Abstract
The oncoprotein BCR-ABL1 triggers chronic myeloid leukemia. It is clear that the disease relies on constitutive BCR-ABL1 kinase activity, but not all the interactors and regulators of the oncoprotein are known. We describe and validate a Drosophila leukemia model based on inducible human BCR-ABL1 expression controlled by tissue-specific promoters. The model was conceived to be a versatile tool for performing genetic screens. BCR-ABL1 expression in the developing eye interferes with ommatidia differentiation and expression in the hematopoietic precursors increases the number of circulating blood cells. We show that BCR-ABL1 interferes with the pathway of endogenous dAbl with which it shares the target protein Ena. Loss of function of ena or Dab, an upstream regulator of dAbl, respectively suppresses or enhances both the BCR-ABL1-dependent phenotypes. Importantly, in patients with leukemia decreased human Dab1 and Dab2 expression correlates with more severe disease and Dab1 expression reduces the proliferation of leukemia cells. Globally, these observations validate our Drosophila model, which promises to be an excellent system for performing unbiased genetic screens aimed at identifying new BCR-ABL1 interactors and regulators in order to better elucidate the mechanism of leukemia onset and progression.
Introduction
Chronic myeloid leukemia (CML) is a clonal myeloproliferative disorder associated with a reciprocal translocation between chromosomes 9 and 22. This process leads to the fusion of the Abelson (ABL1) tyrosine kinase gene with the breakpoint cluster region (BCR) sequences generating a fusion gene encoding the constitutively active protein tyrosine kinase BCR-ABL1. Due to its high frequency in CML patients (95%), the translocation is considered the cytogenetic hallmark of this disease.1,2 Although BCR-ABL1 is one of the most studied oncogenic proteins, some molecular mechanisms leading to cellular transformation are still partially unknown. In particular, positive or negative regulators of BCR-ABL1 have not been completely identified. The fruitfly, Drosophila melanogaster, represents a powerful tool for genome-wide genetic analysis and screens, given the functional conservation and sequence homology between human and Drosophila genes. Genome-wide approaches may allow identification of genetic pathways that contribute to disease onset and/or progression without a priori knowledge of the gene function.3 The high degree of conservation between human and Drosophila Abl (dAbl) proteins and the existence of Drosophila homologs for many proteins that interact functionally with BCR-ABL1 in mammals strongly support the idea that dAbl and presumably BCR-ABL1 signal transduction pathways could be highly conserved from fly to human. The dAbl gene is expressed at high levels in differentiating neurons and plays an important role in central nervous system, eye and epithelia development, mainly regulating cytoskeleton remodeling.4–6 Interestingly, Forgerty and colleagues demonstrated that the neural expression of a chimeric BCR-ABL protein carrying the human BCR fused to dAbl is able to rescue the dAbl mutant phenotype, suggesting that the chimeric BCR-ABL protein can effectively compensate for lack of dAbl.7 To further identify genes and pathways involved in the onset and progression of CML, we developed and validated a genetic model based on transgenic flies that drive inducible human BCR-ABL1 expression under the control of tissue- and stage-specific promoters, providing both an excellent and powerful model to identify novel functional interactors.
Methods
Generation of BCR-ABL1 transgenic flies
The BCR-ABL1 coding sequence was amplified by polymerase chain reactions and cloned into the P-element expression vector pKS69. BCR-ABL1 kinase-dead (BCR-ABL1KD) was obtained through site-directed mutagenesis (Online Supplementary Data). Plasmids were prepared using Qiafilter™ Plasmid Maxi Kit (Qiagen, Venlo, the Netherlands) and injected in Drosophila embryos (The BestGene, Inc, Chino Hills, CA, USA).
Drosophila stocks
Fly stocks were obtained from Bloomington Drosophila Stock Center (Department of Biology, Indiana University, Bloomington, IN, USA). RNA interference (RNAi) lines were obtained from the Vienna Drosophila RNAi Center (Vienna, Austria). domelessGal4 and STATDN flies were kindly provided by A. Giangrande (IGBMC, Illkirch, France) (Online Supplementary Data).
Immunoblotting
Adult heads were dissected and homogenized in a protein extraction buffer. For cell lines, 107 cells were lysed in RIPA buffer. The following primary antibodies were used: c-Abl (sc-23), Dab1 (sc-271136), p-Tyr (sc-7020), GAPDH (sc-137179) (Santa Cruz Biotechnology, Santa Cruz, CA, USA), α-tubulin (CP06; Oncogene Research Products, Merck KGaA, Darmstadt, Germany) mouse monoclonal antibodies, BCR (sc-20707) rabbit polyclonal antibody (Santa Cruz Biotechnology) and mouse 5G2 anti-Enabled supernatant (Developmental Studies Hybridoma Bank - DSHB, University of Iowa, IA, USA). For immunoprecipitation, 1 mg of total protein extract was incubated with anti-Enabled supernatant and subsequently with protein A sepharose (Amersham Bioscience, GE Healthcare, Waukesha, WI, USA) (Online Supplementary Data).
Fluorescent Immunolabeling
Fly eye primordium
Eye imaginal discs were dissected from third instar larvae, fixed in 4% paraformaldehyde, permeabilized with 0.3% Triton X-100, labeled with the rat anti-Elav 7E8A10 supernatant (DSHB), incubated with a Cy3-conjugated anti-rat secondary antibody (Jackson Immunoresearch, Newmarket, UK) and exposed to HOECHST (Sigma-Aldrich Corp., St. Louis, MO, USA) before mounting in Fluormount-G (Electron Microscopy Sciences, Hatfield, PA, USA) (Online Supplementary Data).
Primary cells
The protocol was approved by the local ethics committee (approval n. 212/2015). White blood cells (105) were obtained from peripheral blood. Immunofluorescence was performed as previously described8. Mouse anti-Dab1 and anti-Dab2 primary antibodies (sc-271136 and sc-136963, Santa Cruz BIotechnology) and anti-mouse Alexa Fluor 568 secondary antibody (Molecular Probes-Invitrogen, ThermoFisher Scientific, Waltham, MA, USA) were used (Online Supplementary Data).
Genetic analysis
Eye
Flies carrying gmrGal4 or sevGal4 driver constructs were crossed to the UAS-BCR-ABL1 transgenic lines. To analyze the phenotype, flies from a recombinant line carrying both gmrGal4 and UAS-BCR-ABL1 on the third chromosome (gmrGal4,UAS-BCR-ABL1 4M/TM3) were crossed to lines carrying single gene mutations, deficiencies or RNAi constructs. Fifteen to 30 F1 flies from three independent crosses were classified into three phenotypic classes described in the Results section.
