Sleep is integral to life1, and its insufficiency or disruption increases the risk of multiple pathological conditions, including cardiovascular disease2. Despite these associations, we know little about the cellular and molecular mechanisms by which sleep maintains cardiovascular health. Here we report that sleep regulates hematopoiesis and protects against atherosclerosis. Mice subjected to sleep fragmentation produce more Ly-6Chi monocytes, develop larger atherosclerotic lesions, and produce less hypocretin, a stimulatory and wake-promoting neuropeptide, in the lateral hypothalamus. Hypocretin controls myelopoiesis by restricting CSF1 production by hypocretin-receptor expressing pre-neutrophils in the bone marrow. Consequently, hypocretin-null and hematopoietic hypocretin-receptor-null mice develop monocytosis and accelerated atherosclerosis, which can be mitigated in sleep-fragmented mice via hypocretin supplementation. Together, these results identify a neuro-immune axis that links sleep causally to hematopoiesis and atherosclerosis.
Poor or insufficient sleep is an increasingly significant public health issue3, as nearly half of adults in the United States sleep fewer than the recommended seven to eight hours per day4. Lack of sleep increases risk of obesity5, diabetes6, cancer7 and cardiovascular disease2, but we know little about the underlying mechanisms that link sleep to disease.
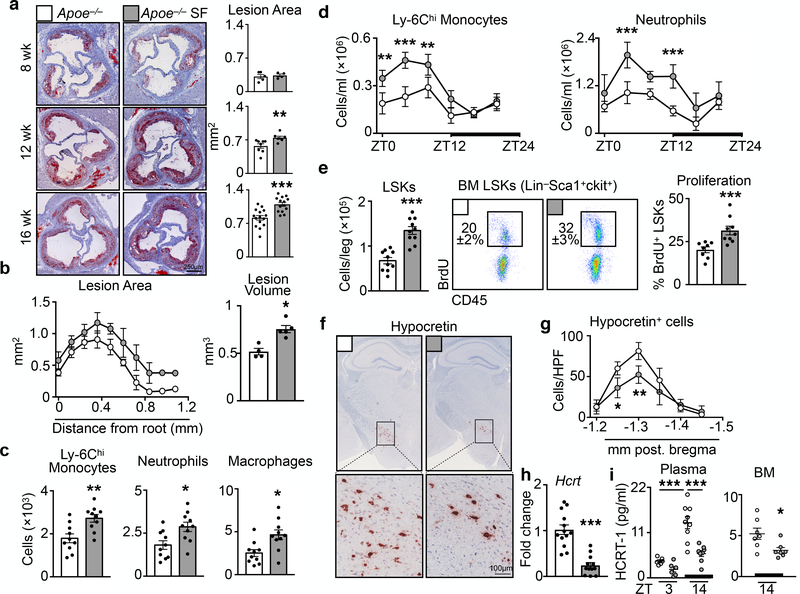
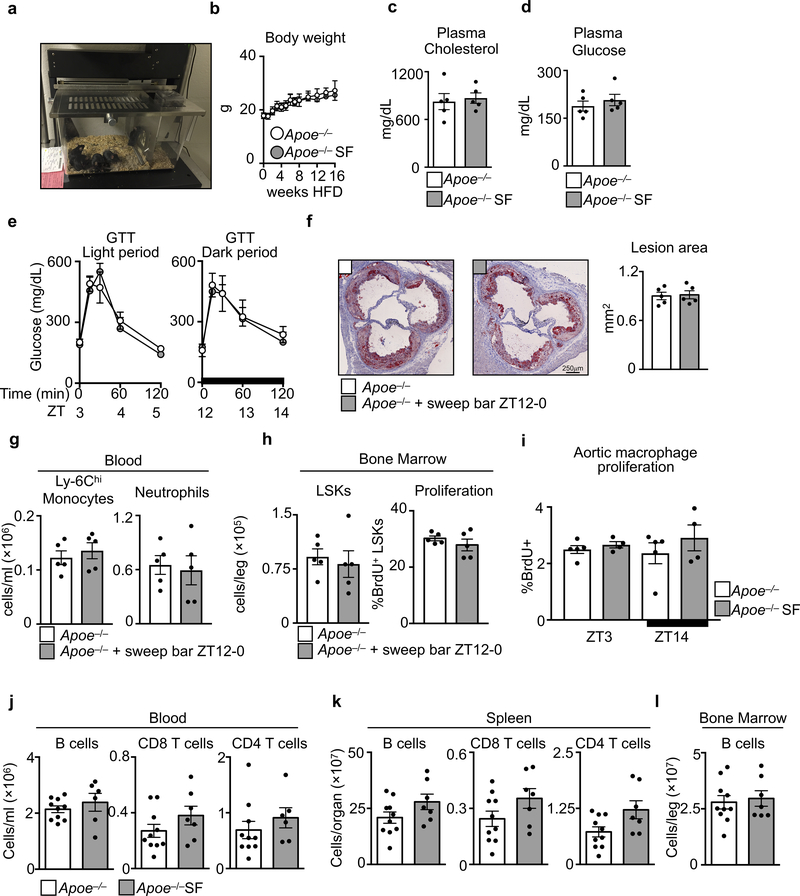
To investigate how sleep might protect against cardiovascular disease, we subjected atherosclerosis-prone Apoe−/− mice to chronic sleep fragmentation (SF)8 (Extended Data Fig. 1a and Video 1). We found no changes in body weight, plasma cholesterol, or glucose tolerance (Extended Data Fig. 1b-e), but the mice developed progressively larger atherosclerotic lesions compared to controls (Fig. 1a and Extended Data Fig. 1f-h). Not only did lesion volume increase in SF mice (Fig. 1b), but aortas from SF mice contained more Ly-6Chi monocytes, neutrophils, and macrophages (Fig. 1c), a change that did not result from increased aortic macrophage proliferation (Extended Data Fig. 1i).
Figure 1. Sleep fragmentation aggravates atherosclerosis, increases hematopoiesis and decreases hypothalamic hypocretin production.
Assessment of sleep fragmentation (SF) in Apoe−/− mice fed a high fat diet (HFD). a, Cross sections of aortic roots stained with oil-red-o and quantification of atherosclerotic lesion area after varying lengths of SF (n=5 8wk Apoe−/−; n=4 8wk Apoe−/−SF; n=8 12wk Apoe−/−; n=7 12wk Apoe−/−SF; n=15 16wk Apoe−/−; n=14 16wkApoe−/−SF). b, Measurement of lesion volume after 16 weeks of SF (n=4 Apoe−/−; n=5 Apoe−/−SF). c, Enumeration, by flow cytometry, of aortic neutrophils, macrophages and Ly-6Chi monocytes in the aorta of Apoe−/− mice and Apoe−/− mice having undergone SF for 16 weeks (n=10 per group). d, Quantification of circulating Ly-6Chi monocytes and neutrophils over 24 hours after 16 weeks of SF (ZT0=lights on, ZT12=lights off, n=4 per group) **p<0.01, ***p<0.001, Two-way ANOVA. e, Quantification of bone marrow Lin−Sca1+cKit+ (LSK) cells and BrdU incorporation after 16 weeks of SF (for LSKs/leg n=10 per group; for proliferation n=8 Apoe−/− and n=9 Apoe−/−SF). f, Immunohistochemical staining for hypocretin in the hypothalamus after 16 weeks of SF. g, Enumeration of hypocretin+ cells per high power field (HPF) in the hypothalamus after 16 weeks of SF (n=4 Apoe−/−; n=5 Apoe−/−SF, of 2 independent experiments) **p<0.01, Two-way ANOVA. h, Transcript expression of hypocretin (Hcrt) in the hypothalamus after 16 weeks of SF (n=12 per group). i, Measurement of hypocretin-1 (HCRT-1) protein in plasma and bone marrow fluid after 16 weeks of SF (n=6 per group plasma ZT3; n=9 plasma Apoe−/− ZT14; n=8 plasma Apoe−/−SF ZT14; n=7 per group BM ZT14) ***p<0.001 One-way ANOVA. Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests unless otherwise indicated.
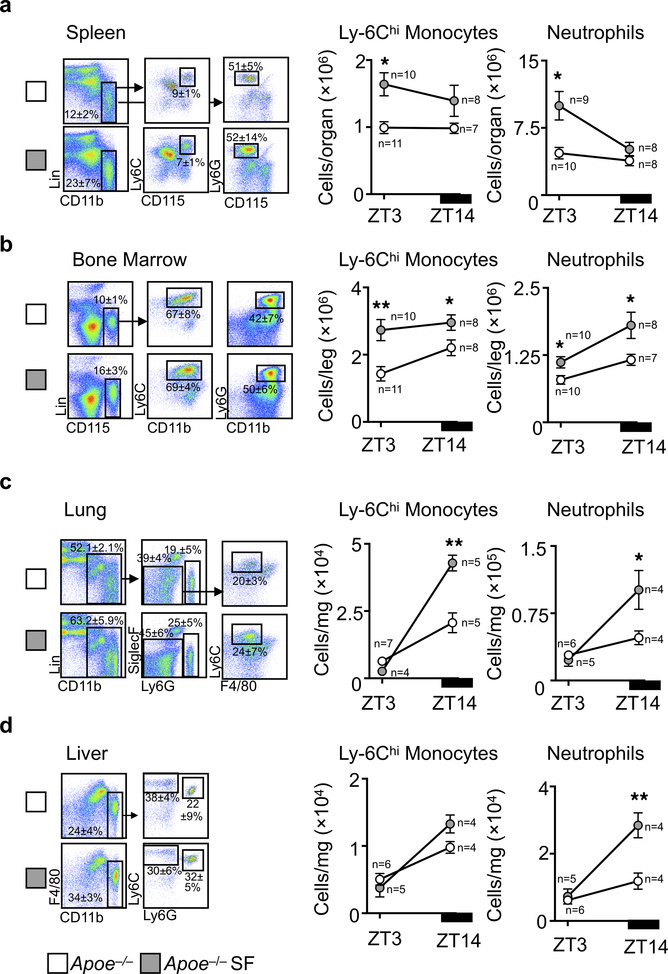
Leukocytosis predicts for cardiovascular disease9. Although myeloid cell numbers in blood of mice fluctuated according to a circadian pattern with a peak at Zeitgeber (ZT)5 and a nadir at ZT14 (Fig. 1d), mice subjected to SF had significantly more circulating Ly-6Chi monocytes and neutrophils during the light period. Rhythmicity analysis revealed that the circadian amplitude was increased (0.7 ± 0.16×105 vs. 1.5 ± 0.15×105, p = 0.02, for Ly-6Chi monocytes and 3.6 ± 0.17×105 vs. 5.1 ± 0.11×105, p=0.13, for neutrophils), but the period and phase were unaltered. SF did not change lymphocyte numbers (Extended Data Fig. 1j-l). To understand sleep’s impact on circadian leukocyte migration to tissue10,11, we profiled leukocytes in various organs at ZT3 and ZT14. Both control and SF mice had elevated Ly-6Chi monocyte and neutrophil levels in various tissues during the dark period, and these increases were higher in SF mice (Extended Data Fig. 2). These observations align with human studies that linked sleep curtailment or interruption with leukocyte numbers12,13.
