Cancer development is an evolutionary process driven by the acquisition of stochastic mutations, some of which increase cellular fitness in a co-evolving microenvironment.1 Evidence across multiple cancer types demonstrates that this process is highly protracted, likely beginning years or even decades prior to the development of an overt lesion, as early as in utero.2 While recurring gene mutations are a feature of many cancers, each tumor is the realization of its own unique evolutionary trajectory and it is usually not possible to determine the variation in cancer evolution. Cases where both the donor and recipient of a hematopoietic stem cell transplant develop the same tumor are a rare exception that occur when, at transplant, the precursor tumor population is transmitted from donor to recipient, and thereafter the two tumor lineages evolve independently.3 The similarities and differences between these ‘evolutionary reruns’ offer a serendipitous natural experiment to assay the repeatability of cancer evolution and study the molecular ontogeny and antecedent genetic steps that occur prior to overt clinical disease. The handful of donor-derived lymphoid malignancies reported thus far have predominantly involved indolent lymphomas and have yielded important insights into the genetic nature of a common ancestral cell with stem cell-like properties that is able to initiate lymphoma after a prolonged latency period.3–7 In aggressive B-cell lymphomas, no parallel examples exist with the molecular pathogenesis demonstrating a complex, multistep process that ultimately results in the transformation and expansion of a malignant clone that has either a germinal or post-germinal center cell of origin.8 This report, to the best of our knowledge, is the first of diffuse large B-cell lymphoma (DLBCL), where the presumed malignant clone was transferred at the time of transplantation, and where both the donor and recipient tumors were genetically profiled, to reveal important insights into the early stages of lymphomagenesis and tumor evolution.
A seven-year-old Caucasian boy was initially diagnosed with acute myeloid leukemia (AML). Bone marrow cytogenetic analysis and mutation screening revealed an abnormal karyotype (47,XY,+8,t(11;12)(q23;p13)[7]/49,XY,+X,+8,del(12)(p12 p13),+19[12]) with no evidence of a clonal KMT2A (11q23) rearrangement as determined by fluorescence in situ hybridisation (FISH), or of FLT3 internal tandem duplication (ITD) or NPM1 mutations. The patient was treated with standard induction chemotherapy and obtained a clinical remission. He relapsed 11 months post treatment and underwent re-induction therapy with a combination of fludarabine and cytarabine (FLA), achieving a second clinical remission that was consolidated by a haploidentical hematopoietic stem cell transplant with his mother as a donor. The post-transplant period was complicated by chronic graft versus host disease, predominantly affecting the skin and mouth, and he remained on immunosuppression until presenting 22 months post transplant with bilateral thigh and buttock pain. Staging investigations revealed an intra-spinal mass extending throughout the sacral canal, a scalp skin lesion and bone marrow involvement. Morphological examination and immunostaining of the spinal mass, bone-marrow trephine and skin lesion confirmed a diagnosis of stage IV DLBCL (CD20+, CD10-, BCL6-, MUM1+, and Ki-67>90%, Epstein-Barr virus-encoded RNA (EBER) negative), and excluded the possibility of relapse of the original AML. A month later the donor presented with an abdominal mass and subsequent investigations confirmed a diagnosis of stage IIa DLBCL (CD20+, CD10-, BCL6+, MUM1+, and Ki67 95%). Apart from discordant BCL6 expression, morphological and IHC findings for the donor and the recipient’s DLBCL were similar, with the same non-germinal-centre B-cell (non-GCB) of origin assigned to both in accordance with the Hans criteria (Figure 1A).9 The recipient’s clinical condition deteriorated soon after diagnosis and he died prior to receiving treatment for DLBCL. The donor was treated with 6 cycles of rituximab, cyclophosphamide, vincristine, doxorubicin and prednisolone (R-CHOP) immuno-chemotherapy and achieved a clinical remission, but relapsed 3 years after initial diagnosis and is currently receiving second-line chemotherapy.