Melanotic nodules
domelessGal4-driven BCR-ABL1 expression was controlled with the TARGET system9,10 (Online Supplementary Data). We performed conditional expression in the medullary zone of the lymph gland starting at different stages during larvae development by moving the animals from 18°C to 29°C. Analysis of the melanotic nodule phenotype and temperature shift experiments were performed as previously described.11
RNA extraction and quantitative analysis
RNA was extracted using standard procedures. Expression levels of Dab1 and Dab2 were evaluated by real-time polymerase chain reaction using specific on-demand kits (Hs00245445_m1 for ABL1, Hs00221518_m1 for Dab1, Hs00184598_m1 for Dab2, Applied Biosystems, ThermoFisher Scientific) according to published methods.12
Results
Expression of human BCR-ABL1 affects eye cell differentiation
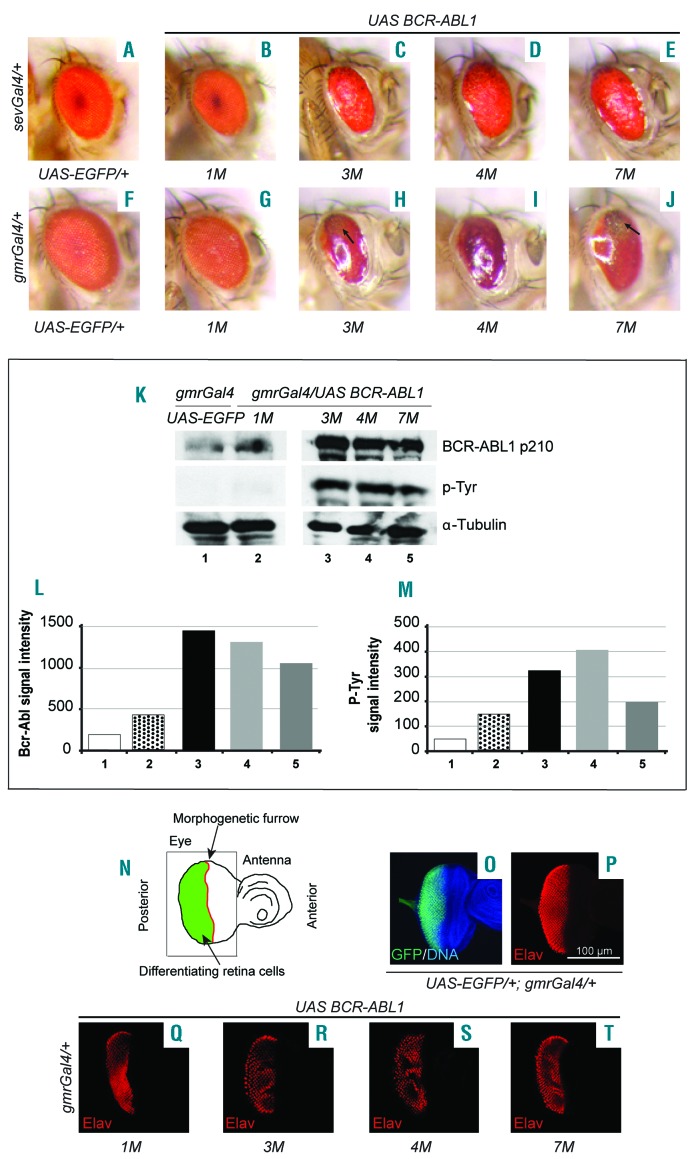
The aim of this work was to set up a CML Drosophila model based on the expression of a completely human BCR-ABL1 fusion protein. Available Drosophila genetic tools allow expression of proteins of interest in developing eye cells, often inducing viable and visible phenotypic traits that can be used as a bait in genetic screening. The Drosophila eye differentiates during the third instar larva (L3) from the eye imaginal disc, a monolayer epithelium that is accessible to dissection. We generated several stable transgenic fly lines to express BCR-ABL1 protein using the yeast Gal4/UAS (Upstream Activating Sequence) transcriptional regulation system controlled by a gene promoter active in specific tissues and stages (Gal4 drivers).13 BCR-ABL1 expression was first triggered with the sevenlessGal4 (sevGal4) construct that drives high levels of expression in some but not all photoreceptors,14 producing a mild rough eye similar to the one observed by Fogerty7 (Figure 1A-E). This suggests that BCR-ABL1 interferes with eye development as described for the human/fly chimera. To drive BCR-ABL1 expression in more eye cells, we used the glass multimer reporterGal4 (gmrGal4) driver, active in all cells committed to differentiation and located posteriorly to the morphogenetic furrow,15 the cell indentation crossing the eye primordium from posterior to anterior (Figure 1N,O). BCR-ABL1 expression in these cells produced a severe “glazed” eye phenotype (Figure 1F-J, Online Supplementary Figure S1A,B,H,I). The regular structure of the eye was almost completely lost: ommatidia, the functional units of the eye, failed to differentiate and were no longer distinguishable. The eye was smaller, bar-shaped and misplaced extra sensory bristles appeared in the dorsal region (Figure 1H-J). Western blot analysis demonstrated that the severity of the phenotype correlated with the amount and phosphorylation of BCR-ABL1 protein (Figure 1K-M): indeed the low level of BCR-ABL1 expression observed in line 1M (Figure 1K-M) resulted in a very mild phenotype (Figure 1G). To better understand the origin of the phenotype, we analyzed the expression of the pan-neuronal and eye photoreceptor marker Elav16 in eye imaginal discs expressing BCR-ABL1. The typical Elav+ photoreceptor clusters (Figure 1P) were reduced in number and altered in BCR-ABL1-expressing flies and this correlated with the described defects of the eye’s ordered structure (Figure 1P-T). To assess whether the phenotype depends on BCR-ABL1 kinase activity, we generated transgenic flies to express a kinase-dead mutant BCR-ABL1. gmrGal4-driven expression of the mutant protein did not affect eye development, indicating that the BCR-ABL1 phenotype requires the enzymatic activity of the oncoprotein (Online Supplementary Figure S1A-C,H).
Figure 1.
BCR-ABL1 expression in the developing eye cells affects photoreceptor differentiation. (A-E) Adult eyes expressing EGFP (A) or BCR-ABL1 in four independent transgenic lines, 1M (B), 3M (C), 4M (D), or 7M (E), in a subset of differentiating photoreceptor cells under the control of the sevenlessGal4 driver construct. (C-E) High levels of BCR-ABL1 induce a “rough” eye phenotype due to impairment of cell differentiation. (F-J) Adult eyes expressing EGFP (F) or BCR-ABL1 (G-J) in all differentiating eye cells under the control of the gmrGal4 driver construct. (H-J) BCR-ABL1 expressed at high level in all differentiating eye cells profoundly disrupts ommatidia development inducing a “glazed” phenotype, depigmented area and the appearance of extra bristles (black arrows). (K-M) Quantification of BCR-ABL1 expression (K,L) and tyrosine-phosphorylation (K,M) in protein extracts from adult heads of flies expressing either EGFP (lane 1) or BCR-ABL1 in independent transgenic fly lines (lanes 2–5). The protein extracts were probed with antibodies raised against BCR, phosphorylated tyrosine residues (p-Tyr) or α–tubulin as the loading control. (N) Schematic of the eye-antenna imaginal disc from a late third instar larva; the positions of the eye and antenna primordia and of the morphogenetic furrows are indicated. The eye imaginal disc area posterior to the morphogenetic furrow, made of cells committed to terminal differentiation, is indicated in green. The thin black square indicates the region of interest shown in panels O-T. (O,P) Eye imaginal disc from wild-type late third instar larvae expressing EGFP under the control of the gmrGal4 driver in cells posterior to the morphogenetic furrow and expressing the pan-neuronal marker Elav in cells committed to terminal differentiation. (Q-T) Elav expression in eye imaginal discs from third instar larvae of the four independent transgenic lines that express BCR-ABL1 under the control of the gmrGal4 driver construct. BCR-ABL1 expression reduces the number of differentiated photoreceptors as indicated by the decrease of Elav-expressing cells.
