gsyn in RNA forms a base-phosphate HB. Inner G-G mismatches cause local unwinding of the helix. Similar results have been found experimentally for the trinucleotide repeats CGG, where crystallographic studies for the sequences 5′-G-(CGG)2-C-3′ (PDB ID 3R1C, ref 64) and 5′-UU-GGGC-(CGG)3-GUCC-3′ (PDB ID 3JS2, ref 65), found that the RNA helices exhibit G-G pairs in a typical anti-syn conformation, with two hydrogen bonds between the Watson–Crick edge of Ganti and the Hoogsteen edge of Gsyn. In particular, all the Gs in 3R1C are in C3′-endo (except for one that is in C4′-exo), and in our simulations for the GGGGCC repeats, RNA Gs are also mainly in C3′-endo, with an occasional C4′-exo.
To further test the reasons why RNA does not favor antiparallel quadruplexes, we assembled the initial RNA quadruplexes in the same sugar conformation as DNA, mainly C2′-endo with a few exceptions (original to the PDB structures used to model the quadruplexes) and allowed the conformations to freely evolve. Sugar puckering is a relatively fast degree of freedom that can be sampled by regular MD, as shown, for instance, in Figure S3. We found that as RNA recovers its preferred sugar conformation of C3′-endo, it triggers the instability of the antiparallel quadruplex, via the coupling to twist and buckle displacemet (Figure 7). The unfolding then proceeds by unwinding and by buckling up and down of the quartet planes. By contrast, the homoduplexes in previous studies accommodate the Gsyn conformation through backbone rotations: in CGG repeats, the Gsyn α and γ backbone torsion angles do not adopt the usual – sc and + sc conformations but rotate around 180° and 120°, resulting in a local straightening of the sugar–phosphate backbone,64 which in turns leads to local unwinding of the helical duplex. These rotations are precluded by the calibrated interlocking of strands required by the quadruplex structure. It is noted that antiparallel RNA quadruplexes, although rare, can be obtained when other components are factored in. Thus, by replacing a guanosine by an 8-bromoguanosine, r(GC(BrG)GCGGC), a tetramolecular, antiparallel architecture of the RNA G-quadruplexes was obtained.66 Another antiparallel RNA G-quadruplex motif was described in an in vitro RNA aptamer (known as Spinach) that binds a GFP-like ligand activating its green fluorescence.67,68 The G-quadruplex exhibits a three-quartet core, composed of two G-quartets stacked on top of a mixed-sequence quartet. The quartets are linked by several loops of various lengths. The guanines in the G-quartets exhibit an equal mix of C3′-endo and C2′-endo sugar puckers, and even Gsyn accepts a C2′-endo conformation.68 In this work we show that the presence of two diagonal loops in the RNA AQ-1-L model is enough to stabilize an antiparallel G-quadruplex, at least in the 1 μs time scale (which is relatively long compared to the quick unraveling of the other antiparallel RNA quadruplexes).
We found that the parallel G-quadruplex stabilizes the adjacent C bases into a C-quartet in both DNA and RNA (thus effectively becoming a mixed quadruplex of 5 layers). As Figure 8 clearly shows, the C-quartet is stabilized by the stacking interactions with the G bases in the preceding G-quartet, and by hydrogen bonds C(N4)-C(O2), whose population is ≃88% for both DNA and RNA PQ models. In addition, there is an ion between the G-quartets and the C-quartet. The end C layer, on the other hand, is unstable, with a total hydrogen bond population of ≃40% for both DNA and RNA. To the best of our knowledge, there are only two experimental structures with atomic resolution that exhibit stable C-tetrads. One is a crystal structure of the DNA decamer, d(CCACNVKGCGTGG) (CNVK, 3-cyanovinylcarbazole), which forms a G-quadruplex structure in the presence of Ba2+. The structure displays a C-tetrad where water molecules mediate contacts between the divalent cations and the C-tetrad, allowing Ba2+ ions to occupy adjacent steps in the central ion channel.69 In this sense, this C-tetrad is rather different to the C-tetrad reported here. The other structure is a NMR structure of the DNA sequence d(TGGGCGGT) in Na+ solution at neutral pH, containing the repeat sequence (GGGCGG) from the SV40 viral genome. The C-tetrad in this structure corresponds exactly to the C-tetrad found in this work, including the environment conditions (neutral pH, monovalent ions, unmodified nucleotides) and the structural features: parallel quadruplex, array of hydrogen bonds between the C bases, and good stacking with the preceding G-quartet. With the right “thumb” in the 5′→3′ direction, the hydrogen bonds form a C(O2)→C(N4) right-handed circular arrangement both for the experimental NMR structure and these simulations. The experimental study was carried out only for DNA; our study shows that the C-tetrad is stable for both DNA and RNA parallel quadruplexes. C-tetrads (and mixed G-C tetrads) were also proposed (but not directly seen) in an experimental study that combined CD, UV and electrophoresis for both (GGGGCC) and (GGCCCC) DNA HRs.70,71
