Abstract
In chronic lymphocytic leukemia, TP53 mutations and deletion of chromosome 17p are well-characterized biomarkers associated with poor progression-free and overall survival following chemoimmunotherapy. Patients harboring low burden TP53 mutations with variant allele frequencies of 0.3-15% have been shown to have similar dismal outcome as those with high burden mutations. We here describe a highly sensitive deep targeted next-generation sequencing assay allowing for the detection of TP53 mutations as low as 0.2% variant allele frequency. Within a consecutive, single center cohort of 290 newly diagnosed patients with chronic lymphocytic leukemia, deletion of chromosome 17p was the only TP53 aberration significantly associated with shorter overall survival and treatment-free survival. We were unable to demonstrate any impact of TP53 mutations, whether high burden (variant allele frequency >10%) or low burden (variant allele frequency ≤10%), in the absence of deletion of chromosome 17p. In addition, the impact of high burden TP53 aberration (deletion of chromosome 17p and/or TP53 mutation with variant allele frequency >10%) was only evident for patients with IGHV unmutated status; no impact of TP53 aberrations on outcome was seen for patients with IGHV mutated status. In 61 patients at time of treatment, the prognostic impact of TP53 mutations over 1% variant allele frequency could be confirmed. This study furthers the identification of a clinical significant limit of detection for robust TP53 mutation analysis in chronic lymphocytic leukemia. Multicenter studies are needed for validation of ultra-sensitive TP53 mutation assays in order to define and implement a technical as well as a clinical lower limit of detection.
Introduction
Chronic lymphocytic leukemia (CLL) is a clonal B-cell malignancy characterized by a heterogeneous clinical course. Prognostic and predictive markers of survival and treatment outcome are essential in management of the disease.1 Deletion of chromosome 17p [del(17p)] and TP53 mutation (TP53mut) remain the most important risk factors for progression-free survival (PFS) and overall survival (OS) following chemo- and chemoimmunotherapy (CIT).2–6 In recent years, B-cell receptor pathway inhibitors (idelalisib, ibrutinib and acalabrutinib) and Bcl-2 inhibitors (venetoclax) have demonstrated remarkable response rates and durable remissions in both treatment naïve and previously treated CLL patients with TP53 aberration (TP53ab: either del(17p) or TP53mut).7–11 Randomized clinical trials comparing CIT directly to targeted therapy in a TP53-aberrated population are still awaited. Thus, assessment of TP53ab is recommended prior to any treatment decision.12
Approximately 80% of patients with del(17p) also harbor TP53muts on the remaining allele, while a subset of patients have TP53muts without del(17p).5 However, Sanger sequencing and fluorescence in situ hybridization (FISH) fail to detect 4–5% of newly diagnosed and untreated patients with CLL harboring low burden TP53muts (Sanger negative) without concomitant del(17p).13,14 Deep-targeted next-generation sequencing (NGS) of TP53 has shown that low burden TP53muts with a variant allele frequency (VAF) as low as 0.3% have similar outcome to patients with high burden TP53muts (Sanger positive).13,14 However, recent data from the UK CLL4 trial indicated that low burden TP53muts impacted neither OS nor PFS for patients treated with chemotherapy.15 For newly diagnosed patients harboring only one TP53ab, better OS is demonstrated compared to patients with both del(17p) and TP53mut. Similarly, patients with del(17p) and additional low burden TP53mut show better OS compared to patients with additional high burden TP53mut.16,17 Thus, the impact of additional TP53ab warrants further investigation.
Upon therapy, the prevalence and size of TP53 clones increase due to clonal evolution and acquisition of new TP53muts.18,19 Targeted therapy is established as the standard of care for patients with TP53 aberrated CLL.20,21 Whether patients with low burden TP53ab may benefit more from targeted therapy compared to standard CIT still remains open for investigation, as the evidence available so far does not allow definitive guidelines to be formulated.12 Thus, studies to elucidate a technical and a clinically significant limit of detection (LOD) for TP53mut are warranted to guide clinical decisions for these patients.
We here describe a robust NGS assay for detection of TP53mut as low as 0.2% VAF. In order to investigate a clinically relevant LOD for low burden TP53mut, we assessed the impact of TP53mut at diagnosis and at time of treatment in a single center cohort of patients with CLL.
Methods
Patients and materials
All consecutive patients diagnosed with CLL from a single center sampled between January 2007 and October 2014 were included in the study (Online Supplementary Figure S1). Samples from patients obtained within 200 days of the diagnostic flow cytometry were considered newly diagnosed.21 To assess the clinical impact of TP53ab at time of treatment, samples obtained up to 200 days before treatment were included for a separate analysis. All available samples considered newly diagnosed and/or sampled at time of treatment were sequenced. Due to the retrospective nature of the study, TP53 analysis was performed on peripheral blood mononuclear cells (PBMCs) and not on purified CLL cells. For 244 patients (81% of the newly diagnosed cohort) with available flow cytometry data at time of sampling, 197 patients (81%) had CLL populations more than 70% of the PBMCs (see Online Supplementary Methods), thus we report VAFs based on PBMCs.