Expression of human BCR-ABL1 interferes with eye development by altering dAbl signaling
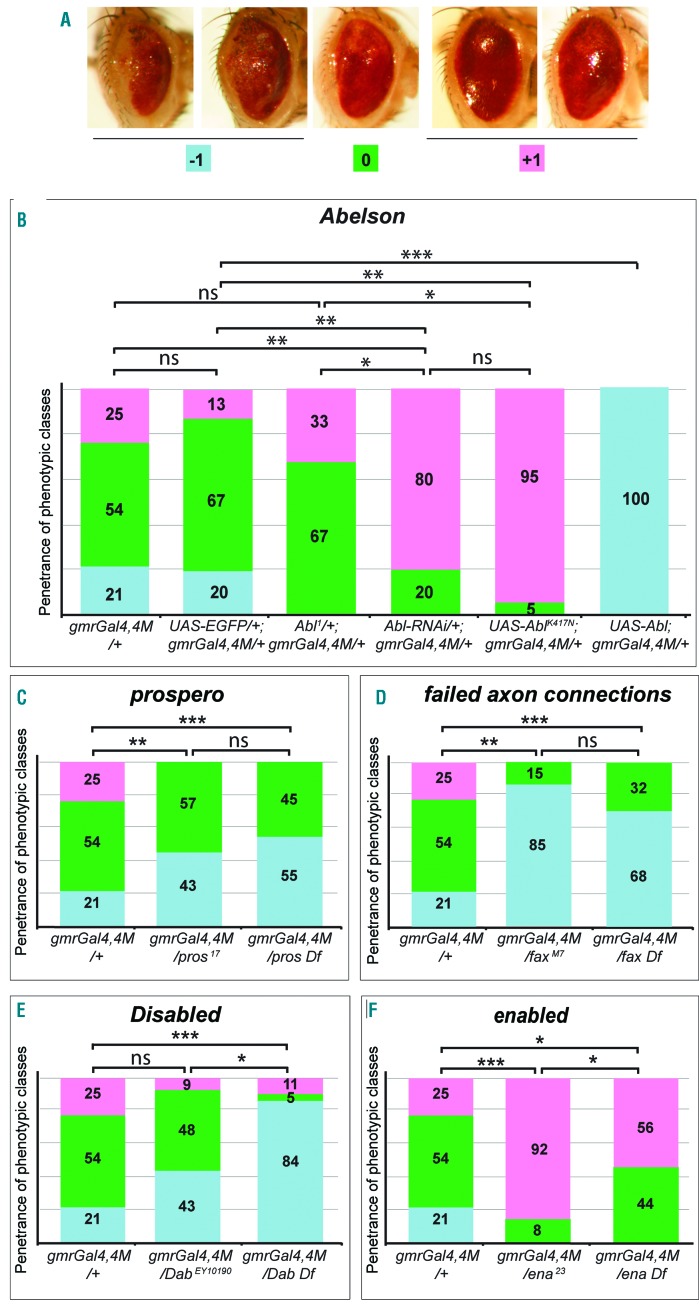
To better understand the consequences of BCR-ABL1 overexpression in the eye, we investigated whether the human oncoprotein could activate the endogenous pathway regulated by the Drosophila Abl kinase (dAbl). To quantify the phenotype we classified BCR-ABL1 eyes (line 4M) into three phenotypic classes. Class 0 represents the most frequent “glazed” phenotype. Class +1 is less severe: the eye is bigger and more prominent, and some ommatidia can be observed. Class −1 is more severe, being characterized by a less differentiated eye with evident lack of pigmentation in the most posterior region (Figure 2A). Interestingly, phenotype expressivity did not change comparing gmrGal4,UAS-BCR-ABL1 4M animals with gmrGal4,UAS-BCR-ABL1 4M;UAS-EGFP (Figure 2B, Online Supplementary Figure S1H,I) indicating that a single gmrGal4 copy does not express a Gal4 limiting amount that could be titrated by increasing the number of UAS sequences. Since overexpression of dAbl (UAS-Abl) induces a very mild rough eye phenotype (Online Supplementary Figure S1A,B,G), we investigated whether it could enhance the BCR-ABL1 phenotype. We observed a worsening of the phenotype: all of the eyes belonged to class −1, showing smaller eyes and more evident loss of pigmentation (Figure 2B, Online Supplementary Figure S1G,H,N). We then investigated whether dAbl loss of function (LOF) could suppress the BCR-ABL1 phenotype. gmrGal4,UAS-BCR-ABL1 4M animals heterozygous for a dAbl hypomorphic recessive lethal allele (Abl1/+) showed a very mild phenotypic suppression but were not statistically different from controls (Figure 2B, Online Supplementary Figure S1E,H,L). However, dAbl downregulation through RNAi (Abl-RNAi) or expression of a dominant negative kinase-defective dAbl (UAS-AblK417N) induced a significant suppression of the BCR-ABL1 phenotype (Figure 2B, Online Supplementary Figure S1D,F,H,I,K,M). Interestingly, we observed that animals expressing either Abl-RNAi or UAS-Abl or UAS-AblK417N showed a similar mild disorganization of the ommatidia (Online Supplementary Figure S1A,B,D,F,G) suggesting that the pathway activated by dAbl is indeed implicated in eye development. Furthermore, the genetic interactions between BCR-ABL1 expression and dAbl loss or gain of function suggest that dAbl, dAblK417N and overexpressed BCR-ABL1 could compete for common binding targets. To confirm that BCR-ABL1 overexpression affects eye development by altering dAbl signaling cascade, we analyzed whether BCR-ABL1 could functionally interact with components of the dAbl pathway. In detail, we focused on four genes whose LOF mutations genetically interact with a dAbl mutant phenotype. Mutations of prospero (pros), a transcription factor that regulates neuronal differentiation17, failed axon connections (fax), implicated both in neurogenesis and axonogenesis18 and Disabled (Dab) that regulates cellular localization of dAbl19, enhance the mutant dAbl phenotype. Moreover, enabled (ena) gene mutations suppress a dAbl mutant phenotype.20 Interestingly, we found that either a deletion or a mutant allele of pros (Figure 2C, Online Supplementary Figure S2A-C) and fax (Figure 2D, Online Supplementary Figure S2A,D,E) was able to enhance the BCR-ABL1 phenotype. Moreover, although the insertional DabEY10190 allele did not change the BCR-ABL1 phenotype significantly, a deletion uncovering the Dab locus enhanced it (Figure 2E, Online Supplementary Figure S2A,F,G), confirming that BCR-ABL1 expression alters eye development likely by interacting with components of the dAbl pathway.
Figure 2.
BCR-ABL1 expression affects ommatidia development by altering the dAbl signaling pathway. (A) Adult eyes showing the phenotypic classes used to quantify the severity of the variable phenotype due to BCR-ABL1 expression in eye cells committed to terminal differentiation. Posterior is on the left. Class 0 (green) corresponds to the average phenotype shown by gmrGal4, UAS-BCR-ABL1 4M flies; the ommatidia are almost totally absent, the eye depigmented region is very small and the eye appears flatter than that of wild-type eyes. Class −1 (pale blue) corresponds to more severe phenotypes: the eyes are even flatter than class 0 eyes and the depigmented area is enlarged including a dorso-ventral sector in the most posterior region of the eye. Class +1 (pink) corresponds to less severe phenotypes: a few ommatidia are visible, the eyes are more bulging and the depigmented area is absent indicating better eye cell differentiation. (B-F) Adult eyes from flies of the indicated genotypes were classified to evaluate the frequency of the three phenotypic classes. (B) A piled histogram chart showing the frequencies of the different phenotypic classes in flies expressing BCR-ABL1 (gmrGal4,4M/+), co-expressing BCR-ABL1 and the EGFP (UAS-EGFP/+;gmrGal4,4M/+), expressing BCR-ABL1 but having a partial loss of the endogenous Abl gene through the heterozygous Abl1 mutation (Abl1/+;gmrGal4,4M/+), RNAi targeting Abl (Abl-RNAi/+;gmrGal4,4M/+), expression of a dominant negative dAbl mutant (UAS-AblK417N;gmrGal4,4M/+) or overexpression of the wild-type Abl protein (UAS-Abl;gmrGal4,4M/+). (C-F) Piled histogram charts showing the frequencies of the three phenotypic classes in flies expressing BCR-ABL1 (gmrGal4,4M) and heterozygous for a loss of function allele or for a deletion of genes that behave as genetic modifiers of the embryonic lethality due to Abl LOF: prospero (C), failed axon connection (D), Disabled (E) and enabled (F). The statistical comparisons were conducted using a Mann-Whitney test (*P<0.05, **P<0.01, ***P<0.001).
BCR-ABL1 expression increases phosphorylation of the dAbl substrate Ena
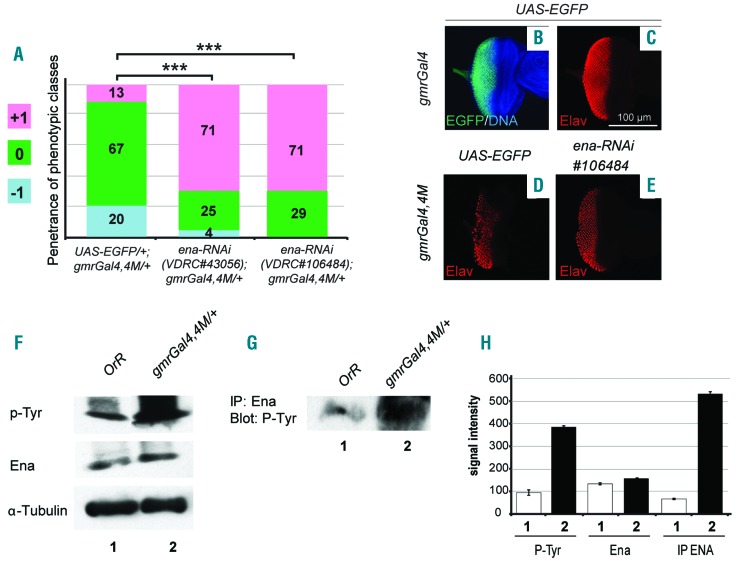
A genetic screen had previously identified an ena LOF allele as a suppressor of the recessive lethality due to dAbl LOF mutations.20 Ena is a cytoskeletal regulator that facilitates actin polymerization.21 Its cellular localization depends on dAbl5,20,22 and it is phosphorylated by both human and Drosophila Abl.23,24 Heterozygosis of a LOF ena allele or of an ena deletion suppressed the BCR-ABL1 phenotype (Figure 2F, Online Supplementary Figure S2A,J,K). ena silencing with two independent constructs (ena-RNAi), induced a size increase and strong decrease of depigmented tissue in eyes expressing BCR-ABL1 (Figure 3A, Online Supplementary Figure S2A,L,M). Consistently, the analysis of Elav expression highlighted a more correct organization of photoreceptor clusters (Figure 3B-E). Furthermore, we looked at tyrosine-phosphorylation of the endogenous Ena. Flies expressing BCR-ABL1 showed increased levels of Ena tyrosine-phosphorylation (Figure 3F,H) even after Ena immunoprecipitation (Figure 3G,H) suggesting that Ena might be phosphorylated by BCR-ABL1. Taken together our data indicate that alteration of several components of the dAbl pathway could be important for the mechanism by which BCR-ABL1 overexpression affects eye development, likely phosphorylating conserved targets in fly eye cells.