Interestingly, ordered C bases are also present in the antiparallel DNA models AQ-1 and AQ-2 (but not in antiparallel RNA), in spite of the fact that the topology of the “hanging” C-strands in these models effectively precludes the formation of quartets. In model AQ-1, there are two strands, each formed by two linked cytidines, across the diagonal in each end of the duplex (Figure 1). Via bending of the backbone the hanging cytidines attempt to form a quartet, with three bases managing a distinct layer and a fourth one flipping freely, as shown in Figure 8. Even model AQ-2 (with two, two-cytidine strands on one end, but only one, twocytidine strand on the other) favors the formation of ordered layers. This stabilizing of the C layers is driven entirely by the stacking with the guanines in the G-quartets. From the point of view of the conformations enabled by the (GGGGCC) HR in C9FTD/ALS, these results clearly indicate that the Gquadruplexes can easily accommodate C-tetrads in order to form ordered, extended mixed-layer quadruplexes.
An important consideration concerning these structures is how strongly the stability of the C-layers are affected by their immediate environment. The two experimental structures reported on previously69 are characterized by internal C-layers in which the C-residues are strongly stabilized by the Gquartets on either sides of the layers. However, the structures shown in Figure 8 are characterized by terminal C’s, and these, for instance, are likely to protonate in a low pH environment. Hence, one can expect that these types of C-layers to be more readily observable experimentally at low pH conditions. We note that our simulations have been carried out in an unprotonated environment. We do not expect protonation to seriously effect the stability of these C-layers since their stability is driven primarily by the stacking interaction with the underlying G-quartet guanines.
Finally, we compared the stability of the quadruplexes as informed by the ion type, K+ and Na+ ions, for 1 μs simulations for PQ-1, AQ-1, and AQ-2 in DNA form. Both K+ and Na+ ions favor a stable twist distribution, with Na+ yielding in general slightly smaller twist angles. Larger buckle displacements for Na+ with respect to K+ provide a corroboration that quadruplex stabilization by K+ is better than by Na+. Another set of simulations comprised these same three quadruplexes but without internal ions, only external K+ ions. The ions were observed to diffuse into the central quadruplex channel within the first few nanoseconds. The ions entered through the nearest channel opening, moved one or two layers, and remained there, blocking just one middle step in the tunnel during the 1 μs time scale.
DNA and RNA G-quadruplexes represent a significant noncanonical nucleic acid structural motif, as in addition of being involved in many crucial biological processes, such as genetic instability, gene and telomere regulation, RNA translation regulation, splicing, and so forth,33,34 they hold great promise as therapeutical targets not only for cancer therapy and aging research but also for the treatment of G-rich SSR diseases.29,72–76 In the context of C9FTD/ALS diseases, knowledge of the RNA secondary structure of GGGGCC repeats (or of GGGCCT in the case of SCA36) may aid in the design of antisense oligonucleotide therapeutic approaches. This approach as been tried, for instance, to inhibit deleterious interactions of proteins with pathogenic RNAs in myotonic dystrophy MD1, caused by expanded CUG repeats.77 At present, there is mounting evidence39 of the structural polymorphism at both DNA and RNA levels of pathological SSRs. Knowledge of the various molecular structures at the atomic level may ultimately prove extremely important for a mechanistic model of SSR neurodegenerative diseases.
METHODS
The simulations were carried out with the AMBER 16 simulation package78 with force field OL1579 for DNA and ff99BSC0+χOL3 80–82 for RNA. The TIP3P water model 83 was used for the explicit solvent simulations with periodic boundary conditions in truncated octahedron water boxes with more than 5000 waters each. Electrostatics were handled by the PME method,84 with a direct space cutoff of 9 Å, and with an average mesh size of approximately 1 Å for the lattice calculations. We used Langevin dynamics with a coupling parameter γ = 2.0 ps–1. The NPT simulations were carried out at 300 K and 1 atm with the Berendsen barostat85 with an isothermal compressibility of β = 44.6 × 10–6 bar–1 and pressure relaxation time τp = 1.0 ps. The length of the simulation was 1 μs for each model, and the coordinate information was collected every 5 ps.