Patients’ characteristics and clinical data were obtained from medical records and registries; CLL-International Prognostic Index (CLL-IPI) factors in terms of age (≤65 vs. >65 years), Binet stage (A vs. B or C), beta-2-microglobulin (β2M) (<4.0 mg/L vs. ≥4.0 mg/L), IGHV mutational status (germline identity <98% vs. ≥98%), and TP53ab only by FISH [no del(17p) vs. del(17p)] were included.3,22 Del(17p) was considered positive if present in at least 10% of 200 interphases. The study was approved by the Danish National Committee on Health Research Ethics, the Data Protection Agency and the Health Authorities involved.
TP53 mutational analysis by deep-targeted sequencing
A high sensitivity TP53 assay was established based on serial 10-fold dilutions of DNA from patient samples with donor DNA. By including a dilution step for each sequenced sample, background noise was filtered and an LOD was established at 0.2% VAF (Online Supplementary Methods, Online Suppplementary Table S1 and Online Suppplementary Figure S2). For each patient, DNA extracted from PBMCs was analyzed undiluted and diluted 20% (dilution factor 5) in DNA derived from the SU-DHL4 cell line. A known TP53mut (p.Arg273Cys) harbored in the cell line DNA acted as internal control of dilution grade. Using 100 ng gDNA per reaction, TP53 exons 2–10 incl. 2 bp intronic overlap for splice sites were amplified with 30 cycles of PCR using Phusion® HSII High-Fidelity DNA polymerase (Life Technologies, Waltham, MA, USA). A list of the primers used is provided in Online Supplementary Table S2. In brief, library preparation was performed following manufacturer protocol KAPA DNA Library Preparation (Nimblegen). Using SeqCap Adapter Kit A and B (Roche NimbleGen) or NEXTflex™ DNA Barcodes 96 (Bioo Scientific, Austin, TX, USA), libraries were pooled (24 or 96 samples per lane) and sequenced as paired-end on a HiSeq2500 using HiSeq® SBS Kit v.4 (2x125 base PE, Illumina) to obtain a minimum target read depth of 20,000x.
Bioinformatic workflow
A workflow for detection of low burden variants was developed in CLC Biomedical Genomics Workbench 3.0 (CLC BGW, Qiagen, Hilden, Germany) as described in the Online Supplementary Methods. Achieving a median coverage of 144,158 reads (99% of region > 26,217x), we applied both a dilution match algorithm (DMA) and a stereotypic error model (SEM) described in detail in the Online Supplementary Methods. In brief, only variants that diluted correctly were called TP53mut by DMA (Online Supplementary Figure S3), while SEM identified outliers from the position-specific and nucleotide-specific background noise as true TP53mut based on the distribution of stereotypic errors (Online Supplementary Figures S4 and S5).13 Results from both DMA and SEM were compared using contingency tables, and only true positive variants were considered true mutations and used in subsequent analyses (Online Supplementary Table S3 and Online Supplementary Figure S6).
Validation by droplet digital PCR and Capture based targeted next-generation sequencing
Droplet digital PCR (ddPCR) was used to validate initial low burden variants. Allele specific Prime Assay™ probes (Bio-Rad, Hercules, CA, USA) were applied for triplicate analyses using QX200™ Droplet Digital™ PCR System and QuantaSoft™ 1.7 (Bio-Rad) according to instructions from the manufacturer. A custom made SeqCap EZ Choice gene panel (Roche Nimblegen) containing TP53 exons 2–10 was used to validate mutations with a VAF of 1% or over, as described in the Online Supplementary Methods.