Figure 3.
Ena loss of function suppresses the eye phenotype due to BCR-ABL1 expression which increases phosphorylation of the dAbl target Ena. (A) Piled histogram chart showing the frequencies of the three phenotypic classes in flies co-expressing BCR-ABL1 (gmrGal4,4M) and EGFP (UAS-EGFP/+;gmrGal4,4M/+), or one of two independent ena-RNAi lines (VDRC#43056 and VDRC#106484). (B,C) Eye imaginal discs from wild-type late third instar larvae expressing EGFP under the control of the gmrGal4 driver in cells posterior to the morphogenetic furrow (B) and expressing the pan-neuronal marker Elav in cells committed to terminal differentiation (C). (D,E) Elav expression in eye imaginal discs from late third instar larvae expressing BCR-ABL1 (D) or larvae co-expressing BCR-ABL1 and ena-RNAi (E) under the control of the gmrGal4 driver construct. BCR-ABL1 expression reduces the number of differentiated photoreceptors, as indicated by a decrease of Elav-expressing cells, and ena downregulation suppresses this phenotypic trait. (F-H) Quantification of Ena expression and tyrosine-phosphorylation in protein extracts (F) or Ena-immunoprecipitated proteins (G) from the heads of adult flies expressing either EGFP (lane 1) or BCR-ABL1 (lane 2). Independent loads of equal amount of protein extracts or Ena-immunoprecipitated proteins were probed with antibodies raised against phosphorylated tyrosine residues (p-Tyr), Ena or α–tubulin as loading control. (H) Average signal intensity from replica of the experiment shown in (F) and (G). Ena immunoprecipitation and probing for tyrosine-phosphorylation confirmed the increase of Ena tyrosine-phosphorylation in animals expressing BCR-ABL1. The statistical comparisons in (A) were conducted using the Mann-Whitney test (*P<0.05, **P<0.01, ***P<0.001).
A component of the BCR-ABL1-activated pathway in human leukemia modulates the eye phenotype in Drosophila
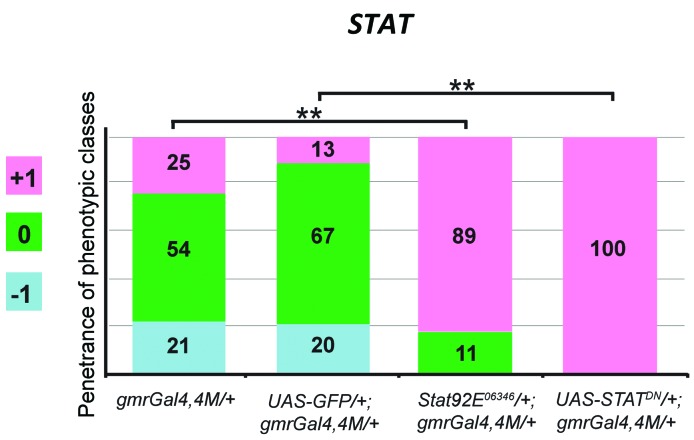
To further assess the effectiveness of the model, we investigated whether a Drosophila homolog of a gene known to be involved in BCR-ABL1 signaling in human leukemia was also able to modulate the BCR-ABL1 phenotype. Signal transducer and activator of transcription 5 (STAT5) is a transcription factor activated in response to cytokines and its role in malignant transformation is well established.25 Several studies showed that BCR-ABL1 induces phosphorylation and constitutive activation of STAT5, hindering apoptosis in leukemic cells.26 The JAK/STAT pathway is required during Drosophila eye morphogenesis and larval hematopoiesis.27,28 Interestingly, loss of STAT92E function (STAT92E06346), the fly counterpart of STAT5, induced strong suppression of the BCR-ABL1 phenotype (Figure 4, Online Supplementary Figure S3A,B). Flies coexpressing a STAT dominant negative allele (STATDN) and BCR-ABL1 showed an even weaker phenotype (Figure 4A, Online Supplementary Figure S3A,C) confirming that STAT is involved in the BCR-ABL1-activated pathway in the Drosophila eye.
Figure 4.
A component of the BCR-ABL1-activated pathway in human leukemia modulates the eye phenotype in Drosophila. Piled histogram chart showing the frequencies of the three phenotypic classes in flies expressing BCR-ABL1 (gmrGal4,4M) and heterozygous for a loss of function STAT92E06346 allele or over-expressing a dominant negative allele STATDN. Reduction of the function of STAT, a gene encoding the homolog of the STAT5 protein involved in the BCR-ABL1-activated pathway in human leukemia cells, suppresses the BCR-ABL1-dependent phenotype in the fly eye. The statistical comparisons were conducted using the Mann-Whitney test (*P<0.05, **P<0.01, ***P<0.001).