The equilibration processes took place in four steps. First, we carried out a steepest descent simulation followed by conjugate gradient minimization keeping the nucleic acid atoms fixed at their initial positions. Then we carried out unrestrained steepest descent followed by conjugate gradient minimizations. This was followed by short MD runs under constant volume while the system was heated from 0K to 300 K with weak restraints on the nucleic acid atoms. After this, Langevin dynamics at constant temperature (300 K) and constant pressure (1 atm) were applied for 2 ns, after which the density of the system was found to be stable around 1.0 g/cm3. Finally, each model was run for 1 μs, and the second half of each run (500 ns) was used for data analysis. Definitions or comments for quadruplex twist angle, buckle displacement, and ion position are presented in the SI.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Extreme Science and Engineering Discovery Environment (XSEDE) TG-MCB160064 for computing support.
Funding
This work has been supported by the National Institute of Health (NIH) Grant R01GM118508 and the National Science Foundation (NSF) Grant SI2-SEE-1534941. We also thank the XSEDE Grant TG-MCB160064 for computational support.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschemneuro.7b00476.
Further details on the analysis, plots of pseudorotation angles and torsion angle distributions for various quadruplex structures (PDF)
REFERENCES
- (1).Ellegren H (2004) Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet 5, 435–445. [DOI] [PubMed] [Google Scholar]
- (2).Oberle I, Rouseau F, Heitz D, Devys D, Zengerling S, and Mandel J (1991) Molecular-basis of the fragile-X syndrome and diagnostic applications. Am. J. Hum. Genet 49, 76.1829582 [Google Scholar]
- (3).Giunti P, Sweeney M, Spadaro M, Jodice C, Novelletto A, Malaspina P, Frontali M, and Harding AE (1994) The trinucleotide repeat expansion on chromosome 6p (SCA1) in autosomal dominant cerebellar ataxias. Brain 117, 645–649. [DOI] [PubMed] [Google Scholar]
- (4).Campuzano V, et al. (1996) Friedreich’s ataxia: Autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427. [DOI] [PubMed] [Google Scholar]
- (5).Wells RD, and Warren S Genetic instabilities and neurological diseases; Academic Press, San Diego, CA, Elsevier, 1998. [Google Scholar]
- (6).Pearson C, and Sinden R (1998) Slipped strand DNA (S-DNA and SI-DNA), trinucleotide repeat instability and mismatch repair: A short review. In Structure, Motion, Interaction and Expression of Biological Macromolecules, Vol 2, pp 191–207, 10th Conversation in Biomolecular Stereodynamics Conference, SUNY, JUN 17–21, 1997. [Google Scholar]
- (7).McMurray C (1999) DNA secondary structure: A common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. U. S. A 96, 1823–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pearson C, Edamura K, and Cleary J (2005) Repeat instability: Mechanisms of dynamic mutations. Nat. Rev. Genet 6, 729–742. [DOI] [PubMed] [Google Scholar]
- (9).Mirkin SM (2006) DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol 16, 351–358. [DOI] [PubMed] [Google Scholar]
- (10).Mirkin S (2007) Expandable DNA repeats and human disease. Nature 447, 932. [DOI] [PubMed] [Google Scholar]
- (11).Wells R, Dere R, Hebert M, Napierala M, and Son L (2005) Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucleic Acids Res 33, 3785–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Orr H, and Zoghbi H (2007) Trinucleotide repeat disorders. Annu. Rev. Neurosci 30, 575. [DOI] [PubMed] [Google Scholar]
- (13).Kim JC, and Mirkin SM (2013) The balancing act of DNA repeat expansions. Curr. Opin. Genet. Dev 23, 280–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Cleary J, Walsh D, Hofmeister J, Shankar G, Kuskowski M, Selkoe D, and Ashe K (2005) Natural oligomers of the amyloidprotein specifically disrupt cognitive function. Nat. Neurosci 8, 79–84. [DOI] [PubMed] [Google Scholar]
- (15).Dion V, and Wilson JH (2009) Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet 25, 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).McMurray CT (2008) Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair 7, 1121–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).DeJesus-Hernandez M, et al. (2011) Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Renton AE, et al. (2011) A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lillo P, and Hodges JR (2009) Frontotemporal dementia and motor neurone disease: Overlapping clinic-pathological disorders. J. Clin. Neurosci 16, 1131–1135. [DOI] [PubMed] [Google Scholar]