Statistical analysis
Time to event was calculated from date of diagnosis for treatment-free survival (TFS), and from date of diagnosis or first date of treatment for OS. Patients were followed until initiation of CLL-specific treatment or death or end of follow up, which ever came first, defined as TFS, and until death or end of follow up, whichever came first, defined as overall survival (OS). Analyses were performed using the Kaplan-Meier method, and log-rank test was used to compare outcome. TP53mut were stratified into high and low burden mutations (VAF>10% and VAF ≤10%, respectively) including minor TP53muts (VAF<1%).12,18 Since allogeneic stem cell transplantation is considered the only curative treatment in CLL, follow up was censored upon allogeneic stem cell transplantation for the cohort analyzed at time of treatment. FISH was not repeated at time of treatment in 5 patients for whom the baseline FISH were extrapolated. All analyses downstream of CLC BGW were performed with R version 3.4.1.23
Results
Patient cohorts and impact of baseline characteristics
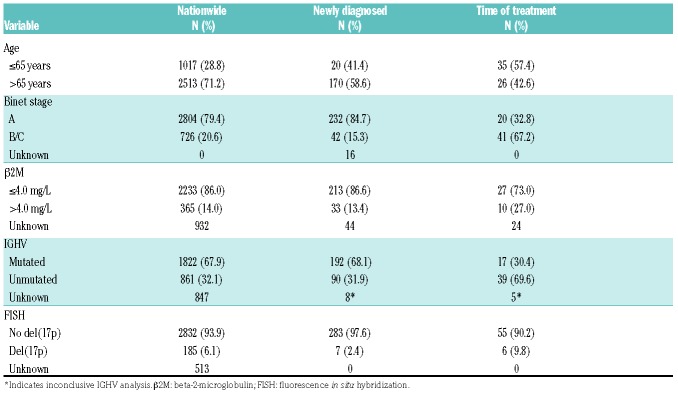
A total of 446 patients were included in our study. We excluded 44 patients with unavailable material and 92 patients who were neither newly diagnosed nor sampled at time of treatment. The two final cohorts included 290 newly diagnosed patients and 61 patients sampled at time of treatment, including 50 patients at time of first-line treatment and 11 at time of later lines of treatment (Online Supplementary Figure S1). Median time from date of diagnosis to sample collection was two days [interquartile range (IQR): 1.25-2.00). During a median follow up of 6.0 years (IQR: 3.9-7.9), 97 (33%) patients received treatment and 81 (28%) deaths were registered among newly diagnosed patients. Compared to the Danish nation-wide cohort, fewer patients were older than 65 years (58.6% vs. 71.2%) and a lower prevalence of Binet stage B/C disease (15.3% vs. 20.6%) as well as a lower prevalence of del(17p) (2.4% vs. 6.1%) were seen in this cohort. Except for age, there were more high-risk features in the 61 patients at time of treatment compared to the newly diagnosed patients (Table 1).
Table 1.
Patients’ characteristics of the Danish nationwide cohort and study cohorts for newly diagnosed patients and patients at time of treatment.
Improved robustness of low burden TP53 mutation detection
Robust detection of low burden TP53muts was ensured by combining a DMA and an SEM. For DMA, the ratio of variants called in both undiluted and diluted samples from the same patient (dilution ratio, DR) and the reference allele frequency of a known cell line mutation used for dilution (dilution grade, DG) were plotted (Online Supplementary Figure S3). The proximity to a line with a slope of one between the DG and adjusted DR defined true mutations for DMA. For SEM, we modeled frequent variants (observed ≥20) according to each unique genomic position and nucleotide change, while infrequent variants (observed <20) were modeled according to each unique nucleotide change only. VAFs were fitted to gamma distributions allowing for identification of true mutations using Bonferroni correction (Online Supplementary Figures S4 and S5). For the full study cohort of 308 patients, 98 and 116 TP53muts were called by DMA and SEM, respectively (Online Supplementary Tables S4 and S5, and Online Supplementary Figure S6). Using an LOD of 1% VAF, we obtained 100% consistency between DMA and SEM for determination of true mutations (Online Supplementary Table S3C). Between 0.3-1% VAF, 32 true positive TP53muts were further identified while excluding four variants only detected by either DMA or SEM (Online Supplementary Table S3B). Reporting TP53muts as low as 0.2% VAF, 10 additional true positive TP53muts could be identified, while 26 mutations only identified by either DMA or SEM were excluded (Online Supplementary Tables S3A and S4). Validating the first 30 low burden TP53muts identified, all were confirmed by ddPCR with high correlation between VAF by ddPCR and deep targeted next-generation sequencing (tNGS) (r2=0.999) (Online Supplementary Table S6). Consequently, one TP53mut (0.2% VAF) only identified by DMA was excluded based on SEM (Online Supplementary Figure S7C) but was in fact validated by ddPCR. However, the TP53 status remained unchanged as this patient harbored additional high and low burden mutations.
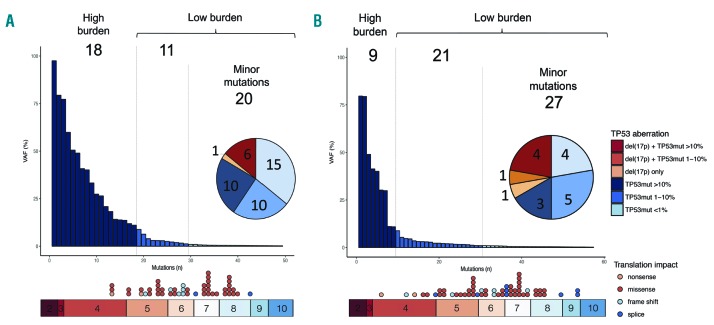
Among the 290 newly diagnosed patients, 41 patients (14%) harbored 18 high burden (VAF >10%) and 31 low burden (VAF ≤10%) mutations, including 20 minor TP53muts (VAF <1%). This resulted in 6 patients with both del(17p) and TP53mut, one with del(17p) only, 10 with high burden TP53mut only, and 25 patients with low burden TP53mut only (Figure 1A and Online Supplementary Table S4). Patients harboring only minor TP53mut were mainly older patients (>65 years) but were otherwise characterized as low risk according to the CLL-IPI (Online Supplementary Table S7A), whereas the distribution of high and low burden TP53muts was similar among patients stratified based on IGHV mutational status (Online Supplementary Table S7B). All mutations were located within exons 4–8 and 80% were missense mutations. Multiple high burden TP53muts were seen in 2 patients, while multiple low burden TP53muts were seen in 5 patients.