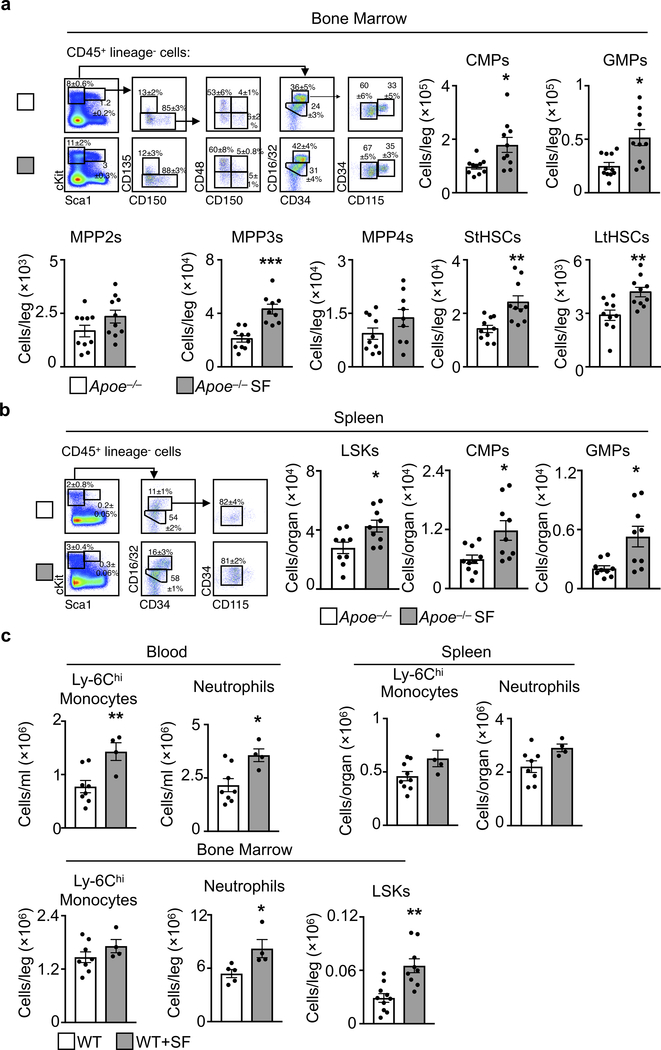
Next, we focused on hematopoiesis. In Apoe−/− mice, SF increased proliferation of Lineage− cKit+ Sca1+ (LSK) hematopoietic progenitors in the bone marrow (BM), and this increase corresponded with a ~two fold higher number of BM LSK cells (Fig. 1e) and other progenitor subsets (Extended Fig. 3a). The spleens of SF mice contained more LSKs and GMPs, indicating heightened extramedullary hematopoiesis (Extended Data Fig. 3b). SF promoted myelopoiesis not only in Apoe−/− HFD-fed mice but also in C57BL/6 mice fed a chow diet (Extended Data Fig. 3c). Together, these data show that SF boosts myeloid-biased hematopoiesis.
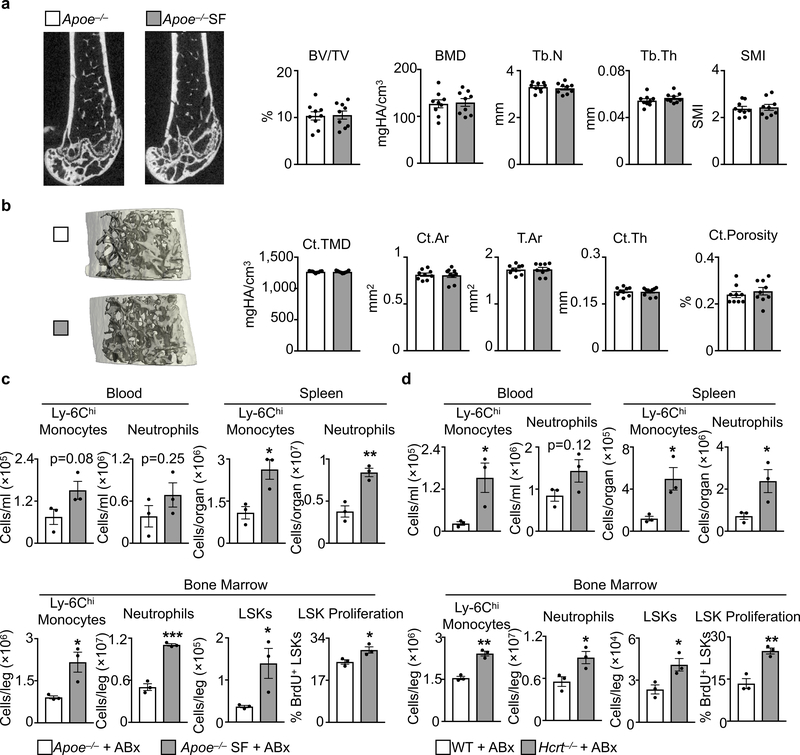
Mice subjected to SF had normal bone structure (Extended Data Fig. 4a, b), and leukocytosis persisted even after prolonged antibiotic treatment (Extended Data Fig. 4c), suggesting that enhanced myelopoiesis was not driven by either physical alterations to the bone or the microbiome, respectively. Because stress activates the sympathetic nervous system (SNS), which can heighten hematopoiesis14, we wondered whether SF-induced myelopoiesis likewise depends on SNS activation, but found no evidence for such a mechanism (Extended Data Fig. 5a-d). Nevertheless, SF mice were more anxious (Extended Data Fig. 5e-g), demonstrating that mice do not easily habituate to sleep fragmentation.
We then focused on the hypothalamus, and specifically on expression of transcripts that encode sleep-regulating proteins (Extended Data Fig. 5h-j). SF decreased hypothalamic hypocretin (Hcrt, also known as orexin) expression (Fig. 1f-h), correlating with reduced levels of the isoform hypocretin-1 in plasma and bone marrow (Fig 1i). Hypothalamic hypocretin reduced gradually and correlated inversely with leukocytosis (Extended Data Fig. 6a). We did not see alterations in hypocretin’s co-transmitter dynorphin15 (Extended Data Fig. 6b-e) or any evidence for death of hypocretin-producing neurons (Extended Data Fig. 6f), suggesting specific repression of hypocretin, potentially mediated by neuropeptide-Y16 (Extended Data Fig. 5h). Furthermore, sleep recovery following SF restored hypothalamic hypocretin content and normalized myelopoiesis (Extended Data Fig. 6g-i). These data intrigued us because hypocretin mediates metabolism, sleep, and appetite17, while autoimmune destruction of hypocretin neurons causes narcolepsy18 and a pro-inflammatory immune signature19. In keeping with hypocretin’s function in appetite promotion, SF mice consumed less food than controls (Extended Data Fig. 5k).
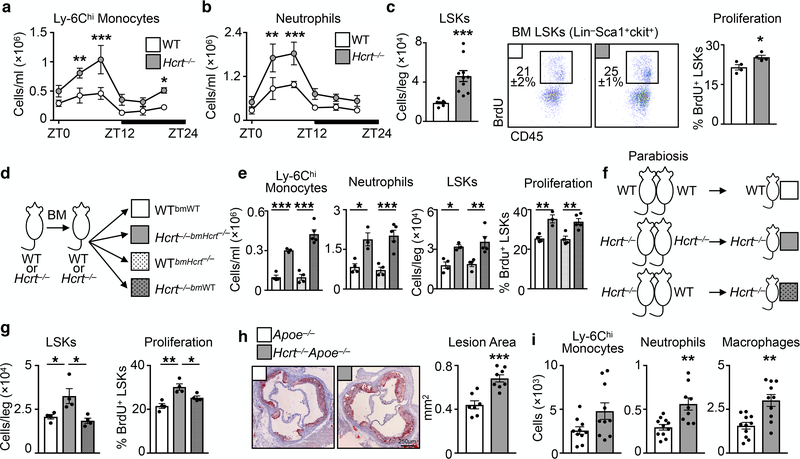
In humans, reduced plasma hypocretin associates with risk of myocardial infarction (MI)20, heart failure21, and obesity22. Likewise, studies suggest narcoleptic patients have heightened risk of heart disease23. In mice, deleting hypocretin severely fragments sleep/wake cycles, causes cataplexy, and promotes diet-induced obesity24,25. Moreover, deleting hypocretin receptor-2 worsens post-MI healing21. We therefore profiled leukocytes in hypocretin-null (Hcrt−/−) mice, and noted more Ly-6Chi monocytes and neutrophils in the blood, spleen, and bone marrow relative to WT controls (Fig. 2a-b and Extended Data Fig. 7). Rhythmicity analysis of Hcrt−/− mouse blood revealed that Ly-6Chi monocytes and neutrophils had elevated circadian amplitudes (1.8 ± 0.32×105 vs. 3.7 ± 0.55×105, p=0.02, for Ly-6Chi monocytes and 4.1 ± 0.69×105 vs.8.1 ± 0.11×105, p=0.03, for neutrophils), but the period and phase were unchanged. Hcrt−/− mouse bone marrow had more hematopoietic progenitors along with heightened LSK proliferation (Fig. 2c and Extended Data Fig. 7). As in the SF mice, accelerated hematopoiesis in hypocretin-deficient mice did not appear to depend on the microbiome (Extended Data Fig. 4d). These results suggest that sleep regulates hematopoiesis via hypocretin.
Figure 2. Hypocretin suppresses hematopoiesis and atherosclerosis.
Assessment of hematopoiesis in hypocretin-deficient (Hcrt−/−) mice. Enumeration of a, Ly-6Chi monocytes and b, neutrophils in the blood of Hcrt−/− and wild type (WT) mice over 24 hours (n=3 per group) **p<0.01, ***p<0.001, Two-way ANOVA. c, Enumeration of bone marrow LSK cells and BrdU incorporation in WT and Hcrt−/− (for LSKs/leg n=8 WT and n=10 Hcrt−/−; for proliferation n=4 per group). d, Schematic of chimeric models. e, Assessment of blood Ly-6Chi monocytes and neutrophils, bone marrow LSK cells and BrdU incorporation in chimeric mice (n=4 WTbmWT mice; n=3 Hcrt−/−bmHcrt−/− mice; n=4 WTbmHcrt−/− mice; n=5 Hcrt−/−bmWT mice) *p<0.05, **p<0.01, ***p<0.001 One-way ANOVA. f, Schematic of parabiosis models. g, Enumeration of LSKs and BrdU incorporation in the bone marrow of parabiosis mice (n=4 per group) *p<0.05, One-way ANOVA. h, Cross section images of aortic roots stained with oil-red-o and quantification of atherosclerotic lesion area in Apoe−/− and Hcrt−/−Apoe−/− mice fed a high fat diet for 16 weeks (n=7 Apoe−/−; n=8 Hcrt−/−Apoe−/−). i, Aortic myeloid cells in Apoe−/− and Hcrt−/−Apoe−/− mice (for Ly-6Chi monocytes n=10 per group; for neutrophils n=11 Apoe−/− and n=9 Hcrt−/−Apoe−/−; for macrophages n=11 Apoe−/− and n=10 Hcrt−/−Apoe−/−). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests unless otherwise indicated.