The human homologs of Disabled, Dab1 and Dab2, are altered in patients with chronic myeloid leukemia
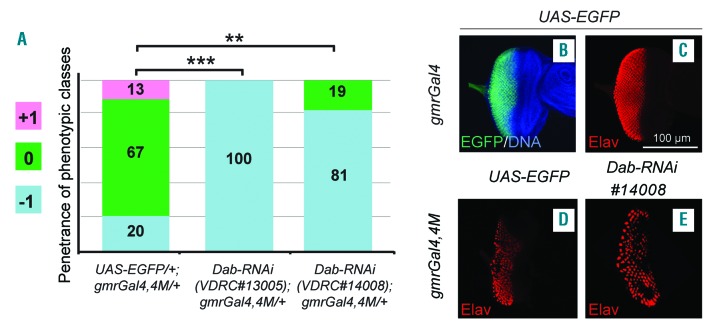
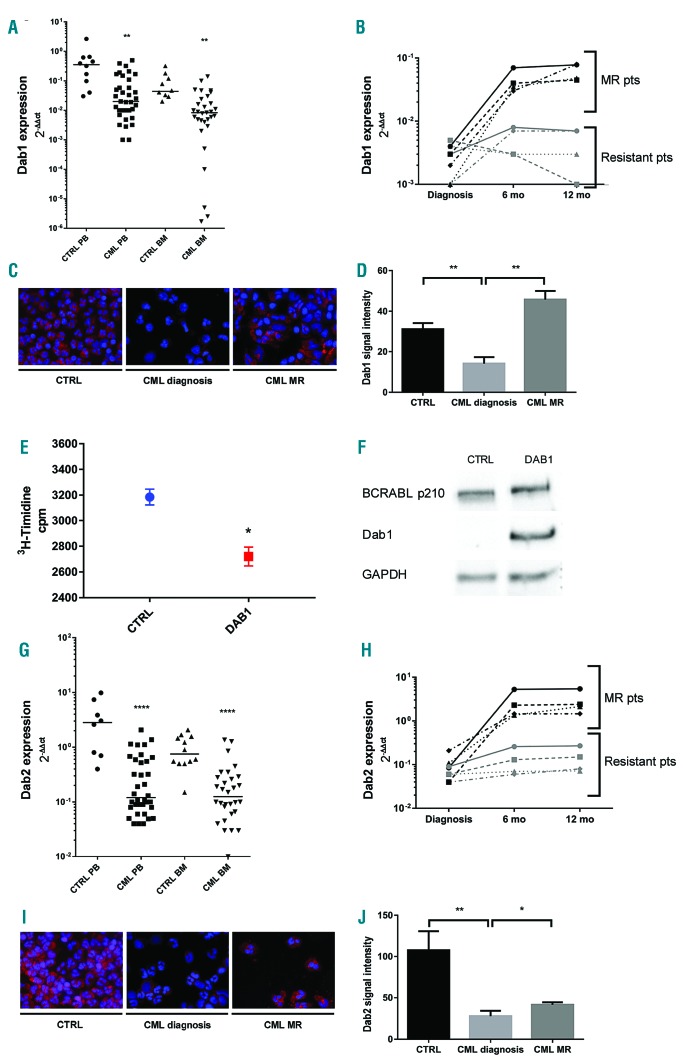
To better explore the efficacy of the model we analyzed the Disabled gene that encodes for an adaptor protein acting downstream of many receptor tyrosine kinases.17,29 In the embryo Dab LOF disrupts the intracellular localization of dAbl and consequently that of phosphorylated Ena and F-Actin accumulation.30 In the fly eye we observed an enhancement of the BCR-ABL1 phenotype in animals heterozygous for a Dab deletion. Thus, we further reduced Dab function by gene silencing. Interestingly, two independent RNAi lines worsened the BCR-ABL1 phenotype more than the Dab deletion did (Figures 2E and 5A, Online Supplementary Figure S2A,F-I): most of the eyes were smaller and showed depigmented scar-like tissue (Online Supplementary Figure S2A,F,H,I). Consistently, alterations of the ommatidia clusters, revealed by Elav expression, worsened compared to those of the control (Figure 5B-E). To establish whether Dab might have a role in CML we analyzed the two human counterparts of Disabled, Dab1 and Dab2 in human primary cells. Dab1 is a large, common fragile site gene and the Dab1 protein acts as a signal transducer that interacts with many receptor tyrosine kinase pathways.31 Dab2 encodes for an adaptor protein implicated in growth factor signaling, endocytosis, cell adhesion, hematopoietic cell differentiation and cell signaling of various receptor tyrosine kinases.32 The expression of both genes is often decreased in many human solid cancers, suggesting their possible role in oncogenesis.31,33 Interestingly, quantitative real-time polymerase chain reaction analysis revealed a significant downregulation of both genes in CML patients at diagnosis compared to controls in peripheral blood or bone marrow samples (Figure 6A,G). Analysis of bone marrow samples from CML patients during molecular remission showed increased levels of expression of both Dab1 (Figure 6B) and Dab2 (Figure 6H) with respect to the levels in treatment-resistant patients. Moreover, immunofluorescence assays demonstrated a significant down-modulation of both proteins in peripheral blood samples at diagnosis compared to the levels in controls or patients in molecular remission (Figure 6C,D,I,J). Finally, transfection experiments in K562 cells using a plasmid carrying the whole Dab1 coding sequence demonstrated that reactivation of Dab1 expression reduced cell proliferation (Figure 6E,F).
Figure 5.
Dab downregulation enhances the eye phenotype due to BCR-ABL1. (A) Piled histogram chart showing the frequencies of the three phenotypic classes in flies co-expressing BCR-ABL1 (gmrGal4,4M) and EGFP (UAS-EGFP/+;gmrGal4,4M/+), or one of two independent Dab-RNAi constructs (VDRC#13005, VDRC#14008). (B,C) Eye imaginal discs from wild-type late third instar larvae expressing EGFP under the control of the gmrGal4 driver in cells posterior to the morphogenetic furrow and expressing the pan-neuronal marker Elav in cells committed to terminal differentiation. (D,E) Elav expression in eye imaginal discs from late third instar larvae expressing BCR-ABL1 (D) or larvae co-expressing BCR-ABL1 and Dab-RNAi (E) under the control of the gmrGal4 driver construct. BCR-ABL1 expression reduces the number of differentiated photoreceptors, as indicated by a decrease of Elav-expressing cells, and Dab downregulation enhances this phenotypic trait. The statistical comparisons were conducted using a Mann-Whitney test (*P<0.05, **P<0.01, ***P<0.001).
Figure 6.
Altered pattern of expression of the human Disabled homologs, Dab1 and Dab2, in patients with chronic myeloid leukemia. (A) Downregulation of Dab1 RNA expression in patients with chronic myeloid leukemia (CML) compared to the expression in healthy donors (CTRL). In particular we found a 1 log reduction of Dab1 expression in both peripheral blood (PB) (P<0.01) and bone marrow (BM) (P<0.01) (median values 2−ΔΔct: 0.02 versus 0.3 in PB and 0.008 versus 0.04 in BM). (B) Expression pattern of Dab1 in CML patients during molecular remission (MR) compared to that in treatment-resistant patients. (C) Immunofluorescence staining of Dab1 protein (red) in PB samples of healthy donors, CML patients at diagnosis and CML patients during MR. Nuclei are stained in blue. (D) Quantification of Dab1 protein expression in the immunofluorescence assay. (E) A 3H-thymidine proliferation assay showing a 20% reduction of cell proliferation in K562 cells transfected with Dab1 plasmid compared to control. (F) Western blot of protein extracts from K562 cells transfected with an empty vector (lane 1) and transfected with a Dab1 expression vector (lane 2), showing detectable expression of Dab1 only in K562 cells transfected with the Dab1 vector. Independent loads of equal amounts of protein extract were probed with antibodies raised against BCR, Dab1 and GAPDH as a loading control. (G) Downregulation of Dab2 RNA expression in CML patients compared to the expression in healthy donors. In particular Dab2 expression was found to be statistically decreased (P<0.0001 and P<0.0001 in PB and BM, respectively) with median values of 0.12 versus 2.8 and 0.12 versus 0.7 in PB and BM, respectively. (H) Pattern of expression of Dab2 in CML patients during MR compared to that in treatment-resistant patients. (I) Immunofluorescence staining of Dab2 protein (red) in PB samples of healthy donors, CML patients at diagnosis and CML patients during MR. Nuclei are stained in blue. (J) Quantification of Dab2 protein expression in an immunofluorescence assay. The statistical comparisons were conducted using a Student t test (*P<0.05, **P<0.01, ****P<0.0001). Bars indicate the standard error.