Figure 1.
Characterization of TP53 mutations. Number of mutations indicated in bar plots [regardless of del(17p)] and the number of patients according to TP53 status indicated in pie charts]. (A) Located within exons 4–8, 49 mutations were detected in 41 of 290 newly diagnosed patients; 6 of 7 del(17p) patients also harbored TP53 mutations. Eighteen (37%) and 31 (63%) mutations classified as high and low burden, respectively. (B) Fifty-seven TP53 mutations within exons 4–9 were detected in 17 of 61 patients at time of treatment; 5 of 6 del(17p) patients also harbored TP53 mutations. Nine (16%) and 48 (84%) mutations classified as high and low burden, respectively. Primarily missense mutations were detected. All percentages indicate variant allele frequencies (VAF). TP53 mutation without del(17p) (TP53mut).
Among 61 patients at time of treatment, we identified 57 mutations in 17 patients (28%): 7 patients with high burden and 10 with low burden TP53muts, including 4 with minor TP53muts (Figure 1B and Online Supplementary Table S4). Forty-three mutations (75%) were observed in 6 out of 11 previously treated patients with a median VAF of 1.01% (IQR: 0.46-2.93). Five of the 6 patients with del(17p) also harbored TP53muts on the remaining allele at time of treatment (Online Supplementary Table S8A). All mutations were located within exons 4–9 and 72% were missense mutations (Figure 1B). Seven patients harbored multiple TP53muts. Patients’ characteristics are summarized in Online Supplementary Table S8.
Prognostic impact on newly diagnosed patients
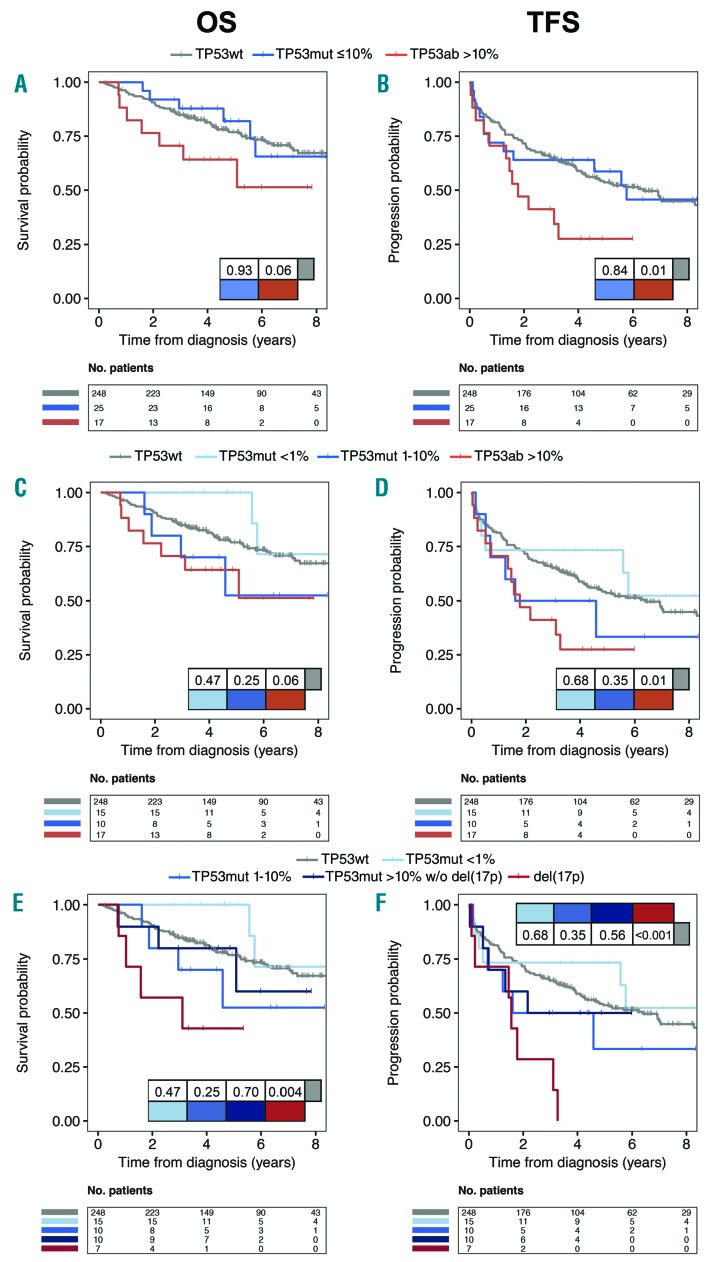
Stratifying TP53 aberrated patients into high and low burden (TP53wt vs. VAF ≤10% vs. VAF >10%), only high burden TP53ab [including del(17p)] patients showed a trend for worse OS and significantly worse TFS compared to TP53wt (Figure 2A and B) (P=0.06 and P=0.01, respectively). Further stratifying low burden TP53mut patients (VAF <1% vs. VAF 1–10%), still no impact on OS or TFS was seen for either group (Figure 2C and D), whereas combining the group of patients with a VAF above 1% could demonstrate a significant impact on OS and TFS compared to TP53wt (Online Supplementary Figure S8A and B). However, this was fully dependent on del(17p) patients (P=0.004 and