Next, we tested whether hypocretin can affect hematopoiesis and atherosclerosis. We found that the hypothalamus produced nearly all the hypocretin (Extended Fig. 8a, b). Sixteen weeks of sleep fragmentation did not alter hypocretin production in the bone and bone marrow (Extended Fig. 8c), indicating that the hypothalamus was the relevant source affected by sleep. Indeed, we found substantial concentrations of the neuropeptide in the cerebrospinal fluid, plasma, and bone marrow fluid of WT but not Hcrt−/− mice, and we detected high HCRT-1 levels in the plasma and bone marrow fluid of Hcrt−/− mice after injecting HCRT-1 into the cerebrospinal fluid of the cisterna magna (Extended Fig. 8d, e). Consequently, we generated chimeric mice lacking hypocretin production in either the non-hematopoietic compartment (including the hypothalamus) or hematopoietic cells (Fig. 2d), and found heightened hematopoiesis in Hcrt−/− mice containing wild-type (WT) BM cells (Fig. 2e). We also placed Hcrt−/− mice in parabiosis with WT mice (Fig. 2f), noting that WT-partnered Hcrt−/− mice had suppressed hematopoiesis compared to Hcrt−/−-partnered Hcrt−/− mice (Fig. 2g). These data suggest that hypothalamus-produced hypocretin can enter the circulation to affect hematopoiesis in the distal bone marrow26,27. To determine if hypocretin deficiency aggravates atherosclerosis, we generated Hcrt−/−Apoe−/− mice, which had larger lesions with more aortic leukocytes than Apoe−/− controls (Fig. 2h, i).
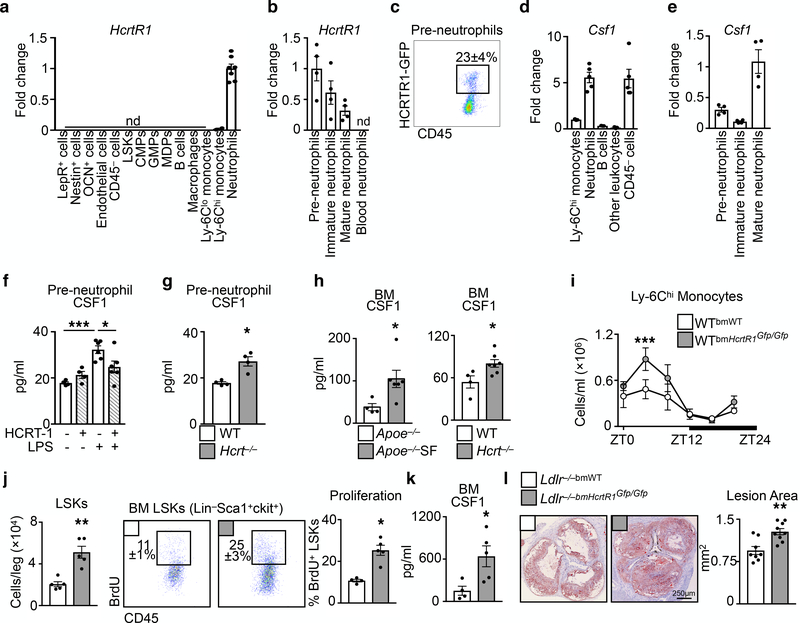
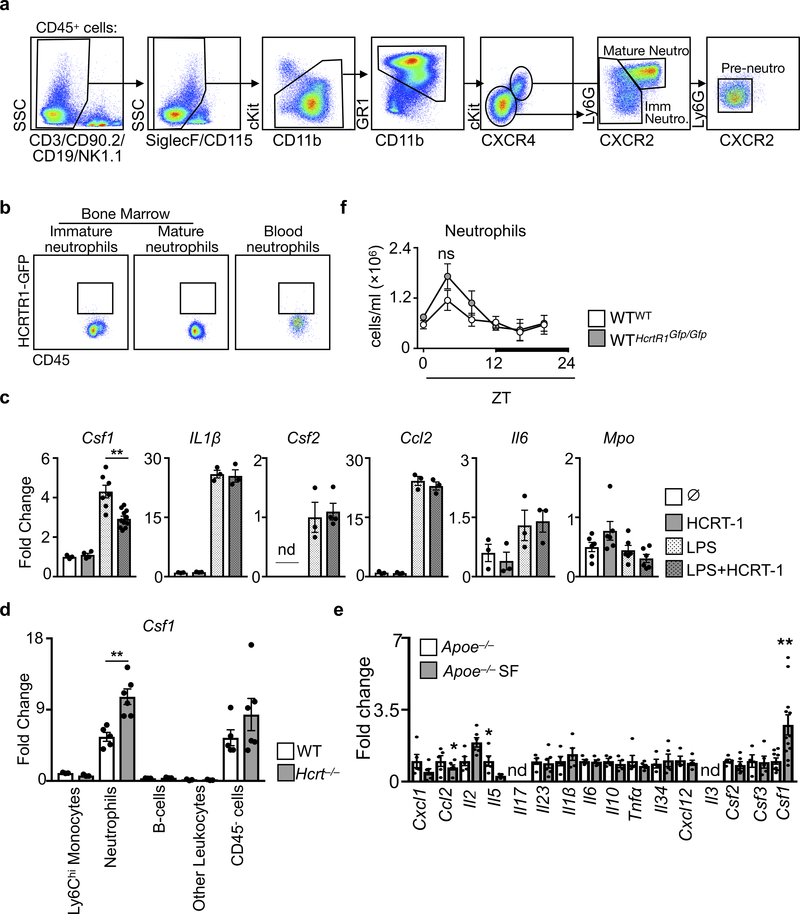
Having identified a link between hypocretin, hematopoiesis, and atherosclerosis, we examined the underlying mechanism. Although hypocretin receptor-1 (HcrtR1) and HcrtR2 expression was highest in the hypothalamus, we detected HcrtR1 in multiple tissues, including the bone marrow (Extended Data Fig. 8f,g). However, adding hypocretin to granulocyte-macrophage colony-forming unit (CFU-GM) cultures had no effect on hematopoiesis in vitro (Extended Data Fig. 8h). We therefore sorted 14 different cell types from the bone marrow and found that neutrophils, but not other cells, expressed HcrtR1 transcript (Fig. 3a) and protein (Extended Data Fig. 8i). We next sorted neutrophils by maturation stage28 (Extended Data Fig. 9a) and found that pre-neutrophils expressed HcrtR1 most abundantly (Fig. 3b). Using knockout-reporter HcrtR1Gfp/Gfp mice29 we identified a subset of bone marrow-resident pre-neutrophils as GFP+ and thus able to express the receptor (Fig. 3c and Extended Data Fig. 9b)
Figure 3. Hypocretin controls pre-neutrophil CSF1 production in the bone marrow.
Hypocretin receptor-1 (Hcrtr1) mRNA in cells sorted from bone marrow (n=4 except neutrophils n=7). b, HcrtR1 mRNA expression in bone marrow and blood neutrophil populations (n=4). c, Flow cytometry plot of HCRTR1+ pre-neutrophils in the bone marrow of WT mice transplanted with HcrtR1Gfp/Gfp BM. d, Colony stimulating factor-1 (Csf1) expression in sorted bone marrow cells (for Ly-6Chi monocytes, B cells, and other leukocytes n=3; for neutrophils and CD45− cells n=5). e, Csf1 expression in sorted bone marrow neutrophil populations (n=4). f, CSF1 production by pre-neutrophils sorted from WT mice exposed to LPS and/or HCRT-1 (for untreated and HCRT-1 n=4 per group; for LPS and LPS+HCRT-1 n=6 per group) *p<0.05, ***p<0.001, One-way ANOVA. g, CSF1 production by pre-neutrophils sorted from WT and Hcrt−/− mice (n=4 per group). h, BM CSF1 concentration in Apoe−/− SF and Hcrt−/− mice (n=4 Apoe−/−; n=6 Apoe−/−SF; n=4 WT; n=7 Hcrt−/−). i, Enumeration of blood Ly-6Chi monocytes over 24 hours in WTbmHcrtR1Gfp/Gfp mice (n=3 per group) ***p<0.001, Two-way ANOVA. j, BM LSKs and LSK proliferation in WT mice transplanted with WT or HcrtR1Gfp/Gfp BM cells (for LSKs/leg n=5 per group; for proliferation n=4 WTbmWT and n=5 WTbmHcrtR1Gfp/Gfp). k, BM CSF1 concentration in WT mice transplanted with WT or HcrtR1Gfp/Gfp BM cells (n=4 WTbmWT; n=5 WTbmHcrtR1Gfp/Gfp ). l, Cross section images of aortic roots stained with oil-red-o and quantification of atherosclerosis in Ldlr−/− mice transplanted with WT or HcrtR1Gfp/Gfp BM cells and fed a high cholesterol diet for 12 weeks (n=8 Ldlr−/−bmWT; n=9 Ldlr−/−bmHcrtR1Gfp/Gfp ). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests unless otherwise indicated.
The observation that BM CXCR4+CXCR2− pre-neutrophils expressed HcrtR1 was intriguing because these cells reside in close proximity to hematopoietic progenitors28. Moreover, neutrophils produce substantial colony stimulating factor-1 (CSF1), which promotes BM myeloid-biased hematopoiesis30 (Fig. 3d). In part because pre-neutrophils sorted from WT mice expressed less Csf1 compared to mature neutrophils (Fig. 3e), we tested whether hypocretin can control hematopoiesis through HcrtR1+ pre-neutrophil-derived CSF1. In vitro, hypocretin limited pre-neutrophils’ capacity to produce CSF1 in response to LPS (Fig. 3f and Extended Data Fig. 9c). Ex vivo, pre-neutrophils and neutrophils sorted from Hcrt−/− mice contained more Csf1 mRNA and secreted more CSF1 protein than cells sorted from WT mice (Fig. 3g and Extended Data Fig 9d). In vivo, we found heightened CSF1 in the bone marrow of Hcrt−/− mice and Apoe−/− mice subjected to SF (Fig. 3h). Indeed, SF increased Csf1 production without substantially changing other mediators (Extended Data Fig. 9e).
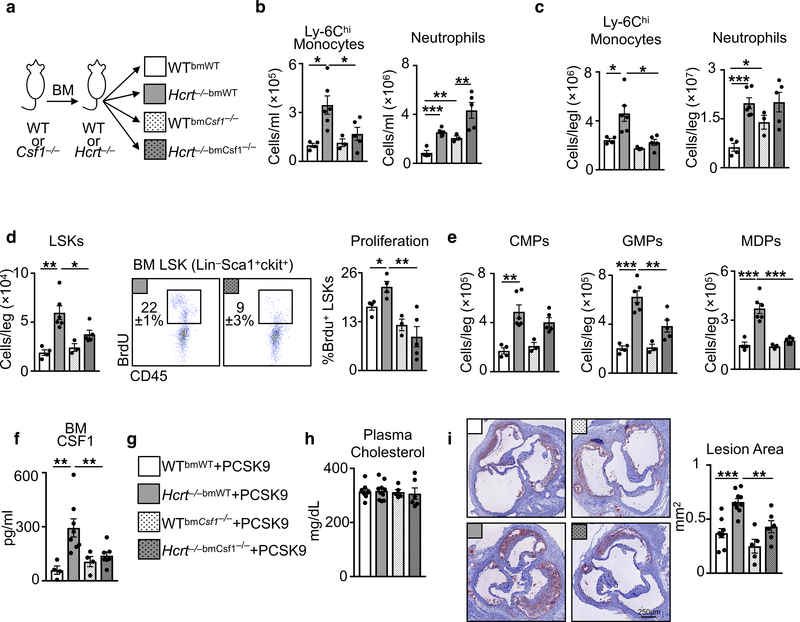
Because BM non-hematopoietic cells do not express HcrtR1 (Fig. 3a) but produce CSF1 (Fig. 3d), we asked whether hypocretin-mediated control of leukocyte-derived CSF1 is important. First, we lethally irradiated WT mice and transplanted them with BM cells from either WT or HcrtR1 knockout-reporter, HcrtR1Gfp/Gfp, mice. Chimeras with HcrtR1Gfp/Gfp bone marrow developed monocytosis (Fig. 3i), along with increased LSK numbers, LSK proliferation (Fig. 3j), and CSF1 in the bone marrow (Fig. 3k). Second, we transplanted Ldlr−/− mice with HcrtR1Gfp/Gfp bone marrow and noted augmented atherosclerosis (Fig. 3l). Third, we lethally irradiated WT or Hcrt−/− mice and transplanted them with BM cells from either WT or Csf1−/− mice (Extended Data Fig. 10a). As expected, WT mice had relatively few Ly-6Chi monocytes regardless of BM source. However, while Hcrt−/− mice reconstituted with WT BM cells developed leukocytosis, Hcrt−/− mice reconstituted with Csf1−/− BM cells had relatively few Ly-6Chi monocytes, limited hematopoietic progenitors and proliferation, and reduced CSF1 concentrations (Extended Data Fig. 10b-f). Curiously, neutrophil numbers remained high in Hcrt−/− mice reconstituted with Csf1−/− BM, indicating that neutrophilia in Hcrt−/− mice was not directly related to Hcrt-leukocyte control of CSF1, an observation we confirmed in WTbmHcrtR1Gfp/Gfp chimeras (Extended Data Fig. 9f). Fourth, we induced atherogenesis in WT and Hcrt−/− chimeras reconstituted with either WT or Csf1−/− BM by over-expressing PCSK9 and feeding the mice a high cholesterol diet. We found that Hcrt−/−bmWT mice developed larger lesions than Hcrt−/−bmCsf1−/− (Extended Data Figure 10g-i). These data agree with the hypothesis that hypocretin controls monocytosis and atherosclerosis by mediating neutrophil CSF1 production.