- (20).Kobayashi H, Abe K, Matsuura T, Ikeda Y, Hitomi T, Akechi Y, Habu T, Liu W, Okuda H, and Koizumi A (2011) Expansion of intronic GGCCTG hexnucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvment. Am. J. Hum. Genet 89, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Gendron TF, et al. (2013) Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng S-M, Schludi MH, van der Zee J, Cruts M, Van Broeckhoven C, Kremmer E, Kretzschmar HA, Haass C, and Edbauer D (2013) Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893. [DOI] [PubMed] [Google Scholar]
- (23).Zu T, et al. (2011) Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl. Acad. Sci. U. S. A 108, 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, DeJesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, Boylan KB, Dickson DW, and Petrucelli L (2013) Unconventional Translation of C9ORF72 GGGGCC Expansion Generates Insoluble Polypeptides Specific to c9FTD/ALS. Neuron 77, 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, and Edbauer D (2013) The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science 339, 1335–1338. [DOI] [PubMed] [Google Scholar]
- (26).Henderson E, Hardin C, Walk S, Tinoco I, and Blackburn E (1987) Telomeric DNA oligonucleotides from novel intramolecular structures containing guanine-guanine base pairs. Cell 51, 899. [DOI] [PubMed] [Google Scholar]
- (27).Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps H, and Pluckthun A (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U. S. A 98, 8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Lipps H, and Rhodes D (2009) G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 19, 414. [DOI] [PubMed] [Google Scholar]
- (29).Patel D, Phan A, and Kuryavyi V (2007) Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 35, 7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Lee J, Yoon J, Kihm H, and Kim D (2008) Structural diversity and extreme stability of unimolecular Oxytricha nova telomeric G-quadrupex. Biochemistry 47, 3389. [DOI] [PubMed] [Google Scholar]
- (31).Lee JY, and Kim DS (2009) Dramatic effect of single-base mutation on the conformational dynamics of human telomeric Gquadruplex. Nucleic Acids Res 37, 3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Santoro MR, Bray SM, and Warren ST (2012) Molecular mechanisms of Fragile X Syndrome: a twenty-year perspective. Annu. Rev. Pathol.: Mech. Dis 7, 219. [DOI] [PubMed] [Google Scholar]
- (33).Maizels N (2015) G4-associated human diseases. EMBO Rep. 16, 910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wu Y, and Brosh RM (2010) G-quadruplex nucleic acids and human disease. FEBS J. 277, 3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Sket P, Pohleven J, Kovanda A, Stalekar M, Zupunski V, Zalar M, Plavec J, and Rogelj B (2015) Characterization of DNA Gquadruplex species forming from C9ORF72 G4C2-expanded repeats associated with amyotrophic lateral sclerosis and frontotemporal lovar degeneration. Neurobiol. Aging 36, 1091–1096. [DOI] [PubMed] [Google Scholar]
- (36).Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EMC, Parkinson G, and Isaacs AM (2012) C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci. Rep 2, 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Reddy K, Zamiri B, Stanley SYR, Macgregor RB Jr., and Pearson CE (2013) The Disease-associated r(GGGGCC)n Repeat from the C9orf72 Gene Forms Tract Length-dependent Uni- and Multimolecular RNA G-quadruplex Structures. J. Biol. Chem 288, 9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Su Z, et al. (2014) Discovery of a Biomarker and Lead Small Molecules to Target r(GGGGCC)-Associated Defects in c9FTD/ALS. Neuron 83, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim M-S, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, Rothstein JD, and Wang J (2014) C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Huppert J (2008) Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev 37, 1375. [DOI] [PubMed] [Google Scholar]
- (41).Malgowska M, Czajczynska K, Gudanis D, Tworak A, and Gdaniec Z (2016) Overview of RNA G-quadruplex structures. Acta Biochim. Pol 63, 609. [DOI] [PubMed] [Google Scholar]
- (42).Tran PLT, de Cian A, Gros J, Moriyama R, and Mergny J-L (2012) Tetramolecular quadruplex stability and assembly. Top. Curr. Chem 330, 243–73. [DOI] [PubMed] [Google Scholar]