P<0.001, respectively) (Figure 2E and F), as TP53 mutated patients without del(17p) demonstrated similar OS and TFS compared to TP53wt patients (P>0.25) (Figure 2E and F and Online Supplementary Figure S8C and D). Multiple TP53muts were observed in 7 newly diagnosed patients without impact on OS or TFS (1 vs. >1 TP53mut; P>0.2) (data not shown), while multiple TP53ab including del(17p) resulted in a significant impact on OS and a trend for TFS (1 vs. >1 TP53ab; P=0.036 and P=0.051, respectively) (data not shown).
Figure 2.
Overall (OS) and treatment-free survival (TFS) in newly diagnosed patients. Kaplan-Meier curves comparing OS (panels A, C, E) and TFS (panels B, D, F) based on (A and B) TP53 aberrations stratified based on variant allele frequencies (VAF) including del(17p) with 10% cut-off or (C and D) 1% and 10% cutoff. (E and F) Del(17p) and subgroups with TP53 mutations without del(17p) [(TP53mut w/o del(17p)] analyzed separately. P-values are indicated in the tables within the panels.
Prediction of treatment outcome
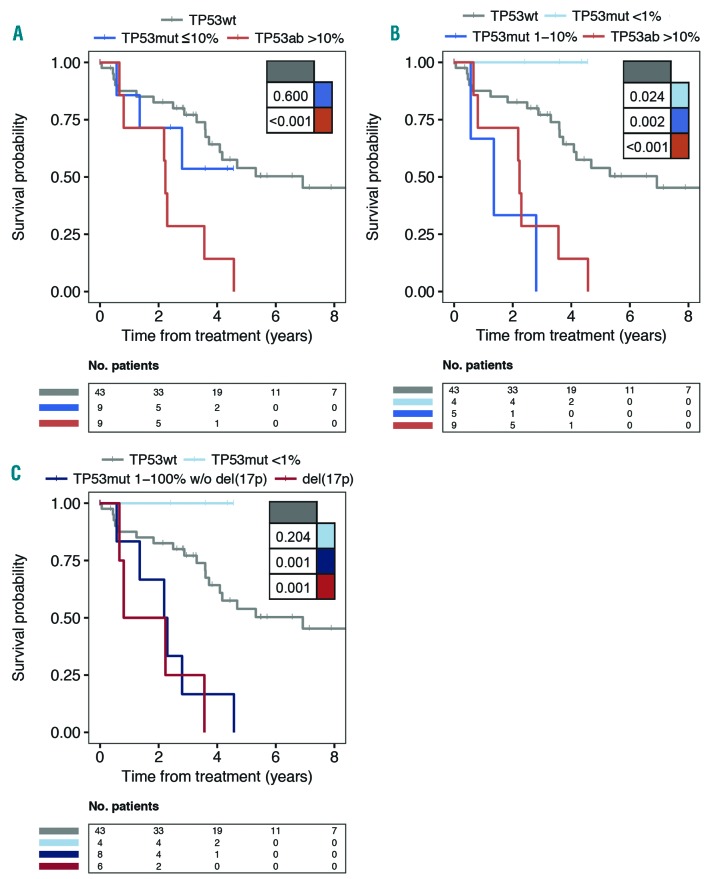
At time of treatment, TP53ab was significantly associated with a poor OS compared to patients with TP53wt (P=0.005) (Figure 3A and data not shown). Stratifying TP53-aberrated patients into high and low burden, only high burden TP53ab patients demonstrated poor OS compared to TP53wt (P<0.001) (Figure 3A). Further stratifying low burden TP53 mutated patients (VAF <1% vs. VAF 1–10%), OS was significantly worse for patients with TP53mut with VAF 1–10% compared to TP53wt (P=0.002) (Figure 3B). Combining the group of patients with TP53ab with VAF 1–100%, the association persisted when omitting del(17p) patients (P≤0.001) (Figure 3C). Four patients with minor TP53muts were still alive and in complete remission at end of follow up (Figure 3B and C). There was no difference in survival between patients harboring one TP53mut with a VAF greater than 1% (VAF 1–100%) and patients with more than one TP53mut (P=0.85) (data not shown). Although both patients receiving first-line treatment (n=50) and patients receiving later lines of treatment (n=11) were included in the cohort at time of treatment, TP53 status demonstrated a similar negative impact on OS in both subcohorts (data not shown).