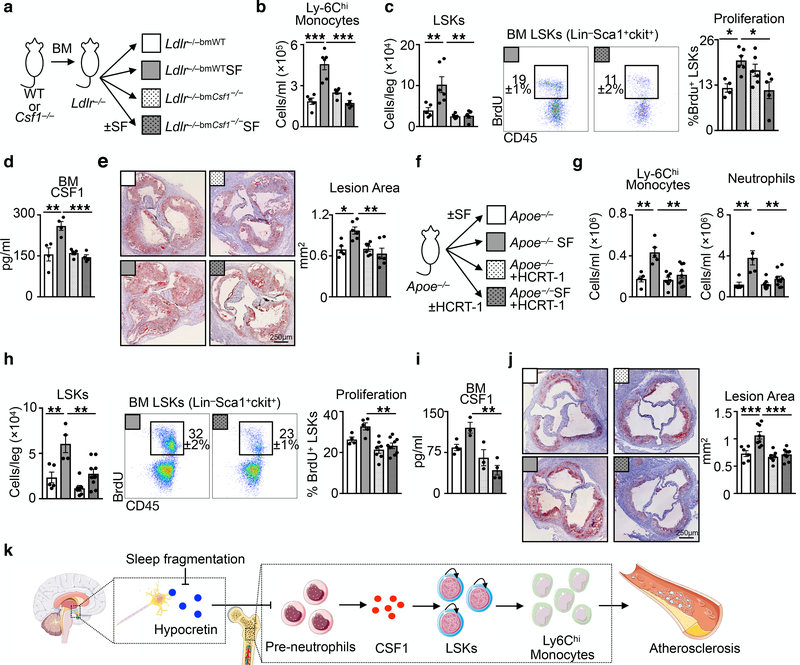
Finally, we sought to explore hypocretin’s role in atherosclerosis and sleep fragmentation. We generated Ldlr−/− mice with either WT or Csf1−/− bone marrow, and subjected the chimeras to SF (Fig. 4a). Whereas Ldlr−/− mice with WT bone marrow subjected to SF developed monocytosis via increased hematopoiesis, LSK proliferation, and more CSF1 in the bone marrow, leading to bigger atherosclerotic lesions, absence of BM-derived Csf1 reduced these parameters in SF mice (Fig. 4b-e). Moreover, we delivered hypocretin to the periphery of Apoe−/− mice subjected to SF (Fig. 4f). Compared to controls, Apoe−/− mice subjected to SF and receiving hypocretin had fewer monocytes and neutrophils in the blood (Fig. 4g), reduced BM LSKs numbers and proliferation (Fig. 4h), lower CSF-1 levels in the bone marrow (Fig. 4i), and smaller lesions (Fig. 4j). These results demonstrate that hypocretin loss during SF aggravates hematopoiesis and atherosclerosis.
Figure 4. Hematopoietic CSF1 deletion or hypocretin supplementation protects against SF-induced hematopoiesis and atherosclerosis.
a, Schematic of chimeric Ldlr−/− mice with WT or CSF1−/− hematopoietic cells subjected to SF. b, Enumeration of blood Ly-6Chi monocytes in chimeric mice (n=5 LDLr−/−bmWT; n=6 LDLr−/−bmWTSF; n=6 Ldlr−/−bmCsf1−/−; n=5 Ldlr−/−bmCsf1−/−SF ). c, Quantification of bone marrow LSKs and BrdU incorporation in chimeric mice (for LSKs/leg n=5 LDLr−/−bmWT; n=6 LDLr−/−bmWTSF; n=6 Ldlr−/−bmCsf1−/−; n=5 Ldlr−/−bmCsf1−/−SF; for proliferation n=4 LDLr−/−bmWT; n=6 LDLr−/−bmWTSF; n=6 Ldlr−/−bmCsf1−/−; n=5 Ldlr−/−bmCsf1−/−SF). d, BM CSF1 concentration in chimeric mice (n=4 LDLr−/−bmWT; n=4 LDLr−/−bmWTSF; n=5 Ldlr−/−bmCsf1−/−; n=4 Ldlr−/−bmCsf1−/−SF). e, Cross section images of aortic roots stained with oil-red-o and assessment of atherosclerosis in chimeric mice after 16 weeks of high cholesterol diet (n=5 LDLr−/−bmWT; n=6 LDLr−/−bmWTSF; n=6 Ldlr−/−bmCsf1−/−; n=6 Ldlr−/−bmCsf1−/−SF). f, Schematic of mice subjected to SF and receiving hypocretin-1 or saline via osmotic mini-pumps for 8 weeks. g, Enumeration of blood Ly-6Chi monocytes and neutrophils (n=5 Apoe−/−; n=5 Apoe−/−SF; n=7 Apoe−/−+HCRT-1; n=9 Apoe−/−SF+HCRT-1). h, LSKs and BrdU incorporation in the bone marrow of SF mice with HCRT-1 supplementation (n=5 Apoe−/−; n=4 Apoe−/−SF; n=7 Apoe−/−+HCRT-1; n=9 Apoe−/−SF+HCRT-1). i, BM CSF1 levels (n=4 Apoe−/−; n=3 Apoe−/−SF; n=3 Apoe−/−+HCRT-1; n=4 Apoe−/−SF+HCRT-1). j, Cross section images of aortic roots stained with oil-red-o and quantification of atherosclerotic lesion area (n=6 Apoe−/−; n=8 Apoe−/−SF; n=7 Apoe−/−+HCRT-1; n=9 Apoe−/−SF+HCRT-1). k, Model of sleep’s role in regulating hypocretin production, hematopoiesis and atherosclerosis. The illustration was modified from Servier Medical Art (http://smart.servier.com/), licensed under a Creative Common Attribution 3.0 Generic License. Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, One-way ANOVA.
Our data indicate that sleep protects against atherosclerosis. Undisturbed sleep maintains proper hypothalamic release of hypocretin, which limits pre-neutrophil CSF1 in the bone marrow, thereby curtailing hematopoiesis and atherosclerosis. This neuro-immune axis directly connects sleep to immune function and cardiovascular disease (Fig. 4k).
Methods
Animals.
C57BL/6J (wild type, WT), Apoetm1Unc (Apoe−/−), Ldlrtm1Her/J (Ldlr−/−) and Csf1op (Csf1−/−) mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). Hcrt−/− mice31 were kindly provided by Dr. Thomas Scammell (Division of Sleep Science, Harvard Medical School) and bred in-house. HcrtR1Gfp/Gfp mice29,32 were kindly provided by Dr. Anne Vassalli (Department of Physiology, University of Lausanne). Stromal cell reporter mice Nestin-GFP33,34, LeptinRcre-R26-EYFP35,36, and OCN-GFPtopaz−37 were kindly provided by Dr. David Scadden (Department of Stem Cell and Regenerative Biology, Harvard University and the Center for Regenerative Medicine, Massachusetts General Hospital). Genotyping for each strain was performed as described on the Jackson Laboratory website. Age- and sex-matched animals were used starting at 8–12 weeks of age. For experiments involving Apoe−/− and Ldlr−/− mice, females mice were used. For all other experiments, both male and female mice were used. All mice were group housed on a 12:12-h light-dark cycles at 22°C with free access to food and water. Where appropriate, animals were randomly assigned to interventions. All protocols were approved by the Animal Review Committee at Massachusetts General Hospital (Protocol No. 2011N000035 and 2015N000044) and were in compliance with relevant ethical regulations.
In vivo interventions.
Diet:
Apoe−/− mice were fed a high-fat diet (HFD, Harlan Teklad, TD.88137) and Ldlr−/− mice were fed a high-cholesterol diet (HCD, Research Diets D12331). 5 × 1011U of adv-PCSK9 virus was injected i.v and mice began consuming a high-cholesterol diet (HCD, Research Diets D12331) immediately after virus injection. Unless otherwise indicated, mice were fed a regular chow diet.
Sleep fragmentation:
For sleep fragmentation studies, mice were placed in a sleep fragmentation chamber (Lafayette Instrument, Lafayette, IN) at the same time as initiation of special diet feeding. The sweep bar moved along the bottom of the cage every 2 minutes during the light cycle (ZT0–12). The sweep bar automatically shut off and was stationary during the dark cycle (ZT12–24). Control mice that received undisturbed sleep were placed in sleep fragmentation chambers with stationary sweep bars. For the control experiment in Extended Data Figure 1f-h, the sweep bar moved along the bottom of the cage every 2 minutes during the dark cycle (ZT12–0) and automatically shut off and was stationary during the light cycle (ZT0–12)
Interventions:
For studies on microbiota, mice were treated with an antibiotic cocktail (0.1% Ampicillin, 0.1% Metronidazole, 0.05% Vancomycin, and 0.1% Neomycin) in drinking water for 4 weeks. Antibiotic-containing drinking water was changed weekly. For hypocretin supplementation studies, HFD fed Apoe−/− control and SF mice were implanted subcutaneously with osmotic mini-pumps (Alzet, Cupertino, CA) 8 weeks after experiment initiation. Mice were randomly assigned a pump containing either saline or Hypocretin-1 (Sigma Aldrich, St. Louis, MO) delivered at 50nmol/hr/kg. Pumps were replaced after 4 weeks.
Parabiosis:
The procedure was conducted as previously described38–40. Briefly, age-, sex- and weight-matched animals were used and housed together for at least 14 days prior to surgery. Each mouse’s corresponding lateral aspects were shaved, incisions were made from the fore-limb joint to the hind-limb joint and the subcutaneous fascia was bluntly dissected to create 0.5 cm of free skin. Fore- and hind-limb joints were joined and the dorsal and ventral skins were approximated by continuous suture using mononylon 5.0 (Ethicon).
Bone marrow transplantation:
Mice were lethally irradiated (950cGy) and reconstituted with 6×106 BM cells injected i.v. to generate chimera groups. Mice were allowed to recover for 8 weeks prior to further manipulation.