- (43).Zhang Y, Roland C, and Sagui C (2017) Structure and dynamics of DNA and RNA double helices obtained from the GGGGCC and CCCCGG hexanucleotide repeats that are the hallmark of C9FTD/ALS diseases. ACS Chem. Neurosci 8, 578–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Pan F, Zhang Y, Man VH, Roland C, and Sagui C (2017) E-motif formed by extrahelical cytosine bases in DNA homoduplexes of trinucleotide and hexanucleotide repeats. Nucleic Acids Res, DOI: 10.1093/nar/gkx1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Pan F, Man VH, Roland C, and Sagui C (2017) Structure and dynamics of DNA and RNA double helices of CAG and GAC trinucleotide repeats. Biophys. J 113, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Pan F, Man VH, Roland C, and Sagui C Structure and dynamics of DNA and RNA double helices obtained from CCG and GGC trinucleotide repeats. Biophys. J, 2018, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zoghbi HY, and Orr HT (2000) Glutamine Repeats and Neurodegeneration. Annu. Rev. Neurosci 23, 217–247. [DOI] [PubMed] [Google Scholar]
- (48).Delot E, King LM, Briggs MD, Wilcox WR, and Cohn DH (1999) Trinucleotide Expansion Mutations in the Cartilage Oligomeric Matrix Protein (Comp) Gene. Hum. Mol. Genet 8, 123–128. [DOI] [PubMed] [Google Scholar]
- (49).Brcic J, and Plavec J (2015) Solution structure of a DNA quadruplex containing ALS and FTD related GGGGCC repeat stabilized by 8-bromodeoxyguanosine substitution. Nucleic Acids Res. 43, 8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and Development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Burge S, Parkinson G, Hazel P, Todd A, and Neidle S (2006) Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 34, 5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Wang Y, de los Santos C, Gao X, Greene K, Live D, and Patel DJ (1991) Multinuclear nuclear magnetic resonance studies of Na cation-stabilized complex formed by d(GGTTTTCGG) is solution. Implications for G-tetrad structures. J. Mol. Biol 222, 819. [DOI] [PubMed] [Google Scholar]
- (53).Wang Y, Jin R, Gaffney B, Jones R, and Breslauer K (1991) Characterization of 1H NMR of glycosidic conformations in the tetramolecular complex formed by d(GGTTTTTGG). Nucleic Acids Res. 19, 4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Schultze P, Macaya RF, and Feigon J (1994) Threedimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol 235, 1532. [DOI] [PubMed] [Google Scholar]
- (55).Mao X, Marky LA, and Gmeiner WH (2004) NMR structure of the thrombin-binding DNA aptamer stabilized by Sr2+. J. Biomol. Struct. Dyn 22, 25. [DOI] [PubMed] [Google Scholar]
- (56).Marathias VM, Wang KY, Kumar S, Pham TQ, Swaminathan S, and Bolton PH (1996) Determination of the number and locations of the manganese binding sites of DNA quadruplexes in solution by EPR and NMR in the presence and absence of thrombin. J. Mol. Biol 260, 378. [DOI] [PubMed] [Google Scholar]
- (57).Marathias VM, and Bolton PH (2000) Stuctures of the potassium-saturated 2:1 and intermediate 1:1 forms of a quadruplex DNA. Nucleic Acids Res. 28, 1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tang CF, and Shafer RH (2006) Engineering the quadruplex fold: nucleoside conformation determines both folding topology and molecularity in quanine quadruplexes. J. Am. Chem. Soc 128, 5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Cang X, Sponer J, and Cheatham TE (2011) Explaining the varied glycosidic conformational, G-tract length and sequence preferences for anti-parallel G-quadruplexes. Nucleic Acids Res. 39, 4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Hall K, Cruz P, Tinoco I, Jovin T, and van de Sande J (1984) Z-RNA - a left-handed RNA double helix. Nature 311, 584. [DOI] [PubMed] [Google Scholar]