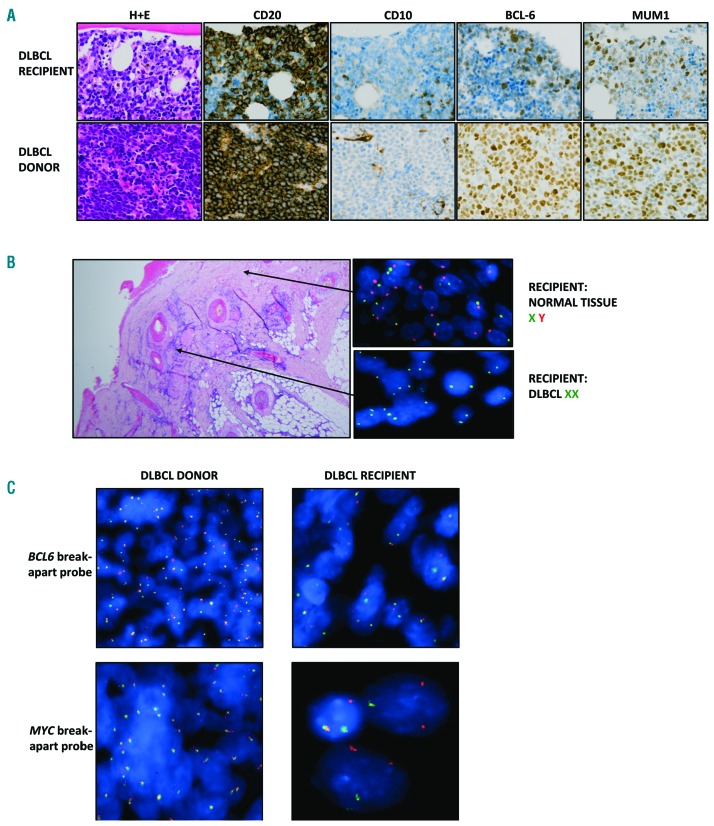
Figure 1.
Histologic findings. (A) Representative histologic and IHC images demonstrating DLBCL in the recipient and donor. All images are shown at the original magnification of x400. Both tumors express CD20 and are negative for CD10. The donor tumor demonstrates strong expression of BCL6 and MUM1, whereas the recipient tumor has weaker expression. Using the immunohistochemical cutoffs described by Hans et al to assign cells of origin,9 BCL6 is classed as positive in the donor and negative in the recipient and MUM1 is positive in both the donor and the recipient. The Hans cells-of-origin classification for both cases is non-GCB DLBCL. (B) Interphase FISH on the recipient skin biopsy demonstrating the presence of both the X and Y chromosome centromeres in normal tissue and two copies of the X chromosome centromere but no Y chromosome in the DLBCL cells, confirming the donor-derived nature of the tumor cells. (C) Interphase FISH using break-apart probes for BCL6 and MYC. The BCL6 probe showed a split signal pattern, consistent with a gene rearrangement, in both tumors whilst the MYC probe showed a split signal pattern, consistent with a MYC rearrangement, only in the recipient’s tumor.
Interphase FISH analysis of the recipient’s scalp lesion demonstrated the lymphoma cells to be of female (donor) origin with two copies of the X-chromosome centromere and absence of a Y-chromosome centromere, indicating that the tumor was donor-derived (Figure 1B). The donor and recipient tumors shared an identical VDJ rearrangement (IGHV3-7*01, D1-26*01, J4*02) with evidence of shared and discordant somatic hypermutation (SHM) changes, indicating that the DLBCL precursor cell transferred at the time of transplant had experienced the germinal center (Online Supplementary Figure S1). Interphase FISH analysis on both the recipient’s spinal mass and the donor’s abdominal mass demonstrated the presence of a BCL6 (3q27) rearrangement, with a standard split signal pattern in the majority of the cells in both tumors. A MYC (8q24) rearrangement, with a standard split signal pattern and unknown partner gene (no evidence of IGH (14q32) involvement), was unique to the recipient’s spinal biopsy and therefore likely to have occurred within the lymphoma clone after transfer into the recipient (Figure 1C).