Intra-cisterna magna injection:
The procedure was conducted as previously described41. Mice were anesthetized and the skin of the neck was shaved and disinfected with 70% ethanol. Mice were placed in a stereotactic frame (Stoelting, Wood Dale,IL) to secure their heads. A skin incision was made at the back of the neck and muscle layers were retracted to expose the cisterna magna. Using a Hamilton syringe 2μg of Hypocretin-1 dissolved in 5μl of PBS was injected into the CSF-filled cisterna magna compartment. After injection the needle was left in place for 5 minutes before removal to prevent back-flow. The skin of the neck was sutured closed and mice were allowed to recover.
Cells.
Cell collection:
Peripheral blood was collected by retro-orbital bleeding and red blood cells were lysed in RBC lysis buffer (Biolegend, San Diego, CA). Aortas, lungs, livers, and hearts were excised after PBS (Thermo Fisher Scientific, Waltham, MA) perfusion, minced and digested with 450 U/ml collagenase I, 125 U/ml collagenase XI, 60 U/ml DNase I and 60 U/ml hyaluronidase (Sigma-Aldrich, St. Louis, MO) in PBS for 20 minutes (liver), 40 minutes (aorta) or 1hr (heart and lung) at 37°C. Spleens were crushed through a 40μm cell strainer and red blood cells were lysed with RBC lysis buffer. Bone marrow cells were collected by flushing bones with PBS after which a single cell suspension was created by passing cells through a 26-gage needle and red blood cells were lysed with RBC lysis buffer. Total viable cell numbers were counted using trypan blue (Cellgro, Mediatech, Inc., Manassas, VA) or counting beads (Thermo Fisher Scientific, Waltham, MA).
Cell sorting:
BM cell suspensions were stained to identify indicated cell population and cells were sorted on a FACS Aria II cell sorter (BD Biosciences, Billerica, MA) directly into collecting medium.
Flow Cytometry:
Single cell suspensions were stained in PBS supplemented with 2% FBS and 0.5% BSA. The following monoclonal antibodies were used for flow cytometric analysis: anti-CD45 (BioLegend, clone30-F11, Cat#103147, Lot#B243834), anti-CD45.1 (BioLegend, clone A20, Cat#110708), anti-CD45.2 (BioLegend, clone 104, Cat#109802), anti-CD3 (BioLegend, clone 17A2, Cat#100206), anti-CD90.2 (BioLegend, clone 53–2.1, Cat#105308, Lot#B260050), anti-CD19 (BioLegend, clone 6D5, Cat#115508, Lot#B226581), anti-B220 (BD Biosciences, clone RA3–6B2, Cat#553089, Lot#6012954), anti-NK1.1 BioLegend, clone PK136, Cat#108708), anti-Ly6G (BioLegend, clone 1A8, Cat127614#, Lot#B259670), anti-Ly6C (BioLegend, AL-21, Cat#128006, Lot#B247728), anti-MHCII (BioLegend, clone M5/114.152, Cat#107602, Lot#B217859), anti-F4/80 (Biolegend, clone BM8, Cat#123114, Lot#B237342), anti-CD11b (BioLegend, clone M1/70, Cat#101226, Lot#B238268), anti-CD115 (BioLegend, clone AFS98, Cat#135517, Lot#B265220), anti-Ter119 (BioLegend, clone TER-119, Cat#116208, Lot#B220899), anti-CD34 (eBioscience, clone RAM34, Cat#11–0341-85, Lot#E00265–1634), anti-CD49b (BioLegend, clone DX5, Cat#1089008, Lot#B258302), ant-CD11c (BioLegend, clone N418, Cat#117310, Lot#B206713), anti-IL7Rα (BioLegend, clone SB/199, Cat#121112, Lot#B189668), anti-CD16/32 (BioLegend, clone 93, Cat#101324, Lot#B250025), anti-CD150 (BioLegend, clone TC15–12F12.2, Cat#115922, Lot#B220585), anti-cKit (BioLegend, clone 2B8, Cat#105814, Lot#B252918), anti-CD135 (BioLegend, clone A2F10, Cat#135310, Lot#B234045), anti-CD48 (BioLegend, clone HM48–1, Cat#103426, Lot#B236445), anti-Sca1 (BioLegend, clone D7, Cat#108126, Lot#B234288), anti-CD8 (BD Bioscience, clone 53–6.7, Cat#553035, Lot#2296946), anti-CD4 (BioLegend, clone GK1.5, Cat#100428, Lot#B237336), anti-SiglecF (BD Pharmingen, clone E50–2440, Cat#562680, Lot#7054789), anti-CXCR4 (Invitrogen, clone 2B11, Cat#12–9991-81, Lot#B251481), anti-CXCR2 (BioLegend, clone SA044G4, Cat#149307, Lot#B251481), anti-BrdU (eBioscience, clone BU20A, Cat#17–5071-42, Lot#4319920). All antibodies were used in a 1:700 dilution.Viable cells were identified as unstained with Zombie Aqua (Biolegend, CA). Cells were identified as (i) Ly6Chigh monocytes (CD45+Lin1−CD11b+CD115+F4/80−Ly-6Chigh), (ii) neutrophils (CD45+Lin1−CD11b+Ly-6G+F4/80−), (iii) macrophages (CD45+Lin1−CD11b+F4/80+Ly-6Clow), (iv) B cells (CD45+B220+CD19+F4/80−CD11b−), (v) CD4 T cells (CD45+CD3+CD90+CD4+CD11b−F4/80−), (vi) CD8 T cells (CD45+CD3+CD90+CD8+CD11b−F4/80−), (vii) LSKs (CD45+Lin2−cKit+Sca1+), (viii) MPP4 (CD45+Lin2−cKit+Sca1+CD135+CD150−), (ix) MPP3 (CD45+Lin2−cKit+Sca1+CD135+CD150−CD48+), (x) MPP2 (CD45+Lin2−cKit+Sca1+CD135+CD150+CD48+), (xi) short-term hematopoietic stem cell (StHSC, CD45+Lin2−cKit+Sca1+CD135+CD150−CD48−), (xii) LtHSC (CD45+Lin2−cKit+Sca1+CD135+CD150+CD48−), (xiii) common myeloid progenitor (CMP, CD45+Lin2−cKit+Sca1−CD34+CD16/32mid), (xiv) granulocyte/macrophage progenitor (GMP, CD45+Lin2−cKit+Sca1−CD34+CD16/32highCD115−), (xv) monocyte-dendritic cell progenitor (MDP, CD45+Lin2−cKit+Sca1−CD34+CD16/32highCD115+), for neutrophils populations (CD45+CD3−CD90.2−CD19−NK1.1−SiglecF−CD115−cKitlowCD11b+Gr-1+) (xvi) pre-neutrophils were then separated as cKitintCXCR4+CXCR2−, (xvii) immature neutrophils as ckit−CXCR4−CXCR2−, and (xviii) mature neutrophils as ckit−CXCR4−CXCR2+Ly6G+. Lineages were defined as Lin1: CD3, CD90.2, CD19, NK1.1, Ter119 and Lin2: B220, CD19, CD49b, Ter119, CD90.2, CD11b, CD11c, Ly-6G, IL7Rα. Osteolineage cells (OCN+) cells were identified as CD45−Ter119−CD31−GFP+ from the bone fraction of OCN-GFP mice. Leptin receptor (LepR+) cells were identified as CD45−Ter119−CD31−YFP+ form the bone marrow fraction of LeptinRcre-R26-EYFP. Nestin+ cells were identified as CD45−Ter119−CD31−GFP+ form the bone and bone marrow fraction of Nestin-GFP mice. Endothelial cells were identified as CD45−Ter119−CD31+ of the bone marrow fraction of LeptinRcre-R26-EYFP mice. Data were acquired on a LSRII (BD Biosciences, Billerica, MA) and analyzed with FlowJo (Tree Star, Ashland, OR).
BrdU incorporation.
To assess cell proliferation, 1mg BrdU was injected intraperitoneally 2 hours prior to sacrifice. BrdU flow kit (BD Biosciences, Billerica, MA) was used to stain BrdU+ cells.
RNA and protein assays.
PCR:
Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Venlo, Netherlands) or the NucleoSpin RNA XS kit (Takara Bio) according to the manufacturer’s instructions. RNase-free DNase Set (Quiagen) was used for DNase digestion during RNA purification. RNA quantity and quality were assessed by nano drop for RNA isolated from tissues and with the Aglient RNA 6000 Pico kit (Aglient Technologies) on the Aglient 2100 Bioanalyzer for RNA of FACS-purified cells. cDNA was generated from 1μg of total RNA per sample using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Quantitative real-time TaqMan PCR was performed using the following TaqMan primers: (Applied Biosystems: Actb (Mm00607939_s1), Hcrt (Mm01964030_s1), HcrtR1 (Mm01185776_m1), HcrtR2 (Mm01179312_m1), Csf1 (Mm00432686_m1), Pmch (Mm01242886_g1), Tph2 (Mm00557715_m1), Agrp (Mm00475829_g1), Galp (Mm00626135_m1), Ghrl (Mm00612524_m1), Gad1 (Mm04207432_g1), Npy (Mm01410146_m1), Pomc (Mm00435874_m1), LepR (Mm00440181_m1), Clock (Mm00455950_m1), Arntl2 (Mm05549497_m1), NR1d2 (Mm01310356_g1), Csf2 (Mm01290062_m1), IL-1β (Mm00434228_m1), CCL2 (Mm00441242_m1), IL-6 (Mm00446190_m1), MPO (Mm01298424_m1), Dpny (Mm00457573_m1). PCR was run on a 7500 thermal cycler (Applied Biosystems, Foster City, CA). Gene expression was normalized to βactin and quantified with the 2ΔΔCT method.
ELISA:
Hypocretin-1 levels were measured by ELISA kit (Fujifilm Wako Chemicals, Richmond, VA)42 and CSF1 (M-CSF) levels were measured by ELISA kit (Boster Biologial Technology, Pleasanton, CA) according to the manufacturers’ instructions. BM ELISAs were performed on BM fluid. Briefly, a small hole was made at the tips of long bones and the marrow was spun out of the bones by centrifugation at 10,000 rpms for 10 minutes. The supernatant fluid was collected.
Western Blot:
Total protein was extracted from cells in RIPA Lysis buffer with protease and phosphatase inhibitor cocktail. Total protein amount was quantified by Bradford assay. 15μg of protein was subjected to electrophoresis using NuPAGE Novex Cel System (Life Technologies, Carlsbad, CA) and transferred to nitrocellulose membrane using iBlot Gel Transfer System (LifeTechnologies, Carlsbad, CA) according to manufacturer’s instructions. Anti-hypocretin receptor-1 antibody (ab68718, Abcam, Cambridge, MA) and anti-βactin antibody (clone 13E5, Cell Signaling, Danvers, MA) were used.
Cell Culture.
For cell culture experiments, cells were cultured in complete medium (RPMI-1640 supplemented with 10% FBS, 2mM L-glutamine, 100 U/ml penicillin and streptomycin, 10mM Hepes, 50μM 2-mercaptoethanol, 1mM sodium pyruvate, and 1xnonessential amino acids) and kept in a humidified 5% CO2 incubator at 37°C. Neutrophils were sorted with a FACS Aria II and 5×105 cells were seeded into 48 well plates in 0.5ml medium. Cells were then pre-incubated with 0.1μM hypocretin-1 for 3 hours followed by co-treatment with 20ng/ml LPS for 3 hours as indicated. Cells and medium were collected for downstream analysis.