Figure 3.
Overall survival (OS) in patients from time of treatment. Stratifying patients with TP53 aberrations including del(17p) based on (A) variant allele frequencies (VAF) with 10% cut-off or (B) 1% and 10% cut-off. (C) Del(17p) versus subgroups with TP53 mutations without del(17p) [TP53mut w/o del(17p)] with 1% VAF cut-off is shown. Del(17p) status may reflect baseline if a second fluorescence in situ hybridization (FISH) was not performed at time of treatment. P-values indicated in tables within the panels.
TP53 status may predict outcome in newly diagnosed patients with unmutated IGHV status
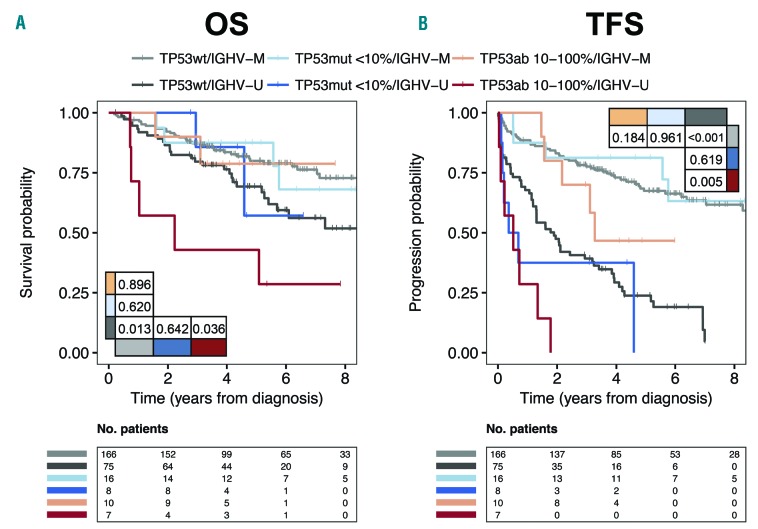
As most patients with TP53ab at time of treatment were also IGHV unmutated (IGHV-U) (Online Supplementary Table S8), we explored the synergy between TP53ab and IGHV mutational status for newly diagnosed patients. For patients with mutated IGHV (IGHV-M), TP53ab [whether low burden or high burden, including 4 patients with del(17p)] did not impact OS or TFS. However, for patients with IGHV-U status, high burden TP53ab [including 3 patients with del(17p)] significantly impacted both OS and TFS (P=0.036 and P=0.005, respectively) (Figure 4).
Figure 4.
Stratified analysis in newly diagnosed patients based on TP53 and IGHV status. Kaplan Meier curves for (A) overall survival (OS) and (B) treatment-free survival (TFS) in newly diagnosed patients stratified for IGHV status and for TP53 aberrations based on variant allele frequencies (VAF) ≤10% and >10%. Four and three del(17p) are included for mutated (IGHV-M) and unmutated IGHV (IGHV-U), respectively. P-values indicated in tables within the panels.
Discussion
This study demonstrates that neither high nor low burden TP53muts at time of CLL diagnosis independently influenced OS or TFS in a consecutive cohort of newly diagnosed patients. However, patients with del(17p) at time of diagnosis had an inferior outcome. In addition, the subgroup of patients with TP53ab over 10% VAF among patients with IGHV-U status demonstrated inferior OS and TFS. At time of treatment, patients with sole TP53muts over 1% VAF had shorter OS, as had patients with del(17p).
In our study, del(17p) in newly diagnosed CLL was rare (2.4%), although still demonstrating a negative prognostic impact in accordance with our previous validation of CLL-IPI3 in a Danish nation-wide cohort.24 The majority of del(17p) patients were, as expected, also TP53 mutated.5 Although we demonstrate a similar prevalence of TP53 mutated patients and a similar distribution of variant allele frequencies, TP53muts without concomitant FISH positive for del(17p) were more frequent in newly diagnosed patients (10.7% using an LOD of 0.3%) compared to previous publications.13,14,17 In particular, sole low burden TP53muts (7.2%) was highly prevalent, whereas the prevalence of patients with sole high burden TP53muts (3.4%) was similar to previous reports.13,14,17 Despite a high prevalence, and in contrast to reports by Rossi et al.,13 we could not demonstrate impact on OS of neither high nor low burden TP53muts without del(17p) in newly diagnosed patients. Similar to our results, Stengel et al. demonstrated a better OS in newly diagnosed patients with TP53mut only compared to concomitant del(17p) and TP53mut, which may support the lack of impact on OS in our smaller cohort.17 Furthermore, Nadeu et al. reported no impact on time to treatment among newly diagnosed TP53 mutated patients compared to TP53wt patients.14 More prevalent high-risk factors with impact on early need of treatment observed across previous studies may also contribute to the different impact of TP53muts.13,14,17 For example, the previous studies included older patients with higher Binet stage and more frequent del(17p), resulting in lower frequencies of TP53-mutated patients without del(17p) than in our cohort. Furthermore, patients in our cohort had a lower CLL-IPI score compared to the Danish nation-wide CLL cohort, probably due to varying regional referral patterns.