BCR-ABL1 expression impairs Drosophila blood cell homeostasis
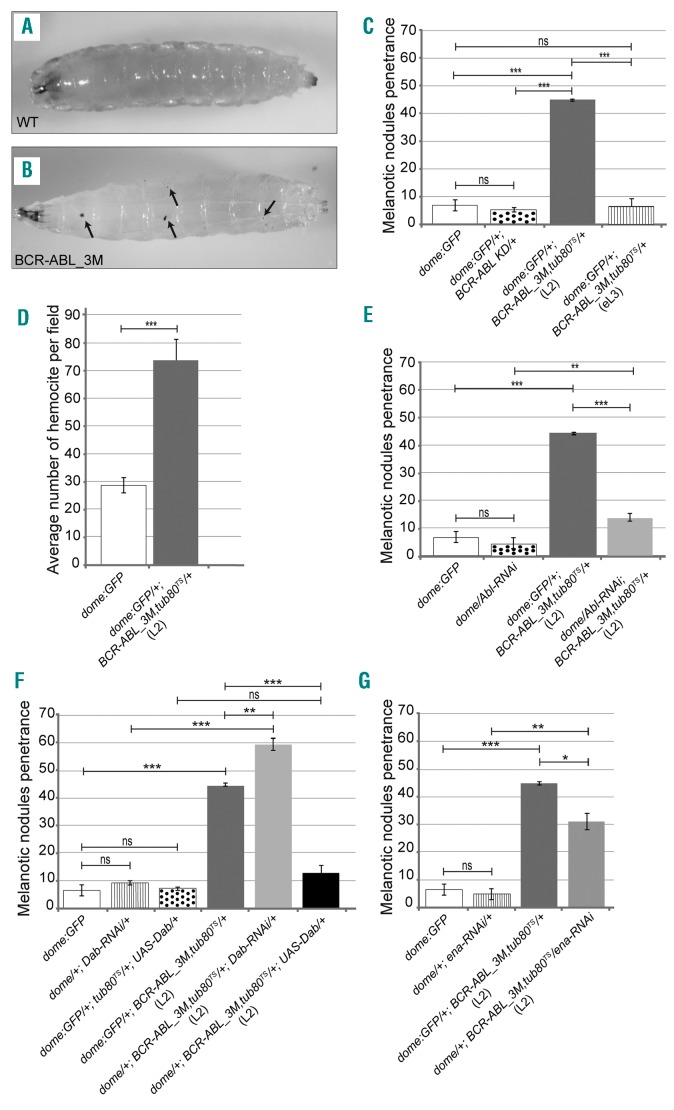
To further confirm the efficacy of the model, we investigated the effects of BCR-ABL1 expression in the lymph gland, the hematopoietic organ of the larva. The lymph gland begins to develop in the embryo34 and grows up from multipotent progenitor cells (prohemocytes) that proliferate and enter a quiescent phase during the second instar (L2). During the third instar (L3) some prohemocytes start to proliferate again and differentiate. The lymph gland breaks apart at the beginning of metamorphosis releasing differentiated blood cells (hemocytes) into the hemolymph, the Drosophila blood.35,36 During the L3, three functional regions can be distinguished in the lymph gland:37 the medullary zone, populated by prohemocytes; the posterior signaling center that regulates the exit of prohemocytes from quiescence; and the cortical zone, made up of differentiating hemocytes.38,39 The lymph gland can break up prematurely in late-L3 if the number of differentiating hemocyte increases. As a reaction to excessive hematopoiesis, the hemocytes aggregate and a spontaneous process of melanization takes place inducing the formation of melanotic nodules.11,36,40,41 Constitutive BCR-ABL1 expression under the control of the domelessGal4 (domeGal4) driver, active in the medullary zone of the lymph gland,11,42 is lethal (data not shown). To overcome this problem, we repressed expression of BCR-ABL1 by co-expressing a heat-sensitive mutant of the Gal4 repressor Gal80 (tubGal80TS) until larvae reached the desired instar (TARGET system).10 While BCR-ABL1 expression from the first instar (L1) induced lethality (data not shown), expression from the L2 allowed larvae to survive and to develop melanotic nodules at L3 (Figure 7A,B). This suggests that BCR-ABL1 expression in the medullary zone precursors might induce an increase of circulating hemocytes (Figure 7C). When compared to controls (Figure 7A), 45% of domeGal4,BCR-ABL1 3M,tubGal80TS larvae showed two to three small melanotic nodules (Figure 7B,C). This correlates with an increased number of circulating hemocytes in hemolymph preparations (Figure 7D). BCR-ABL1 expression starting from the early L3 did not show any significant phenotype (Figure 7C), indicating that only when BCR-ABL1 is expressed when prohemocytes enter quiescence is it able to increase hematopoiesis. Consistently, constitutive expression of the kinase-dead mutant BCR-ABL1KD did not induce any significant phenotype (Figure 7C). Since dAbl, like Dab and ena, is expressed in the lymph gland,43 we assessed whether decreased dAbl function is able to rescue the phenotype. We co-expressed BCR-ABL1 and Abl-RNAi, and observed a significant decrease of the phenotype penetrance (Figure 7E). We then investigated whether Dab or ena downregulation interacts genetically with BCR-ABL1 expression during hematopoiesis as well. Dab-RNAi in the medullary zone starting from L2 was able to enhance the melanotic nodule phenotype, inducing a significant increase of the penetrance (Figure 7F). Consistently, larvae co-expressing Dab (UAS-Dab) and BCR-ABL1 in the medullary zone starting from L2 showed phenotypic suppression (Figure 7F). Moreover, ena-RNAi weakly suppressed the BCR-ABL1 phenotype (Figure 7G), decreasing the phenotype penetrance. As a control, we did not observe any phenotype due to Dab or ena downregulation or Dab overexpression in prohemocytes (Figure 7F,G).
Figure 7.
BCR-ABL1 expression in the hematopoietic precursor cells of the lymph gland impairs Drosophila blood cell homeostasis increasing the number of circulating blood cells. (A) A w1118 mid-L3 instar larva used as the wild-type control. (B) A mid-L3 larva conditionally expressing BCR-ABL1 in the hematopoietic precursors of the lymph gland medullary zone under the control of the domelessGal4 driver construct (dome:GFP/+;BCR-ABL1_3M,tub80TS/+). BCR-ABL1 expression was induced in stage L2 or early-L3 larvae by exposing the animals to 29°C during the indicated larval instars to disrupt the ability of the temperature-sensitive Gal80 mutant to inhibit Gal4 transactivation activity. The black arrows in (B) point to melanotic nodules. Anterior is on the left. (C) Penetrance of the melanotic nodule phenotype in mid-L3 control larvae expressing GFP under the control of the domelessGal4 driver (domeGFP), in larvae constitutively expressing a kinase-dead BCR-ABL1 mutant protein (dome:GFP/+;BCR-ABL1 KD/+) and in larvae in which BCR-ABL1 (dome:GFP/+;BCR-ABL1_3M,tub80TS/+) expression was induced starting from the L2 (L2) or from the early-L3 (eL3) instars. (D) Evaluation of the average number of hemocytes per field after bleeding of dome:GFP/+;BCR-ABL1_3M,tub80TS/+ and dome:GFP/+ larvae. BCR-ABL1 expression induces the appearance of melanotic nodules and this correlates with an increase of circulating hemocytes. (E) Penetrance of the melanotic nodule phenotype in mid-L3 control larvae (dome:GFP), in larvae expressing Abl-RNAi (dome/Abl-RNAi), in larvae in which BCR-ABL1 alone (dome:GFP/+;BCR-ABL1_3M,tub80TS/+) or together with Abl-RNAi (dome/Abl-RNAi;BCR-ABL1_3M,tub80TS/+) is expressed from the L2 instar. (F) Penetrance of the melanotic nodule phenotype in mid-L3 control larvae (dome:GFP), in larvae expressing Dab-RNAi (dome/+;Dab-RNAi/+), in larvae conditionally expressing the Dab protein (dome:GFP/+;tub80TS/+;UAS-Dab/+), and in larvae in which BCR-ABL1 alone (dome:GFP/+;BCR-ABL1_3M,tub80TS/+) or together with either Dab-RNAi (dome/+;BCR-ABL1_3M,tub80TS/+;Dab-RNAi/+) or UAS-Dab (dome/+;BCR-ABL1_3M,tub80TS/+;UAS-Dab/+) is expressed from the L2 instar. (G) Penetrance of the melanotic nodule phenotype in mid-L3 control larvae (dome:GFP), in larvae expressing ena-RNAi (dome/+;ena-RNAi/+), and in larvae in which BCR-ABL1 alone (dome:GFP/+;BCR-ABL1_3M,tub80TS/+) or together with ena-RNAi (dome/+;BCR-ABL1_3M,tub80TS/ena-RNAi) is expressed from the L2 instar. The average phenotype penetrance is calculated from three independent experiments, each involving 15-95 larvae. The statistical comparisons were conducted using a Student t test (*P<0.05, **P<0.01, ***P<0.001, ns=not significant). Bars indicate the standard error.