Myeloid colony forming unit assay:
Single cell suspension from bone marrow was prepared and 2 × 104 cells were plated in triplicates in complete methylcellulose medium (MethoCult GF M3534) with or without hypocretin-1 according to manufacturer’s instructions. Colony forming units were counted after 11 days in a 5% CO2 incubator at 37°C.
Histology and μCT.
Aortas:
Aortic roots were dissected and embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, CA), frozen in 2-Methylbutane (Fisher Scientific, Fair Lawn, NJ) cooled with dry ice, and sectioned into 6μm slices. To compare lesion size among the groups, sections that captured the maximum lesion area were used. To measure lesion volume, sections were collected beginning at the first appearance of the aortic valves and continuing until lesions were no longer visible. Oil-red-O (Sigma Aldrich, St. Louis, MO) staining was performed to visualize lipid content and measure lesion size. Lesion size was measured using Nanozoomer 2.0RS (Hamamatsu, Japan). Briefly, lesion area was quantified by measuring the atherosclerotic plaque of the intima from the endothelial layer to the healthy media.
Brain:
Whole brains were fixed in 10% formalin and embedded in paraffin for histological sectioning. Brains were sectioned coronally and 6μm slices were collected. After deparaffinization and rehydration of the sections, antigen retrieval was performed and endogenous peroxidase activity was blocked by 3% H2O2 solution in dH2O. Immunohistochemistry was done to stain for hypocretin+ cells using an anti-hypocretin antibody (clone D6G9T, Cell Signaling, Danvers, MA) diluted in SignalStain Antibody Diluent (Cell Signaling, Danvers, MA) followed by biotinylated goat anti-Rabbit IgG antibody (Vector Laboratories, Burlingame, CA). The hypocretin+ cells were visualized using avidin/biotin-based peroxidase system, VECTASTAIN Elite ABC-HRP Kit (Vector Laboratories, Burlingame, CA) and AEC Substrate Chromogen (Dako/Agilent, Santa Clara, CA). The tissues were counterstained with Harris Hematoxylin (Sigma-Aldrich, St. Louis, MO) and all images were captured using Nanozoomer 2.0RS (Hamamatsu, Japan). Dynorphin+ cells were identified by staining with Dynorphin A antibody (ab82509, Abcam Cambridge, MA) and AF488 conjugated Hypocretin antibody (clone D6G9T, Cell Signaling, Danvers, MA). Dynorphin+ cells were detected using a biotinylated goat anti-rabbit IgG antibody and streptavidin DyLight 594 (Vector Laboratories, Burlingame, CA). The fluorescent images were captured using a motorized fluorescence microscope Olympus BX63 (Tokyo, Japan) and processed by FIJI/ImageJ software. Apoptotic cells were investigated using In Situ Cell Death Detection Kit, TMR Red (Sigma-Aldrich, St. Louis, MO), and nuclei were counterstained with DAPI (ThermoFisher Scientific, Waltham, MA).
Bone:
Femurs were harvested and fixed in 4% paraformaldehyde overnight and then scanned by μCT at the MGH Center for Skeletal Research Core (NIHP30AR066261). After scanning, femurs were decalcified in 0.375M EDTA in PBS for 10 days prior to paraffin embedding. Sections were cut and stained with anti-tyrosine hydroxylase antibody (Millipore, AB152).
Behavior Phenotyping.
Open Field Test.
Mice were individually placed in a white box (50cm × 50cm) for 5 minutes with ambient lighting and video recorded. The time the mouse spent within 5cm of the box edge was quantified.
Light-dark box test.
Mice were placed in a box in which half the box (25cm × 25cm) was black and blocked from light, while the other half was white and exposed to ambient lighting. A dividing wall with a small hole separated the two halves of the box but allowed mice to move freely between the two halves. Mice were video recorded for 5 minutes and the time spent in the white box quantified.
Y-maze.
Mice were allowed to explore two arms of the Y-maze for 5 minutes before being returned to their home cage. Twenty minutes later, mice were returned to the Y-maze and allowed to explore all three arms for 5 minutes while being video recorded. The time spent in the new arm was quantified. Quantification of behavior phenotype was done with ImageJ software.
Statistics.
Results are shown as mean ± s.e.m. Statistical tests included unpaired, 2-tailed nonparametric Mann-Whitney tests (when Gaussian distribution was not assumed). For multiple comparisons, nonparametric multiple comparisons test comparing mean rank of each group (when Gaussian distribution was not assumed), 1 or 2-way ANOVA followed by Turkey’s test. To analyze circadian rhythmicity, cosinor analysis was performed with the following equation: Y=Baseline+Amplitude*cos(Frequency*X+Phaseshift)43. P values of 0.05 or less were considered to denote significance.
Data availability
All relevant data are included in the paper and its Extended Data Figures.
Extended Data
Extended Data Figure 1. Effects of sleep fragmentation on metabolic and cellular parameters.
a, Image of a sleep fragmentation cage. b, Body weight (n=10 per group). c, Plasma cholesterol at ZT3 (n=5 per group). d, Plasma glucose at ZT3 (n=5 per group). e, Glucose tolerance test (GTT) beginning at ZT3 and ZT12 (n=4 per group). f-h, Apoe−/− mice were placed in sleep fragmentation chambers where the sweep bar operated during the dark period (ZT12–0) when mice are normally awake. Control mice were maintained in SF chambers with a stationary sweep bar. f, Assessment of atherosclerosis and lesion area (n=5 per group). g, Assessment of blood Ly-6Chi monocytes and neutrophils (n=5 per group). h, Assessment of bone marrow LSKs and proliferation (n=5 per group). i, Aortic macrophage proliferation in Apoe−/− and Apoe−/− SF mice after 16 weeks of SF at ZT3 and ZT14 (n=5 Apoe−/−; n=4 Apoe−/− SF). j, Quantification at ZT3 in Apoe−/− and Apoe−/−SF mice of B cells, CD4+ T cells and CD8+ T cells in blood (n=10 Apoe−/−; for B and CD4 T cells n=6 Apoe−/− SF for CD8 T cells n=7 Apoe−/− SF), k, spleen (n=10 Apoe−/−; n=7 Apoe−/− SF) and l, B cells in bone marrow (n=10 Apoe−/−; n=7 Apoe−/− SF). Data presented as mean ± s.e.m.
Extended Data Figure 2. Sleep and circadian leukocyte migration.
Enumeration of Ly-6Chi monocytes and neutrophils at ZT3 and ZT14 in a, spleen, b, bone marrow, c, lung, and d, liver of Apoe−/− mice and Apoe−/− mice after 16 weeks of SF. Group sizes are indicated within the figure. Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Two-way ANOVA.
Extended Data Figure 3. Sleep mediated hematopoiesis and extramedullary hematopoiesis.
a, Gating strategy and quantification of hematopoietic progenitor cells at ZT3 in the bone marrow (n=10 Apoe−/− except GMPs n=11; n=10 Apoe−/− SF except MPP3 and MPP4 n=9) and b, spleen (n=9 Apoe−/− except CMP n=10; n=9 Apoe−/−SF). c, C57BL/6 wildtype (WT) mice consuming a regular chow diet were subjected to SF for 16 weeks after which Ly-6Chi monocytes, neutrophils and LSKs were enumerated at ZT3 (n= 8 WT except n=9 spleen Ly-6Chi monocytes, n=5 BM neutrophils, n=10 BM LSKs; n=4 WT+SF except BM LSKs n=9). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests.
Extended Data Figure 4. Sleep fragmentation does not alter bone structure and does not depend on the microbiome.
μCT analysis of a, tribeculae and b, cortical bone structure of Apoe−/− mice and Apoe−/− mice after 16 weeks of SF. Bone Volume Fraction (BV/TV), Bone Mineral Density (BMD), Trabecular Number (Tb.N), Trabecular Thickness (Tb.Th), Structural Model Index (SMI), Cortical Tissue Mineral Density (Ct.TMD), Cortical Area (Ct.Ar), Total Area (T.Ar), Cortical Thickness (Ct.Th), Cortical Porosity (Ct.Porosity). (n=9 per group) Analysis of leukocytosis in c, SF and d, Hcrt−/− mice at ZT3 after receiving 4 weeks of antibiotic cocktail in drinking water (n=3 per group). Data presented as mean ± s.e.m., *p<0.05,**p<0.01,***p<0.001, Mann-Whitney two-tailed tests.
Extended Data Figure 5. Sleep fragmentation does not activate the peripheral sympathetic nervous system but has effects on hypothalamic gene transcription and food consumption.
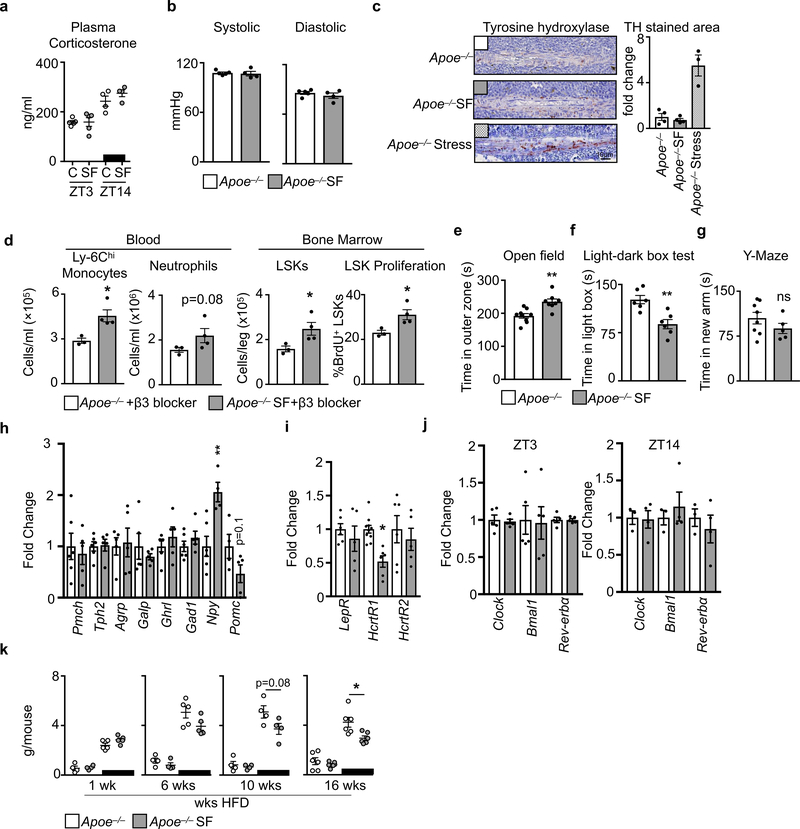
a, Plasma corticosterone levels in Apoe−/− mice and Apoe−/− mice after 16 weeks of SF at ZT3 and ZT14 (n=4 per group except n=5 Apoe−/−ZT3). b, Systolic and diastolic blood pressure at ZT3 (n=4 per group). c, Immunohistochemical analysis and quantification of tyrosine hydroxylaze (TH) staining in the bone marrow of Apoe−/− mice, Apoe−/−SF mice, and Apoe−/− mice subjected to 3 weeks of chronic variable stress (n=4 Apoe−/−; n=4 Apoe−/−SF; n=3 Apoe−/−stress). d, Enumeration at ZT3 of blood Ly-6Chi monocytes and neutrophils, bone marrow LSKs and proliferation in Apoe−/− mice and Apoe−/−SF mice after antagonism of the β3 receptor for 4 weeks (n=3 Apoe−/−+β3 blocker; n=4 Apoe−/−SF+β3 blocker). e, Quantification of time in outer zone during open field test (n=9 Apoe−/−; n=8 Apoe−/−SF). f, Quantification of time spent in light box during light/dark box test (n=6 per group). g, Quantification of time in new arm during Y-maze test (n=8 Apoe−/−; n=5 Apoe−/−SF). h, Analysis of neuropeptide expression in the hypothalamus at ZT3 (n=5 Apoe−/− except n=6 for Pmch, Tph2, Gad1,and Npy; n=5 Apoe−/−SF except n=4 for Npy). i, Neuropeptide receptor expression in the hypothalamus at ZT3 (n=6 Apoe−/− except HcrtR1 n=10; n=5 Apoe−/−SF except HcrtR2 n=6). j, Circadian gene expression in the hypothalamus at ZT3 and ZT14 (n=3 Apoe−/−; n=4 Apoe−/−SF). k, Mouse food consumption during the course of SF (n=4 Apoe−/−ZT3 except n=6 16wksApoe−/−ZT3; n=4 Apoe−/−SFZT3 except n=6 16wksApoe−/−ZT3; n=5 Apoe−/−ZT14 except n=4 10wksApoe−/−ZT14 and n=6 16wksApoe−/−ZT14; n=5 Apoe−/−SFZT14 except n=4 10wksApoe−/−SFZT14 and n=6 16wksApoe−/−SFZT14). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests.