In contrast to previously published data,25 IGHV-U status was not higher in newly diagnosed TP53 mutated patients, which may in part explain the indolence of our cohort. In our study, synergy was demonstrated for IGHV mutational status and TP53ab.25 For patients with IGHV-U status, high burden TP53ab correlated with poor outcome among newly diagnosed patients. However, TP53ab (whether high or low burden) had no negative prognostic impact on the more indolent disease course for patients with IGHV-M status, in accordance with previous studies.26–29 Thus, the less aggressive phenotype in our cohort may diminish any independent impact of TP53muts, especially due to the proportion of IGHV-M status among TP53 mutated patients. High cell proliferation, shorter time to treatment, and a distinct pattern of nucleotide shifts in patients with IGHV-U may contribute to the mechanisms causing this interaction between IGHV mutational status and TP53ab.18,30
Like the majority of NGS studies investigating the clinical impact of TP53muts,13,14,16,18,31,32 we confirm the negative impact on OS of TP53muts over 1% VAF at time of treatment. A recent study, however, was unable to show this association for patients harboring low burden TP53muts only.15
In our study, minor TP53muts were common among pretreated patients. However, minor TP53muts were observed exclusively as the only TP53ab in treatmentnaïve patients. These newly diagnosed patients with only a single minor TP53mut were mainly older patients with an otherwise favorable risk profile and outcome. Even the 4 patients with minor TP53mut requiring initial treatment (3 with IGHV-U status) were still alive and in complete remission at end of follow up. This may indicate that minor TP53muts as the sole TP53ab is an age-related phenomenon of a more benign character, similar to reports on clonal hematopoiesis in myeloid malignancies.33 In accordance with this, a recent study found TP53muts enriched among older CLL patients.17
Current guidelines for assessment of TP53muts prior to treatment recommend an LOD at 10% VAF for clinical decisions, with the option to report low burden mutations down to 5% VAF by NGS as long as the unresolved clinical significance of such mutations is stated.12 The reason for a caveat when reporting TP53muts below 10% VAF results from: 1) low reproducibility between different NGS platforms in this range; and 2) an uncertain clinical significance of low burden mutations.12 To address the technical question of reproducibility, we here report a SEM of both nucleotide and position specific variants from deep tNGS in combination with an algorithm based on the dilution of patient DNA. By this approach, we have developed a technically robust method for detection of TP53mut that could easily be transferred across different platforms and laboratories. For clinical use, we do, however, recommend using a cell line harboring a rare TP53mut predicted to encode functional p53, such as BRG-A (TP53:c.1060C>G), to avoid both risk of contamination and risk of omitting significant low burden mutations.34 We successfully achieved an LOD of 0.3% VAF, applied in previous studies of minor TP53muts,13,14,18 and could even lower the LOD to 0.2% VAF. As we were unable to prove any impact on newly diagnosed patients with IGHV-M, our results support the current guidelines recommending TP53 assessment only prior to treatment.12
This study furthers the identification of a clinically significant LOD for TP53muts in CLL. The method proposed here for analysis of minor TP53muts warrants validation across laboratories for a standard technical LOD for TP53muts. Subsequent validation and standardization of TP53 mutation assays within networks such as the European Research Initiative on CLL (ERIC, http://ericll.org) may provide the platform needed for collaborative multicenter analyses seeking to define a validated clinical LOD for TP53muts.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/4/789
References
- 1.Parikh SA, Shanafelt TD. Prognostic factors and risk stratification in chronic lymphocytic leukemia. Semin Oncol. 2016; 43(2):233–240. [DOI] [PubMed] [Google Scholar]
- 2.Pflug N, Bahlo J, Shanafelt T, et al. Development of a comprehensive prognostic index for patients with chronic lymphocytic leukemia. Blood. 2014;124(1):49–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International CLLIPIwg An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): a meta-analysis of individual patient data. Lancet Oncol. 2016;17(6):779–790. [DOI] [PubMed] [Google Scholar]
- 4.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. [DOI] [PubMed] [Google Scholar]
- 5.Zenz T, Eichhorst B, Busch R, et al. TP53 Mutation and Survival in Chronic Lymphocytic Leukemia. J Clin Oncol. 2010; 28(29):4473–4479. [DOI] [PubMed] [Google Scholar]
- 6.Stilgenbauer S, Schnaiter A, Paschka P, et al. Gene mutations and treatment outcome in chronic lymphocytic leukemia: results from the CLL8 trial. Blood. 2014;123(21):3247–3254. [DOI] [PubMed] [Google Scholar]