- (61).Popenda M, Milecki J, and Adamiak RW (2004) High salt solution structure of a left-handed RNA double helix. Nucleic Acids Res. 32, 4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Pan F, Roland C, and Sagui C (2014) Ion distributions around left- and right-handed DNA and RNA duplexes: a comparative study. Nucleic Acids Res. 42, 13981–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Moradi M, Babin V, Roland C, and Sagui C (2013) Reaction path ensemble of the B-Z-DNA transition: a comprehensive atomistic study. Nucleic Acids Res. 41, 33–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Kiliszek A, Kierzek R, Krzyzosiak WJ, and Rypniewski W (2011) Crystal structures of CGG RNA repeats with implications for fragile X-associated tremor ataxia syndrome. Nucleic Acids Res. 39, 7308–7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Kumar A, Fang P, Park H, Guo M, Nettles KW, and Disney MD (2011) A Crystal Structure of a Model of the Repeating r(CGG) Transcript Found in Fragile Syndrome. ChemBioChem 12, 2140–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Malgowska M, Gudanis D, Kierzek R, Wyszko E, Gabelica V, and Gdaniec Z (2014) Distinctive structural motifs of RNA Gquadruplexes composed of AGG, CGG and UGG trinucleotide repeats. Nucleic Acids Res. 42, 10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Huang H, Suslov NB, Li NS, Shelke SA, Evans ME, Koldobskaya Y, Rice PA, and Piccirilli JA (2014) A Gquadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol 10, 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Warner KD, Chen MC, Song W, Strack RL, Thorn A, Jaffrey SR, and Ferre-D’Amare AR (2014) Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nat. Struct. Mol. Biol 21, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Zhang D, Huang T, Lukeman PS, and Paukstelis PJ (2014) Crystal structure of a DNA/BA2+ G-quadruplex containing a water-mediated C-terad. Nucleic Acids Res. 42, 13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Patel PK, Bhavesh NS, and Hosur RV (2000) NMR observation of a novel C-tetrad in the structure of the SV40 repeat sequence GGGCGG. Biochem. Biophys. Res. Commun 270, 967. [DOI] [PubMed] [Google Scholar]
- (71).Zamiri B, Mirceta M, Bomsztyk K, Macgregor R, and Pearson CE (2015) Quadruplex formation by both G-rich and Crich DNA strands of the C9orf72 (GGGGCC)8*(GGCCCC)8 repeat: effect of CpG methylation. Nucleic Acids Res. 43, 10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Han H, and Hurley L (2000) G-quadruplex DNA: a potential target for anti-cancer drug design. Trends Pharmacol. Sci 21, 136. [DOI] [PubMed] [Google Scholar]
- (73).Kerwin S (2000) G-quadruplex DNA as a target for drug design. Curr. Pharm. Des 6, 441. [DOI] [PubMed] [Google Scholar]
- (74).Hurley L, Wheelhouse R, Sun D, Kerwin S, Salazar M, Fedoroff M, Han FX, Han H, Izbicka E, and Von Hoff DD (2000) G-quadruplexes as targets for drug design. Pharmacol. Ther 85, 141. [DOI] [PubMed] [Google Scholar]
- (75).Bryan T, and Baumann P (2010) G-quadruplexes: from guanine gels to chemotherapeutics. Methods Mol. Biol 608, 1. [DOI] [PubMed] [Google Scholar]
- (76).Neidle S (2009) The structure of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol 19, 239. [DOI] [PubMed] [Google Scholar]
- (77).Wheeler TM, Sobczak K, Lueck j. D., Osborne RJ, Lin X, Dirksen RT, and Thornton CA (2009) Reveral of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science 325, 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Case DA et al. (2016) AMBER 16, University of California, San Francisco. [Google Scholar]
- (79).Zgarbová M, Šponer J, Otyepka M, Cheatham TE, Galindo-Murillo R, and Jurečka P (2015) Refinement of the Sugar- Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J. Chem. Theory Comput 11, 5723–36. [DOI] [PubMed] [Google Scholar]
- (80).Banas P, Hollas D, Zgarbova M, Jurecka P, Orozco M, Cheatham TE, Sponer J, and Otyepka M (2010) Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins. J. Chem. Theory Comput 6, 3836–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE, and Jurecka P (2011) Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J. Chem. Theory Comput 7, 2886–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Besseova I, Banas P, Kuhrova P, Kosinova P, Otyepka M, and Sponer J (2012) Simulations of A-RNA Duplexes. The Effect of Sequence, Solute Force Field, Water Model, and Salt Concentration. J. Phys. Chem. B 116, 9899–9916. [DOI] [PubMed] [Google Scholar]
- (83).Jorgensen WL, Chandrasekhar J, Madura J, Impey RW, and Klein M (1983) Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys 79, 926–935. [Google Scholar]
- (84).Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, and Pedersen LG (1995) A smooth particle mesh Ewald method. J. Chem. Phys 103, 8577–8593. [Google Scholar]
- (85).Berendsen HJC, Postma JPM, van Gunsteren WF, Di Nola A, and Haak JR (1984) Molecular Dynamics with coupling to an external bath. J. Chem. Phys 81, 3684–3690. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.