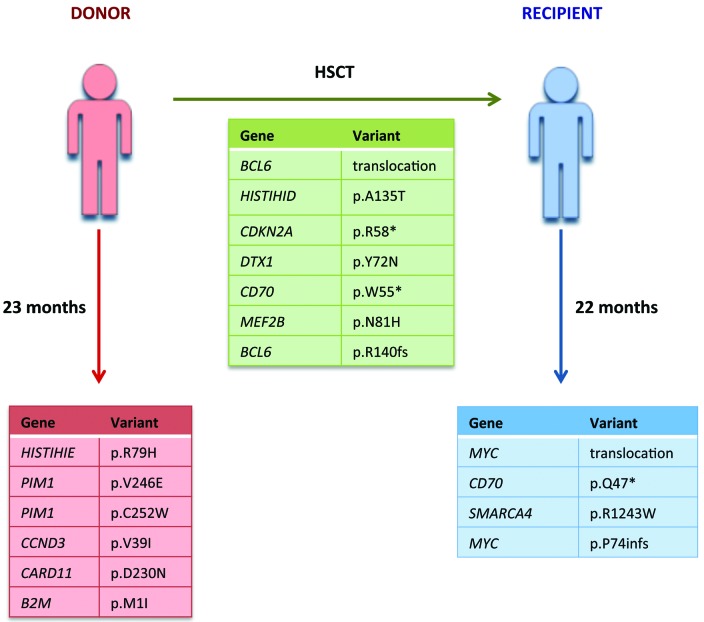
Targeted sequencing performed on the tumor and germ-line DNA from both recipient and donor (mean coverage donor 200x, mean coverage recipient 330x) (Online Supplementary Information) identified five non-synonymous single nucleotide variants (SNV) and one coding insertion shared by both tumors, six SNVs unique to the donor and two SNVs and one deletion specific to the recipient’s tumor (Figure 2, Online Supplementary Table S2). The shared variants included a nonsense mutation in CDKN2A (p.R58*) and missense mutations in DTX1 (p.Y72N) and MEF2B (p.N81H) impacting regions previously shown to be mutated in DLBCL.10–11 SNVs unique to the donor’s tumor, included non-synonymous mutations in PIM1, the coiled-coil domain of CARD11 (p.D230N) and the initiator codon of B2M (p.M1l). The recipient’s tumor had a lower mutational burden compared to the donor’s tumor with a 6-base-pair (bp) in-frame deletion in MYC and a missense mutation in SMARCA4 (p.R1243W). In addition, truncating mutations in CD70 predicted to result in the loss of the extracellular domain, were both shared (p.W55*) and unique to the recipient (p.Q47*), possibly representing an immune escape mechanism due to its physiological role in T-cell activation.10
Figure 2.
Summary of shared and discordant variants identified in the donor and recipient. Shared (green), donor-specific (red) and recipient-specific (blue) variants detected by targeted sequencing. Coverage details and variant allele frequencies are listed in Online Supplementary Table S2.
Donor-derived tumors are rare, underreported complication following stem-cell transplantation yet they present a unique window to trace the temporal evolution of premalignant clones. In indolent lymphomas, temporal and donor-derived cases have firmly established the notion of a long-lived ancestral cell population capable of initiating disease, evading chemotherapy and driving subsequent disease episodes.12–13 In contrast, in aggressive lymphomas the evidence for an analogous population of cells or premalignant phase is more restricted. Recently, temporal studies on DLBCL have suggested two distinct patterns of “early/branched” and “late” clonal evolution based on the degree of genetic semblance between paired diagnostic and relapse tumors.14 In the “early/branched” evolutionary pattern, both the diagnostic and relapse tumors demonstrated significant divergence suggesting that the relapse had emerged from an ancestral cell population. Our case, via a unique approach, provides confirmatory evidence for the existence of an ancestral or precursor population with a latency of almost two years, implying that these cells are long-lived with self-renewal capacity.