Extended Data Figure 6. Hypothalamic expression of hypocretin and dynorphin.
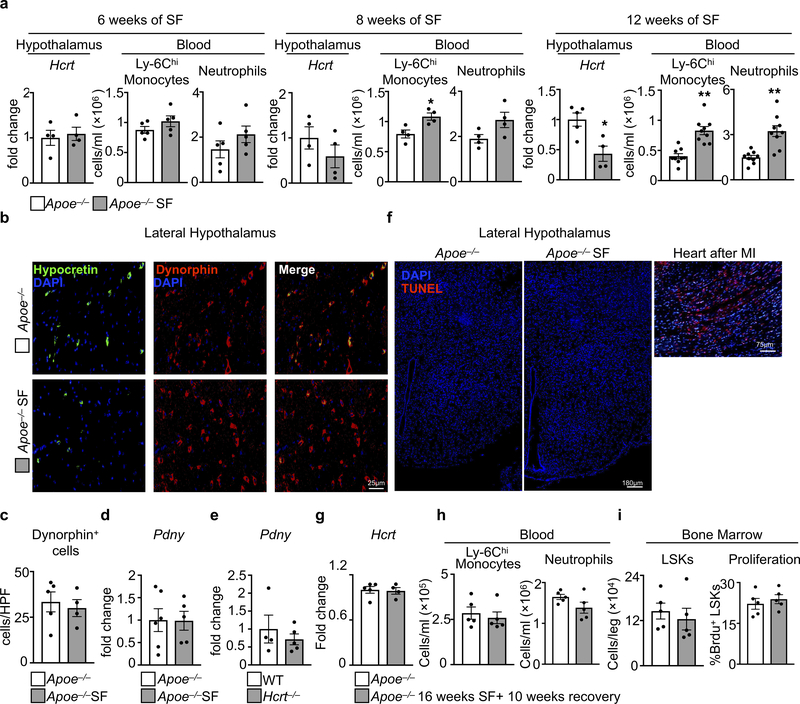
a, Hypothalamic expression of hypocretin and blood Ly-6Chi monocyte and neutrophil enumeration in Apoe−/− mice after 6, 8, and 12 weeks of SF. (for Hcrt n=4 Apoe−/− except n=5 for 12wks Apoe−/−; for Hcrt n=4 Apoe−/−SF; for blood cells at 6wks n=5 per group; for blood cells at 8 wks n=4 per group; for blood cells at 12 wks n=9 per group). b, Sections of the hypothalamus probed for dynorphin and hypocretin. c, Quantification of hypothalamic dynorphin+ cells per high powered field of view (HPV) (n=5 Apoe−/−; n=4 Apoe−/−SF, of 2 independent experiments). Dynorphin (Pdny) mRNA expression in hypothalamus of d, SF mice (n=6 Apoe−/−; n=5 Apoe−/−SF) and e, Hcrt−/− mice (n=4 WT; n= 5 Hcrt−/−). f, TUNEL staining of hypothalamic sections from Apoe−/− and Apoe−/− SF mice (representative of 4 biological replicates) along with a positive control of TUNEL stained myocardium 1 day after myocardial infarction (n = 1). g-i, Apoe−/− mice were sleep fragmented for 16 weeks then allowed to recover and sleep normally for 10 weeks. Control mice slept normally for 26 weeks. g, Analysis of hypothalamic hypocretin expression (n=5 Apoe−/−; n=4 Apoe−/−SF). h, Blood Ly-6Chi monocytes and neutrophils (n=5 per group). i, Bone marrow LSKs and LSK proliferation. (n=5 per group). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests.
Extended Data Figure 7. Hematopoiesis in hypocretin-deficient mice.
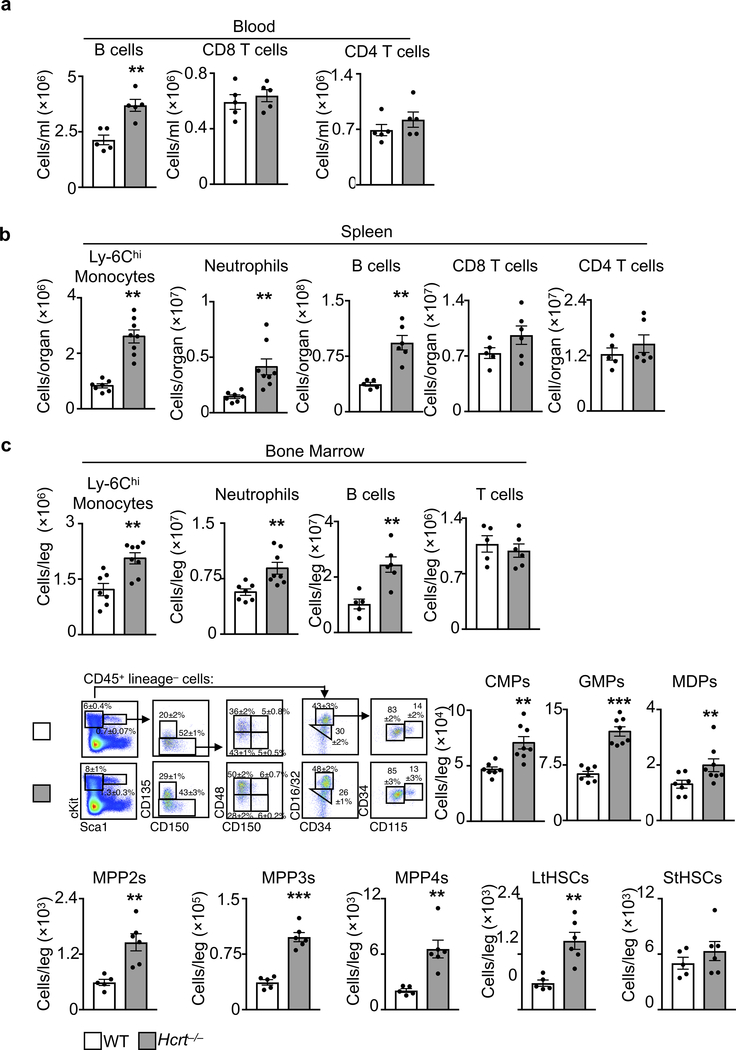
Leukocyte enumeration in WT and Hcrt−/− mice at ZT3 in a, blood (n=5 per group), b, spleen (for Ly-6Chi monocytes and neutrophils n=7 WT and n=8 Hcrt−/−; for B, CD8 T, and CD4 T cells n=5 WT and n=6 Hcrt−/−) and c, bone marrow (for Ly-6Chi monocytes and neutrophils n=7 WT and n=8 Hcrt−/−; for B and T cells n=5 WT and n=6 Hcrt−/−; for CMPs, GMPs and MDPs n=7 WT and n=8 Hcrt−/−; for LSK populations n=5 WT and n=6 Hcrt−/−). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, Mann-Whitney two-tailed tests.
Extended Data Figure 8. Hypocretin and hypocretin receptor-1 expression and production.
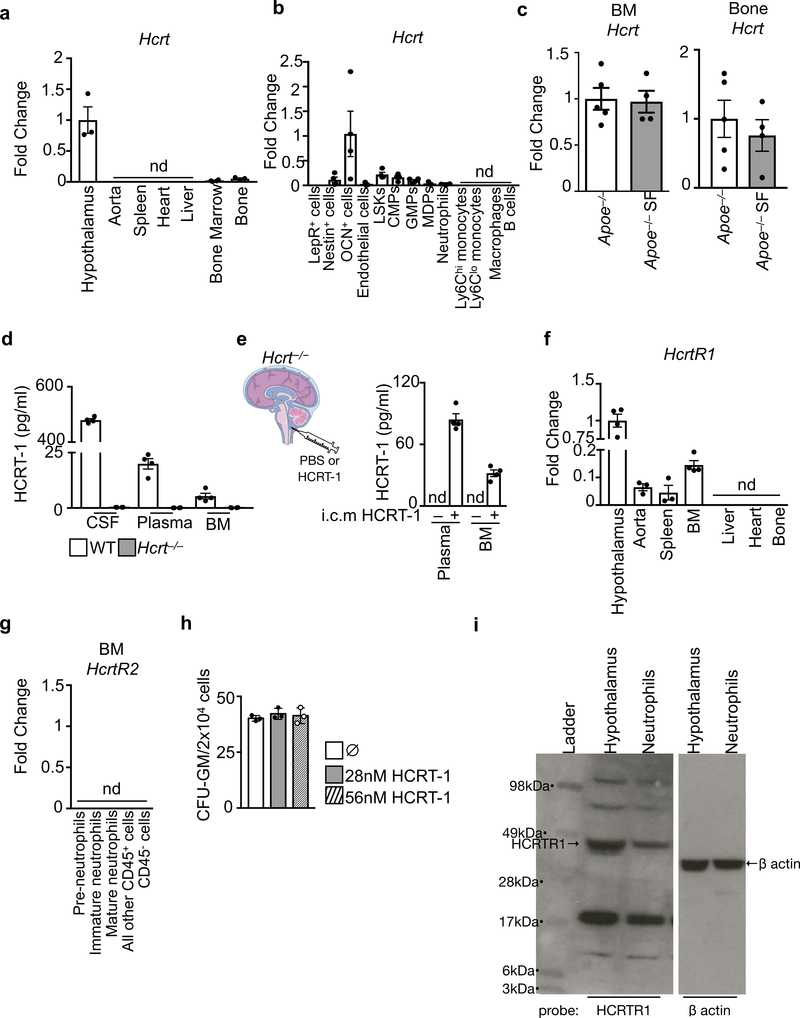
Relative Hcrt mRNA expression in a, tissues (n=3) and b, sorted bone marrow cells (n=4). c, Hcrt expression in the bone marrow and bone in Apoe−/− mice and in Apoe−/− mice subjected to sleep fragmentation for 16 weeks (n=5 Apoe−/−; n=4 Apoe−/−SF). d, Hypocretin-1 protein levels in cerebrospinal fluid (CSF), plasma and bone marrow (BM) fluid of WT and Hcrt−/− mice (n=4 per group). e, Hypocretin-1 protein levels in the plasma and BM fluid of Hcrt−/− mice 3 hours after intra-cisterna magna (i.c.m.) injection of HCRT-1 or PBS. (n=4 per group). f, Relative HcrtR1 mRNA expression in tissues (n=4 except aorta and spleen n=3). g, HcrtR2 expression in sorted BM cells (n=4). h, Granulocyte-macrophage colony forming units (CFU-GM) from bone marrow cells of WT mice exposed to hypocretin-1 (HCRT1) ex vivo in culture media (n=3 per group). i, Assessment of hypocretin receptor-1 (HCRTR1) protein in hypothalamus and sorted bone marrow neutrophils by western blot. Data presented as mean ± s.e.m.