- 7.Byrd JC, O’Brien S, James DF. Ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(13):1278–1279. [DOI] [PubMed] [Google Scholar]
- 8.Farooqui MZ, Valdez J, Martyr S, et al. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: a phase 2, single-arm trial. Lancet Oncol. 2015;16(2):169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furman RR, Sharman JP, Coutre SE, et al. Idelalisib and Rituximab in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med. 2014;370(11):997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts AW, Davids MS, Pagel JM, et al. Targeting BCL2 with Venetoclax in Relapsed Chronic Lymphocytic Leukemia. N Engl J Med. 2016;374(4):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Byrd JC, Furman RR, Coutre SE, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369(1):32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malcikova J, Tausch E, Rossi D, et al. ERIC recommendations for TP53 mutation analysis in chronic lymphocytic leukemia-update on methodological approaches and results interpretation. Leukemia. 2018; 32(5):1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123(14):2139–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadeu F, Delgado J, Royo C, et al. Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia. Blood. 2016;127(17):2122–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blakemore S. The Contribution of Gene Mutations to Long-Term Clinical Outcomes: Data from the Randomised UK LRF CLL4 Trial. Blood. 2017;130(Suppl 1):259. [Google Scholar]
- 16.Yu L, Kim HT, Kasar SN, et al. Survival of Del17p CLL Depends on Genomic Complexity and Somatic Mutation. Clin Cancer Res. 2016;23(3):735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31(3):705–711. [DOI] [PubMed] [Google Scholar]
- 18.Malcikova J, Stano-Kozubik K, Tichy B, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015; 29(4):877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Landau DA, Carter SL, Stojanov P, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152(4):714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown JR, Kay N. Chemoimmunotherapy Is Not Dead Yet in Chronic Lymphocytic Leukemia. J Clin Oncol. 2017;35(26):2989–2992. [DOI] [PubMed] [Google Scholar]
- 21.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745–2760. [DOI] [PubMed] [Google Scholar]
- 22.da Cunha-Bang C, Geisler CH, Enggaard L, et al. The Danish National Chronic Lymphocytic Leukemia Registry. Clin Epidemiol. 2016;8(1):561–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Team RC R: A Language and Environment for Statistical Computing. 2017. [Google Scholar]
- 24.da Cunha-Bang C, Christiansen I, Niemann CU. The CLL-IPI applied in a population- based cohort. Blood. 2016;128(17):2181–2183. [DOI] [PubMed] [Google Scholar]
- 25.Sutton LA, Ljungstrom V, Mansouri L, et al. Targeted next-generation sequencing in chronic lymphocytic leukemia: a high-throughput yet tailored approach will facilitate implementation in a clinical setting. Haematologica. 2015;100(3):370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delgado J, Salaverria I, Baumann T, et al. Genomic complexity and IGHV mutational status are key predictors of outcome of chronic lymphocytic leukemia patients with TP53 disruption. Haematologica. 2014;99(11):e231–e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeromin S, Haferlach C, Dicker F, Haferlach T, Kern C. Patients with TP53 disruption and IGHV Mutated Status Show Indolent Clinical Course: A Study on 1,327 Treatment-Naive CLL Cases. Blood. 2016; 128(22):4378. [Google Scholar]
- 28.Best OG, Gardiner AC, Davis ZA, et al. A subset of Binet stage A CLL patients with TP53 abnormalities and mutated IGHV genes have stable disease. Leukemia. 2009; 23(1):212–214. [DOI] [PubMed] [Google Scholar]
- 29.Sutton LA, Hadzidimitriou A, Baliakas P, et al. Immunoglobulin genes in chronic lymphocytic leukemia: key to understanding the disease and improving risk stratification. Haematologica. 2017;102(6):968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy EJ, Neuberg DS, Rassenti LZ, et al. Leukemia-cell proliferation and disease progression in patients with early stage chronic lymphocytic leukemia. Leukemia. 2017;31(6):1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landau DA, Tausch E, Taylor-Weiner AN, et al. Mutations driving CLL and their evolution in progression and relapse. Nature. 2015;526(7574):525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puente XS, Bea S, Valdes-Mas R, et al. Non-coding recurrent mutations in chronic lymphocytic leukaemia. Nature. 2015; 526(7574):519–524. [DOI] [PubMed] [Google Scholar]
- 33.Wong TN, Ramsingh G, Young AL, et al. Role of TP53mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;2015(518):552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campomenosi P, Fronza G, Ottaggio L, et al. Heterogeneous p53 mutations in a Burkitt lymphoma from an AIDS patient with monoclonal c-myc and VDJ rearrangements. Int J Cancer. 1997;73(6):816–821. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.