Although no samples from the stem-cell donation were available for analysis, the female genotype of the tumor confirmed the donor origin and the identical VDJ gene rearrangements demonstrated a clonal relationship between tumors. After hematopoietic stem cell transplant, the two tumours followed divergent paths as evidenced by the different IHC, cytogenetic and mutational findings with MYC a notable example of discordance. MYC translocations are associated with adverse prognosis and commonly co-occur with either BCL2 or BCL6 rearrangements,15 with the BCL6 rearrangement clearly occurring first in these donor-recipient lymphomas. The precise extent to which the contrasting immune states and chemotherapy-induced cellular damage in the recipient contributed to the evolution of the premalignant clone is unclear although it is noteworthy that a damaging variant in B2M, a component of the MHC class I machinery, was preferentially selected for in the immuno-competent donor but was not observed in the recipient. Strikingly both the donor and recipient tumors occurred virtually simultaneously despite significant differences in host age and immune micro-environment, in line with previous reports of donor-derived follicular lymphoma (7 years)3 and mantle cell lymphoma (12 years),6 suggesting tumor-intrinsic factors may be more important in driving the disease than extrinsic/host characteristics.
This unique case supports other findings suggesting the existence of a premalignant tumor-initiating population of cells in DLBCL that can occur several months or years prior to clinical detection and that harbors genetic lesions in known driver genes. The ability of these long-lived cells to evade chemotherapy is indicative of the “early/branched” pattern of relapse observed in a subset of patients and suggests that future efforts should focus on effectively targeting these cells.
Supplementary Material
Acknowledgements
We are indebted to the patients for donating tumor specimens as part of this study. The authors thank Queen Mary University of London Genome Centre for Illumina Miseq sequencing.
Footnotes
Funding: this work was supported by grants from Cancer Research UK (15968 awarded to JF and Clinical Research Fellowship awarded to SA) and Bloodwise through funding of the Precision Medicine for Aggressive Lymphoma (PMAL) consortium (15002).
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ford AM, Ridge SA, Cabrera ME, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. Nature. 1993;363(6427):358–360. [DOI] [PubMed] [Google Scholar]
- 3.Weigert O, Kopp N, Lane AA, et al. Molecular ontogeny of donor-derived follicular lymphomas occurring after hematopoietic cell transplantation. Cancer Discov. 2012;2(1):47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlotti E, Wrench D, Matthews J, et al. Transformation of follicular lymphoma to diffuse large B-cell lymphoma may occur by divergent evolution from a common progenitor cell or by direct evolution from the follicular lymphoma clone. Blood. 2009;113(15):3553–3557. [DOI] [PubMed] [Google Scholar]
- 5.Hart J, Turner AR, Larratt L, et al. Transmission of a follicular lymphoma by allogeneic bone marrow transplantation - evidence to support the existence of lymphoma progenitor cells. Br J Haematol. 2007;136(1):166–167. [DOI] [PubMed] [Google Scholar]
- 6.Christian B, Zhao W, Hamadani M, et al. Mantle cell lymphoma 12 years after allogeneic bone marrow transplantation occurring simultaneously in recipient and donor. J Clin Oncol. 2010;28(31):e629–632. [DOI] [PubMed] [Google Scholar]
- 7.Berg KD, Brinster NK, Huhn KM, et al. Transmission of a T-cell lymphoma by allogeneic bone marrow transplantation. N Engl J Med. 2001;345(20):1458–1463. [DOI] [PubMed] [Google Scholar]
- 8.Shaffer AL, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275–282. [DOI] [PubMed] [Google Scholar]
- 10.de Miranda NF, Georgiou K, Chen L, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124(16):2544–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying CY, Dominguez-Sola D, Fabi M, et al. MEF2B mutations lead to deregulated expression of the oncogene BCL6 in diffuse large B cell lymphoma. Nat Immunol. 2013;14(10):1084–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okosun J, Bödör C, Wang J, et al. Integrated genomic analysis identifies recurrent mutations and evolution patterns driving the initiation and progression of follicular lymphoma. Nat Genet. 2014;46(2):176–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green MR, Alizadeh AA. Common progenitor cells in mature B-cell malignancies: implications for therapy. Curr Opin Hematol. 2014;21(4):333–340. [DOI] [PubMed] [Google Scholar]
- 14.Juskevicius D, Lorber T, Gsponer J, et al. Distinct genetic evolution patterns of relapsing diffuse large B-cell lymphoma revealed by genome-wide copy number aberration and targeted sequencing analysis. Leukemia. 2016;30(12):2385–2395. [DOI] [PubMed] [Google Scholar]
- 15.Horn H, Ziepert M, Becher C, et al. MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121(12):2253–2263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.