Extended Data Figure 9. Hypocretin, bone marrow neutrophils, and HcrtR1.
a, Flow cytometry gating strategy for bone marrow pre-neutrophils, immature neutrophils and mature neutrophils. b, HCRTR1 (GFP) in bone marrow and blood neutrophils from WTHcrtR1Gfp/Gfp mice. c, mRNA expression in cultured bone marrow pre-neutrophils exposed to LPS and/or HCRT-1 (for untreated n=3 except Mpo n=6; for HCRT-1 n=3 expect Csf1 n=4 and Mpo n=6; for LPS n=3 except Csf1 n=7 and Mpo n=6; for LPS+HCRT-1 n=3 except Csf1 n=11, CSf2 n=4, and Mpo n=6). d, Csf1 expression in sorted bone marrow cells of WT and Hcrt−/− mice (n=5 WT; n=6 Hcrt−/−). e, Analysis of mRNA transcript expression in bone marrow leukocytes of Apoe−/− mice after 16 weeks of SF. (for Apoe−/− n=5 except Il10, IL34, CXCL12, CSF3 n=4 and Csf1 n=9; for Apoe−/− SF n=6 except IL5, IL1β, IL6, IL34, CXCL12, CSF3 n=5, IL10 n=5, and Csf1 n=12). f, Blood neutrophils in WTHcrtR1Gfp/Gfp mice over 24 hours (n=3 per group). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, One-way ANOVA.
Extended Data Figure 10. Hematopoietic CSF1 deletion protects against hematopoiesis and atherosclerosis in hypocretin deficient mice.
a, Schematic of chimeric models. Enumeration of Ly-6Chi monocytes and neutrophils in b, blood (n=4 WTbmWT; n=6 Hcrt−/−bmWT; n=3 WTbmCsf1−/−; n=5 Hcrt−/−bmCsf1−/−) and c, bone marrow (n=4 WTbmWT; n=6 Hcrt−/−bmWT; n=3 WTbmCsf1−/−; n=5 Hcrt−/−bmCsf1−/−). d, LSKs enumeration (n=4 WTbmWT; n=6 Hcrt−/−bmWT; n=3 WTbmCsf1−/−; n=5 Hcrt−/−bmCsf1−/−) and proliferation (n=4 WTbmWT; n=4 Hcrt−/−bmWT; n=3 WTbmCsf1−/−; n=5 Hcrt−/−bmCsf1−/−) in bone marrow. e, Enumeration of CMPs, GMPs and MDPs in chimeric mice (n=4 WTbmWT; n=6 Hcrt−/−bmWT; n=3 WTbmCsf1−/−; n=5 Hcrt−/−bmCsf1−/−). f, Bone marrow CSF1 levels (n=4 WTbmWT; n=8 Hcrt−/−bmWT; n=4 WTbmCsf1−/−; n=7 Hcrt−/−bmCsf1−/−). g, Schematic of chimeric models receiving Adv-PCSK9 and fed a high cholesterol diet for 12 weeks. h, Plasma cholesterol levels (n=7 WTbmWT; n=10 Hcrt−/−bmWT; n=5 WTbmCsf1−/−; n=6 Hcrt−/−bmCsf1−/−). i, Cross section images of aortic roots stained with oil-red-o and quantification of atherosclerosis in chimeric mice (n=7 WTbmWT; n=9 Hcrt−/−bmWT; n=5 WTbmCsf1−/−; n=6 Hcrt−/−bmCsf1−/−). Data presented as mean ± s.e.m., *p<0.05, **p<0.01,***p<0.001, One-way ANOVA.
Supplementary Material
Acknowledgements.
This work was supported in part by NIH grants R35 HL135752, R01 HL128264, P01 HL131478, an American Heart Association Established Investigator Award, and the Patricia and Scott Eston MGH Research Scholar (to F.K.S.); NIH grant R35 HL139598 (to M.N.); Swiss National Science Foundation grants 31003A_125323 and 31003A_144282 (to A.V.); a CIHR postdoctoral fellowship and a Banting postdoctoral fellowship (to C.S.M.); the doctoral program Cell Communication in Health and Disease (CCHD) funded by the Austrian Science Fund (to M.G.K); a Swedish Research Council postdoctoral fellowship (to S.R.); an American Heart Association postdoctoral fellowship (to S.H.); and a postdoctoral fellowship from the Fondation pour la Recherche Medicale (to C.V.); the German Research Foundation (DFG;331536185 to F.K. and DFG; 398190272 to W.C.P.); and a Boehringer-Ingelheim-Fonds MD fellowship (to L.H.). We thank Dr. A. Lichtman for providing the PCSK9 adenovirus, Dr. D. Scadden for providing stromal cell reporter mice, and K. Joyes for editing the manuscript.
Footnotes
Competing Financial Interests
None.
References
- 1.Hublin C, Partinen M, Koskenvuo M & Kaprio J Sleep and mortality: a population-based 22-year follow-up study. Sleep 30, 1245–1253 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cappuccio FP, Cooper D, D’Elia L, Strazzullo P & Miller MA Sleep duration predicts cardiovascular outcomes: a systematic review and meta-analysis of prospective studies. Eur Heart J 32, 1484–1492 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Hafner M, Stepanek M, Taylor J, Troxel WM & van Stolk C Why Sleep Matters-The Economic Costs of Insufficient Sleep: A Cross-Country Comparative Analysis. Rand Health Q 6, 11 (2017). [PMC free article] [PubMed] [Google Scholar]
- 4.Ford ES, Cunningham TJ & Croft JB Trends in Self-Reported Sleep Duration among US Adults from 1985 to 2012. Sleep 38, 829–832 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cappuccio FP et al. Meta-analysis of short sleep duration and obesity in children and adults. Sleep 31, 619–626 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan Z et al. Sleep duration and risk of type 2 diabetes: a meta-analysis of prospective studies. Diabetes Care 38, 529–537 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Blask DE Melatonin, sleep disturbance and cancer risk. Sleep Med Rev 13, 257–264 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Carreras A et al. Chronic sleep fragmentation induces endothelial dysfunction and structural vascular changes in mice. Sleep 37, 1817–1824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swirski FK & Nahrendorf M Leukocyte behavior in atherosclerosis, myocardial infarction, and heart failure. Science 339, 161–166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheiermann C et al. Adrenergic nerves govern circadian leukocyte recruitment to tissues. Immunity 37, 290–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He W et al. Circadian Expression of Migratory Factors Establishes Lineage-Specific Signatures that Guide the Homing of Leukocyte Subsets to Tissues. Immunity 49, 1175–1190.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lasselin J, Rehman JU, Åkerstedt T, Lekander M & Axelsson J Effect of long-term sleep restriction and subsequent recovery sleep on the diurnal rhythms of white blood cell subpopulations. Brain Behav Immun 47, 93–99 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Geovanini GR et al. Association between Obstructive Sleep Apnea and Cardiovascular Risk Factors: Variation by Age, Sex, and Race. The Multi-Ethnic Study of Atherosclerosis. Ann Am Thorac Soc 15, 970–977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidt T et al. Chronic variable stress activates hematopoietic stem cells. Nat Med 20, 754–758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Marchant NJ & Shaham Y Opposing roles of cotransmission of dynorphin and hypocretin on reward and motivation. Proc Natl Acad Sci U S A 111, 5765–5766 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu LY, Acuna-Goycolea C & van den Pol AN Neuropeptide Y inhibits hypocretin/orexin neurons by multiple presynaptic and postsynaptic mechanisms: tonic depression of the hypothalamic arousal system. J Neurosci 24, 8741–8751 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scammell TE, Arrigoni E & Lipton JO Neural Circuitry of Wakefulness and Sleep. Neuron 93, 747–765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Latorre D et al. T cells in patients with narcolepsy target self-antigens of hypocretin neurons. Nature 562, 63–68 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Hartmann FJ et al. High-dimensional single-cell analysis reveals the immune signature of narcolepsy. J Exp Med 213, 2621–2633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibrahim NE et al. Circulating Concentrations of Orexin A Predict Left Ventricular Myocardial Remodeling. J Am Coll Cardiol 68, 2238–2240 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Perez MV et al. Systems Genomics Identifies a Key Role for Hypocretin/Orexin Receptor-2 in Human Heart Failure. J Am Coll Cardiol 66, 2522–2533 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam JA et al. Decreased plasma orexin-A levels in obese individuals. Int J Obes Relat Metab Disord 26, 274–276 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Ohayon MM Narcolepsy is complicated by high medical and psychiatric comorbidities: a comparison with the general population. Sleep Med 14, 488–492 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Mochizuki T et al. Behavioral state instability in orexin knock-out mice. J Neurosci 24, 6291–6300 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellayah D, Bharaj P & Sikder D Orexin is required for brown adipose tissue development, differentiation, and function. Cell Metab 14, 478–490 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Aspelund A et al. A dural lymphatic vascular system that drains brain interstitial fluid and macromolecules. J Exp Med 212, 991–999 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louveau A et al. Corrigendum: Structural and functional features of central nervous system lymphatic vessels. Nature 533, 278 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Evrard M et al. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Li S, Franken P & Vassalli A Bidirectional and context-dependent changes in theta and gamma oscillatory brain activity in noradrenergic cell-specific Hypocretin/Orexin receptor 1-KO mice. Sci Rep 8, 15474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mossadegh-Keller N et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature 497, 239–243 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chemelli RM et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell 98, 437–451 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Vassalli A, Li S & Tafti M Comment on “Antibodies to influenza nucleoprotein cross-react with human hypocretin receptor 2”. Sci Transl Med 7, 314le2 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Mignone JL, Kukekov V, Chiang AS, Steindler D & Enikolopov G Neural stem and progenitor cells in nestin-GFP transgenic mice. J Comp Neurol 469, 311–324 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Méndez-Ferrer S et al. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature 466, 829–834 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeFalco J et al. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science 291, 2608–2613 (2001). [DOI] [PubMed] [Google Scholar]
- 36.Ding L, Saunders TL, Enikolopov G & Morrison SJ Endothelial and perivascular cells maintain haematopoietic stem cells. Nature 481, 457–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilic-Curcic I et al. Visualizing levels of osteoblast differentiation by a two-color promoter-GFP strategy: Type I collagen-GFPcyan and osteocalcin-GFPtpz. Genesis 43, 87–98 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Swirski FK et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science 325, 612–616 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robbins CS et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med 19, 1166–1172 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anzai A et al. The infarcted myocardium solicits GM-CSF for the detrimental oversupply of inflammatory leukocytes. J Exp Med 214, 3293–3310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Da Mesquita S et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ono T, Kanbayashi T, Yoshizawa K, Nishino S & Shimizu T Measurement of cerebrospinal fluid orexin-A (hypocretin-1) by enzyme-linked immunosorbent assay: A comparison with radioimmunoassay. Psychiatry Clin Neurosci 72, 849–850 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Refinetti R, Lissen GC & Halberg F Procedures for numerical analysis of circadian rhythms. Biol Rhythm Res 38, 275–325 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are included in the paper and its Extended Data Figures.