Discussion
In order to identify candidate genes and pathways involved in the onset and progression of CML we developed and validated a CML genetic model based on transgenic Drosophila expressing BCR-ABL1. In order to build and characterize a human functional model that could be sensitive to pharmacological inhibition and suitable for studying the effects of BCR-ABL1 mutations identified in patients with CML, we chose to express a completely human p210-BCR-ABL1 protein, in contrast what has been done previously.7 The expression of the oncoprotein in all eye cells committed to differentiation as photoreceptors or accessory cells (gmrGal4 driver) induces a strong phenotype characterized by altered differentiation of the ommatidia cells.44 The lack of phenotype in flies expressing a BCR-ABL1 kinase-dead mutant supports the role of kinase activity in the eye phenotype. Moreover, BCR-ABL1 expression and phosphorylation levels correlate with the severity of the phenotype. Consistently, BCR-ABL1 expression under the control of gmrGal4 induces a decrease of photoreceptors expressing Elav in eye imaginal discs and this correlates with the disruption of the adult eye. Interestingly, partial loss of dAbl function also slightly reduces the number of eye cells expressing Elav at L3, and to a much greater extent at later stages of development. This suggests that dAbl is implicated in the maintenance of neuronal commitment45,46 and confirms that loss or gain of function of dAbl/BCR-ABL1 can alter eye cell development.7 We have shown that human BCR-ABL1 interacts and interferes with the dAbl signaling pathway. Animals expressing BCR-ABL1 and heterozygous for the recessive Abl1 allele or coexpressing either Abl-RNAi or a kinase-dead dominant negative Abl (AblK417N) showed a weaker phenotype, suggesting that BCR-ABL1 and dAbl proteins most likely share binding sites and/or targets of the kinase activity. Consistently, co-expression of human BCR-ABL1 and dAbl synergizes and the phenotype becomes more severe. Notably, dAbl overexpression per se induces a weak “rough” eye phenotype but the differentiation program is not severely disrupted. We cannot exclude that this is due to a level of dAbl expression below a critic threshold but it could also suggest that excessive dAbl might be still, at least partially, negatively regulated. This possible negative regulation seems to be overcome by BCR-ABL1 since all animals co-expressing dAbl and BCR-ABL1 showed a severe class −1 phenotype. Consistently, LOF or downregulation of genes known to interact genetically with dAbl LOF mutations interact in the same way with BCR-ABL1 expression. Namely, pros and fax alleles or deletions enhance the phenotype and this is consistent with their roles in neuronogenesis and neuronal differentiation. Moreover, ena LOF suppresses and Dab LOF enhances the dAbl LOF phenotype19,20 and we observed that both ena and Dab LOF and downregulation through RNAi also modify the BCR-ABL1 phenotype in the same way. Ena belongs to the ENA/VASP protein family involved in regulation of the actin cytoskeleton.47,48 dAbl regulates Ena by modulating its localization, most likely through its phosphorylation. It is known that both dAbl and the human/Drosophila BCR-ABL chimera phosphorylate Ena7 in vitro and we established that human BCR-ABL1 expression in the eye also increases Ena phosphorylation. This conservation of phosphorylation targets significantly increases the reliability of our model for identifying relevant BCR-ABL1 functional interactors. In this view the observation that decreased Ena function suppresses phenotypes due to both dAbl mutations24 and BCR-ABL1 expression suggests that both phenotypes can be due to Ena mislocalization and consequently actin cytoskeleton alterations can be suppressed if Ena expression decreases. In Drosophila, Abl and Dab are often co-expressed and the phenotype due to Dab mutations mimics the dAbl phenotype. Epistasis experiments have shown that Dab functions upstream of both dAbl and Ena, controlling their localization and thus the actin cytoskeleton, and Dab LOF does indeed enhance the phenotype due to dAbl mutations.30 Interestingly, Dab deletion or downregulation has the same effect on the BCR-ABL1 phenotype. These findings could be explained if Dab is able to regulate, at least partially, BCR-ABL1 localization. This interaction might mitigate more severe BCR-ABL1-dependent effects when Dab is expressed at a physiological level but not if Dab is downregulated or its gene dosage is halved. Furthermore, our study showed that Dab human homologs are less expressed in both peripheral blood and bone marrow of CML patients at diagnosis compared to their expression in controls and are re-expressed in patients during molecular remission. Moreover, Dab1 expression in transfected K562 cells significantly decreases cell proliferation, confirming that Dab activity might alleviate the pathogenic effects of BCR-ABL1. We then assessed whether our model could help to fish-out homologs of leukemia-relevant genes in an ongoing dosage-sensitive genetic screen of the whole Drosophila genome. To this aim we considered STAT5, a transcription factor phosphorylated and activated by BCR-ABL1. Interestingly, LOF conditions of STAT92E, encoding the fly homolog of various human STAT, led to suppression of the BCR-ABL1 phenotype. In order to discover a tissue that could be a reliable second read-out for identifying BCR-ABL1 interactors relevant for hematopoiesis and leukemia, we moved to the larval hematopoietic organ, the lymph gland. We conditionally expressed human BCR-ABL1 in the lymph gland medullary zone where quiescent prohemocytes reside. Only BCR-ABL1 expression during L2 induces the appearance of melanotic nodules, which correlates with an increase of circulating hemocytes. This phenotype can be suppressed by dAbl downregulation, confirming that dAbl is expressed in the lymph gland medullary zone43 where it contributes to BCR-ABL1 pathway activation and to induction of the hematopoietic phenotype. It is worth noting that both Dab and ena interact functionally with BCR-ABL1 during hematopoiesis. In fact, while Dab downregulation enhances the melanotic nodule phenotype and Dab overexpression suppresses it, ena downregulation decreases the penetrance of this phenotype, confirming that ena and Dab are also expressed in the lymph gland medullary zone43 and modulate BCR-ABL1 activity. This phenotype is visible if BCR-ABL1 is expressed from the L2, when prohemocytes become quiescent, but not if it is expressed from the early L3, when the quiescent prohemocytes are still present in the medullary zone of the lymph gland. We are tempted to speculate that the dAbl pathway, activated by BCR-ABL1, could be involved in the mechanisms that regulate entry of prohemocytes into the quiescent state rather than maintenance of this state. This seems consistent with the observation that the lymph glands in mid-L3 larvae expressing BCR-ABL1 from L2 are very small compared to those in controls and do not show any clear partition (Giordani and Bernardoni, unpublished data). This suggests that, upon BCR-ABL1 expression, most of the prohemocytes could undertake the differentiation pathway and leave the lymph gland prematurely without becoming quiescent. We did not test all pathways interacting with BCR-ABL1, for example the Tyr-receptor/Ras pathway, which is known to compete with BCR-ABL1 for binding with the Grb2/Drk proteins1 and is likely involved in the eye phenotype since the Sevenless Tyr-receptor has an established role in eye differentiation.49,50 Nevertheless, we present here a new and efficient CML model based on Drosophila transgenic for human BCR-ABL1. This model could be a powerful tool for identifying new genes and pathways involved in the pathogenesis and progression of CML.
Supplementary Material
Acknowledgments
We thank L. Giardino, V.A. Baldassarro, C. Mangano, and L. Calzà (Fondazione IRET, Ozzano dell’Emilia-Bologna, Italy) for assistance with the confocal microscopy analysis; D. Manzoni and M. Voltattorni for excellent technical help; M. Capovilla and the Trans-FlyER, Startup Company, Ferrara, Italy for generating the BCR-ABL1 kinase-dead transgenic lines. Drosophila lines were obtained from the Bloomington Drosophila Stock Center-BDSC (NIH P40OD018537) and primary antibodies from the Developmental Studies Hybridoma Bank (created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA, USA). We thank the Italian Association for Research on Cancer (AIRC) for funding D. Cilloni (IG10005) and G. Perini (IG11400, IG15182) and for supporting this work.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/4/717
References
- 1.Chereda B, Melo JV. Natural course and biology of CML. Ann Hematol. 2015;94(Suppl 2):S107–121. [DOI] [PubMed] [Google Scholar]
- 2.Quintas-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009;113(8):1619–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez C. Drosophila melanogaster: a model and a tool to investigate malignancy and identify new therapeutics. Nat Rev Cancer. 2013;13(3):172–183. [DOI] [PubMed] [Google Scholar]
- 4.Bashaw GJ, Kidd T, Murray D, Pawson T, Goodman CS. Repulsive axon guidance: Abelson and Enabled play opposing roles downstream of the roundabout receptor. Cell. 2000;101(7):703–715. [DOI] [PubMed] [Google Scholar]
- 5.Grevengoed EE, Loureiro JJ, Jesse TL, Peifer M. Abelson kinase regulates epithelial morphogenesis in Drosophila. J Cell Biol. 2001;155(7):1185–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liebl EC, Forsthoefel DJ, Franco LS, et al. Dosage-sensitive, reciprocal genetic interactions between the Abl tyrosine kinase and the putative GEF trio reveal trio’s role in axon pathfinding. Neuron. 2000;26(1):107–118. [DOI] [PubMed] [Google Scholar]
- 7.Fogerty FJ, Juang JL, Petersen J, Clark MJ, Hoffmann FM, Mosher DF. Dominant effects of the bcr-abl oncogene on Drosophila morphogenesis. Oncogene. 1999;18(1):219–232. [DOI] [PubMed] [Google Scholar]
- 8.Cilloni D, Messa F, Arruga F, et al. The NF-kappaB pathway blockade by the IKK inhibitor PS1145 can overcome imatinib resistance. Leukemia. 2006;20(1):61–67. [DOI] [PubMed] [Google Scholar]
- 9.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004 (220):p16. [DOI] [PubMed] [Google Scholar]
- 10.McGuire SE, Roman G, Davis RL. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 2004;20(8):384–391. [DOI] [PubMed] [Google Scholar]
- 11.Giordani G, Barraco M, Giangrande A, et al. The human Smoothened inhibitor PF-04449913 induces exit from quiescence and loss of multipotent Drosophila hematopoietic progenitor cells. Oncotarget. 2016;7(34):55313–55327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cilloni D, Carturan S, Bracco E, et al. Aberrant activation of ROS1 represents a new molecular defect in chronic myelomonocytic leukemia. Leuk Res. 2013;37(5):520–530. [DOI] [PubMed] [Google Scholar]
- 13.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. [DOI] [PubMed] [Google Scholar]
- 14.Basler K, Yen D, Tomlinson A, Hafen E. Reprogramming cell fate in the developing Drosophila retina: transformation of R7 cells by ectopic expression of rough. Genes Dev. 1990;4(5):728–739. [DOI] [PubMed] [Google Scholar]
- 15.Freeman M. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell. 1996;87(4):651–660. [DOI] [PubMed] [Google Scholar]
- 16.Robinow S, White K. The locus elav of Drosophila melanogaster is expressed in neurons at all developmental stages. Dev Biol. 1988;126(2):294–303. [DOI] [PubMed] [Google Scholar]
- 17.Gertler FB, Hill KK, Clark MJ, Hoffmann FM. Dosage-sensitive modifiers of Drosophila abl tyrosine kinase function: prospero, a regulator of axonal outgrowth, and disabled, a novel tyrosine kinase substrate. Genes Dev. 1993;7(3):441–453. [DOI] [PubMed] [Google Scholar]
- 18.Hill KK, Bedian V, Juang JL, Hoffmann FM. Genetic interactions between the Drosophila Abelson (Abl) tyrosine kinase and failed axon connections (fax), a novel protein in axon bundles. Genetics. 1995;141(2):595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gertler FB, Bennett RL, Clark MJ, Hoffmann FM. Drosophila abl tyrosine kinase in embryonic CNS axons: a role in axonogenesis is revealed through dosage-sensitive interactions with disabled. Cell. 1989;58(1):103–113. [DOI] [PubMed] [Google Scholar]
- 20.Gertler FB, Doctor JS, Hoffmann FM. Genetic suppression of mutations in the Drosophila abl proto-oncogene homolog. Science. 1990;248(4957):857–860. [DOI] [PubMed] [Google Scholar]
- 21.Krause M, Bear JE, Loureiro JJ, Gertler FB. The Ena/VASP enigma. J Cell Sci. 2002;115(Pt 24):4721–4726. [DOI] [PubMed] [Google Scholar]
- 22.Grevengoed EE, Fox DT, Gates J, Peifer M. Balancing different types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J Cell Biol. 2003;163(6):1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Comer AR, Ahern-Djamali SM, Juang JL, Jackson PD, Hoffmann FM. Phosphorylation of Enabled by the Drosophila Abelson tyrosine kinase regulates the in vivo function and protein-protein interactions of Enabled. Mol Cell Biol. 1998;18(1):152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gertler FB, Comer AR, Juang JL, et al. enabled, a dosage-sensitive suppressor of mutations in the Drosophila Abl tyrosine kinase, encodes an Abl substrate with SH3 domain-binding properties. Genes Dev. 1995;9(5):521–533. [DOI] [PubMed] [Google Scholar]
- 25.Nieborowska-Skorska M, Wasik MA, Slupianek A, et al. Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J Exp Med. 1999;189(8):1229–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Groot RP, Raaijmakers JA, Lammers JW, Jove R, Koenderman L. STAT5 activation by BCR-Abl contributes to transformation of K562 leukemia cells. Blood. 1999;94(3):1108–1112. [PubMed] [Google Scholar]
- 27.Bina S, Wright VM, Fisher KH, Milo M, Zeidler MP. Transcriptional targets of Drosophila JAK/STAT pathway signalling as effectors of haematopoietic tumour formation. EMBO Rep. 2010;11(3):201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeidler MP, Bach EA, Perrimon N. The roles of the Drosophila JAK/STAT pathway. Oncogene. 2000;19(21):2598–2606. [DOI] [PubMed] [Google Scholar]
- 29.Le N, Simon MA. Disabled is a putative adaptor protein that functions during signaling by the sevenless receptor tyrosine kinase. Mol Cell Biol. 1998;18(8):4844–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song JK, Kannan R, Merdes G, Singh J, Mlodzik M, Giniger E. Disabled is a bona fide component of the Abl signaling network. Development. 2010;137(21):3719–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAvoy S, Zhu Y, Perez DS, James CD, Smith DI. Disabled-1 is a large common fragile site gene, inactivated in multiple cancers. Genes Chromosomes Cancer. 2008;47(2):165–174. [DOI] [PubMed] [Google Scholar]
- 32.Finkielstein CV, Capelluto DG. Disabled-2: A modular scaffold protein with multifaceted functions in signaling. Bioessays. 2016;38(Suppl 1):S45–55. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Z, Chen Y, Tang J, Xie X. Frequent loss expression of dab2 and promotor hyper-methylation in human cancers: a meta-analysis and systematic review. Pak J Med Sci. 2014;30(2):432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holz A, Bossinger B, Strasser T, Janning W, Klapper R. The two origins of hemocytes in Drosophila. Development. 2003;130(20): 4955–4962. [DOI] [PubMed] [Google Scholar]
- 35.Evans CJ, Hartenstein V, Banerjee U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell. 2003;5(5):673–690. [DOI] [PubMed] [Google Scholar]
- 36.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230(2):243–257. [DOI] [PubMed] [Google Scholar]
- 37.Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132(11):2521–2533. [DOI] [PubMed] [Google Scholar]
- 38.Krzemien J, Dubois L, Makki R, Meister M, Vincent A, Crozatier M. Control of blood cell homeostasis in Drosophila larvae by the posterior signalling centre. Nature. 2007;446(7133):325–328. [DOI] [PubMed] [Google Scholar]
- 39.Lebestky T, Jung SH, Banerjee U. A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes Dev. 2003;17(3):348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo H, Hanratty WP, Dearolf CR. An amino acid substitution in the Drosophila hopTum-l Jak kinase causes leukemia-like hematopoietic defects. EMBO J. 1995;14(7):1412–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minakhina S, Steward R. Melanotic mutants in Drosophila: pathways and phenotypes. Genetics. 2006;174(1):253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bourbon HM, Gonzy-Treboul G, Peronnet F, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech Dev. 2002;110(1–2):71–83. [DOI] [PubMed] [Google Scholar]
- 43.Irving P, Ubeda JM, Doucet D, et al. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 2005;7(3):335–350. [DOI] [PubMed] [Google Scholar]
- 44.Ellis MC, O’Neill EM, Rubin GM. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119(3): 855–865. [DOI] [PubMed] [Google Scholar]
- 45.Bennett RL, Hoffmann FM. Increased levels of the Drosophila Abelson tyrosine kinase in nerves and muscles: subcellular localization and mutant phenotypes imply a role in cell-cell interactions. Development. 1992;116(4): 953–966. [DOI] [PubMed] [Google Scholar]
- 46.Xiong W, Morillo SA, Rebay I. The Abelson tyrosine kinase regulates Notch endocytosis and signaling to maintain neuronal cell fate in Drosophila photoreceptors. Development. 2013;140(1):176–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barzik M, Kotova TI, Higgs HN, et al. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280(31): 28653–28662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bear JE, Svitkina TM, Krause M, et al. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 2002;109(4):509–521. [DOI] [PubMed] [Google Scholar]
- 49.Olivier JP, Raabe T, Henkemeyer M, et al. A Drosophila SH2-SH3 adaptor protein implicated in coupling the sevenless tyrosine kinase to an activator of Ras guanine nucleotide exchange, Sos. Cell. 1993;73(1):179–191. [DOI] [PubMed] [Google Scholar]
- 50.Simon MA, Dodson GS, Rubin GM. An SH3-SH2-SH3 protein is required for p21Ras1 activation and binds to sevenless and Sos proteins in vitro. Cell. 1993;73(1): 169–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.